Abstract
Circulating tumor cells (CTCs) are shed into the bloodstream from primary tumors, but only a small subset generates metastases. We conducted an in vivo genome-wide CRISPR activation screen in breast cancer patient-derived CTCs to identify genes that promote their distant metastasis in mice. Genes coding for ribosomal proteins and regulators of translation were enriched in this screen. Overexpression of RPL15, which encodes a component of the large ribosomal subunit, increased metastatic growth in multiple organs and selectively enhanced translation of other ribosomal proteins and cell cycle regulators. RNA-sequencing of freshly-isolated CTCs from breast cancer patients revealed a subset with strong ribosome and protein synthesis signatures; these CTCs expressed proliferation and epithelial markers and correlated with poor clinical outcome. Therapies targeting this aggressive subset of CTCs may merit exploration as potential suppressors of metastatic progression.
Circulating tumor cells (CTCs) shed into the bloodstream from primary tumors sustain physical, oxidative and other environmental stresses before disseminating to distant organs, where only a small subset may be competent to generate metastatic tumors (1, 2). For hormone receptor-positive (HR+) breast cancer, which may recur at distal sites many years after tumor resection and adjuvant therapy (3), defining the mechanisms that regulate survival and proliferation of CTCs presents an opportunity to suppress such delayed metastatic recurrence. Using a microfluidic platform to enrich viable CTCs from the blood of patients with HR+ metastatic breast cancer, we generated a panel of ex vivo CTC cultures that are highly tumorigenic when injected into the mammary fat pad of immunodeficient NSG mice (4-7). After direct intravenous tail vein injection, however, CTCs that are trapped in the lung fail to generate metastatic tumors for up to 6 months, in contrast to standard human breast cancer cell lines, such as MDA-MB-231-LM2 (Fig. 1A).
Fig. 1. In vivo genome-wide CRISPR activation screen.
(A) Left panel: Whole body bioluminescence monitoring of NSG mice injected via tail vein with GFP-luciferase tagged CTC lines or MDA-MB-231-LM2 cells (n = 3 mice per cell line). Right panel: Representative images of the bioluminescent signal at day 22 and corresponding lung histologic sections stained with anti-GFP to identify tumor cells. Scale bar: 100μm. (B) Diagram of in vivo CRISPR activation screening in CTCs. (C) Classification of known functions of the top 250 genes identified in the combined screen ranking. (D) Distribution of combined screen scores, demonstrating that only the top few hundred genes are enriched. Top genes related to translation are highlighted. (E) Crystal structure of the large and small subunits of the eukaryotic ribosome (11) highlighting the locations of RPL13, RPL15, and RPL35 as well as direct RPL13:RPL35 and RPL15:RPL35 interactions. Ribosome structural features indicated include the central protuberance (CP), exit channel and L1 stalk.
The absence of blood-borne lung metastasis by patient-derived, tumorigenic breast cancer cells provides an experimental system to identify genes in CTCs that promote metastases. We therefore conducted a genome-wide CRISPR activation (CRISPRa) screen using the synergistic activation mediator system; this system combines modified single guide RNAs (sgRNAs) and a catalytically inactive Cas9 (dCas9) to localize protein transactivators to the promoter of a target gene, resulting in gene-specific transcriptional activation (8). The screen included 70,290 sgRNAs, covering all 23,430 human coding isoforms in the RefSeq database. Two different patient-derived breast cancer CTC lines (Brx-82, Brx-142) were each infected with the lentiviral library of sgRNAs and injected into mice via tail vein (Fig. 1B). Two months after tail vein injection, the mice were sacrificed, and, because of the low background of metastatic burden in lungs in this model, bulk lung tissue from each mouse was sequenced to identify enriched sgRNAs. The majority of sgRNAs exhibited reduced or no representation, while a small set showed significant enrichment compared to input (Supplemental Data S1). Genes were scored using an algorithm calculating the level of enrichment of corresponding sgRNAs in the two independent breast CTC lines, allowing for the identification of genes broadly capable of enhancing metastatic potential.
