Abstract
Objective
There is a need for early detection of colorectal cancer (CRC) at precancerous-stage adenoma. Here, we identified novel faecal bacterial markers for diagnosing adenoma.
Design
This study included 1012 subjects (274 CRC, 353 adenoma and 385 controls) from two independent Asian groups. Candidate markers were identified by metagenomics and validated by targeted quantitative PCR.
Results
Metagenomic analysis identified ‘m3’ from a Lachnoclostridium sp., Fusobacterium nucleatum (Fn) and Clostridium hathewayi (Ch) to be significantly enriched in adenoma. Faecal m3 and Fn were significantly increased from normal to adenoma to CRC (p<0.0001, linear trend by one-way ANOVA) in group I (n=698), which was further confirmed in group II (n=313; p<0.0001). Faecal m3 may perform better than Fn in distinguishing adenoma from controls (areas under the receiver operating characteristic curve (AUROCs) m3=0.675 vs Fn=0.620, p=0.09), while Fn performed better in diagnosing CRC (AUROCs Fn=0.862 vs m3=0.741, p<0.0001). At 78.5% specificity, m3 and Fn showed sensitivities of 48.3% and 33.8% for adenoma, and 62.1% and 77.8% for CRC, respectively. In a subgroup tested with faecal immunochemical test (FIT; n=642), m3 performed better than FIT in detecting adenoma (sensitivities for non-advanced and advanced adenomas of 44.2% and 50.8% by m3 (specificity=79.6%) vs 0% and 16.1% by FIT (specificity=98.5%)). Combining with FIT improved sensitivity of m3 for advanced adenoma to 56.8%. The combination of m3 with Fn, Ch, Bacteroides clarus and FIT performed best for diagnosing CRC (specificity=81.2% and sensitivity=93.8%).
Conclusion
This study identifies a novel bacterial marker m3 for the non-invasive diagnosis of colorectal adenoma.
Keywords: colonic bacteria, colorectal adenomas, colorectal cancer screening
Significance of this study.
What is already known on this subject?
Early detection of colonic adenomas and cancer can facilitate the successful treatment and significantly reduces the incidence of colorectal cancer (CRC).
Molecular markers for adenoma, especially non-advanced adenoma, is limited.
What are the new findings?
A new gene marker from a Lachnoclostrium sp., labelled as m3, was identified to be enriched in faecal samples of patients with adenoma by metagenomic analysis.
m3 showed the best performance in diagnosing adenoma in two independent Asian groups of 1012 subjects by quantitative PCR, which is superior to currently available stool-based tests.
Combination of m3 with faecal immunochemical test (FIT) improved diagnostic sensitivity from 50.8% to 56.8% (specificity 79.6%) for advanced adenoma, while combination of m3 with other bacterial markers (Fn, Ch, Bc) and FIT showed good diagnostic performance for CRC (specificity=81.2% and sensitivity=93.8%).
How might it impact on clinical practice in the foreseeable future?
m3 is a novel stool-based non-invasive biomarker for patients with adenoma and CRC.
Introduction
Colorectal cancer (CRC) is one of the most common malignancies worldwide.1 A higher incidence of CRC has been observed in more developed regions than less developed regions, and an increased incidence of CRC is believed to have attributed to changes in diet.1 2 Recent evidences have shown that an altered microbiome environment in the gut is associated with colorectal tumourigenesis. Abnormality in the composition of the gut microbiota has been implicated as a potentially important aetiological factor in the initiation and progression of CRC.3 With the widespread application of metagenomic analyses in the investigation of intestinal microbiota, an increasing number of bacteria have been identified to be positively associated with CRC.4–7 Recent basic research has established a critical function for the intestinal microbiota8 and specific bacterial species, such as Fusobacterium nucleatum (Fn)9–11 and Peptostreptococcus anaerobius,12 in promoting colorectal tumourigenesis. Bacteria such as Fn,13 Clostridium symbiosum 14 and species within the genera Parvimonas, Porphyromonas and Parabacteroides 15 have been shown to be potential markers for the diagnosis of patients with CRC. However, current knowledge on biomarkers for colorectal adenoma detection is limited.
Early detection of cancer can facilitate successful treatment. Endoscopic removal of colorectal adenomas, precursors of most CRCs, significantly reduces the risk of CRC. Early detection of adenomas is thus important for decreasing CRC morbidity and mortality. The most widely used non-invasive stool test is the faecal immunochemical test (FIT), which shows unsatisfying sensitivities for CRC (0.79 (95% CI 0.69 to 0.86); differed greatly among various studies) and is not sensitive for adenoma.16 Sensitivity of FIT for advanced adenoma varied from 6% to 56%, with screening studies involving cohort sizes over 8000 all showing sensitivities of less than 28%.17 Therefore, identification of molecular markers that improve the diagnostic sensitivity for adenoma is warranted.
Using metagenomic analysis to compare the faecal microbiome of patients with CRC and healthy subjects, we identified 20 bacterial gene marker candidates that may serve as non-invasive biomarkers for CRC.4 We further showed that stool-based bacteria could serve as non-invasive diagnostic biomarkers for CRC by targeted quantification using quantitative PCR (qPCR).13 We showed that Fn was a good marker for CRC, and combination with three others (Clostridium hathewayi (Ch), undefined ‘m7’ and Bacteroides clarus (Bc)) could further improve the diagnostic performance of Fn. However, the diagnostic performance of these bacterial gene markers for adenoma was limited. In this study, we identified and evaluated the utility of a new Lachnoclostridium gene marker (labelled as ‘m3’) for the diagnosis of colorectal adenoma. The diagnostic performance of m3, comparing with and in combination with other bacterial gene markers and FIT, was tested in 1012 subjects from two independent groups.
