Significance
Human genes homozygous for apparent loss of function (LoF) variants are increasingly reported in a sizeable proportion of individuals without overt clinical phenotypes. Here, we found 166 genes with 179 predicted LoF variants for which the frequency of homozygous individuals exceeds 1% in at least one of the populations present in databases ExAC and gnomAD. These putatively dispensable genes showed relaxation of selective constraints, suggesting that a considerable proportion of these genes may be undergoing pseudogenization. Eight of these LoF variants displayed robust signals of positive selection, including two variants in genes involved in resistance to infectious diseases. The identification of dispensable genes will facilitate the identification of functions that are now redundant, or possibly even advantageous, for human survival.
Keywords: redundancy, pseudogenization, loss of function, positive selection, negative selection
Abstract
Humans homozygous or hemizygous for variants predicted to cause a loss of function (LoF) of the corresponding protein do not necessarily present with overt clinical phenotypes. We report here 190 autosomal genes with 207 predicted LoF variants, for which the frequency of homozygous individuals exceeds 1% in at least one human population from five major ancestry groups. No such genes were identified on the X and Y chromosomes. Manual curation revealed that 28 variants (15%) had been misannotated as LoF. Of the 179 remaining variants in 166 genes, only 11 alleles in 11 genes had previously been confirmed experimentally to be LoF. The set of 166 dispensable genes was enriched in olfactory receptor genes (41 genes). The 41 dispensable olfactory receptor genes displayed a relaxation of selective constraints similar to that observed for other olfactory receptor genes. The 125 dispensable nonolfactory receptor genes also displayed a relaxation of selective constraints consistent with greater redundancy. Sixty-two of these 125 genes were found to be dispensable in at least three human populations, suggesting possible evolution toward pseudogenes. Of the 179 LoF variants, 68 could be tested for two neutrality statistics, and 8 displayed robust signals of positive selection. These latter variants included a known FUT2 variant that confers resistance to intestinal viruses, and an APOL3 variant involved in resistance to parasitic infections. Overall, the identification of 166 genes for which a sizeable proportion of humans are homozygous for predicted LoF alleles reveals both redundancies and advantages of such deficiencies for human survival.
The human genome displays considerable DNA sequence diversity at the population level. One of its most intriguing features is the homozygosity or hemizygosity for variants of protein-coding genes predicted to be loss-of-function (LoF) found at various frequencies in different human populations (1–3). An unknown proportion of these reported variants are not actually LoF, instead being hypomorphic or isomorphic, because of a reinitiation of translation, readthrough, or a redundant tail, resulting in lower, normal, or even higher than normal levels of protein function. Indeed, a bona fide nonsense allele, predicted to be LoF, can actually be gain-of-function (hypermorphic), as illustrated by IκBα mutations (4). Moreover, the LoF may apply selectively to one isoform or a subset of isoforms of a given gene, but not others (e.g., if the exon carrying the premature stop is spliced out for a specific set of alternative transcripts) (5). Finally, there are at least 400 discernible cell types in the human body (6), and the mutant transcript may be expressed in only a limited number of tissues. Conversely, there are also mutations not predicted to be LoF, such as in-frame insertions−deletions (indels), missense variants, splice region variants not affecting the essential splice regions, and even synonymous or deep intronic mutations, that may actually be LoF but cannot be systematically identified as such in silico.
Many predicted LoF variants have nevertheless been confirmed experimentally, typically when they are associated with a clinical phenotype. Of the 229,161 variants reported in the Human Gene Mutation Database (HGMD) (7), as many as 99,027 predicted LoF alleles in 5,186 genes have been found to be disease causing in association and/or functional studies. For example, for the subset of 253 genes implicated in recessive forms of primary immunodeficiencies (8, 9), 12,951 LoF variants are reported in HGMD. Conversely, a substantial proportion of genes harboring biallelic null variants have no discernible associated pathological phenotype, and several large-scale sequencing surveys in adults from the general population have reported human genes apparently tolerant to homozygous LoF variants (10–15). Four studies in large bottlenecked or consanguineous populations detected between 781 and 1,317 genes homozygous for mutations predicted to be LoF (11, 12, 14, 15). These studies focused principally on low-frequency variants (minor allele frequency < 1 to 5%), and associations with some traits, benign or disease-related, were found for a few rare homozygous LoF variants. Two studies provided a more comprehensive perspective of the allele frequency spectrum of LoF variants. A first systematic survey of LoF variants, mostly in the heterozygous state, was performed with whole-genome sequencing data from 185 individuals of the 1000 Genomes Project; it identified 253 genes with homozygous LoF variants (10). In a larger study of more than 60,000 whole exomes, the Exome Aggregation Consortium (ExAC) database project identified 1,775 genes with at least one homozygous LoF variant, with a mean of 35 homozygous potential LoF variants per individual (13).
These studies clearly confirmed the presence of genes with homozygous LoF variants in apparently healthy humans, but no specific study of such variants present in the homozygous state in a sizeable proportion (>1%) of large populations has yet been performed. In principle, these variants may be neutral (indicating gene redundancy) (16), or may even confer a selective advantage (the so-called “less is more” hypothesis) (3, 17). Indeed, a few cases of common beneficial LoF variants have been documented, including some involved in host defense against life-threatening microbes (18, 19). Homozygosity for LoF variants of DARC (now ACKR1), CCR5, and FUT2 confer resistance to Plasmodium vivax (20, 21), HIV (22–24), and norovirus (25, 26), respectively. We hypothesized that the study of common homozygous LoF variants might facilitate the identification of the set of dispensable protein-coding genes in humans and reveal underlying evolutionary trends. Unlike rare LoF variants, bona fide common homozygous LoF variants are predicted to be enriched in neutral alleles (1–3). They also provide indications concerning genes undergoing inactivation under positive selection (18, 19). The availability of large public databases, such as ExAC (13) and its extended version, Genome Aggregation Database (gnomAD, https://gnomad.broadinstitute.org/), which includes data from more than 120,000 individuals, is making it possible to search for such variants with much greater power, across multiple populations.
Results
Definition of the Set of Dispensable Protein-Coding Genes in Humans.
We used two large exome sequence databases: ExAC (13) and gnomAD (Methods), which have collected 60,706 and 123,136 exome sequences, respectively. For this study, we focused on the 20,232 protein-coding genes, and we excluded the 13,921 pseudogenes (Methods and ref. 27). We defined protein-coding genes as dispensable if they carry variants that 1) are computationally predicted to be LoF with a high degree of confidence, including early stop-gains, indel frameshifts, and essential splice site-disrupting variants (i.e., involving a change in the 2-nt region at the 5′ or 3′ end of an intron; Methods) and 2) have a frequency of homozygous individuals (hemizygous for genes on the X chromosome in males) exceeding 1% in at least one of the five population groups considered in these public databases, i.e., Africans (AFR), including African Americans, East Asians (EAS), South Asians (SAS), Europeans (EUR), and Admixed Latino Americans (AMR). As we focused on exome data, only small indels (<50 base pairs [bp]) were considered in this analysis. Common quality filters for calls and a minimum call rate of 80% were applied to each reference database (ExAC and gnomAD; Methods). Only LoF variants affecting the principal isoform (28) were retained (Methods). The application of these filters led to the detection of 208 (ExAC) and 228 (gnomAD) genes, 190 of which were common to the two databases, and are referred to hereafter as the set of dispensable genes (Table 1 and Dataset S1). No genes on the X or Y chromosomes fulfilled these criteria. Relaxing the thresholds on the single-nt polymorphism (SNP) and insertions and deletions (indel) call quality filters (variant quality score recalibration [VQSR] score; Methods), the variant call rate, or the nonrestriction to the principal isoform did not substantially increase the number of putatively dispensable genes (SI Appendix, Figs. S1 and S2). The frequency of homozygous individuals at which a gene is defined as dispensable appeared to be the criterion with the largest impact on the number of dispensable genes identified, thereby justifying the use of the stringent threshold (>1% homozygotes in at least one specific population) described above (SI Appendix, Figs. S1 and S2).
Table 1.
Number of dispensable genes identified depending on the adopted filtering criteria
| No. of genes | No. of variants | No. of genes with multiple variants | |
| Set of dispensable genes | 190 | 207 | 13 |
| Set of dispensable genes after manual curation (step 1) | 168 | 181 | 9 |
| Set of dispensable genes after manual curation (step 2) | 166 | 179 | 9 |
| No OR genes | 125 | 137 | 8 |
| OR genes | 41 | 42 | 1 |
Manual Curation of the LoF Variants.