Many of the highest scoring genes were established oncogenes or genes involved in processes critical to cancer progression, including transcriptional and translational regulation, cellular motility, and cell cycle progression (Fig. 1C), thus supporting the validity of the screen. Among the protein synthesis-related hits were MTOR and RPS6KA5 and several structural components of the large subunit of the ribosome, RPL15 (eL15), RPL35 (uL29), and RPL13 (eL13) [revised ribosomal protein nomenclature (9)], with RPL15 ranking tenth across the entire genome (Fig. 1D). These three structural ribosomal proteins (RP) cluster on the solvent-exposed surface near the exit tunnel on the back of the 60S ribosome, and they physically interact with each other (10, 11) (Fig. 1E, fig. S1). Given their unexpected link to metastasis, we chose these RPs for further study.
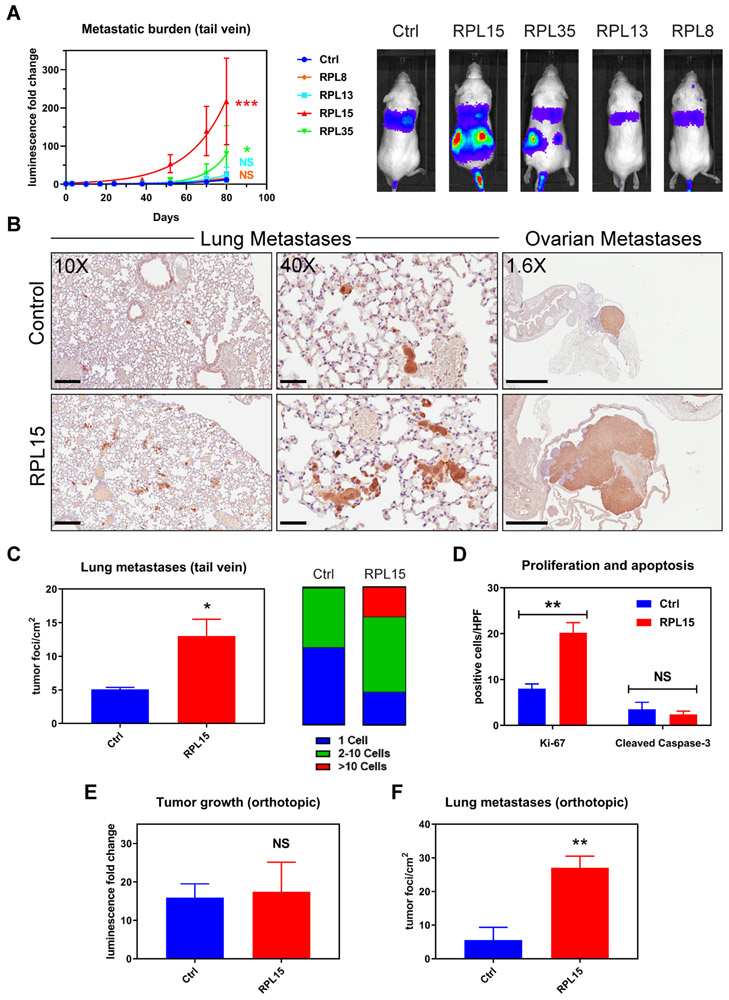
To exclude sgRNA- or CRISPR-specific effects, we first validated the effects of RP expression on metastasis using a cytomegalovirus (CMV) promoter-driven, lentiviral encoded vector to ectopically express these proteins in CTCs, followed by tail vein injection of the cells (fig. S2). Three-fold ectopic expression of RPL15 in a cultured CTC line (RPL15-CTCs) dramatically increased metastatic burden in mice as determined by total body bioluminescence (Fig. 2A). Consistent with its lower sgRNA enrichment, RPL35 also increased metastatic burden, but to a lesser extent than RPL15. In contrast, RPL8 (uL2), which was not a top hit in our screen, did not increase metastatic burden, indicating that the pro-metastatic phenotype is not a general property of all RPs. Histological analysis of the lungs from RPL15-CTC inoculated mice showed an increase in the number of metastatic lesions, with a strong shift from single cells to multicellular foci (p = 0.0299) (Fig. 2, B and C). Mice injected with RPL15-CTCs also developed massive ovarian metastases, compared with small nodular ovarian lesions in control animals (Fig. 2B). A similar increase in the number and size of metastatic lesions was observed in mice injected with RPL35-CTCs (fig. S3). The increased tumor burden correlated with increased proliferation (Ki-67), rather than reduced apoptosis (cleaved caspase-3) (Fig. 2D and fig. S4). To extend these observations beyond direct intravascular inoculation of CTCs, we tested the effect of RPL15 overexpression after orthotopic tumor formation in the mammary fat pad. RPL15-CTCs generated primary tumors of similar size as control, but their spontaneous lung metastases were strikingly increased in number (average 8.2-fold increase in the number of metastatic foci, p = 0.0088), consistent with strongly enhanced metastatic potential (Fig. 2, E and F). Together, these findings establish RPL15 as a potent positive regulator of in vivo metastatic potential in breast cancer CTCs.
