Significance
Bacterial recombineering allows researchers to interrogate microbes by modifying their genomic DNA. Improvements to the efficiency of recombineering have allowed many simultaneous edits to be made at once. Here we describe "serial enrichment for efficient recombineering" (SEER), a method for identifying efficient single-stranded DNA-annealing proteins (SSAPs) in a microbe of interest. We use SEER to identify two SSAPs: 1) CspRecT doubles editing efficiency over Redβ, the state-of-the-art in Escherichia coli recombineering; and 2) PapRecT achieves high efficiency in Pseudomonas aeruginosa, a widely studied human pathogen. We show that these SSAPs work effectively across a broad range of Gammaproteobacteria, demonstrate vastly improved performance in multiplex applications, and provide broad host-range plasmid resources.
Keywords: recombineering, genome engineering, synthetic biology, RecT, MAGE
Abstract
Exploiting bacteriophage-derived homologous recombination processes has enabled precise, multiplex editing of microbial genomes and the construction of billions of customized genetic variants in a single day. The techniques that enable this, multiplex automated genome engineering (MAGE) and directed evolution with random genomic mutations (DIvERGE), are however, currently limited to a handful of microorganisms for which single-stranded DNA-annealing proteins (SSAPs) that promote efficient recombineering have been identified. Thus, to enable genome-scale engineering in new hosts, efficient SSAPs must first be found. Here we introduce a high-throughput method for SSAP discovery that we call “serial enrichment for efficient recombineering” (SEER). By performing SEER in Escherichia coli to screen hundreds of putative SSAPs, we identify highly active variants PapRecT and CspRecT. CspRecT increases the efficiency of single-locus editing to as high as 50% and improves multiplex editing by 5- to 10-fold in E. coli, while PapRecT enables efficient recombineering in Pseudomonas aeruginosa, a concerning human pathogen. CspRecT and PapRecT are also active in other, clinically and biotechnologically relevant enterobacteria. We envision that the deployment of SEER in new species will pave the way toward pooled interrogation of genotype-to-phenotype relationships in previously intractable bacteria.
The increasing accessibility of whole-genome sequencing to microbiologists has created a gap between researchers’ ability to read vs. their ability to write/edit genetic information. As the availability of sequencing data increases, so too does its hypothesis-generating capacity, which motivates the development of genome-editing tools that can be employed to create user-defined genetic variants in a massively parallelizable manner. Currently, most techniques for editing microbial genomes that meet these criteria exploit bacteriophage-derived homologous recombination processes (1–3). Molecular tools derived from phages enable recombineering (recombination-mediated genetic engineering), which uses short homology arms to efficiently direct the integration of double-stranded DNA cassettes and single-stranded DNA (ssDNA) into bacterial genomes (4–6). Improvements to recombineering in Escherichia coli enabled multiplex, genome-scale editing and the construction of billions of genetic variants in a single experiment (7–9). Two of these recombineering-based methods, multiplex automated genome engineering (MAGE) and directed evolution with random genomic mutations (DIvERGE), have been used for a variety of high-value applications (3, 10–14), but at their core they offer the ability to generate populations of bacteria that can contain billions of precisely targeted mutations. However, while these techniques function well in E. coli and some closely related enterobacteria, efforts to reproduce these results in other bacterial species have been sporadic and stymied by low efficiencies (SI Appendix, Table S1) (15–31).
The incorporation of genomic modifications via oligonucleotide annealing at the replication fork, called oligo-mediated recombineering, is the molecular mechanism that drives MAGE and DIvERGE (4, 32, 33). This method was first described in E. coli, and is most commonly promoted by the expression of bet (here referred to as Redβ) from the Red operon of Escherichia phage λ (5, 6, 34). Redβ is an ssDNA-annealing protein (SSAP) whose role in recombineering is to anneal ssDNA to complimentary genomic DNA at the replication fork. Although improvements to recombineering efficiency have been made (7–9), the core protein machinery has remained constant, with Redβ representing the state-of-the-art in E. coli and enterobacterial recombineering. Redβ additionally does not adapt well to use outside of E. coli, displaying host tropism, presumably toward hosts that are targets of infection for Escherichia phage λ. To enable recombineering in organisms in which Redβ does not work efficiently, most often Redβ homologs from prophages of genetically similar bacteria are screened (15, 16, 21, 23, 30), but high levels of recombineering efficiency, as seen in E. coli, remain elusive.
We hypothesized that the identification of an optimal SSAP is currently the limiting factor to improved recombineering efficiency and to developing multiplex genome engineering tools (i.e., MAGE) in new bacterial species. To provide a solution, herein we present a high-throughput method for isolating SSAP homologs that efficiently promote recombineering, which we call “serial enrichment for efficient recombineering” (SEER) (Fig. 1). SEER allowed us to screen two SSAP libraries in a matter of weeks, each of which contains more members than, to our knowledge, the sum total from all previous SSAP-screening efforts (SI Appendix, Table S1). We first used SEER to screen a library of 122 SSAPs from seven different families (RecT, Erf, Sak4, Gp2.5, Sak, Rad52, and RecA), finding that proteins of the RecT and Erf families are the most promising candidates for future screening. In a follow-up screen of 107 RecT variants, we then identified CspRecT (from a Collinsella stercoris phage), which doubles recombineering efficiency in E. coli over Redβ. Next, by focusing on another enriched variant, PapRecT (from a Pseudomonas aeruginosa phage), we demonstrate high efficiency genome editing in its native host P. aeruginosa, which has long lacked good genetic tools. We then broadly profile the activity of CspRecT and PapRecT across diverse Gammaproteobacteria. Following on our successful demonstration in this work, we believe that SEER should be an easily adaptable method for identifying proteins that enable efficient recombineering across a wide range of bacterial species.
Fig. 1.
SEER workflow. The SEER workflow is depicted across the top from left to right, with libraries being first assembled, then moved into a chassis organism, enriched over 3 to 10 cycles for efficient recombineering proteins, and finally analyzed by deep sequencing. This process can then be iterated on by learning from results and making improvements to the library design. The specific selective enrichment strategy that we designed for E. coli is shown in a gray callout bubble. Five successive antibiotic selections were applied to a library of E. coli cells expressing SSAP variants, and after the selective handles were exhausted the plasmid library was extracted and retransformed into the naïve SEER chassis for five further cycles of selection.
Results and Discussion
Identification of SSAPs.
Previous analyses of phylogenetic data suggest that there are seven principal families of phage-derived SSAPs: RecT (Pfam family: PF03837), Erf (Pfam family: PF04404), Rad52 (Pfam family: PF04098), Sak4 (Pfam family: PF13479), Gp2.5 (Pfam family: PF10991), Sak (Pfam family: PF06378), and RecA (Pfam family: PF00154) (18, 35). These have been further grouped into three superfamilies, wherein RecT, Erf, and Sak have been proposed to adopt Rad52-like folds, and Sak4 and RecA cluster together into a Rad51-like superfamily (18). Furthermore, Redβ homologs are best classified as a part of the larger RecT family, and UvsX homologs are best classified in the RecA protein family (36). To generate a library that widely sampled SSAP diversity, we used a hidden Markov model to search metagenomic databases beginning with an ensemble of SSAPs that have demonstrated activity in E. coli (19, 37) (Materials and Methods). A library of 131 proteins was identified and codon-optimized for expression in E. coli, 121 of which were synthesized without error (Dataset S1) and cloned into a plasmid vector with a standard arabinose-inducible expression system. This assembled group of SSAP variants, which we refer to here as the Broad SSAP Library, has members from all seven families of SSAPs (Fig. 2A) and is phylogenetically diverse (Fig. 2B). To facilitate tracking the library over multiple selective cycles, we added a 12-nt barcode 22 nts downstream of the stop codon of each gene (SI Appendix, Fig. S1), which enabled us to identify each SSAP variant by PCR-amplification of the barcoded locus and targeted next-generation sequencing (NGS).
Fig. 2.
