Abstract
BACKGROUND:
Elevated Anoctamin 7 (ANO7) expression is associated with poor survival in prostate cancer patients.
OBJECTIVE:
The aim was to discover proteins that interact with ANO7 to understand its functions and regulatory mechanisms.
METHODS:
The proximity-dependent biotin identification (BioID) method was utilized. ANO7 fused to biotin ligase was transiently transfected into LNCaP cells, and the biotinylated proteins were collected and analysed by mass spectrometry. Four identified proteins were stained with dual fluorescent immunostaining and visualized using Stimulated emission depletion microscopy (STED).
RESULTS:
After bioinformatic filtering steps, 64 potentially ANO7-interacting proteins were identified and analysed with the GO enrichment analysis tool. One of the most prominently enriched cellular components was cellular vesicle. Co-localization was showed for staphylococcal nuclease and tudor domain containing 1 (SND1), heat shock protein family A (Hsp70) member 1A (HSPA1A), adaptor related protein complex 2 subunit beta 1 (AP2B1) and coatomer protein complex subunit gamma 2 (COPG2).
CONCLUSIONS:
This is the first study in which ANO7 interacting proteins have been identified. Although further studies are needed, the findings reported here expand our understanding of the role and regulation of ANO7 in prostate cancer cells. Furthermore, these results are likely to introduce new targets for the novel cancer therapies.
Keywords: ANO7, AP2B1, BioID, COPG2, HSPA1A, interactome, prostate cancer, proteomics
1. Introduction
Prostate cancer (PrCa) is the most common cancer in men, and heritable factors account for 57% of the risk [1]. Although there are many well-characterized pathways involved in PrCa progression, new biomarkers able to predict aggressive disease outcome are needed. In our recent study, we linked the ANO7 gene to the development of aggressive PrCa [2]. Our data showed that high ANO7 expression predicts poor patient survival and that certain recurrent ANO7 single-nucleotide polymorphisms (SNPs) are associated with both prostate cancer susceptibility and the risk of aggressive PrCa [2]. Moreover, since ANO7 expression is prostate tissue-specific [3, 4], ANO7 is a compelling target for novel PrCa therapies.
Of interest, we recently showed that one of the SNPs predisposing to aggressive PrCa clearly associates with favourable response to the first-line docetaxel chemotherapy for metastatic castration-resistant PrCa (mCRPC) [5]. Together these findings strongly suggest that ANO7 possesses a functional role in PrCa progression. However, more detailed studies are needed to investigate the role of ANO7 in normal vs. cancerous prostate.
Certain Anoctamin protein family members have been associated with various cancer types [6]. For example, ANO6 has membrane scramblase function and its activity is required for cell death in epithelial cells and macrophages [7]. Additionally, expression of alternative transcript of ANO6 predicts poor survival of breast cancer patients [8]. Moreover, ANO1 is upregulated in prostate cancer and has been linked to PrCa tumorigenesis [9]. The Anoctamin 7 (ANO7) gene encodes two isoforms: a long form that consists of 25 exons (ENST00000274979.12) and a short form (ANO7S) containing the first four exons (ENST00000402530.7). The protein product of the long form (ANO7L) localizes in the plasma membrane (PM) [3, 10], endoplasmic reticulum (ER) [11] or Golgi apparatus [10]. The structure of ANO7L is unknown, but based on its primary structure, ANO7L is predicted to have eight membrane spanning regions, and its N- and C-terminal regions are predicted to reside inside the cell [12]. The ANO7L is suggested to act as a calcium-activated chloride ion channel [13, 14] or a membrane scramblase [15].
In this study, since there are no reports on the localization of ANO7S, we chose to elucidate solely the interacting partners of the ANO7L protein and determine the cellular components and processes in which ANO7L is involved. To identify ANO7-interacting proteins, we used the well-established BioID method [16, 17]. We chose this method since the strong hydrophobicity of ANO7 and its membrane integration are likely to introduce severe methodological obstacles in the use of traditional co-purification methods. The BioID method allows recognition of protein-protein interactions without a need to purify the intact protein-protein complexes [16]. In more detail, the biotin ligase (BirA) is fused to the bait-protein. The fusion protein (in our case ANO7L-BirA) introduces a biotin label to proteins residing in close (less than 10 nm) proximity of the fusion protein bait. Subsequently, all biotin-labelled proteins are collected with streptavidin-coated beads and analysed with LCI-MS/MS (liquid chromatography-tandem mass spectrometry). This method allows effective identification of proteins from complex biological mixtures. We performed the bioinformatic functional enrichment analysis for the identified proteins with Gene Ontology (GO) knowledgebase (http://geneontology.org/). The most prominent GO terms identified were related to cellular vesicles. For further validation we selected five proteins having high enrichment scores. To visualize the co-localization of selected proteins and ANO7, we performed dual immunofluorescence analysis of ANO7 and the endogenous proteins of interest. The imaging was done with super-resolution stimulated emission depletion (STED) microscope, which is able to separate signals 20 nm apart allowing visualization of protein-protein interactions.
2. Materials and methods
2.1. Plasmids
The pcDNA3.1 MCS-BirA(R118G)-HA plasmid was provided by Professor Johanna Ivaska (University of Turku, Turku, Finland) and was originally a gift from Kyle Roux (Addgene plasmid #36047; http://n2t. net/addgene:36047; RRID:Addgene_36047) (Addgene, Watertown, MA, USA). ANO7 was PCR-amplified from the pNGEP-L plasmid (a gift from Professor Karl Kunzelmann, University of Regensburg, Regensburg, Germany). This construct carries the variant allele rs7590653 (NC_000002.12:g.241223944G A), which causes the E912K amino acid change. The cloning primers used were BirA (R118G) HA_ANO7_ FRW_1: GTCTGCAGAATTCATGCGAATGGCTG and BirA (R118G)-HA_ANO7L_REV_1: GATGTTCGAATTCCTGCTGCAGCTG. The final ANO7L- BirA-HA construct and empty BirA-HA vector were transformed into MAX Efficiency DH5 competent cells (Thermo Fisher Scientific, Waltham, MA, USA), and the insert was verified by Sanger sequencing. Since LNCaP cell line express relatively low levels of endogenous ANO7 we chose to overexpress ANO7 carrying C-terminal V5-His-identification tags (also a gift from Professor Karl Kunzelmann) to carry out the co-localization analyses.
