Abstract
Transposable elements constitute about half of the mammalian genome, and can be divided into two classes: the class I (retrotransposons) and the class II (DNA transposons). A few hundred types of retrotransposons, which are dynamic and stage specific, have been annotated. The copy numbers and genomic locations are significantly varied in species. Retrotransposons are active in germ cells, early embryos and pluripotent stem cells (PSCs) correlated with low levels of DNA methylation in epigenetic regulation. Some key pluripotency transcriptional factors (such as OCT4, SOX2, and NANOG) bind retrotransposons and regulate their activities in PSCs, suggesting a vital role of retrotransposons in pluripotency maintenance and self-renewal. In response to retrotransposons transposition, cells employ a number of silencing mechanisms, such as DNA methylation and histone modification. This review summarizes expression patterns, functions, and regulation of retrotransposons in PSCs and early embryonic development.
Keywords: Retrotransposon, Pluripotent stem cells, Pluripotency, Epigenetic regulation
Background
Pluripotent stem cells (PSCs) are cells that have potential to differentiate into various cells and tissues. The most important properties of PSCs are self-renewal and differentiation into adult stem cells or multiple somatic cells during development (He et al., 2009; Hackett & Surani, 2014). Most researches were focused on cell differentiation, induced PSC (iPSC) reprogramming efficiency, and disease modeling with emphasis on clinical therapies, thus conditions of PSCs are important (Ding et al., 2013; Avior et al., 2016; Mora et al., 2017; Bernareggi et al., 2019). In PSC culture, heterogeneity is a key issue that remains to be unresolved (Hayashi et al., 2019). Apart from researches about gene expression and epigenetic modifications that are specific to PSCs (Bar-Nur et al., 2011; Kim et al., 2012; Stelzer et al., 2013), more and more scientists have begun to focus on transposon elements, which make up almost half of the mammalian genome and were considered “junk” DNA (Chen et al., 2008). Retrotransposons are the most mobile class of transposon elements and studies have demonstrated that they are transcribed more in early embryos and PSCs than somatic cells (Kigami et al., 2003; Zhou & Smith, 2019).
Early mammalian embryos undergo widespread epigenetic reprogramming accompanied by chromatin re-establishment and organization (chromatin interaction) (Xu & Xie, 2018). Studies have shown that cis-regulatory elements (such as promoters, enhancers, and super-enhancers) and trans-factors (such as transcriptional factors and epigenetic effectors) can drive embryonic development. It is worth mentioning that some pluripotency transcriptional factors (such as OCT4, SOX2, and NANOG) control stem cells fate by regulating pluripotency maintenance and lineage differentiation repression, and contributing to the open chromatin architecture of stem cells (Ahmed et al., 2010; Mulas et al., 2018; Bakhmet et al., 2019). Recently, more and more studies have identified that retrotransposons also play important roles in PSCs and embryos, and some factors regulating the elements have been identified (Coluccio et al., 2018; Percharde et al., 2018). Combined with the fact that pluripotency transcriptional factors bind retrotransposons and regulate their activities in PSCs, it is logical to assume that retrotransposons have functions in embryogenesis, maintaining PSCs pluripotency and so on. In this review, we report on recent advances in the study of retrotransposons in mice and humans with a focus on the expression patterns, specific roles in distinct development stages, and important regulation mechanisms.
Retrotransposons in genomes
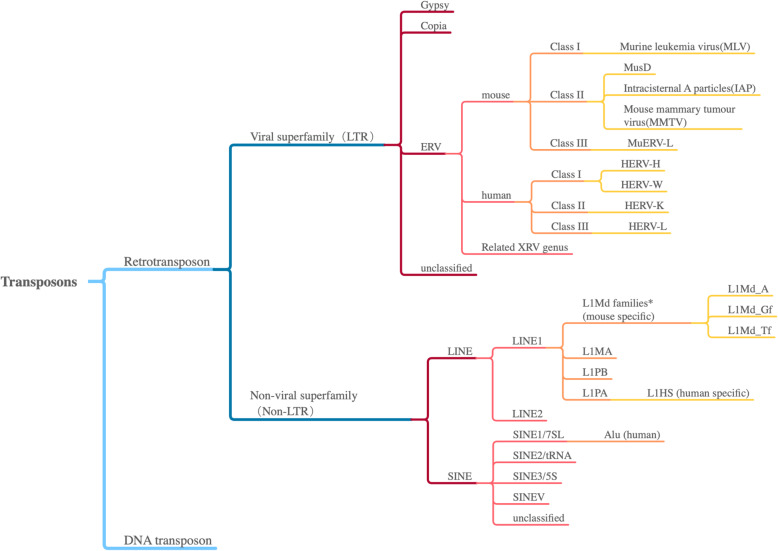
Transposable elements (TE), also known as transposons, were first discovered in 1948 by geneticist Barbara McClintock (Mc, 1948; Mc, 1950). She found certain chromosomes can change their position from one generation to the next and can even be turned on or off in different cell culture stages and environment conditions. In the 1960s to 1970s, this kind of “jumping” DNA sequences were also found in bacteria. As reported in whole genome sequencing projects, almost half of mammalian genomes and approximately 90% of the maize genome was made up of TEs (Lander et al., 2001; Schnable et al., 2009). There are two classes of TEs: the class I (retrotransposons) and the class II (DNA transposons). Class I TEs are transcribed to RNA, then reverse transcribed to cDNA, and integrated in the genome by a “copy and paste” mechanism. Class II TEs can encode transposase enzymes to “cut and paste” directly. The detailed classification is shown in Fig. 1.
Fig. 1.
Retrotransposon classification. A tree diagram of transposons. Well-known classes and subclasses are listed, and mouse/human specific families are highlighted. Transposons are divided into two classes by the type of transposition. Retrotransposons are classified into two types according to the encoding transposase. ERVs are the most popular elements studied in LTRs. In LINE1 subfamilies, L1MA4–1 and L1PB3–1 are already extinct, and L1PA evolved as a single lineage
Retrotransposons are commonly classified into two subclasses with variant characters and functions. Subclass I consists of viral retrotransposons characterized by long terminal repeats (LTRs) on both terminals. They encode reverse transcriptase and change position like retroviruses, but lack a functional envelope gene (Env). They constitute about 8% of the human genome and approximately 10% of the mouse genome (Mameli et al., 2007). The most abundant and more active subclass in human called endogenous retroviruses (ERVs), a type of LTR retrotransposon, may originate from retroviruses and constitute 5–8% of human genome (Nelson et al., 2004). Another endogenous retroviral-like LTR element was identified in mice, such as intracisternal A particle (IAP), which repeated 1–2000 times in mouse genome, and has been found to transpose in mouse tumor cells and germ cells of a few mouse strains (Kuff & Lueders, 1988).