The Broad SSAP Library. (A) Circles of various sizes represent the number of variants present in the Broad SSAP Library from different protein families, as categorized by the Pfam database. The seven principle families of SSAPS are grouped into three clusters based on structural and phylogenetic information as proposed by Lopes et al. (18). (B) The phylogenetic distribution of the Broad SSAP Library is represented as vertical bar charts. For each phylogenetic level, any group that represents more than 5% of the total library is called out to the right of the bar. (C) The enrichment of each Broad SSAP Library member is plotted over 10 successive rounds of selection. Enrichment of each library member was calculated by dividing the average frequency across selective replicates by the frequency of the nonselective control. Frequency data are measured by amplification of the barcoded region of the plasmid library and NGS. (D) Total enrichment is plotted for each protein family over the 10 rounds of SEER. (E) Frequency is plotted against enrichment for each Broad SSAP Library member after the 10th round of selection. The bold line is a linear least-squares fit and the dashed lines represent the 95% confidence bounds of the fit. Five candidate proteins, which are shaded in a yellow box, were selected for further characterization, including SR011 and SR016, which are specifically called out. (F) Five top candidates were tested for their efficiency at incorporating a single base pair silent mutation at a nonessential gene, ynfF. Efficiency was read out by NGS. Significance values are indicated for a parametric two-tailed t test between two groups, where ns indicates *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001; ns, not significant.
SEER.
Next, to evaluate libraries of computationally identified SSAP variants, we established a high-throughput assay to select for SSAPs that promote efficient recombineering. We hypothesized that the iterative, serial integration of easily selectable mutations into the bacterial genome, coupled to subsequent allelic selection, could allow us to select SSAP variants en masse, and thus enrich best performers from diverse SSAP libraries (Fig. 1). This method, which we term SEER, proceeds via 1) the identification and cloning of SSAP libraries into expression vectors, 2) transformation of such libraries into an organism-of-interest, 3) enrichment for SSAPs efficient at recombineering, and finally 4) analysis by deep sequencing to read out library composition. Step 3 comprises the successive transformation of oligos, which upon successful integration into the host genome will confer a resistant phenotype, followed by antibiotic selection for the resistant allele across the entire library in liquid culture. SSAP variants that incorporate mutations most effectively will thereby be enriched in the population that survives antibiotic treatment. Finally, if needed, after the selective handles are exhausted, the SSAP-plasmid library can be extracted and retransformed into a naïve chassis for further cycles of enrichment.
To perform the selections efficiently, we engineered an E. coli chassis to both improve recombineering efficiency and to introduce genetic handles with which we could apply selective pressure against nonedited alleles. We used as our parental strain, EcNR2 (3), a derivative of E. coli K-12 MG1655 that has its methyl-directed mismatch repair (MMR) machinery disabled (ΔmutS::cat) to improve recombineering efficiency (see Materials and Methods for details) (7). EcNR2 was modified to 1) improve recombineering efficiency (DnaG Q576A) (8) and 2) introduce genetic handles for antibiotic selection. To this latter aim, stop codons were introduced into both cat (in the mutS locus) and tolC, making the modified strain sensitive to chloramphenicol (CHL) and sodium dodecyl sulfate (SDS). We refer to this modified organism as the “SEER chassis.” Next, to identify selectable markers beyond cat and tolC, and thus allow multiple SEER selection cycles, we tested a group of resistant alleles that we gathered from an analysis of antibiotic resistance literature (see Materials and Methods for details). We found GyrA_S83L to confer robust resistance to ciprofloxacin (CIP), RpoB_S512P to rifampicin (RIF), and RpsL_K43R to streptomycin (STR) (SI Appendix, Fig. S2). We then verified that these three antibiotic selections, in addition to the CHL and SDS selections described above, are orthogonal and that they could be performed serially on our engineered E. coli SEER chassis. This enabled us to run 10 rounds of SEER with only a single extraction and retransformation of the plasmid libraries into the naïve chassis (Fig. 1).
Following the optimization of the SEER workflow, we performed 10 successive cycles of selection on the Broad SSAP Library, representing all major SSAP families, in the E. coli SEER chassis. Four replicate populations were run: One control population underwent induction of protein expression and oligo transformation, but was not subjected to antibiotic selection. The three experimental replicates underwent antibiotic selections in the following order: SDS → STR → CIP → CHL → RIF (Fig. 1C). To track the enrichment of each library member, we amplified a locus that included the 12-nt barcode region after each round of selection and sequenced by NGS (Dataset S1). Clear winners emerged relatively quickly, with top variants increasing in frequency distinctly after only a few rounds of selection (SI Appendix, Fig. S3). The nonselected control population, however, also displayed significant enrichment effects, which indicated that the overexpression of certain SSAP variants may impair fitness. Thus, to normalize for fitness and the cost of protein expression, we next calculated an enrichment score for each library member: We divided the average frequency of each variant within the selected populations by its frequency in the nonselected control population (Fig. 2C). Based on NGS data, we found that the RecT family was the most enriched family by a large margin (85-fold over the nonselected control), followed by the ERF family (18-fold over the nonselected control), and no other SSAP family showed significant enrichment (Fig. 2D). However, it is important to note that our codon-optimized Redβ (SR085 in our library) was not significantly enriched through the 10 rounds of selection. To investigate this issue further, we compared the performance of Redβ expressed off of its wild-type codons against the codon-optimized version that was included in the Broad SSAP Library. This revealed significantly decreased efficiency for the codon-optimized version of Redβ (SI Appendix, Fig. S4), which indicates that codon choice is an important consideration for library design, and that negative results for individual SSAPs from a large screen of this sort do not necessarily indicate that the protein itself is not functional.
Finally, we chose a set of five library members that exhibited both high frequency and enrichment for further analysis (Fig. 2E). We tested their recombineering efficiency against Redβ expressed off of its wild-type codons on the same plasmid system used for the SEER selections. To ensure an accurate measurement we queried the efficiency of each SSAP by NGS after performing a silent, noncoding genetic mutation at a nonessential gene, ynfF (“silent mismatch MAGE oligo 7”; see SI Appendix, Table S2). Broad SSAP Library member SR016, which we introduced earlier as PapRecT (UniParc ID: UPI0001E9E6CB), demonstrated the highest efficiency of recombineering (31 ± 2%) (Fig. 2F).
Screening Diverse RecT Homologs Identifies a Highly Efficient SSAP.
Of the seven principal families of phage-derived SSAPs, our first SEER screen suggested the RecT family (Pfam family: PF03837) as the most abundant source of efficient recombineering proteins for E. coli. Importantly, previous screens have also found efficient SSAPs from the RecT protein family (SI Appendix, Table S1). Therefore, we hypothesized that by screening additional RecT variants, again exploiting the increased throughput of SEER compared to previous efforts, we might discover recombineering proteins further improved over Redβ and PapRecT. To this aim we constructed a second library, identifying a maximally diverse group of 109 RecT variants, 106 of which were synthesized successfully, which we call the Broad RecT Library (see Materials and Methods for more details). Next, as previously described, we performed 10 rounds of SEER selection on the Broad RecT Library (SI Appendix, Fig. S5), and upon plotting frequency against enrichment after the final selection, a clear winner emerged (Fig. 3A). This protein, which we introduced earlier as CspRecT (UniParc ID: UPI0001837D7F), originates from a phage of the gram-positive bacterium C. stercoris.
Fig. 3.
Broad RecT Library and CspRecT. (A) Frequency is plotted against enrichment for each Broad RecT Library member after the 10th round of selection. One candidate protein, CspRecT (shaded box), was the standout winner. In all subsequent panels, Redβ, PapRecT, and CspRecT are compared when expressed from a pORTMAGE-based construct (SI Appendix, Fig. S1) in wild-type MG1655 E. coli. Significance values are indicated for a grouped parametric two-tailed t test, where not significant (ns) indicates P > 0.05 and ****P < 0.0001. Editing efficiency was measured by blue/white screening at the LacZ locus for (B) eight different single-base mismatches (n = 3) and (C) 18-base and 30-base mismatches (n = 3). (D) MAGE editing targeting 1, 5, 10, 15, or 20 genomic loci at once in triplicate, was read out by NGS. The solid lines represent the mean editing efficiency across all targeted loci, while the dashed lines represent the sum of all single-locus efficiencies, which we refer to as aggregate efficiency. (E) A 130-oligo DIvERGE experiment using oligos that were designed to tile four different genomic loci that encode the drug targets of fluoroquinolone antibiotics and are known hotspots for CIP resistance. The oligos contained 1.5% degeneracy at each nucleotide position along their entire length. All 130 oligos were mixed and transformed together into cells (n = 3). CFUs were measured at three different CIP concentrations after plating 1/100th of the final recovery volume, and “nd” is “none detected.”