Fig. 2. Validation of pro-metastatic effect of RPL15 overexpression.
(A) Left panel: Whole body luminescence monitoring of NSG mice injected via tail vein with CTCs overexpressing RPL8, RPL13, RPL15, or RPL35 (n = 4 mice per group). Curve fit by least squares method. P values calculated by the extra sum-of-squares F test. Right panel: Representative images of the bioluminescent signal one month after injection. (B) Representative sections of lung (left and middle panels) and ovarian (right panel) histology after staining with anti-GFP antibody (brown) and counter-stained with hematoxylin. Average long axis diameter of ovarian metastases in mice injected with RPL15-CTCs vs control: 9.1mm vs 2.4 mm respectively. Scale bars: Left panel 200μm; Middle panel 50μm; Right panel 2mm. (C) Quantitation of the number and size of tumor foci per cm2 identified by anti-GFP staining of lung histologic sections from mice injected with RPL15-CTCs or control. (D) Quantitation of the number of cells positive for Ki-67 or cleaved caspase-3 per high powered field by immunohistochemical staining of ovarian histologic sections. (E) Fold change in tumor bioluminescence after mammary fat pad injections of RPL15-CTCs or control at the terminal time point of day 78 (n = 4 mice per group, 2 tumors per mouse). (F) Quantitation of the number of tumor foci per cm2 identified by anti-GFP staining of lung histologic sections from mice after orthotopic injection of RPL15-CTCs or control. Error bars represent SEM. P values calculated by two-tailed unpaired Student’s t test. ***: p<0.001, **: p<0.01 *: p<0.05, NS: p>0.05.
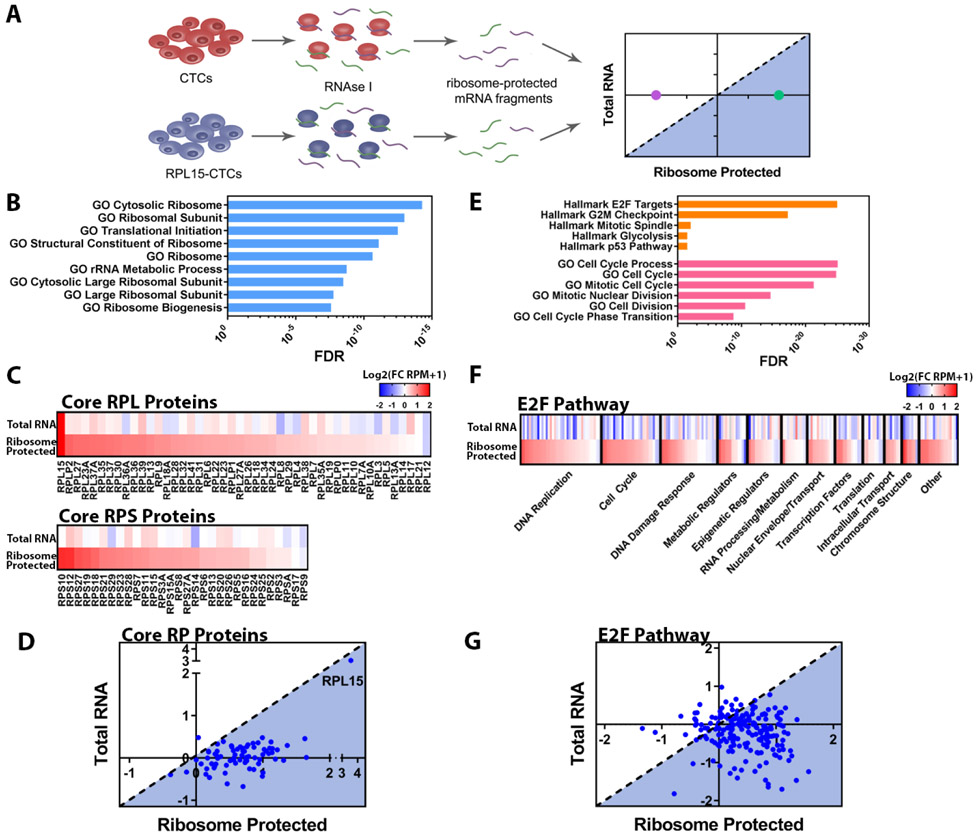
As components of large and complex structures, RPs are reported to have highly coordinated expression to ensure the fidelity of ribosome subunit biogenesis and assembly (12). Altered expression of an individual component, such as RPL15, may alter ribosome translational efficiency, either globally or for specific subsets of mRNAs (13). We performed ribosomal profiling in control and RPL15-CTCs to determine the global ribosome occupancy of mRNA transcripts, identify changes in translational preference, and calculate the translational efficiency for any given transcript (ratio of ribosome-bound mRNA to total mRNA) (14) (Fig. 3A and fig. S5). Consistent with ectopic expression, RPL15-CTCs showed an 8-fold increase in total RPL15 transcripts