2.2. Cell lines
The LNCaP cell line endogenously expressing ANO7 mRNA [3, 18] was used in all experiments. The cell line was authenticated using PCR-single-locus-technology by Eurofins Genomics Europe Applied Genomics (Ebersberg, Germany). The cells were cultured in RPMI-1640 medium (Lonza, Basel, Switzerland) with inactivated 10% foetal bovine serum, 100 U/ml penicillin, 0.1 mg/mL streptomycin and 2 mM ultraglutamine in 5% CO at 37C.
2.3. Transient transfection and biotin administration
To isolate biotinylated proteins, two 10 cm Petri dishes were plated with 1.5 million cells in parallel. Sterile glass cover slips were included and processed for immunofluorescence and biotin detection as described below. For dual immunofluorescence detection, the cells were plated on sterile glass coverslips coated with poly-L-lysine (Sigma-Aldrich, St. Louis, MI, USA) in 6-well plates (100,000 cells/well).
One day after plating, the cells were transiently transfected using the Lipofectamine 3000 reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to Thermo Fisher’s optimized protocol for LNCaP cells. Twenty-four hours after transfection, 50 M biotin was added to the cells, which were further cultured for 24 h before analysis.
2.4. Cell lysis and purification of the proximity-labelled biotinylated proteins
Twenty-four hours after biotin treatment, the cells were rinsed twice with cold PBS, and 800 l of lysis buffer (50 mM Tris (pH 7.5), 150 mM NaCl, 0.2% SDS (w/v), 5 mM EDTA, 1% Triton X-100, 0.5% deoxycholate (w/v), 20 mM NaF, 1 mM DTT and protease inhibitors) was added to the cells. Subsequently, the cells were scraped with a rubber policeman, and the cell suspension was transferred into a 1.5 ml Eppendorf tube. The cells were rotated for 30 minutes at 4C. Five hundred microliters of 50 mM Tris-HCl (pH 7.5) was added, and the lysates were sonicated with a Sonopuls ultrasonic homogenizer (Bandelin, Berlin, Germany) for 2 minutes per sample (2 s pulses with 0.5 s breaks, 50% amplitude). The cell lysates were centrifuged at 12,000 g for 15 minutes at 4C, and the supernatant was collected into a new tube. The pelleted cellular debris was resuspended in Laemmli lysis buffer for Western blot analysis. Streptavidin beads (MyOne Streptavidin C1 Dynabeads, 10 mg/ml, Invitrogen, Carlsbad, CA, USA) were added to the supernatant (200 l/sample), and the samples were incubated with rotation overnight at 4C. The next day, the beads were washed three times with 1 ml of RT washing buffer (1% Triton X-100, 0.2% SDS (w/v), 0.2% deoxycholate (w/v), 1 mM EDTA, 150 mM NaCl and 10 mM Tris-HCL (pH 7.5)). The proteins were then eluted into 100 l of 50 mM Tris-HCl. Twenty percent of the bead volume was mixed with Laemmli buffer and prepared for Western blot analysis. The remaining beads were dried, frozen and sent to the Turku Proteomics Facility (Turku Bioscience Centre, Turku, Finland) for LC-ESI-MS/MS analysis. Five replicates of LNCaP cells at passage numbers 11, 13 and 18 after thawing were processed.
2.5. Western and streptavidin blotting
For Western blot analysis, the resuspended pellets and eluates from the protein purification step were run on 10% SDS-PAGE gels, and the proteins were transferred onto an Immuno-Blot PVDF Membrane (Bio-Rad, CA, USA). Biotinylated proteins were detected according to the methods of Roux et al. [16]. For HA tag staining, we used a rabbit anti-HA tag primary antibody (H6908, Sigma-Aldrich, St. Louis, MI, USA) in 5% BSA-TBST and a HRP-conjugated anti-rabbit IgG secondary antibody (sc-2054, Santa Cruz Biotechnology, Dallas, TX, USA) in 5% BSA-TBST.
2.6. Identification of interacting proteins by LC-ESI-MS/MS
The samples were digested on beads and dissolved in 0.1% formic acid. Then, the samples were analysed with an LC-ESI-MS/MS nanoflow HPLC system (Easy-nLC 1200, Thermo Fisher Scientific, Waltham, MA, USA) coupled to a Q Exactive mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) equipped with a nanoelectrospray ionization source. Peptides were first loaded on a trapping column and subsequently separated inline on a 15 cm C18 column (75 m 15 cm, ReproSil-Pur, 5 m, 200 Å C18-AQ, Dr. Maisch HPLC GmbH, Ammerbuch-Entringen, Germany). The mobile phases were water with 0.1% formic acid (solvent A) or acetonitrile/water (80:20 v/v) with 0.1% formic acid (solvent B). A 20 min linear gradient from 8% to 43% B was used. PAGE-MS data were acquired automatically with Thermo Xcalibur 4.0 software (Thermo Fisher Scientific, Waltham, MA, USA). An information-dependent acquisition method consisted of an Orbitrap MS survey scan with a mass range of 300–2000 m/z followed by HCD fragmentation for the 10 most intense peptide ions.
Data files were searched for protein identification using Proteome Discoverer 2.2 software (Thermo Fisher Scientific, Waltham, MA, USA) connected to an in-house server running Mascot 2.6.1 software (Matrix Science) against the SwissProt (SwissProt_2018_4) protein sequence database with the taxonomy filter ‘homo sapiens’. The following database search parameters were used: Enzyme, Trypsin; Static modifications, Carbamidomethyl (C); Variable modifications, Oxidation (M); Biotin-tyramide (Y), Acetyl (Protein N-term); Peptide Mass Tolerance, 5 ppm; Fragment Mass Tolerance, 0.02 Da; Max Missed Cleavages, 2; Instrument type, ESI-TRAP) Methionine oxidation is a common modification during sample processing and is therefore normally included in the search parameters. Data were filtered by peptide spectrum matches per protein (PSM) (minimal number of PSMs: 2). The required number of PSMs was set to two to reduce false positive identifications. Percolator was applied for statistical evaluation and rescoring of the search results.