Results
Identification of ‘m3’ from a Lachnoclostridium species as a potential biomarker for colorectal neoplasm
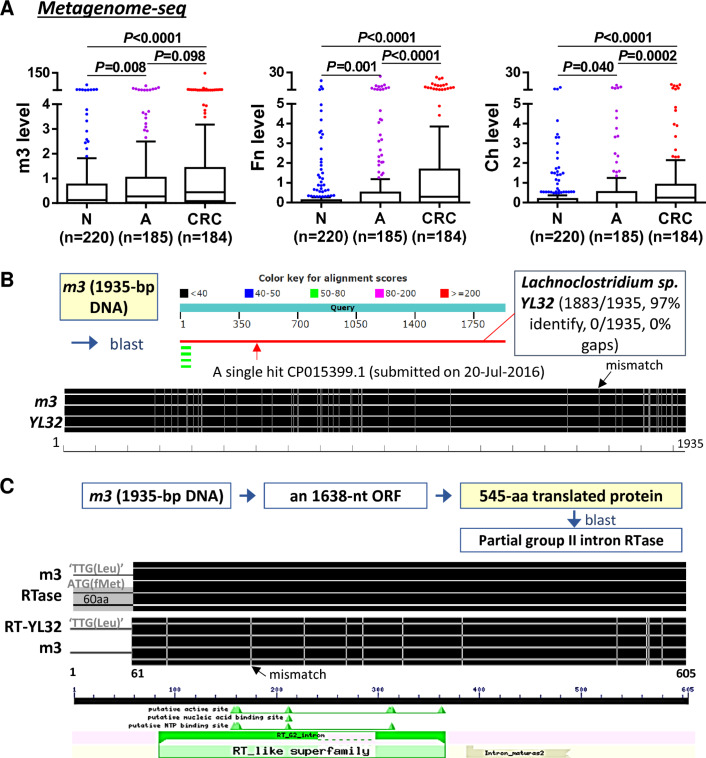
To investigate whether our previously identified 20 bacterial gene markers for CRC4 may also serve as biomarkers for adenoma, we analysed their abundances in our in-house metagenomics data from 589 Asian subjects (184 CRC, 185 adenoma and 220 control subjects) (online supplementary table S1). Among them, the marker labelled as ‘m3’, which was not assignable to any known species at the time of the previous discovery study,4 was found to be significantly enriched in patients with CRC and adenoma as compared with control subjects, as well as two previously verified CRC markers (Fn and Ch) (figure 1A; other gene markers in online supplementary figure 1). A blast search for the 1935 nt m3 sequence in the non-redundant nucleotide collection of NCBI identified Lachnoclostridium sp. YL32, a new species with genome sequence deposited in GenBank in July 2016 (accession no. CP015399). m3 and Lachnoclostridium sp. YL32 shared 97% (1883/1935) DNA sequence similarity (figure 1B). The m3 DNA contains a 1638 nt open reading frame (nt 298–1935), encoding a putative 545 aa protein with 100% sequence similarity to a group II RTase (GenBank accession no. WP_055650193). Although m3 protein lacks the first 60 aa due to ‘TTG’ codon instead of ‘ATG’ at the corresponding translation start site, it retains the RTase conserved domain (figure 1C). The corresponding sequence in Lachnoclostridium sp. YL32 genome also encodes a partial group II RTase, showing 98% (534/545) sequence similarity with m3-RTase and the group II RTase. We further analysed the abundance of Lachnoclostridium sp. YL32 genome based on the Prokka-annotated protein coding gene sequences in our in-house metagenomics data. The result showed that Lachnoclostridium sp. YL32 was significantly increased in adenoma but to a less extent in CRC as compared with control subjects (online supplementary figure 2A). Therefore, the candidate gene marker m3 may belong to Lachnoclostridium species close to Lachnoclostridium sp. YL32.
Figure 1.
Identification and characterisation of m3. (A) Metagenome sequencing identified m3, as well as Fusobacterium nucleatum (Fn) and Clostridium hathewayi (Ch), to be significantly increased in faecal samples of patients with adenoma. (B) DNA sequence of m3 showed high similarity to Lachnoclostridium sp. YL32. (C) m3 encodes a putative reverse transcriptase (RTase) that maps to a group II intron RTase, lacking the first 60 amino acids but retaining the RTase conserved domain. A, adenoma; CRC, colorectal cancer; N, normal control.
gutjnl-2019-318532supp001.pdf (1.1MB, pdf)
gutjnl-2019-318532supp002.pdf (592.7KB, pdf)
Validation of m3 as a novel faecal biomarker for colorectal adenoma by qPCR
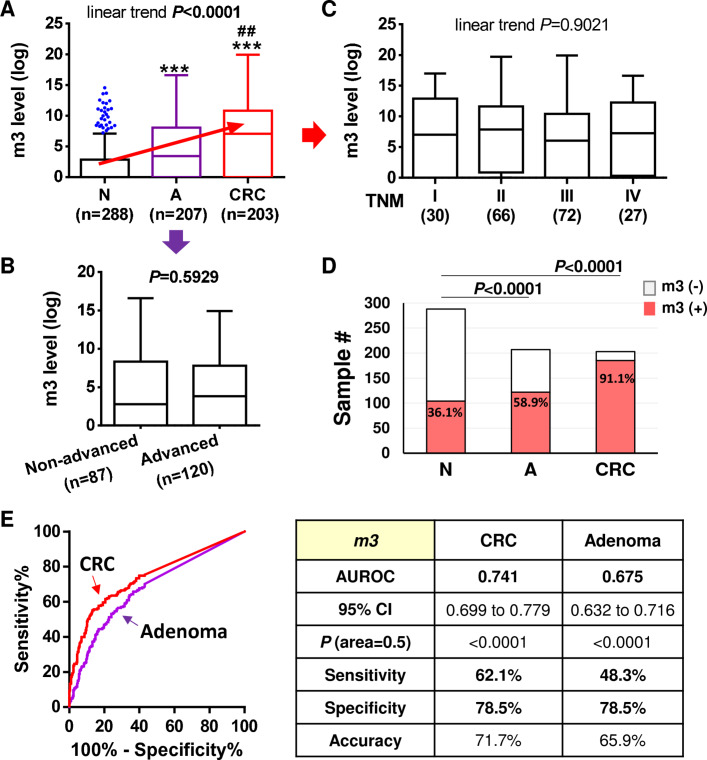
We further quantitatively examined the abundance of m3 in an enlarged group of stool samples from Hong Kong Chinese by using our previously established duplex-qPCR platform.13 The results showed that faecal m3 level was significantly higher in patients with adenoma (n=207) versus control subjects (n=288), and was significantly higher in patients with CRC (n=203) versus control subjects or patients with adenoma (all p<0.0001 by multiple comparison). There was a significant linear trend of m3 increasing from control to adenoma to cancer (p<0.0001, one-way ANOVA) (figure 2A). Interestingly, m3 level was similar between non-advanced and advanced adenomas (figure 2B). Similarly, m3 level showed no significant change across tumour-node-metastasis staging in patients with CRC (figure 2C). The occurrence rate of faecal m3 was significantly higher in patients with adenoma as compared with control subjects, and highest in patients with CRC (both p<0.0001 vs control subjects; figure 2D). Receiver operating characteristic (ROC) curve analysis showed that m3 could significantly discriminate CRC and adenoma patients from control subjects, with areas under ROC (AUROCs) of 0.741 and 0.675 for CRC and adenoma, respectively (both p<0.0001; figure 2E). At specificity of 78.5%, m3 showed sensitivities of 62.1% for CRC and 48.3% for adenoma; the accuracies were 71.7% and 65.9% for distinguishing patients with CRC and adenoma from control subjects, respectively. These results demonstrated that m3 may serve as a new stool-based biomarker to assist the non-invasive diagnosis of CRC and adenoma.
