Significance
Exocytosis is of vital importance for life, as it allows cells to communicate with each other and with their environment. This dynamic process is highly regulated with a set of both positive and negative regulators. While promotors of exocytosis are well studied, negative regulators are poorly understood. We discovered that a small SNARE protein amisyn (STXBP6) acts as a vertebrate-specific competitor of synaptobrevin-2, a key player in exocytosis. Amisyn contains an N-terminal pleckstrin homology domain that mediates its transient association with the plasma membrane by binding to phospholipid PI(4,5)P2. Both the pleckstrin homology and SNARE domains are needed to inhibit exocytosis. Understanding the mechanisms by which amisyn regulates exocytosis will advance our comprehension of pathological processes in diabetes and autism.
Keywords: SNARE complex; exocytosis inhibition; PI(4,5)P2; tomosyn; autism spectrum disorders
Abstract
The functions of nervous and neuroendocrine systems rely on fast and tightly regulated release of neurotransmitters stored in secretory vesicles through SNARE-mediated exocytosis. Few proteins, including tomosyn (STXBP5) and amisyn (STXBP6), were proposed to negatively regulate exocytosis. Little is known about amisyn, a 24-kDa brain-enriched protein with a SNARE motif. We report here that full-length amisyn forms a stable SNARE complex with syntaxin-1 and SNAP-25 through its C-terminal SNARE motif and competes with synaptobrevin-2/VAMP2 for the SNARE-complex assembly. Furthermore, amisyn contains an N-terminal pleckstrin homology domain that mediates its transient association with the plasma membrane of neurosecretory cells by binding to phospholipid PI(4,5)P2. However, unlike synaptrobrevin-2, the SNARE motif of amisyn is not sufficient to account for the role of amisyn in exocytosis: Both the pleckstrin homology domain and the SNARE motif are needed for its inhibitory function. Mechanistically, amisyn interferes with the priming of secretory vesicles and the sizes of releasable vesicle pools, but not vesicle fusion properties. Our biochemical and functional analyses of this vertebrate-specific protein unveil key aspects of negative regulation of exocytosis.
The complex, highly controlled cascade of protein–protein and lipid–protein interactions, Ca2+-triggered exocytosis, leads to the externalization of secretory molecules and neurotransmitters. The machinery mediating and regulating exocytosis has been studied in detail over the past three decades. Numerous proteins were identified and characterized, including the SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) protein superfamily, central to the later steps of regulated exocytosis and bilayer fusion. All members of the SNARE family have a characteristic conserved homologous stretch of 60 to 70 amino acids, referred to as a SNARE motif (1, 2).
Four SNARE motifs assemble spontaneously into a thermostable, sodium dodecyl sulfate (SDS) and protease resistant coiled-coil bundle termed the SNARE core complex (3, 4). Heptad repeats in components of the core complex form 16 conserved layers of interacting amino acid side chains that are arranged perpendicular to the axis of the complex. All layers except one contain hydrophobic amino acids; the unique central layer, termed the “0-layer,” is hydrophilic and consists of three glutamine (Q) and one arginine (R) residue that is stabilized by ionic interactions. Based on this characteristic, SNARE proteins are classified into four subfamilies: Qa, Qb, Qc, and R-SNAREs (4). Three or four SNARE proteins bundle together to form a QaQbQcR complex; this arrangement contributes to specific Q- and R-SNARE pairing.
Among the large number of SNARE proteins, the members of the neuronal SNARE complex are most intensely studied. This complex, first purified in Söllner et al. (5), comprises a minimal fusion machinery consisting of three proteins: Syntaxin-1 (6), synaptosome-associated protein of 25 kDa (SNAP-25) (7), and synaptobrevin2/vesicle associated membrane protein 2 (VAMP2) (8, 9). Yet, fusion mediated by SNARE proteins alone is slow and uncoordinated, which conflicts with the physiology of neuronal and neuroendocrine cells that require fast, spatially coordinated secretion. Consequently, accessory factors are needed to modulate the SNARE-driven exocytosis, like synaptotagmins that sense the rise in calcium (Ca2+) levels and Sec1/Munc18 proteins that are essential for several exocytic steps (10, 11). Additional regulators bind to SNARE complexes and to neuronal SNARE proteins: For example, complexin, tomosyn (also known as syntaxin binding protein 5, STXBP5) and amisyn (known as syntaxin binding protein 6, STXBP6) to promote or interfere with the SNARE complex formation (12–14).
Little is known about amisyn, a 24-kDa protein reported to contain an uncharacterized N-terminal domain and a C-terminal SNARE motif (14). This brain-enriched protein coimmunoprecipitated with syntaxin-1a and syntaxin-4 (14). It was mostly cytosolic, but a fraction of amisyn cosedimented with membranes (14, 15). Like tomosyn’s SNARE motif, the recombinant SNARE motif of amisyn could form a ternary complex with the neuronal SNARE proteins syntaxin-1a and SNAP-25 (14). Addition of the amisyn-SNARE domain is reported to inhibit secretion from cultured neuroendocrine PC12 cells (14). Amperometry-based experiments revealed that overexpression of full-length amisyn in rat chromaffin cells had no effect on the basic characteristics of the amperometric spikes, but it reduced the number of spikes elicited, and increased the lifetime of the prespike foot (15). Yet, the inhibition of secretion was independent of amisyn’s interaction with syntaxin-1 (15), and the mechanisms of amisyn action in exocytosis are not yet known.
Overexpression of amisyn also inhibited secretion from human insulin-secreting β-cells (16). In addition, the cAMP-sensor Epac2 is reported to restrict fusion pore expansion by acutely recruiting amisyn and GTPase dynamin-1 to the exocytic site in insulin-secreting β-cells (17). Of note, amisyn is linked to several diseases, including diabetes (16, 17), autism (18, 19) and cancer (20, 21). Consequently, this poorly studied protein necessitates detailed characterization of its biochemical, structural, and functional roles.
Here, we report that amisyn transiently associates with the plasma membrane by binding phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] through its pleckstrin homology (PH) domain. Full-length amisyn formed stable SNARE complexes with syntaxin-1a and SNAP-25, yet both PH and SNARE domains were needed to mediate its role in exocytosis. Elevated amisyn levels resulted in a reduced number of fusion events in neuroendocrine cells, most likely since amisyn works as a vertebrate-specific competitor of synaptobrevin-2, but they did not alter the fusion pore properties.
Results
Amisyn Is a Conserved Vertebrate-Specific Protein that Forms a Ternary SNARE Complex with Neuronal SNAP-25 and Syntaxin-1.
We first used the full-length coding region of human amisyn to identify orthologs in the animal kingdom with moderate stringency conditions (Methods): Orthologs of amisyn were identified in all classes of vertebrates (SI Appendix, Table S1), but not in other orders of animal kingdom, in agreement with Kloepper et al. (22). The comparative protein sequences revealed that amisyn is evolutionarily conserved (SI Appendix, Table S1).
Amisyn contains the SNARE motif at its C terminus (14), while the structural and functional nature of the N-terminal region is unclear. We aligned the SNARE motifs of syntaxin-1, SNAP-25, and synaptobrevin-2 with that of amisyn, and detected the characteristic heptad repeats (layers −7 to +8) essential for the putative amisyn–SNARE complex formation (SI Appendix, Fig. S1A; layers −7 to +8 are shaded gray and SNARE motifs are boxed). Notably, amisyn contains an R-SNARE motif like synaptobrevin, implying that amisyn may participate in the neuronal SNARE complex formation, possibly instead of synaptobrevin-2.
Given a suggested role for the SNARE motif of amisyn in the SNARE complex assembly (14), supported by the sequence alignment (SI Appendix, Fig. S1A), we examined the biochemical properties of full-length amisyn protein in the context of its interactions with other neuronal SNAREs. Full-length amisyn was reported as difficult to express heterologously and poorly soluble (14). We confirmed that the full-length protein expresses poorly under commonly used expression conditions, but it was nevertheless soluble. By subsequent optimization of the expression conditions (Methods), we achieved high expression levels, and succeeded to purify soluble amisyn protein (∼5 mg amisyn/L medium). The amisyn R-SNARE motif (amisyn-SNARE) was expressed and purified to even higher concentrations. Both proteins were >95% pure, as assessed by SDS/polyacrylamide gel electrophoresis (PAGE) (SI Appendix, Fig. S2). We noted, however, that both proteins were inactivated by freeze–thawing, and were active only when kept at 4 °C for up to 3 to 4 d.
Analysis by circular dichroism spectroscopy revealed the change in the secondary structure when syntaxin-1a and SNAP-25a were added to the purified SNARE motif of amisyn (SI Appendix, Fig. S1B). Specifically, the characteristic α-helical spectrum showed minima at 208 and 222 nm. The structural change occurred after the addition of the SNARE motif of amisyn: Similar observations were made with the SNARE motif of synaptobrevin-2 (23), and the SNARE motif of tomosyn (24, 25). Our data further revealed that the SNARE motif of amisyn interacted with syntaxin-1a and SNAP-25a to form an SDS-resistant ternary complex (SI Appendix, Fig. S1C), as originally reported (14). Also, the full-length amisyn protein formed a stable, SDS-resistant ternary complex with the neuronal Q-SNAREs in vitro (SI Appendix, Fig. S1D; note that the complexes dissociated after heating at 95 °C).
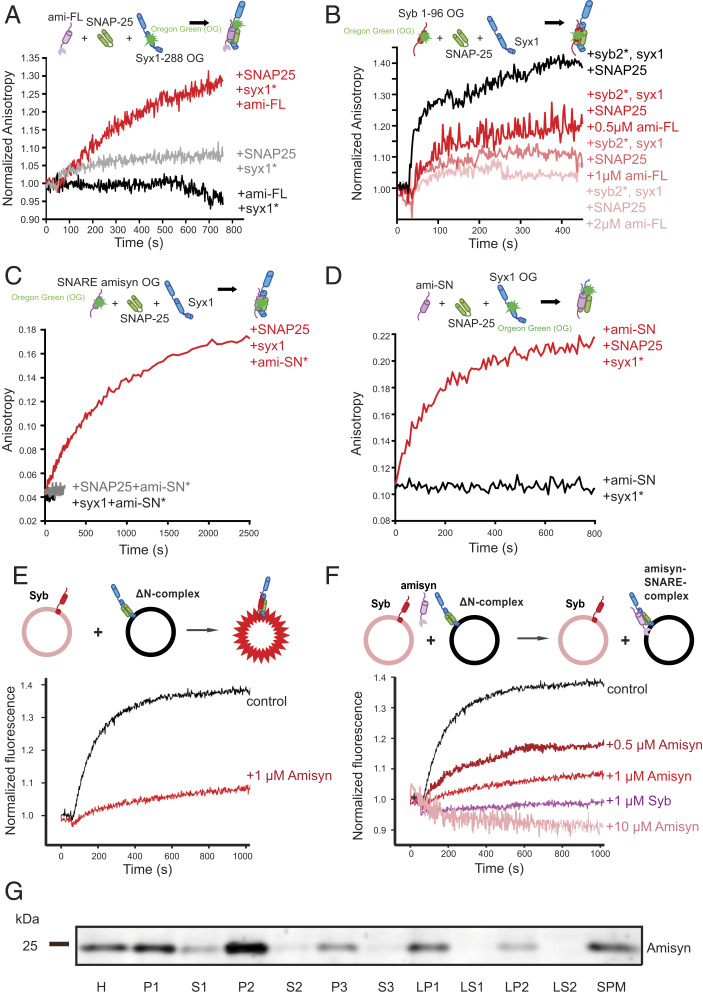
Detailed characterization of amisyn-containing SNARE complex formation was performed by a comprehensive set of fluorescence anisotropy experiments using recombinant purified SNARE proteins (Methods). First, fluorescence anisotropy spectra of syntaxin-1a (syx11−288OG; 1 μM) revealed that its interaction with full-length amisyn (1 μM) occurred only in the presence of SNAP-25a (1.5 μM) (Fig. 1A). Second, fluorescence anisotropy spectra of synaptobrevin-1 (syb1−96OG) showed that full-length amisyn protein (0.25 μM to 2 μM) competes for the SNARE complex formation in the presence of SNAP-25a (1.5 μM) and syntaxin-1a (1 μM) in a concentration-dependent manner (Fig. 1B). Third, when the cysteine residue at position 210 of amisyn’s C terminus was labeled with Oregon green dye (amisyn-SNARE210OG), we observed unchanged fluorescence anisotropy spectra of amisyn-SNARE210OG after addition of Q-SNARE motifs of SNAP-25a (1.5 μM) or syntaxin-1a (1 μM) (Fig. 1C). In contrast, a robust increase in fluorescence anisotropy was observed when both SNAP-25a and syntaxin-1a were present (Fig. 1C). Similarly, the fluorescence anisotropy of syntaxin-1a, labeled at position 197 with Oregon green (syx1-H3197OG, ∼0.5 μM), did not change after the addition of the SNARE motif of amisyn (amisyn-SNARE, 1 μM) (Fig. 1D). Yet, an increase in anisotropy was evident when both amisyn-SNARE (1 μM) and SNAP-25a (1 μM) were added to syntaxin-1a197OG (Fig. 1D). From the complex-forming and fluorescence anisotropy experiments, we concluded that full-length amisyn can form stable ternary complexes with syntaxin-1 and SNAP-25.