Subclass II consists of non-viral retrotransposons, also known as non-LTR TEs (Feng et al., 1996). They can be further divided into two subgroups by the length of elements. The first subgroup is long interspersed nuclear elements (LINE), which has an average length of 7000 base pairs and is widespread in many eukaryotes, comprising 21% of the human genome. The structure of LINEs contains a 5’UTR, two non-overlapping open reading frames (ORF), a 3’UTR, and a 3′ poly (A) tail. The first ORF encodes a nucleic acid binding protein (Holmes et al., 1992) and the second ORF of LINEs may encode reverse transcriptase (Sakaki et al., 1986), and there is an endonuclease domain at the ORF2 N-terminus, which helps to cleave the target site for insertion (Moran et al., 1996). Most LINE elements are 5′ truncated, suggesting that 5′ sequence is not indispensable while 3′ poly (A) tail is critical in transposition (Feng et al., 1996). LINE-1(L1), a subgroup of LINEs, can be found in all mammals and is the only activated autonomous element in human genome with an estimated 500,000 copies (Belancio et al., 2008). The second subgroup of non-LTR retrotransposons short interspersed nuclear elements (SINE) is 50–500 base pairs in length. They are non-coding transposons and utilize other TE reverse transposases for reposition. There are over 500,000 copies of Alu SINE elements in human genome and suspected to cause various diseases by disrupting gene sequences.
Retrotransposons in early embryo development
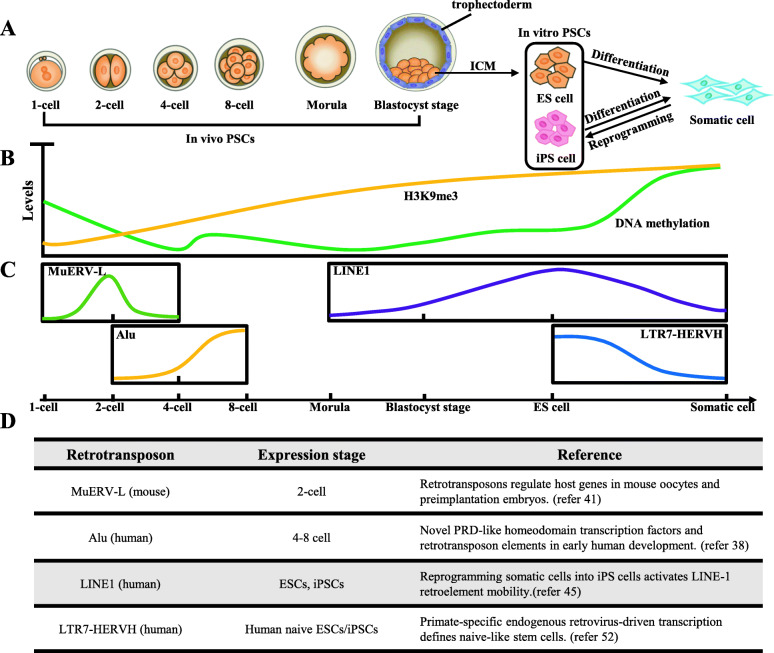
There is a remarkable change in DNA methylation from sperm to the zygote. DNA methylation level increases progressively during embryo development and finally reaches normal level in post-implantation embryos (Fig. 2a and b). This process is accompanied by changes in retrotransposon expression.
Fig. 2.
Epigenetic modification and retrotransposon expression changes in embryogenesis. a Different stages of embryonic development process are showed (from left to right): 1-, 2-, 4-, 8-cells, morula, and blastocyst stages. Embryonic stem cells (ESCs) are derived from the inner cell mass (ICM) of the early blastocysts, and have similar developmental potential as ICM. Inducible pluripotent stem cells (iPSCs) come from differentiated somatic cells and together with ESCs comprise in vitro PSCs; b DNA methylation (green) and H3K9me3 (yellow) levels decrease after fertilization, then increase to normal levels in somatic cells; c The expression of retrotransposon subclasses changes in the embryonic development process, and some typical elements are shown such as MuERV-L(green), Alu(yellow), LINE1(purple), and LTR7-HERVH(blue); d The summary of expression patterns of retrotransposons demonstrated in (c) and related references
For non-LTR elements, studies have shown that L1 and IAP retrotransposons become reactivated from both parental genomes after fertilization (Fadloun et al., 2013). In mice, over 85% LINEs have significant translational changes and about 18% LINEs display a change in methylation levels (more than 45%), which are concentrated in Mus musculus domesticus L1 retrotransposons (L1Md_Tf and L1Md_Gf classes), suggesting specific roles during fertilization (Smith et al., 2012). L1Md families can express at the onset of germ cell meiotic prophase I (MPI) and are the most abundant and active TEs in the mouse genome (Gaysinskaya et al., 2018). Further research also indicates that each of the three L1Md classes (A, Gf, and Tf) has much higher expression in 2-cell embryonic stage than in mouse ESCs (mESCs). Notably, L1Md_A can even be detected at 8-cells stage and progressively decreases till 16-cells stage (Jachowicz et al., 2017). Most of the newly generated L1 DNA copies are unstably integrated into the genome, while some of them can stably insert to the genome leading to genetic diseases, such as aberrant splicing of glycine receptor beta subunit mRNA and exon skipping caused by the L1 element retrotransposition (Mulhardt et al., 1994; Takahara et al., 1996; van den Hurk et al., 2007; Vitullo et al., 2012). When L1 elements in zygotes are repressed immediately after fertilization, pre-implantation development is arrested at the two- and four-cells stages, leading to 50% reduction the rates of the blastocyst development and embryo fragmentation. It has shown that the L1 contributes to early embryo development and exits from the two-cells stage (Percharde et al., 2018; Jachowicz et al., 2017; Beraldi et al., 2006). Furthermore, researchers have found that overexpression of L1 causes abnormal chromatin decondensation, thus suggesting the another role of L1 transcript independent (Jachowicz et al., 2017).
Meanwhile, Alu elements, which belong to SINE family, are the most abundant repeats in the human genome, participating in regulating gene expression in early embryos. The studies have shown that 32 and 129 genes are unregulated in the transition from oocyte to four-cells stage and from four- to eight-cells stage, respectively (Tohonen et al., 2015) (Fig. 2c). Most of these genes have similar regulatory motifs that overlapped with Alu elements and these sequences appear in the 5’UTR, promoter, and around the transcription start site (TSS), which demonstrates that active Alu elements may regulate certain gene expression in human embryos.