2.7. Enrichment analyses
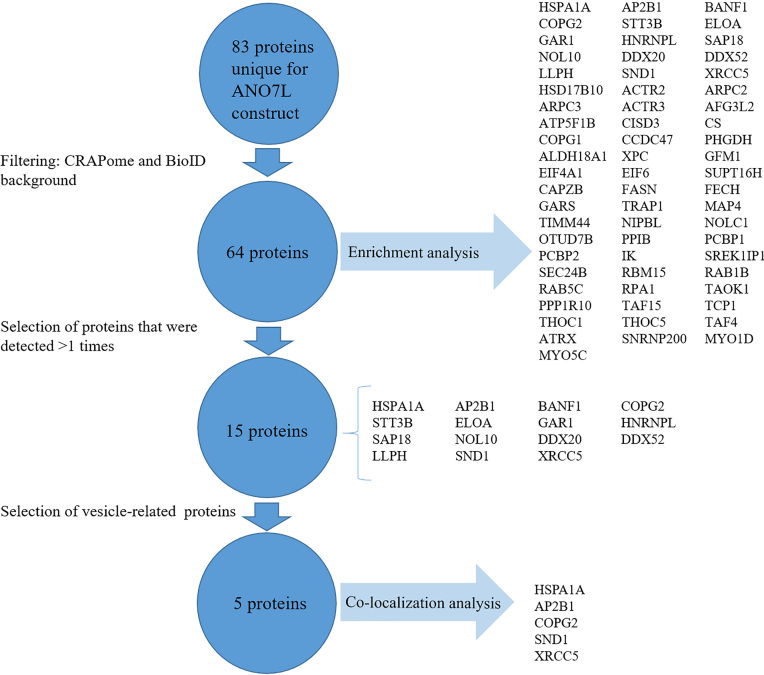
The process of LC-ESI-MS/MS result filtration is presented in Fig. 1. Eighty-three proteins uniquely identified with the ANO7 construct but not with the control construct were filtered against the list of the known BioID background proteins and CRAPome database version 1.0 (https://www.crapome.org/) to filter out contaminants. Proteins with average spectral counts 20 or those that were identified in 50% of screens in the database were excluded from the list. The remaining 64 proteins were analysed with the Gene Ontology (GO) ‘cellular component’ enrichment tool. A false discovery rate (FDR) 0.05 indicated statistical significance.
Figure 1.
Proteins selected for validation. A schematic diagram showing a brief description of how proteins were selected for the final analysis. Eighty-three proteins were unique to the ANOL construct and not found in Bir-HA controls. After filtering against the CRAPome database and a known BioID background, 64 proteins remained. Among the 15 proteins detected in at least 2 replicates, those with vesicle-related functions were selected for validation with co-localization analysis (5 proteins).
2.8. Immunofluorescence and detection of biotinylated proteins
Biotinylated proteins were detected according to the methods of Roux et al. [16] using a streptavidin-Alexa Fluor 488 conjugate (S32354, Thermo Fisher Scientific, Waltham, MA, USA). Bir*A fusion proteins were detected with anti-HA tag antibody (H6908, Sigma-Aldrich, St. Louis, MI, USA) and anti-rabbit IgG secondary antibody conjugated to Alexa Fluor 568 (A-11036, Thermo Fisher Scientific, Waltham, MA, USA). The cells were visualized with the Nikon Eclipse Ni-U upright fluorescence microscope (Nikon Instruments, Inc. Shinagawa, Tokyo, Japan).
For co-localization analyses, the cells were washed 2 times with PBS, fixed with 4% PFA-PBS and permeabilized with 0.4% Triton X-100 in PBS. After permeabilization, the cells were washed three times with PBS and incubated with primary antibodies overnight at 4C. The following primary and secondary antibodies were used: anti-COPG2 (Novus Biologicals, Colorado, USA NBP2-55178), anti-HSPA1A (ab79852), anti-AP2B1 (ab220778), anti-SND1 (Abcam ab65078, Boston, USA), anti-XRCC5 (Abcam ab80592, Boston, USA), anti-6 His tag (Abcam ab18184), anti-HA tag (Abcam ab130275), (Abcam, Boston, USA), STAR RED anti-rabbit IgG (Abberior 2-0012-011-9), and STAR 580 anti-mouse IgG (Abberior 2-0002-005-1) (Abberior Instruments GmbH, Göttingen, Germany). The coverslips were mounted with Mount Solid Antifade (Abberior Instruments GmbH, Göttingen, Germany) and dried overnight. The stained cells were visualized by STED super-resolution microscope allowing the separation of proteins at the distance of 20 nm. Co-localization and fluorescence intensity profiles were analysed with ImageJ (NIH, version 18.0).
