Significance
Knowledge of how thermal tolerances are distributed across major clades and biogeographic regions is important for understanding biome formation and climate change responses. However, most research has concentrated on animals, and we lack equivalent knowledge for other organisms. Here, we compile global data on heat and cold tolerances of plants, showing that many, but not all, broad-scale patterns known from animals are also true for plants. Importantly, failing to account simultaneously for influences of local environments, and evolutionary and biogeographic histories, can mislead conclusions about underlying drivers. Our study unravels how and why plant cold and heat tolerances vary globally and highlights that all plants, particularly at mid-to-high latitudes and in their nonhardened state, are vulnerable to ongoing climate change.
Keywords: macrophysiology, cold and heat, hardening, temperature, latitude
Abstract
Thermal macrophysiology is an established research field that has led to well-described patterns in the global structuring of climate adaptation and risk. However, since it was developed primarily in animals, we lack information on how general these patterns are across organisms. This is alarming if we are to understand how thermal tolerances are distributed globally, improve predictions of climate change, and mitigate effects. We approached this knowledge gap by compiling a geographically and taxonomically extensive database on plant heat and cold tolerances and used this dataset to test for thermal macrophysiological patterns and processes in plants. We found support for several expected patterns: Cold tolerances are more variable and exhibit steeper latitudinal clines and stronger relationships with local environmental temperatures than heat tolerances overall. Next, we disentangled the importance of local environments and evolutionary and biogeographic histories in generating these patterns. We found that all three processes have significantly contributed to variation in both heat and cold tolerances but that their relative importance differs. We also show that failure to simultaneously account for all three effects overestimates the importance of the included variable, challenging previous conclusions drawn from less comprehensive models. Our results are consistent with rare evolutionary innovations in cold acclimation ability structuring plant distributions across biomes. In contrast, plant heat tolerances vary mainly as a result of biogeographical processes and drift. Our results further highlight that all plants, particularly at mid-to-high latitudes and in their nonhardened state, will become increasingly vulnerable to ongoing climate change.
As our global climate continues to change, there is a need to increase understanding of the ecological and evolutionary processes that cause variation in temperature tolerances across organisms and biomes. Improved knowledge of how individuals cope with novel extreme thermal conditions can lead to better predictions of how species and communities will respond to climate change and aid development of mitigation strategies (1, 2). Moreover, knowledge of how thermal tolerances are distributed geographically and phylogenetically sheds light on the fundamental biogeographic and evolutionary processes that shape inherent physiological limits (3, 4), with important implications for how and why species’ range limits and biodiversity gradients are formed (5, 6). As a consequence, the past decade has seen a reinvigoration of the field of macrophysiology (4, 7–9), and several global analyses of physiological thermal limits have been conducted for different animal groups (3, 10–12). However, equivalent in-depth studies for nonanimal systems are lacking, limiting the generality of our understanding and ability to predict biotic responses to climate change.
Previous work on the global distribution of thermal tolerances in animals has led to the recognition of several major patterns, including (i) cold tolerances are more phenotypically variable and exhibit greater acclimation response than heat tolerances, for a similar set of organisms (11, 13), (ii) cold tolerances exhibit stronger latitudinal clines than heat tolerances (3, 10), (iii) the extent to which acclimation improves thermal tolerance increases with latitude (4, 14), and (iv) signatures of local adaptation in thermal tolerances are stronger under more extreme conditions (i.e., under strong directional selection; refs. 15–18).
Multiple hypotheses have been developed to explain these patterns, in particular the lower latitudinal variability and acclimation capacity of heat tolerances in comparison to cold tolerances. Hypotheses include lower evolvability of heat tolerance (11, 13), lower spatial variability in extreme heat than extreme cold environmental conditions themselves, limiting the magnitude of divergence in local adaptation for heat tolerance (3), and/or stronger mechanistic or scaling-related associations between metabolic optima and heat tolerance (19, 20). However, biogeographic processes, such as range shifts and endemism, may also play critical roles in driving the global distribution of heat or cold tolerances, both because limited dispersal between speciation events can constrain the phylogenetically determined rate of thermal tolerance evolution (21) and because large-scale dispersal events, e.g., during postglacial and contemporary range shifts, can transport thermal tolerance limits far from where they evolved (18). Species movement processes can also produce asymmetrical variability in heat vs. cold tolerances, depending on whether net migration is to colder or warmer regions (18).
Here, we test the diverse patterns and hypotheses developed for animals in a previously overlooked group: land plants. Plant thermal tolerances have been extensively studied in a mechanistic context (22, 23), but they have rarely been used to test fundamental macrophysiological hypotheses (but see discussions in refs. 24–26). Latitudinal gradients have been discovered for several other ecologically important plant traits (e.g., refs. 27–29), but latitudinal gradients in plant thermal tolerances remain undescribed (but see ref. 25). This is surprising given that plants cover every terrestrial surface of Earth, and that their distribution is strongly spatially and climatically structured, with temperature being considered one of the strongest determinants of plant distribution patterns globally (30, 31). The spatial structuring of plants is reflected in the major biomes of the world (e.g., broadleaf forest, coniferous forest, and grassland) and is the result of biogeographical and evolutionary processes over thousands to millions of years (32–34). Contemporary range shifts in response to changing climates have been documented for plants (35, 36), but migration through anthropogenically fragmented landscapes may be too slow for many species to keep pace with geographically shifting climate niches (37); the already elevated rates of plant extinction in the Anthropocene (38) are therefore likely to increase.
To increase understanding of global patterns of plant thermal tolerances, and how such patterns evolve, we compiled a database of thermal tolerances from the literature (Dataset S1), examined latitudinal patterns, and tested for the importance of local climate, phylogeny, and geographic distance in explaining those patterns, taking into account hardening status and method, measurement method, and hemisphere. We further fitted phylogenetic trait evolution models to test for a potential constraint in heat and cold tolerance evolution. Given the large variation in lifespan, growth form, and dispersal ability across land plants, the associated myriad ways in which they avoid or tolerate thermal stress might lead to new patterns, and confirm or refute existing macrophysiological hypotheses developed for (ectothermic) animals.
Results
Geographic and Taxonomic Coverage of Plant Thermal Tolerance Data.