Figure 2.
Quantitative detection of faecal m3 in the diagnosis of patients with colorectal cancer (CRC) and adenoma. (A) Relative abundance of m3 in faecal samples differed significantly between healthy control subjects (N, n=288), patients with adenoma (A, n=207) and patients with CRC (n=203). ***p<0.0001 as compared with N; ##p<0.001 as compared with A. (B) No significant difference in faecal abundance of m3 was observed between non-advanced and advanced adenomas. (C) No difference in faecal abundance of m3 was observed among patients with CRC of different tumour-node-metastasis (TNM) stages. (D) Occurrence rates of m3 was significantly higher in patients with adenoma compared with control subjects, and highest in patients with CRC. (E) Receiver operating characteristic (ROC) curves and diagnostic performance of m3 in discriminating patients with CRC and adenoma from control subjects, respectively. AUROC, area under ROC.
m3 performs better than other bacterial markers (Fn and Ch) in diagnosing colorectal adenoma
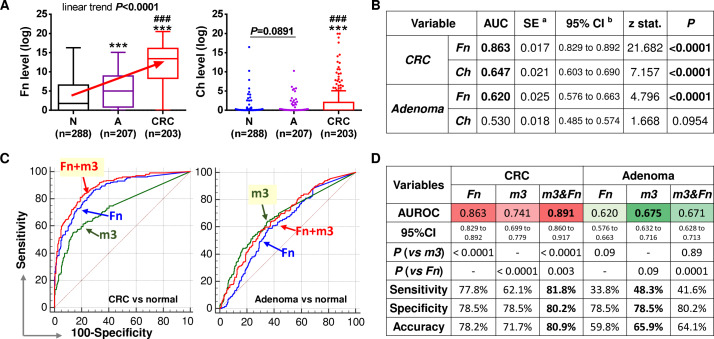
As Fn and Ch were also identified to be significantly increased in patients with adenoma by metagenome sequencing, we further examined the levels of Fn and Ch by qPCR and compared their diagnostic performances with m3. Results confirmed that, similar to m3, the relative faecal abundance of Fn was significantly elevated in patients with adenoma compared with control subjects, and highest in patients with CRC, with a significant linear trend of increase during CRC development (all p<0.0001; figure 3A). However, faecal Ch was not significantly increased in patients with adenoma as compared with control subjects by qPCR (p>0.05). Ch was significantly enriched in patients with CRC compared with patients with adenoma and control subjects (both p<0.0001, figure 3A). ROC curve analyses also showed that both Fn and Ch performed well in diagnosing CRC, but only Fn could significantly distinguish patients with adenoma from control subjects (p<0.0001; figure 3B). The abundances of both Fn and m3 were not associated with gender, CRC staging, lesion location or body mass index. Multivariate analysis showed that Fn and m3 were significantly associated with CRC and adenoma diagnosis, as well as Fn with age (table 1). Although Fn performed better than m3 in CRC diagnosis (AUROC of Fn=0.863 vs m3=0.741; p<0.0001), m3 may work better than Fn for adenoma diagnosis as shown by comparison of ROC curves (AUROCs: m3=0.675 vs Fn=0.620, p=0.09; figure 3C, D). As single diagnostic factors at specificity of 78.5%, Fn discriminated patients with CRC from control subjects with a sensitivity of 77.8% and accuracy of 78.2% (vs 62.1% and 71.7%, respectively, by m3). At specificity of 78.5%, m3 discriminated patients with adenoma from control subjects with a sensitivity of 48.3% and accuracy of 65.9% (vs 33.8% and 59.8%, respectively, by Fn) (figure 3C, D). Interestingly, combination of Fn and m3 by a logistic regression model significantly improved their individual diagnostic performances for CRC (AUROC=0.891, p=0.003 vs Fn) but not for adenoma (AUROC=0.671, p=0.89 vs m3). These results demonstrate that m3 is a potential good diagnostic biomarker especially for adenoma.
Figure 3.
Comparison and combination of bacterial markers for non-invasive diagnosis of colorectal cancer (CRC) and adenoma. (A) Relative abundances of Fusobacterium nucleatum (Fn), Clostridium hathewayi (Ch) in faecal samples of control subjects, patients with adenoma and patients with CRC. N, normal control; A, adenoma; ***p<0.0001 as compared with N; ###p<0.0001 as compared with A. (B) ROC curve analyses showed Fn could discriminate adenoma and CRC from controls, while Ch could discriminate CRC but not adenoma from controls. (C) Comparison of ROC curves of Fn, m3 and their combination. (D) Diagnostic performances of Fn, m3 and their combination. Fn performed better than m3 in diagnosing CRC, and m3 was superior to Fn in diagnosing adenoma. Combination with Fn improved the diagnostic performance of m3 for CRC but not for adenoma.
Table 1.
Correlations between faecal marker abundances and clinical characteristics
| Univariate | Multivariate | |||||||
| Markers | Fn | m3 | Fn | m3 | ||||
| Variable | Coef | P value | Coef | P value | Coef | P value | Coef | P value |
| Age | 0.217 | <0.0001 | 0.111 | <0.0001 | 0.064 | 0.007 | 0.025 | 0.269 |
| Gender | −0.348 | 0.443 | −0.143 | 0.706 | ||||
| Diagnosis | 2.768 | <0.0001 | 1.535 | <0.0001 | 2.597 | <0.0001 | 1.472 | <0.0001 |
| Pre/post-colonoscopy | −0.181 | 0.729 | −0.735 | 0.095 | ||||
| BMI | 0.052 | 0.761 | −0.069 | 0.633 | ||||
| CRC staging | 0.173 | 0.689 | −0.227 | 0.620 | ||||
| Lesion location | −0.153 | 0.862 | −0.201 | 0.830 | ||||
BMI, body mass index; CRC, colorectal cancer.
