Significance
Arp2/3 complex is an ATPase that binds to the side of a preexisting actin filament and nucleates an actin filament branch. Growing branched networks experience variable resistance and respond by adapting growth speed, power, and architecture. How force influences the dissociation of actin filament branches was not known. We used microfluidics to show that mechanical force promotes the dissociation of actin filament branches and that Arp2/3 complex adopts two distinct mechanical states with different responses to force. Phosphate release from Arp2/3 complex increases the sensitivity to both force and the debranching protein GMF. Thus, phosphate release from Arp2/3 complex may regulate debranching by force and debranching proteins.
Keywords: actin, Arp2/3 complex, branched filament, debranching, force
Abstract
Networks of branched actin filaments formed by Arp2/3 complex generate and experience mechanical forces during essential cellular functions, including cell motility and endocytosis. External forces regulate the assembly and architecture of branched actin networks both in vitro and in cells. Considerably less is known about how mechanical forces influence the disassembly of actin filament networks, specifically, the dissociation of branches. We used microfluidics to apply force to branches formed from purified muscle actin and fission yeast Arp2/3 complex and observed debranching events in real time with total internal reflection fluorescence microscopy. Low forces in the range of 0 pN to 2 pN on branches accelerated their dissociation from mother filaments more than two orders of magnitude, from hours to <1 min. Neither force on the mother filament nor thermal fluctuations in mother filament shape influenced debranching. Arp2/3 complex at branch junctions adopts two distinct mechanical states with different sensitivities to force, which we name “young/strong” and “old/weak.” The “young/strong” state 1 has adenosine 5′-diphosphate (ADP)−Pi bound to Arp2/3 complex. Phosphate release converts Arp2/3 complex into the “old/weak” state 2 with bound ADP, which is 20 times more sensitive to force than state 1. Branches with ADP−Arp2/3 complex are more sensitive to debranching by fission yeast GMF (glia maturation factor) than branches with ADP−Pi−Arp2/3 complex. These findings suggest that aging of branch junctions by phosphate release from Arp2/3 complex and mechanical forces contribute to disassembling “old” actin filament branches in cells.
Arp2/3 complex forms networks of branched actin filaments that generate and sustain mechanical forces that power cell motility, endocytosis, and vesicle trafficking (1, 2). Membrane-bound proteins, called nucleation-promoting factors, such as WASP activate Arp2/3 complex, which then nucleates a branch when it binds to the side of a preexisting “mother” filament (3). The new “daughter” filament elongates and pushes against the membrane until it is capped. All of the filaments, including branches formed by Arp2/3 complex, must disassemble for recycling to form new filaments and branches. Similar to actin, Arp2/3 complex is an ATPase (4–6). Hydrolysis of bound ATP and subsequent phosphate release have been implicated in controlling branched network dynamics (5, 7–10), but mechanistic details are lacking.
The assembly and architecture of branched actin networks are sensitive to force in vitro and in cells (11, 12). Under load, branched actin networks assembled from purified proteins grow more slowly and with a higher branch density (13, 14), but how these mechanical forces directly affect the biochemical interactions of branched actin network protein components has not been firmly established. Similar to measurements with purified protein components, branched actin networks in cells respond to external load by increasing density of branched filaments while also reorganizing relative to the membrane (15).
Networks of branched actin filaments turn over much more rapidly in cells than in vitro when assembled from purified proteins (16–19). The regulatory proteins cofilin and glia maturation factor (GMF) accelerate debranching and have been implicated in accelerating branched network remodeling and turnover (20–24). Although not investigated previously, mechanical forces may also affect network disassembly through debranching.
We report that mechanical forces promote dissociation of branches formed by Arp2/3 complex and that phosphate bound to Arp2/3 complex regulates the sensitivity to force. Phosphate release from Arp 2/3 complex at branch junctions also regulates debranching by GMF. Thus, phosphate release from the Arp2/3 complex could target “older” adenosine 5′-diphosphate (ADP)−Arp2/3 branches for dissociation while sparing “younger” branches with ADP−Pi−Arp2/3 complex.
Results
Microfluidics Assay to Measure Dissociation of Branches Formed by Arp2/3 Complex under Force.
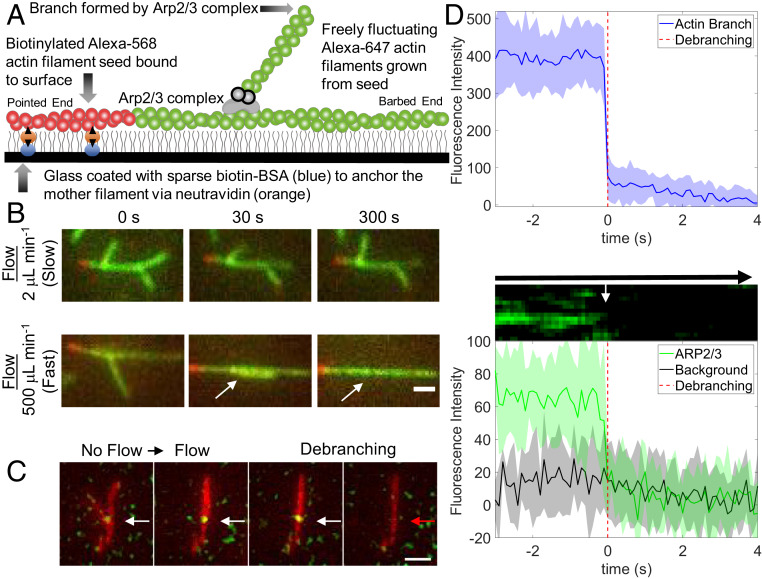
We used fluid flowing through a microfluidics apparatus to apply force to actin filament branches formed by purified fission yeast Arp2/3 complex and muscle actin monomers (Fig. 1A) as we observed the dissociation of the branches by fluorescence microscopy. Starting with short filament seeds tethered to the surface of the slide, we assembled branched filaments from purified ATP-actin monomers and Arp2/3 complex for 2 min to 4 min. After washing out the soluble proteins, the filaments were allowed to “age” for an additional, variable time with very slow fluid flow. Thereafter, we started the debranching process under constant force by flowing buffer over the surface at a rate of 2 μL⋅min−1 to 500 μL⋅min−1 until the end of the experiment, while we recorded a series of images. The seeds were immobilized on the slide, but both the mother filaments and branches were free to fluctuate (Fig. 1A).
Fig. 1.
Microfluidics assay to measure Arp2/3 complex debranching under force. (A) Diagram showing a short segment of an actin filament containing 10% biotinylated and 15% Alexa 568-labeled (red) actin subunits immobilized on the neutravidin-coated surface. The surface is passivated with 0.2% tween (illustrated with gray vertical lines). This seed was elongated at its barbed end with 1.5 µM 15% Alexa 647-labeled Mg-ATP-actin (green), and Arp2/3 complex formed a branch with Alexa 647-labeled Mg-ATP-actin. The green filaments fluctuate freely and are subject to viscous drag forces applied by fluid flow. (B) TIRF microscopy images of representative branched filaments under slow flow (2 μL⋅min−1, ∼0.004 pN of force for a 1.5-μm branch; Top) and fast flow (500 μL⋅min−1, ∼1.02 pN of force for a 1.5-μm branch; Bottom). Branches are aligned in the direction of flow. (Scale bar, 1 μm.) (C) The Arpc5 subunit of the Arp2/3 complex was labeled with Alexa 488 via snap tag and tracked during debranching. Time-lapse images with the actin filaments represented in red and the Alexa 488−Arp2/3 complex located at the junction of the daughter branch and mother filament represented in green. (Scale bar, 1 μm.) (D) Top shows the spatially integrated fluorescence intensity of actin at a branch junction as a function of time, used to determine the observed debranching event time (t = 0). Middle shows a kymograph measured across a branched actin filament. Bottom shows the time course of spatially integrated fluorescence intensity of Arpc5 subunit at a branch junction with time aligned to its corresponding actin frame. The fluorescence intensity from Arp2/3 complex reproducibly decreased in a single step for all 12 debranching events observed.
Rapid flow rates flattened branches against the mother filaments until they dissociated (Fig. 1B and Movies S1–S3). Neither the angle of the applied force (relative to mother filament orientation), the tension in the mother filament, nor fluctuations in mother filament shape had a strong influence on debranching (SI Appendix, Fig. S1 and Movies S6 and S7).
We added a snap tag to the Arpc5 subunit of Arp2/3 complex for labeling with Alexa 488 and simultaneous viewing with Alexa 647-labeled actin. Force dissociated Arp2/3 complex and the daughter filament concurrently, within the 0.1-s time resolution of our imaging (Fig. 1D and Movies S4 and S5). Buffer flowed across the sample at 500 µm⋅s−1 dissociated both the daughter filament and Arp2/3 complex from the field of view by the next frame, making it impossible to determine whether the labeled Arp2/3 complex remained bound to the dissociated daughter filament.
Piconewton Forces Decreased the Time for Branch Dissociation from Hours to <1 Min.