3. Results
3.1. Validation of the expression and functionality of the ANO7-BirA-HA and BirA-HA fusion proteins
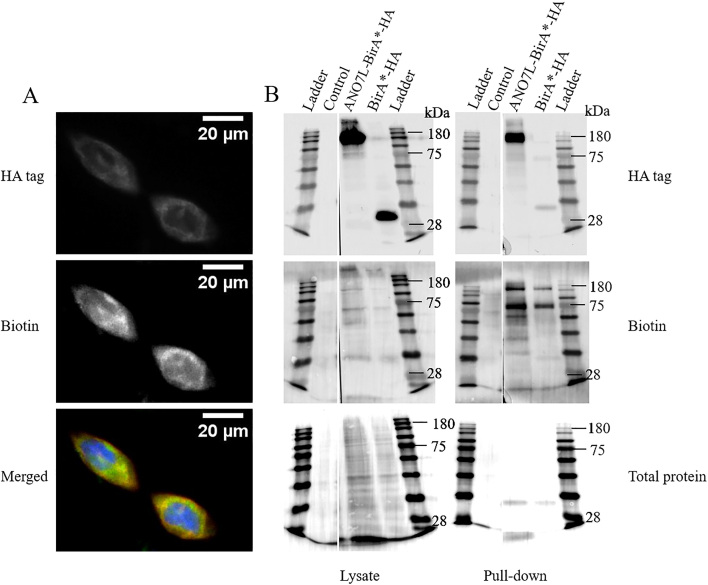
To confirm that biotin ligase was able to biotinylate proteins fused to ANO7, we performed dual staining of overexpressed ANO7L-BirA-HA and biotinylated proteins in LNCaP cells. ANO7L-BirA-HA was localized close to the membrane and biotinylated proteins detected with Alexa Fluor 488-conjugated streptavidin co-localized with ANO7L-BirA-HA (Fig. 2a). In contrast, the BirA-HA produced from the empty vector was distributed evenly inside the cells. In Western blot analyses, the ANO7L-BirA-HA protein product migrated at 150 kDa, while the calculated size of the fusion protein was 141 kDa. The difference between the expected and detected protein sizes is most likely because of post-translational glycosylation of ANO7L -BirA-HA [12]. Expression of both ANO7L-BirA-HA and BirA-HA increased the abundance of biotinylated proteins in the total lysates of transfected cells compared to those of untransfected cells demonstrating the functional activity of the cloned biotin ligase (BirA-HA).
Figure 2.
Biotinylation of proteins in ANO7L-Bir-HA-transfected LNCaP cells. A. Immunohistochemical staining of HA-tagged ANO7L-Bir (red) and biotinylated proteins with Alexa Fluor 488-tagged streptavidin (green) and nuclear staining with DAPI (blue). B. The insoluble cell lysate fraction is on the left side, and streptavidin bead pull-down samples are on the right side. The control lane contains untransfected LNCaP cells treated with biotin. Expression of the fusion proteins was detected with anti-HA, biotinylated proteins were detected with streptavidin-HRP, and total protein were detected with Ponceau S staining. The staining was visualized with Nikon Eclipse Ni-U upright fluorescence microscope (Nikon Instruments, Inc. Shinagawa, Tokyo, Japan).
3.2. Identification of interacting proteins
Mass spectrometry analysis detected 442 proteins (Supporting Information Table 1). At least two unique peptides mapped to the protein were required for identification. Eighty-three interactions were unique to ANO7L-Bir-HA, 67 interactions were observed with only BirA-HA and 292 interactions were found with both constructs. After filtering against common BioID background proteins [14] and likely contaminants listed in the CRAPome database (https://www.crapome. org/), 64 proteins that were detected as interacting uniquely with ANO7 remained. Fifteen of these proteins were detected in at least two replicates.
3.3. Vesicular and nuclear proteins are enriched among the ANO7 interacting proteins
GO cellular component analysis for the filtered 64 protein interactions showed strong enrichment in the ‘vesicles’ (fold enrichment 2.54, FDR 9.26E-06) and ‘extracellular exosomes’ (fold enrichment 3.77, FDR 2.57E-07) GO terms. According to the analysis, 6 of the 15 proteins detected in at least two replicates (HSPA1A, AP2B1, COPG2, HNRNPL, SND1, XRCC5) were associated with vesicles. HSPA1A, XRCC5 and SND1 were also associated with extracellular exosomes. Additionally, among other enriched pathways with a fold enrichment 2 and an FDR value 1E-04 were the ‘site of DNA damage’ and ‘chromosome’ (fold enrichment 28.78, FDR 1.15E-06 and fold enrichment 5.5, FDR 2.67E-07, respectively) GO terms, in which one (XRCC5) and four (BANF1, LLPH, GAR1, XRCC5) of the proteins, respectively, were detected in at least two replicates by LCI-MS/MS analysis. The GO term ‘Arp2/3 protein complex’ had a fold enrichment of 100 and FDR 1.72E-05, but none of the proteins involved were among those detected in at least two replicates. Interestingly, eleven of the proteins detected in at least two replicates were enriched in the ‘nuclear lumen’ (fold enrichment 2.37, FDR 5.51E-05) GO term. The cellular components with a fold enrichment 2 and FDR 1E-04 are listed in Table 1. The complete list of the statistically significantly enriched cellular components are presented in Supporting Information Table 2.
Table 1.
Results of the GO cellular component enrichment analysis (fold enrichment 2 and FDR 1E-04)
| GO cellular component | Fold | FDR | |
| enrichment | |||
| Protein-containing complex | 2. | 43 | 2.18E-08 |
| Membrane-enclosed lumen | 2. | 37 | 2.56E-07 |
| Extracellular exosome | 3. | 77 | 2.57E-07 |
| Chromosome | 5. | 5 | 2.67E-07 |
| Extracellular organelle | 3. | 72 | 2.67E-07 |
| Intracellular organelle lumen | 2. | 37 | 2.77E-07 |
| Extracellular vesicle | 3. | 73 | 2.82E-07 |
| Organelle lumen | 2. | 37 | 3.02E-07 |
| Site of DNA damage | 28. | 78 | 1.15E-06 |
| Intracellular non-membrane-bounded | 2. | 49 | 2.60E-06 |
| organelle | |||
| Non-membrane-bounded organelle | 2. | 48 | 2.60E-06 |
| Vesicle | 2. | 54 | 9.26E-06 |
| Nuclear chromosome, telomeric region | 19. | 19 | 1.14E-05 |
| Arp2/3 protein complex | 100 | 1.72E-05 | |
| Chromosomal part | 4. | 91 | 2.74E-05 |
| Nucleoplasm | 2. | 53 | 4.86E-05 |
| Nucleoplasm part | 4. | 35 | 4.88E-05 |
| Chromosome, telomeric region | 14. | 59 | 5.39E-05 |
| Nuclear lumen | 2. | 37 | 5.51E-05 |
FDR false discovery rate.