We searched the literature for published estimates of georeferenced physiological thermal limits for land plants, focusing on both heat tolerance (Tmax) and cold tolerance (Tmin). These estimates represent a set of measures for assessing the environmental temperatures under which plants lose function physiologically. We found 70 books, monographs, and articles, which provided n = 1,732 thermal tolerance data points with geographical information for n = 1,028 plant species (Dataset S1). The thermal tolerance data were gathered from 246 unique locations (149 for cold and 138 for heat tolerance; Fig. 1 A and B). In addition, n = 806 records included confirmed information on hardening or acclimation status.
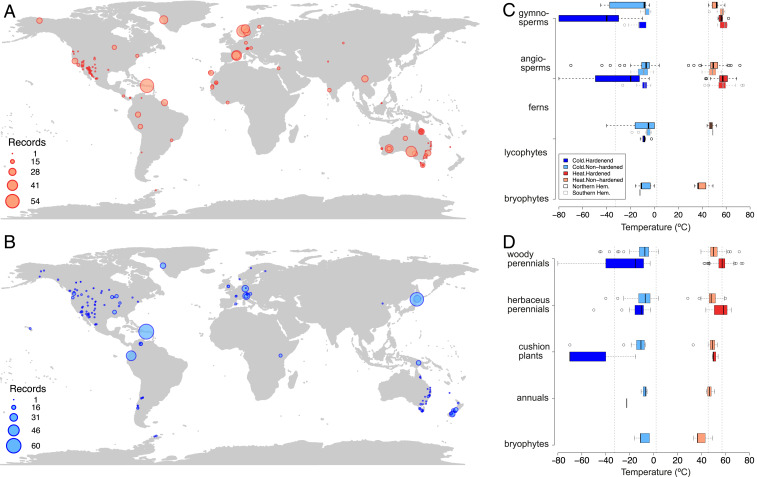
Fig. 1.
Global variation in plant thermal tolerances—distribution of data. (A and B) Geographic distribution of heat (A) and cold (B) tolerance measurements (n = 966 for Tmax, n = 769 for Tmin). The size of the circles is proportional to the number of data points it represents and ranges from 1 to 114 measurements at the same location for heat and cold tolerance together. Color hues are used for visibility but do not indicate hardening status (c.f. C and D). Most thermal tolerance data are from North America, Europe, Australia, and New Zealand with very few records from Africa or Asia. (C and D) Variation in thermal tolerance among major groups of land plants (C) and growth forms (D). Maximum temperatures (Tmax, heat tolerance) are plotted in reds and minimum temperatures (Tmin, cold tolerance) in blues. In C and D, measurements on hardened plants are shown in dark hues and nonhardened (including those with no information on hardening status) in light hues. In C, data for Northern (black frame) and Southern (gray frame) Hemispheres are plotted separately. Vertical dashed lines denote the SD across all data for each of heat and cold tolerance, which is wider for cold than heat tolerance.
Across all data, there is more variation in cold than heat tolerance (Tmin: mean −15.4 ± 17.4 °C SD, Tmax: 51.3 ± 5.8 °C). Most of the variation in cold tolerance comes from hardened plants in the Northern Hemisphere, especially cushion plants and gymnosperms (Fig. 1 C and D). There are very few data for Southern Hemisphere bryophytes, lycophytes, and ferns. (See SI Appendix, sections i–v, Figs. S1–S3, and Table S1 for further analysis and discussion of thermal tolerances in the context of taxonomic group, growth form, experimental approach, other plant traits, and plant thermal tolerance and avoidance strategies.)
Phylogenetic Signal and Evolutionary Mode of Heat and Cold Tolerance.
To estimate phylogenetic signal and test how cold and heat tolerances are evolving across land plants, we obtained phylogenetic information (39) for n = 653 and 455 species for heat and cold tolerance, respectively, representing 95% and 89% of the total dataset, respectively, with a bias against retention of nonvascular plants (SI Appendix, Fig. S4). Heat and cold tolerances exhibited similar phylogenetic signal, being significantly different from both 0 and 1 (cold: λ = 0.67, difference in corrected akaike information criterion (ΔAICc) ≥ 55; heat: λ = 0.65, ΔAICc ≥ 100; SI Appendix, Table S2). Further, we tested whether there was support for heat tolerance being evolutionarily constrained, as expressed by an Ornstein–Uhlenbeck (OU; ref. 40) model, in which species’ heat tolerances are pulled to an optimal value, and whether there was support for punctuated evolution for cold tolerance, as expressed by a “kappa” (κ) model (41), and as expected if extreme cold tolerance is conferred by hardening ability and that ability evolves only rarely (ref. 42 and SI Appendix, sections vi–viii, Tables S2 and S3, and Figs. S4–S5).
The OU model could not be rejected in any of the analyses for either heat or cold tolerance (based on ΔAICc ≥ 3.0; SI Appendix, Table S2); it was the best model in all cases except nonhardened heat, where the λ model had a slightly better fit (ΔAICc = 1.83). Nevertheless, parameter estimates suggest that the OU model is a better explanation for change in heat tolerances than cold tolerances: The stationary variance (σ2/2α), which measures the rate of stochastic change (or “drift,” as described by Brownian motion [BM], σ2) relative to the strength of the adaptive pull (α) toward the optimal value, is much higher for cold tolerance (344.3 [overall], 566.5 [hardened-only]) than heat tolerance (33.7 [overall], 26.4 [hardened-only]; SI Appendix, Table S3). This suggest a much weaker pull toward a globally optimal thermal state for cold tolerance than for heat tolerance.
Spatial Autocorrelation.
Both heat and cold tolerance exhibited significant spatial autocorrelation, calculated using Moran’s I, particularly at short to moderate spatial scales (i.e., within 50° latitude or longitude, corresponding to ∼5,000 km; SI Appendix, Fig. S6), both within and across taxonomic groups (SI Appendix, section ix). There is a clearer distance–decay relationship in cold tolerance than in heat tolerance. For cold tolerance, spatial autocorrelation is stronger in hardened than in nonhardened individuals, whereas for heat tolerance, hardened and nonhardened individuals show similar levels of spatial autocorrelation (SI Appendix, Fig. S6).
Global Variation in Thermal Tolerances: Latitudinal Trends.