To maximize the phylogenetic reach and applicability of these new tools, we characterized CspRecT alongside Redβ and PapRecT subcloned into the pORTMAGE plasmid system (SI Appendix, Fig. S1) [CspRecT and PapRecT were cloned in place of Redβ into pORTMAGE311B (38): Addgene accession no. 120418]. This plasmid contains a broad-host-range RSF1010 origin of replication (39), establishes tight regulation of protein expression with an m-toluic acid-inducible expression system (40), and disables MMR by transient overexpression of a dominant-negative mutant of E. coli MutL (EcMutL_E32K). Nyerges et al. (41) found that off-target mutations were greatly reduced when MMR was disabled periodically by expressing EcMutL_E32K only transiently alongside other recombineering proteins, when compared to permanent disruption of MMR with a knockout of MutS. Fewer than three mutations accumulated over 24 rounds of MAGE in E. coli that transiently overexpressed EcMutL_E32K, whereas 84 off-target mutations were seen in a mutS-inactivated strain. Measured with a standard lacZ recombineering assay, wild-type E. coli MG1655 expressing CspRecT off of the pORTMAGE plasmid exhibited editing efficiency of 35 to 51% for various single-base mismatches, averaging 43% or more than double the efficiency of cells expressing Redβ or PapRecT off of the same plasmid system (Fig. 3B). This pORTMAGE plasmid expressing CspRecT we refer to as pORTMAGE-Ec1 (Addgene accession no. 138474). To our knowledge the efficiency of CspRecT single-locus genome editing reported here is unique in significantly exceeding 25%, the theoretical maximum for a single incorporation event (42), implying that editing occurs either at multiple forks or over successive rounds of genome replication. To investigate the background mutation rate of this method, we measured the accumulation of off-target mutations in E. coli MG1655 expressing CspRecT, PapRecT, or Redβ off of the pORTMAGE plasmid. LacZ(−) colonies were whole-genome–sequenced after one round of recombineering for incorporation of a LacZ-inactivating oligo. We found elevated numbers of off-target mutations (P < 0.01 by two-tailed t test) in cells expressing PapRecT and CspRecT when compared with Redβ (Table 1 and SI Appendix, Table S3). This effect deserves further investigation and should be kept in mind for applications in which genome fidelity is essential.
Table 1.
Number of off-target mutations
| Plasmid | n | Off-target mutations per colony |
| pORTMAGE311B (Redβ) | 5 | 1.0 ± 0.7 |
| pORTMAGE312B (PapRecT) | 3 | 3.3 ± 0.6 |
| pORTMAGE-Ec1 (CspRecT) | 4 | 4.0 ± 1.4 |
The number of off-target mutations were calculated from whole genome sequencing of single E. coli MG1655 colonies after successful incorporation of an oligonucleotide that disrupts lacZ through a single nucleotide mutation (LacZ_TT).
As CspRecT displayed high efficiency at editing single sites, we next tested the protein in a variety of more complex genome-editing tasks. For longer strings of consecutive mismatches, which are lower-efficiency events, CspRecT was again about twice as efficient as Redβ. Wild-type E. coli MG1655 expressing CspRecT displayed 6% or 3% efficiency (vs. 3% or 1% for Redβ) for the insertion of oligos conferring 18-bp or 30-bp consecutive mismatches into the lacZ locus, respectively (Fig. 3C). To investigate the performance of CspRecT at complex, highly multiplexed genome-editing tasks, we designed a set of 20 oligos spaced evenly around the E. coli genome, each of which incorporates a single-nucleotide synonymous mutation at a nonessential gene. Next, while expressing Redβ, PapRecT, and CspRecT separately from the corresponding pORTMAGE plasmid in E. coli MG1655, we performed a single cycle of genome editing with equimolar pools of 1, 5, 10, 15, and 20 oligos and assayed editing efficiency at each locus by PCR amplification coupled to targeted NGS. NGS analysis revealed a general trend: As the number of parallel edits grew, the degree of overperformance by CspRecT also grew (Fig. 3D). For example, when making 19 simultaneous edits (1 oligo from the pool of 20 could not be read out due to inconsistencies in allelic amplification), CspRecT averaged 10.0% editing efficiency at all loci, whereas PapRecT averaged 4.0% and Redβ averaged 1.9%. Importantly, despite keeping total oligo concentration fixed across all pools, aggregate editing efficiency increased as more oligos were present in each pool. For example, when using CspRecT with a 19-oligo pool, aggregate editing efficiency was more than 200%, implying that across the total recovered population of E. coli there averaged more than two edits per cell.
Finally, because of CspRecT's efficiency at multiplexed genome editing tasks, we tested its performance in a DIvERGE experiment (13). DIvERGE uses large libraries of soft-randomized oligos that have a low basal error rate at each nucleotide position along their entire sequence to incorporate mutational diversity into a targeted genomic locus. To compare the performance of Redβ, PapRecT, and CspRecT, we performed one round of DIvERGE mutagenesis by simultaneously delivering 130 partially overlapping DIvERGE oligos designed to randomize all four protein subunits of the drug targets of CIP (gyrA, gyrB, parC, and parE) in E. coli MG1655. Following library generation, cells were subjected to 250, 500, and 1,000 ng/mL CIP on Lysogeny-broth (LB) agar plates. Variant libraries that were generated by expressing CspRecT produced more than 10 times as many colonies at 250 ng/mL CIP as Redβ and PapRecT, while at 1,000 ng/mL CIP, which requires the simultaneous acquisition of at least two mutations (usually at gyrA and parC) to confer a resistant phenotype, only the use of CspRecT produced resistant colonies (Fig. 3E). Because gyrA and parC mutations are usually necessary to confer high-level CIP resistance, we performed sequence analysis of gyrA and parC from 11 randomly selected CIP-resistant colonies and found many different mutations, in combinations of up to three, most of which have been described in resistant clinical isolates (13, 43) (SI Appendix, Table S4). In sum, in both MAGE and DIvERGE experiments, which require multiplex editing, CspRecT provided about an order-of-magnitude improvement to editing efficiency over Redβ, the current state-of-the-art recombineering tool.
Improved Genome Editing in Diverse Gammaproteobacteria.
SSAPs frequently show host tropism (15, 25, 26), but there are also indications that within bacterial clades certain SSAPs may function broadly (21, 41, 44). Therefore, we next investigated the functionality of PapRecT and CspRecT in selected Gammaproteobacteria and compared their efficiency to that of Redβ. We chose to focus our efforts on two enterobacterial species: Citrobacter freundii ATCC 8090 and Klebsiella pneumoniae ATCC 10031, along with the more distantly related P. aeruginosa PAO1. Pathogenic isolates of K. pneumoniae and P. aeruginosa are among the most concerning clinical threats due to widespread multidrug resistance (45). In these species, oligo-recombineering–based multiplexed genome editing (i.e., MAGE and DIvERGE) holds the promise of enabling rapid analysis of genotype-to-phenotype relationships and predicting future mechanisms of antimicrobial resistance (13, 46). In contrast, C. freundii is an intriguing biomanufacturing host in which the optimization of metabolic pathways has remained challenging (47, 48).