and a 12-fold increase in ribosome-protected RPL15 transcripts. In total, 183 genes were significantly enriched for ribosome-protected fragments in RPL15-CTCs, while 250 genes were significantly depleted (fold change > 2.0; FDR < 0.05). Gene Set Enrichment Analysis (GSEA) of these ribosome-protected transcripts identified other RP genes and regulators of translation as the most highly enriched ribosome-protected signatures in RPL15-CTCs (Fig. 3B).
Fig. 3. RPL15 overexpression promotes translation of core ribosomal proteins and E2F pathway proteins.
(A) Schema illustrating ribosome profiling of control CTCs or RPL15-CTCs. (B) Gene Set Enrichment Analysis of transcripts preferentially bound by ribosomes in RPL15-CTCs. The most enriched ribosomal/translational GO gene sets and associated FDR values are shown. (C) Heat map representing the fold change of RPL15-CTCs relative to control for each RP gene for total RNA sequencing and ribosome profiling. Upper panel represents RPL proteins and lower panel represents RPS proteins. (D) Scatter plot representing the translational efficiency of individual RP gene transcripts. The y-axis represents the log2(fold change in RNA-seq), and the x-axis represents the log2(fold change in ribosome profiling). The shaded region represents transcripts that have increased translational efficiency relative to the level of the transcript. (E) Gene Set Enrichment Analysis of transcripts preferentially bound by ribosomes in RPL15-CTCs. The most enriched Hallmarks of Cancer gene sets from the Broad Molecular Signatures Database and most enriched cell cycle related Gene Ontology (GO) gene sets, and associated FDR values are shown. (F) Heat map representing fold change of RPL15-CTCs relative to control for each gene within the Hallmark E2F Targets gene set for total RNA sequencing and ribosome profiling. Genes were categorized according to their function and ordered based on the fold change in the ribosome profiling. (G) Scatter plot representing the translational efficiency of individual transcripts within the Hallmark E2F Targets gene set. The y-axis represents the log2(fold change in RNA-seq), and the x-axis represents the log2(fold change in ribosome profiling). The shaded region represents transcripts that have increased translational efficiency relative to the level of the transcript. Ribosome profiling was performed in duplicate.