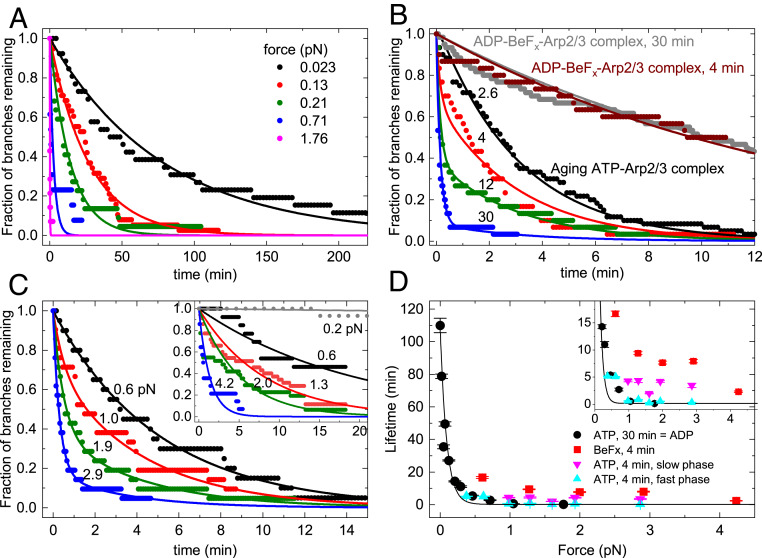
Branches formed by ATP−Arp2/3 complex were stable for many minutes without buffer flow (Fig. 2A) but had a higher probability of dissociating when subjected to the forces produced by the range of buffer flow rates in our experiments (Fig. 2A). The following sections explain how the time course of debranching depends on the applied force and how long the newly formed branches were aged after assembly. At a given flow rate, the force on a branch scales with its length. In some experiments, we show the force on individual branches (SI Appendix, Fig. S2), but, in most experiments, we report flow rates from which we calculate the average force on branches (e.g., Fig. 2A).
Fig. 2.
Effects of mechanical force and nucleotide bound to Arp2/3 complex on the time course of dissociation of actin filament branches. Arp2/3 complex branches were assembled in the flow chamber before applying flow as described in Materials and Methods. In A and C, the force on each observed branch was calculated from its length and the flow rate, and then binned at the indicated average forces values (see Materials and Methods). (A) The effect of force on the time course of dissociation of actin filament branches formed by ATP-actin monomers and ATP−Arp2/3 complex and aged for 30 min, when most branches had ADP bound to Arp2/3 complex. The fraction of branches remaining is plotted. Smooth curves are the best fits of single exponentials to the data. Each trace includes at least 14 branches. (B) Dependence of the time course of dissociation of branches formed with ATP-actin monomers and ATP−Arp2/3 complex with different aging times (2.6, 4, 12, and 30 min) and the presence of BeFx. For all time courses, 500 μL⋅min−1 of buffer flow was applied to the branches for debranching, producing a force of ∼1 pN for a branch of 1.5 µm. The force on each branch was not calculated for the data shown. Smooth curves are the best global fits of double exponentials to the data for aging branches and yielded two shared rate constants for debranching: slow ks,F = 0.32 (±0.005) min−1 and fast kf,F = 6.67 (±0.44) min−1. Smooth curves are the best single exponential fits to the data for branches aged for 4 and 30 min with BeFx with lifetimes of 13.9 (±0.2) min−1 for the sample aged for ∼4 min and 14.8 (±0.09) min−1 for the sample aged for 30 min. Fig. 4D presents the fractional amplitudes obtained from the double exponential fits. (C) The effect of force on the time course of the dissociation of actin filament branches formed by ATP-actin monomers and ATP−Arp2/3 complex and aged for ∼4 min using the same data collection and analysis methods as in A. The smooth curves are best fits of the data to single (F ≤ 0.6 pN) or double (F > 0.6 pN) exponentials. The fractional amplitudes of the slow phase in the time courses that follow the double exponentials are 0.71 ± 0.05 (F = 0.98 pN), 0.39 ± 0.06 (F = 1.29 pN), 0.81 ± 0.05 (F = 1.60 pN), 0.38 ± 0.02 (F = 1.93 pN), and 0.17 ± 0.02 (F = 2.86 pN). Each trace includes at least 19 branches. (Inset) Time courses of branch dissociation under a range of forces for branches formed from ATP−Arp2/3 complex in the presence of 2 mM BeFx and aged for ∼4 min. Each trace includes at least 13 branches. The smooth curves are single exponential fits to the time courses. Branches dissociate slowly under 0.2 pN of force, so the debranching time course cannot be reliably fitted to obtain the branch lifetime. (D) Dependence of branch lifetimes on force for four different conditions: (filled black circles) branches formed from ATP−Arp2/3 complex and aged for 30 min to form branches with ADP−Arp2/3 complex (time courses in A); (filled red squares) branches formed from ADP−BeFx−Arp2/3 complex and aged for ∼4 min (time courses in C, Inset); (filled pink triangles) slow debranching phase of branches formed from ATP−Arp2/3 complex and aged for ∼4 min (ADP−Pi branch population; time courses in C); and (filled blue triangle) fast debranching phase of branches formed from ATP−Arp2/3 complex and aged for ∼4 min (ADP branch population; time courses in C). The uncertainty bars for all data represent the SDs from the fits to exponentials shown in A and C. The smooth black curve is the best single exponential fit (Eq. 1) to the ADP−Arp2/3 complex debranching data points, yielding a half-force (F1/2) of 0.054 (±0.008) pN and branch lifetime in the absence of force (τ0) of 106 (±8) min (observed rate constant = τ0−1 = 0.01 (±0.007) min−1). Inset shows that 1) fast phase lifetimes (blue triangles; 0.3 min to 0.8 min at F > 1 pN) differ from the slow phase lifetimes (pink triangles; 2 min to 4 min at F > 1 pN; t = 6.85, one-tail tcritical = 2.13 and P = 0.001 by Welch’s unequal variances t test); 2) the slow phase lifetimes (pink triangles; 2 min to 4 min at F > 1 pN) differ from the debranching lifetimes with BeFx (red squares; 8 min to 9 min at F > 1 pN; t = 6.65, one-tail tcritical = 2.02 and P = 0.0006 by Welch’s unequal variances t test); and 3) the fast phase lifetimes (blue triangles) do not differ significantly from the debranching with ATP aged for 30 min lifetimes (black circles) at F > 1 pN (t = −0.86, two-tail tcritical = 2.45 and P = 0.42 by Welch’s unequal variances t test).
In samples of branches formed by ATP−Arp2/3 complex and aged for 30 min, the time course of debranching followed a single exponential that depended on the applied force (Fig. 2A). The observed lifetimes of these branches decreased with force (Fig. 2D), suggesting a slip bond behavior, so we used the Bell’s equation (25) (Eq. 1) to estimate the force sensitivity,
| [1] |
Here kB is the Boltzmann constant, T is the absolute temperature, F is force, τ0 is the branch lifetime in the absence of force, and d is the characteristic distance to the transition state (26). The value of d is typically considered a force sensitivity parameter for bond rupture. An alternative estimate of force dependence is the force that reduces the branch lifetime by half (half-force, F1/2 = 0.693kBT/d), which we estimate to be 0.054 (±0.008) pN.
Without force, the branch lifetime (τ0), estimated from extrapolation of the fit of the force dependence (Fig. 2D), was 106 (±8) min [Table 1; k2 ≈ 1/τ0 = 0.01 (±0.0007) min−1], indicating that sustained pN forces reduce τobs more than two orders of magnitude to <30 s. The force dependence of individual branch lifetimes (SI Appendix, Fig. S2) yielded a half-force (F1/2) of 0.054 (±0.008) pN and τ0 = 96 (±6) min, comparable to the values estimated from exponential fits of debranching time courses (Fig. 2 A and D).
Table 1.
Rate constants for branch formation and dissociation in the absence of force and related figures
| Conversionkconv (min−1) | State 1 debranchingk1* (min−1) | State 2 debranchingk2† (min−1) | Branch formationkform* (μM−1⋅s−1) |
| 0.14 ± 0.03 | 0.012 | 0.01 ± 0.007 | 0.02 |
| Fig. 4 | Fig. 4 | Fig. 2D | Fig. 4 |
The equations used to determine these parameters are approximations, so uncertainties are not reported.
From the fit of force-dependent debranching data to Eq. 1; k2 = 1/τ0. The error was calculated from the SD τ0 in the fit.
Branches Assembled from ATP−Arp2/3 Complex Dissociate Faster under Force as They Age.
As they age, branches assembled from ATP−Arp2/3 complex dissociated progressively faster under force produced by a buffer flow rate of 500 μL⋅min−1 (Fig. 2B). Immediately after the 2.6-min assembly reaction, when the dominant nucleotide bound to both Arp2/3 complex and the actin filaments is expected to be ADP−Pi, the time course of branch dissociation followed a single exponential with a slow observed lifetime (τs,F, where the subscript F indicates under flow force) of 3.08 (±0.05) min corresponding to a first-order rate constant ks,F of 0.32 (±0.005) min−1. After aging for 30 min, when both Arp2/3 complex and the subunits in the actin filaments are expected to have bound ADP, the time course of debranching also followed a single exponential with a 20-fold shorter lifetime (τf,F) of 0.15 (±0.01) min (kf,F = 6.67 (±0.44) min−1). At intermediate aging times, the time courses followed double exponentials (Fig. 2B) with distinguishable fast and slow phases, indicating that (at least) two reactions contributed to debranching.