3.4. Co-localization analysis of ANO7 and the interacting proteins
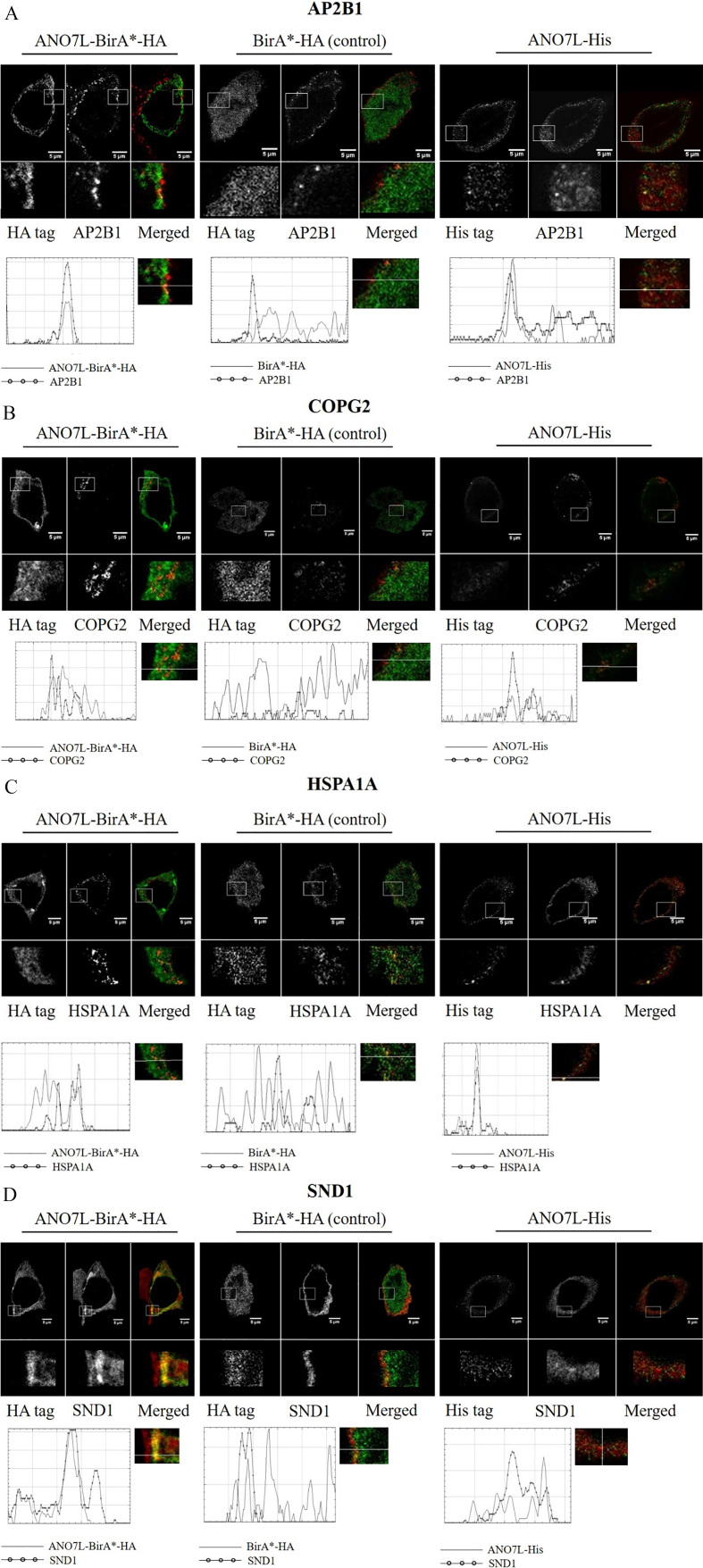
The putative interactions between ANO7 and the identified binding partners were visualized with dual immunofluorescence and STED microscopy. ANO7 was detected with anti-HA or anti-His following transfection of LNCaP cells with ANO7L-BirA-HA or ANO7 with C-terminal His-tag, while the putative endogenous ANO7 interacting proteins were detected with their respective polyclonal antibodies. ANO7L-BirA-HA was used to verify the results obtained by LCI-MS/MS analysis, while ANO7L-V5-His was used to verify that the observed interactions were not due to the proteins interacting with the biotin ligase. ANO7L-BirA-HA and His-tagged ANO7 both localised to plasma membrane in bright spotted structures (Fig. 3). No nuclear staining was observed.
Figure 3.
Co-localization of ANO7 and the identified interaction partners. Double staining of ANO7L-BirA-HA, BirA-HA or His-tagged ANO7 shown in green and the interaction partners (AP2B1/COPG2/HSPA1A/SND1) shown in red. Co-localization was visualized with the fluorescence intensity profiles below the microscope figures. The intensity profile allows visualisation of the intensity of red and green staining along the line added into the immunofluorescence figure. In the microscope figures, yellow spots indicate co-localization, which are also indicated by overlapping peaks in the signal intensity curves. A. AP2B1. B. COPG2. C. HSPA1A. D. SND1.
Since ANO7 is predicted to be localized in membrane structures, we chose to validate the five putative binding partners that, according to GO enrichment analysis, are associated with vesicle structures and were uniquely detected with ANO7-BirA-HA in at least two replicate experiments: HSPA1A (heat shock protein family A [Hsp70] member 1A), SND1 (staphylococcal nuclease and tudor domain containing 1), AP2B1 (Adaptor Related Protein Complex 2 Subunit Beta 1), COPG2 (Coatomer subunit gamma-2) and XRCC5 (X-Ray Repair Cross-Complementing Protein 5).
Both AP2B1 and HSPA1A formed bright spots at the plasma membrane, and their co-localization with ANO7 was partial. COPG2 staining showed granular structures in the cytosol, likely in the Golgi apparatus, and COPG2 was partly co-localized with ANO7. SND1 once again co-localized to the cell surface with ANO7, but in a more diffuse manner than ANO7 (Fig. 3). We did not detect XRCC5 in the cytoplasm with the antibody used in this work; thus, XRCC5 did not co-localize with ANO7.