Combination with other bacterial markers (Fn, Bc and Ch) increases the diagnostic performance of m3 for CRC, while m3 alone works best for adenoma
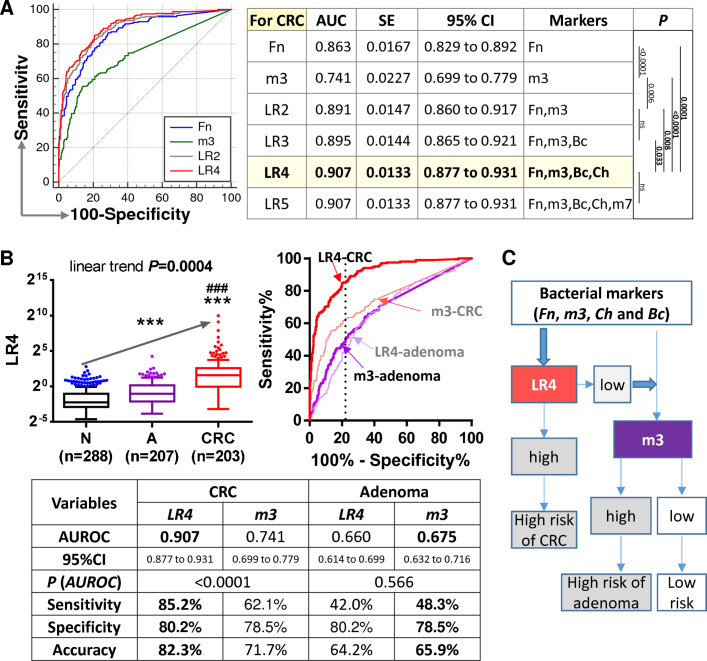
As we have previously reported bacterial markers Fn, Ch, Bc and m7 for diagnosis of CRC, we further tested the performance of the combination of these markers with m3 for CRC. The results showed that combination of Fn, m3, Ch and Bc by a logistic regression model performed best in diagnosing CRC, with an AUROC of 0.907 (all p<0.05 as compared with combinations of fewer markers by comparison of ROC curves; figure 4A). At specificity of 80.2%, combination of Fn, m3, Ch and Bc showed a sensitivity of 85.2% and accuracy of 82.3% for CRC. Although combination of Fn, m3, Ch and Bc showed significantly increased score in patients with adenoma as compared with control subjects, its diagnostic performance for adenoma was not better than m3 (AUROC=0.660 vs m3=0.675, p=0.086; figure 4B). Therefore, combination of Fn, m3, Ch and Bc may serve as a novel tool for the non-invasive diagnosis of CRC, while m3 alone may be applied for further detection of adenoma (figure 4C).
Figure 4.
Combination of four markers for olorectal cancer (CRC) and m3 alone for adenoma. (A) Receiver operating characteristic (ROC) curve analysis of combination of the five bacterial markers of interest showed that combination of Fn, m3, Ch and Bc by a logistic regression (LR) model worked best for CRC diagnosis. Shown p values are by comparison ROC curves. (B) Level of the combination of Fn, m3, Ch and Bc (LR4) in faecal samples and comparison of its diagnostic performance with m3. N, normal control; A, adenoma; ***p<0.0001 as compared with N; ###p<0.0001 as compared with A. (C) Proposed strategy for the application of Fn, m3, Ch and Bc in the diagnosis of CRC and adenoma.
Verification of diagnostic performance of bacterial markers in a second independent group
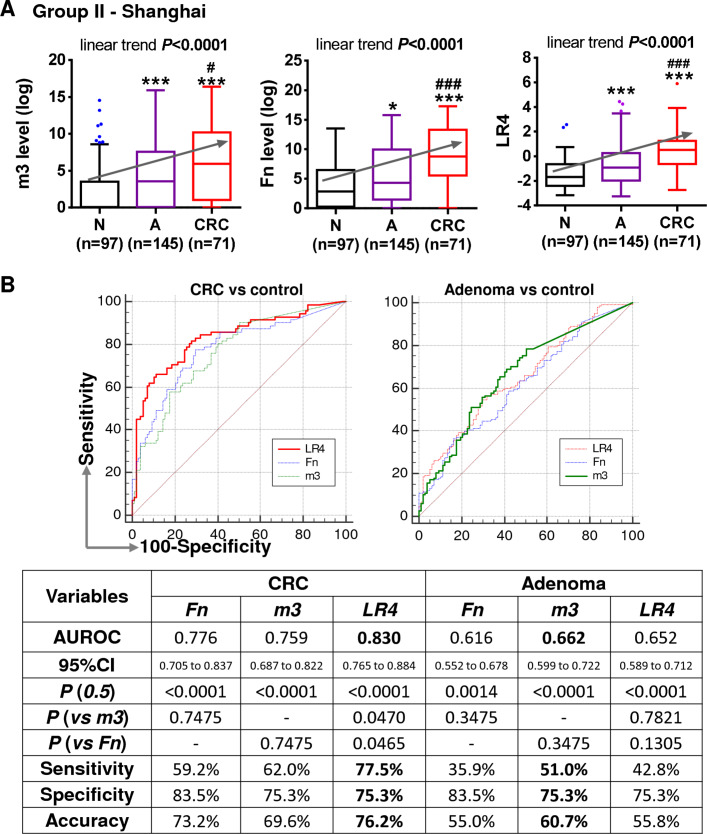
We further tested the bacterial markers in a second independent group of 313 samples from Shanghai China (71 CRC, 145 adenoma and 97 controls). Results showed that both Fn and m3 were significantly increased in patients with adenoma compared with control subjects, and further increased in patients with cancer, with a significant linear trend of increase from control to adenoma to cancer by multiple comparison (p<0.0001; figure 5A). Using the same logistic regression model established in the Hong Kong group, the combined score of the four markers (Fn, m3, Ch and Bc) also showed a significant increase during the normal-adenoma-CRC sequence (p<0.0001). Fn performed better than m3 in discriminating CRC from controls although not significantly (AUROCs for CRC: Fn=0.776 vs m3=0.759), and the four-marker combination showed the best performance in diagnosing CRC (AUROC=0.830, sensitivity=77.5% and specificity=75.3%; figure 5B). m3 performed better than Fn and the four-marker combination in distinguishing patients with adenoma from controls although not significantly (AUROCs: m3=0.662, Fn=0.616 and four-marker=0.652), and m3 showed a sensitivity of 51.0% and specificity of 75.3% for adenoma (figure 5B). These results further confirm the diagnostic values of the four bacterial markers for CRC and m3 for adenoma.
Figure 5.
Validation of bacterial markers in diagnosing colorectal cancer (CRC) and adenoma in a second independent group of faecal samples. (A) Relative faecal abundances of Fn and m3 and level of the combination of Fn, m3, Ch and Bc (LR4) in patients with CRC and adenoma compared with control subjects of the second group. N, normal control; A, adenoma; *p<0.05 and ***p<0.0001 as compared with N; #p<0.05 and ##p<0.001 as compared with A. (B) Comparison of ROC curves and diagnostic performances of Fn, m3 and LR4.