To evaluate how force affects slow and fast debranching, we aged samples for a short time (∼4 min), so the sample would include mixtures of branches with Arp2/3 complex with bound ADP−Pi and ADP and thus exhibit both slow and fast phases of dissociation over a range of forces applied with different flow rates. However, the actual aging times prior to making observations varied. This uncertainty influenced the observed amplitudes but not the observed lifetimes, so we only analyzed the lifetimes. The debranching time courses at forces of >1 pN followed double exponential decays (Fig. 2C), yielding the force dependence of the slow and fast phase lifetimes (Fig. 2 D and Inset; triangles). The lifetimes of the slow phase (2- to 4-min range) differed from the fast phase (0.3 min to 0.8 min) in this force range (>1 pN), but neither depended strongly on the applied force. At low forces of <0.6 pN, dissociation of branches formed from ATP−Arp2/3 complex followed single exponentials, with lifetimes similar to branches with ADP−Arp2/3 complex (Fig. 2D).
The debranching model presented below accounts for these different time courses. The fast phase lifetime behaved similar to ADP−Arp2/3 complex branches (i.e., assembled from ATP−Arp2/3 complex and aged for 30 min), consistent with fast debranching population corresponding to ADP−Arp2/3 complex branches. We discuss the force dependence of the slow phase below.
The Nucleotide Bound to Arp2/3 Complex Influences the Sensitivity of Branches to Force.
ATP−Arp2/3 complex hydrolyzes the bound nucleotide upon or soon after branch formation (5, 7) followed by dissociation of the γ-phosphate with an unknown rate constant. Therefore, branches formed by ATP−Arp2/3 complex rapidly transition to the ADP−Pi state for an unknown duration before the release of Pi. Actin filament branches formed by Arp2/3 complex with mutations that slow ATP hydrolysis are more stable than those formed by native Arp2/3 complex, so Martin et al. (10) proposed that the hydrolysis of ATP bound to Arp2/3 complex destabilizes branches.
We performed a series of experiments to determine whether the nucleotide state of Arp2/3 complex could explain the slow and fast debranching states (Fig. 2 B and C). Like Acanthamoeba Arp2/3 complex (5), Schizosaccharomyces pombe ADP−Arp2/3 complex did not form branches from ATP-actin monomers nor did the ADP−Arp2/3 complex form branches in the presence of 20 mM phosphate (SI Appendix, Fig. S3). A likely interpretation of this behavior is that the transient intermediate ADP−Pi−Arp2/3 complex is competent to form branches, but ADP−Arp2/3 complex binds Pi very weakly (Kd > 20 mM). On the other hand, S. pombe ADP−Arp2/3 complex formed branches with 2 mM beryllium fluoride (BeFx) in the buffer (SI Appendix, Fig. S3), as originally shown for Acanthamoeba Arp2/3 complex (5). ATP−Arp2/3 complex also forms branches with 2 mM BeFx in the buffer.
Branches with ADP−BeFx−Arp2/3 complex dissociate with indistinguishable time courses whether assembled from ATP−Arp2/3 complex or ADP−Arp2/3 complex in the presence of 2 mM BeFx and aged for ∼4 min (SI Appendix, Fig. S4). We assume that, in both cases, BeFx binds to the ADP−Arp2/3 complex and stabilizes conformations similar to ADP−Pi, as established for actin (27, 28), so we used ADP−BeFx−Arp2/3 complex branches made with either method. ADP−BeFx−Arp2/3 complex branches aged for 4 or 30 min in the presence of 2 mM BeFx dissociate with similar time courses and follow single exponentials (Fig. 2B). Thus, branches did not convert from the slowly to the rapidly dissociating state when aged with BeFx.
Force is required to dissociate, within our experimental observation period, ADP−BeFx−Arp2/3 complex branches formed from ATP−Arp2/3 complex in the presence of 2 mM BeFx (Fig. 2 C, Inset). Under low forces (i.e., <<0.6 pN), <20% of branches with ADP−BeFx−Arp2/3 complex dissociated within 5 h, so branching lifetimes could not be measured reliably. With forces >0.6 pN, branches dissociated with single exponential time courses and observed rate constants (inverse of lifetimes) that depended on the applied force (Fig. 2 C, Inset). Therefore, branches with ADP−BeFx−Arp2/3 complex were more stable under force than branches with ADP−Arp2/3 complex (formed from ATP−Arp2/3 complex and aged for 30 min; Fig. 2D).
Under a given force, the lifetimes of ADP−BeFx−Arp2/3 complex branches were longer (∼twofold) than the slow debranching phase of ATP−Arp2/3 complex branches that had been aged for ∼4 min (Fig. 2D). The observed rate constant of the slow phase debranching reflects the sum of the ADP−Pi state debranching rate constant and the rate constant for conversion of the ADP−Pi−Arp2/3 complex to the ADP state (see Discussion).
It is therefore expected to be faster than ADP−BeFx−Arp2/3 complex debranching, which converts more slowly to ADP−Arp2/3 complex (Fig. 2B).
The Nucleotide State of the Actin Filaments Does Not Influence Debranching.
We assembled branches from ATP−Arp2/3 complex with ADP or ADP−Pi bound to the subunits in the actin filaments and observed that all had similar time courses of debranching after aging for a given time (SI Appendix, Fig. S5). Samples with ADP-actin filaments were prepared by assembly from ATP-actin monomers and ATP-actin Arp2/3 complex followed by aging for 30 min. Samples with ADP−Pi actin filaments were prepared by assembly from ATP-actin monomers and ATP−Arp2/3 complex in buffer containing 20 mM phosphate followed by aging for 4 or 30 min (SI Appendix, Fig. S5). This concentration of phosphate is well above the Kd for binding ADP-actin subunits (29), so subunits in the filaments likely had ADP−Pi in the active site, assuming that Arp2/3 complex binding does not dramatically change the affinity of actin for phosphate. Samples assembled from ATP-actin monomers and ATP−Arp2/3 complex followed by ∼4 min of aging had a mixture of ADP and ADP−Pi nucleotide states for both actin subunits and Arp2/3 complex. Samples with AMPPNP actin (a nonhydrolyzable analog of ATP) were assembled from AMPPNP actin monomers and ATP-actin Arp2/3 complex in buffer containing 2 mM AMPPNP followed by aging for ∼4 min. Since AMPPNP−Arp2/3 complex does not form branches (ref. 5 and SI Appendix, Fig. S3), all of the branches formed from Arp2/3 complex with bound ATP, while the actin filaments had bound AMPPNP. A previous study of bovine Arp2/3 complex (16) reported more branches on mother filament segments with ADP−Pi than segments with ADP. This difference was attributed to slower dissociation of branches aged in buffer with 25 mM phosphate allowing time for them to be stabilized by binding to the slide coated with N-ethylmaleimide (NEM)-myosin anchors. However, that study did not measure time course of the dissociation of branches, and we did not compare the rate of branch formation on ADP and ADP−Pi mother filaments, so we do not know whether the sources of Arp2/3 complex, the presence of NEM-myosin anchors on the surface, or other factors account for the apparent difference.
Since the debranching kinetics are independent of the nucleotide (i.e., ADP, ADP−Pi, or AMPPPNP) bound to the mother and daughter filaments, the stabilization of branches by BeFx (Fig. 2 B–D) is likely due to BeFx bound to Arp2/3 complex in the branch junction. This suggests that the ADP−Pi−Arp2/3 complex intermediate has different mechanical properties than the ADP−Arp2/3 complex after phosphate dissociation. The lack of an effect of 20 mM phosphate on debranching (SI Appendix, Fig. S5) is consistent with the affinity of S. pombe ADP−Arp2/3 complexes in branch junctions for phosphate being very weak (Kd > 20 mM).
Force Promotes but BeFx Inhibits Debranching by GMF.
GMF (20–22) promotes the dissociation of actin filament branches without applied force, so we measured how the concentration of fission yeast GMF influences the rates of dissociation of actin filament branches with ADP−BeFx−Arp2/3 complex or ADP−Arp2/3 complex at a low buffer flow rate (15 µL⋅min−1). This flow rate exerts very little force on branches and had little or no effect on debranching (Fig. 2).