We then investigated the potential molecular consequences of the 207 putative LoF variants retained, collectively associated with the 190 genes, by manually curating the annotation of these LoF variants. An examination of the sequencing reads in gnomAD showed that 18 variants—10 single-nt variants (SNVs) and 8 indels initially annotated as stop-gains and frameshift variants, respectively—were components of haplotypes with consequences other than the initial LoF annotation (Dataset S1). Thus, for each of the 10 SNVs, a haplotype encompassing a contiguous second variant led to the creation of a missense rather than a stop variant allele. For each of the eight frameshift variations, a haplotype with a nearby second indel (observed in the same sequencing reads) collectively led to an in-frame indel allele. In addition, six essential splice site-disrupting variants caused by indels resulted in no actual modification of the splice receptor or acceptor site motif. Overall, annotation issues occurred in about 13% of the initial set, indicating that there is a need for annotation methods to take the underlying haplotype inferred from sequencing reads into account. We also analyzed the common putative HLA-A LoF frameshift variant rs1136691 in more detail, as HLA-A null alleles are known to occur very rarely in large transplantation databases (http://hla.alleles.org/alleles/nulls.html). An analysis of the sequencing reads corresponding to this variant revealed an alternative haplotype of several variants, and a Blast analysis of this sequence yielded a perfect match with an alternative unmapped contig of chromosome 6 (chr6_GL000256v2_alt in GRCh38). This observation suggests that the alternative haplotype may have been wrongly mapped to the closest sequence in the reference genome, resulting in an artifactual frameshift in the HLA-A gene. This hypothesis is consistent with the known genomic complexity and high level of polymorphism of the HLA region, which can lead to incorrect mapping (29). Finally, we noted that one variant, rs36046723 in the ZNF852 gene, had an allele frequency >0.9999, suggesting that the reference genome carries the derived, low-frequency variant at this position. After curation, we retained 181 predicted LoF variants from 168 genes (Table 1).
Characteristics of the LoF Variants.
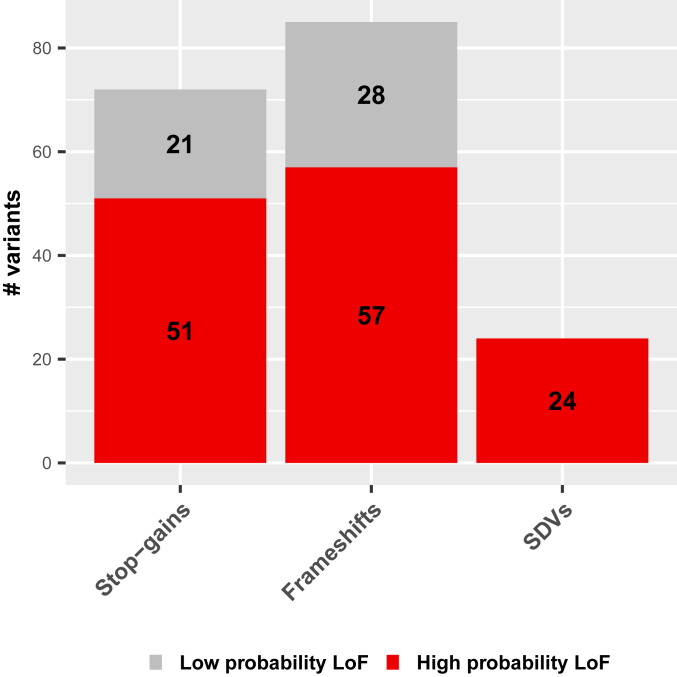
The set of 181 predicted LoF variants defining the set of dispensable genes included premature stop-gains (40%), frameshifts (47%), and splice site variants (13%; Fig. 1). The variants could be classified further into those with a high or low predicted probability of being LoF (Methods): 27% presented features consistent with a low probability of LoF, whereas the remaining 73% were predicted to have more severely damaging consequences and were therefore considered to have a high probability of LoF (Fig. 1 and SI Appendix, Fig. S3). Despite possible differences in their impact, low- and high-probability LoF variants had similarly distributed global allele frequencies (SI Appendix, Fig. S4; two-tailed Wilcoxon test P value = 0.3115 based on ExAC allele frequencies, and 0.3341 based on gnomAD allele frequencies). Only 30 of the LoF variants finally retained were reported in at least one PubMed publication in the dbSNP database (30) (SI Appendix, Table S1). Manual inspection of these studies revealed that only 11 LoF variants had been experimentally demonstrated to abolish gene function (Dataset S1). Focusing on the overlap between the 181 predicted LoF variants and the genome-wide association study (GWAS) hits, we found that only two were reported in the GWAS catalog (31) as being significantly associated with a phenotypic trait: rs41272114 (LPA), associated with plasma plasminogen levels, lipoprotein A levels, and coronary artery disease; and rs601338 (FUT2), associated with the levels of certain blood proteins, such as fibroblast growth factor 19. Finally, only 27 out of the 181 predicted LoF presented annotations in ClinVar, including 25 of them labeled as benign/likely benign, one as protective/risk factor (rs5744168 in TLR5, discussed in later sections of this manuscript), and another as pathogenic (rs17147990 in HTN3) (Dataset S1). Pathogenicity for rs17147990 is only based on a publication reporting this mutation in the Histatin 3 gene (p.Tyr47Ter) (32), without any evidence of causality for a clinical phenotype. Overall, this analysis shows that most of the common LoF considered here present features consistent with severely damaging variants for gene function, although experimental characterization is largely lacking.
Fig. 1.
Functional consequences of LoF variants defining the set of dispensable protein-coding genes. Bar plots show the distribution of LoF variants defining the set of dispensable genes according to their molecular consequences (stop-gains, frameshifts, and splice-disrupting variants [SDVs]) and the predicted severity of their functional impact: low probability (light gray) and high probably LoF (dark red; Methods).
Overlap of Dispensable Genes with Disease-Causing and Essential Genes.
We then explored the features characterizing the list of 168 putatively dispensable genes, and searched for those previously shown to be 1) associated with Mendelian diseases (n = 3,622, Online Mendelian Inheritance in Man database [OMIM]) (33), or 2) essential in human cell lines (n = 1,920) or knockout mice (n = 3,246) (34) (Methods). We focused on LoF variants predicted to be severely damaging, as described above (Fig. 1). We found that three LoF variants from our list affected OMIM genes (CLDN16, TMEM216, GUF1; Table 2), whereas none affected essential genes (Fisher’s exact P value = 7.727e-06 and 6.249e-10 for human cell lines and knockout mice essential genes, respectively). It has been suggested that the common LoF variant rs760754693 of CLDN16—a gene associated with a renal disorder known as familial hypomagnesemia with hypercalciuria and nephrocalcinosis—affects the 5′-untranslated region of the gene rather than its coding sequence (35). A second methionine residue downstream from the affected position in CLDN16 could potentially serve as the actual translation start site. The TMEM216 variant rs10897158 is annotated as benign in ClinVar (36). TMEM216 is required for the assembly and function of cilia, and pathogenic mutations of this gene cause Joubert, Meckel, and related syndromes (37). The canonical transcript of TMEM216 encodes a 145-amino acid protein. The rs10897158 splice variant (global frequency of homozygous individuals of >70%) results in the synthesis of a longer protein (148 amino acids) corresponding to the most prevalent isoform (37), which could probably be considered to be the reference protein in humans. There is currently little evidence to support the third variant, rs141526764 (GUF1), having any pathogenic consequences. The association of GUF1 with Mendelian disease (early infantile epileptic encephalopathy) is reported in OMIM as provisional and based solely on the finding of a homozygous missense variant (A609S) in three siblings born to consanguineous parents (38). For all subsequent analyses, we filtered out the two variants considered highly likely not to be LoF (rs760754693, rs10897158), resulting in a final list of 179 predicted LoF variants corresponding to 166 genes (Table 1). The absence of common LoF variants predicted with a high degree of confidence in disease genes or essential genes is consistent with these genes being dispensable.
Table 2.
Common high-probability LoF variants in OMIM genes
| Gene | rs ID | Consequence | Global allele Frequency (gnomAD) | Percent of homozygous individuals per population (gnomAD) |
| CLDN16 | rs760754693 | Frameshift | 0.194 | AFR (0.68) |
| AMR (1.50) | ||||
| EAS (0.05) | ||||
| EUR (6.07) | ||||
| SAS (3.18) | ||||
| GUF1 | rs141526764 | Splice donor variant | 0.015 | AFR (0.00) |
| AMR (1.35) | ||||
| EAS (0.00) | ||||
| EUR (0.00) | ||||
| SAS (0.00) | ||||
| TMEM216 | rs10897158 | Splice acceptor variant | 0.830 | AFR (12.83) |
| AMR (74.35) | ||||
| EAS (92.64) | ||||
| EUR (72.84) | ||||
| SAS (70.05) |
Features of the Set of Putative Dispensable Protein-Coding Genes.