Fig. 1.
Amisyn forms SNARE complex with syntaxin-1 and SNAP-25 and inhibits liposome fusion in vitro. (A) Fluorescence anisotropy of syntaxin-1 (syx11−288OG; 1 μM) revealed interaction with full-length amisyn (ami-FL; 1 μM) in the presence of SNAP-25 (1.5 μM) (red), versus SNAP-25 (gray), and amisyn alone (black trace). (B) Fluorescence anisotropy of synaptobrevin-2 (syn21−96OG) revealed interaction with syntaxin-1 in the presence of SNAP-25 (1.5 μM) and full-length amisyn (0.5, 1, or 2 μM). Competition between amisyn and synaptobrevin-2 is demonstrated by decrease in anisotropy upon adding amisyn in a concentration-dependent manner. (C) Anisotropy of the SNARE motif of amisyn (100 nM), labeled at cysteine-210 with Oregon green-488, did not change by addition of the H3-syntaxin-1a (syx1, 1 μM) or SNAP-25 (SNAP25; 1.5 μM). In contrast, increased anisotropy was observed when both syx1 and SNAP-25 were added. (D) Anisotropy of H3-syntaxin-1a (syx1), labeled at position 197 with Oregon green-488 (syx1197OG), did not change by addition of the SNARE motif of amisyn (Ami-SN). Increased anisotropy was evident when both ami-SN and SNAP-25 (SNAP25) were added to syx1197OG. (E) In the presence of amisyn, liposome fusion is severely impaired. Schematic representation of the liposome fusion assay is shown above the graph. Donor liposomes contain Syb-2 (1–116) and two fluorophores, nitrobenzoxadiazole (NBD) and Rhodamine, coupled to lipids, thereby quenching their fluorescence. Acceptor liposomes contain ∆N ternary complexes of SNAP-25, syntaxin-1a, and a C-terminal fragment of Syb-2. When donor (200 nM) and acceptor (200 nM) liposomes are mixed, they fuse due to trans-SNARE complex formation, which allows de-quenching of the NBD fluorescence and kinetic and quantitative measurements of the fusion process. Amisyn caused less dequenching, indicative of inhibition of the fusion process. (F) Inhibition of liposome fusion by amisyn is concentration-dependent. Monitoring of NBD-dequenching fluorescence after mixing liposomes revealed that higher concentrations of amisyn blocked liposome fusion more efficiently. Soluble synaptobrevin-2, which competes with synaptobrevin-2 on the liposomes for the SNARE complex formation, was added as a control. (G) Fractionation of mouse brain to define the subcellular distribution of amisyn. See also SI Appendix, Fig. S1F. Abbreviations: H, mouse brain homogenate; P1, 1,400 × g pellet; S1, supernatant, further centrifuged (13,800 × g, 10 min) to yield pellet P2: crude synaptosomes. P2 was lysed and centrifuged (32,800 × g, 20 min) to yield pellet LP1: crude synaptic plasma membranes and supernatant LS2: synaptic vesicles and synaptosomal cytosol. S2: crude cytosol further centrifuged (165,000 × g, 1 h) to obtain P3: membranes and S3: cytosol. Fraction LS1 was centrifuged (165,000 × g, 1 h) to yield pellet containing crude synaptic vesicles (LP2) and supernatant containing synaptosomal cytosol (LS2). SPM contains isolated synaptic membranes after sucrose gradient centrifugation. Amisyn was predominantly found in membrane fractions.
Taken together, our observations conclusively show that full-length amisyn interacts with syntaxin-1 and SNAP-25, two key neuronal Q-SNAREs, through its SNARE domain. Furthermore, amisyn interacts with the Q-SNARE proteins in a similar manner as synaptobrevin-2 (Fig. 1 A–D) and the SNARE motif of tomosyn (25). We thus propose that amisyn, with the arginine residue in the central layer position (SI Appendix, Fig. S1A), is a competitor of synaptobrevin-2 for a ternary SNARE complex formation (SI Appendix, Fig. S1E).
Amisyn Inhibits Liposome Fusion In Vitro.
A common approach to investigate the function of a SNARE protein is by reconstituting complementary SNARE proteins into liposomes and examining liposomal fusion by Förster resonance energy transfer (FRET) between two fluorophore-labeled lipid analogs (26, 27). We used an established liposome-based fusion assay (28) to examine if exogenously added amisyn interferes with liposome fusion. Addition of purified full-length amisyn inhibited liposome fusion (Fig. 1E), and the effect was concentration-dependent (Fig. 1F). The lack of a detectable transmembrane domain in amisyn implied that the amisyn-containing SNARE complex was “fusion-inactive,” supporting the hypothesis that amisyn acts as a negative regulator of exocytosis.
Amisyn Interacts with the Plasma Membrane through an N-Terminal PH Domain.
We started studying the subcellular distribution of native amisyn protein by an affinity-purified, custom-made polyclonal antibody (Methods). This antibody could specifically recognize amisyn, both native and recombinant forms (SI Appendix, Fig. S3), and was thus used to investigate the distribution of amisyn in the mouse brain subcellular fractions. Notably, in the mouse brain, amisyn was conspicuously absent from supernatant fractions S3, LS1, and LS2, but was enriched in synaptosomal membranes (LP1) and in crude synaptosomal vesicles (LP2) (Fig. 1G and SI Appendix, Fig. S1F). The evident implication is that the majority of amisyn associates with the membranes in nerve terminals of mouse brains. These data are in agreement with refs. 14 and 15, who reported that a fraction of amisyn is membrane-bound.
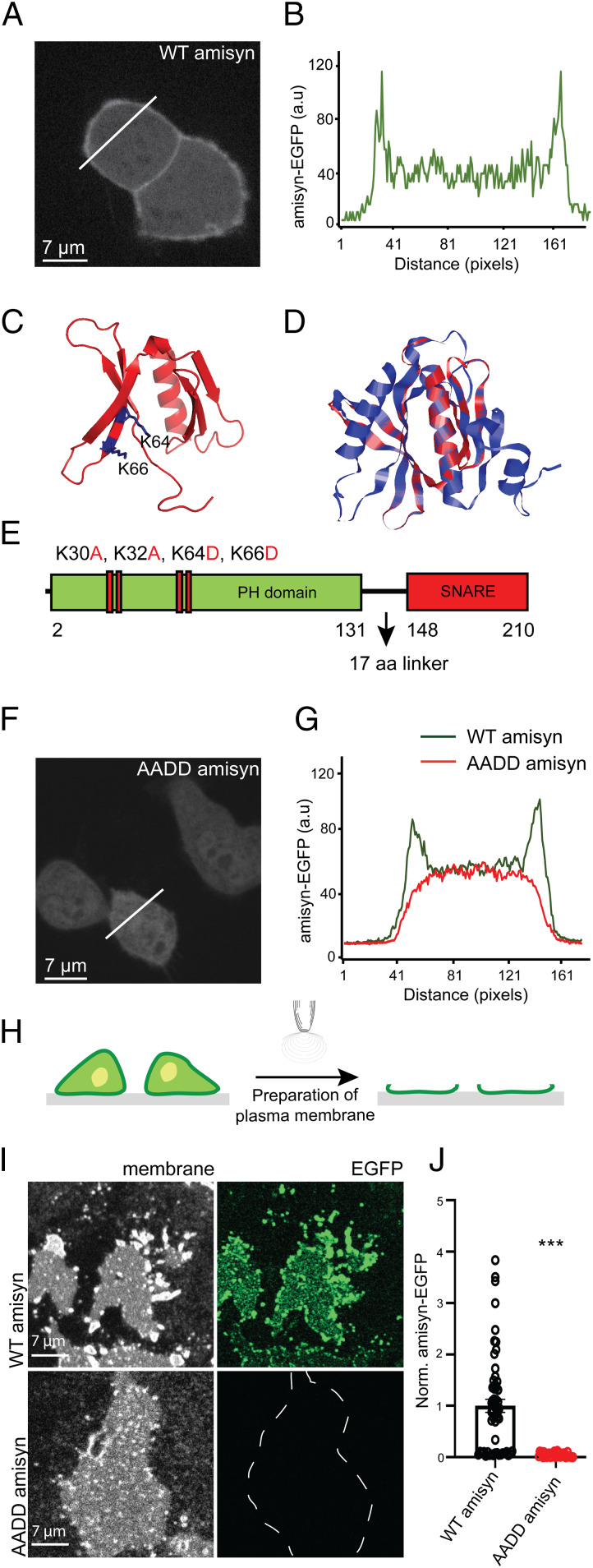
We next cloned human amisyn (amino acids 1 to 210) fused to enhanced green fluorescent protein (EGFP) and expressed this construct in pheochromocytoma PC12 cells, neuroendocrine cells isolated from rat adrenal medulla. We noted, besides cytosolic localization, the significant association of amisyn-EGFP with the plasma membrane (Fig. 2 A and B). The stable binding of amisyn to the plasma membrane was also seen by an in vitro assay in which cultured cells are unroofed by a single sonication pulse, exposing the inner leaflet of the plasma membrane to the extracellular environment (29, 30), as shown in Fig. 2 H–J. In summary, amisyn was found to associate considerably with the plasma membrane in living cells and isolated membrane fractions despite the absence of membrane anchor sequences. Of note, the N-terminal part of amisyn remains to be explored for as yet unknown functions. This part of amisyn protein showed sequence homology (∼32%) to Sec3-like protein, and we noted that Sec3 contains a PH domain implicated in tethering. Furthermore, homology was noted with the Caenorhabditis elegans homolog Uso1p, also known as a tethering protein. We consequently scrutinized the uncharacterized N-terminal domain of amisyn for potential lipid-binding sequences and activity.
Fig. 2.