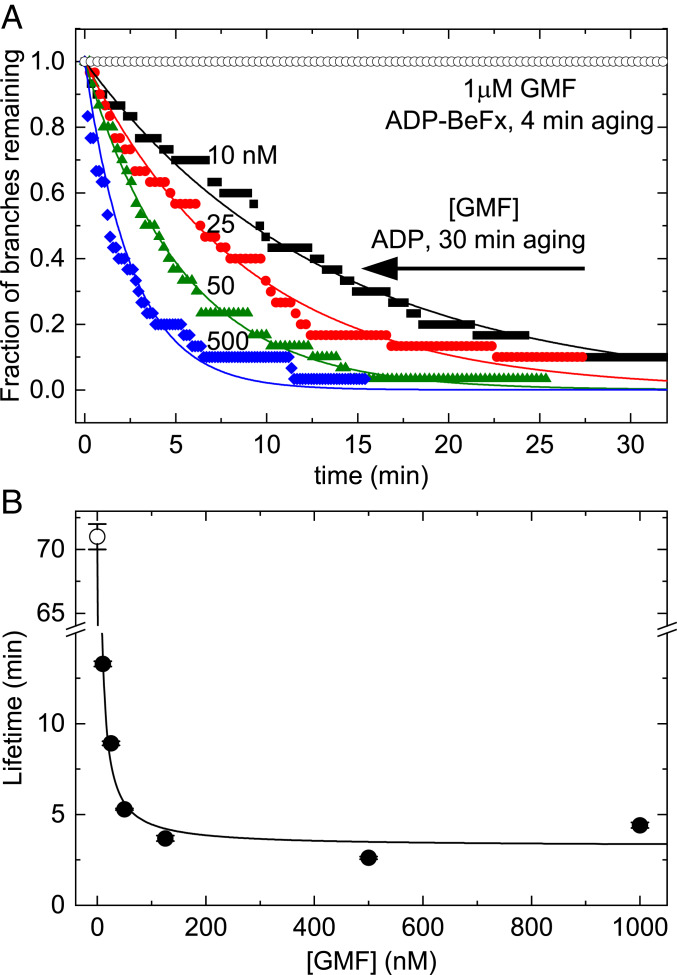
Concentrations of GMF up to 1 µM did not dissociate branches with ADP−BeFx−Arp2/3 complex (Fig. 3A), but nanomolar concentrations of GMF promoted dissociation of branches with ADP−Arp2/3 complex (Fig. 3). Time courses of dissociation of branches with ADP−Arp2/3 complex followed single exponentials (Fig. 3A) with lifetimes (τobs) that depended hyperbolically on the concentration of GMF (Fig. 3B).
Fig. 3.
BeFx inhibits debranching by GMF. (A) Dependence of the time course of dissociation of branches with ADP−BeFx−Arp2/3 complex or ADP−Arp2/3 complex on the concentration of GMF. Branches with ADP−BeFx−Arp2/3 complex were assembled in buffer containing 0.2 mM ATP, 2 mM BeSO4, and 10 mM NaF and aged for ∼4 min. Branches with ADP−Arp2/3 complex were assembled in buffer with 0.2 mM ATP and aged for 30 min to allow for ATP hydrolysis and phosphate dissociation (Fig. 2B). Debranching was initiated by flowing buffer with GMF at 15 μL⋅min−1 and continued throughout the debranching measurements. The smooth curves are the best fits of single exponentials to the data, yielding the (average) branch lifetimes; n = 30 branches for each trace. A concentration of 1 μM GMF did not dissociate branches with ADP−BeFx−Arp2/3 complex assembled from ATP-actin and ATP−Arp2/3 complex with BeFx and aged for ∼4 min (open black circles). (B) Dependence of the lifetimes of branches with ADP−Arp2/3 complex on the concentration of GMF at a buffer flow rate of 15 μL⋅min−1. The line is the best fit of Eq. 3 to the data, yielding a GMF binding affinity (Kd,GMF) of 40 (±10) nM and a maximum debranching rate constant (kdiss,GMF) of 0.31 (±0.05) min−1. At this low flow rate, branches with ADP−Arp2/3 complex (without GMF) dissociated with a rate constant (kdiss,0 = 0.014 ± 0.0002 min−1, indicated by an open circle) similar to that under zero force (Fig. 2B and Table 1). The uncertainty bars are within the data points and represent the SDs of lifetimes in the best single exponential fits of time traces in A.
Force increased the rate at which GMF dissociated branches with ADP−Arp2/3 complex (SI Appendix, Fig. S6), but dissociation was slower than predicted if GMF and force increased the rate of dissociation independently (i.e., their energetic contributions to debranching were additive). This raises the possibility that the reactions catalyzed by force and GMF are coupled and/or that debranching follows different pathways in the presence absence of force.
Discussion
Quantitative Analysis of the Two-State Model for Dissociation of Actin Filament Branches.
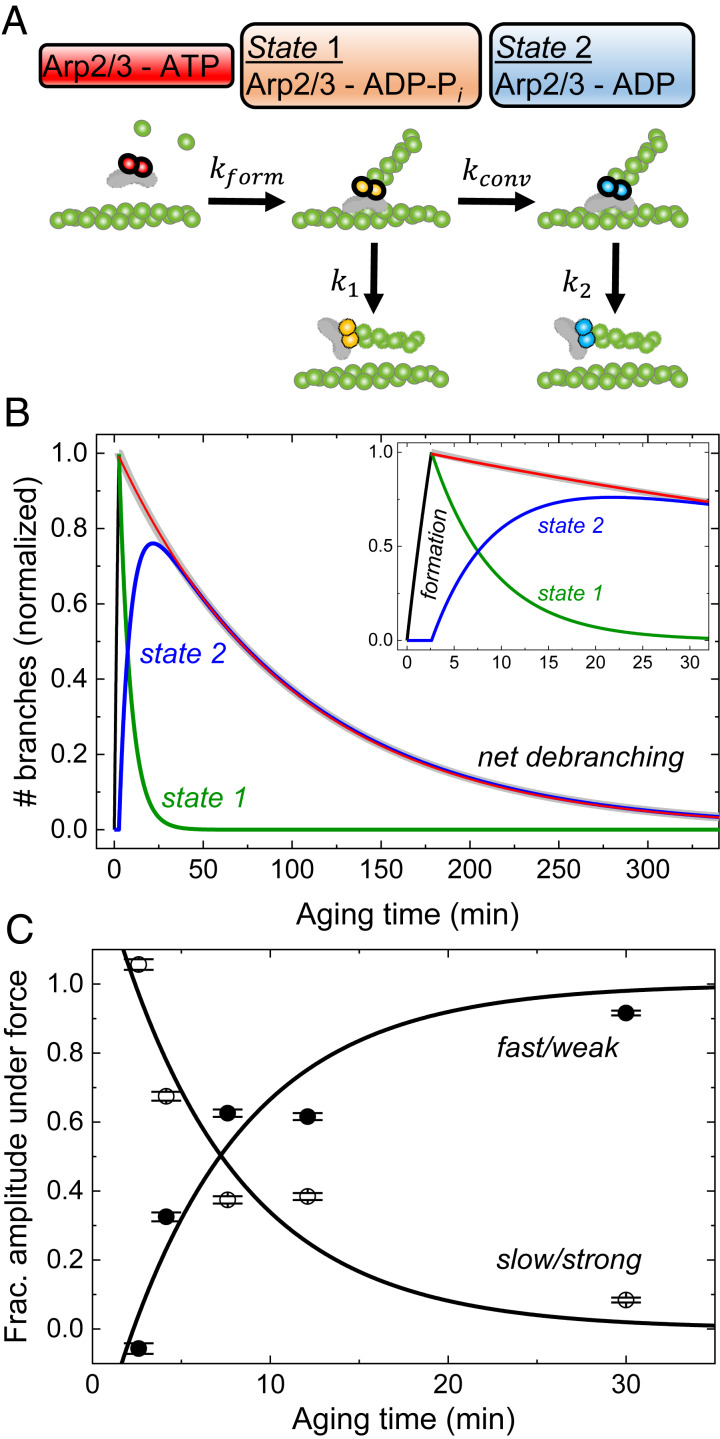
Our experiments show that the nucleotide bound to Arp2/3 complex determines the sensitivity of branches to dissociation by physical force. We use a formal description of a simple two-state model (Fig. 4A) to analyze our data and estimate the rate constants for the three reactions and their sensitivities to force.
Fig. 4.
Model and simulations of the pathways of branch formation, aging, and debranching. (A) Schematic of our hypothesis. Formation of a branch by ATP−Arp2/3 complex (red) is coupled to hydrolysis of ATP bound to the Arps (5, 7) with rate constant kform, yielding ADP−Pi−Arp2/3 complex (orange) in state 1. Irreversible phosphate dissociation with rate constant kconv converts state 1 to ADP−Arp2/3 complex (blue) in state 2. Branches dissociate from mother filaments with rate constants k1 for state 1 and k2 for state 2, both sensitive to force. (B) Simulated time course of the model showing how the populations of state 1 branches, state 2 branches, and dissociated branches evolve over time in the absence of force. We assumed that Mg-ATP-actin monomers and Mg-ATP−Arp2/3 complex formed branches for 2.6 min, when the free proteins were removed, and the reactions continued without additional branch formation. The red line represents the best single exponential fit to the observed debranching (i.e., combined from both states) starting with a normalized value of 1 at the end of branch formation (2.6 min). The experimentally determined or estimated rate constants used in the simulation are kform[Arp] = 0.12 min−1, kconv = 0.14 min−1, k1 = 0.012 min−1, and k2 = 0.01 min−1. (C) Aging time dependence of the fractional the slow and fast phase amplitudes in the debranching time courses under 500 µL⋅min−1 of buffer flow, obtained from double exponential fits to the time courses (Fig. 2B). These data were used for extraction of fundamental rate constants in Table 1. The best global fits of the two-state model (Eq. 2) to the fractional amplitude data gave rate constants for conversion kconv of 0.14 min−1, state 1 branch dissociation k1 of 0.012 s−1, and branch formation kform, of 0.02 μM−1⋅s−1 without force (Table 1). The one negative fast phase amplitude at 2.6 min results from a net increase in the state 2 branch population during debranching under force after 2.6 min aging time. The state 2 branch population is the net sum of depletion from debranching (negative contribution to population, exponential decay) and gain from conversion of state 1 branches (positive contribution, exponential rise). For short aging times, little or no state 2 branches exist for depletion, and the conversion from state 1 branches, represented by an exponential rise (negative amplitude), dominates the time course (54, 55). The uncertainty bars are SDs of the fractions of branches from the global double exponential fits of debranching time courses with different aging times in Fig. 2B.