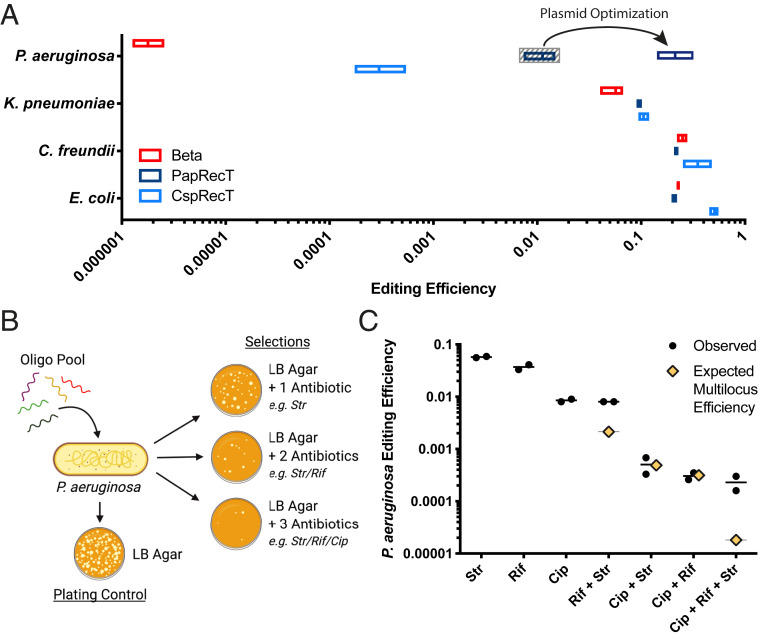
To test the activity of PapRecT and CspRecT in these three organisms, we built on the broad host-range pORTMAGE system (41) described above (SI Appendix, Fig. S1). For experiments in C. freundii and K. pneumoniae we used the same pORTMAGE311B-based plasmid system that we had used in E. coli. In P. aeruginosa the plasmid architecture remained constant, except that we replaced the RSF1010 origin of replication and KAN resistance cassette, instead using the broad host-range pBBR1 origin, which was shown to replicate in P. aeruginosa (49), and a gentamicin resistance marker (SI Appendix, Fig. S1). Next we tested these constructs (see Materials and Methods for details), and in all three species, PapRecT and CspRecT displayed high editing efficiencies (Fig. 4A). In C. freundii and K. pneumoniae, just as in E. coli, we found CspRecT to be the optimal choice of protein, whereas in P. aeruginosa PapRecT performed the best. We further compared PapRecT to two recently reported Pseudomonas putida SSAPs (Rec2 and Ssr) (26, 50), and found that PapRecT, isolated from a large E. coli screen, performed equal to or better than proteins found in smaller screens run through P. putida (SI Appendix, Fig. S6). We found, however, that the efficiency of our plasmid construct was lower in P. aeruginosa than in the enterobacterial species for which pORTMAGE was optimized. Therefore, to increase editing efficiency in P. aeruginosa, we next 1) optimized ribosomal binding sites (RBS) for PapRecT and EcMutL, 2) replaced EcMutL_E32K with its equivalent homologous mutant from P. aeruginosa (PaMutL_E36K), and 3) incorporated the native P. aeruginosa coding sequence for PapRecT instead of the E. coli codon-optimized version (SI Appendix, Fig. S7). Together these changes significantly improved the editing efficiency of our best plasmid construct featuring PapRecT in P. aeruginosa, which we call pORTMAGE-Pa1 (Addgene accession no. 138475), to ∼15%.
Fig. 4.
Recombineering in Gammaproteobacteria. (A) Recombineering experiments were run with Redβ, PapRecT, and CspRecT expressed off of the pORTMAGE311B backbone, or with a pBBR1 origin in the case of P. aeruginosa. Editing efficiency was measured by colony counts on selective vs. nonselective plates (n = 3) (see Materials and Methods). Vector optimization resulted in improved efficiency of PapRecT in P. aeruginosa (SI Appendix, Fig. S7). (B) Diagram of a simple multidrug resistance experiment in P. aeruginosa harboring an optimized PapRecT plasmid expression system, pORTMAGE-Pa1. In a single round of MAGE, a pool of five oligos was used to incorporate genetic modifications that would provide resistance to STR, RIF, and CIP (n = 3). These populations were then selected by plating on all combinations of one-, two-, or three-antibiotic agarose plates and compared with a nonselective control. (C) Observed efficiencies were calculated by comparing colony counts on selective vs. nonselective plates. Expected efficiencies for multilocus events were calculated as the product of all relevant single-locus efficiencies.
Virulent strains of P. aeruginosa are a frequent cause of acute infections in healthy individuals, as well as chronic infections in high-risk patients, such as those suffering from cystic fibrosis (51). The rate of antibiotic resistance in this species is growing, with strains adapting quickly to all clinically applied antibiotics (52, 53). The development of multidrug resistance in P. aeruginosa requires the successive acquisition of multiple mutations, but due to the lack of efficient tools for multiplex genome engineering in P. aeruginosa (54, 55), investigation of these evolutionary trajectories has remained cumbersome. Therefore, and to demonstrate the utility of pORTMAGE-Pa1–based MAGE in P. aeruginosa, we simultaneously incorporated a panel of genomic mutations that individually confer resistance to STR, RIF, and fluoroquinolones (i.e., CIP) (56, 57). Importantly, the corresponding genes are also clinical antibiotic targets in P. aeruginosa (58). Following a single cycle of MAGE delivering five mutation-carrying oligos, a single-day experiment with pORTMAGE-Pa1, we were able to isolate all possible combinations of five resistant mutations, with more than 105 cells from a 1-mL overnight recovery attaining simultaneous resistance to STR, RIF, and CIP (Fig. 4B). Interestingly, because rpsL and rpoB, the resistant loci for STR and RIF, respectively, are located only ∼5 kb apart from each other on the P. aeruginosa genome, these two mutations cosegregated much more often than would be expected by independent inheritance, confirming that coselection functions similarly in P. aeruginosa to E. coli (Fig. 4C) (12). By genotyping and characterizing resistant colonies, we could then easily determine the minimum inhibitory concentration (MIC) of CIP for various resistant genotypes (Table 2 and SI Appendix, Fig. S8). The allure of this method is that the entire workflow took only 3 d to complete, in contrast with other genome engineering methods (i.e., CRISPR/Cas9 or base-editor–based strategies) that are either less effective, have biased mutational spectra, or would require tedious plasmid cloning and cell manipulation steps (54, 55).
Table 2.
Minimum inhibitory concentration
| Genotype | CIP MIC (µg/mL) |
| PAO1 wild-type | 0.25 |
| nfxB knockout | 4 |
| GyrA_T83I | 16 |
| GyrA_T83I + ParC_S87L | 32 |
| GyrA_T83I + ParC_S87L + nfxB knockout | >128 |
MIC was measured for various combinations of CIP resistance-conferring alleles. GyrA_T83I displays strong positive epistasis with ParC_S87L, and so clonal populations with mutations to parC but not gyrA were not pulled out of our antibiotic selection (59).
Conclusion
The discovery and subsequent improvement of recombineering in E. coli enabled the development of advanced genome-engineering techniques—such as MAGE (3), trackable multiplex recombineering (60), replicon excision for enhanced genome engineering through programmed recombination (REXER) (1), retron-recombineering (10, 61), and DIvERGE (13)—and applications ranging from genomic recoding (62), biocontainment (11), and viral resistance (62) to the study and prediction of antibiotic resistance (13) and the improved bioproduction of chemicals (12). These powerful techniques all rely on efficient recombineering, but despite the essential role that an optimal SSAP plays, no method had hitherto been developed to rapidly and efficiently screen complex libraries of SSAPs. Here we describe such a method that we term SEER, a technique for isolating efficient SSAPs from a large number of in silico-identified candidates, in which successive rounds of recombineering are followed by allelic selection (Fig. 1). In the present work we have used SEER to screen two large libraries of SSAPs through E. coli and demonstrated the power of this method by isolating a variant, CspRecT that doubles the already high single-locus efficiency in E. coli to around 50%, a standout number and the highest reported gene-editing efficiency of any system that is not nuclease-dependent that we are aware of. We also demonstrate that CspRecT radically improves the multiplexability of oligo recombineering in E. coli, showing 5- to 10-fold improvement over Redβ with methods that rely on multiplex editing such as MAGE and DIvERGE.
Beyond the success of SEER in E. coli, the true promise of the technique is to increase the number of bacterial species in which these powerful genome-engineering tools are available to researchers. To this end we demonstrate that SEER can discover recombineering proteins that work not only in the targeted organism, but in closely related species as well. We show that proteins enriched in two SSAP libraries that were screened through E. coli meet or exceed the efficiency of the best tools available in C. freundii, K. pneumoniae, and P. aeruginosa. We report single-locus editing efficiencies of over 10% in each of these clinically and biotechnologically relevant Gammaproteobacteria, which should readily enable multiplex genome engineering.