Combination with FIT improves the diagnostic ability of bacterial markers for CRC
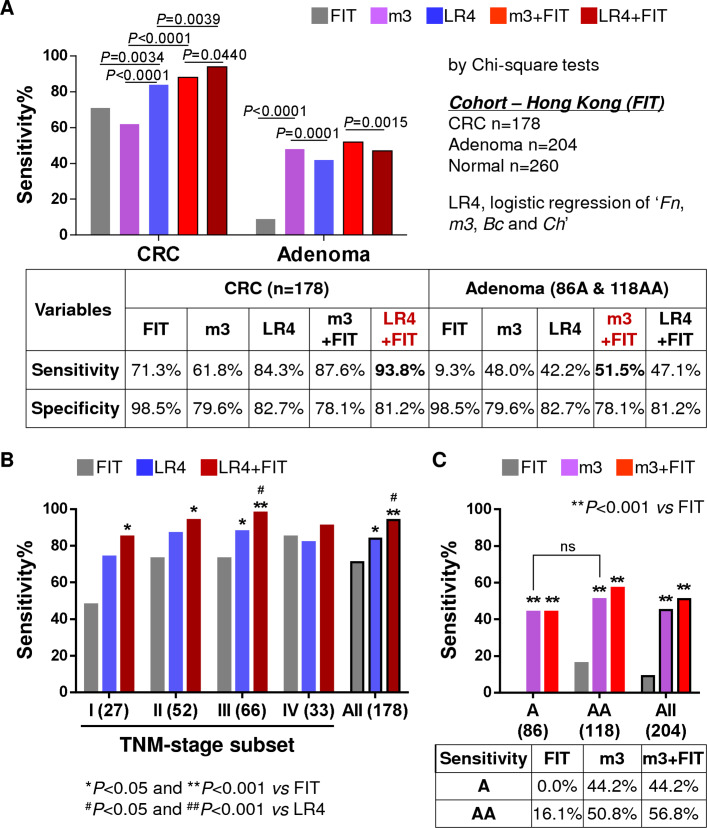
To compare the diagnostic performance of bacterial markers with the most widely used non-invasive stool test in CRC screening, FIT was performed in a subgroup of 642 samples from Hong Kong (178 CRC, 204 adenoma and 260 controls). The CRC detection rate by combination of Fn, m3, Ch and Bc (84.7%) was significantly higher than m3 alone (61.8%) or FIT (71.3%) (both p<0.01). The combination of FIT with four-marker showed the best performance in detecting CRC, with a sensitivity of 93.8% and specificity of 81.2% (figure 6A). The four-marker showed higher sensitivities than FIT for stages I–III cancers but not late stage IV. Combination of the four-marker and FIT showed significantly increased sensitivities than FIT for stages I–III cancers, and also elevated the detection rate for stage IV cancers (figure 6B). These results demonstrate that the bacterial marker panel is superior to FIT for detection of stages I–III CRC, and their combination further improves the non-invasive diagnosis of CRC.
Figure 6.
Comparison and combination of bacterial markers with faecal immunochemical test (FIT). (A) Comparison of sensitivity and specificity of FIT, m3, combination of four makers (Fn, m3, Ch and Bc; LR4) and combination of bacterial markers with FIT in a subgroup of Hong Kong samples. LR4 combined with FIT performed best for colorectal cancer (CRC) detection, while m3 combined with FIT performed best for detecting adenoma. (B) Comparison of the sensitivities of FIT, LR4 and their combination in detecting CRC according to tumour-node-metastasis (TNM) stage subsets. (C) Comparison of the sensitivities of FIT, m3 and their combination in detecting non-advanced and advanced adenomas. All comparison of sensitivities was conducted by χ2 tests. A, non-advanced adenoma; AA, advanced adenoma.
m3 performs significantly better than FIT in diagnosing adenoma
In the subgroup of 204 adenoma (86 non-advanced and 118 advanced) cases with FIT results, m3 alone (sensitivity=48.0%) showed a significantly higher detection rate than FIT (9.3%) and the four-marker (42.2%) (both p<0.001). Combination of m3 with FIT showed the best diagnostic performance for adenoma, with a sensitivity of 51.5% and specificity of 78.1% (figure 6A). FIT failed to detect any non-advanced adenoma (0/86, 0%) and detected 16.1% (19/118) of the advanced adenoma. On the other hand, m3 showed no difference in the detection between non-advanced and advanced adenomas (figures 2B and 6C), and the detection rates of m3 were significantly higher for both non-advanced (44.2%) and advanced (50.8%) adenomas than FIT (both p<0.001; figure 6C). Combination with FIT increased the sensitivity of m3 for advanced adenoma to 56.8%. These results demonstrate that m3 alone shows good performance for stool-based detection of adenoma.
Discussion
In this study, we screened a previously identified panel of CRC-associated gene markers in patients with CRC or adenoma compared with control subjects by metagenomics analysis. Focusing on the candidate gene markers that were significantly changed in patients with adenoma as compared with control subjects, we further validated their application values in non-invasive diagnosis of adenoma and CRC by qPCR. We demonstrated a faecal bacterial marker m3 that is useful for adenoma detection and at the mean time devised a new panel of faecal bacterial markers (m3+Fn+Ch+Bc) to achieve improved diagnostic capacity for CRC as compared with markers reported in our previous studies.4 13
m3 is a well-assembled DNA sequence (1935 bp) from shotgun sequencing data. As m3 DNA is long enough and distinct from DNA polymerase genes of other micro-organisms, with a single specific hit to Lachnoclostridium sp. YL32 with high score (97% identify), it is reliable to conclude that the host bacterium of ‘m3’ belong to the bacterial genus Lachnoclostridium. We further analysed the abundances of known Lachnoclostridium genomes in our in-house metagenomics data, with 160 strains from GenBank and 27 species from ChocoPhlAn pangenome database (online supplementary figure 2A, B). We found stepwise increase from control to adenoma to cancer in some species, such as Clostridium (C.) aldenense, C. bolteae, C. citroniae and C. clostridioforme (online supplementary figure 2C), demonstrating the potential of Lachnoclostridium species in discriminating patients with colorectal neoplasm from control subjects. Lachnoclostridium is a newly defined genus under the highly polyphyletic class Clostridia,18 with an increasing number of new species identified from human gut microbiota in recent few years, such as Lachnoclostridium (L.) edouardi,19 L. pacaense 20 and L. touaregense.21 The Lachnoclostridium species carrying m3 and its roles in colorectal tumourigenesis warrant further characterisation in future studies.