We characterized the set of 166 putatively dispensable protein-coding genes further by performing a Gene Ontology (GO) enrichment analysis (Methods). We found a significant overrepresentation of genes involved in G protein coupled receptor activity and related GO terms (Fisher’s exact P value = 5.50e-25; Dataset S2). Such enrichment was driven by the presence of 41 olfactory receptor (OR) genes, accounting for 25% of the total set of dispensable genes, consistent with previous analyses (10). We then stratified dispensable genes according to their mutational damage, assessed by calculating the gene damage index (GDI) score (39). The GO enrichment analyses conducted separately for the two sets gave very similar results, driven by the OR genes in both cases (Dataset S2). After the removal of ORs, no additional functional categories were identified as displaying significant enrichment (Dataset S2). Thus, a large number of protein-coding genes may be dispensable, but this has no apparent impact at the level of pathways or functional categories. Based on previous findings and the known specific features of OR genes (40, 41), we opted to consider the 41 OR and 125 non-OR dispensable genes separately in subsequent analyses (Table 1). In comparisons with a reference set of 382 OR, and 19,850 non-OR protein-coding genes (Methods), the coding lengths of the 41 OR (median = 945 nt) and the 125 non-OR (median = 1153.5 nt) dispensable genes were not significantly different from those of the corresponding nondispensable OR (median = 945 nt; Wilcoxon test P value = 0.6848) and non-OR (median = 1,275 nt; Wilcoxon test P value = 0.1263) genes. The genomic distribution of dispensable OR genes displayed some clustering on some chromosomes, but did not differ significantly from that of the reference OR genes (SI Appendix, Fig. S5A). The distribution of dispensable non-OR genes across autosomal chromosomes was also similar to that for the reference set, except that no dispensable genes were present on the X and Y chromosomes (SI Appendix, Fig. S5B). This finding suggests that common LoF variants on sexual chromosomes have been more efficiently purged from the population than autosomal variants, presumably, because they have pathogenic effects in hemizygous males.
Organ Expression Patterns of the Dispensable Protein-Coding Genes.
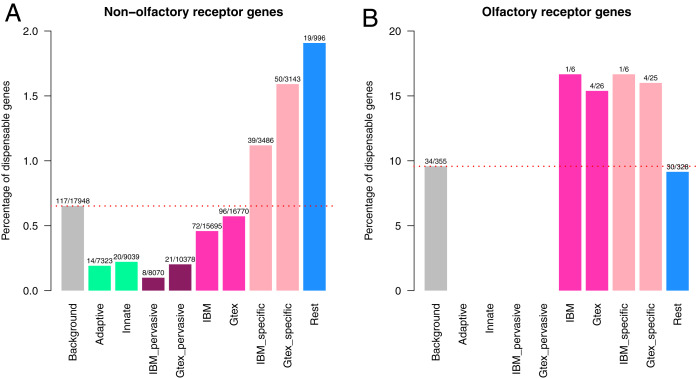
We then investigated the expression patterns of the 166 putatively dispensable genes across organs and leukocyte types. For organs, we used RNA sequencing (RNA-seq) expression data from the Illumina Body Map project (IBM) and the Genotype-Tissue Expression (GTEx) project, and, for leukocytes, we used data from the BluePrint project (Methods and Dataset S3). Most of the dispensable OR (30 of 34, 88%) were not found to be expressed in any of the datasets considered, consistent with the general expression pattern for all OR genes (328 of 355 OR genes not expressed, 92%; Fig. 2). We found that 996 of a reference set of 17,948 non-OR protein-coding genes were not expressed in any of the databases considered (referred to hereafter as nondetected genes). Interestingly, the nondetected genes displayed a significant enrichment in dispensable genes relative to the reference set: odds ratio (OR), 3.34; 95% CI, 1.92 to 5.53; Fisher’s exact test P value, 2.29e-05 (Fig. 2). A similar pattern was observed for organ-specific genes, which were defined as genes expressed in <20% of the organs evaluated in the corresponding dataset: OR of 2.09 (CI 1.38 to 3.11) for IBM, P value = 3.67e-04, and OR of 3.56 (CI 2.41 to 5.22) for GTEx, P value = 1.53e-10. We further characterized the distribution of non-OR dispensable genes among the organ-specific genes for the various organs evaluated. Consistent with previous observations (12), the brain appeared to be the only organ in which the proportion of dispensable genes was significantly lower than that observed for organ-specific genes in both the IBM and the GTEx datasets: OR of 0.23 (CI 0.04 to 0.72), P value = 5.47e-03, and OR of 0.08 (CI 0.002 to 0.45), P value = 2.69e-04, respectively (Dataset S4). Organ-pervasive genes were also found to be significantly depleted of dispensable genes, reflecting a lower degree of redundancy: OR of 0.09 (CI 0.04 to 0.18), P value = 6.47e-20 for IBM, and OR of 0.16 (CI 0.09 to 0.26), P value = 8.04e-19 for GTEx. The number of organs in which a gene is expressed was consistently, and negatively associated with the proportion of dispensable genes (linear regression P values after adjustment for coding sequence length < 2e-16 for both IBM and GTEx). Overall, genes widely expressed or specifically expressed in the brain are less dispensable than those with a more restricted pattern of expression.
Fig. 2.
Distribution of the set of dispensable genes in organ-expressed genes and adaptive and innate leukocytes-expressed genes, defined from gene expression datasets. Relative enrichment in dispensable genes among different gene sets defined from expression datasets based on RNA-seq expression data from IBM, the GTEx project, and the BluePrint project (adaptive and innate leukocytes). Organ-expressed genes are further classified into two subcategories: organ-specific and organ-pervasive genes (see text). The ratio of dispensable genes versus the total size of each category is indicated. Results are presented separately for (A) 17,948 non-OR genes and (B) 355 OR genes for which expression data could be retrieved, from an initial list of 20,232 protein-coding genes (Methods). P values for two-tailed Fisher’s exact tests comparing the fraction of dispensable genes among the gene subsets against the reference background are reported in text.
Expression Patterns for Dispensable Genes in Leukocytes.
We then investigated the expression patterns of the 166 putatively dispensable genes in leukocytes. Human immune genes were classified on the basis of the RNA-seq data generated by the BluePrint project (42). We identified 7,323 adaptive leukocyte-expressed genes on the basis of their expression in B or T cells in at least 20% of the samples considered, and 9,039 innate leukocyte-expressed genes defined on the basis of their expression in macrophages, monocytes, neutrophils, or dendritic cells (DC) in at least 20% of the samples considered (Dataset S3). We are aware that the main function of DCs is to present antigens to T cells, making their classification as “innate” both arbitrary and questionable. These leukocyte-expressed genes included no OR genes, and all of the results therefore correspond to non-OR genes. A significant depletion of dispensable non-OR genes relative to the reference set was observed among the genes expressed in adaptive and innate leukocytes: OR of 0.20 (95% CI 0.10 to 0.34), P value = 8.80e-12, and OR of 0.20 (CI 0.12 to 0.33), P value = 9.98e-14, respectively (Fig. 2). In total, 24 dispensable genes were identified as expressed in adaptive leukocytes (n = 4 genes), innate leukocytes (n = 10), or both (n = 10). Detailed information about the common homozygous LoF variants of these genes is presented in Dataset S5. Sixteen of these 24 genes had variants predicted to have highly damaging consequences, including a well-known stop-gain variant of TLR5 (43). This truncating TLR5 variant, which abolishes cellular responses to flagellin, appears to have evolved under neutrality (44). These genes also included APOL3, which is known to be involved in the response to infection with African trypanosomes (45). Thus, genes widely expressed in leukocytes are generally less dispensable than the reference set. However, specific immune-related genes may become LoF tolerant due to functional redundancy (e.g., TLR5) or positive selection, by increasing protective immunity, for example. This aspect is considered in a subsequent section.
Population Distribution of the Dispensable Genes.
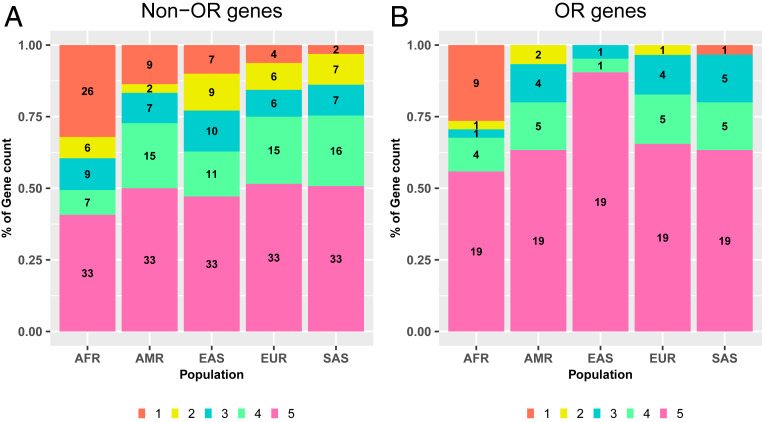
We analyzed the distribution of the dispensable genes across the five specific populations considered: AFR (including African Americans), EAS, SAS, EUR (including Finnish), and AMR (Fig. 3). Of the 125 non-OR genes, we found that 33 were dispensable in all populations, 16 were dispensable in four populations, and 13 were dispensable in three populations (homozygous LoF frequency >1% in each population). Conversely, 48 genes were population specific, with Africans providing the largest fraction in both absolute (26 genes) and relative terms, as a reflection of their greater genetic diversity (46). The remaining 15 genes were found in two populations. Almost half the 41 dispensable OR were common to all five populations (n = 19), 10 were present in four or three populations, two were present in two populations, and 10 were population specific (including nine in Africans). As expected, the number of populations in which a gene was found to be dispensable correlated with the maximum frequency of homozygous individuals in the populations concerned (SI Appendix, Fig. S6). Overall, 49% of the non-OR and 71% of the OR dispensable genes (>90% of the dispensable OR genes in non-African populations) were common to at least three human populations, suggesting a general process of tolerance to gene loss independent of the genomic background of the population.
