Significance
The accurate quantification of the amounts of small oligomeric assemblies formed by the amyloid β (Aβ) peptide represents a major challenge in the Alzheimer’s field. There is therefore great interest in the development of methods to specifically detect these oligomers by distinguishing them from larger aggregates. The availability of these methods will enable the development of effective diagnostic and therapeutic interventions for this and other diseases related to protein misfolding and aggregation. We describe here a single-domain antibody able to selectively quantify oligomers of the Aβ peptide in isolation and in complex protein mixtures from animal models of disease.
Keywords: Alzheimer’s disease, amyloid, protein aggregation, protein design
Abstract
Protein misfolding and aggregation is the hallmark of numerous human disorders, including Alzheimer’s disease. This process involves the formation of transient and heterogeneous soluble oligomers, some of which are highly cytotoxic. A major challenge for the development of effective diagnostic and therapeutic tools is thus the detection and quantification of these elusive oligomers. Here, to address this problem, we develop a two-step rational design method for the discovery of oligomer-specific antibodies. The first step consists of an “antigen scanning” phase in which an initial panel of antibodies is designed to bind different epitopes covering the entire sequence of a target protein. This procedure enables the determination through in vitro assays of the regions exposed in the oligomers but not in the fibrillar deposits. The second step involves an “epitope mining” phase, in which a second panel of antibodies is designed to specifically target the regions identified during the scanning step. We illustrate this method in the case of the amyloid β (Aβ) peptide, whose oligomers are associated with Alzheimer’s disease. Our results show that this approach enables the accurate detection and quantification of Aβ oligomers in vitro, and in Caenorhabditis elegans and mouse hippocampal tissues.
Alzheimer’s disease (AD), which is the most prevalent cause of dementia, affects over 50 million people worldwide (1). A molecular hallmark of AD is the accumulation of insoluble protein deposits, most notably amyloid plaques and neurofibrillary tangles, in specific brain tissues (2–6). Amyloid plaques, in particular, predominantly consist of aggregated forms of the amyloid β peptide (Aβ) (2–6) and are formed through a complex process that involves a variety of structurally different species (7, 8). Among such species, increasing evidence has implicated small, soluble oligomers as major agents responsible for neurotoxicity in AD (8–13). In particular, these oligomers have been shown to trigger various toxic pathways, including synaptic dysregulation, membrane permeabilization, oxidative stress, mitochondrial dysfunction, and activation of proinflammatory response (9, 11).
The conformational heterogeneity, low concentrations, and transient nature of these oligomeric species have made their isolation and characterization very challenging. Antibodies offer a variety of opportunities to overcome this challenge, as they represent powerful and versatile tools, owing to their high binding specificity and affinity and well-established discovery methods (14, 15). These protein molecules have highly successful applications in diagnostics, therapeutics, and targeted drug delivery systems, for infectious diseases, cancer, and metabolic and hormonal disorders (16). In particular, many diagnostic tests routinely used in the clinic are based on antibodies. For this reason, in the last 20 years major efforts have been made to overcome the challenges in isolating and stabilizing oligomeric species for immunization and phage display protocols to develop antibodies that selectively recognize such species in positron emission tomography scans and biological samples from patients (11, 17–20).
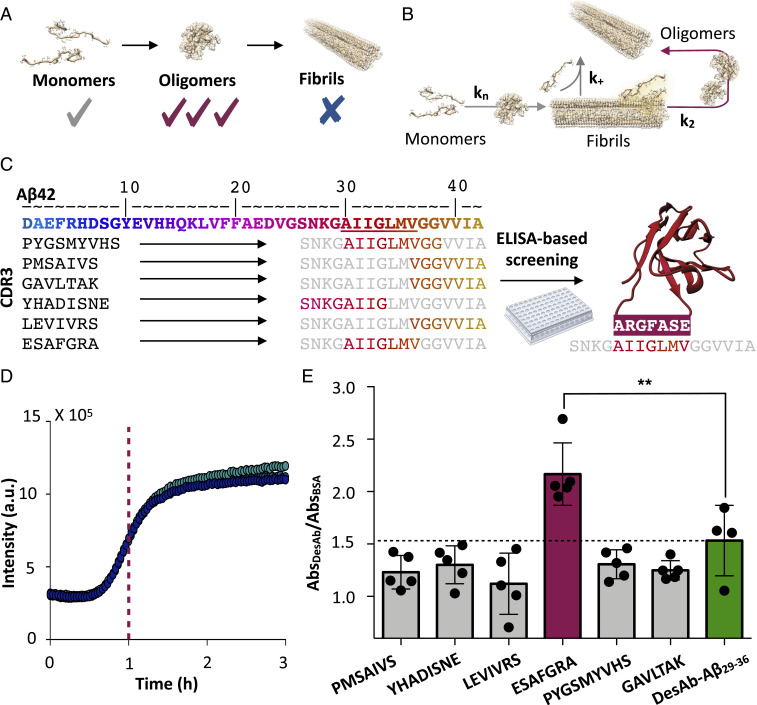
Recently, we introduced a scanning method based on the use of rationally designed, single-domain (VH) antibodies (DesAbs) for sequence-activity relationship studies (21–23). Using this strategy, we found that the antibody DesAb-Aβ29–36, targeting the epitope 29 to 36 of the 42-residue form of Aβ (Aβ42), inhibits the secondary nucleation step during the aggregation of Aβ42 at substoichiometric concentrations (Fig. 1B). This result suggests that DesAb-Aβ29–36 binds to Aβ42 aggregates with higher affinity than to monomers. Furthermore, recent structural characterizations of Aβ42 amyloid fibrils have shown that the C-terminal region of this peptide is buried within the core of the cross-β structure (24), and, accordingly, DesAb-Aβ29–36 shows low binding toward mature fibrils (21). Taken together, these results indicate that DesAb-Aβ29–36 inhibits secondary nucleation by interacting with oligomers and residues 29 to 36 of Aβ42 is likely to be solvent-exposed when the peptide is oligomeric, before becoming buried in amyloid fibrils. Therefore, targeting epitopes in this region could represent a strategy for generating oligomer-specific antibodies.
Fig. 1.
Rational design of a conformation-specific antibody for Aβ42 oligomers. (A) Schematic representation of the target selection strategy used in this work, which aims to generate an antibody with higher affinity for Aβ42 oligomers than monomers or fibrils. (B) Representation of the aggregation mechanism of Aβ42 (8). Primary nucleation (kn), secondary nucleation (k2), and elongation (k+) rate constants are shown. The red arrow indicates secondary nucleation processes, which are primarily responsible for the production of oligomers (21). (C) Sequence of Aβ42; the color gradient provides a visual representation of the results of the antigen scanning phase, where red residues are increasingly more effective targets for binding Aβ42 oligomers; the initial methionine residue is not shown. In the epitope mining phase, six designed complementary peptides (Left) targeting the region of residues 29 to 36 in the Aβ42 sequence were used to generate six single-domain antibodies, whose CDR3 sequences are shown together with their corresponding Aβ42 epitopes (Right). The sequence of the complementary peptide of DesAb-O, the designed antibody with highest binding affinity for Aβ oligomers, is shown in red on the right-hand side. (D) ThT-based in vitro aggregation assay of 3 μM Aβ42 (three replicates are shown). The purple dashed line indicates the time at which samples were collected from the aggregation reaction to perform the ELISA experiment. a.u., arbitrary units. (E) ELISA experiment performed on samples collected from the aggregation reaction shown in B, using the six DesAbs as primary antibodies. For each antibody, absorbance values recorded in the presence of Aβ42 aggregated samples were normalized over background absorbance values (i.e., in the presence of BSA only). The bar corresponding to DesAb-O is colored magenta while the one corresponding to the original DesAb-Aβ29–36 is green. The dashed line corresponds to the value of DesAb-Aβ29–36. Error bars are representative of the SD. Statistical analysis was performed by ANOVA with multiple comparison (95% CI, **P ≤ 0.01).
In this work, we use rational design to generate a battery of DesAbs, targeting epitopes in the region of residues 29 to 36 of Aβ42, and experimental screenings to identify one, DesAb-O, which selectively binds oligomers of Aβ, rather than its monomeric and fibrillar forms (Fig. 1A). We characterize the binding properties of DesAb-O in vitro and in complex mixtures from animal models toward the development of diagnostic tools.
Results
Antibody Mining to Identify An Oligomer-Specific Antibody.