Amisyn binds to membranes by its N-terminal PH-domain. (A) Amisyn-EGFP distribution in PC12 cells revealing amisyn enrichment at the plasma membrane. Cells were fixed and imaged 20 h posttransfection. An identical result was observed in six independent experiments. (B) The fluorescence intensity line profile according to the white line in A. (C) Model of the N-terminal domain of amisyn that assembles into a PH-like domain. The essential Lysine residues (K64 and K66) are marked in blue. (D) Alignment of the identified PH domain of amisyn (red) with that of PLCɗ1 (blue) as template. (E) Schematic representation of amisyn with the PH and SNARE domains separated by the 17-amino acid linker sequence. The mutant AADD-amisyn protein contained four point-mutations in the PH domain: K30A, K32A, K64D, K66D. (F) The mutant AADD-amisyn failed to bind to the plasma membrane. Representative confocal image of AADD amisyn-EGFP expressed in PC12 cells. Cells were fixed and imaged 20 h posttransfection. The experiment was repeated three times. (G) The fluorescence intensity profiles of PC12 cells expressing WT amisyn-EGFP (green trace) and AADD amisyn-EGFP (red trace; from the white line in F). (H) Schematic of isolated PC12 plasma membrane sheet preparation by sonication. (I) Isolated membrane sheets from PC12 cells expressing either WT or AADD amisyn-EGFP (indicated by the captions on the left). Representative images are shown of the membranes (Left) and the associated fluorescence (Right). (J) Quantification of the fluorescence signals (as shown in I) of WT and AADD amisyn-EGFP. Three independent experiments, WT (31 cells) and AADD (33 cells). Unpaired two-sided t test, mean ± SEM, ***P < 0.001.
First, we modeled the N-terminal domain of amisyn using the yeast exocyst subunit Sec3p (PDB ID code 3A58) as a template (Fig. 2 C and D). The 89 residues of the amisyn N-terminal domain (68% coverage) was modeled using the protein threading method with 99.7% confidence (Methods). As evidenced by the structural alignment with the template structure (rmsd 0.56 Å), as well as with the PH domain from the exchange factor ARNO in mouse (PDB ID code 1U27, rmsd 2.23 Å), the N-terminal amisyn sequence represented a typical PH domain, with a distinct fold consisting of an antiparallel β-sheet followed by an α-helix. In addition, the molecular docking simulation revealed a potential binding site for PI(4,5)P2, with the highlighted lysine residues (K64 and K66) located at the putative PI(4,5)P2 binding site. In analogy with homologous proteins where the N-terminal domains is flexibly linked to the SNARE domain, we refer to a 17-residue stretch between the putative N-terminal PH domain and C-terminal SNARE domain in amisyn as a linker region (Fig. 2E).
To experimentally verify the predicted PH-domain in amisyn, we attempted to prepare amisyn crystals, but failed to obtain suitably diffracting crystals. Thus, we experimentally approached our prediction that the PH-domain of amisyn was responsible for its membrane interactions. We prepared a mutated amisyn with the lysine residues predicted to interact with PI(4,5)P2 based on molecular docking and analogy with phospholipase C-ɗ1 and Sec3p as PI(4,5)P2-binding proteins (31). We constructed and expressed several mutants, including the quadruple amisyn mutant: K30A, K32A, K64D, K66D, denoted AADD-amisyn (Fig. 2E). In transfected PC12 cells, we noted that the expressed AADD-amisyn failed to locate at the plasma membrane (Fig. 2 F and G). Moreover, plasma membrane sheets from transfected PC12 cells expressing either WT amisyn-EGFP or AADD-amisyn-EGFP confirmed that WT amisyn was bound to the plasma membrane, while the AADD-amisyn was not (Fig. 2 H–J).
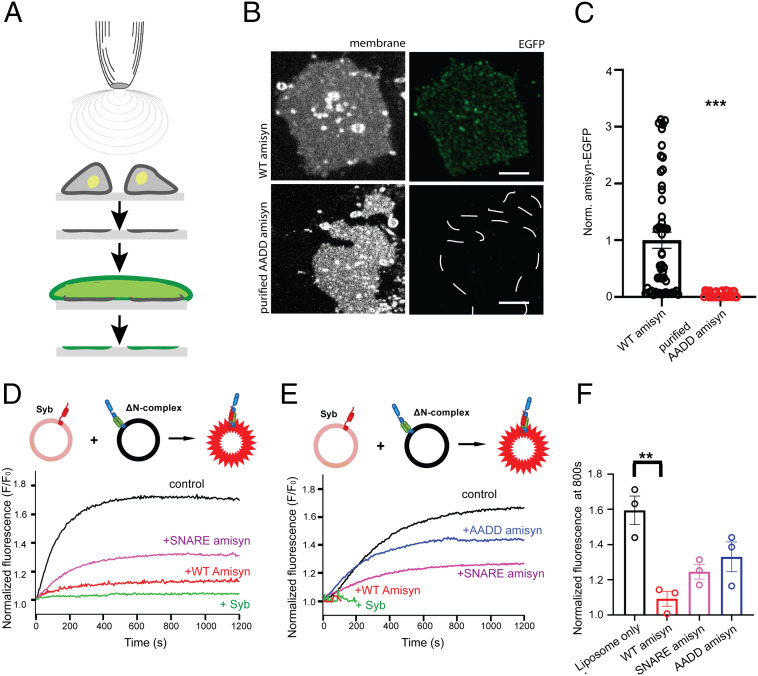
Next, we expressed and purified WT and AADD-amisyn heterologously. The mutant showed similar circular dichroism spectra and gel-filtration profile as WT amisyn (SI Appendix, Fig. S2), suggesting that the mutations did not alter the local and the overall protein fold. Upon incubation with the freshly prepared PC12 plasma membrane sheets, only recombinant WT amisyn-EGFP, and not AADD-amisyn-EGFP, bound to the isolated membranes (Fig. 3 A–C). Notably, fluorescently labeled amisyn was bound to membranes in a nonuniform punctate pattern (Fig. 3 B, Upper). The plasma membrane sheets generated from PC12 cells expressing amisyn-EGFP also showed nonuniform pattern (Fig. 2 I, Upper). We concluded from the combined experiments that amisyn interacts with membranes through its N-terminal PH domain.
Fig. 3.
Recombinant amisyn binds to plasma membranes in vitro. (A) Schematic of PC12 plasma membrane sheet preparation by sonication and subsequent immediate incubation with purified recombinant amisyn protein. (B) Binding of recombinant WT and AADD-amisyn to membranes from PC12 cells. The purified WT amisyn-EGFP (1 μM) or AADD amisyn-EGFP (1 μM) were incubated with freshly prepared membranes at room temperature. Images show WT but not AADD-amisyn-EGFP bound to the membranes (visualized by trimethylamine-diphenylhexatriene [TMA-DPH]). (Scale bar, 5 μm.) (C) Quantitative analysis of data as shown in B. Unpaired two-sided t test, At least 49 sheets/conditions from three experiments; mean ± SEM, ***P < 0.001. (D) Representative experiment showing full-length WT amisyn inhibiting liposome fusion more efficiently than its SNARE domain. Fluorescence dequenching of NBD fluorophore-labeled liposomes containing synaptobrevin-2 (Syb) after mixing with liposomes containing the ΔN complex. Soluble synaptobrevin-2 that competes with synaptobrevin-2 on the liposomes for the SNARE complex formation was added as a control. (E) Representative experiment showing that WT amisyn inhibits liposome fusion more efficiently than AADD amisyn mutant. Fluorescence dequenching of NBD fluorophore-labeled liposomes containing synaptobrevin-2 (Syb) after mixing with liposomes containing the ΔN complex. (F) Fluorescence dequenching of NBD fluorophore labeled liposomes at 800s reveals that WT amisyn inhibits liposome fusion more efficiently than its SNARE domain alone or AADD-amisyn. Three independent experiments; mean ± SEM.
Finally, we used a liposome-based assay fusion assay (as shown in Fig. 1 E and F) to examine whether AADD-amisyn or the SNARE domain of amisyn alone can alter liposome fusion. Addition of either AADD amisyn mutant, or the SNARE domain of amisyn, had a negative effect on liposome fusion, yet neither had an effect as potent as the inhibition observed with full-length WT amisyn (Fig. 3 D and E; a mean of three independent experiments is shown in Fig. 3F).
PI(4,5)P2 Controls Amisyn Binding to Membranes in Living PC12 Cells.
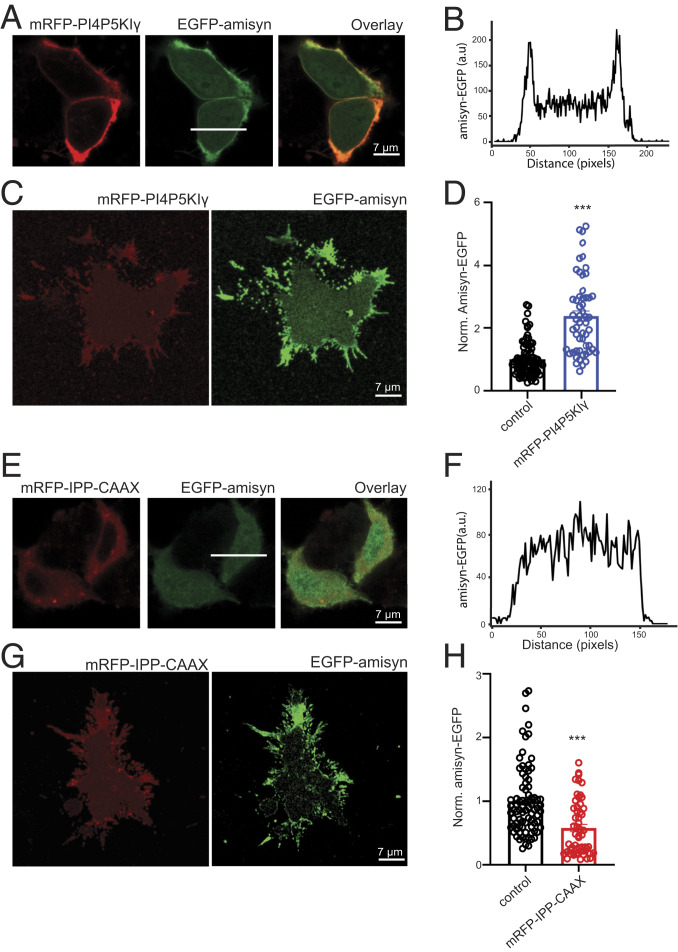
We next performed the series of ex vivo and in vitro experiments to explore if amisyn interaction with membranes by its PH-domain is PI(4,5)P2-dependent. First, we elevated the levels of PI(4,5)P2 in the inner leaflet of the plasma membrane of PC12 cells by transfection and expression of PI4P5KIɣ (29). The result revealed that the vast majority of amisyn, coexpressed in PC12 cells with mRFP-PI4P5KIɣ, became associated with the plasma membrane (Fig. 4 A and B). In addition, more amisyn was bound to plasma membrane sheets isolated from amisyn-EGFP/mRFP-PI4P5KIɣ coexpressing PC12 cells (Fig. 4 C and D). Next, we reduced the PI(4,5)P2 levels in PC12 cells by transfection with the membrane-targeted 5′-phosphatase domain of synaptojanin-1 (IPP-CAAX) (29). Thereby, the amisyn-EGFP coexpressed with mRFP-IPP-CAAX was no longer associated with the plasma membrane (Fig. 4 E and F). Moreover, less amisyn was detected on plasma membrane sheets from amisyn-EGFP/mRFP-IPP-CAAX coexpressing PC12 cells (Fig. 4 G and H). Taken together, by varying the amount of PI(4,5)P2 in the inner leaflet of the plasma membrane in both directions by well-established methods, we demonstrated correlating alterations in plasma membrane-anchored amisyn.