Apart from the non-LTR retrotransposons, LTR elements also have indispensable roles in embryos. Although most LTRs have lost retroviral activity (Lower et al., 1996), stage-specific expression demarcates the different developmental stages and distinct cell populations in early embryos (Goke et al., 2015). Murine endogenous retrovirus-like gene (MuERV-L) which the most quickly transcribed gene from a zygotic genome, begins in the S phase of the first cell cycle and continues to blastula (Kigami et al., 2003). Compared with somatic tissues, the TSSs of two-cell-specific genes show preferential enrichment for MuERV-L and other ERV-L elements. Thus, MuERV-L is a potential marker of two-cell like cells because of the maximal transcription among LTRs. Transposition in cleavage-stage embryos is believed to use MuERV-L reverse transcriptase (Peaston et al., 2004; Macfarlan et al., 2012) (Fig. 2c). Disrupting the transcription of MuERV-L leads to a remarkable decrease in developmental competence after the four-cell stage, confirming its function in embryos (Kigami et al., 2003). In addition, existing researches have confirmed that LTRs not only promote some stage-dependent genes expression (such as Rpl41 and 2610005H11Rik) (Peaston et al., 2004) but also contribute to the accessibility of chromatin landscape in early embryos (Wu et al., 2016).
Retrotransposons in PSCs
PSCs have the potential to self-renew and differentiate into three germ layer cell types (endodermal, mesodermal, and ectodermal cells) until adulthood (He et al., 2009; Hackett & Surani, 2014) (Fig. 2a). Transcription factors specifically expressed in PSCs (such as OCT4, SOX2, KLF4, C-MYC, and NANOG) control pluripotency, and they prefer to bind to functional genome features (such as promoters, enhancers) and open chromatin. Studies in recent years have shown that a large portion of human open chromatin regions (44%) overlap with TEs (Jacques et al., 2013) and transcription factors (such as OCT4, SOX2, and CTCF) are found bind to distinctive classes of TEs (Chen et al., 2008). On the other hand, some TEs work as promoters and enhancers (such as ERV) in PSCs. All the aforementioned evidence illustrates that retrotransposons provide regulation elements for core pluripotency transcription factors and thus may help maintain pluripotency.
It has been demonstrated that L1 full-length mRNA and the ORF1-encoded protein (L1TD1) can be detected in both ESCs and iPSCs, caused by an overall decrease in CpG methylation in the L1 promoter region (Wissing et al., 2012). L1TD1 is a stem-cell-specific RNA binding protein and chromatin immunoprecipitation sequencing (ChIP-seq) results show that pluripotency factors OCT4, SOX2, and NANOG all bind to the promoter of L1TD1, while deleting L1TD1 leads to the immediate down regulation of OCT4 and NANOG (Narva et al., 2012). Further research shows that more than half of de novo LINE-1 insertions are full length and enriched in the specific protein-coding genes of PSCs (Garcia-Perez et al., 2007; Klawitter et al., 2016) (Fig. 2c). Knockdown of L1 RNA inhibits mESC self-renewal (Percharde et al., 2018). Therefore, it is logical to assume that reactivation of LINE1 is required for pluripotency maintenance and self-renewal.
There are two states of pluripotency, naive and primed (Nichols & Smith, 2009). Each state has its own special morphology, gene expression pattern, and epigenetic modification, which can serve as methods of distinction. Some retrotransposons have been found to exhibit dramatic stem-cell-specific expression, and are considered as a marker of the naive state. LTR7-HERVH, a primate-specific endogenous retrovirus, was found to be enriched in the TSS of 127 HERVH-lincRNAs in both human ESCs and iPSCs, and 40% of them are annotated as enhancers in ESCs, whereas only 2.2% are annotated in other cells (Kelley & Rinn, 2012; Lu et al., 2014; Wang et al., 2014) (Fig. 2c). HERVH is considered a promoter because of the enrichment of H3K4me3, and it provides binding sites at 5’UTR for NANOG, with OCT4 and SOX2 (Goke et al., 2015; Wang et al., 2014; Loewer et al., 2010; Santoni et al., 2012). Recently, the study has reported that HERVH influences on chromatin architecture through establishing topologically associating domains (TAD) boundaries (Zhang et al., 2019). When knockout or transcriptional repression of HERVH in hESCs is achieved, corresponding TAD boundaries are eliminated and new chromatin domain boundaries rebuild when HERVH elements are inserted into the genome. In this process, transcription of genes upstream from the HERVH loci are affected, suggesting HERVH participates in hESC-specific gene regulation network (Zhang et al., 2019). Another naive pluripotency transcription factor LBP9 has been confirmed to work with HEVRH to drive hESC-specific pluripotency-modulating long non-coding RNAs, suggesting that HERVH elements also participate in pluripotency regulation and self-renewal development via lincRNA regulation (Kelley & Rinn, 2012; Wang et al., 2014).
Heterogeneity is another important phenotype in PSCs and is a key issue that remains unsolved. iPSCs, which are reprogramed from somatic cells, are highly similar to ESCs in transcription profiles but display a more open chromatin state (Cao et al., 2018). Despite the origin, almost all the repetitive elements have been found to be upregulated in mouse iPSCs compared to differentiated cells, such as L1. ERV MusD remains highly expressed, whereas other elements, such as IAP, HERVK, and S71 (LTR6b) display a fully repressed state, similar to expression patterns in ESCs (Fort et al., 2014; Friedli et al., 2014). Some of the elements are partially expressed in iPSCs, which are completely repressed in ESCs, suggesting a failure to reactivate sequence-specific repressors during reprogramming (Friedli et al., 2014) and shows heterogeneity is also associated with retrotransposon-selective transcription.
Epigenetic regulation of retrotransposon
Although most retrotransposons have become inactive during evolution, some are still active and can insert new copies in the genome. Additionally, active retrotransposons are triggered in specific stages, especially in germlines, early embryos, and PSCs, which have a more open epigenetic environment. However, if somatic mutagenesis occurs as a result of diseases, cancer, or aging, the retrotransposons may be reactivated, and the abnormal activity may cause frequent transposition, breakage of DNA, and genome instability (Tubio et al., 2014). In response, cells have evolved mechanisms to keep retrotransposons elements tightly repressed (Vagin et al., 2006; Tam et al., 2008; Watanabe et al., 2008; Di Giacomo et al., 2013), and the regulation mechanisms can be divided into several levels: transcription, mRNA mature and export, translation, RNP formation, and transposition.