Fig. 3.
Distribution of dispensable genes across the five human populations considered. The numbers of (A) dispensable non-OR genes and (B) dispensable OR genes in each population (>1% homozygous LoF frequency in a given population) are represented across five categories, indicating whether the gene is dispensable in all five populations considered, or four, three, or two of them, or is a population-specific dispensable gene. The five populations considered were AFR, AMR, EAS, EUR, and SAS. The homozygous LoF variant frequencies were taken from the gnomAD dataset for the purposes of this analysis.
Negative Selection of Dispensable Genes.
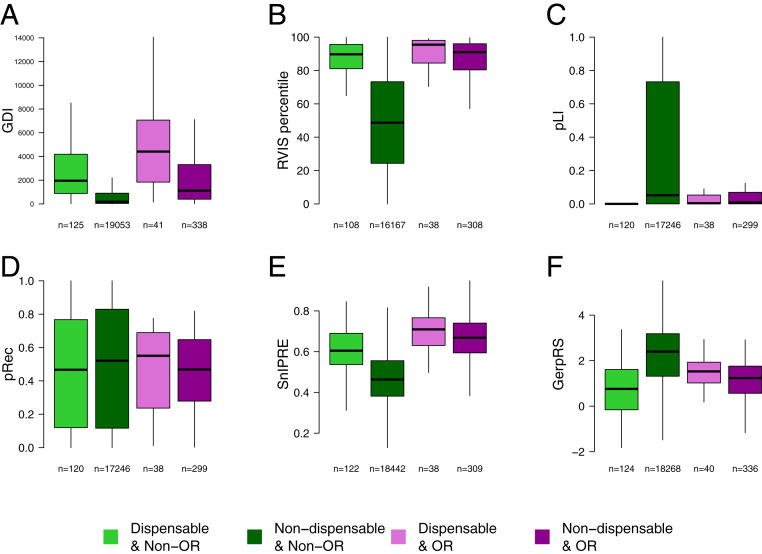
We then investigated the behavior of dispensable genes in terms of the functional scores associated with gene essentiality and selective constraints (Fig. 4). We first evaluated a metric that was designed to assess the mutational damage amassed by a gene in the general population [GDI (39)] and three gene-level scores that measure the extent of recent and ongoing negative selection in humans [residual variation intolerance score (RVIS) (47), gene probability of LoF intolerance (pLI), and The Probability of being intolerant to homozygous LoF variants (pRec) (13); Methods]. Consistent with expectations, dispensable non-OR genes had much higher GDI (two-tailed Wilcoxon test P value = 6.52e-35), higher RVIS (P value = 1.30e-37), and lower pLI (P value = 1.548597e-22) values than nondispensable non-OR genes (Fig. 4), whereas no significant differences were found for pRec values (P value = 0.29). In analyses focusing on OR genes, GDI and RVIS values were also significantly higher for dispensable genes than for nondispensable genes (P values = 3.08e-06 and 1.96e-02, respectively), whereas no differences were found for the pLI and pRec distributions (P value > 0.05). When restricting the assessment of the GDI score to missense variants, the same trends where observed: P value = 1.24e-15 and 1.44e-02, for non-OR and OR comparisons, respectively. We then evaluated the strength of negative selection at a deeper evolutionary level, using two interspecies conservation scores: the estimated proportion f of nonlethal nonsynonymous mutations (48), which was obtained with the selection inference using a Poisson random effects model (SnIPRE), by comparing polymorphism within humans and divergence between humans and chimpanzee at synonymous and nonsynonymous sites (49), and the genomic evolutionary rate profiling rejected substitutions (GerpRS) conservation score, obtained from alignments of sequences from multiple mammalian species (excluding humans) (50). Neither the f nor the GerpRS values obtained differed significantly (P value > 0.01) between dispensable and nondispensable OR genes (Fig. 4). However, dispensable non-OR genes had higher f and GerpRS values than nondispensable non-OR genes (P value = 1.51e-29 and = 9.70e-28, respectively), indicating that dispensable non-OR genes were more tolerant to nonsynonymous variants than nondispensable non-OR genes. Dispensable genes also had more human paralogs than other protein-coding genes (P value = 8.76e-06), suggesting a higher degree of redundancy. For OR genes, the number of paralogs did not differ between dispensable and nondispensable genes, further confirming that the negative selection parameters of dispensable and nondispensable genes OR genes were similar. Overall, these results reveal a relaxation of the selective constraints acting at dispensable non-OR gene loci relative to nondispensable genes, providing further evidence for evolutionary dispensability.
Fig. 4.
Distribution of functional scores relating to gene essentiality/redundancy in the stringent set of dispensable genes. Score distributions presented include (A) GDI; (B) RVIS; (C) pLI; (D) pRec; (E) proportion of nonlethal nonsynonymous mutations, f, estimated by SnIPRE; and (F) interspecies conservation, estimated by GerpRS (Methods). Panels display the distribution of scores across dispensable non-OR genes (light green), nondispensable non-OR genes (dark green), dispensable OR genes (light purple), and nondispensable OR genes (dark purple). Two-tailed Wilcoxon test P values comparing the distribution of dispensable non-OR genes to that of nondispensable non-OR genes are (A) P value = 6.52e-35, (B) P value = 1.30e-37, (C) P value = 1.55e-22, (D) P value = 2.94e-01, (E) P value = 1.51e-29, and (F) P value = 9.70e-28. Two-tailed Wilcoxon test P values comparing the distribution of dispensable OR genes to that of nondispensable OR genes are (A) P value = 3.04e-06, (B) P value = 1.96e-02, (C) P value = 4.64e-01, (D) P value = 7.98e-01, (E) P value = 6.81e-02, and (F) P value = 1.85e-02.
Positive Selection of Common LoF Variants.
We investigated the possibility that the higher frequency of some LoF mutations was due to a selective advantage conferred by gene loss (i.e., the “less-is-more” hypothesis), by searching for population-specific signatures of positive selection acting on these variants (18, 19). We considered two neutrality statistics: FST, which measures between-population differences in allele frequencies at a given locus (51), and integrated haplotype score (iHS) (52), which compares the extent of haplotype homozygosity around the ancestral and derived alleles in a given population. Both statistics could be computed for 68 variants fulfilling the quality control criteria (Methods). Considering a cutoff point of the 95th genome-wide percentile for each statistic (Fig. 5), we detected 39 common LoF alleles in putatively dispensable genes displaying signals suggestive of positive selection, as attested by their low iHS (n = 7), high FST values (n = 24), or both (n = 8; Dataset S6). Seven of these variants in OR genes had only high FST values, suggestive of genetic drift related to the ongoing pseudogenization of ORs (41). We also noted that the LoF mutation (rs2039381) of IFNE displayed significant levels of population differentiation (e.g., FST = 0.25 for Gujarati Indian from Houston [GIH] vs. Utah residents with European ancestry [CEU], Pemp = 0.002). This result is intriguing, as IFNE encodes IFNε, which plays an important role in protective immunity against microbes in the female reproductive tract in mice (53, 54). This nonsense variant (Q71X) is predicted to decrease the length of the encoded protein by two-thirds, but this has not been validated experimentally. The proportion of homozygotes is highest in East Asia (3.5%) and South Asia (2%), is much lower in Europe (0.02%), and does not differ between males and females. The iHS scores were not significant for this variant (SI Appendix, Fig. S7), and neither were other selection scores, such as Tajima’s D, Fu and Li’s D* and F *, and Fay and Wu’s H, previously obtained in a large evolutionary genetic study of human interferons (55). In light of these observations, the most likely explanation for the high FST observed at this locus is genetic drift.
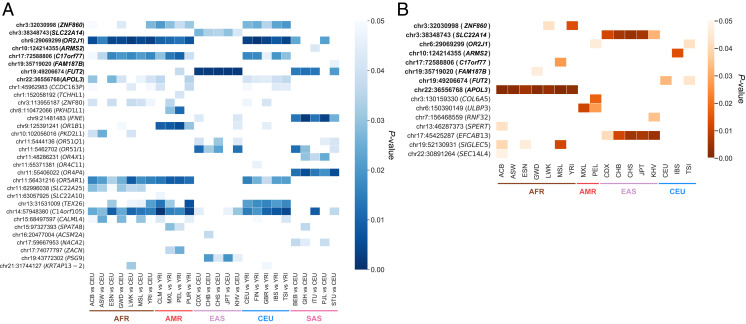
Fig. 5.