However, it is also important to highlight the current limitations to the scope of this work. To expand modern genome-engineering tools to bacteria that are only distantly related to E. coli, SEER will need to be performed in new host organisms in the future. To this end, we detail three important considerations for constructing SSAP libraries for future studies: 1) Based on our screens in E. coli, and in concert with previous efforts (SI Appendix, Table S1), the recT and ERF families are the most likely sources of SSAPs to drive efficient recombineering; 2) phylogenetic diversity is essential, as CspRecT, the best-performing SSAP in E. coli, originates from a phage of C. stercoris, a gram-positive coriobacterium that is phylogenetically distant to E. coli; and 3) future SSAP screening efforts should give careful consideration to codon optimization, as codon choice can have a large impact on apparent recombineering efficiency (as we observed with both Redβ and PapRecT). In short, SEER holds the promise of being a universal screening method that allows for the identification of highly active SSAPs in virtually any target bacterium, with the potential to broaden the applicability of recombineering-based genome-engineering tools toward as yet genetically intractable microorganisms.
Materials and Methods
Strains and Reagents.
All strains used in this study are listed in SI Appendix, Table S5. Unless otherwise noted, bacterial cultures were grown in Lysogeny-Broth-Lennox (LBL) (10 g tryptone, 5 g yeast extract, 5 g NaCl in 1 L H2O). Super optimal broth with catabolite repression (SOC) was used for recovery after electroporation. MacConkey agar (17 g pancreatic digest of gelatin, 3 g peptone, 10 g lactose, 1.5 g bile salt, 5 g NaCl, 13.5 g agar, 0.03 g neutral red, 0.001 g Crystal violet in 1 L H2O) and isfopropyl-β-d-thiogalactopyranoside (IPTG)-X-gal Mueller-Hinton II agar (3 g beef extract, 17.5 g acid hydrolysate of casein, 1.5 g starch, 13.5 g agar in 1 L H2O, supplemented with 40 mg/L X-gal and 0.2 μM IPTG) were used to differentiate LacZ(+) and LacZ(−) mutants. Cation-adjusted Mueller Hinton II Broth (MHBII) was used for antimicrobial susceptibility tests. Antibiotics were ordered from Sigma-Aldrich. Recombineering oligos were synthesized by Integrated DNA Technologies (IDT) or by the DNA Synthesis Laboratory of the Biological Research Centre (Szeged, Hungary) with standard desalting as purification.
Oligo-Mediated Recombineering.
Bacterial cultures (E. coli, K. pneumoniae, C. freundii, or P. aeruginosa) were grown in LBL at 37 °C in a rotating drum. Overnight cultures were diluted 1:100, grown for 60 min or until OD600 ∼ 0.3, whereupon expression of SSAPs was induced for 30 min with 0.2% arabinose or 1 mM m-toluic acid as appropriate. Cells were then prepared for transformation. Briefly, E. coli, K. pneumoniae, and C. freundii cells were put on ice for approximately 10 min, washed three times with cold water, and resuspended in 1/100th culture volume of cold water. This same procedure was followed for P. aeruginosa with the following differences: 1) Resuspension buffer (0.5 M sucrose + 10% glycerol) was used in place of water and 2) there was no preincubation on ice, as competent cell preparation was carried out at room temperature, which we found to be much more efficient than preparation at 4 °C. After competent cell preparation, 9 µL of 100 µM oligo was added to 81 µL of prepared cells for a final oligo concentration of 10 µM in the transformation mixture (2.5 µM final oligo concentration was used for C. freundii and K. pneumoniae). This mixture was transferred to an electroporation cuvette with a 0.1-cm gap and electroporated immediately on a Gene Pulser (Bio-Rad) with the following settings: 1.8 kV (2.2 kV in the case of P. aeruginosa), 200 Ω, 25 µF. Cultures were recovered with SOC media for 1 h and then 4 mL of LB with 1.25× selective antibiotic and 1.25× antibiotic for plasmid maintenance were added for outgrowth.
Engineering of SEER Chassis.
The E. coli strain described in this work as the SEER chassis is engineered from EcNR2 (3). EcNR2 harbors a small piece of λ-phage integrated at the bioAB locus, which allows expression of λ-Red genes, and a knockout of the methyl-directed MMR gene mutS, which improves the efficiency of mismatch inheritance (MG1655 ΔmutS::cat Δ(ybhB-bioAB)::[λcI857 Δ(cro-orf206b)::tetR-bla]). Modifications made to EcNR2 to engineer the SEER chassis include: 1) Improvement of MAGE efficiency by mutating DNA primase (dnaG_Q576A) (8), 2) introduction of a handle for SDS selection (tolC_STOP), 3) introduction of a handle for CHL selection (mutS::cat_STOP), and 4) removal of λ phage with a zeocin resistance marker Δ[λcI857 Δ(cro- orf206b)::tetR-bla]::zeoR. The final strain, which we refer to as the SEER chassis, is therefore: MG1655 Δ(ybhB-bioAB)::zeoR ΔmutS::cat_STOP tolC_STOP dnaG_Q576A.
Selective Allele Testing in the SEER Chassis.
To complement the SEER chassis’ two engineered selective handles, we tested the following native antibiotic resistance alleles: (TMP: FolA P21→L, A26→G, and L28→R) (63), (KAN/GEN: 16S rRNA U1406→A and A1408→G), (SPT: 16S rRNA A1191→G and C1192→U) (64), (RIF: RpoB S512→P and D516→G) (65), (STR: RpsL K4→R and K88→R) (66), and [(CIP: GyrA S83→L)] (67). The 90-bp oligos conferring each mutation, with two PT bonds at their 5′ end and with complementarity to the lagging strand were designed. Two oligos were designed to repair the engineered selective handles: 1) Elimination of a stop codon in the CHL acetyltransferase (cat) to confer CHL resistance, and 2) elimination of a stop codon in tolC to confer SDS resistance. Oligo-mediated recombineering was run with Redβ expressed off of the pARC8 plasmid and the cultures were then plated onto a range of concentrations of the antibiotic to which the oligo was expected to confer resistance. Colony counts were made and compared to a water-blank control. Modifications targeted to provide TMP, KAN, and SPT resistance did not work adequately and so were dropped. RpsL_K43R was chosen for STR selection and RpoB_S512P for RIF selection, although in both cases there was not a significant observable difference between the two tested alleles. An antibiotic concentration was chosen that provided the largest selective advantage for those cultures transformed with oligo (SI Appendix, Fig. S2). The concentrations chosen for the selective antibiotics were: 0.1% (vol/vol) SDS, 25 μg/mL STR, 100 µg/mL RIF, 0.1 µg/mL CIP, and 20 μg/mL CHL.
Identification of SSAP Library Members.
To generate the Broad SSAP Library we used a multiple sequence alignment of eight SSAPs that had been shown to function in E. coli [Redβ, EF2132 from Enterococcus faecalis, OrfC from Legionella pneumophila, S065 from Vibrio cholerae, Plu2935 from Photorhabdus luminescens, Orf48 from Listeria monocytogenes, Orf245 from Lactococcus lactis, and Bet from Prochlorococcus siphovirus P-SS2 (19, 37)] to generate a hidden Markov model that described the weighted positional variance of these proteins. We then queried nonredundant nucleotide and environmental metagenomic databases using a web-based search interface (68). Candidates were filtered based on gene size and annotation. Those that exhibited intrasequence similarity of greater than 98% were removed from the group. We added three eukaryotic SSAP homologs to the library (69). In total, the Broad SSAP Library contains 120 members from the homology search, 8 members from the starting sequence alignment, and 3 eukaryotic members, or a total of 131 SSAP homologs (Dataset S1).
The Broad RecT Library was generated from the full alignment of Pfam family PF03837, containing 576 sequences from Pfam 31.0 (70). Using ETE 3, a phylogenetic tree made by FastTree and accessed from the Pfam31.0 database was pruned, and from it a maximum diversity subtree of 100 members was identified (SI Appendix, Fig. S9) (71). Five members of this group were found in Library S1, and so were excluded, and in their place six RecT variants from Streptomyces phages and eight other RecT variants were added that had previously reported activity or were otherwise of interest (6, 15, 19, 22, 23), bringing the library size to 109 (Dataset S1).
Library Assembly.