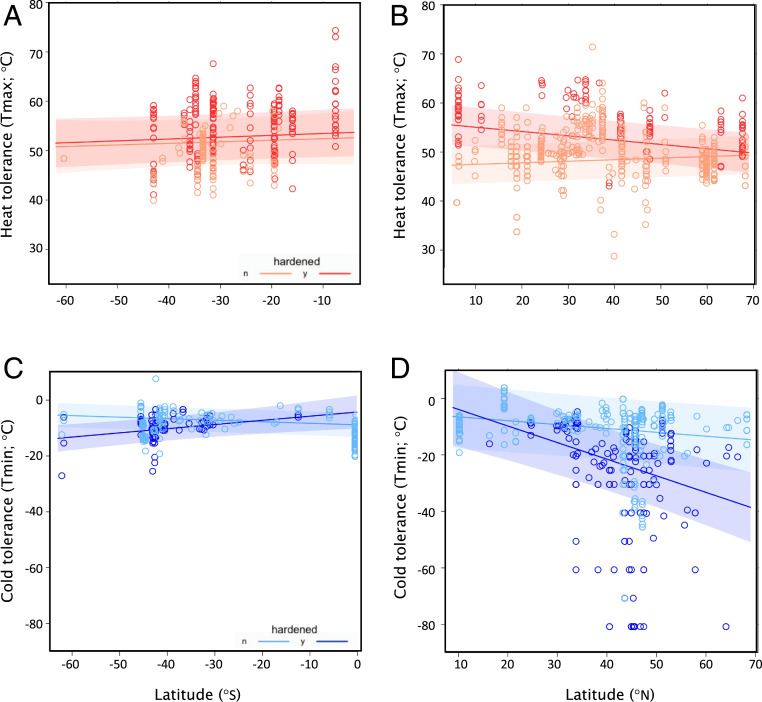
We tested for latitudinal variation in thermal tolerance using a Bayesian mixed modeling approach (43), further testing whether latitudinal effects on Tmin or Tmax were impacted by hemisphere and hardening status, and correcting for effects of phylogeny, sampling location, growth form, and the experimental approach used to assess tolerance. For heat tolerance, the best fit model included a significant three-way interaction among latitude, hemisphere, and hardening status, as well as significant two-way interactions between each of these variables: effect of latitude × hemisphere × hardening status = 0.27 [0.12–0.45 CI], P < 0.005; effect of latitude × hardening status = −0.32 [−0.44 to −0.16 CI], P < 0.005; effect of hemisphere (S) × hardening status = −12.39 [−16.95 to −6.28 CI], P < 0.005; effect of latitude × hemisphere (S) = −0.23 [−0.42 to −0.03], P = 0.01 (Fig. 2 A and B). Heat tolerance declines with latitude, but this is primarily observed in hardened individuals, and the difference in latitudinal patterns between hardened and nonhardened individuals was also driven primarily by Northern Hemisphere plants.
Fig. 2.
Latitudinal clines in plant thermal tolerances. Latitudinal clines are largely driven by Northern Hemisphere plants in the hardened state and likely reflect the combined influences of phylogenetic, biogeographic, and local adaptation processes (see Fig. 3 and SI Appendix, Figs. S4, S6, and S8 for graphical depictions of these contributing factors). Maximum temperatures (Tmax, heat tolerance) (A and B) are plotted in reds and minimum temperatures (Tmin, cold tolerance) (C and D) in blues; measurements on hardened plants are shown in dark hues and nonhardened (including those with no information on hardening status) in light hues. Plotted relationships are marginal effects of climate x hardening status from reported models (see main text).
The best Bayesian mixed model describing latitudinal effects on cold tolerance included significant fixed effect interactions of both latitude and hemisphere with hardening status, but not a three-way interaction among all three of these variables: effect of latitude × hardening status = −0.29 [−0.46 to −0.09 95% CI], P < 0.005; effect of hemisphere (S) × hardening status = 9.96 [6.58–13.95 CI], P < 0.005 (Fig. 2 C and D). In essence, the global distribution of cold tolerance in plants exhibits the predicted latitudinal variation (better tolerance at higher latitudes), but this pattern only holds for hardened individuals. For nonhardened individuals, there is no apparent latitudinal variation in cold tolerance. Moreover, latitudinal variation in hardened individuals is driven largely by Northern Hemisphere plants, as hardening status has negligible effects on cold tolerance in the Southern Hemisphere.
As is typically found in ectothermic animals and has previously been reported in plants (25, 44), Tmax was closest to local environmental heat extremes at mid latitudes and in the Northern Hemisphere, with unhardened heat tolerances often being exceeded by local environmental thermal maxima there (SI Appendix, section x and Fig. S7). In contrast, Tmin was at greatest risk for increasing cold snaps at high latitudes in both hemispheres, where estimated Tmin values, especially unhardened, already often fail to protect individuals against extremes of local environments (SI Appendix, section x and Fig. S7).
Environmental Predictors of Cold and Heat Tolerances.
After correcting for phylogeny, geographic distance, growth form, and experimental approach, the best Bayesian mixed model describing environmental effects on Tmax included significant interactions of mean annual temperature and temperature seasonality with hardening status (effect of mean temperature × hardening status = 0.24 [0.13–0.37 CI], P < 0.005; effect of seasonality × hardening status = 0.53 [0.13–0.90 CI], P < 0.005). Hardened heat tolerance increased at higher values of temperature mean and seasonality, but nonhardened heat tolerance was not positively affected by these environmental variables (SI Appendix, Fig. S8). However, combined fixed effects of environment and hardening status explained very little of the variation in heat tolerance overall (Fig. 3).
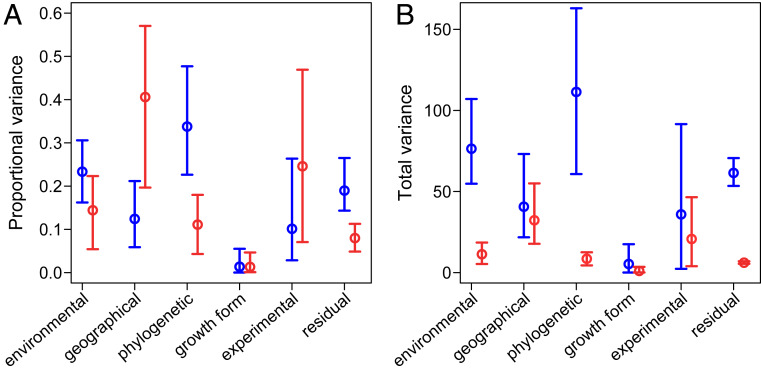
Fig. 3.