Evidence for positive selection on common LoF alleles. (A) Empirical P values for 32 LoF mutations presenting FST scores measuring allele frequency differentiation, in the 95th percentile of highest values genome wide, in at least one population. (B) Empirical P values for 15 LoF mutations presenting iHS below the fifth percentile of the lowest values genome wide (i.e., selection on the LoF allele), in at least one population. FST and iHS values are reported for each of the 26 populations of the 1000 Genomes Project, grouped into five super populations: AFR (brown), AMR (orange), EAS (purple), EUR (blue), and SAS (pink). Eight common LoF variants with both high FST and low iHS are highlighted. The color gradients indicate the significance of the P values, and only polymorphic sites involving common biallelic LoF SNPs from the stringent set predicted to have severe damaging functional consequences were considered.
LoF Mutations of FUT2 and APOL3 Are under Positive Selection.
Eight common LoF variants provided more robust evidence for positive selection, as they had both a high FST and a low iHS (Dataset S6 and Fig. 5). For five of these variants, there was no obvious relationship between the gene concerned and a possible selective advantage. However, one variant with a high FST (FST = 0.25 for Indian Telugu from UK [ITU] vs. Han Chinese in Beijing [CHB]), Pemp = 0.012) and a low iHS (iHS = −2.33, Pemp = 0.004 in Southern Han Chinese [CHS]) was located in SLC22A14, which has been shown to be involved in sperm motility and male infertility in mice (56). Finally, the two remaining variants were in the FUT2 and APOL3 genes, which are known to be involved in defense against infections. Consistent with previous observations, we observed high FST values (FST = 0.54 for CEU vs. CHS, Pemp = 0.002) and extended haplotype homozygosity (iHS = −1.7, Pemp = 0.03 in CEU) around the LoF mutation (rs601338) in FUT2 (SI Appendix, Fig. S7). This gene is involved in antigen production in the intestinal mucosa, and null variants are known to lead to the nonsecretor phenotype conferring protection against common enteric viruses, such as norovirus (25, 26), and rotavirus (57, 58). We also identified a hit at the APOL3 LoF variant rs11089781. This nonsense variant (Q58X) was detected only in African populations (15 to 33% frequency), in which it had a low iHS (iHS = −2.75 in Mende in Sierra Leone [MSL], Pemp = 0.001), indicating extended haplotype homozygosity around the LoF mutation in these populations (SI Appendix, Fig. S7). APOL3 is located within a cluster of APOL genes including APOL1. These two members of the six-member APOL cluster, APOL1 and APOL3, are known to be involved in defense against African trypanosomes (45, 59). These analyses indicate that, although positive selection remains rare in humans, it may have increased the frequency of LoF variants when gene loss represents a selective advantage.
Discussion
We identified 166 putatively dispensable human protein-coding genes. Even after manual curation, it is likely that a certain proportion of the variants retained for these genes are not actually LoF, and the availability of experimental validation for only a small fraction of the predicted LoF variants is one of the limitations of this study. It is also likely that additional dispensable genes could be detected in the general population on the basis of common homozygosity for other types of genetic variants abrogating protein function, notably, those for which whole-exome sequencing is not particularly suitable, such as deletions of more than 50 nts (60). The putatively dispensable human protein-coding genes identified included 120 that overlapped either with the total list of 2,641 genes apparently tolerant to homozygous rare LoF variants reported from bottlenecked or consanguineous populations (11, 12, 14, 15) or with the list of 253 genes initially identified from individuals of the 1000 Genomes Project (10) (SI Appendix, Fig. S8), or both. These partly discordant results are probably due to differences in study designs, in particular, in terms of sample sizes, population structures, and frequencies of the LoF variants studied. Most of the 46 genes specific to our study had a maximum homozygote frequency below 0.05 or had a higher frequency in only one or two of the five studied populations, as for FUT2 and IFNE. Our set of dispensable genes was strongly enriched in OR genes, as previously reported (10, 61). However, dispensable OR genes had no particular features distinguishing them from nondispensable OR genes other than slightly higher GDI and RVIS values. This finding is consistent with the notion that the number of functional ORs has decreased during evolution in humans (62), and provides evidence for ongoing pseudogenization (41, 63). Conversely, dispensable non-OR genes displayed a strong relaxation of selective constraints relative to non-OR nondispensable genes at both the interspecies and intraspecies levels. In addition, the set of dispensable non-OR genes was depleted of genes widely expressed in the panel of organs evaluated, brain-specific genes and genes expressed in leukocytes. This suggests that the redundancy observed for some microbial sensors and effectors (19, 64) does not necessarily translate into higher rates of gene dispensability.
The set of dispensable genes identified here probably largely corresponds to genes undergoing pseudogenization (65, 66) due to present-day superfluous molecular function at the cell, organ, or organism levels, or a redundant function in the genome that may be recovered (e.g., by paralogous genes or alternative pathways). This is consistent with the observation that most of the dispensable genes were common to at least three human populations. The overlap was particularly large for OR genes, which are strongly enriched in dispensable genes, and generally present a strong relaxation of selective constraints and signs of ongoing pseudogenization (41). Another example is provided by TLR5, encoding a cell surface receptor for bacterial flagellin, which harbors a dominant negative stop mutation at high population frequencies (43, 44, 67). This finding is consistent with the notion that a substantial proportion of modern-day humans can survive with complete TLR5 deficiency (43). These observations also suggest that additional mechanisms of flagellin recognition, such as those involving the NAIP-NLRC4 inflammasome (68, 69), may provide sufficient protection in the absence of TLR5. Finally, 45 of our dispensable genes belong to the set of 2,278 Ensembl/GENCODE coding genes recently reported to display features atypical of protein-coding genes (70). Our study may therefore provide additional candidates for inclusion in the list of potential non−protein-coding genes.
High population frequencies of LoF variants may also reflect recent and ongoing processes of positive selection favoring gene loss (i.e., the “less-is-more” paradigm) (18). Two of the eight variants with the most robust signals of positive selection were located in genes involved in resistance to infectious diseases. The FUT2 gene is a well-known example of a gene for which loss is an advantage, as it confers Mendelian resistance to common enteric viruses and has a profile consistent with positive selection. However, nonsecretor status has also been associated with predisposition to Crohn’s disease (71), Behçet’s disease (72), and various bacterial infections (73), including otitis media (74), suggesting an advantage for secretor status. Accordingly, an evolutionary genetics study concluded that FUT2 genetic diversity was compatible with the action of both positive and balancing selection on secretor status (75). An interesting finding from this study is provided by the APOL3 LoF variant (rs11089781), which we found to display signals of recent, positive selection in Africans. APOL3 and APOL1 are known to be involved in the response to African trypanosomes (45). Two variants encoding APOL1 proteins with enhanced trypanolytic activity are present only in African populations, in which they harbor signatures of positive selection despite increasing the risk of kidney disease (76). Interestingly, the APOL3 LoF variant was also recently associated with nephropathy independently of the effect of the two late-onset kidney disease-risk APOL1 variants, which are not in strong linkage disequilibrium with rs11089781 (77). A physical interaction occurs between APOL1 and APOL3 (77), and APOL1 may protect against pathogens more effectively when not bound to APOL3. Similar mechanisms may, therefore, be involved in the positive selection of the APOL1 kidney disease-risk alleles and the APOL3 LoF variant in African populations. Additional common LoF homozygotes could probably be further identified in other unstudied populations, or involving variants not currently predicted to be LoF in silico. Improvements in the high-confidence identification of dispensable genes will make it possible to identify biological functions and mechanisms that are, at least nowadays, redundant, or possibly even advantageous, for human survival.
Methods
Exome Sequencing Data.
Human genetic variants from the ExAC database (13) (https://console.cloud.google.com/storage/browser/gnomad-public/legacy) were downloaded on 9 September 2016, release 0.3.1, non-TCGA subset. Variants from Exome data of gnomAD (https://gnomad.broadinstitute.org/) were obtained from release 2.0.1 of 27 February 2017. For consistency with ExAC and gnomAD pipelines, variants were annotated with the Ensembl Variant Effect Predictor VEP (78) (v81 for ExAC, v85 for gnomAD), with LoF annotations from LOFTEE (13) (https://github.com/konradjk/loftee). Homo sapiens genome build GRCh37/hg19 was used with both databases. Analyses were restricted throughout this study to a background set of 20,232 human protein-coding genes obtained from BioMart Ensembl 75, version Feb 2014 (GRCh37.p13; ref. 79). Protein-coding genes were defined as those containing an open reading frame. By contrast, pseudogenes were typically defined as gene losses resulting from fixations of null alleles that occurred in the human lineage after a speciation event; some of them may actually be human specific, that is, fixed after the human−chimpanzee divergence. However, the definition might be larger, including the so-called processed pseudogenes, corresponding to DNA sequences reverse-transcribed from RNA and randomly inserted into the genome (65, 66). A total of 20,232 protein-coding genes and 13,921 pseudogenes were reported by Ensembl following the Ensembl Genebuild workflow incorporating the HAVANA group manual annotations (27). Among the protein-coding genes, 382 genes were identified with a gene name starting with “Olfactory receptor” (Dataset S7).