Epigenetic regulation occurs at transcriptional and translational levels. Silencing TEs are characterized by heterochromatin which is highly compact chromatin architecture, and are rich in repressive histone modifications, DNA methylation, and co-repressor proteins (such as NuRD complex, HP1, and repressive epigenetic enzymes). DNA methylation levels regulate retrotransposon expression in ESCs and iPSCs. DNA methylation enzymes (DNMT3a, DNMT3b, or DNMT1) are responsible for DNA methylation. Double knockout of DNMT3a/3b causes global loss of DNA methylation, and over 98% of the loss overlaps with repeat sequences (Li et al., 2015). The majority of ERVK subfamily members are sensitive to DNMT3a/3b, while DNMT1-mediated methylation activity is critical for suppressing IAP transcription in the mouse genome (Li et al., 2015). Additionally, the DNA methylation level of LINE1 promoter that contains some CpG sites, affects its transcription, and DNA hypomethylation of the promoter triggers its expression (Li et al., 2015). After fertilization, genome-wide DNA demethylation occurs in mammalian embryos. Thus, PSCs genomes are characterized by their low content of DNA methylation, which correlates with increasing levels of L1 mRNA expression (Gaysinskaya et al., 2018).
Repressive histone modifications are important in silencing retrotransposons. After knockout of H3K9me3 site methyltransferase Setdb1 in mESCs, ERV elements lose H3K9me3 accompanied by unregulated genes as 15% of promoters are proximal to demethylated ERVs (Karimi et al., 2011). Death domain associated protein (Daxx) binding sites are highly enriched in the IAP and ERVK. Daxx/Atrx complex can recruit Suv39h1 (another H3K9me3 site methyltransferase) to promote H3K9me3 and safeguards hypomethylated genome in naive pluripotent state (He et al., 2015). Similarly, histone chaperone CAF-1 mediates the replacement of H3.3 with H3.1/3.2 at retrotransposon regions, and this process is associated with deposition of repressive histone markers (H3K9me3, H3K9me2, H3K27me3, and H4K20me3). Subsequently, CAF-1 protects preimplantation mouse embryos from endogenous retrotransposons. Recently, a genome-wide CRIPSR-Cas9 screening was done in human cells, which aims to find genes that participant in L1 retrotransposition. Protein MORC2 and human silencing hub (HUSH) complex helped each other to repress the endogenous, evolutionarily young L1s in both hESCs and K562 cells, and they finally promoted deposition of H3K9me3 for L1s transcriptional silencing (Liu et al., 2018).
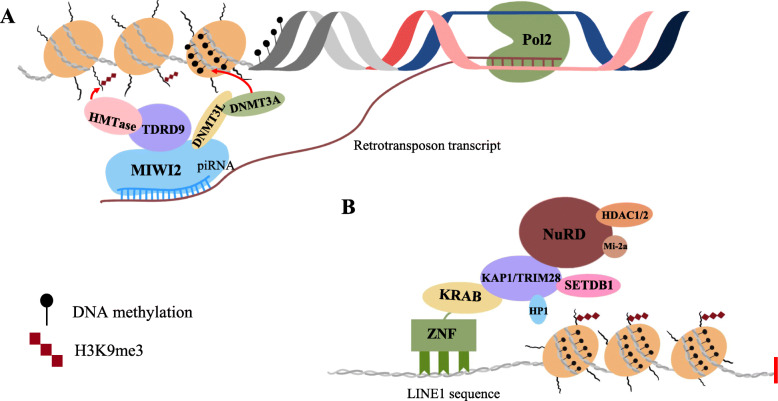
It is worth mentioning that histone and DNA methylation work in tandem and form a typical mechanism. In early embryogenesis and ESCs, KRAB domain-containing zinc finger proteins (KRAB-ZFPs) and KRAB-associated protein 1 (KAP1, also known as tripartite motif containing protein 28, TRIM28)-regulating retrotransposon silencing has been explored extensively. KRAB-ZFPs recognize specific retrotransposon sequences and recruit KAP1 to repress transcription of the targeted retrotransposon and adjacent genes. KAP1 repressed genes by recruiting histone methyltransferase SETDB1, HP1, and the NuRD histone deacetylase complex. KAP1 deletion decreased the level of H3K9me3, and upregulated ERVs, partial IAP elements, and genes such as ZFP575, Prnp, and Serinc3. Therefore, ESCs used KAP1-dependent pathway which resulting in H3K9me3 enrichment on retro-elements (Matsui et al., 2010; Rowe et al., 2010). Through this mechanism, we found that epigenetic silencing could spread from repetitive elements to neighboring genes (Fig. 3a).
Fig. 3.
Epigenetic regulations of retrotransposons by DNA methylation and H3K9me3 modification. a Schematic diagram of different forms in retrotransposon repression. a piRNA-pathway is the most common mechanism to regulate retrotransposons in germ cells; piRNA guides PIWI protein to bind transposon transcripts, which have a matching sequence, and cleave target RNA. In addition, piRNAs work not only at the post-transcriptional level, but also regulate DNA methylation and histone modification. b KAP1/Trim28 complex is important in retrotransposons repression in early embryogenesis and ESCs. In this mechanism, KRAB-ZFPs recruit KAP1 to repress transcription of targeted retrotransposon, and KAP1 represses genes by recruiting histone methyltransferases SETDB1, HP1, and NuRD histone deacetylase complex
In post-transcription regulation, the functions of silencing TEs by microRNA (miRNA), short interfering RNA (siRNA), and the P-element-induced wimpy testis (PIWI)-interacting RNA (piRNA) pathways have been confirmed (Fig. 3b). Tissue-specific siRNA and miRNA pathways are active in all tissues while the piRNA pathway is predominantly active in the germline (Slotkin & Martienssen, 2007; Ghildiyal & Zamore, 2009; Heras et al., 2013; Hamdorf et al., 2015). Genome-wide DNA demethylation occurs in mammalian embryos after fertilization. However, piRNA/Dnmt3L pathway is not responsible for suppressing TEs in this time (Rougier et al., 1998; Hajkova et al., 2002; Bourc'his & Bestor, 2004). In MII oocytes, piRNA is present in almost 50% of transcripts, reduces to about 25% (similar to miRNA), and finally drops to undetectable levels in ICM (Ohnishi et al., 2010). One-cell embryos injecting with mDicer-siRNA or long mDicer-dsRNA show a 50% increase in IAP and MuERV-L transcript abundance at 8-cells stage, suggesting the function of piRNA is progressively replaced by other small RNAs (Svoboda et al., 2004).With the development of pre-implantation mouse embryos, small RNA transcription changes from siRNA/piRNA to miRNA, but the post-transcriptional mechanism continues to be effective in repressing retrotransposons (Ohnishi et al., 2010; Russell et al., 2016).