Broad SSAP Library and Broad RecT Library variants with a DNA barcode 22 nucleotides downstream of the stop codon were codon-optimized for E. coli and synthesized by Gen9 (Broad SSAP Library) or Twist (Broad RecT Library). Synthesized DNA was amplified by PCR (New England Biolabs Q5 polymerase) and cloned into pDONR/Zeo (Thermo) by Gibson Assembly (New England Biolabs HiFi DNA Assembly Master Mix) and then moved into pARC8-DEST for arabinose-inducible expression. pARC8-DEST was engineered from a pARC8 plasmid (72) that shows good inducible expression in E. coli by moving Gateway sites (attR1/attR2), a CHL marker, and a ccdB counter selection marker downstream of the pBAD-araC regulatory region (SI Appendix, Fig. S1). This enabled easy, one-step cloning of the entire library into pARC8-DEST by Gateway cloning (Thermo). The Gateway reaction was transformed into E. cloni Supreme electrocompetent cells (Lucigen), providing >10,000× coverage of both libraries in total transformants.
SEER.
Libraries were miniprepped (New England Biolabs Monarch Kit) and electroporated into the SEER chassis with more than 1,000-fold coverage. Five cycles of oligo-mediated recombineering followed by antibiotic selection were then conducted (Fig. 1B). Next, 5 µL of the 5-mL recovery from the recombineering step was immediately plated onto LBL + selective antibiotic plates to estimate the total throughput of the selective step. This allowed us to ensure that the library was never bottlenecked; the first round of selection was the most stringent, but we ensured that there was >500× coverage at this stage. Following five rounds of selection, the plasmid library was miniprepped and transformed back into the naïve parent strain, followed by five further rounds of selection (10 in total). After each selective step a 100-µL aliquot of the antibiotic-selected recovery was frozen down at −80 °C in 25% glycerol for analysis by NGS.
NGS of Libraries.
Primers were designed to amplify a 215-bp product containing the barcode region of the SSAP libraries from the pARC8 plasmid and to add on Illumina adaptors. PCR amplification was done with Q5 polymerase (New England Biolabs) performed on a LightCycler 96 System (Roche), with progress tracked by SYBR Green dye and amplification halted during the exponential phase. Barcoding PCR for Illumina library preparation was performed as just described, but with NEBNext Multiplex Oligos for Illumina Dual Index Primers Set 1 (New England Biolabs). Barcoded amplicons were then purified with AMPure XP magnetic beads (Beckman Coulter), pooled, and the final pooled library was quantified with the NEBNext Library Quant Kit for Illumina (New England Biolabs). The pooled library was diluted to 4 nM, denatured, and a paired end read was run with a MiSeq Reagent Kit v3, 150 cycles (Illumina). Sequencing data were downloaded from Illumina, sequences were cleaned with Sickle (73), and analyzed with custom scripts written in Python. Briefly, reads were sorted by selection step, protein barcodes were recognized by the presence of a preceding 12-nt DNA sequence, and frequencies were calculated for each library member. An average of 50,000 individual reads were analyzed for each replicate, or about 500× coverage of each initial library.
Measuring Recombineering Efficiency in E. coli by NGS.
To measure single locus editing, a recombineering cycle was run with an oligo that confers a single base pair noncoding mismatch in a nonessential gene (SI Appendix, Table S2, silent mismatch MAGE oligo 7). The allele was then amplified by PCR and editing efficiency was measured by NGS as described above. To test multiplex editing, the concentration of oligo was held fixed (10 µM in the final electroporation mixture), but the total number of oligos in the mixture was varied. Pools of oligos to test editing at 5, 10, 15, or 20 alleles simultaneously were designed so as to space the edits relatively evenly around the genome. The five-oligo pool contained oligo nos. 3, 7, 11, 15, 17; the 10-oligo pool added oligo nos. 1, 5, 9, 13, 19; the 15-oligo pool added oligo nos. 4, 8, 12, 16, 18; and the final 20-oligo pool contained all of the silent mismatch MAGE oligos listed in SI Appendix, Table S2. One locus (locus 8) showed major irregularities when sequenced, and so we eliminated it from our analyses.
Whole-Genome Sequencing and Alignment to Measure Off-Target Mutagenesis.
Prior to sequencing, gDNA was isolated from each isolate and the corresponding parental strain by using the MasterPure Complete DNA and RNA Purification Kit (Lucigen). Extracted gDNA was sent to the Microbial Genome Sequencing Center (Pittsburgh, PA) for 2 × 150-bp paired-end shotgun sequencing on an Illumina NextSeq 550 (Illumina). Variants were called against E. coli MG1655 genome (accession no. U00096.3) with breseq (74).
DIvERGE-Based Simultaneous Mutagenesis of gyrA, gyrB, parE, and parC.
A single round of DIvERGE mutagenesis was carried out to simultaneously mutagenize gyrA, gyrB, parE, and parC in E. coli MG1655 by the transformation of an equimolar mixture of 130 soft-randomized DIvERGE oligos, tiling the four target genes. The sequences and composition of these oligos were published previously (13). To perform DIvERGE, 4 µL of this 100-µM oligo mixture was electroporated into E. coli K-12 MG1655 cells expressing Redβ from pORTMAGE311B, PapRecT from pORTMAGE312B, or CspRecT from pORTMAGE-Ec1, in five parallel replicates according to our previously described protocol (38). Following electroporation, the replicates were combined into 10 mL fresh TB media. Following recovery for 2 h, cells were diluted by the addition of 10 mL LB and allowed to reach stationary phase at 37 °C, 250 rpm. Library generation experiments were performed in triplicates. Following library generation, 1 mL of outgrowth from each library was subjected to 250, 500, and 1,000 ng/mL CIP stresses on 145-mm-diameter LB-agar plates. Colony counts were determined after 72 h of incubation at 37 °C, and individual colonies were subjected to further genotypic (i.e., capillary DNA sequencing) analysis and phenotypic (i.e., MIC) measurements.
pORTMAGE Plasmid Construction and Optimization.
All plasmids used in the study are listed in SI Appendix, Table S6. Cloning reactions were performed with Q5 High-Fidelity Master Mix and HiFi DNA Assembly Master Mix (New England Biolabs). pORTMAGE312B (Addgene accession no. 128969) and pORTMAGE-Ec1 (Addgene accession no. 138474) were constructed by replacing the Redβ ORF of pORTMAGE311B plasmid (Addgene accession no. 120418) (46) with PapRecT and CspRecT, respectively. pORTMAGE-Pa1 was constructed in many steps: 1) The Kanamycin resistance cassette and the RSF1010 origin-of-replication on pORTMAGE312B with Gentamicin resistance marker and pBBR1 origin-of-replication, amplified from pSEVA631 (75) (gift from Victor de Lorenzo, Consejo Superior de Investigaciones Cientificas, Madrid, Spain); 2) optimization of RBSs in pORTMAGE-Pa1 was done by designing a 30-nt optimal RBS in front of the SSAP ORF and in between the SSAP and MutL ORFs with an automated design program, De Novo DNA (76); 3) PaMutL was amplified from P. aeruginosa genomic DNA and cloned in place of EcMutL_E32K; and finally 4) PaMutL was mutated by site-directed mutagenesis to encode E36K. Ssr and Rec2 were ordered as gblocks from IDT and cloned in place of PapRecT into earlier versions of pORTMAGE-Pa1 for the comparisons in SI Appendix, Fig. S6.
Measuring Recombineering Efficiency in Gammaproteobacteria by Selective Plating.
Oligos were designed to introduce 1) premature STOP codons into lacZ for E. coli, K. pneumoniae, and C. freundii, or 2) RpsL K43→R; GyrA T83→I; ParC S83→L; RpoB D521→V, or a premature STOP codon into nfxB for P. aeruginosa. Oligo-mediated recombineering was performed as described above on all bacterial strains. After recovery overnight, cells were plated at empirically determined dilutions to a density of 200 to 500 colonies per plate. In the case of LacZ screening, plating was assayed on MacConkey agar plates or on X-Gal/IPTG LBL agar plates in the case of K. pneumoniae. In the case of selective antibiotic screening, cultures were plated onto both selective and nonselective plates. Selective antibiotic concentrations used were the same as those described for the selective testing above, except that in P. aeruginosa 100 µg/mL STR and 1.5 µg/mL CIP were used unless otherwise noted. Variants that were resistant to multiple antibiotics were selected on LBL agar plates that contained the combination of all corresponding antibiotics. Nonselective plates were antibiotic-free LBL agar plates. In all cases, allelic-replacement frequencies were calculated by dividing the number of recombinant CFUs by the number of total CFUs. Plasmid maintenance was ensured by supplementing all media and agar plates with either KAN (50 µg/mL) or GEN (20 µg/mL).