Based on our previous results (21), we performed a rational search for complementary peptides binding epitopes in the region of residues 29 to 36. We thus designed a panel of six additional antibodies with the aim of selecting the antibody with highest preferential binding to Aβ42 oligomers with respect to monomers and fibrils (Fig. 1). To do so, first we generated six complementary peptides, using the cascade method (22) (Methods). In this method the complementary peptides are assembled from peptide fragments selected from the Protein Data Bank (PDB), so that such fragments are complementary to the corresponding short peptide fragments of the target sequence because they are found in two opposing strands of a β-sheet. We grafted these peptides into a human VH domain antibody scaffold (20), and we expressed and purified the resulting DesAbs in Escherichia coli as previously reported (21) (SI Appendix, Fig. S1A). Circular dichroism (CD) spectroscopy revealed that all six DesAbs have a secondary structure content compatible with the native conformation of a VH domain (SI Appendix, Fig. S1B).
In order to identify the DesAbs with the strongest binding to Aβ42 oligomers, we developed an enzyme-linked immunosorbent assay (ELISA)-based screening assay (Fig. 1 C–E). We performed our analysis on samples collected directly from aggregation reactions at the half-time of aggregation (Fig. 1D), where the oligomers are present at their highest concentration (25). We immobilized these samples onto an ELISA plate and then used the various DesAbs as primary antibodies in an indirect ELISA setup (Fig. 1 D and E). We found that all of the antibodies in the panel were able to bind to the oligomeric mixtures. In particular, the antibody carrying the complementary peptide ESAFGRA, which we will refer to as DesAb-O (SI Appendix, Fig. S1C), showed the strongest binding (Fig. 1E).
Characterization of the Conformational Specificity of DesAb-O.
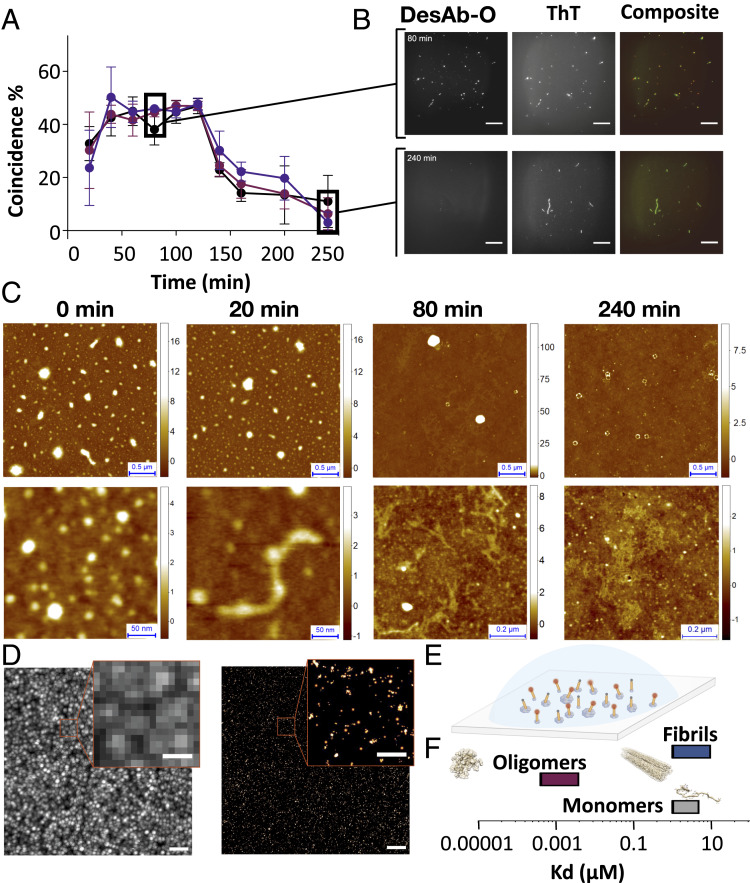
Next, in order to determine whether DesAb-O is specific for oligomers over fibrils, we used total internal reflection fluorescence (TIRF) microscopy to visualize individual aggregates in samples taken at specific incubation times from aggregation reactions of 3 μM Aβ42 (Fig. 2 and SI Appendix, Figs. S2 and S3). We performed a colocalization analysis by depositing these protein samples on glass slides and labeling them with two different fluorescent dyes: 5 μM thioflavin T (ThT), a generic dye for amyloid aggregates, and 1 nM AF647-labeled DesAb-O (AF647-DesAb-O). We then measured how many of the aggregates contained in the samples were recognized by either ThT, AF647-DesAb-O, or both. We found that many aggregates were probed by both ThT and AF647-DesAb-O at early aggregation times, with a percentage of coincidence instances up to 45% between 60 and 100 min (Fig. 2 A and B). In contrast, at later incubation times (beyond 140 min), when mature amyloid fibrils are formed, the percentage of coincidence dramatically drops down to less that 10%.
Fig. 2.
Characterization of the conformational specificity of DesAb-O by TIRF, AFM, and dSTORM. (A) Coincidence of ThT and AF647-DesAb-O signals in the TIRF single-molecule imaging of aggregates from an aggregation reaction of Aβ42. Three independent experiments in three different colors (black, red, and violet) are shown. Each point is the average of 10 fields. Error bars represent the SD. (B) TIRF images at 80 and 240 min of aggregation time are shown. ThT, AF647, and composite channel images are shown. (Scale bars, 20 μm.) (C) Wide (Top) and detailed (Bottom) high-resolution AFM 3D morphology maps of samples at 0, 20, 80, and 240 min. (D) Super-resolution dSTORM imaging of an aggregation reaction of Aβ42 after 40 min of incubation. (Left) Diffraction limited image. (Right) dSTORM image. (Scale bars, 2 μm in the lower-magnification images and 500 nm in the zoomed images.) (E) Representation of the experimental setup for super-resolution imaging. (F) Estimation of the Kd values of binding of DesAb-O with different aggregated species of Aβ42.
To prove that, in these conditions, DesAb-O binds oligomers in the presence of many other aggregated species, we further investigated the content of the samples at the single aggregate nanoscale by acquiring high-resolution, three-dimensional (3D) morphology maps using phase-controlled atomic force microscopy (AFM) (26). We found that samples at 20, 80, and 240 min were all highly heterogeneous and contained different types of aggregated species (Fig. 2C). Starting from 80 min, we also observed the presence of ring-shaped toroidal and prefibrillar oligomers, whose size matches those recognized by DesAb-O and observed by TIRF.
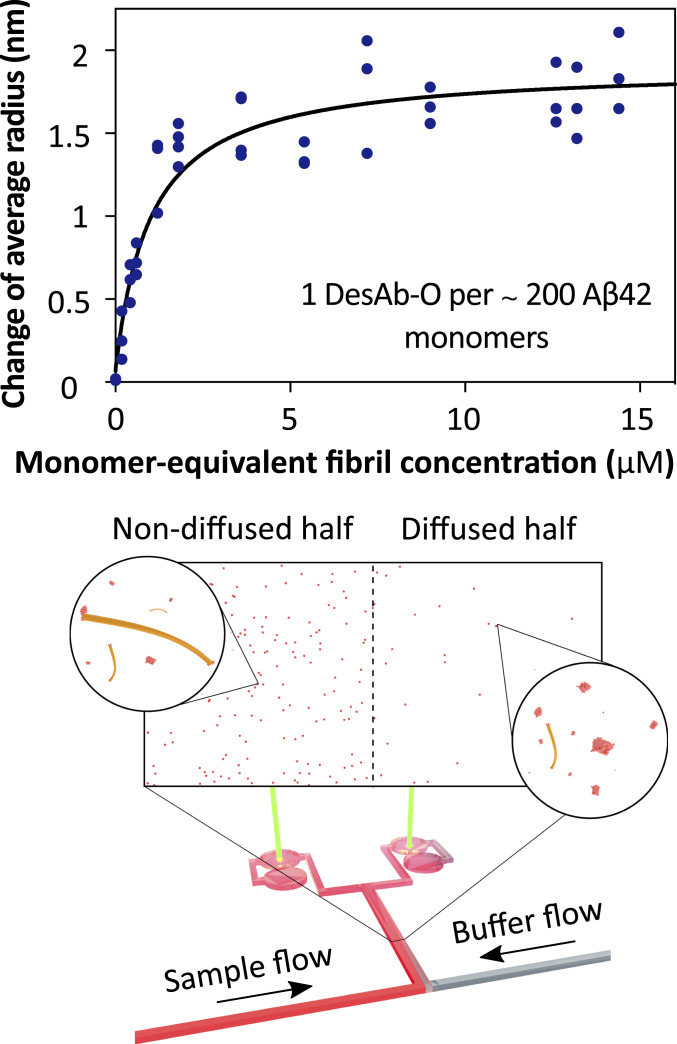
In order to determine the sizes and shapes of the Aβ42 aggregates recognized by DesAb-O, we performed super-resolution imaging by direct stochastic optical reconstruction microscopy (dSTORM) using AF647-DesAb-O on samples collected at 40 min of aggregation. These data show that DesAb-O binds Aβ42 aggregates that are significantly smaller than the diffraction limit (∼230 nm; Fig. 2 D and E and Movie S1). On the contrary, DesAb-O does not bind to late-stage (140 min) aggregates (Movie S2), further confirming the specificity of the DesAb to oligomeric species. Furthermore, DesAb-O was able to bind oligomers with an apparent subnanomolar binding affinity, compared to the low micromolar affinity for fibrils (Fig. 2F), as derived by microfluidic diffusional sizing (27) (Fig. 3).