Conclusions and perspectives
TEs comprised a great part of the genome, but were previously considered useless. With more and more researches, we have recognized that few retrotransposons still work even though a majority of them have been truncated during genome evolution. Although retrotransposons are usually regulated by small RNAs, DNA methylation, and histone modifications, the global genome is in a more open state in PSCs compared to somatic cells, providing opportunities for repeat sequence transcription, and retrotransposons take part in gene regulation by actively reshaping chromatin structure or providing transcription factor binding sites.
The vast majority of retrotransposon functions are not very clear, but specific expression patterns of each family and subfamily and their effect on the genome can be observed. It is worth mentioning that different iPS cell lines have different transcription levels in some transposon classes, indicating heterogeneity of cell lines. Moreover, although ESCs and iPSCs share similar repetitive element expression, differences may exist due to variations in epigenetic modifications. As of yet, we still do not have a detailed protocol to evaluate a good iPS cell line. Retrotransposon transcription level may be considered as a criterion if it proves to work well in cell therapy. Furthermore, active or repressive types of transposon expression may be a new way to induce cells into the desired stage or condition, thus having potential in model construction and tissue differentiation.
Despite having benefits, the random insertion of transposons may cause genome instability and possibility of affecting promoter function, gene transcription, and pre-mRNA slicing, which happens more frequently in germ cells and early embryos because of the low level of repressive epigenetic regulation. LINE1 is associated with tumorigenesis in several cancers and human genetic diseases caused by Alu element homologous recombination (reach up to 0.3%) (Ahmed et al., 2010; Deininger & Batzer, 1999). By evaluating the expression of LINE1 elements, it was found that high levels of ORF1 occur more frequently in breast and ovarian cancer (Chen et al., 2012; Rodic et al., 2014), whereas ORF2 expression often occurs earlier in the tumorigenesis process (De Luca et al., 2016), indicating that retrotransposons may be a useful early diagnostic marker of certain cancers. However, translation of transposons can also be used as a tool. Transformation efficiency is improved by using hyperactive PiggyBac transposase in insect (Eckermann et al., 2018), and CRISPR/Cas9 gene editing system also can be stably transformed in multiple human cells, including iPSCs (Ishida et al., 2018; Schertzer et al., 2019), providing a new platform for genetic disease treatment.
Aberrant transposon activation leads to genome instability and carcinogenesis, thus, the mechanism of transposons regulation needs to be analyzed in detail. Apart from the classic regulation mechanism, more and more epigenetic modifications have been discovered, such as histone succinylation, crotonylation, 6 mA DNA and m6A mRNA modifications. As there are no detailed studies about the relationship between these new modifications and TE regulation, more regulation mechanisms need to be elucidated.
To date, many TEs are still not recognized, and the sequences may have changed with genome evolution. Functions of enhancers are not easy to detect because of the complex spatial structures of chromatin. TEs are a large family and have multiple functions and with the development of new technologies, we will be one step closer to understanding them and their regulation.
Acknowledgements
This work was supported by the National Key R&D Program of China (2017YFA0102801 and 2017YFC1001901), the National Natural Science Foundation (31671540and 31971365), the Natural Science Foundation of Guangdong Province (2017A030313093 and 2017A030313491), the Guangdong Basic and Applied Basic Research Foundation (2019A1515011422) and the Guangzhou Science and Technology Project (201803010020).
Abbreviations
- ESCs
Embryonic stem cells
- iPSCs
Induced pluripotent stem cells
- hPSCs
Human pluripotent stem cells
- LTRs
Long terminal repeats
- ERVs
Endogenous retroviruses
- IAP
Intracisternal A particle
- ORF
Open reading frames
- L1
LINE1–1
- SINEs
Short interspersed nuclear elements
- LINEs
Long interspersed nuclear elements
- L1Md
Mus musculus domesticus L1 retrotransposon
- ICM
Inner cell mass
- TE
Transposable element
- ChIP-seq
Chromatin immunoprecipitation sequencing
- piRNAs
P-element-induced wimpy testis (PIWI)-interacting RNAs
- TSS
Transcription start site
- MuERV-L
Murine endogenous retrovirus-like gene
- NHEJ
Non-homologous end joining
- Daxx
Death Domain Associated Protein
- KRAB-ZFPs
KRAB domain-containing zinc finger proteins
- KAP1
KRAB-associated protein 1
- siRNA
short interfering RNA
- miRNA
microRNA
Authors’ contributions
JW, JH and GS conceived and wrote the paper. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Contributor Information
Junjiu Huang, Email: hjunjiu@mail.sysu.edu.cn.
Guang Shi, Email: shguang@mail.sysu.edu.cn.
References
- Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One. 2010;5:e10531. doi: 10.1371/journal.pone.0010531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avior Y, Sagi I, Benvenisty N. Pluripotent stem cells in disease modelling and drug discovery. Nat Rev Mol Cell Biol. 2016;17:170–182. doi: 10.1038/nrm.2015.27. [DOI] [PubMed] [Google Scholar]
- Bakhmet EI, Nazarov IB, Gazizova AR, Vorobyeva NE, Kuzmin AA, Gordeev MN, Sinenko SA, Aksenov ND, Artamonova TO. Khodorkovskii MA, Alenina N, Onichtchouk D, Wu G, Scholer HR. Tomilin AN: hnRNP-K targets open chromatin in mouse embryonic stem cells in concert with multiple regulators. Stem Cells. 2019;37:1018–1029. doi: 10.1002/stem.3025. [DOI] [PubMed] [Google Scholar]
- Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- Beraldi R, Pittoggi C, Sciamanna I, Mattei E, Spadafora C. Expression of LINE-1 retroposons is essential for murine preimplantation development. Mol Reprod Dev. 2006;73:279–287. doi: 10.1002/mrd.20423. [DOI] [PubMed] [Google Scholar]