Variance partitioning of heat and cold tolerance among environmental effects (including hardening status), geographical distance, phylogenetic distance, growth form, experimental protocol, and residual variance (dots and whiskers represent mean values ± HPD from the reported Bayesian analyses). (A) Proportional variance in heat (red) and cold (blue) tolerance explained by each factor. (B) Total variance in heat (red) and cold (blue) tolerance explained by each factor. Geographical, experimental, and growth form effects account for similar amounts of the total variance in heat and cold tolerance, with the higher total variance in cold tolerance (Fig. 1) largely being explained by environmental and phylogenetic effects (plus a higher residual variance). However, a significantly higher proportion of the overall variance in heat tolerance is explained by geography, with a significantly higher proportion of the variance in cold tolerance being explained by phylogeny (plus residual variance). Other factors account for a similar proportion of the variance in both heat and cold tolerance. Thus, the single most important factor for explaining global variation in heat tolerance among land plants is geographical proximity, while the single most important factor for explaining variation in cold tolerance is phylogenetic relatedness.
The best Bayesian mixed model describing environmental effects on Tmin included significant interactions between fixed effects of mean annual temperature and temperature seasonality of the site with hardening status (effect of mean temperature × hardening status = 0.88 [0.50–1.31 CI], P < 0.005; effect of seasonality × hardening status = −0.26 [−0.32 to −0.19 CI], P < 0.005; SI Appendix, Fig. S8). In effect, these environmental factors predicted variation in hardened cold tolerance (hardened Tmin was positively correlated with mean temperature, and negatively correlated with temperature seasonality), but, as for Tmax, hardening and environmental variation explained only a small proportion of the overall variance in Tmin (Fig. 3), and none at all for nonhardened Tmin, which exhibited less correlation with environmental variables (no correlation with mean temperatures, shallower correlation with temperature seasonality; SI Appendix, Fig. S8).
Global Variation in Thermal Tolerances: Intrinsic, Biogeographic, and Environmental Drivers.
In the context of our Bayesian mixed models, we further partitioned the variance in cold and heat tolerance among fixed effects of local climate variables × hardening status, geographic and phylogenetic distances, growth form, and experimental method (Fig. 3). The total variance in heat tolerance (Tmax) explained by the model was 92% [36–149% highest posterior density (HPD) interval], with spatial distance having the largest effect (41% [20–57%]), followed by measurement method (25% [7–47%]), fixed effects of local environment and acclimation (14% [5–22%]), phylogeny (11% [4–18%]), and growth form (only 1% [0.01–5%]; Fig. 3A).
The total variance explained for cold tolerance (Tmin) was 81% [48–126% HPD], with the largest proportion of the total variance attributed to phylogeny (34% [23–48% HPD]), followed by the fixed effects of local environmental variables and hardening status (23% [16–31%]), geographic distance (12% [6–21%]), measurement method (10% [3–26%]), and very little variance explained by growth form (1% [0–5%]; Fig. 3A).
Differences in the proportional contribution of each of these factors to heat vs. cold tolerance arise in part due to differences in the total variance in these traits (greater for cold than for heat, see above and Fig. 1). The total variance explained by environmental factors is 11 [5–18] for heat tolerance vs. 76 [55–107 HPD] for cold; geographic distance: 32 [18–55] for heat vs. 41 [22–73] for cold; phylogeny: 9 [4–13] heat vs. 111 [61–163] cold; growth form: 1 [0.1–4] for heat vs. 5 [1–18] cold; measurement method: 21 [4–47] heat vs. 36 [2–92] cold; leaving a residual variance of 6 [5–7] for heat vs. 62 [53–71] for cold (Fig. 3B). It is evident that geographical distance, measurement method, and growth form explain similar amounts of the absolute variance for heat and cold tolerance. The larger total variance in cold tolerance is in addition explained by phylogenetic distance and local environmental factors (as well as a larger residual variance). Thus, the higher phenotypic variance in cold tolerance is largely explained by phylogenetic distance and more extreme acclimation processes.
Discussion
Our knowledge of the thermal tolerance of plants is extremely limited; we found data for 1,028 land plant species overall, which amounts to a mere 0.31% of the ca. 330,200 species recognized (45). This acute lack of information on the intrinsic thermal tolerances of most plants implies we have limited ability to incorporate such information in realistic predictions about how specific plant lineages will fare under future climates and how plant distributions might be altered. However, the dataset is taxonomically and geographically broad (SI Appendix, Table S5) and spans a large latitudinal range (Fig. 1), allowing for analysis of how thermal tolerances vary globally and what might be driving this variation.
The Generality of Macrophysiological Rules Developed in Animals.
Overall, we found several expected macrophysiological patterns, including: greater overall and latitudinal variability in cold than heat tolerance (Janzen’s rule; Figs. 1 and 2 and refs. 4, 11, 13, and 46); greater acclimation potential at higher latitudes (Vernberg’s rule; Fig. 2 and refs. 4 and 14); greater effect of acclimation on cold than heat tolerance and greater acclimation ability under more extreme climatic conditions (Payne’s rule; SI Appendix, Fig. S8 and refs. 4, 47, and 48). These patterns are in agreement with previous macrophysiological “rules” primarily generated from the study of ectothermic animals.
However, we also found significant departures from the expected macrophysiological patterns. We found similar phylogenetic signal in heat and cold tolerance, but higher variance explained by phylogeny for cold than heat tolerance (Fig. 3 and see SI Appendix, Table S5 for how this compares to results from animals). Variation in heat tolerance in plants was instead better explained by geographic distance, a finding that has received mixed support in animals (refs. 7, 8, and 16 and SI Appendix, section xii and Table S5). In addition, we found a stronger hemisphere effect on macrophysiological patterns and weaker (often nonexistent) evidence for the macrophysiological drivers of unhardened thermal tolerances of plants compared to animals (10, 13) (Fig. 3). We discuss our findings in detail below but, overall, differences among studies (SI Appendix, Table S5) suggest more work is required to understand what aspects of ecology, physiology, and biogeography result in different phylogenetic, spatial, or hemispherical signals in heat or cold tolerance distributions across major divisions of life, as well as to establish the robustness of these differences to varying geographic and/or phylogenetic scales (and modeling approaches) of different study systems (SI Appendix, sections iv and xii).
Evolutionary, Ecological, and Biogeographical Drivers of Global Variation in Plant Thermal Tolerances.