Fig. 3.
Determination of the binding affinity of DesAb-O for Aβ42 amyloid fibrils. We used microfluidic diffusion measurements to determine the affinity of DesAb-O for Aβ42 amyloid fibrils. The analysis yielded an effective Kd of 1 μM, and the binding ratio of about 1 to 200 (DesAb-O:Aβ42) suggests that DesAb-O binds the fibril ends, where the epitope Aβ4229–36 may not be buried in the structural core of the fibrils.
Taken together, these data indicate that DesAb-O is able to preferentially bind oligomers over monomers and fibrils of Aβ42.
ELISA-Based Real-Time Oligomer Quantification Using DesAb-O.
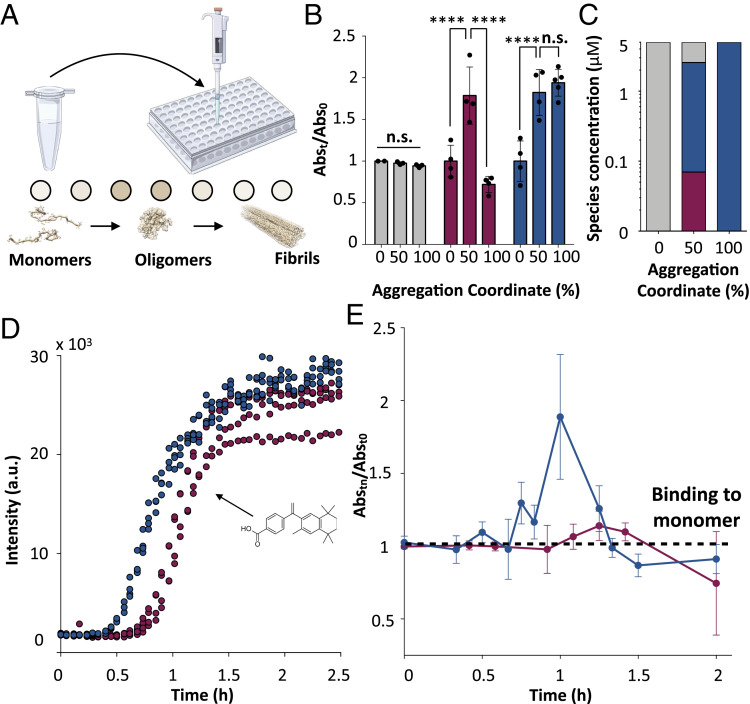
Given the high oligomer specificity of DesAb-O, we developed an assay to monitor the formation, conversion, and inhibition by anti-amyloid molecules of oligomeric populations during the aggregation of Aβ42. First, we examined Aβ42 oligomers formed during in vitro aggregation assays (25). We used aggregation reactions of 5 μM Aβ42, from which we extracted 20-μL samples at specific incubation times. These samples were then analyzed by an indirect ELISA set up using DesAb-O as primary antibody (Fig. 4A and Methods).
Fig. 4.
Development and validation of a real-time ELISA using DesAb-O for the quantification of Aβ42 oligomers in an aggregation reaction. (A) Illustration of the experimental setup of the time-course ELISA. Samples from 5 μM Aβ42 aggregation reactions were collected at specific incubation times and loaded onto an ELISA plate. The amounts of oligomers were determined from absorbance measurements upon incubation with DesAb-O and a commercial HRP-conjugated anti-His tag antibody. (B) ELISA measurements taken at 0 h, 0.5 h (50% of the total aggregation time), and 2 h (plateau, 100% of the total aggregation time) from a 5 μM Aβ42 aggregation reaction using DesAb-O (red), DesAb-Aβ18–24 (blue), and the commercial antibody 6E10 (gray). Data of each antibody were normalized for the corresponding average value at t = 0. Error bars are representative of the SD. Statistical analysis was performed by ANOVA with multiple comparisons (CI 95%, ****P < 0.0001; n.s., not significant). (C) Concentrations of monomers (gray), oligomers (red), and fibrils (blue) at varying time points for a 5 μM Aβ42 aggregation reaction. (D) ThT aggregation assays of 3 μM Aβ42 alone (blue) or in the presence of fivefold molar excess bexarotene (red). (E) Time course ELISA of 3 μM Aβ42 alone (blue) or in the presence of fivefold molar excess bexarotene (red). Error bars are representative of the SD. Individual data points are shown in SI Appendix, Fig. S4B.
In order to assess whether this antibody was able to detect Aβ42 oligomers under our experimental conditions, we compared its performance to that of a commercial antibody (6E10), which targets the N-terminal region of Aβ42, and of a previously described DesAb (DesAb-Aβ18–24) (21), which binds to all aggregated species, oligomers, and fibrils with no particular preference, at the beginning (0 h), at the half-time (0.5 h), and at the plateau (2 h) of aggregation (Fig. 4B). As expected, we observed that 6E10 leads to similar signals at the three incubation times (Fig. 4B, gray bars). As this antibody binds the N terminus of Aβ42, which is a portion of the peptide exposed in most aggregated conformations, this result proves that the total amount of Aβ42 in the wells is similar for all three time points and the presence of aggregates does not affect the absorption of the samples on the ELISA wells. In contrast, the DesAb-Aβ18–24 signal increased with the aggregation time, demonstrating that this antibody preferentially binds aggregated species but cannot distinguish between oligomers and fibrils (Fig. 4B, blue bars, and SI Appendix, Fig. S4). Finally, DesAb-O showed the highest signal at the half-time of the aggregation reaction (Fig. 4B, red bars), indicating that this antibody specifically binds oligomers, which transiently form during the aggregation of Aβ42. In particular, the signal of DesAb-O was twofold higher at the half-time than at the beginning of the aggregation. Notably, it has been shown that the concentration of monomers is about 35-fold higher than the concentration of oligomers (2.4 μM vs. 70 nM) at this time point under the same aggregation conditions (8) (Fig. 4C).
We then performed a time-course experiment (Fig. 4 D and E) to test whether we could characterize the time evolution of the Aβ42 oligomer population during the aggregation process. To do so, we prepared solutions of 3 μM Aβ42, and we monitored the aggregation of Aβ42 by ThT (Fig. 4D) and by using DesAb-O as primary antibody in ELISA experiments on different incubation times of the aggregation reaction (Fig. 4E). We found that, in the ELISA experiments, the absorbance from DesAb-O progressively increased, reaching the highest value at 1 h of incubation, approximately at the half-time of aggregation as determined by ThT (Fig. 4D), where oligomers are at their highest concentrations. This value was approximately twofold higher than that initial aggregation time points. After 1 h of incubation, the absorbance progressively decreased to values even lower than the initial ones, further indicating that DesAb-O does not bind fibrillar species (Fig. 4E and SI Appendix, Fig. S4B).
Using DesAb-O to Assess Aβ42 Oligomer Inhibition by Drug Candidates.
Next, we evaluated the capability of the DesAb-O assay to capture changes in the oligomeric population in the presence of anti-amyloid compounds. To do so, we performed an aggregation assay in the presence of the small molecule bexarotene (Fig. 4 D and E), which has been shown to inhibit primary nucleation and thus to delay the formation of Aβ42 oligomers (28). In this case, the DesAb-O–based ELISA detected a shift of 30 min in the peak of the oligomers (Fig. 4E), which matches the shift in half-time of aggregation observed by ThT assay (Fig. 4D). These results indicate that our antibody-based assay is able to detect oligomeric populations formed during in vitro aggregation of Aβ42 and that this technique can be used to probe the effects of anti-amyloid compounds on these populations.
DesAb-O Detects Aβ42 Oligomers in a Caenorhabditis elegans Model of AD.