The localization of all the ANO7-interacting proteins was similar between ANO7L-BirA-HA and BirA-HA constructs, indicating that the overexpression of ANO7 does not affect the localization of these proteins.
4. Discussion
Based on our previous data, the ANO7 gene is a promising new biomarker for PrCa since changes in its gene expression predicts disease aggressiveness [2]. We have also reported two ANO7 SNPs which are linked to the aggressive progression of PrCa [2]. These findings prompted us to study ANO7-interacting proteins to obtain insights into the regulatory milieu of ANO7 in the prostate and its functions in the development of PrCa.
In this study, we used a proximity-dependent labelling technique (BioID) for identifying the ANO7 protein interactions. The BioID method recognizes not only the strong direct interactions but also weak and transient interactions or parts of protein complexes that are closer than 10 nm to the studied fusion protein [19].
Whereas the enrichment of the ANO7 interacting proteins in Arp2/3 complex and nuclear proteins is a novel finding, the connection of Anoctamins with vesicles and exosomes has been described earlier [20, 21]. Interestingly, Anoctamin family member ANO6 regulates the formation of a subset of extracellular vesicles (EVs) via phospholipid scrambling [22]. Furthermore, it has been shown that the chemical induction of giant plasma membrane vesicles (GPMVs) is dependent on ANO6 [23]. ANO7 has also been shown to act as a scramblase [15], and could thus play a similar role in vesicle formation as ANO6. ANO7 peptides have been detected in patient-derived prostasomes, i.e., prostate-specific extracellular vesicles (EVs) [24, 25], which further indicates a role of ANO7 in vesicle maturation and/or trafficking. In normal prostate tissue, the luminal cells of the prostatic ducts secrete prostasomes into the seminal plasma, where they play a role in sperm cell motility and immunosuppression. During cancer progression, the luminal cells lose their polarization, and consequently, the prostasomes are secreted into stromal tissue, where they can support tumour growth [26, 27]. Interestingly, Corcoran et al. have showed that docetaxel chemotherapy resistance between cells spreads via exosomes [28]. Accordingly, reduced exosome maturation in ANO7 mutant PrCa cells would provide a mechanistic explanation to our previous finding indicating that a possibly pathogenic SNP in ANO7 is associated with favourable docetaxel response in mCRPC patients [5].
Similarly to some other Anoctamin family members, ANO7 was demonstrated to interact with proteins having known functions in vesicle formation. This indicates that the methodology used was able to identify biologically meaningful protein interactions.
Of the identified protein interactions, COPG2 and AP2B1 participate in intracellular vesicle trafficking, whereas HSPA1A [29, 30, 31], SND1 [29, 30] and XRCC5 [30, 32] have been identified in extracellular vesicles. Four of the ANO7-interacting proteins, COPG2, AP2B1, HSPA1A and SND1, were shown to co-localize with ANO7, suggesting their functional interaction. Although XRCC5 was only detected in the nucleus under these conditions, earlier studies have indicated that XRCC5 can also locate in the cytoplasm and to play a role in cell adhesion and invasion [33].
COPG2 is a subunit of the COPI coatomer complex that participates in retrograde transport of intracellular vesicles from the Golgi to the ER by coating the vesicles. Remarkably, COPB1 directly interacts with ANO1 and has been shown to downregulate ANO1 expression on cell surfaces [34]. ANO7 could also be regulated by the COPI coatomer complex. AP2B1 together with three other subunits form the AP2 complex, which mediates endocytosis and regulates receptor trafficking [35]. In addition, AP2B1 has been detected in prostasomes [24].
HSPA1A is a multifunctional chaperone. Interestingly, in prostate cancer, HSPA1A interacts with the androgen receptor (AR) and regulates its activation [36]. HSPA1A is localized in both the cytosol and the membrane, and membrane-associated HSPA1A expression has been linked to cancer aggressiveness and resistance to cancer therapy [37, 38]. According to the work of Dubois et al., HSPA1A is present in the lipid rafts of prostasomes [24].
SND1 was first identified as a transcription coactivator [39] but has also been shown to have several functions related to RNA metabolism [40]. The expression of SND1 is increased in many cancers, including PrCa. Similar to HSPA1A, SND1 is associated with development of resistance to cancer therapies [41, 42].
This is the first study in which ANO7 interactions have been investigated. ANO7 has been linked to PrCa, and understanding its role in the development of the disease may provide new prognostic as well as therapeutic targets. ANO7 is expressed almost exclusively in prostate tissue [3, 4], making it a promising target for PrCa therapy. The ANO7 protein has indicated to be immunogenic, and ANO7-targeted T cells have been shown to specifically kill PrCa cells [43]. More studies are warranted to reveal the wider functions of ANO7.
Supplementary Material
Acknowledgments
Mass spectrometry analysis was performed at the Turku Proteomics Facility, University of Turku and Åbo Akademi University. The facility is supported by Biocenter Finland. This study was financially supported by Worldwide Cancer Research (#14-0089), the Sigrid Juselius Foundation, the Academy of Finland (#310105), State Research Funding of Turku University Hospital, Cancer Foundation Finland sr, and Jane & Aatos Erkko Foundation.
Conflict of interest
The authors declare no potential conflicts of interest.