VEP annotations were done against the set of Ensembl Transcript IDs associated with Ensembl protein-coding genes, focusing on the canonical transcript as described in the ExAC flagship paper (13). This was done through VEP option “–canonical”, which provides the variant consequence for what is considered to be the “canonical transcript” of a gene. As reported in ref. 80 and further detailed in Ensembl documentation (http://www.ensembl.org/info/website/glossary.html) for human, the canonical transcript for a gene is set according to the following hierarchy: 1) Longest CCDS translation with no stop codons. 2) If no item 1, choose the longest Ensembl/HAVANA merged translation with no stop codons. 3) If no item 2, choose the longest translation with no stop codons. 4) If no translation, choose the longest non−protein-coding transcript. However, as acknowledged in the Ensembl documentation, the canonical transcript does not necessarily reflect the most biologically relevant transcript of a gene. For the purpose of this study, we thus further focused on variants affecting canonical transcripts when these are considered, in turn, the “principal isoform” of the associated gene, that is, its most functionally important transcript. To that aim, we used transcript annotations from the APPRIS database (29) (downloaded on 2 March 2017, using Gencode19/Ensembl74). APPRIS is a system to annotate principal isoforms based on a range of computational methods evaluating structural information, presence of functionally important residues, and conservation evidence from cross-species alignments.
LoF Variants and Definition of the Set of Dispensable Genes.
LoF variants were considered here as those predicted to lead to an early stop-gain, indel frameshift, or essential splice site disruption (i.e., splice site donor and splice site acceptor variants). Following the criteria used in the ExAC flagship paper (13), only variants with a genotype quality ≥ 20, depth ≥ 10, and a call rate > 80%, mapped to canonical isoforms and labeled as “high-confidence” LoF variants by LOFTEE, were retained. LOFTEE (13) (https://github.com/konradjk/loftee) is a VEP plugin allowing flagging and filtering of variants according to quality control criteria characteristic of falsely considered LoF variants. Thus, the following low-confidence LoF variants flagged by LOFTEE were filtered out: variants for which the purported LoF allele is the ancestral state (across primates); stop-gain and frameshift variants in the last 5% of the transcript, or in an exon with noncanonical splice sites around it (i.e., intron does not start with GT and end with AG); and splice site variants in small introns (<15 bp), in an intron with a noncanonical splice site or rescued by nearby in-frame splice sites. In a previous work, we showed that features associated with low-confidence LoF variants are enriched in common (AF > 5%) and in homozygous LoF in the general population (5). Consistent with such observations, putatively false LoF variants filtered by LOFTEE are enriched in common variants (81), and would confound the detection of dispensable genes as defined above unless they are filtered out. Following https://macarthurlab.org/2016/03/17/reproduce-all-the-figures-a-users-guide-to-exac-part-2/, variants in the top 10 most multiallelic kilobases of the human genome were filtered out, that is, Chr14:106,330,000 to 106,331,000; Chr2:89,160,000 to 89,161,000; Chr14:106,329,000 to 106,330,000; Chr14:107,178,000 to 107,179,000; Chr17:18,967,000 to 18,968,000; Chr22:23,223,000 to 23,224,000; Chr1:152,975,000 to 152,976,000; Chr2:89,161,000 to 89,162,000; Chr14:107,179,000 to 107,180,000; Chr17:19,091,000 to 19,092,000. For gnomAD variants, a heterozygote genotype allele balance of >0.2 was further required. In addition to the previous criteria, a set of filters for LoF variants was adopted in this work, retaining variants with a VQSR equal to “PASS” both for SNPs and frameshifts and affecting the APPRIS principal isoform (28) as described above. Dispensable genes were defined as those presenting a LoF variant with a frequency of homozygous individuals higher than 1% in at least one of the five main populations considered, that is, AFR (including African Americans), EAS, SAS, EUR (Finnish and non-Finnish), and AMR.
Impact Prediction of LoF Variants.
The messenger RNA region capable of triggering transcript degradation by nonsense mediated decay (NMD) upon an early stop-gain was defined as in a previous study (5), according to HAVANA annotation guidelines (v.20). Specifically, the NMD target region of a transcript was defined as those positions more than 50 nts upstream from the 3′-most exon−exon junction. Transcripts bearing stop-gain variants in these regions are predicted to be degraded by NMD (82). Molecular impact prediction of splice-disrupting variants was performed with Human Splicing Finder (HSF) software (83) (online version 3.1 available at http://www.umd.be/HSF/ with default parameters). HSF classifies splicing variants into five categories: unknown impact, no impact, probably no impact, potential alternation, and most probably affecting variant. LoF variants were classified into those predicted to be LoF with low and high probability. Low-probability LoF arbitrarily include 1) stop-gains and frameshift variants truncating the last 15% of the protein sequence, which might translate into small truncations in the final protein, or mapping into the first 100 nts of the transcript, which has been reported as a window length in which LoF variants are often recovered by means of alternative start site usage (84); and 2) essential splice site variants with an unknown or low computationally predicted impact on splicing motifs based on position weight matrices, maximum entropy, and motif comparison methods (Methods). High-probability LoF variants include 1) stop-gains and frameshifts truncating more than 15% of the protein sequence or occurring in a region prone to transcript degradation by NMD, which probably result in the complete abolition of protein production (Methods); and 2) putative splice site variants with an intermediate or high computationally predicted impact (Methods). All associations of LoF variants reported in the GWAS catalog (version v1.0, date 11 January 2019) were downloaded from ftp://ftp.ebi.ac.uk/pub/databases/gwas/. As for Clinvar annotations, the 2 March 2020 release was used (36).
GO Enrichment Analysis.
GO enrichment analysis was performed with the database for annotation, visualization and integrated discovery (DAVID) functional annotation tool (85). Only terms with a Benjamini-corrected Fisher’s exact test P value < 0.05 were retained.
Gene Expression Patterns across Human Organs and Immune Cell Types.
Two lists of organ- and tissue-expressed genes were defined on the basis of the RNA-seq expression data from IBM (15,688 expressed genes), and the GTEx project (16,762 expressed genes). First, RNA-seq expression data from a panel of 11 human organs and tissues (one sample each) from IBM were extracted from the Expression Atlas database (EBI accession E-MTAB-513; 1 February 2017 release; https://www.ebi.ac.uk/gxa/experiments/E-MTAB-513/Results). The list of organs and tissues included adipose tissue, brain, breast, colon, heart, kidney, liver, lung, ovary, skeletal muscle tissue, and testis. It should be noted that leukocyte, lymph node, adrenal, prostate, and thyroid gland data were removed from these datasets. A total of 15,688 organ-expressed genes were defined as being expressed in more than three transcripts per million (TPM; according to ref. 86) in at least one of the IBM samples considered. Second, RNA-seq expression data from a panel of 24 human organs (multiple samples per organ) from the GTEx project (87) were extracted from https://gtexportal.org/home/tissueSummaryPage (version V6p). Blood, blood vessel, salivary gland, adrenal gland, thyroid, pituitary, and bone marrow data were removed from these datasets. A total of 16,762 organ-expressed genes were defined as being expressed in more than three TPM in at least 20% of the samples from at least one GTEx organ. The organ-expressed set of genes previously defined was further classified into a set of “organ-specific genes” and “organ-pervasive genes,” depending on whether the gene was defined as expressed in <20% (organ-specific) or >80% of the organs and tissue types evaluated in the corresponding dataset (i.e., IBM or GTEx).
In addition, the RNA-seq dataset generated by the BluePrint project (42) was used as a reference set for gene expression for the different immune cell types from human venous blood (19 August 2017 release, data available at http://dcc.blueprint-epigenome.eu/#/files). Only cell types with more than two samples were considered. In total, 85 libraries were retained, including 9 B cell and 17 T cell samples (collectively considered as adaptive immune cell types), and 15 monocyte, 25 macrophage, 6 DC, and 13 neutrophil samples (collectively considered as innate immune cell types). A total of 7,346 adaptive immune cell-expressed genes were detected as those expressed in more than three TPM in B cells or T cells in at least in 20% of the corresponding samples collectively considered. Similarly, 9,069 innate immune cell-expressed genes were defined here as expressed in more than three TPM in macrophages, monocytes, neutrophils, or DCs in at least 20% of the corresponding samples collectively considered. Full details about the libraries used, as provided by the BluePrint project, are reported in Dataset S8. The set of genes not found to be expressed in any of the previous lists was determined from the complement of the reference list of 20,120 protein-coding genes defined as Ensembl Biomart, release 75 (79).