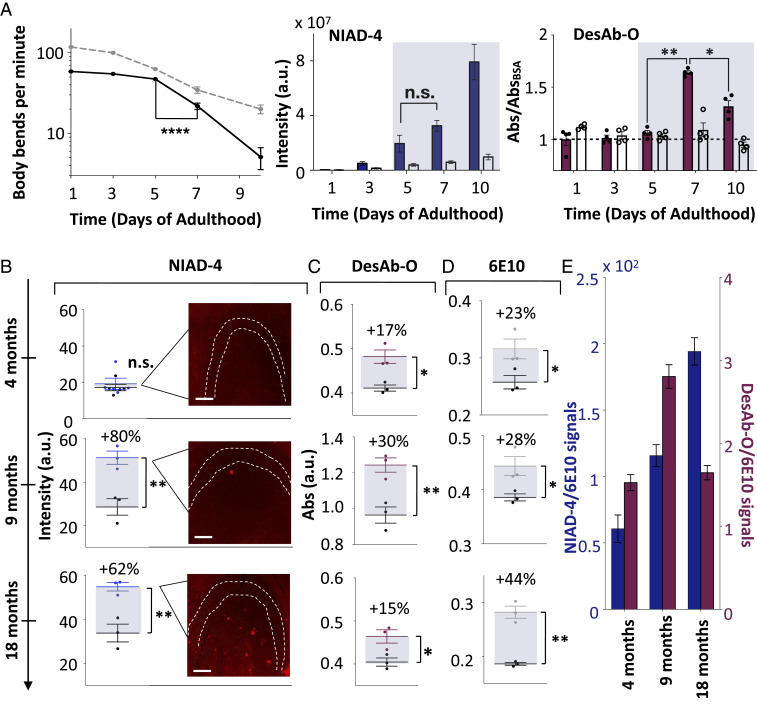
In order to verify whether DesAb-O specifically detects Aβ42 oligomers formed in vivo, we used a C. elegans model of Aβ42-mediated dysfunction, called GMC101, in which human Aβ42 is expressed in body wall muscle cells where it forms aggregates and results in severe age-progressive paralysis (29). The analysis was performed on protein extracts from 500 GMC101 worms at days 0, 3, 5, 7, and 10 of adulthood (Fig. 5 A and B). We first assessed the fitness of the worms for 10 d starting at 24 h of induction of aggregation (Methods). We found that the GMC101 worms had the most dramatic drop of mobility between days 5 and 7, which indicates that the toxic species likely reach their highest concentrations at that time. Then, we monitored the formation of the amyloid aggregates using the amyloid-specific dye NIAD-4, whose fluorescence increases with the concentration of amyloid aggregates. We found no significant NIAD-4 fluorescence change able to capture the pathological behaviors observed between days 5 and 7, suggesting that this compound is not selective for toxic aggregates. Then, we verified whether we were able to specifically detect these species using DesAb-O. To do so, we analyzed protein samples from worms at different days of adulthood by means of ELISA (Methods) and compared the result of this experiment with a quantification of the aggregates using NIAD-4. In particular, we coated the ELISA wells with the protein extracts and used DesAb-O as a primary antibody. As a control, we performed the same procedure on protein extracts from the wild-type C. elegans model N2 and used these signals as a reference (Fig. 5A). We found that DesAb-O was able to specifically recognize toxic Aβ42 species in the GMC101 protein extracts formed between days 5 and 7 that NIAD-4 is unable to detect.
Fig. 5.
Quantification of Aβ42 oligomers in C. elegans and mouse hippocampal tissue. (A) From left to right: plots showing body bends per minute of N2 (dashed gray line) and GMC101 (continuous black line) worms (n = 500 individual worms), NIAD-4 fluorescence intensities of GMC101 (blue bars) and N2 (white bars) worms (n = 30 individual worms), and ELISA absorbance of DesAb-O of GMC101 (red bars) and N2 (white bars) worms at different days of adulthood. NIAD-4 fluorescence intensity was calculated as corrected total cell fluorescence using the ImageJ software (Methods). (B–E) Comparison of different aggregate-detection assays for the hippocampus CA3 area from J20 (blue) and control wild-type (black) mice at 4, 9, and 18 mo of age. (B) NIAD-4 fluorescence from J20 (blue) and control wild-type (black) mice; representative fluorescence images are shown in the insets. The dashed line marks the borders of the cell body layer in the CA3 region of the hippocampus. (Scale bars, 100 μm.) (C) Absorbance of an ELISA using DesAb-O from J20 (red) and control wild-type (black) mice. (D) Absorbance of an ELISA using the monoclonal antibody 6E10 from J20 (gray) and control wild-type (black) mice. (E) Bar plot of the NIAD-4 and DesAb-O signals of J20 mice from B and C divided by the 6E10 signals of D; all error bars represent SEs. Statistical analysis in A was performed by ANOVA with multiple-comparison and in B–D with a t test (CI 0.95; *P **P, ****P; n.s., not significant).
DesAb-O Quantifies Aβ42 Oligomers in Hippocampal Tissue in a Mouse Model of AD.
To further validate the ability of DesAb-O to detect and quantify Aβ42 oligomers formed in vivo, we performed similar analyses using a J20 mouse model of AD (PDGF-APPSw,Ind), which overexpresses a variant of human amyloid precursor protein carrying the Swedish and Indiana familiar mutations (30). We performed ELISAs on hippocampus extracts using DesAb-O and 6E10, and quantified NIAD-4 fluorescence from tissue slices from mice at 4, 9, and 18 mo of age. We then normalized these signals using a control wild-type mouse. We were able to detect positive signals of DesAb-O at 4 mo of age, when the mice show the initial signs of memory impairment (31) and neuroinflammation and synaptic deficit (32), but before amyloid deposits are significantly detected with NIAD-4. Furthermore, the DesAb-O signal increases between 4 and 9 mo and then decreases at 18 mo, while both 6E10 and NIAD-4 signals progressively increase (Fig. 5 B–E). The decrease of DesAb-O signal observed at 18 mo suggests that the amounts of oligomers may decrease at this age. These studies demonstrate that DesAb-O detects oligomers early in the pathogenesis in this mouse model of AD and may provide a valuable tool to assess oligomer load in the AD brain.
DesAb-O Detects Zinc-Stabilized Aβ40 Oligomers but Not Aβ40 Monomers and α-Synuclein Oligomers.
Given that different types of Aβ oligomers (33) are likely to exist upon in vivo aggregation, we also tested the ability of DesAb-O to detect other types of Aβ oligomers. To this end, we carried out a fluorescence-based screening assay in combination with Zn2+-stabilized Aβ40 oligomers (33). This assay uses the fluorescent probe 8-anilinonaphthalene-1-sulfonic acid (ANS), whose fluorescence emission increases upon binding to exposed hydrophobic regions on the surface of oligomers. We thus monitored the change in ANS fluorescence in the presence of a 1:1 ratio of DesAb:Aβ40 (SI Appendix, Fig. S5). We found that the addition of the DesAbs decreased ANS fluorescence, suggesting it prevents ANS molecules from binding the surface of the oligomers as a result of the antibody binding. Among all of the antibodies produced in this work, the antibody DesAb-O showed the greatest binding in this experiment (SI Appendix, Fig. S6), significantly higher than that of DesAb-Aβ29–36 (SI Appendix, Fig. S5). As a complementary measurement, we performed an ELISA (SI Appendix, Fig. S6) monitoring the binding of immobilized DesAbs in the presence of Aβ40 oligomer solution. We thus found that, while the majority of the antibodies had a binding similar to the original antibody DesAb-Aβ29–36, DesAb-O showed significantly stronger binding to Aβ40 oligomers (SI Appendix, Fig. S6).
We further confirmed these results with a dot blot analysis. To do so, we immobilized the antibodies DesAb-O and the antibody containing a complementary peptide (PYGSMYVHS), the latter being the least reactive antibody in the ANS quenching experiments on a nitrocellulose membrane (SI Appendix, Fig. S6). We then incubated the membrane in the presence of a solution of stabilized Aβ40 oligomers and revealed the bound oligomers using 6E10, a commercial primary antibody targeting Aβ. In agreement with the previous experiments, DesAb-O was the antibody showing the strongest binding to Aβ40 oligomers. Furthermore, we tested the anti-aggregation proprieties of this antibody, as we previously did for DesAb29–36 (21). We found that DesAb-O mainly affects the secondary nucleation of Aβ42 aggregation (SI Appendix, Fig. S7). This is in line with what we observed for DesAb29–36, as both antibodies target the same epitope of Aβ42. In the case of DesAb-O, however, the inhibitory effect is less strong, probably as a consequence of the lower affinity for monomers and fibrils.