References
- [1]. Mucci L.A., Hjelmborg J.B., Harris J.R., Czene K., Havelick D.J., Scheike T., Graff R.E., Holst K., Möller S., Unger R.H., McIntosh C., Nuttall E., Brandt I., Penney K.L., Hartman M., Kraft P., Parmigiani G., Christensen K., Koskenvuo M., Holm N.V., Heikkilä K., Pukkala E., Skytthe A., Adami H.O., Kaprio J. and Collaboration N.T.S.o.C.N., Familial risk and heritability of cancer among twins in nordic countries, JAMA 315 (2016), 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Kaikkonen E., Rantapero T., Zhang Q., Taimen P., Laitinen V., Kallajoki M., Jambulingam D., Ettala O., Knaapila J., Boström P.J., Wahlström G., Sipeky C., Pursiheimo J.P., Tammela T., Kellokumpu-Lehtinen P.L., Fey V., Maehle L., Wiklund F., Wei G.H., Schleutker J. and Consortium P., ANO7 is associated with aggressive prostate cancer, Int J Cancer 143 (2018), 2479–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Bera T.K., Das S., Maeda H., Beers R., Wolfgang C.D., Kumar V., Hahn Y., Lee B. and Pastan I., NGEP, a gene encoding a membrane protein detected only in prostate cancer and normal prostate, Proc Natl Acad Sci U S A 101 (2004), 3059–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. Kiessling A., Weigle B., Fuessel S., Ebner R., Meye A., Rieger M.A., Schmitz M., Temme A., Bachmann M., Wirth M.P. and Rieber E.P., D-TMPP: A novel androgen-regulated gene preferentially expressed in prostate and prostate cancer that is the first characterized member of an eukaryotic gene family, Prostate 64 (2005), 387–400. [DOI] [PubMed] [Google Scholar]
- [5]. Kaikkonen E., Ettala O., Nikulainen I., Taimen P., Lehtinen I., Boström P.J., Kellokumpu-Lehtinen P.L. and Schleutker J., rs77559646 is associated with first-line docetaxel treatment response in metastatic castration-resistant prostate cancer, Anticancer Res 39 (2019), 5353–5359. [DOI] [PubMed] [Google Scholar]
- [6]. Wanitchakool P., Wolf L., Koehl G.E., Sirianant L., Schreiber R., Kulkarni S., Duvvuri U. and Kunzelmann K., Role of anoctamins in cancer and apoptosis, Philos Trans R Soc Lond B Biol Sci 369 (2014), 20130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Ousingsawat J., Schreiber R. and Kunzelmann K., TMEM16F/anoctamin 6 in ferroptotic cell death, Cancers (Basel) 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Dutertre M., Lacroix-Triki M., Driouch K., de la Grange P., Gratadou L., Beck S., Millevoi S., Tazi J., Lidereau R., Vagner S. and Auboeuf D., Exon-based clustering of murine breast tumor transcriptomes reveals alternative exons whose expression is associated with metastasis, Cancer Res 70 (2010), 896–905. [DOI] [PubMed] [Google Scholar]
- [9]. Liu W., Lu M., Liu B., Huang Y. and Wang K., Inhibition of Ca(2+)-activated Cl(-) channel ANO1/TMEM16A expression suppresses tumor growth and invasiveness in human prostate carcinoma, Cancer Lett 326 (2012), 41–51. [DOI] [PubMed] [Google Scholar]
- [10]. Das S., Hahn Y., Nagata S., Willingham M.C., Bera T.K., Lee B. and Pastan I., NGEP, a prostate-specific plasma membrane protein that promotes the association of LNCaP cells, Cancer Res 67 (2007), 1594–1601. [DOI] [PubMed] [Google Scholar]
- [11]. Duran C., Qu Z., Osunkoya A.O., Cui Y. and Hartzell H.C., ANOs 3-7 in the anoctamin/tmem16 Cl-channel family are intracellular proteins, Am J Physiol Cell Physiol 302 (2012), C482–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Das S., Hahn Y., Walker D.A., Nagata S., Willingham M.C., Peehl D.M., Bera T.K., Lee B. and Pastan I., Topology of NGEP, a prostate-specific cell: Cell junction protein widely expressed in many cancers of different grade level, Cancer Res 68 (2008), 6306–6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13]. Schreiber R., Uliyakina I., Kongsuphol P., Warth R., Mirza M., Martins J.R. and Kunzelmann K., Expression and function of epithelial anoctamins, J Biol Chem 285 (2010), 7838–7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Tian Y., Schreiber R. and Kunzelmann K., Anoctamins are a family of Ca2+-activated Cl- channels, J Cell Sci 125 (2012), 4991–4998. [DOI] [PubMed] [Google Scholar]
- [15]. Suzuki J., Fujii T., Imao T., Ishihara K., Kuba H. and Nagata S., Calcium-dependent phospholipid scramblase activity of TMEM16 protein family members, J Biol Chem 288 (2013), 13305–13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Roux K.J., Kim D.I. and Burke B., BioID: A screen for protein-protein interactions, Curr Protoc Protein Sci 74 (2013), Unit 19.23. [DOI] [PubMed] [Google Scholar]
- [17]. Varnaitė R. and MacNeill S.A., Meet the neighbors: Mapping local protein interactomes by proximity-dependent labeling with BioID, Proteomics 16 (2016), 2503–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18]. Mohsenzadegan M., Tajik N., Madjd Z., Shekarabi M. and Farajollahi M.M., Study of NGEP expression in androgen sensitive prostate cancer cells: A potential target for immunotherapy, Med J Islam Repub Iran 29 (2015), 159. [PMC free article] [PubMed] [Google Scholar]
- [19]. Kim D.I., Birendra K.C., Zhu W., Motamedchaboki K., Doye V. and Roux K.J., Probing nuclear pore complex architecture with proximity-dependent biotinylation, Proc Natl Acad Sci U S A 111 (2014), E2453–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Kunzelmann K., Ousingsawat J., Benedetto R., Cabrita I. and Schreiber R., Contribution of anoctamins to cell survival and cell death, Cancers (Basel) 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Whitlock J.M. and Hartzell H.C., Anoctamins/TMEM16 proteins: Chloride channels flirting with lipids and extracellular vesicles, Annu Rev Physiol 79 (2017), 119–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22]. Fujii T., Sakata A., Nishimura S., Eto K. and Nagata S., TMEM16F is required for phosphatidylserine exposure and microparticle release in activated mouse platelets, Proc Natl Acad Sci U S A 112 (2015), 12800–12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Han T.W., Ye W., Bethel N.P., Zubia M., Kim A., Li K.H., Burlingame A.L., Grabe M., Jan Y.N. and Jan L.Y., Chemically induced vesiculation as a platform for studying TMEM16F activity, Proc Natl Acad Sci U S A 116 (2019), 1309–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Dubois L., Ronquist K.K., Ek B., Ronquist G. and Larsson A., Proteomic profiling of detergent resistant membranes (lipid rafts) of prostasomes, Mol Cell Proteomics 14 (2015), 3015–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Poliakov A., Spilman M., Dokland T., Amling C.L. and Mobley J.A., Structural heterogeneity and protein composition of exosome-like vesicles (prostasomes) in human semen, Prostate 69 (2009), 159–167. [DOI] [PubMed] [Google Scholar]