- Bernareggi D, Pouyanfard S, Kaufman DS. Development of innate immune cells from human pluripotent stem cells. Exp Hematol. 2019;71:13–23. doi: 10.1016/j.exphem.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Cao ST, Yu SY, Li DW, Ye J, Yang XJ, Li C, Wang XS, Mai YB, Qin Y, Wu J, He JP, Zhou CH, Liu H, Zhao BT, Shu XD, Wu CM, Chen RP, Chan WY, Pan GJ, Chen JK, Liu J, Pei DQ. Chromatin Accessibility Dynamics during Chemical Induction of Pluripotency. Cell Stem Cell. 2018;22:529. doi: 10.1016/j.stem.2018.03.005. [DOI] [PubMed] [Google Scholar]
- Chen L, Dahlstrom JE, Chandra A, Board P, Rangasamy D. Prognostic value of LINE-1 retrotransposon expression and its subcellular localization in breast cancer. Breast Cancer Res Treat. 2012;136:129–142. doi: 10.1007/s10549-012-2246-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang WW, Jiang JM, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan YJ, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Coluccio A, Ecco G, Duc J, Offner S, Turelli P, Trono D. Individual retrotransposon integrants are differentially controlled by KZFP/KAP1-dependent histone methylation, DNA methylation and TET-mediated hydroxymethylation in naive embryonic stem cells. Epigenet Chromatin. 2018;11:1–18. [DOI] [PMC free article] [PubMed]
- De Luca C, Guadagni F, Sinibaldi-Vallebona P, Sentinelli S, Gallucci M, Hoffmann A, Schumann GG, Spadafora C, Sciamanna I. Enhanced expression of LINE-1-encoded ORF2 protein in early stages of colon and prostate transformation. Oncotarget. 2016;7:4048–4061. doi: 10.18632/oncotarget.6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA. Alu repeats and human disease. Mol Genet Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- Di Giacomo M, Comazzetto S, Saini H, De Fazio S, Carrieri C, Morgan M, Vasiliauskaite L, Benes V, Enright AJ, O'Carroll D. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell. 2013;50:601–608. doi: 10.1016/j.molcel.2013.04.026. [DOI] [PubMed] [Google Scholar]
- Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, Kuperwasser N, Motola DL, Meissner TB, Hendriks WT, Trevisan M, Gupta RM, Moisan A, Banks E, Friesen M, Schinzel RT, Xia F, Tang A, Xia Y, Figueroa E, Wann A, Ahfeldt T, Daheron L, Zhang F, Rubin LL, Peng LF, Chung RT, Musunuru K, Cowan CA. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckermann KN, Ahmed HMM, KaramiNejadRanjbar M, Dippel S, Ogaugwu CE, Kitzmann P, Isah MD, Wimmer EA. Hyperactive piggyBac transposase improves transformation efficiency in diverse insect species. Insect Biochem Mol Biol. 2018;98:16–24. doi: 10.1016/j.ibmb.2018.04.001. [DOI] [PubMed] [Google Scholar]
- Fadloun A, Le Gras S, Jost B, Ziegler-Birling C, Takahashi H, Gorab E, Carninci P, Torres-Padilla ME. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat Struct Mol Biol. 2013;20:332–338. doi: 10.1038/nsmb.2495. [DOI] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Fort A, Hashimoto K, Yamada D, Salimullah M, Keya CA, Saxena A, Bonetti A, Voineagu I, Bertin N, Kratz A, Noro Y, Wong CH, de Hoon M, Andersson R, Sandelin A, Suzuki H, Wei CL, Koseki H, Consortium F, Hasegawa Y, Forrest AR, Carninci P. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet. 2014;46:558–566. doi: 10.1038/ng.2965. [DOI] [PubMed] [Google Scholar]
- Friedli M, Turelli P, Kapopoulou A, Rauwel B, Castro-Diaz N, Rowe HM, Ecco G, Unzu C, Planet E, Lombardo A, Mangeat B, Wildhaber BE, Naldini L, Trono D. Loss of transcriptional control over endogenous retroelements during reprogramming to pluripotency. Genome Res. 2014;24:1251–1259. doi: 10.1101/gr.172809.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez JL, Marchetto MCN, Muotri AR, Coufal NG, Gage FH, O'Shea KS, Moran JV. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- Gaysinskaya V, Miller BF, De Luca C, van der Heijden GW, Hansen KD, Bortvin A. Transient reduction of DNA methylation at the onset of meiosis in male mice. Epigenet Chromatin. 2018;11:1–15. [DOI] [PMC free article] [PubMed]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goke J, Lu X, Chan YS, Ng HH, Ly LH, Sachs F, Szczerbinska I. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell. 2015;16:135–141. doi: 10.1016/j.stem.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Surani MA. Regulatory principles of Pluripotency: from the ground state up. Cell Stem Cell. 2014;15:416–430. doi: 10.1016/j.stem.2014.09.015. [DOI] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Hamdorf M, Idica A, Zisoulis DG, Gamelin L, Martin C, Sanders KJ. Pedersen IM: miR-128 represses L1 retrotransposition by binding directly to L1 RNA. Nat Struct Mol Biol. 2015;22:824–831. doi: 10.1038/nsmb.3090. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Ohnuma K, Furue MK. Pluripotent Stem Cell Heterogeneity. Adv Exp Med Biol. 2019;1123:71–94. doi: 10.1007/978-3-030-11096-3_6. [DOI] [PubMed] [Google Scholar]
- He Q, Kim H, Huang R, Lu W, Tang M, Shi F, Yang D, Zhang X, Huang J, Liu D, Songyang Z. The Daxx/Atrx complex protects tandem repetitive elements during DNA Hypomethylation by promoting H3K9 Trimethylation. Cell Stem Cell. 2015;17:273–286. doi: 10.1016/j.stem.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He SH, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Bi. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- Heras SR, Macias S, Plass M, Fernandez N, Cano D, Eyras E, Garcia-Perez JL, Caceres JF. The Microprocessor controls the activity of mammalian retrotransposons. Nat Struct Mol Biol. 2013;20:1173. doi: 10.1038/nsmb.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes SE, Singer MF, Swergold GD. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J Biol Chem. 1992;267:19765–19768. [PubMed] [Google Scholar]
- Ishida K, Xu H, Sasakawa N, Lung MSY, Kudryashev JA, Gee P, Hotta A. Site-specific randomization of the endogenous genome by a regulatable CRISPR-Cas9 piggyBac system in human cells. Sci Rep. 2018;8:310. doi: 10.1038/s41598-017-18568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachowicz JW, Bing XY, Pontabry J, Boskovic A, Rando OJ, Torres-Padilla ME. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat Genet. 2017;49:1502. doi: 10.1038/ng.3945. [DOI] [PubMed] [Google Scholar]