We then quantified the sequence and conformational specificity of DesAb-O binding. In particular, we determined whether this antibody was selective for stabilized Aβ40 oligomers over monomers of Aβ40, and similarly whether it was specific for Aβ oligomers over those formed by another amyloid protein, α-synuclein. To do so we performed isothermal titration calorimetry (ITC) experiments (SI Appendix, Fig. S8), where we measured the heat change over time upon injections of the DesAbs into solutions containing Zn2+-stabilized oligomers or monomers of Aβ40. We found that DesAb-O binds oligomers with a dissociation constant (Kd) of 440 ± 1.5 nM (SI Appendix, Fig. S8). We observed the binding to be exothermic and enthalpically driven [∆H = −1.27E4 ± 9.4E2 cal/mol; ∆S = −13.4 ± 9.4E2 cal/(mol⋅deg)]. The stoichiometry of binding, DesAb-O:Aβ40 molecules (about 1:5), suggests a binding of about one antibody per oligomer, in agreement with a previous report, which suggest that these oligomers typically contain five Aβ40 monomers (33). The binding of DesAb-O to monomers of Aβ40 was extremely weak and not quantifiable, as well as the binding of DesAb-O to stabilized α-synuclein oligomers (12), indicating that DesAb-O is both conformation- and sequence-specific (SI Appendix, Fig. S8).
Discussion and Conclusions
Sequence-specific and conformation-specific antibodies capable of binding neurotoxic protein oligomers are powerful tools for basic research, as well as for diagnostic and therapeutic applications. These antibodies enable the quantification of the amounts of oligomers present in given samples, and the characterization of their properties and behavior. In particular, in combination with fluorescence microscopy and with immunoassays such as ELISA and related methods, antibodies offer unique possibilities to assess the presence and abundance of oligomers in cells and tissues.
The discovery of oligomer-specific antibodies, however, is very challenging because of the transient nature of the oligomers produced during protein aggregation reactions. These oligomers are characteristically not sufficiently stable to be used as antigens in standard in vivo or in vitro antibody discovery methods. To overcome this challenge and contribute to the development of oligomer-specific antibodies, we have presented a rational design method that does not require previous knowledge of the structure of the target oligomers. The method involves two phases, whereby an initial panel of antibodies is designed to bind different epitopes covering the entire sequence of a target protein (scanning phase). Analysis of the antibodies in this panel enables the identification of epitopes exposed in the oligomers but mostly inaccessible in the fibrils. In the second step, a second panel of antibodies is designed to target the epitopes identified in the first step (mining phase). Further binding assays enable the identification of the oligomer-specific antibody in the panel with the highest binding affinity.
We have illustrated the application of this rational design method for the determination of the oligomeric populations formed during the aggregation of Aβ42 both in vitro and in vivo using C. elegans and mouse models of AD. We anticipate that this technology will create novel opportunities for the detection and accurate quantification of oligomers of amyloidogenic proteins for diagnostic and therapeutic applications.
Methods
Rational Design of the Antibodies.
We summarize the rational method for the identification of complementary peptides that bind to specific linear epitopes in target proteins of interest, which we graft onto the CDR loops of domain antibodies. A detailed description of the method is provided in ref. 22. The complementary peptide design procedure consists of two steps. First, given a target linear epitope, we collect from the PDB all protein fragments that face in a β-strand any subsequence of at least three residues in which the target epitope can be fragmented. Second, complementary peptides predicted to bind the target epitope are built by merging together these fragments using the cascade method (22). In essence, in the cascade method fragments are linked using three rules: 1) fragments can be joined together only if found in β-strands of the same type (i.e., parallel or antiparallel), 2) all fragments making up a complementary peptide must partly overlap with their neighboring fragments, and 3) the overlapping regions must be identical both in the sequence and in the backbone hydrogen-bond pattern that is extracted from the β-strand where each fragment is found. Since the identification of the complementary peptides is based on the analysis of amino acid sequences facing each other in β-strands in the PDB, the interaction with the target sequence is already shown to be viable in a biological context. In addition, given this design strategy, the resulting complementary peptides are expected to bind the target epitope by enforcing a β-strand–like conformation. Therefore, such complementary peptides will be particularly effective in binding to solvent-exposed regions of protein sequences that do not form persistent hydrogen bonds with other parts of the protein molecule, such as in the case of disordered regions (22).
Protein Expression and Purification.
The various complementary peptides were grafted into the CDR3 of the DesAb scaffold by means of mutagenic PCR with phosphorylated oligonucleotides (21). The different DesAb constructs were then expressed and purified using Ni2+ chromatography, as previously described (21). Imidazole was finally removed using size-exclusion chromatography with a HiLoad 16/600 Superdex 75 pg column (GE Healthcare). Protein concentration was determined by absorbance measurement at 280 nm using theoretical extinction coefficients calculated with ExPASy ProtParam (34). Aβ40 (M1–40) and Aβ42 (M1–42) peptides were expressed in E. coli BL21 (DE3) Gold Strain (Agilent Technologies) and purified as described previously (8). Aliquots of purified Aβ42 and Aβ40 were lyophilized and stored at −80 °C.
CD.
Far-ultraviolet (UV) CD spectra of the DesAbs were recorded using a Jasco J-810 spectropolarimeter equipped with a Peltier holder (Jasco UK Ltd), using a 0.1-cm-pathlength cuvette. Samples contained 10 μM protein in 20 mM Tris pH 7.4, 100 mM NaCl. The far-UV CD spectra of all DesAbs were recorded from 200 to 250 nm at 20 °C, and the spectrum of the buffer was subtracted from the spectra of all DesAbs.
Aggregation Assays of Aβ42.
The lyophilized Aβ42 peptide was dissolved in 6 M guanidinium, pH 8, and incubated for 3 h on ice. This solution was then injected into a Superdex 75 10/300 Increase size-exclusion column (GE Healthcare), and the peak corresponding to the monomeric Aβ42 peptide was isolated and collected in low-binding test tubes (Corning) on ice (8). Ninety micromoles of each aggregation sample were then pipetted into black polystyrene 96-well half-area plates with clear bottoms and polyethylene glycol coating (Corning). Plates were sealed to prevent evaporation. Aggregation assays were performed at 37 °C under quiescent conditions using a CLARIOstar plate reader (BMG Labtech). Fluorescence emissions at 480 nm were recorded upon excitation at 440 nm through the bottom of the plate every 2 min. Aggregations samples were prepared with 2 μM monomeric Aβ42 in the presence of different molar equivalents of DesAb-O in 20 mM sodium phosphate, pH 8, 200 μM ethylenediaminetetraacetic acid (EDTA), 0.02% NaN3, and 20 μM ThT. For seeded experiments, aggregation samples were prepared in the same manner with the addition of 0.4 μM preformed Aβ42 fibrils.
ELISA-Based Binding Screening of the Antibodies.
Monomeric Aβ42 peptides were aggregated at a protein concentration of 5 μM in 20 mM sodium phosphate buffer (pH 8) and 200 μM EDTA under quiescent conditions. Twenty-microliter aliquots were taken at the t50 (i.e., half-time) of aggregation from aggregation reactions. Samples were then immobilized on a 96- or 384-well Maxisorp ELISA plate (Nunc) with no shaking for 1 h at room temperature. The plate was then washed three times with 20 mM Tris, pH 7.4, and 100 mM NaCl and incubated in 20 mM Tris, pH 7.4, 100 mM NaCl, and 5% bovine serum albumin (BSA) under constant shaking overnight at 4 °C. The day after the plate was washed six times with 20 mM Tris, pH 7.4, and 100 mM NaCl and then incubated with 30 μL solutions of 5 µM DesAb-O under constant shaking either for 1 h at room temperature or overnight at 4 °C. At the end of this incubation, the plate was washed six times with 20 mM Tris, pH 7.4, and 100 mM NaCl and incubated with 30 μL solutions of rabbit polyclonal 6x His tag horseradish peroxidase (HRP) conjugated (Abcam) at a dilution of 1:4,000 in 20 mM Tris, pH 7.4, 100 mM NaCl, and 5% BSA under shaking for 1 h at room temperature. Finally, the plate was washed three times with 20 mM Tris, pH 7.4, and 100 mM NaCl, then twice with 20 mM Tris, pH 7.4, 100 mM NaCl, and 0.02% Tween-20 and again three times with 20 mM Tris, pH 7.4, and 100 mM NaCl. Finally, the amount of bound DesAb-O was quantified by using 1-Step Ultra TMB-ELISA Substrate Solution (Thermo Fisher Scientific), according to the manufacturer’s instructions, and the absorbance was measured at 450 nm by means of a CLARIOstar plate reader (BMG Labtech).