- [26]. Lundholm M., Schröder M., Nagaeva O., Baranov V., Widmark A., Mincheva-Nilsson L. and Wikström P., Prostate tumor-derived exosomes down-regulate NKG2D expression on natural killer cells and CD8+ T cells: Mechanism of immune evasion, PLoS One 9 (2014), e108925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Ronquist K.G., Ronquist G., Larsson A. and Carlsson L., Proteomic analysis of prostate cancer metastasis-derived prostasomes, Anticancer Res 30 (2010), 285–290. [PubMed] [Google Scholar]
- [28]. Corcoran C., Rani S., O’Brien K., O’Neill A., Prencipe M., Sheikh R., Webb G., McDermott R., Watson W., Crown J. and O’Driscoll L., Docetaxel-resistance in prostate cancer: Evaluating associated phenotypic changes and potential for resistance transfer via exosomes, PLoS One 7 (2012), e50999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29]. Bijnsdorp I.V., Geldof A.A., Lavaei M., Piersma S.R., van Moorselaar R.J. and Jimenez C.R., Exosomal ITGA3 interferes with non-cancerous prostate cell functions and is increased in urine exosomes of metastatic prostate cancer patients, J Extracell Vesicles 2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Soekmadji C., Riches J.D., Russell P.J., Ruelcke J.E., McPherson S., Wang C., Hovens C.M., Corcoran N.M., Hill M.M., Nelson C.C. and BioResource A.P.C.C., Modulation of paracrine signaling by CD9 positive small extracellular vesicles mediates cellular growth of androgen deprived prostate cancer, Oncotarget 8 (2017), 52237–52255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Hosseini-Beheshti E., Pham S., Adomat H., Li N. and Tomlinson Guns E.S., Exosomes as biomarker enriched microvesicles: Characterization of exosomal proteins derived from a panel of prostate cell lines with distinct AR phenotypes, Mol Cell Proteomics 11 (2012), 863–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32]. Duijvesz D., Burnum-Johnson K.E., Gritsenko M.A., Hoogland A.M., Vredenbregt-van den Berg M.S., Willemsen R., Luider T., Paša-Tolić L. and Jenster G., Proteomic profiling of exosomes leads to the identification of novel biomarkers for prostate cancer, PLoS One 8 (2013), e82589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33]. Muller C., Paupert J., Monferran S. and Salles B., The double life of the Ku protein: Facing the DNA breaks and the extracellular environment, Cell Cycle 4 (2005), 438–441. [DOI] [PubMed] [Google Scholar]
- [34]. Lee Y.S., Bae Y., Park N., Yoo J.C., Cho C.H., Ryoo K., Hwang E.M. and Park J.Y., Surface expression of the Anoctamin-1 (ANO1) channel is suppressed by protein-protein interactions with β-COP, Biochem Biophys Res Commun 475 (2016), 216–222. [DOI] [PubMed] [Google Scholar]
- [35]. Collins B.M., McCoy A.J., Kent H.M., Evans P.R. and Owen D.J., Molecular architecture and functional model of the endocytic AP2 complex, Cell 109 (2002), 523–535. [DOI] [PubMed] [Google Scholar]
- [36]. Dong J., Wu Z., Wang D., Pascal L.E., Nelson J.B., Wipf P. and Wang Z., Hsp70 binds to the androgen receptor N-terminal domain and modulates the receptor function in prostate cancer cells, Mol Cancer Ther 18 (2019), 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37]. Multhoff G. and Hightower L.E., Distinguishing integral and receptor-bound heat shock protein 70 (Hsp70) on the cell surface by Hsp70-specific antibodies, Cell Stress Chaperones 16 (2011), 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38]. Shevtsov M., Huile G. and Multhoff G., Membrane heat shock protein 70: A theranostic target for cancer therapy, Philos Trans R Soc Lond B Biol Sci 373 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39]. Tong X., Drapkin R., Yalamanchili R., Mosialos G. and Kieff E., The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE, Mol Cell Biol 15 (1995), 4735–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40]. Ochoa B., Chico Y. and Martínez M.J., Insights into SND1 oncogene promoter regulation, Front Oncol 8 (2018), 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Blanco M.A., Alečkoviéc M., Hua Y., Li T., Wei Y., Xu Z., Cristea I.M. and Kang Y., Identification of staphylococcal nuclease domain-containing 1 (SND1) as a Metadherin-interacting protein with metastasis-promoting functions, J Biol Chem 286 (2011), 19982–19992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42]. Fu X., Zhang C., Meng H., Zhang K., Shi L., Cao C., Wang Y., Su C., Xin L., Ren Y., Zhang W., Sun X., Ge L., Silvennoinen O., Yao Z., Yang X. and Yang J., Oncoprotein Tudor-SN is a key determinant providing survival advantage under DNA damaging stress, Cell Death Differ 25 (2018), 1625–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43]. Cereda V., Poole D.J., Palena C., Das S., Bera T.K., Remondo C., Gulley J.L., Arlen P.M., Yokokawa J., Pastan I., Schlom J. and Tsang K.Y., New gene expressed in prostate: A potential target for T cell-mediated prostate cancer immunotherapy, Cancer Immunol Immunother 59 (2010), 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.