- Jacques Pierre-Étienne, Jeyakani Justin, Bourque Guillaume. The Majority of Primate-Specific Regulatory Sequences Are Derived from Transposable Elements. PLoS Genetics. 2013;9(5):e1003504. doi: 10.1371/journal.pgen.1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, Lorincz MC. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley David, Rinn John. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biology. 2012;13(11):R107. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kigami D, Minami N, Takayama H, Imai H. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol Reprod. 2003;68:651–654. doi: 10.1095/biolreprod.102.007906. [DOI] [PubMed] [Google Scholar]
- Kim H, Kim D, Jang MJ, Han YM. Variations in the epigenetic regulation of lineage-specific genes among human pluripotent stem cell lines. Biochem Biophys Res Commun. 2012;424:331–337. doi: 10.1016/j.bbrc.2012.06.122. [DOI] [PubMed] [Google Scholar]
- Klawitter S, Fuchs NV, Upton KR, Munoz-Lopez M, Shukla R, Wang JC, Garcia-Canadas M, Lopez-Ruiz C, Gerhardt DJ, Sebe A, Grabundzija I, Merkert S, Gerdes P, Pulgarin JA, Bock A, Held U, Witthuhn A, Haase A, Sarkadi B, Lower J, Wolvetang EJ, Martin U, Ivics Z, Izsvak Z, Garcia-Perez JL, Faulkner GJ, Schumann GG. Reprogramming triggers endogenous L1 and Alu retrotransposition in human induced pluripotent stem cells. Nat Commun. 2016;7:1–15. [DOI] [PMC free article] [PubMed]
- Kuff EL, Lueders KK. The intracisternal A-particle gene family: structure and functional aspects. Adv Cancer Res. 1988;51:183–276. doi: 10.1016/s0065-230x(08)60223-7. [DOI] [PubMed] [Google Scholar]
- Lander ES, Consortium IHGS, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang HM, Yu J, Wang J, Huang GY, Gu J, Hood L, Rowen L, Madan A, Qin SZ, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan HQ, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JGR, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang WH, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz JR, Slater G, Smit AFA, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, Conso IHGS: Initial sequencing and analysis of the human genome. Nature 2001;409: 860–921. [DOI] [PubMed]
- Li ZG, Dai HZ, Martos SN, Xu BS, Gao Y, Li T, Zhu GJ, Schones DE, Wang ZB. Distinct roles of DNMT1-dependent and DNMT1-independent methylation patterns in the genome of mouse embryonic stem cells. Genome Biol. 2015;16. [DOI] [PMC free article] [PubMed]
- Liu NA, Lee CH, Swigut T, Grow E, Gu B, Bassik MC, Wysocka J. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature. 2018;553:228. doi: 10.1038/nature25179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, Manos PD, Datta S, Lander ES, Schlaeger TM, Daley GQ, Rinn JL. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower R, Lower J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci U S A. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XY, Sachs F, Ramsay L, Jacques PE, Goke J, Bourque G, Ng HH. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol. 2014;21:423–U168. doi: 10.1038/nsmb.2799. [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mameli G, Astone V, Arru G, Marconi S, Lovato L, Serra C, Sotgiu S, Bonetti B, Dolei A. Brains and peripheral blood mononuclear cells of multiple sclerosis (MS) patients hyperexpress MS-associated retrovirus/HERV-W endogenous retrovirus, but not human herpesvirus 6. J Gen Virol. 2007;88:264–274. doi: 10.1099/vir.0.81890-0. [DOI] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–U149. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- Mc CB. Mutable loci in maize. Carnegie Inst Wash Year Book. 1948;47:155–169. [Google Scholar]
- Mc CB. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci U S A. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora C, Serzanti M, Consiglio A, Memo M, Dell'Era P. Clinical potentials of human pluripotent stem cells. Cell Biol Toxicol. 2017;33:351–360. doi: 10.1007/s10565-017-9384-y. [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Mulas Carla, Chia Gloryn, Jones Kenneth Alan, Hodgson Andrew Christopher, Stirparo Giuliano Giuseppe, Nichols Jennifer. Oct4 regulates the embryonic axis and coordinates exit from pluripotency and germ layer specification in the mouse embryo. Development. 2018;145(12):dev159103. doi: 10.1242/dev.159103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulhardt C, Fischer M, Gass P, Simon-Chazottes D, Guenet JL, Kuhse J, Betz H, Becker CM. The spastic mouse: aberrant splicing of glycine receptor beta subunit mRNA caused by intronic insertion of L1 element. Neuron. 1994;13:1003–1015. doi: 10.1016/0896-6273(94)90265-8. [DOI] [PubMed] [Google Scholar]
- Narva E, Rahkonen N, Emani MR, Lund R, Pursiheimo JP, Nasti J, Autio R, Rasool O, Denessiouk K, Lahdesmaki H, Rao A, Lahesmaa R. RNA-binding protein L1TD1 interacts with LIN28 via RNA and is required for human embryonic stem cell self-renewal and Cancer cell proliferation. Stem Cells. 2012;30:452–460. doi: 10.1002/stem.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PN, Hooley P, Roden D, Davari Ejtehadi H, Rylance P, Warren P, Martin J, Murray PG. Molecular immunology research G: human endogenous retroviruses: transposable elements with potential? Clin Exp Immunol. 2004;138:1–9. doi: 10.1111/j.1365-2249.2004.02592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y, Totoki Y, Toyoda A, Watanabe T, Yamamoto Y, Tokunaga K, Sakaki Y, Sasaki H, Hohjoh H. Small RNA class transition from siRNA/piRNA to miRNA during pre-implantation mouse development. Nucleic Acids Res. 2010;38:5141–5151. doi: 10.1093/nar/gkq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Percharde M, Lin CJ, Yin YF, Guan J, Peixoto GA, Bulut-Karslioglu A, Biechele S, Huang B, Shen XH, Ramalho-Santos M. A LINE1-Nucleolin Partnership Regulates Early Development and ESC Identity. Cell. 2018;174:391. doi: 10.1016/j.cell.2018.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodic N, Sharma R, Sharma R, Zampella J, Dai L, Taylor MS, Hruban RH, Iacobuzio-Donahue CA, Maitra A, Torbenson MS, Goggins M, Shih Ie M, Duffield AS, Montgomery EA, Gabrielson E, Netto GJ, Lotan TL, De Marzo AM, Westra W, Binder ZA, Orr BA, Gallia GL, Eberhart CG, Boeke JD, Harris CR, Burns KH. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am J Pathol. 2014;184:1280–1286. doi: 10.1016/j.ajpath.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougier N, Bourc'his D, Gomes DM, Niveleau A, Plachot M, Paldi A, Viegas-Pequignot E. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12:2108–2113. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, Spitz F, Constam DB, Trono D. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Russell SJ, Stalker L, Gilchrist G, Backx A, Molledo G, Foster RA, LaMarre J. Identification of PIWIL1 isoforms and their expression in bovine testes, oocytes, and early embryos. Biol Reprod. 2016;94:1–11. [DOI] [PubMed]