Fig. 4.
PI(4,5)P2 levels control membrane binding of amisyn in PC12 cells. (A) Representative confocal images of PC12 cells expressing either amisyn-EGFP or mRFP-PI4P5KIγ, or both to increase PI(4,5)P2 in inner leaflet of membrane, resulting in more amisyn associated with the plasma membrane. (B) Fluorescence profile of amisyn-EGFP expressing cell through a white line in A. (C) Plasma membrane sheets isolated from cotransfected PC12 cells as in A contain more amisyn-EGFP. (D) Quantitation of amisyn-EGFP fluorescence on the plasma membrane sheets isolated from transfected PC12 cells. At least 50 sheets/conditions, two independent experiments; mean ± SEM, unpaired two-sided t test, ***P < 0.001. (E) Confocal images of PC12 cells expressing either amisyn-EGFP or mRFP-IPP-CAAX or both, to decrease levels of PI(4,5)P2 in the inner leaflet of the plasma membranes, resulting in less amisyn-EGFP bound to the plasma membrane. (F) Fluorescence profile through an amisyn-EGFP–expressing cell through a white line in E. (G) Membrane sheets isolated from cotransfected PC12 cells as in E contain less amisyn-EGFP. (H) Quantitative determination of amisyn-EGFP fluorescence intensity on membrane sheets from transfected PC12 cells. At least 50 sheets/conditions, three experiments. Mean ± SEM, unpaired two-sided t test, ***P < 0.001.
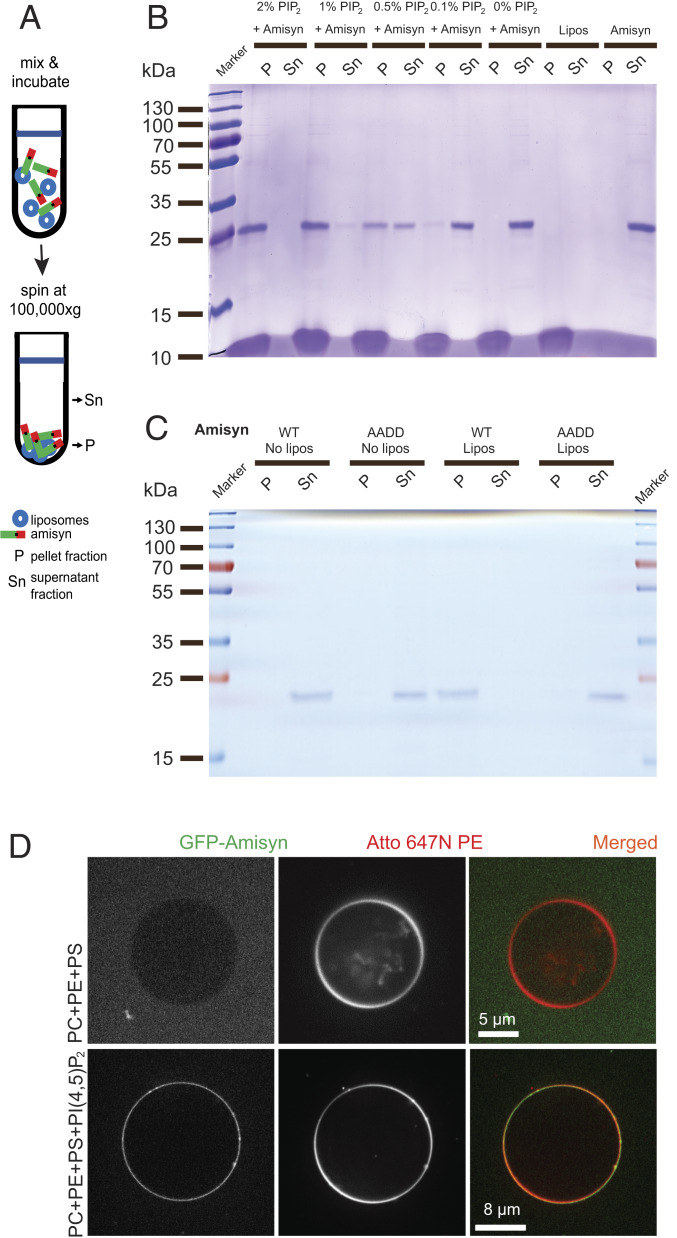
To further test if binding of amysin to the membrane requires PI(4,5)P2, we used purified recombinant full-length WT amisyn in a cosedimentation assay and in a liposome-binding assay. We prepared unilamellar liposomes composed of phosphatidylcholine (PC), phosphatidylserine (PS), and phosphatidylethanolamine (PE), supplemented with PI(4,5)P2, various phosphatidylinositol phosphates (PIPs) or PC only as control. We assayed the ability of amisyn to interact with the liposome’s outer membrane. First, using cosedimentation assays (Fig. 5A) with different levels of PI(4,5)P2 incorporated into the liposomes, we observed a close positive correlation between PI(4,5)P2 levels and amisyn cosedimentation (Fig. 5B). Remarkably, WT amisyn cosedimented with PI(4,5)P2-containing liposomes, but not with liposomes without PI(4,5)P2 (Fig. 5B), showing that amisyn interaction with the liposomal membrane is mediated by negatively charged PI(4,5)P2. In contrast to WT amysin, amisyn AADD mutant did not cosedimented with PI(4,5)P2-containing liposomes (Fig. 5C). Using the same assay, we observed that amisyn, in addition to binding PI(4,5)P2-liposomes, could also bind to liposomes containing PI(3,4)P2, PI(3,5)P2, or PI(3,4,5)P3, but not phosphatidylinositol (PI), suggesting that PH domain of amisyn has high affinity but low specificity for PIPs (SI Appendix, Fig. S4).
Fig. 5.
Liposome binding of amisyn is dependent on PI(4,5)P2. (A) Schematic representation of a cosedimentation assay. (B) Representative cosedimentation of amisyn with liposomes depends on their PI(4,5)P2 content (shown in captions over each lane). Representative SDS/PAGE gel (12%) shows that PI(4,5)P2 levels in liposomes correlate with more amisyn bound to liposomes cosedimenting in the pellets (P) relative to the supernatant (Sn). Three independent experiments were performed. (C) Representative cosedimentation assay shows that AADD amisyn mutant does not bind well to PI(4,5)P2-containing liposomes (2%). Representative SDS/PAGE gel (12%) of sedimentation assay. P, pellet; Sn, supernatant; three independent experiments were performed. (D) Recombinant amisyn-EGFP bound only to liposomes containing PI(4,5)P2. Representative confocal images of liposomes without or containing PI(4,5)P2 (captions on the left). Two experiments with independently purified amisyn-EGFP were performed, each time with several technical replicates.
We next performed imaging-based membrane-binding experiments using giant unilamellar vesicles (GUVs). Here, we tested the binding of purified amisyn-EGFP fusion protein to fluorescently labeled GUVs with different lipid compositions (Methods). Amisyn was thereby found to bind only to PI(4,5)P2-containing vesicles (Fig. 5D), because replacing PI(4,5)P2 with PS or PC did not lead to any appreciable amisyn binding.
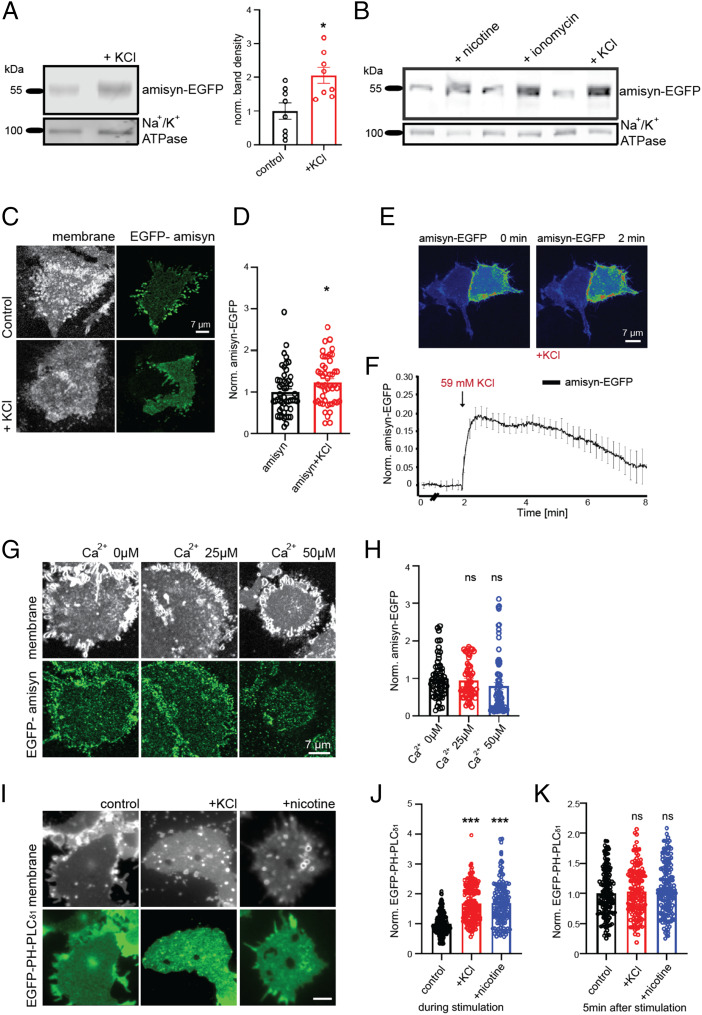
Stimulation of PC12 Cells Recruited Amisyn-EGFP to the Plasma Membrane.
Depolarization of neurosecretory chromaffin cells was shown to increase PI(4,5)P2 levels rapidly and transiently (32). Therefore, we analyzed whether more amisyn was bound to the plasma membranes of stimulated PC12 cells, depolarized by addition of 59 mM KCl. Fractionation of the stimulated cells (Methods) revealed more amisyn to be present in the plasma membrane fractions compared to nondepolarized cells (Fig. 6A; quantification shown, Right). PC12 cells stimulated by either nicotine or ionomycin, which both increase cytosolic Ca2+ levels, provoked a similar increase in amisyn membrane binding (Fig. 6B). Finally, plasma membrane sheets prepared from KCl-stimulated amisyn-EGFP–expressing PC12 cells contained more amisyn-EGFP than plasma membrane sheets prepared from nondepolarized amisyn-EGFP–expressing cells (Fig. 6 C and D).
Fig. 6.