Our comprehensive mixed modeling approach led to a number of important conclusions. First, our models explained almost all variation in thermal tolerance for plants (81% for cold tolerance; 92% for heat tolerance). This suggests plant thermal tolerances can be understood with just a few parameters, making predictions more straightforward. Second, our findings are not an artifact of measurement method (cf. ref. 48). Third, our models show that phylogeny, geography, and the local environment are all needed to explain global variation in thermal tolerances (Fig. 3). Failure to incorporate one or more of these variables decreased the explanatory power of the models overall and overestimated the importance of the factors included (SI Appendix, section xi and Table S4). For example, including only phylogenetic or geographic information inflated the importance of the included random effect, while models including neither phylogenetic nor geographic information enormously inflated the apparent importance of the local environment and acclimation status (fixed effect; SI Appendix, Table S4). This occurs partly because of spatial autocorrelation in climates, meaning that environmental effects can be confounded with effects of spatial or phylogenetic processes, if gene flow or biogeographic events produce patterns of trait variance that correlate with, but are not caused by, environmental gradients (18, 49). Our results can also be explained by the tendency of closely related lineages to occur in greater spatial proximity to each other and, thus, may also inhabit more similar environments by chance, compared to more distantly related species; failing to account for direct environment effects and spatial distance can therefore inflate the phylogenetic signal in thermal traits (21, 50, 51). Our results clearly demonstrate that incomplete models that do not account for all potential drivers simultaneously can yield erroneous conclusions about the importance of local adaptation, evolutionary legacies, or biogeographical drivers of global variation in thermal tolerances.
At first glance, our resulting models were strikingly similar for heat and cold tolerances: similar phylogenetic signal; similar support for an evolutionary model with a central tendency (OU model); similar importance of hardening and a Northern Hemisphere distribution; similar levels of spatial autocorrelation; similar relationships with latitude (direction, not magnitude); similar environmental temperature variables ranking highest in importance; and a conspicuous lack of any relationship with any precipitation variable (SI Appendix, section v). However, there were major differences in the relative importance of each factor for explaining variation in heat versus cold tolerance (Fig. 3), and the parameter estimates of the OU models suggest different underlying evolutionary processes (SI Appendix, Table S3). Taken together, these results suggest that evolutionary history, particularly transitions to and within cold hardening capacity, strongly structure how plant cold tolerances are distributed globally. This is consistent with evolutionary innovations in hardened cold tolerances playing a critical role in determining plant distributions across biomes, and tropical-to-temperate transitions being key evolutionary events (30, 42). In contrast, plant heat tolerances and nonhardened cold tolerances are primarily structured spatially, likely reflecting effects of gene flow or colonization history. The magnitude of spatial drift in heat tolerance and nonhardened cold tolerance may, however, be limited (13, 18), as indicated by the relatively strong strength of the pullback to ancestral values for these traits (SI Appendix, section vii and Table S3). Thus, our results suggest strongly divergent underlying processes structuring global variation in heat and (hardened) cold tolerances of plants.
Implications for Climate Change.
The importance of hardening in our data has implications for plant responses to climate change. Throughout, hardening status was found to be a significant mediator of patterns of both heat and cold tolerances; we found no relationship of nonhardened thermal tolerances with latitude (Fig. 2), and weaker or no relationships with the local environment (SI Appendix, Fig. S8). Yet, nonhardened tolerances may become increasingly important under less predictable temperature fluctuations, which increase the exposure of unhardened plants to extreme weather. Particular risks suggested by our data are unseasonal cold snaps at high latitudes and unseasonal heat waves at midlatitudes (SI Appendix, Fig. S7 and section x); such events are predicted to increase under future climate scenrios (52).
An inability of plants to cope with higher or unseasonable temperatures under future warming can affect the functioning of entire ecosystems (53, 54). These changes have consequences not only for the future survival and distributions of plants, but for the animals and people that depend on them too. For example, substantial losses to winegrowing areas have been predicted from even modest warming (55), and freezing damage to grain crops due to changing weather patterns is already a serious economic problem (56). Understanding the ecological role of thermal safety margins must therefore focus on thermal tolerance traits, as well as how they interact with and trade-off against traits that influence other aspects of plant survival, productivity, and reproduction.
Methods
Dataset Construction.
Suitable literature was identified using Web of Science and Google Scholar, employing search strings including combinations of: “heat,” “cold,” “temperature,” “limits,” “tolerance,” “metabolic,” “respiration,” “photosynthesis,” “physiological,” “chill,” “freeze,” “critical,” and “lethal.” We also searched citing and cited references of relevant articles. The search was carried out between October 2017 and January 2018. For reviewing articles, e.g., ref. 24, we referred to the original study where possible. For articles not written in English, we used Google Translate (https://translate.google.com) to extract relevant methodological details. Across studies, thermal tolerances were estimated on a variety of scales, but mostly included LT50 under heat or cold stress, assessed visually via stain uptake or electrolyte leakage assessments (n = 439 heat, n = 512 cold), LT100 (n = 8 heat, n = 37 cold), Tcrit (n = 177 heat), Tmax (n = 340 heat), freezing resistance (n = 183 cold), freezing tolerance (n = 22 cold), or unknown, i.e., where the methods were insufficiently recorded (n = 14). We also recorded hardening status (heat/cold acclimation; n = 594 hardened vs. n = 212 nonhardened and n = 928 records where no information was provided). Where stated, we also separated whether hardening was induced in the laboratory (n = 51 heat, n = 249 cold), field (warming: n = 6 heat), or greenhouse (n = 36 cold) or was the result of natural seasonal variation (n = 356 heat, n = 106 cold). Additional data exploration with respect to experimental approach is provided in SI Appendix, section iii.
Environmental Variables.
Climatic data for the spatial coordinates of the collection localities for each thermal tolerance estimate were extacted using the bioclim environmental layers (57) at the 10′ resolution using the raster package for R (58, 59). Elevational data for each point were extracted from the US Geological Survey GMTED2010 digital elevation model at the 30″ resolution (60). Where the elevation of a sampling location was reported in the original report, we used this value. We further extracted distance from the nearest coastline from the National Aeronautics and Space Administration (NASA) oceancolor dataset, at the 0.01° resolution (61).
Growth Form, Taxonomic, and Phylogenetic Information.