Preparation for ThT/AF647–DesAb-O Fluorescence Imaging.
Aliquots were taken from aggregation reactions at 20-min intervals and imaged immediately. Borosilicate coverslips (22 × 22 mm, 630-2186; VWR) were cleaned for 1 h using argon plasma (PDC-002; Harrick Plasma). Frame-Seal slide chambers (9 × 9 mm, SLF-0201; Bio-Rad) were affixed to the glass, and then washed three times with filtered (0.2-µm syringe filter, 6780-1302; Whatman) phosphate-buffered saline (PBS) buffer. A working solution containing AF647-DesAb-O (1 nM) and ThT (5 μM) in filtered PBS (pH 7.4) was prepared 5 min prior to imaging. Aβ aliquots were diluted (500 nM), incubated on the poly-l-lysine–coated coverslip for 5 min and then washed twice with filtered PBS. The DesAb-ThT working solution (50 μL) was added to the cover slide and incubated for 2 min prior to imaging.
DesAb–ThT Colocalization Imaging.
Imaging experiments were carried out with bespoke TIRF inverted microscope (Eclipse TE2000-U; Nikon) fitted with a Perfect Focus unit. Excitation of ThT and AF647 was achieved with either a 405-nm laser (LBX-405-50-CIR-PP; Oxxius) or 641-nm laser (Cube, 1150205; Coherent), respectively. The beams were aligned parallel to the optical axis and directed into an oil immersion objective lens (1.49 numerical aperture [N.A.], 60×, Plan Apo, TIRF; Nikon) above the critical angle to ensure TIR at the coverslip/sample (glass/water) interface. Fluorescence emission was also collected by the same objective and selected by the presence of a dichroic (Di01-R405/488/561/635; Semrock) and subsequently passed through appropriate emission filters (BLP01-488R-25, FF01-480/40-25, and FF01-676/37-25; Semrock). Image stacks of the AF647 and ThT emission channels were collected by sequential excitation of AF647 followed by ThT. Images were recorded by an electron multiplying charge-coupled device (Evolve delta 512; Photometrics) with an electron multiplication gain of 250 analog-to-digital units per photon running in frame transfer, clear presequence mode. Each pixel on the image was 237 nm. Images from 27 different fields of view were recorded at 50 ms for 200 frames in each emission channel using a custom beanshell script through Micromanager software (v. 1.4).
DesAb–ThT Colocalization Image Analysis.
Colocalization data were analyzed with a bespoke ImageJ macro. Separate average intensity z-projections of ThT and AF647 channels were created which results in single-frame images representing the mean pixel intensities calculated for the total image stack. Following this, points of intensity above a background threshold were located, counted, and binarized. Pixels with a value of 1 in both the AF647 and ThT images were identified as coincident points. Chance coincident spots were extracted by performing a 90° rotation of the AF647 binary image and subtracted from the total coincidence value. Percentage coincidence was calculated as
AFM.
We performed in air AFM measurements of the sample deposited on glass, where TIRF measurements had been acquired. High-resolution images (1,024 × 1,024 pixels) and phase-controlled maps (26) were collected using an NX10 Atomic Force Microscope (Park Systems) under ambient conditions and in amplitude modulation noncontact (NC-AM) mode. We imaged square areas of 2 × 2 µm and 4 × 4 µm. We performed all of the measurements using sharp cantilevers (PPP-NCHR; Park Systems) with resonance frequency of 330 kHz and a typical apical radius of 8 nm. The raw images were flattened using the built-in software (XEI; Park Systems). To maintain consistency in the subsequent statistical analysis, all images were processed using the same parameters. The images were first flattened by a plane and then line-by-line in first regression order. This second step was repeated until a flat baseline in the line profile of the image was reached. During the process of flattening of the images, the aggregates were masked from the calculation to avoid modification and underestimation of their heights.
Sample Preparation for dSTORM Imaging.
Glass cover slides (631-1570; VWR) for single-aggregate fluorescence imaging were cleaned for 1 h using an argon plasma (PDC-002; Harrick Plasma). Frame-Seal slide chambers (9 × 9 mm, SLF-0201; Bio-Rad) were attached to the cover slide. The slide was then washed three times with filtered PBS prior to imaging. Gold nanoparticles in PBS (100 nm, 753688; Sigma-Aldrich) were used as fiducial markers and were diluted to 2 μM and incubated on the plasma-cleaned glass cover slides for 5 min followed by two gentle washes with PBS. Aβ42 aggregates were diluted to 2 μM in PBS + EDTA [(ethylenedinitrilo)tetraacetic acid] and incubated on the cover slide for 5 min followed by two washes with PBS + EDTA. dSTORM experiments were carried out in dSTORM buffer of PBS-Tris (50 mM), glucose (0.5 mM), glucose oxidase (1.3 μM), catalase (2.2 μΜ), and mercaptoethylamine (MEA) (50 mM). MEA was added to buffer immediately prior to imaging. DesAb-AF647 (70 μM) was diluted into the STORM buffer to a concentration of 100 pM and 50 μL added to the Aβ42-coated cover slide.
dSTORM Imaging.
Imaging was performed on a bespoke TIRF microscope using a 641-nm excitation laser (Obis, 0.5 kW⋅cm−2; Coherent). The beam was circularly polarized using a quarter-wave plate and then expanded, collimated, and aligned parallel to the optical axis at the edge of an objective lens (UPlanSApo, 100×, 1.4 N.A.; Olympus) mounted on an inverted optical microscope (Ti2-E, Eclipse; Nikon Corporation). Emission was collected by the same objective lens and separated from excitation light using a dichroic mirror (Di01-R405/488/561/635; Semrock) and passed through appropriate emission filters (FF01-692/40-25; Semrock). The fluorescence was then expanded and focused on an electron-multiplying charge-coupled device (Evolve 512 Delta; Photometrics) for imaging. Image stacks of 10,000 frames were recorded in frame-transfer mode at 50 frames per second with an exposure time of 20 ms. Each pixel on the final image was equal to 106 nm. Super-resolution images were reconstructed using the Drift Calculator and Peak Fit package (GDSC SMLM; University of Sussex) in ImageJ.
Microfluidic Diffusional Sizing.
The binding affinity of DesAb-O for Aβ42 fibrils was quantified in 20 mM sodium phosphate buffer (pH 8) by microfluidic diffusional sizing, which measures the hydrodynamic radius of fluorescently labeled species in their native state in solution (27). Aβ42 were sonicated (3/3 s on/off cycles) for 1 min on ice with 50% power before use in the binding experiments. Samples containing 0.54 nM AF47-DesAb-O and various concentrations of Aβ42 fibrils in PBS were equilibrated at 22 °C for at least 17 h. The hydrodynamic radius of AF647-DesAb-O at various concentrations of Aβ42 fibrils was recorded using a Fluidity One-W instrument (Fluidic Analytics Ltd) between 27 and 28 °C.
The dissociation constant Kd was calculated by direct nonlinear regression based on the fractional saturation as a function of the concentration of total ligand added:
with Δrtot, ro, and r being the maximum change in the DesAb-O radius between the bound and unbound state, the radius in the absence of any ligand, and the measured radius at fibril mass concentration [Ltot], respectively. Furthermore, [Btot] and n are the DesAb-O concentration used and the number of fibril binding sites per DesAb-O molecule. Since the binding mode of DesAb-O with fibrils is not known, the effective binding affinity was determined through fitting the molar stoichiometry of DesAb-O and fibrils, finding that on average about 200 Aβ monomers within the fibrils bind each DesAb-O molecule, with low micromolar affinity.
ELISA with Aliquots from Aggregation Reactions.
Twenty-microliter aliquots were taken at specific incubation times and immobilized on a 96- or 384-well Maxisorp ELISA plate (Nunc) with no shaking for 1 h at room temperature. Experiments were then performed as described above (ELISA-Based Screening of the Antibodies). Experiments in the presence of the compound bexarotene were performed using the same protocol on aggregation reactions supplemented with 5 molar excess bexarotene.
Strains of C. elegans.
The following strains of C. elegans were used: dvIs100 [unc-54p::A-beta-1-42::unc-54 3′-UTR + mtl-2p::GFP] (GMC101), which produces constitutive expression of green fluorescent protein (GFP) in intestinal cells, and unc-54p::A-beta-1-42, which expresses full-length human Aβ42 in body-wall muscle cells that aggregates in vivo; shifting L4 or young adult animals from 20 °C to 25 °C causes paralysis (29). C. elegans var Bristol (N2) was used as a control strain. Generation time is about 3 d. The control strain was isolated from mushroom compost near Bristol, England (35).