Stimulation recruits amisyn-EGFP to the plasma membrane of PC12 cells. (A) Membranes isolated from PC12 cells after stimulation (59 mM KCl, 5 s) contain more amisyn-EGFP, relative to membranes isolated from naive cells. Na+/K+ ATPase is used as internal loading control and membrane marker. (Left) Representative Western blots from eight experiments. (Right) Quantification. Mean ± SEM, unpaired two-sided t test, *P < 0.1. (B) Representative Western blot of membranes isolated from naive PC12 cells and after stimulation as shown in the captions (100 μM nicotine, 1 μM ionomycin, 59 mM KCl, 5 s). Na+/K+ ATPase was used as the internal loading control and membrane marker. (C) Membrane sheets generated from stimulated PC12 cells (59 mM KCl, 5 s) contain more amisyn-EGFP than membrane sheets from naive cells. (D) Fluorescence quantified from samples as in C (n = 41 sheets/conditions, 3 experiments). Mean ± SEM; unpaired two-sided t test, *P < 0.1. (E) Representative confocal images of PC12 cells before and after stimulation (59 mM KCl) demonstrating that stimulation recruited amisyn to plasma membranes. (F) Time-course of amisyn-EGFP fluorescence on the plasma membrane in living PC12 cells stimulated with 59 mM KCl. Mean of 15 cells from 3 experiments ± SEM (G) Plasma membrane sheets from naive PC12 cells were incubated with recombinant amisyn-EGFP (3 μM) and different calcium ion concentrations (captions). (Upper) TMA-DPH–stained isolated plasma membranes; (Lower) isolated plasma membranes with bound recombinant amisyn-EGFP. (H) Quantification of amisyn fluorescence (as on samples in G) revealed that recruitment of amisyn-EGFP to the plasma membrane was not mediated by calcium ions. At least 46 sheets/condition from 3 experiments. Mean ± SEM; one-way ANOVA with Tukey’s post hoc test; ns, not significant. (I) Plasma membrane sheets isolated from stimulated PC12 cells (59 mM KCl, 5 s) were incubated with recombinant EGFP-PH-PLCɗ1 (3 μM) for 60 s, washed and fixed. (Upper) TMA-DPH dye stains isolated plasma membranes; (Lower) isolated plasma membranes with bound recombinant EGFP-PH-PLCɗ1. (Scale bar, 5 µm.) (J) Quantification of EGFP-PH-PLCɗ1 fluorescence (as on samples in I) revealed elevated PI(4,5)P2 levels in the plasma membranes of stimulated cells. At least 144 sheets/conditions from 3 experiments. Mean ± SEM; one-way ANOVA with Tukey’s post hoc test, ***P < 0.001. (K) Plasma membrane sheets were isolated from PC12 cells 5 min poststimulation (59 mM KCl), immediately incubated with recombinant EGFP-PH-PLCɗ1 (3 μM) for 60 s, washed and fixed. Quantification of EGFP-PH-PLCɗ1 fluorescence revealed no change in the levels of PI(4,5)P2 in the plasma membranes between nonstimulated and stimulated cells. At least 144 sheets/conditions from 3 experiments. Mean ± SEM; one-way ANOVA with Tukey’s post hoc test; ns, not significant.
The kinetics of amisyn recruitment to the plasma membrane upon depolarization was analyzed in a series of live-imaging experiments with PC12 cells expressing amisyn-EGFP. We noted the rapid rearrangement of amisyn distribution in PC12 cells expressing amisyn-EGFP, concomitantly with the elevation in plasma membrane-bound amisyn, almost immediately upon the addition of KCl (Fig. 6 E and F). The association of amisyn with the plasma membrane was transient, as the majority of amisyn dissociated from the membrane and returned to cytosol within 6 min after stimulation. We concluded that depolarization recruited amisyn-EGFP to the plasma membrane of PC12 cells, rapidly and transiently.
Upon stimulation/depolarization, the levels of intracellular calcium ions also increase rapidly and transiently. To address whether recruitment of amisyn to the plasma membrane was mediated by an increase in calcium concentration, we incubated plasma membrane sheets (isolated from nondepolarized PC12 cells) with recombinant amisyn-EGFP protein and various concentrations of calcium ions in the specified buffers (prepared as described in ref. 30). Imaging revealed that similar amounts of amisyn-EGFP became bound to the membranes regardless of the calcium ion concentration, and even in the complete absence of externally added Ca2+ (Fig. 6 G and H). We also examined the PI(4,5)P2 levels in the plasma membrane upon stimulation of PC12 cells. Using recombinant EGFP-PH-PLCɗ1 as a specific PI(4,5)P2-probe and plasma membrane sheet assay (29), we detected elevated PI(4,5)P2 levels in the isolated plasma membranes of stimulated PC12 cells (Fig. 6 I and J). When the same experiment was done with PC12 cells 5 min poststimulation, the difference in PI(4,5)P2 levels was no longer observed (Fig. 6K). These PC12 cell-based data are in agreement with a previous report showing increased levels of PI4P and PI(4,5)P2 in whole-cell lysates prepared from chromaffin cells (32). Given a correlation between rapid and transient amisyn recruitment and a transient increase in the plasmalemmal PI(4,5)P2 levels, we concluded that the recruitment and binding of amisyn to the plasma membrane is mediated, at least in part, by PI(4,5)P2 concentrations in the inner leaflet of the plasma membrane, and not by calcium ions.
Addition of Amisyn, but Not Amisyn’s SNARE Domain, Reduced Secretion of Adrenal Chromaffin Cells.
PC12 cells were instrumental for the characterization of membrane binding of amisyn, while for physiological studies primary chromaffin cells from adrenal medulla that also secrete adrenaline and noradrenaline were preferred. The secretion in adrenal chromaffin cells has been well characterized, which make these cells an ideal system to study the effects of amisyn on releasable vesicle pools and release kinetics (33).
First, we attempted to express amisyn from an internal ribosome entry site-EGFP construct in bovine adrenal chromaffin cells using Semliki Forest virus. We noticed that the infected chromaffin cells became stressed, were detaching from the glass coverslips, and eventually died. We therefore opted for acute direct delivery of the recombinant amisyn protein through the patch pipette. Purified amisyn (5 µM) was included in the pipette solution, and each cell was loaded for at least 90 s before combined electrophysiological and electrochemical experiments were performed. As a control, purified EGFP (5 µM) was included into the pipette solution, and each cell was loaded and recorded, like the experiments with amisyn. This approach allowed the acute studies of amisyn function in the fast-secreting primary chromaffin cells. In addition to purified amisyn, chromaffin cells were loaded via the patch pipette with the photolabile Ca2+ chelator nitrophenyl-EGTA and with two Ca2+-sensitive dyes that enabled accurate Ca2+ measurements during the whole-cell patch-clamp experiments (29, 34). Flash photo-release of caged calcium increases the intracellular calcium ion concentration, [Ca2+]i, resulting in robust secretion that was assayed by the increase in membrane capacitance measurements. Notably, exocytosis of chromaffin cells loaded with full-length amisyn protein was strongly inhibited compared to control cells loaded with EGFP, as measured by the lesser increase in capacitance (Fig. 7A). Acute amisyn addition robustly reduced both an exocytotic burst (secretion during the first second after stimulation) and a sustained component of release (secretion during the next 4 s). A second flash stimulation, applied to the same cells after 90-s recovery, also triggered a smaller response in amisyn-loaded cells compared to control (SI Appendix, Fig. S5A).
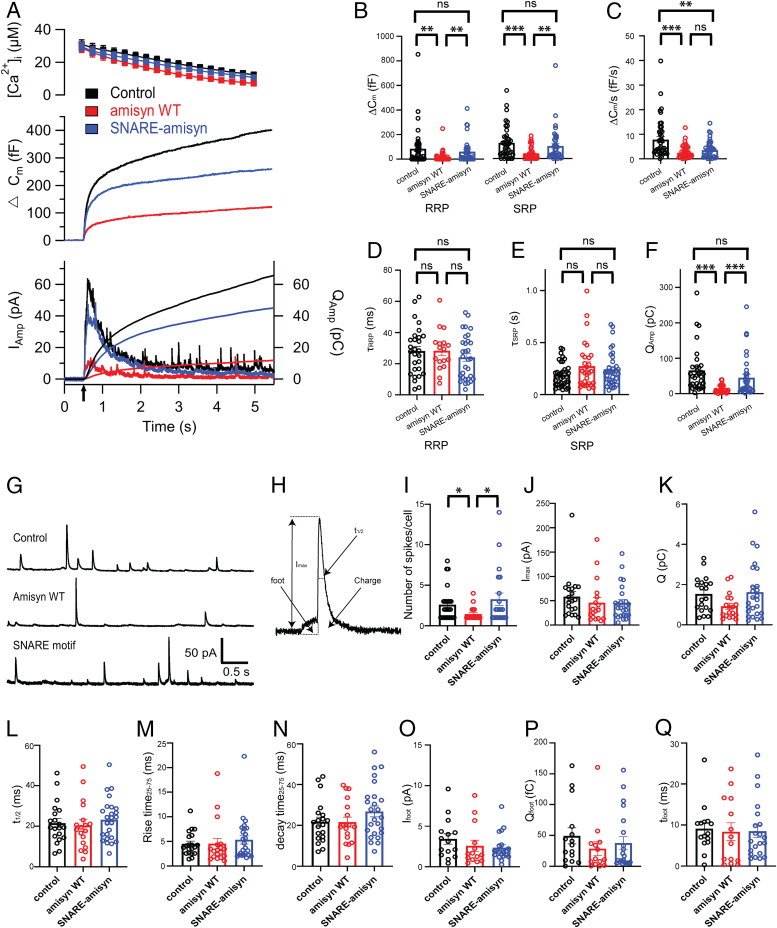
Fig. 7.
Amisyn, but not amisyn-SNARE domain, reduced number of released vesicles but did not alter rates of vesicle fusion in bovine chromaffin cells. (A–F) Exocytosis induced by UV-flash photolysis of caged calcium ions (stimulus #1, arrow) was reduced in amisyn-loaded chromaffin cells (red trace) compared to control cells (black trace). Cells loaded with amisyn-SNARE protein (blue trace) did not differ significantly from control cells (with an exception of sustained release): 42 control cells, (black); 38 amisyn-loaded cells (red); 38 amisyn-SNARE–loaded cells (blue) from 5 independent experiments. Kruskal–Wallis test with Dunn’s multiple comparison test; ns, non significant; **P < 0.01, ***P < 0.001. (A, Top) Intracellular calcium level increase induced by flash photolysis (at t = 0.5 s). (Middle) Averaged traces of membrane capacitance changes upon Ca2+-induced exocytosis. (Bottom) Mean amperometric current (Iamp; left axis) and cumulative charge (right axis). (B and C) Exponential fitting of the capacitance traces revealed changes in RRP and SRP size and sustained phase of release (C). Note the marked reduction in exocytosed vesicles in amisyn-loaded cells. (D and E) Fusion kinetics of vesicles from RRP and SRP pools were not altered. (F) Reduced detection of catecholamines in chromaffin cells loaded with amisyn by amperometry: Cumulative charge during 5 s after stimulation. (G–Q) Single-spike amperometry analysis revealed problems in vesicle fusion, but no alterations in the stability of the fusion pore. (G) Exemplary traces from single-spike amperometric recordings of control, amisyn WT, and amisyn-SNARE domain-injected adrenal chromaffin cells. (H) Schematic of analyzed amperometric spike parameters. (I) Number of fusion events per cell was significantly reduced in chromaffin cells injected with full-length amisyn. Each spot in the dot plot represents a mean from an analyzed cell. Four experiments and four independent cell preparations, total of control (20 cells), amisyn WT (18 cells), and amisyn-SNARE domain (25 cells). Mean ± SEM. One-way ANOVA with Tukey’s post hoc test, *P < 0.05. (J–N) Single-spike amplitude (J), charge (K), and the kinetics of single fusion events, such as duration at half-maximal amplitude (L), rise time (M), and decay time (N), were unchanged. For number of cells and statistics, see I. (O–Q) The stability of the fusion pore was not altered, as revealed by unchanged foot amplitude (O), foot charge (P), and foot duration (Q). Four experiments and four independent cell preparations, total of control (15 cells), amisyn WT (14 cells), and amisyn-SNARE domain (21 cells). Mean ± SEM. One-way ANOVA with Tukey’s post hoc test, *P < 0.05.