Taxonomic designations at the family, genus, and species levels were updated using the taxize package for R, based on the Taxonomic Name Resolution Service (TNRS) and National Center for Biotechnology Information (NCBI) databases (62–64). The taxonomy was further verified using the World Checklist of Selected Plant Families (WCSP, http://wcsp.science.kew.org), Tropicos (https://www.tropicos.org), and The Plant List (http://www.theplantlist.org). Broad classifications were then assigned as follows: bryophytes (liverworts and mosses), lycophytes, “ferns” (ferns and horsetails), gymnosperms, and angiosperms. Using online floras and the WCSP, we further recorded the growth form of each species as woody perennial (including trees [>10 m height] and shrubs [<10 m]), cushion plant (herbaceous or woody), herbaceous perennial (including facultative angiosperm annuals, ferns, horsetails, and lycophytes), herbaceous annual, or bryophyte (including liverworts and mosses).
Phylogenetic information was obtained from Slik et al. (39) using the Phylomatic query tool (65). For fitting phylogenetic trait evolution models, branch lengths were set to 1. For fitting phylogenetic mixed models, an ultrametric tree of unit height was generated with a default smoothing parameter of 1, under a correlated substitution model, using the chronos() function in ape (66).
Statistical Analyses.
Phylogenetic signal and trait evolution analyses for heat and cold tolerance.
Phylogenetic signal and the best evolutionary model for cold and heat tolerances were assessed using several models founded in BM (SI Appendix, sections vi–viii). Models were fitted on the complete dataset and separately for hardened and nonhardened subsets using Geiger (67) and compared using AICc (68). To visualize the phylogenetic distribution of each trait (SI Appendix, Fig. S4), each tree was rescaled with the estimated phylogenetic signal (λ) in Geiger and then ancestral states were reconstructed on the rescaled tree using “fastAnc” in Phytools (69).
Spatial autocorrelation in heat and cold tolerance.
Spatial autocorrelation was tested using Moran’s I and a randomization test to determine the significance of spatial autocorrelation at each distance class, using ncf (70). We evaluated spatial autocorrelation separately for each broad taxonomic group, as well as a combined estimate across all of our data. We also examined spatial autocorrelation of hardened vs. nonhardened tolerances separately (SI Appendix, section ix). Significance of spatial autocorrelation at each distance class was assessed using a Bonferroni correction for the number of distance classes tested (n = 10 distance classes, α = 0.005). Further testing for effects of geographic distance on thermal tolerance was conducted in a mixed model framework simultaneously accounting for phylogeny and local environments (see below).
Global variation in thermal tolerances: Latitudinal trends.
Bayesian linear mixed effects models were fit using MCMCglmm (43), fitting either Tmin or Tmax as the response variable, and including latitude, hemisphere, hardening status (and hardening method; SI Appendix, section iii and Fig. S3), and all interactions as fixed effects. An inverse phylogenetic similarity matrix was fit as a random effect to account for autocorrelation due to phylogenetic distance, and additional random effects were included to account for growth form, effects of shared sampling locations (concatenated Lat/Long), and the methodological approach used to estimate Tmin or Tmax. We used an Inverse Wishart prior for random and residual terms with V = 1 and nu = 1.002, and a normal prior for fixed effects. We also assessed model outputs for qualitatively similar outcomes after specifying a prior to account for potential correlations among fixed effects and using parameter-expanded priors for random effects. All Markov-chain Monte Carlo (MCMC) chains were run with a length of 1,000,000, burnin of 50,000, and thinning interval of 5,000. This was sufficient to achieve model convergence and avoid temporal autocorrelation among the posteriors. Deviance information criterion (DIC) was used to select the best combination of fixed effects.
To address the lack of nonvascular plants in the Slik et al. phylogeny, we replicated these analyses on the full dataset using a maximum likelihood approach in lme4 and lmerTest R packages (71, 72). A taxonomic correction was applied, with separate random effects for higher-order taxonomic group (e.g., class or unranked higher clade), family, genus, and species, and additional random effects for growth form, location, and thermal tolerance assessment methodology as described above. AICc was used for model comparison. Results of these models were similar to those from the Bayesian models (SI Appendix, section x).
We further plotted latitudinal variation in Tmin and Tmax against local values of maximum and minimum environmental temperatures (BioClim Bio5 and Bio6) to visually assess latitudinal variation in tolerance to climate extremes in plants (SI Appendix, Fig. S7).
Global variation in thermal tolerances: Intrinsic, biogeographic, and environmental drivers.
We next partitioned the variation in each of heat and cold tolerance among factors representing the local environment versus phylogenetic or spatial distance. Models were run in MCMCglmm, implementing fixed effects of environmental variables and hardening status, plus phylogenetic and geographic similarity matrices, and additional random effect terms for tolerance measurement method and growth form. For the geographic similarity matrix, we calculated great circle distances using geosphere (73). We then constructed a Gaussian spatial kernel of the distances, K = e−h*distance^2 (74), with the value of the tuning parameter (h) determined via optimization (75). For both heat and cold tolerance, the optimal value for h was 9e-13. To identify climatic drivers of thermal tolerances, we included temperature (bio1, mean annual temperature; bio2, diurnal temperature range; bio4, temperature seasonality) and precipitation (bio12, annual precipitation; bio15, precipitation seasonality) as fixed effect variables, as well as effects for elevation and distance from the coast, which might capture elements of alpine or maritime climates not reflected in extracted temperature and precipitation qualities. Interactions of these with hardening status were also tested. The best combination of these environmental covariates was determined using DIC and significance of effects. Priors and chain lengths were established as described above. The proportion of variance in heat or cold tolerance explained by fixed effects versus each random effect in the final models was calculated using the Nakagawa and Schielzeth approach (76).
Data Availability.
The global dataset of plant thermal tolerances generated for and analyzed within this study is appended to Supplementary Information (Dataset S1).
Supplementary Material
Acknowledgments
We thank Mark Adams for providing code to optimise spatial distance kernels, and Jake Alexander, Per-Ola Karis, members of the British Ecological Society Macroecology special interest group, and members of the Bolin Centre for Climate Research (Research Area 8) for helpful discussions.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918162117/-/DCSupplemental.