Description of the model.
The model (Fig. 4A) assumes that branches form when ATP−Arp2/3 complex binds to the side of a mother filament and nucleates a daughter filament growing at its barbed end. Hydrolysis of ATP in the active sites of Acanthamoeba Arp2/3 complex is closely associated with nucleation of the daughter filament (5, 7), so we assume that state 1 branches have Arp2/3 complex with bound ADP−Pi. Dissociation of the γ-phosphate with a rate constant kconv converts branch state 1 to branch state 2 with ADP bound to Arp2/3 complex. Branches in either state can dissociate from the mother filament. The model predicts that, overall, net debranching time courses follow double exponentials with two rate constants. One observed rate constant is the sum of the rate constants for state 1 debranching and conversion (kobs,1(F) = k1(F) + kconv(F); (F) indicates optional force). The second observed rate constant corresponds to debranching of state 2 (kobs,2(F) = k2(F); SI Appendix, Eqs. S25 and S29). Force accelerates the dissociation of branches with either ADP−Arp2/3 and ADP−Pi−Arp2/3 complex, but branches with ADP−Pi or ADP−BeFx−Arp2/3 (mimicking the ADP−Pi state) complex are much more resistant to force than branches with ADP−Arp2/3 complex. As branches age, phosphate release converts slowly dissociating, “young and strong” state 1 branches with ADP−Pi−Arp2/3 complex into rapidly dissociating, “old and weak” state 2 branches with ADP−Arp2/3 complex.
Formulation of the model.
We analyzed the experimental data using a parallel debranching pathway (Fig. 4A and SI Appendix, Part 2 and Scheme S1) where “young” (strong, state 1) branches convert to “old” (weak, state 2) branches as they age (Fig. 4A). We fit the aging time dependence of the fractions of slowly and rapidly dissociating branches observed under force as the amplitudes (As,F and Af,F, respectively; Fig. 4C) of double exponential fits to the following functions (SI Appendix, Eqs. S34 and S37):
| [2] |
where t1 is the 2.6 min during which branches formed from ATP−Arp2/3 complex and ATP-actin monomers, tage is aging time, the intrinsic debranching rate constants are k1 for state 1 and k2 for state 2 in the absence of force, and kconv is the rate constant for conversion of state 1 to state 2 in the absence of force (Fig. 4A and SI Appendix, Part 2 and Scheme S1). The constants a and b are unitless composites of rate constants and initial branch concentrations defined in SI Appendix, Part 2 and Scheme S1 (SI Appendix, Eqs. S39 and S40). The constant a is force independent and determined only by intrinsic debranching in the absence of an applied force (SI Appendix, Eq. S39). The constant b is a function of debranching and conversion under force (SI Appendix, Eq. S40), yielding force-dependent amplitudes (As,F and Af,F) when debranching and/or conversion are force dependent.
Estimation of the rate constants for branch formation and phosphate dissociation.
Global analysis of the data (Figs. 2B and 4C and SI Appendix, Part 2) yielded an overall pseudo−first-order association rate constant for branch formation of 0.12 min−1 (SI Appendix, Part 2). Given 100 nM Arp2/3 complex used in our experiments, we estimate the second-order association rate constant for Arp2/3 complex binding to mother filaments and subsequent activation (kform) to be ∼0.02 μM−1⋅s−1 (Table 1), consistent with published reports (30–32).
The best fit of the fractional amplitudes to Eq. 2 (Fig. 4C) yielded a rate constant (kconv) of 0.14 (±0.03) min−1 (corresponding to a lifetime of >7 min) for conversion of state 1 branches to state 2 branches. We interpret the conversion reaction as the release of Pi from the Arp2/3 complex, and thus interpret state 2 branches as ADP−Arp2/3 complex branches. This rate constant inferred for Pi release from ADP-Pi Arp2/3 complex is similar to the rate constant of ∼0.18 min−1 for Pi release from actin filaments (29).
We can place a limit on the rate constant for phosphate binding to ADP-Arp2/3 complex in a branch junction (k+Pi). We interpret conversion to reflect Pi release, so k−Pi = kconv = 0.14 (±0.03) min−1, and know that Pi binds Arp2/3 complex in branches with a low affinity (Kd > 20 mM; SI Appendix, Fig. S5A). Therefore, the second order association rate constant of k+Pi is less than 7 M−1 min−1 = 0.12 M−1 s−1. This value is two orders of magnitude slower than Pi binding to ADP-actin subunits in the interior of filaments [600 M−1min−1 = 10 M−1s−1 (29)].
Estimation of the rate constants for branch dissociation at low force.
The rate constant for dissociation of state 2 branches at zero force is k2 ≈ 0.01 min−1, based on the intercept of the force-dependence of ADP-Arp2/3 complex (state 2) branch lifetimes (Fig. 2D). The rate constant for debranching from state 1 at zero force (k1) is ∼0.012 min−1, from the analysis of the aging time-dependence of time course of dissociation of branches (Figs. 2B and 4C, Table 1, and SI Appendix, Part 2 and Eq. S42). As a result of these two rate constants being similar (k1 ≈ k2), debranching without force follows a single exponential with a rate constant of ∼k2 (SI Appendix, Part 2 and Eq. S25 and Fig. 4B), even though three reactions (conversion and debranching from state 1 and state 2) occur simultaneously.
Time courses also follow single exponentials at low forces (<0.6 pN), independent of the aging time (Fig. 2 Cand D). This behavior arises because dissociation is much slower than conversion (k1 << kconv), so ADP-Pi branches convert to ADP before dissociating. At forces >1 pN, the observed dissociation time courses follow double exponentials because kconv < k1 and debranching occurs from both ADP-Pi and ADP branches.
Estimation of the force sensitivity of the branch dissociation rate constants.
Force increases the dissociation rate constants of both branch states, but has a larger effect on state 2 branches with ADP-Arp2/3 complexes than state 1 branches with ADP-Pi-Arp2/3 complexes (Fig. 2D). The best global fits of the double exponential time courses at intermediate aging times yielded a ∼20-fold difference in the lifetimes of the species that dissociated slowly (ks,F = 0.32 (±0.005) min−1) and rapidly (kf,F = 6.67 (±0.44) min−1) at a buffer flow rate of 500 μL min−1 (Fig. 2B). Under this force, the observed fast rate constant is the rate constant for the state 2 branch dissociation (i.e., kf,F = k2,F) and the observed slow rate constant is the sum of the rate constants for dissociation for state 1 branches and conversion from state 1 to state 2 (i.e., ks,F = k1,F + kconv,F; SI Appendix, Part 2 and Eqs. S29 and S41).
Force produced by a flow of 500 μL min−1 increased the observed dissociation of branches ∼670-fold (from k2 = 0.01 to k2,F = 6.67 min−1) for those with ADP-Arp2/3 complex but only 32-fold (from k1 = 0.012 to k1,F = ks,F − kconv,F < ks,F = 0.32 min−1; Fig. 2B and SI Appendix, Fig. S5) for those with ADP-Pi-Arp2/3 complex. This difference in force sensitivity of the two states explains the dramatic acceleration in overall debranching with aging (Fig. 2B). Furthermore, we estimate that force produced by 500 μL min−1 flow increased kconv only two-fold from kconv = 0.14 min−1 in the absence of force to kconv,F = ks,F − k1,F < ks,F = 0.32 min−1 in the presence of force (SI Appendix, Fig. S5).
Implications of the Force Sensitivity of Branches for Their Turnover in Cells.
Implications for debranching by myosin motor proteins.
A sustained flow force of ∼1 pN dissociates ADP-Arp2/3 complex branches in <30 s (Fig. 2D), raising the possibility that pN contractile forces generated by myosin motors could rapidly debranch and reorganize Arp2/3 complex networks. Myosin accelerates network disassembly and reorganization in both biomimetic systems (33, 34) and in cells (35, 36). We note, however, that the work (i.e., energy term) from the applied force determines the overall debranching acceleration, not the force per se.
Implications of Nucleotide-Dependent Force Sensitivity of Arp2/3 Complex Debranching.
“Young” branches with ADP−Pi−Arp2/3 complex are more resistant to dissociation by force than “old” branches with ADP−Arp2/3 complex. Nucleation-promoting factors associated with membranes (plasma membrane, vesicles, intracellular bacteria) activate Arp 2/3 complex, so “young” branches are located closer to these surfaces than “old” branches. If force is uniformly distributed across the filament network, old branches farthest from the surface with nucleation-promoting factors would be preferentially debranched.