We complemented the membrane capacitance measurements, showing the net change in exocytotic and endocytotic activity, with simultaneous recordings by carbon fiber amperometry. The amperometric data provide a direct measure of catecholamine release, not influenced by endocytosis. The observed net (cumulative) amperometric signal in amisyn-loaded cells revealed a strong decrease in the number of fused catecholamine-filled secretory vesicles (Fig. 7A and SI Appendix, Fig. S5 A, Lower; quantified in panels Fig. 7F and SI Appendix, Fig. S5F). These results are consistent with an earlier study that shows a smaller cumulative number of exocytic events by amperometry in chromaffin cells expressing amisyn (15), and with the membrane capacitance measurements described in the previous section. Furthermore, these data are also in agreement with another study where the reduction in secretion in insulin-secreting β-cells expressing amisyn was reported (16). Taken together, our data demonstrate that increasing the levels of amisyn caused a significant decrease in secretory vesicle secretion, and that this decrease is attributable to a smaller number of fused secretory vesicles.
Remarkably, the inclusion of purified recombinant SNARE domain of amisyn (5 µM) into the pipette solution did not affect secretion robustly, except the sustained component of release (Fig. 7A). This became even more obvious by the second stimulation, applied 90 s later (SI Appendix, Fig. S5A). The same result was obtained from amperometry measurements during the first and second stimulation (Fig. 7A and SI Appendix, Fig. S5 A, Lower; quantified in panels Fig. 7F and SI Appendix, Fig. S5F). These data match results obtained with the liposome fusion assay (Fig. 3 D–F). Nonetheless, this was a surprising observation since the SNARE domain of amisyn was previously shown to inhibit exocytosis of noradrenaline in cracked PC12 cells (14), and to form a SNARE complex with soluble SNAP-25 and syntaxin-1 (Fig. 1) (14). In conclusion, when injected into live chromaffin cells, only full-length amisyn with both PH and SNARE domains could potently inhibit exocytosis.
Amisyn Inhibited Exocytosis by Attenuating Secretory Vesicle Docking/Priming and Fusion.
Chromaffin cell secretion elicited by flash photolysis stimulation consists of an exocytotic burst (comprising of two pools of release-competent vesicles: The readily and slowly releasable pools [RRP and SRP, respectively], corresponding to fast [∼30 ms] and slow release [∼200 ms], respectively), followed by the sustained phase that represents vesicle recruitment and subsequent fusion (33–35). We analyzed which of these distinct phases of the exocytotic burst was affected by acute addition of amisyn to determine the vesicle pool sizes by the amplitudes of the exponential fittings, and the fusion kinetics by the time constants of the exponential fittings (29). The results revealed that the amplitudes (sizes) of RRP in amisyn-loaded cells were reduced by about 80%, and that SRP were similarly affected, compared to control cells (Fig. 7B). The time constant of the vesicle release from RRP was not affected (Fig. 7D). Of note, in most amisyn-loaded cells (24 of 38) the fast component was absent, reducing the number of observations. The time constant of the vesicle release from SRP was also not significantly affected by acute addition of amisyn (Fig. 7E). The sustained component of release, which measures the refilling of the RRP and SRP pools, was reduced (Fig. 7C). Altogether, the results suggested that acute amisyn addition substantially inhibited the docking/priming and fusion of vesicles into the releasable pools but did not alter the kinetics of vesicle fusion. The results obtained with a second stimulation were almost identical to the first (SI Appendix, Fig. S5 B–E).
Notably, the inclusion of purified recombinant amisyn-SNARE domain into the pipette solution did not significantly affect the sizes of RRP and SRP, while the sustained component was reduced (Fig. 7 B and C, first stimulation, and SI Appendix, Fig. S5 B and C, second stimulation). Kinetics of vesicle release from RRP and SRP were not significantly affected either during first or second stimulation (Fig. 7 D and E and SI Appendix, Fig. S5 D and E). These data, in addition to the liposome fusion assay data, show that only the full-length amisyn protein can alter docking/priming and fusion of secretory vesicles.
Amisyn Addition Reduced the Number of Fusion Events, but did Not Affect Fusion Properties.
To further explore the putative role of amisyn in the regulation of the fusion pore stability, as reported earlier (15, 17), we performed a thorough examination of single amperomteric spikes in bovine adrenal chromaffin cells acutely loaded with amisyn (5 µM) and SNARE domain of amisyn (5 µM) (36). Representative amperometric traces for WT, amisyn, and amisyn-SNARE–loaded cells are shown in Fig. 7G. Significant differences were observed in the number of detected events per cell (Fig. 7I), in agreement with Constable et al. (15). Single-spike charge (reflecting the amount of catecholamines oxidized at the electrode) (Fig. 7H) and amplitude were not changed (Fig. 7 J and K), as well as the kinetic features of single spikes (Fig. 7 L–N). Amperometric spikes are often preceded by a prespike foot that reflects catecholamine leakage through the forming fusion pore (37). The prespike foot duration and its amplitude were not altered in chromaffin cells loaded with amisyn, or the amisyn-SNARE domain (Fig. 7 O–Q), suggesting that amisyn and its SNARE domain do not alter fusion pore stability when acutely added to adrenal chromaffin cells.
Altogether, our electrophysiological and electrochemical data show that upon acute addition of recombinant proteins, only full-length amisyn and not amisyn’s SNARE domain can strongly inhibit exocytosis by attenuating secretory vesicle docking/priming and fusion, while the release time constants and fusion pore properties remain unaffected.
Discussion
The intracellular mechanisms governing exocytosis are regulated by numerous proteins, yet aspects of negative regulation of this complex process are poorly understood. Amisyn has been implied to play a role in several disorders, including diabetes, autism and cancer, by way of its regulatory functions in exocytosis (15–17, 38). Our research on this functionally largely unknown protein was motivated by its inhibitory role in membrane fusion and a presence of the SNARE domain in amisyn. We first explored the biochemical properties of full-length amisyn with respect to SNARE complex formation in vitro. Such studies were required since the SNARE domain of amisyn alone does not efficiently inhibit exocytosis, unlike the SNARE domain of synaptobrevin-2.
We show that full-length amisyn forms a SNARE complex with SNAP-25 and syntaxin-1 and may act as a vertebrate-specific competitor of synaptobrevin-2. We refer to amisyn-containing SNARE complex as “fusion-inactive” since amisyn does not contain a distinct transmembrane sequence or recognizable lipidation motifs. Nonetheless, we and others (14, 15) have found it to be enriched at the membranes. Our subcellular fractionations of mouse brain indicated that amisyn was largely absent from the soluble fractions. It is still possible that amisyn is indirectly attached to the plasma membrane through its interaction partner syntaxin-1. However, we have not observed a correlation between syntaxin-1a and amisyn levels associated with the plasma membrane. In contrast, we observed that amisyn association with the plasma membrane was dependent on the membrane concentration of PI(4,5)P2 in vitro and ex vivo. This dependence of amisyn on PI(4,5)P2, considerable sequence homology to yeast Sec3p that contains a PH-domain, and a review by Barg and Guček (38), inspired us to explore whether the N-terminal domain of amisyn contained a functional PH-domain. We first attempted to obtain amisyn structure through crystallography but had little success. Thus, we performed a series of mutagenesis-based experiments that led to the construction of an AADD mutant incapable of PI(4,5)P2 binding. Altogether, it was revealed that amisyn’s PH domain mediates its interaction with the plasma membrane and is needed for its regulatory role in vesicle priming and fusion. Indeed, intricate regulation of exocytosis can be achieved by varying the levels and types of phosphatidylinositides, which, in turn, alter the nature and composition of the recruited exocytic machinery (29, 39). The PH domain may allow amisyn to act as a PI(4,5)P2-dependent effector, contributing to the spatial and temporal regulation of exocytosis. Additional research is needed to distinguish if amisyn’s PH domain is just a recruiting device to create a high local concentration of the inhibitory SNARE motif or whether amisyn’s N-terminal domain mediates more than membrane binding.
Of note, the N-terminal part of amisyn corresponds to the N-terminal domain of the exocyst component Sec3p. This yeast protein is targeted to the budding tip through interactions with PI(4,5)P2 and the small GTPase Rho/Cdc42. Consequently, Sec3 was used as a structural model and template, similar to the C. elegans homolog of the tethering protein Uso1 (40), with unknown structure and function. This connection of amisyn to the exocyst complex is intriguing. We envisage that amisyn may have taken over a part of the function of Sec3 (given the interaction of the N-terminal domain of Sec3 with sso2/syntaxin) (41) and could act in concert with other exocyst components as ExoC8, and possibly Exo84. In addition, Kloepper et al. (22) proposed that amisyn evolved with the vertebrates, likely containing the SNARE-motif of tomosyn, the gene of which was possibly duplicated and evolved with a different N-terminal domain. Tomosyn is evolutionarily very old; it belongs to the SNARE repertoire of the last common ancestor of all eukaryotes. Despite the fact that tomosyn and Lgl (a protein that likely arose from tomosyn or a predecessor that lost its SNARE domain) are active in different contexts, we speculate that amisyn is related to tomosyn, and possibly Lgl. Like amisyn, Lgl and tomosyn regulate secretion of vesicles, presumably independently of calcium.
In any case, the role of amisyn in exocytosis goes beyond its SNARE motif; the whole protein (including both the PH and SNARE domains) was needed to block exocytosis efficiently, as seen by in vitro liposome fusion assays, and electrophysiological and electrochemical recordings in live chromaffin cells. Notably, the inhibition of exocytosis by amisyn was directly proportional to its concentration: The higher the levels, the more potent was the exocytic block (high levels of amisyn were detrimental for the cells). Mechanistically, we document that amisyn interfered with the docking/priming and fusion of secretory vesicles, and with the sizes of releasable pools. Fusion kinetics of releasable vesicle pools (RRP and SRP) were not altered upon acute addition of amisyn protein in chromaffin cells, nor were the single spike features [only the number of detected spikes was lower, in agreement with previous studies (15, 16)]. The only difference to an earlier report (15) is that we do not detect a significant difference in foot duration and charge when amisyn was acutely loaded into cells through the patch pipette. Yet, we do not exclude that amisyn may affect fusion pore expansion under different conditions. In our study, the recombinant amisyn was delivered acutely through a patch pipette, which may cause diffusion of small molecules like cAMP (possibly also cAMP effector proteins, e.g., EPAC2) out of the cell and prevent detecting the effect of amisyn on fusion pores, as reported previously (17). Furthermore, amisyn has a strong inhibitory effect on exocytosis, making these experiments difficult; the number of detected amperometic spikes was severely reduced and many amisyn-loaded cells showed little or no secretion.