Media for C. elegans.
Standard conditions were used for the propagation of C. elegans. Briefly, the animals were synchronized by hypochlorite bleaching, hatched overnight in M9 (3 g/L KH2PO4, 6 g/L Na2HPO4, 5 g/L NaCl, and 1 µM MgSO4) buffer, and subsequently cultured at 20 °C on nematode growth medium (NGM) (CaCl2 1mM, MgSO4 1 mM, cholesterol 5 µg/mL, 250 µM KH2PO4, pH 6, agar 17 g/L, NaCl 3 g/L, and casein 7.5 g/L) plates seeded with the E. coli strain OP50. Saturated cultures of OP50 were grown by inoculating 50 mL of LB medium (tryptone 10 g/L, NaCl 10 g/L, and yeast extract 5 g/L) with OP50 and incubating the culture for 16 h at 37 °C. NGM plates were seeded with bacteria by adding 350 µL of saturated OP50 to each plate and leaving the plates at 20 °C for 2 to 3 d. On day 3 after synchronization, the animals were placed on NGM plates containing 5-fluoro-2′deoxy-uridine (FUDR) (6.83 nM, unless stated otherwise) to inhibit the growth of offspring. FUDR plates were seeded with bacteria by adding 350 µL of 10× concentrated OP50 solution to ensure starvation did not occur for the lifespan of the worm. Concentrated OP50 solution was obtained by centrifuging 1 L of saturated OP50 culture at 5,000 rpm for 15 min and suspending the resultant pellet in 100 mL sterile water.
Staining of the Aggregates Using the Fluorescent Probe NIAD-4.
To visualize the amount of aggregates in the worms, live transgenic were incubated with 1 μM NIAD-4 (0.1% dimethyl sulfoxide in M9 buffer) for 4 h at room temperature. After staining, animals were allowed to recover on NGM plates for about 24 h to allow destaining via normal metabolism. Stained animals were mounted on 2% agarose pads containing 40 mM NaN3 as anesthetic on glass microscope slides for imaging. Images were captured using a Zeiss Axio Observer D1 fluorescence microscope (Carl Zeiss Microscopy GmbH) with a 20× objective and a 49004 ET-CY3/TRITC filter (Chroma Technology Corp.). Fluorescence intensity was calculated using ImageJ and then normalized as the corrected total cell fluorescence. Only the head region was considered because of the high background signal in the guts. All experiments were carried out in triplicate, and the data from one representative experiment are shown.
Automated Motility Assay on Agar Plates and Imaging of the Aggregates.
All C. elegans populations were cultured at 20 °C and developmentally synchronized from a 4-h egg lay. At 64 to 72 h after egg lay (time 0), individuals were shifted to 24 °C and transferred to FUDR plates and body movements were assessed over the times indicated. Videos were analyzed using a custom-made tracking code.
ELISA with C. elegans Protein Extracts.
GMC101 and N2 worms were incubated on FUDR plates until days 0, 1, 3, 5, 7, and 10 of adulthood, according to what is reported in the text for each experiment. At a specific time the worms were washed out of the plates and snap-frozen using liquid nitrogen. For sample preparation, around 1,000 animals were thawed and resuspended in 500 μL PBS supplemented with EDTA-free protease inhibitor tablets 1× (Roche), after which they were sonicated (5 × 45 s, 50% cycles, 50% maximum power on ice) using a Bandelin Sonopuls HD 2070. After sonication, the lysates were centrifuged at maximal speed using a bench centrifuge and 20 μL of supernatant (5 μg) were loaded on a 96 Maxisorp ELISA plate (Nunc) with no shaking overnight at 4 °C. ELISAs were conducted as described above (ELISA-Based Screening of the Antibodies).
Use and Care of the Mice.
Experiments were performed according to the Animals Scientific Procedures Act UK (1986). Tg(PDGFB-APPSwInd)20Lms/2Mmjax (J20) heterozygous mice were housed at University College London, maintained on a 12-h light/dark cycle and provided with food and water for ad libitum consumption. Transgenic or nontransgenic female littermates (16 to 18 mo old) were used for this experiment (n = 3 per group).
Immunofluorescence Staining.
J20 mouse brains were collected, immediately fixed overnight at 4 °C with 4% paraformaldehyde in PBS, then washed twice with PBS and immersed in 30% sucrose/PBS for 2 d. Next, brain samples were frozen in precooled isopentane on dry ice, and 50-µm sections were cut on a cryostat and stored in ethylene glycol-based cryoprotectant (30% glycerol and 30% ethylene glycol in 0.1 M sodium/potassium phosphate buffer, pH 7.4). Brain sections were washed twice in PBS and then incubated for 30 min with 10 μM NIAD-4 (0.1% dimethyl sulfoxide [DMSO] and 0.5% Triton in PBS). The samples were washed three times with PBS. Finally, brain slices were mounted in Fluoromount-G (SouthernBiotech) on a glass slide for imaging. Images of the hippocampus area were captured using a Leica SP8 confocal microscope. NIAD-4 fluorescence was excited at 488 nm and emission recorded at 550 to 650 nm. Images were taken using a 20× (N.A. 0.72) dry objective, with 1,024 × 1,024 image resolution and 15 z-steps of 0.5 µm.
ELISA with Mouse Protein Extracts.
Thirty-microliter aliquots (5 μg) were immobilized on a 96 Maxisorp ELISA plate (Nunc) with no shaking overnight at 4 °C. Experiments were then performed as described above (ELISA-Based Screening of the Antibodies).
Preparation of Aβ40 and α-Synuclein Oligomers.
For the oligomers of Aβ40, lyophilized Aβ40 (1 mg/mL) was solubilized overnight in 300 µL hexafluoroisopropanol. Solvent was gently evaporated off with nitrogen and protein resuspended in 100% DMSO. Two sonication steps of 10 min were performed, after which the protein was resuspended at 100 µM in 20 mM sodium phosphate buffer and 200 μM ZnCl2, pH 6.9, for 20 h at 20 °C. Samples were centrifuged (15,000 rcf, 20 °C, 15 min) and the supernatant was removed. Oligomers were resuspended in buffer (20 mM Tris and 100 mM NaCl) with thorough mixing. For the oligomers of α-synuclein, 6 mg of lyophilized protein was resuspended in PBS to give a final concentration of 800 μM and passed through a 0.22-μm cutoff filter immediately before incubation at 37 °C for 20 under quiescent conditions. Small fibrils were removed by ultracentrifugation for 1 h at 90,000 rpm (TLA-120.2 Beckman rotor). The excess of monomeric protein and the low levels of small oligomers were removed by filtration (using 100-kDa cutoff membranes).
ANS Binding Measurements.
Ten-micromolar oligomers (in monomer equivalents) were incubated for 2 h in the absence and presence of an equimolar concentration of the different DesAbs at 20 °C. Subsequently, ANS (Sigma-Aldrich) was added to a final concentration of 30 µM (i.e., threefold excess dye). Emission spectra were recorded using a plate reader (BMG Labtech) with excitation at 380 nm. Duplicate samples are shown representative of three independent experiments that gave consistent results.
ELISA with Aβ40 Oligomers.
Forty microliters of 5 μM DesAb solutions were used to coat the wells of a 96-well Maxisorp ELISA plate (Nunc) with no shaking for 1 h at room temperature. The plate was then washed three times with PBS and incubated in PBS and 5% BSA under constant shaking overnight at 4 °C. The day after the plate was washed six times with PBS and then incubated with 40-μL solutions of 5 µM oligomers in PBS under constant shaking either for 1 h at room temperature or overnight at 4 °C. At the end of this incubation, the plate was washed six times with PBS and incubated with 40-μL solutions of the mouse monoclonal anti-Aβ antibody [6E10] (1:2,000 dilution; Absolute Antibody Ltd). After six additional washes with PBS and 0.02% Tween-20, the plate was incubated with 40 μL of goat anti-mouse IgG (H+L) secondary antibody and HRP conjugate (1:2,000 dilution; Life Technologies) in PBS and 5% BSA for 1 h at room temperature. Finally, the plate was washed six times with PBS and 0.02% Tween-20 and bound oligomers were quantified by using 1-Step Ultra TMB-ELISA Substrate Solution (Thermo Fisher Scientific) according to the manufacturer’s instructions and the absorbance at 450 nm was measured by means of a CLARIOstar plate reader (BMG Labtech).
Dot-Blot with Aβ40 Oligomers.