We have reported that combination of Fn, Bc, Ch and the undefined ‘m7’ showed good diagnostic performance for CRC.13 Comparison of ROC curves showed that combination of ‘Fn, Bc, Ch and m3’ (AUROC=0.907 (0.877 to 0.931)) showed an increased AUROC than the combination of ‘Fn, Bc, Ch and m7’ (AUROC=0.892 (0.856 to 0.921)) in our Hong Kong group and in Shanghai group (AUROCs: 0.830 (0.765 to 0.884) with m3 vs 0.795 (0.705 to 0.867) with m7). These results suggest the impact of m3 on improving other bacterial markers for the non-invasive diagnosis of CRC.
Fn is prevalently detected in human CRC, with important roles in the initiation and progression of CRC. Fn level in colonic adenoma and adenocarcinoma tissues was found to be >10–100 times higher than normal colonic mucosa,10 demonstrating Fn accumulation may occur at an early stage of colonic tumourigenesis. However, there is disagreement about the relationship between Fn and colorectal adenoma.22 Fn was found to be enriched in cancerous versus matched normal tissues, but not significantly higher in adenoma versus normal tissues in a European cohort.23 Similarly, faecal abundance of Fn was found to be strongly associated with CRC but not adenoma in a German cohort.24 Although we observed a significant increase of faecal Fn in patients with adenoma compared with control subjects, the diagnostic value of Fn for adenoma is not as good as m3, and combination with Fn could not improve the diagnostic performance of m3 for adenoma.
We have showed that the gram-negative bacterium Bc 25 was significantly decreased in patients with CRC as compared with healthy subjects and thus could help improve diagnostic specificity. The gram-positive bacterium Ch, which participates in glucose metabolism using carbohydrates as fermentable substrates to produce acetate, ethanol, carbon dioxide and hydrogen,26 was significantly increased in patients with CRC compared with healthy subjects. However, faecal abundances of Bc and Ch showed no differences between patients with adenoma and control subjects.
On the other hand, m3 is superior to other bacterial markers in discriminating patients with adenoma from control subjects according to our results from two independent Chinese groups, although its diagnostic capacity for CRC is not as good as Fn. FIT only detected 16.1% advanced adenoma and none of non-advanced adenoma. The multitarget stool DNA test approved by the US Food and Drug Administration, which combines mutant and methylated DNA markers and a FIT, shows sensitivities of 42.4% for advanced adenoma and 17.2% for non-advanced adenoma.27Although the sensitivity of m3 (48.3%) is still low for adenoma, m3 showed no significant difference in the detection between advanced and non-advanced adenomas, with sensitivities of 50.8% and 44.2%, respectively. Therefore, m3 may outperform all other available stool-based tests in detecting non-advanced adenoma. Moreover, combination with FIT improved the detection rate of m3 for advanced adenoma from 50.8% to 56.8%.
Some of the faecal samples were collected after colonoscopy, with 40.6%, 36.2% and 40.4% in control, adenoma and cancer groups, respectively (p=0.577). However, these post-colonoscopy samples were collected at least 1 month after colonoscopy when gut microbiome should have recovered to baseline.28 Furthermore, we have adjusted for confounding effects of sample collection before/after colonoscopy during marker discovery.4 There was no significant difference in m3 level between pre-colonoscopy and post-colonoscopy adenoma samples by qPCR or metagenome sequencing. There were also no difference in Fn, m3 or the four-marker combination between pre-colonoscopy and post-colonoscopy samples of the control, adenoma or CRC groups (online supplementary figure 3). Therefore, the markers involved in this study may not be affected by colonoscopic/bowel-prep status, given enough time for gut microbiome to recover after colonoscopy. Although age and gender differed significantly among the groups, inclusion of age and gender in the logistic regression model did not affect the ROC curves for CRC and adenoma significantly (online supplementary figure 4).
Recent studies of CRC have identified a large number of faecal microbial markers, and attempts to combine such markers from shotgun metagenomics data showed good diagnostic performance.6 29 30 Our recent meta-analysis of multicohort metagenomics data, covering 526 samples from Chinese, Austrian, American, and German and French cohorts, identified seven CRC-enriched bacterial species showing an AUROC of 0.8 in discriminating patients with CRC from control subjects, which was increased to 0.88 when the clinical data were added.6 Application of direct shotgun metagenomics to diagnosis is not cost-efficient due to cumbersome experimental procedure and heavy computing workload. Targeted detection of identified microbial marker candidates based on shotgun metagenomics for clinical application is a more promising strategy. Based on our bacterial gene markers identified by metagenomics investigation, quantification of four bacterial gene markers by qPCR shows an AUROC of 0.907 for CRC diagnosis in this study. However, as the true performance of the markers cannot be established from these case–control samples, future validation is required in large sample cohorts representative of the CRC screening populations. We have also reported for the first time that faecal CRC-enriched virome and mycobiome biomarkers distinguished CRC from controls with AUROCs of 0.802 and 0.93, respectively.29 30 The application of these viral and fungal markers to non-invasive diagnosis of CRC by targeted quantification needs further exploration.
In conclusion, we identified a novel bacterial marker m3, from a Lachnoclostridium species, for the non-invasive diagnosis of colorectal adenoma. m3 is superior to other bacterial markers and currently available stool-based tests for adenoma detection.
Methods
Metagenomic marker gene sequence analysis
Metagenomic sequencing data from 589 Hong Kong Chinese subjects (184 CRC, 185 adenoma and 220 control subjects) from our previous study were analysed,29 which included the discovery cohort of 74 CRC and 54 controls for the identification of the 20 CRC-related markers.4 Raw faecal shotgun metagenomic sequences were quality-trimmed and decontaminated as described previously.29 Low complexity subsequences of bacterial genes were hard-masked with the DUST program and indexed using the Burrows-Wheeler Aligner (BWA; V.0.7.17) to create the gene database for short read alignment.31 32 Post-quality control sequences in FASTQ format were mapped against the BWA database with maximal exact match (mem) algorithm and default parameters of penalty scoring. Histograms of aligned sequence coverage were reported using the ‘genomecov’ module of BEDTools suite (V.2.27.0).33 Mean sequence coverage table of metagenomic samples was constructed by computing summed products of coverage depth and base-pair fraction of marker gene length for positional features in input BAM files. Multiple group comparison of clinical phenotype was performed by pairwise Wilcoxon’s rank-sum tests, and p values were corrected by Benjamini-Hochberg step-up procedure. We then derived average weighted contribution (AWC) scores to estimate differential genomic enrichment and depletion of Lachnoclostridium species using marker gene sequences originating from the ChocoPhlAn pangenome database (V.293)34 as well as Prokka-annotated protein coding gene sequences representing 160 Lachnoclostridium genomes at all assembly levels from the NCBI GenBank (release 234.0; accessed 16 Oct 2019).35 The AWC of species i with gene set j to phenotype k was computed as follows:
where NEij (or NDij) is the total count of significant enrichment (or depletion) of a genomic sequence in gene set j for species i in a one-versus-all comparative statistical analysis of clinical phenotype k at 5% false discovery rate, respectively. Nij denotes the number of gene sequences of species i.