References
- 1.Kidane Y. O., Steinbauer M. J., Beierkuhnlein C., Dead end for endemic plant species? A biodiversity hotspot under pressure. Glob. Ecol. Conserv. 19, e00670 (2019). [Google Scholar]
- 2.Liancourt P. et al., Plant response to climate change varies with topography, interactions with neighbors, and ecotype. Ecology 94, 444–453 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Addo-Bediako A., Chown S. L., Gaston K. J., Thermal tolerance, climatic variability and latitude. Proc. Biol. Sci. 267, 739–745 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaston K. J. et al., Macrophysiology: A conceptual reunification. Am. Nat. 174, 595–612 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Thuiller W. et al., Consequences of climate change on the tree of life in Europe. Nature 470, 531–534 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Sandel B. et al., The influence of Late Quaternary climate-change velocity on species endemism. Science 334, 660–664 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Diamond S. E., Chick L. D., The Janus of macrophysiology: Stronger effects of evolutionary history, but weaker effects of climate on upper thermal limits are reversed for lower thermal limits in ants. Curr. Zool. 64, 223–230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grigg J. W., Buckley L. B., Conservatism of lizard thermal tolerances and body temperatures across evolutionary history and geography. Biol. Lett. 9, 20121056 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kellermann V., van Heerwaarden B., Sgrò C. M., Hoffmann A. A., Fundamental evolutionary limits in ecological traits drive Drosophila species distributions. Science 325, 1244–1246 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Sunday J. M., Bates A. E., Dulvy N. K., Global analysis of thermal tolerance and latitude in ectotherms. Proc. Biol. Sci. 278, 1823–1830 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Araújo M. B. et al., Heat freezes niche evolution. Ecol. Lett. 16, 1206–1219 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Khaliq I., Hof C., Prinzinger R., Böhning-Gaese K., Pfenninger M., Global variation in thermal tolerances and vulnerability of endotherms to climate change. Proc. R Soc. B Biol. Sci. 281, 20141097 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann A. A., Chown S. L., Clusella-Trullas S., Upper thermal limits in terrestrial ectotherms: How constrained are they? Funct. Ecol. 27, 934–949 (2013). [Google Scholar]
- 14.Vernberg F. J., Compartive physiology: Latitudinal effects on physiological properties of animal populations. Annu. Rev. Physiol. 24, 517–546 (1962). [DOI] [PubMed] [Google Scholar]
- 15.Baudier K. M., D’Amelio C. L., Malhotra R., O’Connor M. P., O’Donnell S., Extreme insolation: Climatic variation shapes the evolution of thermal tolerance at multiple scales. Am. Nat. 192, 347–359 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Kellermann V. et al., Upper thermal limits of Drosophila are linked to species distributions and strongly constrained phylogenetically. Proc. Natl. Acad. Sci. U.S.A. 109, 16228–16233 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lancaster L. T., Dudaniec R. Y., Hansson B., Svensson E. I., Latitudinal shift in thermal niche breadth results from thermal release during a climate-mediated range expansion. J. Biogeogr. 42, 1953–1963 (2015). [Google Scholar]
- 18.Lancaster L. T., Widespread range expansions shape latitudinal variation in insect thermal limits. Nat. Clim. Chang. 6, 618–621 (2016). [Google Scholar]
- 19.Payne N. L., Smith J. A., An alternative explanation for global trends in thermal tolerance. Ecol. Lett. 20, 70–77 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Huey R. B., Kingsolver J. G., Evolution of thermal sensitivity of ectotherm performance. Trends Ecol. Evol. 4, 131–135 (1989). [DOI] [PubMed] [Google Scholar]
- 21.Coelho M. T. P., Rodrigues J. F. M., Diniz‐Filho J. A. F., Rangel T. F., Biogeographical history constrains climatic niche diversification without adaptive forces driving evolution. J. Biogeogr. 46, 1020–1028 (2019). [Google Scholar]
- 22.Franklin K. A., Wigge P. A., Temperature and Plant Development, (John Wiley & Sons, Ames, Iowa, 2014). [Google Scholar]
- 23.Long S. P., Plants and Temperature, Steven P., Woodward F. I., Eds. (Published for the Society for Experimental Biology by the Company of Biologists, 1988). [Google Scholar]
- 24.Bannister P., Godley review: A touch of frost? Cold hardiness of plants in the southern hemisphere. N. Z. J. Bot. 45, 1–33 (2007). [Google Scholar]
- 25.O’sullivan O. S. et al., Thermal limits of leaf metabolism across biomes. Glob. Change Biol. 23, 209–223 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Gauslaa Y., Heat resistance and energy budget in different scandinavian plants. Holarctic Ecol. 7, 5–78 (1984). [Google Scholar]
- 27.Moles A. T. et al., Global patterns in plant height. J. Ecol. 97, 923–932 (2009). [Google Scholar]
- 28.Moles A. T. et al., Global patterns in seed size. Glob. Ecol. Biogeogr. 16, 109–116 (2006). [Google Scholar]
- 29.Swenson N. G., Enquist B. J., Ecological and evolutionary determinants of a key plant functional trait: Wood density and its community-wide variation across latitude and elevation. Am. J. Bot. 94, 451–459 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Wiens J. J., Donoghue M. J., Historical biogeography, ecology and species richness. Trends Ecol. Evol. 19, 639–644 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Humboldt F. W. H. A., Bonpland A. J. A., Essai sur la géographie des plantes, accompagné d’un tableau physique des regions équinoxiales, fondé sur des measures executes, (Schoell, Paris, France, 1805). [Google Scholar]
- 32.Svenning J.-C., Skov F., Ice age legacies in the geographical distribution of tree species richness in Europe. Glob. Ecol. Biogeogr. 16, 234–245 (2007). [Google Scholar]
- 33.Hewitt G. M., Post-glacial re-colonization of European biota. Biol. J. Linn. Soc. Lond. 68, 87–112 (1999). [Google Scholar]
- 34.Crisp M. D. et al., Phylogenetic biome conservatism on a global scale. Nature 458, 754–756 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Parmesan C., Yohe G., A globally coherent fingerprint of climate change impacts across natural systems. Nature 421, 37–42 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Lenoir J., Gegout J. C., Marquet P. A., de Ruffray P., Brisse H., A significant upward shift in plant species optimum elevation during the 20th century. Science 320, 1768–1771 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Miller K. M., McGill B. J., Land use and life history limit migration capacity of eastern tree species. Glob. Ecol. Biogeogr. 27, 57–67 (2018). [Google Scholar]