This differential response to force may contribute to the observed remodeling of cellular branched actin networks under load, including local changes in branch density (13–15). We apply a simple a minimal kinetic model (SI Appendix, Part 3) to analyze how selective, force-sensitive debranching might influence the distribution of branches in actin networks at the leading edge of cells. The model assumes Arp 2/3 complex is activated uniformly at the membrane, such that the branch density distribution is also uniform, on average, along the plane parallel to the cell membrane at the leading edge. The model with the experimental parameters determined here predicts that the density of branches decays exponentially (see also refs. 37–39) along the axis perpendicular to the membrane, toward cell interior in both the absence and presence of force, but preferential debranching of old Arp 2/3 complexes shortens the branch density decay length and can, under some conditions, increase the local branch density near the membrane (SI Appendix, Part 3 and Figs. S9 and S10). Thus, external force on the leading edge of cells favors debranching of old branches that have migrated toward the cell interior over young branches still near the membrane and can thus contribute to spatial and vectorial network turnover. External force may also influence binding of cofilin (40) and GMF (Fig. 3; discussed below), both of which display debranching activity (20, 24).
GMF Selectively Dissociates ADP−Arp2/3 Complex Branches.
Fission yeast GMF increases the rate constant for dissociation of branches with ADP−Arp 2/3 complex up to ∼20-fold, but does not dissociate branches with ADP−BeFx−Arp 2/3 complex (Fig. 3). The following parallel reactions describe GMF-catalyzed dissociation of branches with ADP−Arp2/3 complex:
| Scheme 1 |
where kdiss,0 and kdiss,GMF are the dissociation rate constants of branches formed by Arp2/3 complex alone (Arp) and GMF-bound Arp2/3 complex (Arp-GMF), and Kd,GMF is the affinity of GMF for Arp 2/3 complex in a branch junction. This scheme assumes that GMF binding equilibrates rapidly compared to the rate of debranching (and k+GMF[GMF], k−GMF >> kdiss,0, kdiss,GMF) and that [GMF] >> [Arp2/3 complex].
Debranching from the parallel pathway in Scheme 1 follows a single exponential (41). The dependence of the observed branch lifetime (τobs) on the concentration of GMF can be expressed in two ways, first,
| [3] |
Alternatively, the relationships can also be expressed in terms of observed rate constants (41, 42),
| [4] |
The best fit of Eq. 3 to the dependence of the observed branch lifetime (τobs) on the concentration of GMF (Fig. 3B) yielded an apparent affinity of GMF for ADP−Arp2/3 complex (Kd, GMF) of 40 (±10) nM and a lifetime of GMF bound to ADP−Arp2/3 complex in branches (τGMF) of 3.3 (±0.5) min. This corresponds to a debranching rate constant (kGMF = 1/τGMF) of 0.31 (±0.05) min−1. In Eqs. 3 and 4, k0 is the debranching rate constant and τ0 is the branch lifetime without GMF under very low flow rate (15 μL⋅min−1), that is, very low average force. The best fit yielded k0 = 1/τ0 = 0.014 (±0.0002) min−1, corresponding to τ0 = 71 (±1) min. These values are slightly faster than the intrinsic dissociation rate constant for branches with ADP−Arp2/3 complex in the absence of force (>100 min; Fig. 3) due to the 15 μL⋅min−1 flow rate employed in this experiment. We note, for general readers, that the GMF concentration at half-maximum effect is given by Kd,GMF (τGMF/τ0) in Eq. 3 and Kd,GMF in Eq. 4, so its value differs depending on how the data are plotted.
Reported affinities of GMF for soluble Arp2/3 complex (Kd,GMF) vary widely: >>10 μM for ATP−Arp 2/3 complex (43); 0.7 μM for ADP−Arp2/3 complex (43); and 13 nM (20–22) or ∼1 μM (44) for Arp2/3 complex without a specified nucleotide. We found that the measured affinity of GMF for Arp2/3 complex in branch junctions bound to both a mother and daughter filament depended on the bound nucleotide. Arp2/3 complex used in these studies came from different organisms: Saccharomyces cerevisiae (20–22), bovine brain (43), and S. pombe (this study and ref. 44).
Thus, GMF preferentially dissociates “old” branches with ADP−Arp2/3 complex rather than “young” branches with ADP−Pi−Arp2/3 complex. Given that GMF has a much lower affinity for ATP−Arp2/3 complex in solution than ADP−Arp2/3 complex (43), weak binding of GMF to ADP−Pi−Arp2/3 complex in branches is the most likely explanation for the resistance of young branches and branches with ADP−BeFx−Arp2/3 complex to dissociation by GMF. The nucleotide bound to Arp2/3 complex might also affect the debranching reaction directly.
Saturating GMF destabilizes ADP−Arp2/3 complexes, reducing their lifetime from more than 60 min to ∼3 min (Fig. 3). While GMF strongly promotes debranching, these long lifetimes (3 min) at saturating GMF concentrations place limits on the role of GMF in mediating debranching and network turnover in cells. More-rapid debranching may be achieved by combining GMF with force and/or from contributions from other debranching proteins such as cofilin, which dissociates branches on a second time scale at micromolar concentrations without force (24).
Arp2/3 Complex Likely Dissociates with the Daughter Filament.
A daughter filament and its associated Arp2/3 complex dissociate simultaneously (within the 100 ms time resolution of our experiments) during debranching events. We cannot eliminate a pathway in which the daughter filament detaches first, followed by rapid release of Arp2/3 complex. However, we favor a mechanism where the interface between Arp2/3 complex and the mother filament ruptures and simultaneously releases the daughter filament with Arp2/3 complex on its pointed end. This mechanism is consistent with actin filaments rarely fragmenting under debranching conditions, so the rate constant for rupture of the actin−actin interface is hundreds of times slower than the rate constant for rupturing a branch junction. Given that the interface between Arp2/3 complex and the daughter filament is structurally similar on many levels to an actin−actin interface (45), it is likely to rupture slowly like actin filaments.
Materials and Methods
Protein Purification.
Rabbit skeletal muscle actin was purified from back and leg muscles and labeled on lysine residues with NHS esters of Alexa 568, Alexa 647, or biotin (46, 47) or on Cys-374 with pyrene iodoacetamide (48). Actin monomers with bound ATP were passed twice through a desalting column to exchange into G-Buffer (20 mM Tris pH 8, 2 mM CaCl2, 10 mM NaN3, 0.2 mM ATP, and 0.5 mM dithiothreitol [DTT]) containing 0.2 mM AMPPNP instead of ATP and incubated for 1 h at 25 °C. Remaining free nucleotides were removed with a desalting column in G-Buffer containing AMPPNP instead of ATP prior to polymerization experiments. Arp2/3 complex was stored in ATP buffer solution composed of 10 mM Pipes at pH 6.8, 100 mM KCl, 1 mM MgCl2, 0.25 mM ethylene glycol bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid [EGTA], 0.1 mM ATP, and 1 mM DTT.
Recombinant fission yeast glutathione S-transferase (GST)-VCA was purified from bacteria (49). S. pombe Arp2/3 complex was purified from the TP150 strain, a protease-deleted S. pombe strain (49). Fluorescent Arp2/3 complex was generated by conjugating Alexa 488 to a snap tag on the C terminus of the Arpc5 subunit. Bulk polymerization assays with pyrenyl actin and GST-VCA showed that Arp2/3 complex with 82% labeling of the snap tag nucleated 91% as many actin filaments as unlabeled Arp2/3 complex (4.2 nM vs. 4.6 nM of barbed ends at half-maximal polymerization; SI Appendix, Fig. S6) (refs. 48 and 49 and SI Appendix, Fig. S8).
A complementary DNA for S. pombe GMF was cloned and inserted into the pet28a plasmid that includes GST and Tobacco Etch Virus (TEV) protease sites. Recombinant GMF was expressed in Rosetta2 BL21 Escherichia coli cells (Novagen) and purified using glutathione affinity chromatography. After cleaving off GST with TEV protease, a second round of glutathione chromatography removed the protease and GST. After fast protein liquid chromatography on a MonoQ column (20), the GMF was greater than 95% pure when analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Microscopy and Microfluidics.
A total internal reflection fluorescence (TIRF) microscopy system with a Till iMic digital microscope equipped with a ×100 objective (Olympus) and an Andor iXon897 electron multiplying charge-coupled device (EMCCD) camera was used in this study. Images were acquired at a rate of 5 to 30 frames per second. Coverslips were cleaned with the following solutions, all incubated in a sonicator for 30 min and rinsed intensively with Milli-Q water in between steps: 2% Hellmanex, water, acetone, 1 M HCl, 5 M KOH, and hexane, then silanized with 650 µL of dichlorodimethyl silane in 500 mL of hexane for 30 min, and subsequently rinsed and sonicated in hexane for 3 × 1 min. Coverslips were dried using a stream of nitrogen gas and stored in falcon tubes at −20 °C for up to 2 mo (50).