Human faecal sample collection
Faecal samples (n=1012) were collected from two independent groups of subjects, including group I—Hong Kong (698 subjects: 203 CRC, 207 adenoma and 288 normal controls) at the Prince of Wales Hospital, the Chinese University of Hong Kong between 2009 and 2014 and group II—Shanghai (313 subjects: 71 CRC, 145 adenoma and 97 normal controls) at Renji Hospital, Shanghai Jiaotong University between 2014 and 2018 (detailed clinical characteristics in online supplementary table S2). Subjects recruited for faecal sample collection included individuals presenting symptoms such as change of bowel habit, rectal bleeding, abdominal pain or anaemia, and asymptomatic individuals aged 50 or above undergoing screening colonoscopy as in our previous metagenomic study.4 Samples were collected before or 1 month after colonoscopy, when gut microbiome should have recovered to baseline.28 The exclusion criteria were (1) use of antibiotics within the past 3 months, (2) on a vegetarian diet, (3) had an invasive medical intervention within the past 3 months and (4) had a history of any cancer or inflammatory disease of the intestine. Subjects were asked to collect stool samples in standardised containers at home and store the samples in their home freezer at −20°C immediately. Frozen samples were then delivered to the hospitals in insulating polystyrene foam containers and stored at −80°C immediately until further analysis. Patients were diagnosed by colonoscopic examination and histopathological review of any biopsies taken.
gutjnl-2019-318532supp003.pdf (77.5KB, pdf)
DNA extraction, design of primers and probes and qPCR
DNA extraction, design of primer and probe sequences and qPCR amplifications on an ABI QuantStudio sequence detection system were conducted as our previous description.13 Primer and probe sequences specifically targeting m3 are as following: forward 5′-AATGGGAATGGAGCGGATTC-3′; reverse 5′-CCTGCACCAGCTTATCGTCAA-3′; probe 5′-AAGCCTGCGGAACCACAGTTACCAGC-3′. Primer and probe sequences targeting other bacterial gene markers and 16s rDNA internal control are as in our previous study.13 Each probe carried a 5′ reporter dye FAM (6-carboxy fluorescein) or VIC (4,7,2′-trichloro-7′-phenyl-6-carboxyfluorescein) and a 3′ quencher dye TAMRA (6-carboxytetramethyl-rhodamine). Primers and hydrolysis probes were synthesised by Invitrogen (Carlsbad, CA). PCR amplification specificity was confirmed by direct Sanger sequencing of the PCR products or by sequencing randomly picked TA clones. Relative abundance of each marker was calculated by using delta Cq method as compared with internal control and shown as Log value of ‘*10e6+1’.
Faecal immunochemical test
A subgroup of Hong Kong samples (n=642; 178 CRC, 118 advanced adenoma, 86 non-advanced adenoma and 260 control subjects) were examined by FIT using the automated quantitative OC-Sensor test (Eiken Chemical, Japan). The quantitative OC-Sensor test was performed as our previous description,36 with a positive cut-off value equivalent to a concentration of 100 ng of haemoglobin per millilitre.
Statistical analyses
Values were all expressed as mean±SD or median (IQR) as appropriate. The differences in bacterial abundances were determined by Mann-Whitney U test. One-way ANOVA multiple comparison with test for linear trend was used to evaluate the changes of marker levels during disease progression (from control to adenoma to cancer). Simple and multiple regression analyses were used to estimate the association between marker levels and factors of interest. Occurrence rates between different groups and sensitivities by different markers were analysed using the χ2 test. Combination of multiple biomarkers was performed by applying logistic regression model to obtain values for estimating the incidence of CRC as compared with controls. The scores of the combination of four markers were calculated as follows: LR4=Power (2, (α+β1X1+β2X2+β3X3+β4X4)), where α represented the intercept, β represented the regression coefficients and X represented the levels of the corresponding markers. ROC curves were used to evaluate the diagnostic value of bacterial markers/models in distinguishing CRC/adenoma and controls. Pairwise comparison of ROC curves was performed using a non-parametric approach.37 The best cut-off values were determined by ROC analyses that maximised the Youden index (J=Sensitivity+Specificity−1).38 All tests were done by GraphPad Prism V.5.0 (GraphPad Software, San Diego, CA) or MedCalc Statistical Software V.18.5 (MedCalc Software bvba, Ostend, Belgium; http://www.medcalc.org; 2018). A p value <0.05 was taken as statistical significance.
Footnotes
Contributors: Study conception and design: QL, JY. Development of methodology: QL. Acquisition of data: TL, Y-XC, TOY, GN, EC, SW, SCN, FKLC, Y-YF. Analysis and interpretation of data: QL. Writing, review and/or revision of the manuscript: QL, FKLC, JJYS, JY. Administrative, technical or material support: QL, JY. Study supervision: QL, JJYS, JY.
Funding: This project was supported by HRMF research fellowship scheme (02160037), China MOST fund (2016YFC1303200), Science and Technology Program Grant Shenzhen (JCYJ20170413161534162), National Key R&D Program of China (2017YFE0190700, 2018YFC135000, 2018YFC1315004), National Natural Science Foundation of China (81773000) and Shenzhen Virtual University Park Support Scheme to CUHK Shenzhen Research Institute.
Competing interests: None declared.
Patient consent for publication: Obtained.
Ethics approval: The study was approved by the Clinical Research Ethics Committee of the Chinese University of Hong Kong and the Ethics Committee of Renji Hospital, Shanghai Jiaotong University.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: All data relevant to the study are included in the article or uploaded as online supplementary information.