Gene-Level Annotations.
The following gene-level features associated with natural selection were obtained: GDI scores, a gene-level metric of the mutational damage that has accumulated in the general population, based on the combined annotation dependent depletion (CADD) scores, were taken from ref. 39. High GDI values reflect highly damaged genes. The RVIS percentile, provided in ref. 47, assesses the gene’s departure from the mean number of common functional mutations in genes with a similar mutational burden in humans. High RVIS percentiles reflect genes that are highly tolerant to variation. The pLI (13), estimating the depletion of rare and de novo protein-truncating variants relative to expectations derived from a neutral model of de novo variation on ExAC exomes data, and pRec, estimating gene intolerance to two rare and de novo protein-truncating variants (analogous to recessive genes), were obtained from the ExAC Browser (release 0.3.1, ref. 13). The pLI and pRec values close to 1 indicate gene intolerance to heterozygous and homozygous LoF and to homozygous mutations, respectively. “f” values were obtained through SnIPRE (49) as described in ref. 48, based on a comparison of polymorphism and divergence at synonymous and nonsynonymous sites. GerpRS scores (50) were downloaded from http://mendel.stanford.edu/SidowLab/downloads/gerp/. Data on the number of human paralogs for each gene were collected from the online gene essentiality database (OGEE) database (88). Monogenic Mendelian disease genes were obtained as described in Chong et al. (89): OMIM raw data files were downloaded from ref. 33. Phenotype descriptions containing the word “somatic” were flagged as “somatic,” and those containing “risk,” “quantitative trait locus,” “QTL,” “{,” “[,” or “susceptibility to” were flagged as “complex.” Monogenic Mendelian genes were defined as those having a supporting evidence level of three (i.e., the molecular basis of the disease is known) and not having a “somatic” or “complex” flag. (Dataset S7). Genes essential in human cell lines and in knockout mice were obtained from ref. 34 (Dataset S7).
Genome-wide Scans for Recent Positive Selection at LoF Mutations.
We tested for the occurrence of positive selection of LoF mutations, by calculating two neutrality statistics: the interpopulation FST, which identifies loci displaying high levels of variation in allele frequencies between groups of populations (51), and the intrapopulation iHS (52), which compares the extent of haplotype homozygosity at the ancestral and derived alleles. Positive selection analyses were confined to biallelic SNPs found in the 1000 Genomes Project phase 3 data (90), including 2,504 individuals from 26 populations, assigned to five metapopulations and predicted to have severely damaging consequences (SI Appendix, Table S1). Multiallelic SNPs, SNPs not detected in the 1000 Genomes Project, and indel frameshifts were discarded in the positive selection analysis. For FST calculation, we investigated a total of 75 LoF mutations that passed quality filters, and compared the allele frequencies of these variants in 26 populations to the allele frequencies of the same mutations in the European CEU and African Yoruba in Ibadan (YRI) reference populations (SI Appendix, Table S1). More specifically, we compared allele frequencies in populations from AFR, EAS, and SAS metapopulations to allele frequencies in the CEU groups, and allele frequencies in populations from the AMR and EUR metapopulations to allele frequencies in the YRI group. For the detection of candidate variants for positive selection based on FST values, we used an outlier approach and considered LoF mutations presenting FST values located in the top 5% of the distribution of FST genome wide. We identified 32 LoF mutations presenting high FST values in at least one population, in 32 genes (including 8 mutations located in OR genes). For haplotype-based iHS score calculations, we first defined the derived allele state of each SNP based on the 6-EPO alignment, and retained only SNPs with a derived allele frequency between 10% and 90% to maximize the power of iHS to detect selective signals. These additional filters led to a total of 68 LoF mutations to be investigated. We calculated iHS scores in 100-kb windows with custom-generated scripts and normalized values. For the detection of selection events targeting derived alleles, we considered LoF mutations located in the top 5% most negative iHS values genome wide and found a total of 15 LoF mutations with iHS scores in the lowest 5% of the iHS values genome wide in at least one population (including 1 mutation located in an OR gene).
Data and Materials Availability.
All data used in the paper are present in the main text and SI Appendix.
Supplementary Material
Acknowledgments
We thank the Laboratory of Clinical Bioinformatics and both branches of the Laboratory of Human Genetics of Infectious Diseases, Yuval Itan, Sophie Saunier, and Corinne Antignac for helpful discussions and support. The Laboratory of Clinical Bioinformatics was supported by the French National Research Agency (ANR) “Investissements d’Avenir” Program (Grant ANR-10-IAHU-01) and Christian Dior Couture, Dior. The Laboratory of Human Genetics of Infectious Diseases was supported, in part, by grants from ANR under the “Investissements d’Avenir” Program (Grant ANR-10-IAHU-01), the Fondation pour la Recherche Médicale (Equipe FRM EQU201903007798), the St. Giles Foundation, and the Rockefeller University. The laboratory of L.Q.-M. is supported by the Institut Pasteur, the Collège de France, the French Government’s Investissements d’Avenir program, Laboratoires d’Excellence “Integrative Biology of Emerging Infectious Diseases” (Grant ANR-10-LABX-62-IBEID) and “Milieu Intérieur” (Grant ANR-10-LABX-69-22701), and the Fondation pour la Recherche Médicale (Equipe FRM DEQ20180339214). D.N.C. and P.D.S. acknowledge the financial support of Qiagen Inc. through a license agreement with Cardiff University.
Footnotes
Competing interest statement: L.H. coauthored research papers with J.-L.C. in 2017 and with E.P., J.-L.C., L.Q.-M., and L.A. in 2018.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917993117/-/DCSupplemental.
References
- 1.Alkuraya F. S., Human knockout research: New horizons and opportunities. Trends Genet. 31, 108–115 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Narasimhan V. M., Xue Y., Tyler-Smith C., Human knockout carriers: Dead, diseased, healthy, or improved? Trends Mol. Med. 22, 341–351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casanova J.-L., Abel L., Human genetics of infectious diseases: Unique insights into immunological redundancy. Semin. Immunol. 36, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Courtois G. et al., A hypermorphic IkappaBalpha mutation is associated with autosomal dominant anhidrotic ectodermal dysplasia and T cell immunodeficiency. J. Clin. Invest. 112, 1108–1115 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rausell A. et al., Analysis of stop-gain and frameshift variants in human innate immunity genes. PLOS Comput. Biol. 10, e1003757 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vickaryous M. K., Hall B. K., Human cell type diversity, evolution, development, and classification with special reference to cells derived from the neural crest. Biol. Rev. Camb. Philos. Soc. 81, 425–455 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Stenson P. D. et al., The Human Gene Mutation Database: Towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 136, 665–677 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Picard C. et al., International Union of Immunological Societies: 2017 Primary Immunodeficiency Diseases Committee Report on Inborn Errors of Immunity. J. Clin. Immunol. 38, 96–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tangye S. G., et al. , Human Inborn Errors of Immunity: 2019 Update on the Classification from the International Union of Immunological Societies Expert Committee. J. Clin. Immunol. 40, 24–64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacArthur D. G. et al.; 1000 Genomes Project Consortium , A systematic survey of loss-of-function variants in human protein-coding genes. Science 335, 823–828 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim E. T. et al.; Sequencing Initiative Suomi (SISu) Project , Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 10, e1004494 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sulem P. et al., Identification of a large set of rare complete human knockouts. Nat. Genet. 47, 448–452 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Lek M. et al.; Exome Aggregation Consortium , Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narasimhan V. M. et al., Health and population effects of rare gene knockouts in adult humans with related parents. Science 352, 474–477 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saleheen D. et al., Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature 544, 235–239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nowak M. A., Boerlijst M. C., Cooke J., Smith J. M., Evolution of genetic redundancy. Nature 388, 167–171 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Olson M. V., When less is more: Gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 64, 18–23 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barreiro L. B., Quintana-Murci L., From evolutionary genetics to human immunology: How selection shapes host defence genes. Nat. Rev. Genet. 11, 17–30 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Quintana-Murci L., Clark A. G., Population genetic tools for dissecting innate immunity in humans. Nat. Rev. Immunol. 13, 280–293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller L. H., Mason S. J., Clyde D. F., McGinniss M. H., The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N. Engl. J. Med. 295, 302–304 (1976). [DOI] [PubMed] [Google Scholar]
- 21.Tournamille C., Colin Y., Cartron J. P., Le Van Kim C., Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat. Genet. 10, 224–228 (1995). [DOI] [PubMed] [Google Scholar]
- 22.Samson M. et al., Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382, 722–725 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Liu R. et al., Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86, 367–377 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Dean M. et al., Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 273, 1856–1862 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Lindesmith L. et al., Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 9, 548–553 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Thorven M. et al., A homozygous nonsense mutation (428G–>A) in the human secretor (FUT2) gene provides resistance to symptomatic norovirus (GGII) infections. J. Virol. 79, 15351–15355 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aken B. L. et al., The Ensembl gene annotation system. Database (Oxford) 2016, baw093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez J. M. et al., APPRIS: Annotation of principal and alternative splice isoforms. Nucleic Acids Res. 41, D110–D117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brandt D. Y. C. et al., Mapping bias overestimates reference allele frequencies at the HLA genes in the 1000 Genomes Project phase I data. G3 (Bethesda) 5, 931–941 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherry S. T. et al., dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buniello A. et al., The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabatini L. M., Azen E. A., Two coding change mutations in the HIS2(2) allele characterize the salivary histatin 3-2 protein variant. Hum. Mutat. 4, 12–19 (1994). [DOI] [PubMed] [Google Scholar]
- 33.McKusick-Nathans Institute of Genetic Medicine , Johns Hopkins University, Online Mendelian Inheritance in Man, OMIM. https://omim.org/. Accessed 6 October 2019.