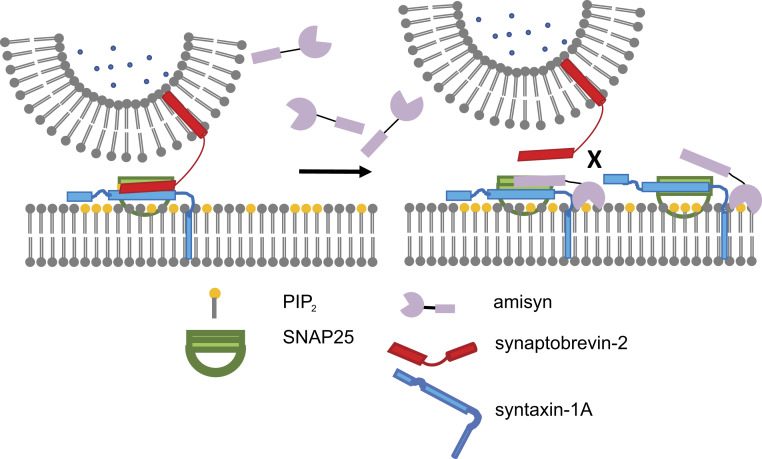
Based on available data, we designed a putative model of amisyn’s role in exocytosis (Fig. 8). Amisyn’s enrichment in the plasma membrane suggests that amisyn exerts its main functions there. The R-SNARE motif of amisyn interacts with the Q-SNAREs syntaxin-1 and SNAP-25 as effectively as synaptobrevin-2. In addition, amisyn acts primarily on the regulation of number of released vesicles from RRP, SRP, and during sustained secretion. Therefore, one could envision that amisyn acts as a negative regulator of SNARE complex assembly by competing with the “fusion-active” synaptobrevin-2 for SNAP-25/syntaxin-1 binding (Fig. 8). Since amisyn does not contain any transmembrane domains, it most likely forms a fusion-inactive SNARE complex, as originally suggested (14). However, in contrast with the earlier study (14), our model does not assume that amisyn holds SNAP-25/syntaxin-1 in a conformation ready for synaptobrevin-2 to replace it before membrane fusion takes place. Instead, we propose that amisyn controls the number of fused vesicles and timing of exocytosis, which may be important for brain development and autism, as well as insulin-secreting β-cells and diabetes. Furthermore, amisyn interacts with the plasma membrane in a PI(4,5)P2-dependent manner through its N-terminal PH domain that may serve as the protein’s recruiting device and, in turn, generate the required high local concentrations of amisyn that are needed to outcompete the secretory vesicle-anchored synaptobrevin-2. PI(4,5)P2-sensitive PH domain is also able to provide temporal and spatial characteristics needed for the regulation of exocytosis. Such a model could elegantly explain key observations in amisyn’s studies, including decreased exocytosis efficiency (refs. 14–16 and this study) and enhance our understanding of membrane fusion timing control.
Fig. 8.
Model of role of amisyn in secretory vesicle exocytosis. Amisyn acts as a negative regulator of SNARE-complex assembly by competing with the fusion-active synaptobrevin-2. Formation of the SNARE complex drives membrane fusion, whereby PI(4,5)P2 recruits amisyn to the plasma membrane to compete with synaptobrevin-2 in formation of the SNARE complex and vesicle exocytosis. Since amisyn does not contain a transmembrane domain, it forms a fusion-inactive SNARE complex.
Given the complexity of the exocytic machinery, it is not surprising that regulatory proteins like amisyn are needed to balance and control exocytosis, and are relevant for various diseases. Further studies are needed to fully comprehend this overlooked protein. Understanding amisyn both structurally and functionally is not only fundamentally important for our knowledge of the physiology of secretory and neuronal cells, but may also help to unravel complex pathological processes, like in diabetes and autism.
Methods
Plasmids.
Cloning of pEGFP(N1)-amisyn WT, pEGFP(N1)-amisyn-SNARE domain, pEGFP(N1)-amisyn AADD, pEGFP(N1)-amisyn PH domain, pGEX-6p1-amisyn WT, pGEX-6p1-amisyn SNARE domain, pGEX-6p1-amisyn AADD, and pGEX-6p1 amisyn PH domain are described in SI Appendix. All constructs were verified by control restriction enzyme digestions and by sequencing. The following additional expression constructs were used: rat syntaxin1a H3 domain (amino acids 180 to 262) (4), rat syntaxin-1 with its transmembrane region (amino acids 183 to 288) (42), rat SNAP-25a (amino acids 1 to 206) (4), soluble rat synaptobrevin-2 (amino acids 1 to 96) (4), and synaptobrevin-2 with transmembrane domain (amino acids 1 to 116) (42). pEGFP-PH-PLCɗ1, pmRFP-PI4P5KI and pmRFP-IPP1-CAAX (29), or empty pEGFP-N1 (Clontech) as control, were used as indicated.
Cell Culture, Cell Transfections, and Cell Stimulations.
The neuroendocrine cell line PC12 (ATCC CRL-1651) was maintained and propagated as detailed in SI Appendix. PC12 cells (until passage 14) were transfected with plasmids expressing amisyn-EGFP, mRFP-PI4P5KI, and mRFP-IPP1-CAAX (29). In some experiments, EGFP expressed from pEGFP-N1 was used as a control. Cell transfection was performed using Lipofectamine (ThermoFisher Scientific) 18 to 30 h before analysis. PC12 cells were either imaged live, used for plasma membrane sheet preparations, or fixed in 3.7% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 30 min at room temperature and further processed for confocal microscopy. Stimulation of PC12 cells was achieved with 59 mM KCl (Sigma), nicotine (Merck, N0267), and ionomycin (Merck, I0634) in Tyrode’s buffer (119 mM NaCl, 5 mM KCl, 25 mM Hepes pH 7.4, 2 mM CaCl2, 1 mM MgCl2, 6 g/L glucose), as indicated. The primary culture of bovine chromaffin cells was prepared as described previously (29), and handled as described in SI Appendix.
Protein Expression, Purification, and Labeling.
Due to the abundance of nonpreferred arginine codons, the full-length amisyn could be expressed only at very low levels under standard conditions. Thus, Escherichia coli BL21-CodonPlus(DE3)-RIL (Stratagene; these cells contain plasmids that encode the nonpreferred tRNAs) or Rosetta (DE3) (Merck 70954-4) competent cells transformed with pGEX-6p1-amisyn constructs were grown in Luria-Bertani medium (LB; tryptone, yeast extract, NaCl, 50 μg/mL ampicillin, 50 μg/mL chloramphenicol) to OD600 0.6 to 0.8 at 37 °C. Amisyn recombinant protein (amisyn WT, amisyn-SNARE, and amisyn-AADD) expression was induced by 10 µM isopropyl-1-thio-β-d-galacto-pyranoside (IPTG) and further culturing for 12 h at 18 °C and 250 rpm. The cells were pelleted by centrifugation (54,000 × g), washed with ice-cold PBS, and resuspended in equilibration buffer (150 mM NaCl, 50 mM Hepes, 2 mM EDTA, 2 mM DTT, pH 7.5) containing 1 mM phenylmethylsulfonyl fluoride (PMSF). The cells were subjected to cell fractionation (Fluidizer; Microfluidics). After centrifugation (25,000 × g, 20 min), the supernatant was loaded to the glutathione column (Protino Glutathione Agarose 4B column; Machery-Nagel; equilibration buffer: 150 mM NaCl, 50 mM Hepes, 2 mM EDTA, 2 mM DTT, pH 7.5). GST-amisyn was eluted with reduced glutathione (10 mM, in 50 mM Tris⋅HCl, 100 mM NaCl, pH 8.0). Next, the eluate was dialyzed before proteolytic cleavage (PreScission, GE Healthcare Life) to remove the GST-tag by incubation overnight at 4 °C. The eluate was again affinity-purified over the glutathione column to obtain the pure recombinant amisyn protein, as confirmed by SDS/PAGE analysis and Western blotting.
EGFP-PH-PLCɗ1 was expressed and purified as in ref. 29. Unless otherwise stated, proteins from other expression constructs were expressed through the pET28a vector (Novagen) in E. coli BL 21 (DE3) competent cells (Merck), purified, and labeled as described in SI Appendix.
Antibodies.
Custom-made polyclonal amisyn antibody was generated against full-length recombinant amisyn protein that was expressed and purified as detailed above. The rabbit was immunized with 300 μg recombinant amisyn emulsified in Freud’s adjuvant complete (Merck, F5881). Booster injections (150 μg recombinant amisyn emulsified in Freud’s adjuvant) were given every 2 to 3 wk for 4 mo. The blood was obtained from the rabbit’s ear veins and processed to remove the blood cells. The serum with polyclonal antiamisyn antibody was then aliquoted, and stored at −80 °C. This antiamisyn antibody (Aminchen #172) was affinity-purified, and its specificity was characterized as described in SI Appendix. The list of commercial antibodies and their dilutions is presented in SI Appendix.
DNA Sequence Analysis and Structural Modeling of Amisyn.
We searched available databases for possible homologs of amisyn using BLAST (43) against the nonredundant dataset with default parameters. We restricted the search to insects, molluscs, arachnids, crustaneans (all invertebrates), as well as all vertebrates, and separately mammals and primates. Homologous protein sequences (e-value < 10−5, and sequence coverage >70%) were collected and multiple sequence alignments were performed using the T-Coffee server (44).
Using the Phyre2 web portal for protein modeling, prediction, and analysis with default parameters (45), the tertiary structure of the N-terminal domain of amisyn and its SNARE motif were modeled. The N-terminal domain of 89 amino acids (68% of the query sequence) was modeled with 99.7% confidence, using the tertiary structure of the yeast Sec3p exocyst subunit (PDB ID code 3A58). The C-terminal domain, previously described as the SNARE domain, was modeled on the tomosyn (PDB ID code 1URQ) as template. The model was obtained with 99.8% confidence on a stretch of 58 amino acids (94% of the query sequence).
The resulting PDB files of models of the N-terminal domain and the SNARE domain of amisyn were used to perform structural alignments using the TM-align server (46). The N-terminal domain was aligned with its template (PDB ID code 3A58), as well as with several typical PH domains (e.g., PH domain from the exchange factor ARNO) (Mus musculus, PDB ID code 1U27). The SNARE domain was aligned with its template (PDB ID code 1URQ). PyMol software was used for protein visualization.
Anisotropy Measurements.
Anisotropy measurements were performed as previously described (28) on a Fluorolog 3 spectrometer with magnetic stirrer and built-in T-configuration equipped for polarization (Model FL322, Jobin Yvon). The experimental details are described in SI Appendix.
GUV Preparation and GUV Assays.
GUVs were prepared as described in Tarasenko et al. (47), with lipid composition as indicted in SI Appendix. GUV visualization experiments were carried out using the spinning-disk confocal set-up (UltraVIEW VoX, Perkin-Elmer) with an inverted microscope (Nikon Ti-E Eclipse) and a 14-bit electron-multiplying charge-coupled device camera (C9100; Hamamatsu) using Volocity 6.3 software for image acquisition. The experiments were carried out using a 250-μL borosilicate chamber (Lab-Tek). To avoid disruption of the GUV on contact, the chamber was coated with bovine serum albumin (BSA) and 50 μL GUVs (300 mM sucrose, 10 mm Hepes, pH 7.4) were added to 150 μL 192 mM NaCl, 10 mM Hepes, pH 7.4. Osmolality of GUV suspension and buffer was matched to that of the protein solutions. Proteins were added to a final concentration of 2.2 mg/mL, and solution was mixed by gently rotating the pipette tip.
For liposome sedimentation assays, freshly prepared liposomes (prepared as described in SI Appendix) were incubated with 10 μM recombinant amisyn for 20 min at 37 °C. The liposomes were then centrifuged for 1 h at 70,000 rpm (Sorvall RC-M120, rotor S120-AT3). Supernatant and pellet were separated, analyzed by SDS/PAGE, and the gel was scanned (EPSON Perfection V700 Photo).
Liposome Fusion Assays.
Liposome fusion was studied using a lipid dequenching assay (27), as described in SI Appendix.
Isolation of Plasma Membranes from PC12 Cells.