Virtually all 76 core ribosomal proteins, from both the large and small subunits, were markedly increased (mean 1.6-fold) among the ribosome-protected transcripts (Fig. 3C), suggesting that overexpression of RPL15 leads to coordinate translational upregulation of the core RPs (Fig. 3D). While a conserved 5’ UTR (untranslated region) element in the RP genes is known to drive the stoichiometric coordination of RPs (15, 16), the ability of a single RP to drive translation of all other RPs is unexpected. To validate these ribosome-protection experiments, we conducted polysome profiling and found that, compared with control CTCs, RPL15-CTCs had an increased proportion of RP transcripts within the polysome fractions (fig. S6A). Polysome profiling also identified a significant increase in the global polysome-to-monosome ratio in RPL15-CTCs (fig. S6, B and C), indicating a global enhancement in the proportion of ribosomes found within polysomes, in addition to the specific enrichment for RP transcripts. Indeed, RPL15-CTCs had increased total RNA, a reflection of increased ribosomal RNA (rRNA) content, which accounts for the majority of cellular RNA (17) (fig. S7A). Finally, RPL15-CTCs had increased global translational activity, as measured by O-propargyl-puromycin (OP-Puro) and L-Azidohomoalanine (L-AHA) incorporation, two orthogonal measures of total cellular translation (Figure S7B-C).
In addition to the RP genes, GSEA of ribosome-protected transcripts from RPL15-CTCs identified cell proliferation pathways, including downstream targets of the transcription factor E2F, as highly enriched among ribosome-protected signatures (Fig. 3E). The absence of E2F pathway enrichment by total cellular RNA-sequencing confirmed that upregulation of this proliferative program represents increased translational efficiency (Fig. 3, F to G). Quantitative proteomic analysis of E2F targets identified a positive correlation between ribosomal occupancy of E2F target mRNAs and protein level, confirming a functional effect of RPL15-induced increased translational efficiency (fig. S8). The RPL15-induced proliferative signature is not correlated with increased ERBB2 expression and therefore is independent of the previously-reported heterogeneity of Her2 protein expression in breast CTCs (7). Together, these findings demonstrate that overexpression of the ribosomal structural protein RPL15 augments global protein translation, with a selectively enhanced impact on translation of transcripts encoding other RPs and proliferation programs.
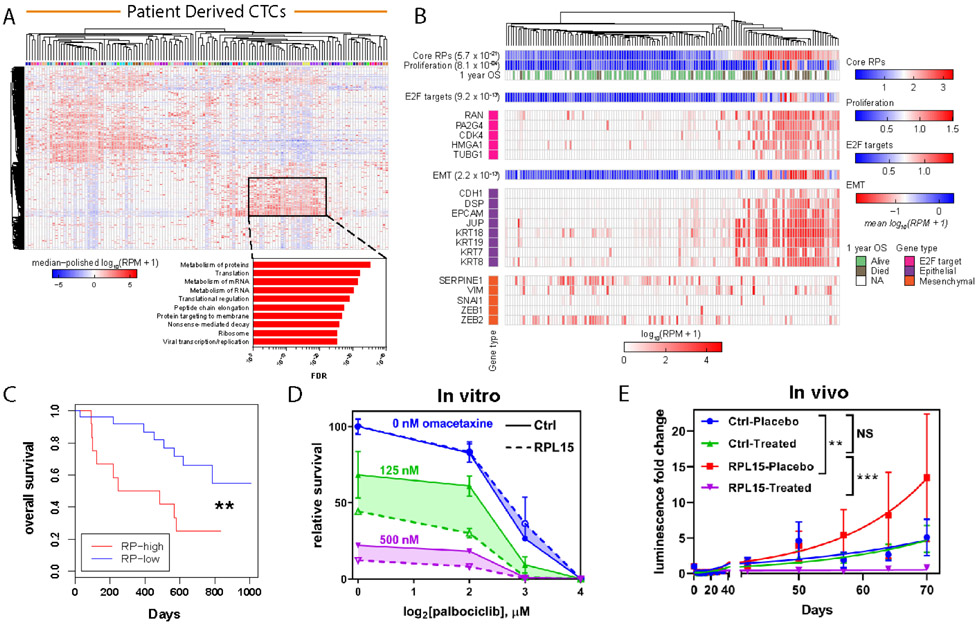
The CTC-iChip microfluidic device (4) enables isolation of rare viable CTCs directly from whole blood specimens of patients with cancer, with RNA quality compatible with single-cell RNA sequencing (RNA-seq) (18-20). We interrogated RNA-seq profiles of 135 freshly isolated single CTCs or CTC-clusters from 45 patients with HR+ metastatic breast cancer: unsupervised clustering based on the 2000 most variant genes revealed a subset of CTCs with strikingly increased expression of multiple RP genes (Fig. 4A). GSEA of the genes driving this subset clustering demonstrated highly significant enrichment for RP genes and genes involved with translational regulation (FDR ranging from 10−25 to 10−35) (Fig. 4A). Supervised clustering of CTCs based on expression of RP genes confirmed coordinate expression of the core RPs, allowing division of the clinical dataset into CTCs with RP-high (33% of all patient-derived CTCs) and RP-low gene expression (67%) (fig. S9). RP-high versus RP-low expression in patient-derived CTCs