- Sakaki Y, Hattori M, Fujita A, Yoshioka K, Kuhara S, Takenaka O. The LINE-1 family of primates may encode a reverse transcriptase-like protein. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):465–469. doi: 10.1101/sqb.1986.051.01.056. [DOI] [PubMed] [Google Scholar]
- Santoni FA, Guerra J, Luban J. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology. 2012;9:1–15. [DOI] [PMC free article] [PubMed]
- Schertzer MD, Thulson E, Braceros KCA, Lee DM, Hinkle ER, Murphy RM, Kim SO, Vitucci ECM, Calabrese JM. A piggyBac-based toolkit for inducible genome editing in mammalian cells. RNA. 2019;25:1047–1058. doi: 10.1261/rna.068932.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei FS, Pasternak S, Liang CZ, Zhang JW, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du FY KK, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen WZ, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M, Kohlberg S, Sgro J, Delgado B, Mead K, Chinwalla A, Leonard S, Crouse K, Collura K, Kudrna D, Currie J, He RF, Angelova A, Rajasekar S, Mueller T, Lomeli R, Scara G, Ko A, Delaney K, Wissotski M, Lopez G, Campos D, Braidotti M, Ashley E, Golser W, Kim H, Lee S, Lin JK, Dujmic Z, Kim W, Talag J, Zuccolo A, Fan C, Sebastian A, Kramer M, Spiegel L, Nascimento L, Zutavern T, Miller B, Ambroise C, Muller S, Spooner W, Narechania A, Ren LY, Wei S, Kumari S, Faga B, Levy MJ, McMahan L, Van Buren P, Vaughn MW, Ying K, Yeh CT, Emrich SJ, Jia Y, Kalyanaraman A, Hsia AP, Barbazuk WB, Baucom RS, Brutnell TP, Carpita NC, Chaparro C, Chia JM, Deragon JM, Estill JC, Fu Y, Jeddeloh JA, Han YJ, Lee H, Li PH, Lisch DR, Liu SZ, Liu ZJ, Nagel DH, McCann MC, SanMiguel P, Myers AM, Nettleton D, Nguyen J, Penning BW, Ponnala L, Schneider KL, Schwartz DC, Sharma A, Soderlund C, Springer NM, Sun Q, Wang H, Waterman M, Westerman R, Wolfgruber TK, Yang LX, Yu Y, Zhang LF, Zhou SG, Zhu Q, Bennetzen JL, Dawe RK, Jiang JM, Jiang N, Presting GG, Wessler SR, Aluru S, Martienssen RA, Clifton SW, McCombie WR, Wing RA, Wilson RK. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu HC, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–U374. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer Y, Ronen D, Bock C, Boyle P, Meissner A, Benvenisty N. Identification of novel imprinted differentially methylated regions by global analysis of human-parthenogenetic-induced pluripotent stem cells. Stem Cell Reports. 2013;1:79–89. doi: 10.1016/j.stemcr.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Anger M, Bernstein E, Hannon GJ, Schultz RM. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev Biol. 2004;269:276–285. doi: 10.1016/j.ydbio.2004.01.028. [DOI] [PubMed] [Google Scholar]
- Takahara T, Ohsumi T, Kuromitsu J, Shibata K, Sasaki N, Okazaki Y, Shibata H, Sato S, Yoshiki A, Kusakabe M, Muramatsu M, Ueki M, Okuda K, Hayashizaki Y. Dysfunction of the Orleans reeler gene arising from exon skipping due to transposition of a full-length copy of an active L1 sequence into the skipped exon. Hum Mol Genet. 1996;5:989–993. doi: 10.1093/hmg/5.7.989. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohonen V, Katayama S, Vesterlund L, Jouhilahti EM, Sheikhi M, Madissoon E, Filippini-Cattaneo G, Jaconi M, Johnsson A, Burglin TR, Linnarsson S, Hovatta O, Kere J. Novel PRD-like homeodomain transcription factors and retrotransposon elements in early human development. Nat Commun. 2015;6:8207. doi: 10.1038/ncomms9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubio JM, Li Y, Ju YS, Martincorena I, Cooke SL, Tojo M. L1 Retrotransposition transduces nonrepetitive DNA throughout Cancer genomes. Cancer Discov. 2014;4:1115. [Google Scholar]
- Vagin VV, Sigova A, Li CJ, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- van den Hurk JAJM, Meij IC, Seleme MDC, Kano H, Nikopoulos K, Hoefsloot LH, Sistermans EA, de Wijs IJ, Mukhopadhyay A, Plomp AS, de Jong PTVM, Kazazian HH, Cremers FPM. L1 retrotransposition can occur early in human embryonic development. Hum Mol Genet. 2007;16:1587–1592. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- Vitullo P, Sciamanna I, Baiocchi M, Sinibaldi-Vallebona P, Spadafora C. LINE-1 retrotransposon copies are amplified during murine early embryo development. Mol Reprod Dev. 2012;79:118–127. doi: 10.1002/mrd.22003. [DOI] [PubMed] [Google Scholar]
- Wang JC, Xie GC, Singh M, Ghanbarian AT, Rasko T, Szvetnik A, Cai HQ, Besser D, Prigione A, Fuchs NV, Schumann GG, Chen W, Lorincz MC, Ivics Z, Hurst LD, Izsvak Z. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014;516:405. doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Wissing S, Munoz-Lopez M, Macia A, Yang ZY, Montano M, Collins W, Garcia-Perez JL, Moran JV, Greene WC. Reprogramming somatic cells into iPS cells activates LINE-1 retroelement mobility. Hum Mol Genet. 2012;21:208–218. doi: 10.1093/hmg/ddr455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JY, Huang B, Chen H, Yin QZ, Liu Y, Xiang YL, Zhang BJ, Liu BF, Wang QJ, Xia WK, Li WZ, Li YY, Ma J, Peng X, Zheng H, Ming J, Zhang WH, Zhang J, Tian G, Xu F, Chang Z, Na J, Yang XR, Xie W. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534:652. doi: 10.1038/nature18606. [DOI] [PubMed] [Google Scholar]
- Xu Q, Xie W. Epigenome in early mammalian development: inheritance, reprogramming and establishment. Trends Cell Biol. 2018;28:237–253. doi: 10.1016/j.tcb.2017.10.008. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li T, Preissl S, Amaral ML, Grinstein JD, Farah EN, Destici E, Qiu Y, Hu R, Lee AY, Chee S, Ma K, Ye Z, Zhu Q, Huang H, Fang R, Yu L, Izpisua Belmonte JC, Wu J, Evans SM, Chi NC, Ren B. Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat Genet. 2019;51:1380–1388. doi: 10.1038/s41588-019-0479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Smith AD. Subtype classification and functional annotation of L1Md retrotransposon promoters. Mobile DNA-Uk. 2019. p. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]