A glass microfluidic chamber was constructed as described in ref. 51. Briefly, the input and output ports for flow solution were formed in polydimethylsiloxane (PDMS) using a flat-tip needle. Holes connecting the PDMS to parallel sample chambers were drilled through the glass with a diamond tip bit. Subsequently, a plasma cleaner was employed to bond PDMS to slide glass. Immediately before use, chambers were assembled using parafilm on the glass slide opposite to PDMS ports, and a coverslip was placed over the parafilm and sealed with heat.
A fixed rate hydrodynamic flow exerted by an automatic syringe pump applied pulling forces on actin filaments. The magnitude of pulling force on a branch joint (Fd; SI Appendix, Fig. S1 A and B) scales with daughter filament branch length and flow rate. These values span a range up to a few piconewtons for the daughter filament lengths and flow rates examined here. Pulling force was calculated according to the Batchelor (52) equation,
| [5] |
Eq. 5 estimates drag force on a cylindrical filament with correction for the height (h) of the filaments from the surface of the flow chamber (53). Solution viscosity (η) is assumed to equal unity; l is the (daughter) filament length and varies for individual branches. The average actin filament radius is r = 4 nm, and v is the linear fluid velocity in the plane of the filament. We assume an average height (h) of 200 nm from the flow chamber surface. The fluid velocity (v) is proportional to bulk flow rate and was determined from the movement of 100 nM TetraSpek beads (Thermo Fisher) through the sample chamber (52). Only beads moving parallel to the surface were employed in our analysis, as the flow velocity profile changes with the height from the chamber surface. Bead flow velocity was measured via ImageJ “Manual Tracking” and used in force calculations (52). In the experiments presented in Fig. 2 A and C, multiple experiments at different flow rates were performed, and the force on each branch was calculated using the flow rate and length of the branch. Then, debranching data from different flow cells with a range of flow rates were binned according to the force on each branch and plotted as survival time courses, given by the fraction of branches remaining at each time point. For example, the branches in Fig. 2A were divided into bins of at least 13 branches. The bin size for each data point is approximately the midpoint between the point itself and its neighboring data points (Fig. 2D). At low forces (0.002 pN to 0.4 pN), the bin sizes were small, for example, 0 pN to 0.04 pN, 0.1 pN to 0.2 pN, etc., and, at higher forces (>∼1 pN), the bin sizes were larger, for example, 0.8 pN to 1.4 pN, 1.4 pN to 2.4 pN, etc.
Some of these parameters are uncertain, so the estimated force values may be offset systematically for all of our experiments. The least certain parameter is the distance of individual filaments from the surface, which may vary by <50% from the average distance of 200 nm assumed here. A 500 μL⋅min−1 flow rate produces forces on a 1.5-µm branch of 0.94 pN at a height of 300 nm, 1.02 pN at 200 nm, and 1.20 pN at 100 nm (SI appendix in ref. 52). Therefore, the absolute forces and parameters calculated from them may be imprecise, but no more than 20%, and the relative forces between experiments can be compared.
Preparation of Branched Actin Filament Networks and Experimental Procedures.
KMIE buffer (6, 23) (10 mM imidazole pH 7.0, 50 mM KCl, 2 mM MgCl2, 1 mM EGTA, 0.2 mM ATP, 2 mM DTT) supplemented with 15 mM glucose, 0.02 mg⋅mL−1 catalase, and 0.1 mg⋅mL−1 glucose oxidase was used for all microscope experiments, unless noted otherwise. Filaments of Alexa 568−labeled actin (Fig. 1A, red filament segment; 15% labeled with Alexa 568 and containing 10% biotinylated actin subunits) were polymerized in KMIE buffer, sheared by vigorous pipetting, and subsequently mixed with 1.5 μM Alexa 647-labeled actin monomers (Fig. 1A, green segment and branch), 0.1 μM Arp2/3 complex, and 0.5 μM GST-VCA in KMIE buffer. This solution was incubated for 1 min to 2 min to initiate branch formation. During the branching reaction, green actin monomers elongated from either the barbed end of short Alexa 568 actin segments or mother filament bound Arp2/3 complex VCA, forming mother or daughter filaments, respectively. This mixture was pipetted into flow chambers where the short, red actin segments bind the neutravidin-coated coverslip surface, and the branching reaction was allowed to continue for 1 min to 2 min. The total time of branch formation and aging for a given experiment (e.g., 2.6 min) is indicated in the text. Branch formation and filament elongation was then terminated by removing untethered proteins (labeled actin, Arp2/3 complex, and GST-VCA) from the sample chamber with gentle flow of KMIE buffer. Tethered filaments were further aged for different time periods after the removal of unbound protein. Low (2 µL⋅min−1) flow applied during aging ensured irreversible debranching. Filament segments with Alexa 568 actin subunits (Fig. 1A, red) were tethered to the surface, while Alexa 647 actin (Fig. 1A, green) mother and daughter filaments were allowed to freely fluctuate and align with flow. Unlabeled actin monomer (0.2 μM) was included in flow buffer solution to prevent filament depolymerization. The “aging time” specified in each experiment is the time between the initial mixing of the proteins and application of debranching flow, including the time for branch formation and further aging. The age times were precisely measured in the experiment in Fig. 2B but are approximate times in all other experiments. Debranching under force was performed by applying flow at a fixed rate (specified in text) throughout the experiment. Since unbound proteins were removed after the branch formation period, actin filament elongation was terminated, and individual branch length (l) and consequently applied force (F) on the branch joint were constant (Eq. 5).
Experiments with BeFx were performed two different ways. 1) For experiments with ATP−Arp2/3 complex (Fig. 2 C, Inset), branches were formed from ATP-actin monomers and ATP−Arp2/3 complex as described above but with 2 mM BeSO4 and 10 mM NaF included in the buffer at all times, and aged for ∼4 min before applying various rates of flow with KMIE buffer supplemented with 2 mM BeSO4 and 10 mM NaF. For the experiment with BeFx and 30-min aging (Fig. 2B, gray), ATP−Arp2/3 complex branches were formed as described above and aged for 30 min with 2 mM BeSO4 and 10 mM NaF before applying flow with KMIE buffer. 2) For experiments with ADP−Arp2/3 complex and BeFx (Fig. 2B, brown), mother filaments with ATP-actin (10% biotinylated) monomers were immobilized on the surface as described above, and the KMIE buffer (which includes ATP) was washed out and replaced with KMIE ADP buffer supplemented with 2 mM BeSO4 and 10 mM NaF. Then ADP-actin monomers, ADP−Arp2/3 complex, and GST-VCA in KMIE buffer with 2 mM BeSO4 and 10 mM NaF and 2 mM ADP instead of 2 mM ATP were gently flowed into the chamber and allowed to form branches. After a ∼4-min branch formation period, all unbound proteins were washed out with KMIE ADP and 2 mM BeSO4 and 10 mM NaF and maintained under flow as noted. ADP-actin and ADP−Arp2/3 complex were prepared by exchanging nucleotide from ATP-actin and ATP−Arp2/3 complex, respectively, using desalting columns. ADP−Arp2/3 complex never formed branches unless 2 mM BeSO4 and 10 mM NaF was present as reported previously and shown here (SI Appendix, Fig. S3) (5, 6).
For experiments with AMPPNP mother filaments, filaments were polymerized from AMPPNP actin monomers and immobilized on the surface as described above. Branch formation was initiated with ATP−Arp2/3 complex as described above in KMIE buffer with 0.2 mM AMPNP instead of 0.2 mM ATP. Since AMPPNP−Arp2/3 complex does not form branches (5), verified in our experiments (SI Appendix, Fig. S3), all branches formed from Arp2/3 complex with bound ATP. Since the flow buffer included 0.2 mM AMPPNP, all actin monomers remained bound to AMPNP. The solution with Arp2/3 complex accounted for less than 1% of the final solution volume, so it introduced a negligible amount of ATP.
In debranching experiments with GMF with or without BeFx, samples were prepared as above except that debranching was initiated by gentle flow of buffer (15 μL⋅min−1, very low force) containing a range of concentrations of GMF, and the flow was maintained throughout the experiment.
Data Analysis.
Analysis of branch dissociation was done by manual tracking with ImageJ (https://imagej.nih.gov/). Branches that were fluctuating at all times and did not stick to the glass surface were selected randomly to minimize bias. These branches were labeled, cataloged, and observed for their entire lifetimes. Origin (https://www.originlab.com/) was used to fit all data and make plots. For the Arp2/3 complex debranching images in Fig. 1, a Gaussian blur with a sigma radius of 1.25 pixels was applied. Then images were background subtracted with a rolling ball radius of 15 pixels and subsequently contrast enhanced.
Data Availability.
All protein purification protocols are referenced in Materials and Methods. Purification of GMF is described in Materials and Methods. Plasmid and the DNA sequence of GMF are provided in SI Appendix, Part 4. Details on movies for microscopy experiments are provided in SI Appendix, Part 1.
Supplementary Material
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. H.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911183117/-/DCSupplemental.