- 38.Humphreys A. M., Govaerts R., Ficinski S. Z., Nic Lughadha E., Vorontsova M. S., Global dataset shows geography and life form predict modern plant extinction and rediscovery. Nat. Ecol. Evol. 3, 1043–1047 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Slik J. W. F. et al., An estimate of the number of tropical tree species. Proc. Natl. Acad. Sci. U.S.A. 112, 7472–7477 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen T. F., Stabilizing selection and the comparative analysis of adaptation. Evolution 51, 1341–1351 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Pagel M., Inferring the historical patterns of biological evolution. Nature 401, 877–884 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Zanne A. E. et al., Three keys to the radiation of angiosperms into freezing environments. Nature 506, 89–92 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Hadfield J. D., MCMC methods for multi-response generalized linear mixed models: The MCMCglmm R package. J. Stat. Softw. 33, 1–22 (2010).20808728 [Google Scholar]
- 44.Sunday J. M. et al., Thermal-safety margins and the necessity of thermoregulatory behavior across latitude and elevation. Proc. Natl. Acad. Sci. U.S.A. 111, 5610–5615 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Christenhusz M. J. M., Byng J. W., The number of known plants species in the world and its annual increase. Phytotaxa 261, 201–217 (2016). [Google Scholar]
- 46.Janzen D. H., Why mountain passes are higher in the tropics. Am. Nat. 101, 233–249 (1967). [Google Scholar]
- 47.Payne N. M., Measures of insect cold hardiness. Biol. Bull. 52, 449–457 (1926). [Google Scholar]
- 48.Sunday J. et al., Thermal tolerance patterns across latitude and elevation. Philos. Trans. R. Soc. B Biol. Sci. 374, 20190036 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lancaster L. T., Host use diversification during range shifts shapes global variation in Lepidopteran dietary breadth. Nat. Ecol. Evol., 10.1038/s41559-020-1199-1 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Cooper N., Freckleton R. P., Jetz W., Phylogenetic conservatism of environmental niches in mammals. Proc. R. Soc. B Biol. Sci. 278, 2384–2391 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Humphreys A. M., Linder H. P., Evidence for recent evolution of cold tolerance in grasses suggests current distribution is not limited by (low) temperature. New Phytol. 198, 1261–1273 (2013). [DOI] [PubMed] [Google Scholar]
- 52.IPCC , Climate Change 2013: The Physical Science Basis, (Cambridge University Press, New York, 2013). [Google Scholar]
- 53.Jeong S. J. et al., Potential impact of vegetation feedback on European heat waves in a 2 × CO2 climate: Vegetation impact on European heat waves. Clim. Change 99, 625–635 (2010). [Google Scholar]
- 54.Menzel A., Seifert H., Estrella N., Effects of recent warm and cold spells on European plant phenology. Int. J. Biometeorol. 55, 921–932 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Hannah L. et al., Climate change, wine, and conservation. Proc. Natl. Acad. Sci. U.S.A. 110, 6907–6912 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baxter R., “Plant acclimation and adaptation to cold environments” in Temperature and Plant Development, K. A. Franklin, P. A. Wigge, Eds. (Wiley-Blackwell, Ames, IA, 2014), pp. 19–42. [Google Scholar]
- 57.Hijmans R. J., Cameron S. E., Parra J. L., Jones P. G., Jarvis A., Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978 (2005). [Google Scholar]
- 58.Hijmans R. J., raster: Geographic Data Analysis and Modeling (R Package Version 2.6-7, 2017). https://cran.r-project.org/web/packages/raster/index.html.
- 59.R Core development Team , R: A language and environment for statistical computing, (R Foundation for Statistical Computing, Vienna, Austria, 2018). [Google Scholar]
- 60.Danielson J. J., Gesch D. B., “Global multi-resolution terrain elevation data 2010 (GMTED2010)”, US Geological Survey Open-File Report 2011–1073 (U.S. Geological Survey, Reston, Virginia, 2011). [Google Scholar]
- 61.Stumpf R., Kuring N., GMT intermediate distance from coast. NASA Ocean Biol Satell Altimetry Lab NOAA Satell Altimetry Lab (2009). https://oceancolor.gsfc.nasa.gov/docs/distfromcoast/. Accessed 24 June 2009.
- 62.Chamberlain S., et al. , Taxize–Taxonomic search and retrieval in R. (2019). https://github.com/ropensci/taxize. Accessed 20 February 2019.
- 63.Taxosaurus organization http://taxosaurus.org/. Accessed 20 February 2019.
- 64.NCBI Taxonomy Database https://www.ncbi.nlm.nih.gov/taxonomy. Accessed 20 February 2019.
- 65.Webb C. O., Donoghue M. J., Phylomatic: Tree assembly for applied phylogenetics. Mol. Ecol. Notes 5, 181–183 (2005). [Google Scholar]
- 66.Paradis E., Schliep K., ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Harmon L. J., Weir J. T., Brock C. D., Glor R. E., Challenger W., GEIGER: Investigating evolutionary radiations. Bioinformatics 24, 129–131 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Burnham K. P., Anderson D. R., Model Selection and Multimodel Inference : A Practical Information-Theoretic Approach, (Springer, New York, 1998). [Google Scholar]
- 69.Revell L. J., phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012). [Google Scholar]
- 70.Bjornstad O. N., ncf: Spatial Covariance Functions (R package Version 1.2-8, 2019). https://CRAN.R-project.org/package=ncf.
- 71.Bates D., Mächler M., Bolker B., Walker S., Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 51 (2014). [Google Scholar]
- 72.Kuznetsova A., Brockhoff P. B., Christensen R. H. B., lmerTest Package: Tests in linear mixed effects models. J. Stat. Softw. 82, 1–26 (2017). [Google Scholar]
- 73.Hijmans R. J., geosphere: Spherical Trigonometry (R package version 1.5-7, 2017). https://cran.r-project.org/package=geosphere.
- 74.de Los Campos G., Gianola D., Rosa G. J. M., Reproducing kernel Hilbert spaces regression: A general framework for genetic evaluation. J. Anim. Sci. 87, 1883–1887 (2009). [DOI] [PubMed] [Google Scholar]
- 75.Adams M. J., Example of Making a Spatial Kernel and Using it as the Inverse G Matrix in a Mixed-Effects Model with MCMCglmm. (2013). https://gist.github.com/mja/6807269#file-geok. Accessed 7 April 2019.
- 76.Nakagawa S., Schielzeth H., A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol. Evol. 4, 133–142 (2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The global dataset of plant thermal tolerances generated for and analyzed within this study is appended to Supplementary Information (Dataset S1).