MIC Measurement in P. aeruginosa.
MICs were determined using a standard serial broth microdilution technique according to the Clinical Laboratory Standards Institute guidelines (ISO 20776-1:2,006, Part 1: Reference method for testing the in vitro activity of antimicrobial agents against rapidly growing aerobic bacteria involved in infectious diseases). Briefly, bacterial strains were inoculated from frozen cultures onto MHBII agar plates and were grown overnight at 37 °C. Next, independent colonies from each strain were inoculated into 1 mL MHBII medium and were propagated at 37 °C, 250 rpm overnight. To perform MIC tests, 12-step serial dilutions using twofold dilution-steps of the given antibiotic were generated in 96-well microtiter plates (Sarstedt 96-well microtest plate). Antibiotics were diluted in 100 μL of MHBII medium. Following dilutions, each well was seeded with an inoculum of 5 × 104 bacterial cells. Each measurement was performed in three parallel replicates. Plates were incubated at 37 °C under continuous shaking at 150 rpm for 18 h in an INFORS HT shaker. After incubation, the OD600 of each well was measured using a Biotek Synergy 2 microplate reader. MIC was defined as the antibiotic concentration which inhibited the growth of the bacterial culture (i.e., the drug concentration where the average OD600 increment of the three replicates was below 0.05).
Data Availability.
All NGS data are available in the SI Appendix and Dataset S1. Plasmids are available with Addgene, and all code and supporting data has been made available on GitHub at https://github.com/churchlab/SEER.
Supplementary Material
Acknowledgments
We thank Erkin Kuru, Max Schubert, Aditya Kunjapur, and Devon Stork for helpful discussions; Dan Snyder at the Microbial Genome Sequencing Center (Pittsburgh, PA) for whole-genome sequencing support; Prof. Victor de Lorenzo (Centro Nacional de Biotecnologia, Consejo Superior de Investigaciones Cientificas) for providing plasmids for the study; Claire O’Callaghan and Verena Volf for providing experimental help in various capacities in closely related work; and John Aach for thinking through conceptual ideas. Funding for this research was graciously provided by the Department of Energy under grant DE-FG02-02ER63445 (to G.M.C.); the European Research Council under grant H2020-ERC-2014-CoG 648364 ‘Resistance Evolution’ (to C.P.); Economic Development and Innovation Operational Programme (GINOP) (MolMedEx TUMORDNS) under grant GINOP-2.3.2-15-2016-00020, GINOP (EVOMER) under grant GINOP-2.3.2-15-2016-00014 (to C.P.); the ‘Lendület’ Program of the Hungarian Academy of Sciences (C.P.); and a European Molecular Biology Organization Long-Term Fellowship ALTF 160-2019 (to A.N.). M.C. was supported by fellowships from the Szeged Scientists Academy under the sponsorship of the Hungarian Ministry of Human Capacities (EMMI: 13725-2/2018/INTFIN), UNKP-18-2 New National Excellence Program of the Hungarian Ministry of Human Capacities, and UNKP 10-2 New National Excellence Program of the Hungarian Ministry for Innovation and Technology.
Footnotes
Competing interest statement: G.M.C. has related financial interests in EnEvolv, GRO Biosciences, and 64-x. G.M.C., C.J.G., M.J.L., and X.R. have submitted a patent application relating to pieces of this work (WO2017184227A2). T.M.W., G.T.F., and G.M.C. have submitted a patent application related to the improved single-stranded DNA-annealing proteins variants referenced here. A.N. and C.P. have submitted a patent application related to directed evolution with random genomic mutations (DIvERGE) (PCT/EP2017/082574 [WO2018108987] Mutagenizing Intracellular Nucleic Acids).
This article is a PNAS Direct Submission.
Database deposition: All next-generation sequencing data are available in SI Appendix and Dataset S1. Plasmids are available with Addgene, and all code and supporting data has been made available on GitHub at https://github.com/churchlab/SEER.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001588117/-/DCSupplemental.
References
- 1.Wang K. et al., Defining synonymous codon compression schemes by genome recoding. Nature 539, 59–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bird A. W. et al., High-efficiency counterselection recombineering for site-directed mutagenesis in bacterial artificial chromosomes. Nat. Methods 9, 103–109 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Wang H. H. et al., Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellis H. M., Yu D., DiTizio T., Court D. L., High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl. Acad. Sci. U.S.A. 98, 6742–6746 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu D. et al., An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 97, 5978–5983 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y., Buchholz F., Muyrers J. P., Stewart A. F., A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20, 123–128 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Costantino N., Court D. L., Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc. Natl. Acad. Sci. U.S.A. 100, 15748–15753 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lajoie M. J., Gregg C. J., Mosberg J. A., Washington G. C., Church G. M., Manipulating replisome dynamics to enhance lambda Red-mediated multiplex genome engineering. Nucleic Acids Res. 40, e170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grogan D. W., Stengel K. R., Recombination of synthetic oligonucleotides with prokaryotic chromosomes: Substrate requirements of the Escherichia coli/lambdaRed and Sulfolobus acidocaldarius recombination systems. Mol. Microbiol. 69, 1255–1265 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Farzadfard F., Lu T. K., Synthetic biology. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science 346, 1256272 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandell D. J. et al., Biocontainment of genetically modified organisms by synthetic protein design. Nature 518, 55–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H. H. et al., Genome-scale promoter engineering by coselection MAGE. Nat. Methods 9, 591–593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nyerges Á. et al., Directed evolution of multiple genomic loci allows the prediction of antibiotic resistance. Proc. Natl. Acad. Sci. U.S.A. 115, E5726–E5735 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wannier T. M. et al., Adaptive evolution of genomically recoded Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 115, 3090–3095 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Z. et al., A high-efficiency recombineering system with PCR-based ssDNA in Bacillus subtilis mediated by the native phage recombinase GP35. Appl. Microbiol. Biotechnol. 99, 5151–5162 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Binder S., Siedler S., Marienhagen J., Bott M., Eggeling L., Recombineering in Corynebacterium glutamicum combined with optical nanosensors: A general strategy for fast producer strain generation. Nucleic Acids Res. 41, 6360–6369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang Y., Wang Q., Su T., Qi Q., The efficiency for recombineering is dependent on the source of the phage recombinase function unit. bioRxiv:10.1101/745448 (24 August 2019). [Google Scholar]
- 18.Lopes A., Amarir-Bouhram J., Faure G., Petit M.-A., Guerois R., Detection of novel recombinases in bacteriophage genomes unveils Rad52, Rad51 and Gp2.5 remote homologs. Nucleic Acids Res. 38, 3952–3962 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datta S., Costantino N., Zhou X., Court D. L., Identification and analysis of recombineering functions from Gram-negative and Gram-positive bacteria and their phages. Proc. Natl. Acad. Sci. U.S.A. 105, 1626–1631 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo T., Xin Y., Zhang Y., Gu X., Kong J., A rapid and versatile tool for genomic engineering in Lactococcus lactis. Microb. Cell Factories 18, 22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Pijkeren J.-P., Britton R. A., High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. 40, e76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Pijkeren J.-P., Neoh K. M., Sirias D., Findley A. S., Britton R. A., Exploring optimization parameters to increase ssDNA recombineering in Lactococcus lactis and Lactobacillus reuteri. Bioengineered 3, 209–217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Kessel J. C., Hatfull G. F., Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152 (2007). [DOI] [PubMed] [Google Scholar]