Crude PC12 cell membranes were prepared as described in Sharma et al. (48) with minor modifications. Cell were resuspended by pipetting in PBS containing 2 mM EDTA at room temperture, and pelleted by centrifugation (300 × g, 5 min, 4 °C). The pellet was resuspended in low ionic strength TEP buffer (20 mM Tris⋅HCl pH 7.4, 1 mM EDTA) containing protease inhibitors (Roche) and homogenized with 20 strokes of a tight-fitting glass-glass homogenizer. Intact cells and nuclei were removed by centrifugation (300 × g, 7 min). The supernatant was centrifuged (50,000 × g, 30 min) and the pelleted membranes resuspended in TEP buffer. Protein content was determined with the bicinchoninic acid method (BCA; Pierce Chemical Co.) using BSA as the standard. Protein electrophoresis and immunoblotting was performed as described in SI Appendix.
Live PC12 Cells and Plasma Membrane Sheet Experiments.
Experiments with live PC12 cells and plasma membrane sheets were performed and imaged as described in SI Appendix. Plasma membrane sheet from stimulated PC12 cells were generated 5 s after the cell stimulation started. Digital image analysis of data shown in Figs. 3H, 4D, 5 D and H, and 7 D and H were performed using ImageJ (49), as stated in the SI Appendix.
Electrophysiology and Electrochemistry.
Capacitance and amperometric measurements on bovine adrenal chromaffin cells were performed concurrently at room temperature (22–24 °C), and as described in SI Appendix. Recombinant amisyn (5 µM) or SNARE-amisyn mutant (5 µM) was added to the intracellular pipette solution (100 mM Cs-glutamate, 8 mM NaCl, 4 mM CaCl2, 32 mM Cs-Hepes pH 7.25, 2 mM Mg-ATP, 0.3 mM GTP, 0.4 mM Fura4F, and 0.4 mM Furaptra, 5 mM NPE, osmolarity ∼295 mOsm/kg), and kept cold until injection into cells through the custom-made glass patch pipette (NPI Electronic, GB150TF-8P, prepared with P-97 Flaming/Brown Micropipette Puller, Sutter Instruments). Analysis of amperometric recordings were performed by IGOR Pro (Wave Metrics) as in Mosharov and Sulzer (50).
Statistical Analysis.
Unless otherwise stated, all statistical analysis was done using Prism (GraphPad). Where indicated, nonparametric one-way ANOVA tests were used with significance set at P < 0.05. Unpaired two-sided t tests with Tukey’s correction were used for comparisons between specific groups. Cumulative frequency distributions were compared using the Kolmogorov–Smirnov test. Electrophysiological data were examined by the Kruskal–Wallis test with Dunn’s multiple comparison test. Unless otherwise stated, data are presented as mean ± SEM.
Data Availability.
All datasets generated and analyzed during the study are included in the main text, SI Appendix, and Dataset S1.
Supplementary Material
Acknowledgments
We thank D. Schwitters and B. Ide for excellent technical assistance; J. Peña del Castillo for help with amperometry analysis; J. Ferreira for protein expression; and Dr. I. Gottfried for initial discussion about amisyn’s pleckstrin homology domain. This work was supported by the Schram-Stiftung T287/25457 and the Deutsche Forschungsgemeinschaft Emmy Noether Young Investigator Award MI-1702/1 and SFB1190/P02 (to I.M.), SFB803/B09 and SFB1190/P12 (to B.K. and M.M.), Israel Science Foundation Grants 2546/16 and 953/16 (to U.A.), and the Engelhorn-Stiftung fellowship (to I.K.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908232117/-/DCSupplemental.
References
- 1.Terrian D. M., White M. K., Phylogenetic analysis of membrane trafficking proteins: A family reunion and secondary structure predictions. Eur. J. Cell Biol. 73, 198–204 (1997). [PubMed] [Google Scholar]
- 2.Weimbs T., Mostov K., Low S. H., Hofmann K., A model for structural similarity between different SNARE complexes based on sequence relationships. Trends Cell Biol. 8, 260–262 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Sutton R. B., Fasshauer D., Jahn R., Brunger A. T., Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Fasshauer D., Eliason W. K., Brünger A. T., Jahn R., Identification of a minimal core of the synaptic SNARE complex sufficient for reversible assembly and disassembly. Biochemistry 37, 10354–10362 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Söllner T., Bennett M. K., Whiteheart S. W., Scheller R. H., Rothman J. E., A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell 75, 409–418 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Bennett M. K., Calakos N., Scheller R. H., Syntaxin: A synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science 257, 255–259 (1992). [DOI] [PubMed] [Google Scholar]
- 7.Oyler G. A., et al. , The identification of a novel synaptosomal-associated protein, SNAP-25, differentially expressed by neuronal subpopulations. J. Cell Biol. 109, 3039–3052 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trimble W. S., Cowan D. M., Scheller R. H., VAMP-1: A synaptic vesicle-associated integral membrane protein. Proc. Natl. Acad. Sci. U.S.A. 85, 4538–4542 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baumert M., Maycox P. R., Navone F., De Camilli P., Jahn R., Synaptobrevin: An integral membrane protein of 18,000 daltons present in small synaptic vesicles of rat brain. EMBO J. 8, 379–384 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Q., et al. , Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis. Nature 525, 62–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prinslow E. A., Stepien K. P., Pan Y. Z., Xu J., Rizo J., Multiple factors maintain assembled trans-SNARE complexes in the presence of NSF and αSNAP. eLife 8, e38880 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMahon H. T., Missler M., Li C., Südhof T. C., Complexins: Cytosolic proteins that regulate SNAP receptor function. Cell 83, 111–119 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Fujita Y., et al. , Tomosyn: A syntaxin-1-binding protein that forms a novel complex in the neurotransmitter release process. Neuron 20, 905–915 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Scales S. J., Hesser B. A., Masuda E. S., Scheller R. H., Amisyn, a novel syntaxin-binding protein that may regulate SNARE complex assembly. J. Biol. Chem. 277, 28271–28279 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Constable J. R., Graham M. E., Morgan A., Burgoyne R. D., Amisyn regulates exocytosis and fusion pore stability by both syntaxin-dependent and syntaxin-independent mechanisms. J. Biol. Chem. 280, 31615–31623 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Collins S. C., et al. , Increased expression of the diabetes gene SOX4 reduces insulin secretion by impaired fusion pore expansion. Diabetes 65, 1952–1961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guček A., et al. , Fusion pore regulation by cAMP/Epac2 controls cargo release during insulin exocytosis. eLife 8, e41711 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castermans D., et al. , Position effect leading to haploinsufficiency in a mosaic ring chromosome 14 in a boy with autism. Eur. J. Hum. Genet. 16, 1187–1192 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Castermans D., et al. , SCAMP5, NBEA and AMISYN: Three candidate genes for autism involved in secretion of large dense-core vesicles. Hum. Mol. Genet. 19, 1368–1378 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Fernandez S. V., et al. , DNA methylation changes in a human cell model of breast cancer progression. Mutat. Res. 688, 28–35 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenka G., et al. , Identification of methylation-driven, differentially expressed STXBP6 as a novel biomarker in lung adenocarcinoma. Sci. Rep. 7, 42573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kloepper T. H., Kienle C. N., Fasshauer D., SNAREing the basis of multicellularity: Consequences of protein family expansion during evolution. Mol. Biol. Evol. 25, 2055–2068 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Fasshauer D., Otto H., Eliason W. K., Jahn R., Brünger A. T., Structural changes are associated with soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor complex formation. J. Biol. Chem. 272, 28036–28041 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Hatsuzawa K., Lang T., Fasshauer D., Bruns D., Jahn R., The R-SNARE motif of tomosyn forms SNARE core complexes with syntaxin 1 and SNAP-25 and down-regulates exocytosis. J. Biol. Chem. 278, 31159–31166 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Pobbati A. V., Razeto A., Böddener M., Becker S., Fasshauer D., Structural basis for the inhibitory role of tomosyn in exocytosis. J. Biol. Chem. 279, 47192–47200 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Fasshauer D., Antonin W., Margittai M., Pabst S., Jahn R., Mixed and non-cognate SNARE complexes. Characterization of assembly and biophysical properties. J. Biol. Chem. 274, 15440–15446 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Weber T., et al. , SNAREpins: Minimal machinery for membrane fusion. Cell 92, 759–772 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Pobbati A. V., Stein A., Fasshauer D., N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science 313, 673–676 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Milosevic I., et al. , Plasmalemmal phosphatidylinositol-4,5-bisphosphate level regulates the releasable vesicle pool size in chromaffin cells. J. Neurosci. 25, 2557–2565 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milosevic I., Spatial and temporal aspects of phosphoinositides in endocytosis studied in the isolated plasma membranes. Methods Mol. Biol. 1847, 147–160 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Pleskot R., Cwiklik L., Jungwirth P., Žárský V., Potocký M., Membrane targeting of the yeast exocyst complex. Biochim. Biophys. Acta 1848, 1481–1489 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Eberhard D. A., Holz R. W., Calcium promotes the accumulation of polyphosphoinositides in intact and permeabilized bovine adrenal chromaffin cells. Cell. Mol. Neurobiol. 11, 357–370 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neher E., A comparison between exocytic control mechanisms in adrenal chromaffin cells and a glutamatergic synapse. Pflugers Arch. 453, 261–268 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Voets T., Dissection of three Ca2+-dependent steps leading to secretion in chromaffin cells from mouse adrenal slices. Neuron 28, 537–545 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Xu T., Binz T., Niemann H., Neher E., Multiple kinetic components of exocytosis distinguished by neurotoxin sensitivity. Nat. Neurosci. 1, 192–200 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Mosharov E. V., Analysis of single-vesicle exocytotic events recorded by amperometry. Methods Mol. Biol. 440, 315–327 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Chow R. H., von Ruden L., Neher E., Delay in vesicle fusion revealed by electrochemical monitoring of single secretory events in adrenal chromaffin cells. Nature 356, 60–63 (1992). [DOI] [PubMed] [Google Scholar]
- 38.Barg S., Guček A., How kiss-and-run can make us sick: SOX4 puts a break on the pore. Diabetes 65, 1791–1793 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Schink K. O., Tan K. W., Stenmark H., Phosphoinositides in control of membrane dynamics. Annu. Rev. Cell Dev. Biol. 32, 143–171 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Nakajima H., et al. , A cytoskeleton-related gene, uso1, is required for intracellular protein transport in Saccharomyces cerevisiae. J. Cell Biol. 113, 245–260 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yue P., et al. , Sec3 promotes the initial binary t-SNARE complex assembly and membrane fusion. Nat. Commun. 8, 14236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Margittai M., Otto H., Jahn R., A stable interaction between syntaxin 1a and synaptobrevin 2 mediated by their transmembrane domains. FEBS Lett. 446, 40–44 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 44.Notredame C., Higgins D. G., Heringa J., T-coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 302, 205–217 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y., Skolnick J., TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarasenko D., et al. , The MICOS component Mic60 displays a conserved membrane-bending activity that is necessary for normal cristae morphology. J. Cell. Biol. 216, 889–899 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma N., D’Arcangelo G., Kleinlaus A., Halegoua S., Trimmer J. S., Nerve growth factor regulates the abundance and distribution of K+ channels in PC12 cells. J. Cell Biol. 123, 1835–1843 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rasband W. S., ImageJ. https://imagej.nih.gov/ij/ (National Institutes of Health, Bethesda, MD, 1997–2018). Accessed 1 February 2020.
- 50.Mosharov E. V., Sulzer D., Analysis of exocytotic events recorded by amperometry. Nat. Methods 2, 651–658 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated and analyzed during the study are included in the main text, SI Appendix, and Dataset S1.