Solutions of DesAb-O (4 and 2 μM) were spotted (3.5 μL) on a 0.2-μm-pore-size nitrocellulose membrane (GE Healthcare). As negative control, samples with same amounts of DesAb-PYGSMYVHS, which showed the weakest binding to Aβ40 oligomers in in the ANS binding measurements, were used. The blots were blocked in PBS and 5% BSA overnight at 4 °C. Then, they were incubated in solutions containing 5 μM Aβ40 oligomers in PBS overnight at 4 °C. Blots with bound oligomers were then probed with the mouse monoclonal anti-Aβ antibody [6E10] (1:2,000 dilution; Absolute Antibody Ltd) and with goat anti-mouse IgG (H+L) secondary antibody and AF488 conjugate (1:5,000 dilution; Life Technologies).
.
Kinetic Analysis.
The time evolution of the total fibril mass, M(t), in the absence of seeds is described by the following integrated rate law:
where the kinetic parameters B+, C+, k, , and have been previously described in detail (8) and are functions of the two combinations of the microscopic rate constants k+kn and k+k2, where kn, k+, and k2 are the primary nucleation, elongation, and secondary nucleation rate constants, respectively.
The above equation was used to identify the nucleation rate constants inhibited by the DesAb-O to describe the macroscopic aggregation profiles shown in SI Appendix, Figs. S5–S7 and comparing the set of microscopic rate constants k+kn and k+k2 to describe the time evolution of the fibril formation in the absence and presence of antibody.
The apparent rate constant k+ was derived as the rate of formation of fibrils within the first 20% of monomer conversion, r = 2k + P0m, where P0 is the number of seeds introduced in the system and m is the initial monomer concentration. The relative decrease in the apparent rate constant k+ was evaluated by dividing the rate in the presence of DesAb-O with the value calculated in its absence. Finally, the decreases in kn and k2 were calculated by dividing the decreases in k+k2 and k+kn obtained under unseeded conditions by the decrease in k+ derived from the seeded aggregation profiles.
ITC.
Measurements were performed on a MicroCal iTC200 (Malvern Panalytical Ltd) at 25 °C. The DesAb-O (100 μM) was injected 16 times into a solution containing 10 μM of either monomers of Aβ40, stabilized Aβ40 oligomers, or stabilized oligomers of α-synuclein. The first injection was 0.2 μL; all other injections were 2 μL. Aβ40 experiments were carried out in 20 mM Tris, pH 7.4, and 100 mM NaCl. Additionally, all solutions for the experiments involving monomeric Aβ40 contained 0.05% DMSO to ensure that the peptide was in its monomeric form. α-Synuclein solutions were carried out in standard PBS. Each injection occurred at 3-min intervals. Heats of dilution, obtained by separately injecting the antibody into the corresponding buffer and the buffer into either the monomer or oligomer solutions, were subtracted from the final data. The corrected heats were divided by the number of moles injected and analyzed using Origin 7.0 software (OriginLab).
Data Availability Statement.
All data are provided in the main text and SI Appendix.
Supplementary Material
Acknowledgments
We thank Ms. Swapan Preet and Ms. Ewa Klimont for the expression and purification of Aβ42, and Dr. Georg Meisl and Ms. Catherine Xu for helpful discussions. F.A.A. thanks UK Research and Innovation (Grant MR/S033947/1) and the Alzheimer’s Society UK (Grants 317 and 511) for support. G.T.H. is supported by the Gates Cambridge Trust and Rosalind Franklin Research Fellowship at Newnham College. P.S. is supported by a Borysiewicz Fellowship from the University of Cambridge.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919464117/-/DCSupplemental.
References
- 1.Patterson C., “World Alzheimer report 2018” (Alzheimer’s Disease International, London, 2018). [Google Scholar]
- 2.Jack C. R. Jr. et al.; Contributors , NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 14, 535–562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardy J., Selkoe D. J., The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 297, 353–356 (2002). [DOI] [PubMed] [Google Scholar]
- 4.De Strooper B., Karran E., The cellular phase of Alzheimer’s disease. Cell 164, 603–615 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Holtzman D. M., Morris J. C., Goate A. M., Alzheimer’s disease: The challenge of the second century. Sci. Transl. Med. 3, 77sr71 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobson C. M., Protein folding and misfolding. Nature 426, 884–890 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Knowles T. P. et al., An analytical solution to the kinetics of breakable filament assembly. Science 326, 1533–1537 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Cohen S. I. et al., Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. U.S.A. 110, 9758–9763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haass C., Selkoe D. J., Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 8, 101–112 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Michaels T. C. et al., Dynamics of oligomer populations formed during the aggregation of Alzheimer’s Aβ42 peptide. Nat. Chem. 12, 445–451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benilova I., Karran E., De Strooper B., The toxic Aβ oligomer and Alzheimer’s disease: An emperor in need of clothes. Nat. Neurosci. 15, 349–357 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Cremades N. et al., Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell 149, 1048–1059 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fusco G. et al., Structural basis of membrane disruption and cellular toxicity by α-synuclein oligomers. Science 358, 1440–1443 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Winter G., Griffiths A. D., Hawkins R. E., Hoogenboom H. R., Making antibodies by phage display technology. Annu. Rev. Immunol. 12, 433–455 (1994). [DOI] [PubMed] [Google Scholar]
- 15.Bradbury A. R., Sidhu S., Dübel S., McCafferty J., Beyond natural antibodies: The power of in vitro display technologies. Nat. Biotechnol. 29, 245–254 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leader B., Baca Q. J., Golan D. E., Protein therapeutics: A summary and pharmacological classification. Nat. Rev. Drug Discov. 7, 21–39 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Sehlin D. et al., Antibody-based PET imaging of amyloid beta in mouse models of Alzheimer’s disease. Nat. Commun. 7, 10759 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kayed R. et al., Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Yang T. et al., Target engagement in an Alzheimer trial: Crenezumab lowers Aβ oligomers in cerebrospinal fluid. Ann. Neurol. 86, 215–224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meli G. et al., Conformational targeting of intracellular Aβ oligomers demonstrates their pathological oligomerization inside the endoplasmic reticulum. Nat. Comm. 5, 3867 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aprile F. A. et al., Selective targeting of primary and secondary nucleation pathways in Aβ42 aggregation using a rational antibody scanning method. Sci. Adv. 3, e1700488 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sormanni P., Aprile F. A., Vendruscolo M., Rational design of antibodies targeting specific epitopes within intrinsically disordered proteins. Proc. Natl. Acad. Sci. U.S.A. 112, 9902–9907 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sormanni P., Aprile F. A., Vendruscolo M., Third generation antibody discovery methods: In silico rational design. Chem. Soc. Rev. 47, 9137–9157 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Colvin M. T. et al., Atomic resolution structure of monomorphic Aβ42 amyloid fibrils. J. Am. Chem. Soc. 138, 9663–9674 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chia S. et al., SAR by kinetics for drug discovery in protein misfolding diseases. Proc. Natl. Acad. Sci. U.S.A. 115, 10245–10250 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruggeri F. S. et al., Nanoscale studies link amyloid maturity with polyglutamine diseases onset. Sci. Rep. 6, 31155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arosio P. et al., Microfluidic diffusion analysis of the sizes and interactions of proteins under native solution conditions. ACS Nano 10, 333–341 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Habchi J. et al., An anticancer drug suppresses the primary nucleation reaction that initiates the production of the toxic Aβ42 aggregates linked with Alzheimer’s disease. Sci. Adv. 2, e1501244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McColl G. et al., Utility of an improved model of amyloid-beta (Aβ1−42) toxicity in Caenorhabditis elegans for drug screening for Alzheimer’s disease. Mol. Neurodegener. 7, 57 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mucke L. et al., High-level neuronal expression of abeta 1-42 in wild-type human amyloid protein precursor transgenic mice: Synaptotoxicity without plaque formation. J. Neurosci. 20, 4050–4058 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright A. L. et al., Neuroinflammation and neuronal loss precede Aβ plaque deposition in the hAPP-J20 mouse model of Alzheimer’s disease. PLoS One 8, e59586 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong S. et al., Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mannini B. et al., Stabilization and characterization of cytotoxic Aβ40 oligomers isolated from an aggregation reaction in the presence of zinc ions. ACS Chem. Neurosi. 19, 2959–2971 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Gasteiger E. et al., “Protein identification and analysis tools on the ExPASy Server” in The Proteomics Protocols Handbook, Walker J. M., Ed. (Springer, 2005), pp. 571–607. [Google Scholar]
- 35.Brenner S., The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are provided in the main text and SI Appendix.