- 24.van Kessel J. C., Hatfull G. F., Efficient point mutagenesis in mycobacteria using single-stranded DNA recombineering: Characterization of antimycobacterial drug targets. Mol. Microbiol. 67, 1094–1107 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Yin J. et al., Single-stranded DNA-binding protein and exogenous RecBCD inhibitors enhance phage-derived homologous recombination in Pseudomonas. iScience 14, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ricaurte D. E. et al., A standardized workflow for surveying recombinases expands bacterial genome-editing capabilities. Microb. Biotechnol. 11, 176–188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aparicio T., de Lorenzo V., Martínez-García E., A broad host range plasmid-based roadmap for ssDNA-based recombineering in Gram-negative bacteria. Methods Mol. Biol. 2075, 383–398 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Aparicio T. et al., Mismatch repair hierarchy of Pseudomonas putida revealed by mutagenic ssDNA recombineering of the pyrF gene. Environ. Microbiol. 22, 45–58 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Corts A. D., Thomason L. C., Gill R. T., Gralnick J. A., A new recombineering system for precise genome-editing in Shewanella oneidensis strain MR-1 using single-stranded oligonucleotides. Sci. Rep. 9, 39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penewit K. et al., Efficient and scalable precision genome editing in Staphylococcus aureus through conditional recombineering and CRISPR/Cas9-mediated counterselection. MBio 9, e00067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aparicio T., Nyerges A., Martínez-García E., de Lorenzo V., High-efficiency multi-site genomic editing (HEMSE) of Pseudomonas putida through thermoinducible ssDNA recombineering. bioRxiv:851576 (21 November 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maresca M. et al., Single-stranded heteroduplex intermediates in lambda Red homologous recombination. BMC Mol. Biol. 11, 54 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mosberg J. A., Lajoie M. J., Church G. M., Lambda red recombineering in Escherichia coli occurs through a fully single-stranded intermediate. Genetics 186, 791–799 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy K. C., Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180, 2063–2071 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iyer L. M., Koonin E. V., Aravind L., Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics 3, 8 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Story R. M., Bishop D. K., Kleckner N., Steitz T. A., Structural relationship of bacterial RecA proteins to recombination proteins from bacteriophage T4 and yeast. Science 259, 1892–1896 (1993). [DOI] [PubMed] [Google Scholar]
- 37.Sullivan M. B. et al., The genome and structural proteome of an ocean siphovirus: A new window into the cyanobacterial “mobilome”. Environ. Microbiol. 11, 2935–2951 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szili P. et al., Rapid evolution of reduced susceptibility against a balanced dual-targeting antibiotic through stepping-stone mutations. Antimicrob. Agents Chemother. 63, e00207-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Honda Y. et al., Functional division and reconstruction of a plasmid replication origin: Molecular dissection of the oriV of the broad-host-range plasmid RSF1010. Proc. Natl. Acad. Sci. U.S.A. 88, 179–183 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gawin A., Valla S., Brautaset T., The XylS/Pm regulator/promoter system and its use in fundamental studies of bacterial gene expression, recombinant protein production and metabolic engineering. Microb. Biotechnol. 10, 702–718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nyerges Á. et al., A highly precise and portable genome engineering method allows comparison of mutational effects across bacterial species. Proc. Natl. Acad. Sci. U.S.A. 113, 2502–2507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pines G., Freed E. F., Winkler J. D., Gill R. T., Bacterial recombineering: Genome engineering via phage-based homologous recombination. ACS Synth. Biol. 4, 1176–1185 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Piddock L. J., Mechanisms of fluoroquinolone resistance: An update 1994–1998. Drugs 58 (suppl. 2), 11–18 (1999). [DOI] [PubMed] [Google Scholar]
- 44.van Kessel J. C., Marinelli L. J., Hatfull G. F., Recombineering mycobacteria and their phages. Nat. Rev. Microbiol. 6, 851–857 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tommasi R., Brown D. G., Walkup G. K., Manchester J. I., Miller A. A., ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discov. 14, 529–542 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Szili P., et al. , Antibiotic usage promotes the evolution of resistance against gepotidacin, a novel multi-targeting drug. bioRxiv:495630 (13 December 2018). [Google Scholar]
- 47.Yang C. et al., Fed-batch fermentation of recombinant Citrobacter freundii with expression of a violacein-synthesizing gene cluster for efficient violacein production from glycerol. Biochem. Eng. J. 57, 55–62 (2011). [Google Scholar]
- 48.Jiang P. X. et al., Pathway redesign for deoxyviolacein biosynthesis in Citrobacter freundii and characterization of this pigment. Appl. Microbiol. Biotechnol. 94, 1521–1532 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Szpirer C. Y., Faelen M., Couturier M., Mobilization function of the pBHR1 plasmid, a derivative of the broad-host-range plasmid pBBR1. J. Bacteriol. 183, 2101–2110 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aparicio T., Jensen S. I., Nielsen A. T., de Lorenzo V., Martínez-García E., The Ssr protein (T1E_1405) from Pseudomonas putida DOT-T1E enables oligonucleotide-based recombineering in platform strain P. putida EM42. Biotechnol. J. 11, 1309–1319 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Marvig R. L., Sommer L. M., Molin S., Johansen H. K., Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat. Genet. 47, 57–64 (2015). [DOI] [PubMed] [Google Scholar]
- 52.AbdulWahab A. et al., The emergence of multidrug-resistant Pseudomonas aeruginosa in cystic fibrosis patients on inhaled antibiotics. Lung India 34, 527–531 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tacconelli E. et al.; WHO Pathogens Priority List Working Group , Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Agnello M., Wong-Beringer A., The use of oligonucleotide recombination to generate isogenic mutants of clinical isolates of Pseudomonas aeruginosa. J. Microbiol. Methods 98, 23–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen W. et al., CRISPR/Cas9-based genome editing in Pseudomonas aeruginosa and Cytidine deaminase-mediated base editing in Pseudomonas species. iScience 6, 222–231 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cabot G. et al., Evolution of Pseudomonas aeruginosa antimicrobial resistance and fitness under low and high mutation rates. Antimicrob. Agents Chemother. 60, 1767–1778 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jatsenko T., Tover A., Tegova R., Kivisaar M., Molecular characterization of Rif(r) mutations in Pseudomonas aeruginosa and Pseudomonas putida. Mutat. Res. 683, 106–114 (2010). [DOI] [PubMed] [Google Scholar]
- 58.PEW Charitable Trust , Antibiotics currently in global clinical development. https://www.pewtrusts.org/en/research-and-analysis/data-visualizations/2014/antibiotics-currently-in-clinical-development. Accessed 29 October 2019.
- 59.Marcusson L. L., Frimodt-Møller N., Hughes D., Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog. 5, e1000541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sandoval N. R. et al., Strategy for directing combinatorial genome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 109, 10540–10545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simon A. J., Morrow B. R., Ellington A. D., Retroelement-based genome editing and evolution. ACS Synth. Biol. 7, 2600–2611 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Lajoie M. J. et al., Genomically recoded organisms expand biological functions. Science 342, 357–360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Novais A. et al., Evolutionary trajectories of beta-lactamase CTX-M-1 cluster enzymes: Predicting antibiotic resistance. PLoS Pathog. 6, e1000735 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Criswell D., Tobiason V. L., Lodmell J. S., Samuels D. S., Mutations conferring aminoglycoside and spectinomycin resistance in Borrelia burgdorferi. Antimicrob. Agents Chemother. 50, 445–452 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Campbell E. A. et al., Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell 104, 901–912 (2001). [DOI] [PubMed] [Google Scholar]
- 66.Okamoto-Hosoya Y., Hosaka T., Ochi K., An aberrant protein synthesis activity is linked with antibiotic overproduction in rpsL mutants of Streptomyces coelicolor A3(2). Microbiology 149, 3299–3309 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Yoshida H., Bogaki M., Nakamura M., Nakamura S., Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34, 1271–1272 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Finn R. D., Clements J., Eddy S. R., HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eisen A., Camerini-Otero R. D., A recombinase from Drosophila melanogaster embryos. Proc. Natl. Acad. Sci. U.S.A. 85, 7481–7485 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.El-Gebali S. et al., The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huerta-Cepas J., Serra F., Bork P., ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Choe W., Chandrasegaran S., Ostermeier M., Protein fragment complementation in M.HhaI DNA methyltransferase. Biochem. Biophys. Res. Commun. 334, 1233–1240 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Joshi N. A., Fass J. N., Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33). Available at https://github.com/najoshi/sickle. Accessed 29 October 2019.
- 74.Deatherage D. E., Barrick J. E., Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martínez-García E., Aparicio T., Goñi-Moreno A., Fraile S., de Lorenzo V., SEVA 2.0: An update of the Standard European Vector Architecture for de-/re-construction of bacterial functionalities. Nucleic Acids Res. 43, D1183–D1189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salis H. M., Mirsky E. A., Voigt C. A., Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 27, 946–950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All NGS data are available in the SI Appendix and Dataset S1. Plasmids are available with Addgene, and all code and supporting data has been made available on GitHub at https://github.com/churchlab/SEER.