- 34.Bartha I., di Iulio J., Venter J. C., Telenti A., Human gene essentiality. Nat. Rev. Genet. 19, 51–62 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Weber S. et al., Novel paracellin-1 mutations in 25 families with familial hypomagnesemia with hypercalciuria and nephrocalcinosis. J. Am. Soc. Nephrol. 12, 1872–1881 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Landrum M. J. et al., ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 44, D862–D868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valente E. M. et al., Mutations in TMEM216 perturb ciliogenesis and cause Joubert, Meckel and related syndromes. Nat. Genet. 42, 619–625 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alfaiz A. A. et al., West syndrome caused by homozygous variant in the evolutionary conserved gene encoding the mitochondrial elongation factor GUF1. Eur. J. Hum. Genet. 24, 1001–1008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Itan Y. et al., The human gene damage index as a gene-level approach to prioritizing exome variants. Proc. Natl. Acad. Sci. U.S.A. 112, 13615–13620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilad Y., Lancet D., Population differences in the human functional olfactory repertoire. Mol. Biol. Evol. 20, 307–314 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Pierron D., Cortés N. G., Letellier T., Grossman L. I., Current relaxation of selection on the human genome: Tolerance of deleterious mutations on olfactory receptors. Mol. Phylogenet. Evol. 66, 558–564 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Stunnenberg H. G., Hirst M.; International Human Epigenome Consortium , The International Human Epigenome Consortium: A blueprint for scientific collaboration and discovery. Cell 167, 1145–1149 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Casanova J.-L., Abel L., Quintana-Murci L., Human TLRs and IL-1Rs in host defense: Natural insights from evolutionary, epidemiological, and clinical genetics. Annu. Rev. Immunol. 29, 447–491 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Barreiro L. B. et al., Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 5, e1000562 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fontaine F. et al., APOLs with low pH dependence can kill all African trypanosomes. Nat. Microbiol. 2, 1500–1506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nielsen R. et al., Tracing the peopling of the world through genomics. Nature 541, 302–310 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrovski S., Wang Q., Heinzen E. L., Allen A. S., Goldstein D. B., Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 9, e1003709 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deschamps M. et al., Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am. J. Hum. Genet. 98, 5–21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eilertson K. E., Booth J. G., Bustamante C. D., SnIPRE: Selection inference using a Poisson random effects model. PLOS Comput. Biol. 8, e1002806 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davydov E. V. et al., Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLOS Comput. Biol. 6, e1001025 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holsinger K. E., Weir B. S., Genetics in geographically structured populations: Defining, estimating and interpreting FST. Nat. Rev. Genet. 10, 639–650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voight B. F., Kudaravalli S., Wen X., Pritchard J. K., A map of recent positive selection in the human genome. PLoS Biol. 4, e72 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fung K. Y. et al., Interferon-ε protects the female reproductive tract from viral and bacterial infection. Science 339, 1088–1092 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stifter S. A. et al., Defining the distinct, intrinsic properties of the novel type I interferon, IFNε. J. Biol. Chem. 293, 3168–3179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manry J. et al., Evolutionary genetic dissection of human interferons. J. Exp. Med. 208, 2747–2759 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maruyama S. Y. et al., A critical role of solute carrier 22a14 in sperm motility and male fertility in mice. Sci. Rep. 6, 36468 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nordgren J. et al., Both Lewis and secretor status mediate susceptibility to rotavirus infections in a rotavirus genotype-dependent manner. Clin. Infect. Dis. 59, 1567–1573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Payne D. C. et al., Epidemiologic association between FUT2 secretor status and severe rotavirus gastroenteritis in children in the United States. JAMA Pediatr. 169, 1040–1045 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vanhamme L. et al., Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature 422, 83–87 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Shigemizu D. et al., IMSindel: An accurate intermediate-size indel detection tool incorporating de novo assembly and gapped global-local alignment with split read analysis. Sci. Rep. 8, 5608 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alsalem A. B., Halees A. S., Anazi S., Alshamekh S., Alkuraya F. S., Autozygome sequencing expands the horizon of human knockout research and provides novel insights into human phenotypic variation. PLoS Genet. 9, e1004030 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nei M., Niimura Y., Nozawa M., The evolution of animal chemosensory receptor gene repertoires: Roles of chance and necessity. Nat. Rev. Genet. 9, 951–963 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Alonso S., López S., Izagirre N., de la Rúa C., Overdominance in the human genome and olfactory receptor activity. Mol. Biol. Evol. 25, 997–1001 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Nish S., Medzhitov R., Host defense pathways: Role of redundancy and compensation in infectious disease phenotypes. Immunity 34, 629–636 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X., Grus W. E., Zhang J., Gene losses during human origins. PLoS Biol. 4, e52 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim H. L., Igawa T., Kawashima A., Satta Y., Takahata N., Divergence, demography and gene loss along the human lineage. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 2451–2457 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Quach H. et al., Different selective pressures shape the evolution of Toll-like receptors in human and African great ape populations. Hum. Mol. Genet. 22, 4829–4840 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ferwerda B. et al., Human dectin-1 deficiency and mucocutaneous fungal infections. N. Engl. J. Med. 361, 1760–1767 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao Y., Shao F., The NAIP-NLRC4 inflammasome in innate immune detection of bacterial flagellin and type III secretion apparatus. Immunol. Rev. 265, 85–102 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Abascal F. et al., Loose ends: Almost one in five human genes still have unresolved coding status. Nucleic Acids Res. 46, 7070–7084 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McGovern D. P. B. et al.; International IBD Genetics Consortium , Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. Hum. Mol. Genet. 19, 3468–3476 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takeuchi M. et al., Dense genotyping of immune-related loci implicates host responses to microbial exposure in Behçet’s disease susceptibility. Nat. Genet. 49, 438–443 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Le Pendu J., Ruvoën-Clouet N., Kindberg E., Svensson L., Mendelian resistance to human norovirus infections. Semin. Immunol. 18, 375–386 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Santos-Cortez R. L. P. et al.; University of Washington Center for Mendelian Genomics (UWCMG) , FUT2 variants confer susceptibility to familial otitis media. Am. J. Hum. Genet. 103, 679–690 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ferrer-Admetlla A. et al., A natural history of FUT2 polymorphism in humans. Mol. Biol. Evol. 26, 1993–2003 (2009). [DOI] [PubMed] [Google Scholar]
- 76.Genovese G. et al., Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329, 841–845 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Skorecki K. L. et al., A null variant in the apolipoprotein L3 gene is associated with non-diabetic nephropathy. Nephrol. Dial. Transplant. 33, 323–330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McLaren W. et al., The Ensembl variant effect predictor. Genome Biol. 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zerbino D. R. et al., Ensembl 2018. Nucleic Acids Res. 46, D754–D761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hubbard T. J. P. et al., Ensembl 2009. Nucleic Acids Res. 37, D690–D697 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karczewski K. J., et al. , Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioArxiv:10.1101/531210 (30 January 2019).
- 82.Nagy E., Maquat L. E., A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem. Sci. 23, 198–199 (1998). [DOI] [PubMed] [Google Scholar]
- 83.Desmet F.-O. et al., Human splicing finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 37, e67 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lindeboom R. G. H., Supek F., Lehner B., The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat. Genet. 48, 1112–1118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 86.Kin K., Nnamani M. C., Lynch V. J., Michaelides E., Wagner G. P., Cell-type phylogenetics and the origin of endometrial stromal cells. Cell Rep. 10, 1398–1409 (2015). [DOI] [PubMed] [Google Scholar]
- 87.Lonsdale J. et al.; GTEx Consortium , The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen W.-H., Lu G., Chen X., Zhao X.-M., Bork P., OGEE v2: An update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res. 45, D940–D944 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chong J. X. et al.; Centers for Mendelian Genomics , The genetic basis of mendelian phenotypes: Discoveries, challenges, and opportunities. Am. J. Hum. Genet. 97, 199–215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.The 1000 Genomes Project Consortium , A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.