References
- 1.Pollard T. D., Cooper J. A., Actin, a central player in cell shape and movement. Science 326, 1208–1212 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janmey P. A., McCulloch C. A., Cell mechanics: Integrating cell responses to mechanical stimuli. Annu. Rev. Biomed. Eng. 9, 1–34 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Pollard T. D., Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 36, 451–477 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Le Clainche C., Didry D., Carlier M. F., Pantaloni D., Activation of Arp2/3 complex by Wiskott-Aldrich Syndrome protein is linked to enhanced binding of ATP to Arp2. J. Biol. Chem. 276, 46689–46692 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Dayel M. J., Holleran E. A., Mullins R. D., Arp2/3 complex requires hydrolyzable ATP for nucleation of new actin filaments. Proc. Natl. Acad. Sci. U.S.A. 98, 14871–14876 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanchoin L., Pollard T. D., Mullins R. D., Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr. Biol. 10, 1273–1282 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Dayel M. J., Mullins R. D., Activation of Arp2/3 complex: Addition of the first subunit of the new filament by a WASP protein triggers rapid ATP hydrolysis on Arp2. PLoS Biol. 2, E91 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ingerman E., Hsiao J. Y., Mullins R. D., Arp2/3 complex ATP hydrolysis promotes lamellipodial actin network disassembly but is dispensable for assembly. J. Cell Biol. 200, 619–633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Clainche C., Pantaloni D., Carlier M. F., ATP hydrolysis on actin-related protein 2/3 complex causes debranching of dendritic actin arrays. Proc. Natl. Acad. Sci. U.S.A. 100, 6337–6342 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin A. C., Welch M. D., Drubin D. G., Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nat. Cell Biol. 8, 826–833 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Fletcher D. A., Mullins R. D., Cell mechanics and the cytoskeleton. Nature 463, 485–492 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blanchoin L., Boujemaa-Paterski R., Sykes C., Plastino J., Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 94, 235–263 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Bieling P. et al., Force feedback controls motor activity and mechanical properties of self-assembling branched actin networks. Cell 164, 115–127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parekh S. H., Chaudhuri O., Theriot J. A., Fletcher D. A., Loading history determines the velocity of actin-network growth. Nat. Cell Biol. 7, 1219–1223 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Mueller J. et al., Load adaptation of lamellipodial actin networks. Cell 171, 188–200.e16 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Mahaffy R. E., Pollard T. D., Kinetics of the formation and dissociation of actin filament branches mediated by Arp2/3 complex. Biophys. J. 91, 3519–3528 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahaffy R. E., Pollard T. D., Influence of phalloidin on the formation of actin filament branches by Arp2/3 complex. Biochemistry 47, 6460–6467 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berro J., Sirotkin V., Pollard T. D., Mathematical modeling of endocytic actin patch kinetics in fission yeast: Disassembly requires release of actin filament fragments. Mol. Biol. Cell 21, 2905–2915 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sirotkin V., Berro J., Macmillan K., Zhao L., Pollard T. D., Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast. Mol. Biol. Cell 21, 2894–2904 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gandhi M. et al., GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Curr. Biol. 20, 861–867 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goode B. L., Sweeney M. O., Eskin J. A., GMF as an actin network remodeling factor. Trends Cell Biol. 28, 749–760 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ydenberg C. A. et al., GMF severs actin-Arp2/3 complex branch junctions by a cofilin-like mechanism. Curr. Biol. 23, 1037–1045 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang H. et al., Site-specific cation release drives actin filament severing by vertebrate cofilin. Proc. Natl. Acad. Sci. U.S.A. 111, 17821–17826 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan C., Beltzner C. C., Pollard T. D., Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr. Biol. 19, 537–545 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell G. I., Models for the specific adhesion of cells to cells. Science 200, 618–627 (1978). [DOI] [PubMed] [Google Scholar]
- 26.Rakshit S., Sivasankar S., Biomechanics of cell adhesion: How force regulates the lifetime of adhesive bonds at the single molecule level. Phys. Chem. Chem. Phys. 16, 2211–2223 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Muhlrad A., Cheung P., Phan B. C., Miller C., Reisler E., Dynamic properties of actin. Structural changes induced by beryllium fluoride. J. Biol. Chem. 269, 11852–11858 (1994). [PubMed] [Google Scholar]
- 28.Ge P., Durer Z. A. O., Kudryashov D., Zhou Z. H., Reisler E., Cryo-EM reveals different coronin binding modes for ADP- and ADP-BeFx actin filaments. Nat. Struct. Mol. Biol. 21, 1075–1081 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlier M. F., Pantaloni D., Binding of phosphate to F-ADP-actin and role of F-ADP-Pi-actin in ATP-actin polymerization. J. Biol. Chem. 263, 817–825 (1988). [PubMed] [Google Scholar]
- 30.Helgeson L. A., Nolen B. J., Mechanism of synergistic activation of Arp2/3 complex by cortactin and N-WASP. eLife 2, e00884 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith B. A., Daugherty-Clarke K., Goode B. L., Gelles J., Pathway of actin filament branch formation by Arp2/3 complex revealed by single-molecule imaging. Proc. Natl. Acad. Sci. U.S.A. 110, 1285–1290 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith B. A. et al., Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. eLife 2, e01008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reymann A. C. et al., Actin network architecture can determine myosin motor activity. Science 336, 1310–1314 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sonal et al., Myosin-II activity generates a dynamic steady state with continuous actin turnover in a minimal actin cortex. J. Cell Sci. 132, jcs219899 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Wilson C. A. et al., Myosin II contributes to cell-scale actin network treadmilling through network disassembly. Nature 465, 373–377 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medeiros N. A., Burnette D. T., Forscher P., Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat. Cell Biol. 8, 215–226 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Vinzenz M. et al., Actin branching in the initiation and maintenance of lamellipodia. J. Cell Sci. 125, 2775–2785 (2012). [DOI] [PubMed] [Google Scholar]
- 38.McMillen L. M., Vavylonis D., Model of turnover kinetics in the lamellipodium: Implications of slow- and fast- diffusing capping protein and Arp2/3 complex. Phys. Biol. 13, 66009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyoshi T. et al., Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: Evidence of frequent filament severing. J. Cell Biol. 175, 947–955 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayakawa K., Tatsumi H., Sokabe M., Actin filaments function as a tension sensor by tension-dependent binding of cofilin to the filament. J. Cell Biol. 195, 721–727 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hannemann D. E., Cao W., Olivares A. O., Robblee J. P., De La Cruz E. M., Magnesium, ADP, and actin binding linkage of myosin V: Evidence for multiple myosin V-ADP and actomyosin V-ADP states. Biochemistry 44, 8826–8840 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Wong E. V. et al., Nup159 weakens Gle1 binding to Dbp5 but does not accelerate ADP release. J. Mol. Biol. 430, 2080–2095 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boczkowska M., Rebowski G., Dominguez R., Glia maturation factor (GMF) interacts with Arp2/3 complex in a nucleotide state-dependent manner. J. Biol. Chem. 288, 25683–25688 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakano K., Mabuchi I., Actin-depolymerizing protein Adf1 is required for formation and maintenance of the contractile ring during cytokinesis in fission yeast. Mol. Biol. Cell 17, 1933–1945 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rouiller I. et al., The structural basis of actin filament branching by the Arp2/3 complex. J. Cell Biol. 180, 887–895 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCullough B. R., Blanchoin L., Martiel J. L., De la Cruz E. M., Cofilin increases the bending flexibility of actin filaments: Implications for severing and cell mechanics. J. Mol. Biol. 381, 550–558 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elam W. A. et al., Phosphomimetic S3D cofilin binds but only weakly severs actin filaments. J. Biol. Chem. 292, 19565–19579 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cooper J. A., Walker S. B., Pollard T. D., Pyrene actin: Documentation of the validity of a sensitive assay for actin polymerization. J. Muscle Res. Cell Motil. 4, 253–262 (1983). [DOI] [PubMed] [Google Scholar]
- 49.Nolen B. J., Pollard T. D., Structure and biochemical properties of fission yeast Arp2/3 complex lacking the Arp2 subunit. J. Biol. Chem. 283, 26490–26498 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hua B. Y. et al., An improved surface passivation method for single-molecule studies. Nat. Methods 11, 1233–1236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson-Chavarria E. M., Agrawal U., Tanyeri M., Kuhlman T. E., Schroeder C. M., Automated single cell microbioreactor for monitoring intracellular dynamics and cell growth in free solution. Lab Chip 14, 2688–2697 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Courtemanche N., Lee J. Y., Pollard T. D., Greene E. C., Tension modulates actin filament polymerization mediated by formin and profilin. Proc. Natl. Acad. Sci. U.S.A. 110, 9752–9757 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brennen C., Winet H., Fluid-mechanics of propulsion by cilia and flagella. Annu. Rev. Fluid Mech. 9, 339–398 (1977). [Google Scholar]
- 54.Robblee J. P., Cao W., Henn A., Hannemann D. E., De La Cruz E. M., Thermodynamics of nucleotide binding to actomyosin V and VI: A positive heat capacity change accompanies strong ADP binding. Biochemistry 44, 10238–10249 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Taylor E. W., Kinetic studies on the association and dissociation of myosin subfragment 1 and actin. J. Biol. Chem. 266, 294–302 (1991). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All protein purification protocols are referenced in Materials and Methods. Purification of GMF is described in Materials and Methods. Plasmid and the DNA sequence of GMF are provided in SI Appendix, Part 4. Details on movies for microscopy experiments are provided in SI Appendix, Part 1.