References
- 1. Allemani C, Matsuda T, Di Carlo V, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018;391:1023–75. 10.1016/S0140-6736(17)33326-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. The Lancet GLOBOCAN 2018: counting the toll of cancer. Lancet 2018;392:985 10.1016/S0140-6736(18)32252-9 [DOI] [PubMed] [Google Scholar]
- 3. Irrazábal T, Belcheva A, Girardin SE, et al. The multifaceted role of the intestinal microbiota in colon cancer. Mol Cell 2014;54:309–20. 10.1016/j.molcel.2014.03.039 [DOI] [PubMed] [Google Scholar]
- 4. Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 2017;66:70–8. 10.1136/gutjnl-2015-309800 [DOI] [PubMed] [Google Scholar]
- 5. Nakatsu G, Li X, Zhou H, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun 2015;6:8727 10.1038/ncomms9727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dai Z, Coker OO, Nakatsu G, et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome 2018;6 10.1186/s40168-018-0451-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tilg H, Adolph TE, Gerner RR, et al. The intestinal microbiota in colorectal cancer. Cancer Cell 2018;33:954–64. 10.1016/j.ccell.2018.03.004 [DOI] [PubMed] [Google Scholar]
- 8. Wong SH, Zhao L, Zhang X, et al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology 2017;153:1621–33. 10.1053/j.gastro.2017.08.022 [DOI] [PubMed] [Google Scholar]
- 9. Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013;14:207–15. 10.1016/j.chom.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013;14:195–206. 10.1016/j.chom.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017;170:548–63. e16 10.1016/j.cell.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tsoi H, Chu ESH, Zhang X, et al. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology 2017;152:1419–33. 10.1053/j.gastro.2017.01.009 [DOI] [PubMed] [Google Scholar]
- 13. Liang Q, Chiu J, Chen Y, et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res 2017;23:2061–70. 10.1158/1078-0432.CCR-16-1599 [DOI] [PubMed] [Google Scholar]
- 14. Xie Y-H, Gao Q-Y, Cai G-X, et al. Fecal Clostridium symbiosum for noninvasive detection of early and advanced colorectal cancer: test and validation studies. EBioMedicine 2017;25:32–40. 10.1016/j.ebiom.2017.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shah MS, DeSantis TZ, Weinmaier T, et al. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut 2018;67:882–91. 10.1136/gutjnl-2016-313189 [DOI] [PubMed] [Google Scholar]
- 16. Lee JK, Liles EG, Bent S, et al. Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta-analysis. Ann Intern Med 2014;160:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robertson DJ, Lee JK, Boland CR, et al. Recommendations on fecal immunochemical testing to screen for colorectal neoplasia: a consensus statement by the US Multi-Society Task force on colorectal cancer. Gastroenterology 2017;152:1217–37. 10.1053/j.gastro.2016.08.053 [DOI] [PubMed] [Google Scholar]
- 18. Yutin N, Galperin MY. A genomic update on clostridial phylogeny: gram-negative spore formers and other misplaced clostridia. Environ Microbiol 2013;140:2631–41. 10.1111/1462-2920.12173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Traore SI, Azhar EI, Yasir M, et al. Description of 'Blautia phocaeensis' sp. nov. and 'Lachnoclostridium edouardi' sp. nov., isolated from healthy fresh stools of Saudi Arabia Bedouins by culturomics. New Microbes New Infect 2017;19:129–31. 10.1016/j.nmni.2017.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pham T-P-T, Cadoret F, Alou MT, et al. 'Urmitella timonensis' gen. nov., sp. nov., 'Blautia marasmi' sp. nov., 'Lachnoclostridium pacaense' sp. nov., 'Bacillus marasmi' sp. nov. and 'Anaerotruncus rubiinfantis' sp. nov., isolated from stool samples of undernourished African children. New Microbes New Infect 2017;17:84–8. 10.1016/j.nmni.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tidjani Alou M, Khelaifia S, La Scola B, et al. "Lachnoclostridium touaregense," a new bacterial species isolated from the human gut microbiota. New Microbes New Infect 2016;14:51–2. 10.1016/j.nmni.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang S, Cai S, Ma Y. Association between Fusobacterium nucleatum and colorectal cancer: progress and future directions. J Cancer 2018;9:1652–9. 10.7150/jca.24048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Flanagan L, Schmid J, Ebert M, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis 2014;33:1381–90. 10.1007/s10096-014-2081-3 [DOI] [PubMed] [Google Scholar]
- 24. Amitay EL, Werner S, Vital M, et al. Fusobacterium and colorectal cancer: causal factor or passenger? Results from a large colorectal cancer screening study. Carcinogenesis 2017;38:781–8. 10.1093/carcin/bgx053 [DOI] [PubMed] [Google Scholar]
- 25. Watanabe Y, Nagai F, Morotomi M, et al. Bacteroides clarus sp. nov., Bacteroides fluxus sp. nov. and Bacteroides oleiciplenus sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 2010;60:1864–9. 10.1099/ijs.0.015107-0 [DOI] [PubMed] [Google Scholar]
- 26. Steer T, Collins MD, Gibson GR, et al. Clostridium hathewayi sp. nov., from human faeces. Syst Appl Microbiol 2001;24:353–7. 10.1078/0723-2020-00044 [DOI] [PubMed] [Google Scholar]
- 27. Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014;370:1287–97. 10.1056/NEJMoa1311194 [DOI] [PubMed] [Google Scholar]
- 28. Jalanka J, Salonen A, Salojärvi J, et al. Effects of bowel cleansing on the intestinal microbiota. Gut 2015;64:1562–8. 10.1136/gutjnl-2014-307240 [DOI] [PubMed] [Google Scholar]
- 29. Nakatsu G, Zhou H, Wu WKK, et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology 2018;155:529–41. 10.1053/j.gastro.2018.04.018 [DOI] [PubMed] [Google Scholar]
- 30. Coker OO, Nakatsu G, Dai RZ, et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 2019;68:654–62. 10.1136/gutjnl-2018-317178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morgulis A, Gertz EM, Schäffer AA, et al. A fast and symmetric dust implementation to mask low-complexity DNA sequences. J Comput Biol 2006;13:1028–40. 10.1089/cmb.2006.13.1028 [DOI] [PubMed] [Google Scholar]
- 33. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26:841–2. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Franzosa EA, McIver LJ, Rahnavard G, et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods 2018;15:962–8. 10.1038/s41592-018-0176-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014;30:2068–9. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 36. Wong SH, Kwong TNY, Chow T-C, et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut 2017;66:1441–8. 10.1136/gutjnl-2016-312766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837–45. 10.2307/2531595 [DOI] [PubMed] [Google Scholar]
- 38. Youden WJ. Index for rating diagnostic tests. Cancer 1950;3:32–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2019-318532supp001.pdf (1.1MB, pdf)
gutjnl-2019-318532supp002.pdf (592.7KB, pdf)
gutjnl-2019-318532supp003.pdf (77.5KB, pdf)