was again highly correlated with expression of E2F targets (p = 9.2 x 10−13) and a proliferation signature (p = 8.1 x 10−4) (21) (Fig. 4B and fig. S10). To validate these findings, we interrogated a second, independent cohort of single-cell RNA-seq data that we had previously derived from 109 CTCs individually isolated from 33 patients with metastatic breast cancer (7). As in our first cohort, expression of RP genes clearly identified a distinct RP-high CTC subset (44% of all CTCs) and an RP-low CTC subset (56%), with high RP expression again correlated with E2F target gene expression (p = 9.4 x 10−13) (fig. S11 and fig. S12).
Fig. 4. Heterogeneity of RP expression in primary patient CTCs correlates with worse clinical outcome and sensitivity to translational and cell cycle inhibition:
(A) Upper panel: RNA-seq from CTCs enriched from whole blood with the iChip microfluidic device and isolated as single cells or clusters. Expression values represent log10(RPM+1), and the dataset was median polished. Dendrogram represents unsupervised clustering of the 2000 most variant genes within the dataset. Color bar identifies individual patients. Highlighted box represents a subset of CTCs with coordinately expressed RP genes. Lower panel: GSEA for KEGG and Reactome gene sets of the genes found in the highlighted box. (B) Heat map of the expression level of selected E2F target genes, epithelial markers and mesenchymal markers. Dendrogram represents supervised clustering of the CTC samples based on RP gene expression. OS bar indicates whether patient was alive one year after CTC sample collection. Color bar illustrates metagene analysis of core RPs, a proliferation signature, E2F targets, and an EMT signature and associated p values. (C) Kaplan-Meier analysis of the overall survival for patients with high average RP gene expression versus low average RP gene expression. The RP-high and RP-low subgroups were determined based on average RP gene expression for each patient blood draw. P value calculated by log rank test. (D) Dose response curves for RPL15-CTCs and control treated with increasing doses of palbociclib and omacetaxine. Shaded region represents the difference between RPL15-CTCs and control across tested concentrations of palbociclib. (E) Whole body luminescence monitoring of NSG mice after intracardiac injection with RPL15-CTCs or control and treatment with placebo or a combination of palbociclib and omacetaxine (n = 4-5 mice per condition). Error bars represent SEM. Curve fit by least squares method. P values calculated by the extra sum-of-squares F test. ***: p<0.001, **: p<0.01, NS: p>0.05.
Primary CTCs exhibit heterogeneous expression of epithelial and mesenchymal markers (22), and among these cells, the RP-high subset had significantly increased epithelial markers and decreased mesenchymal markers (Fig. 4B). To explore the link between epithelial-to-mesenchymal transition (EMT) and protein translation, we assessed translational changes in the canonical EMT model of transforming growth factor-β (TGF-β) treatment in MCF10A breast epithelial cells (23). Indeed, EMT induction by TGF-β in these cells suppressed global translational activity and rRNA expression (fig. S13, A to B). Polysome profiling revealed depletion in gene sets related to RPs and translation (fig. S13C). The coordinated and pronounced suppression in translation of RP transcripts (89% of RPs translationally downregulated, median downregulation of 1.25-fold) was accompanied by a smaller decrease in total RP mRNA content (76% of RPs transcriptionally downregulated, median downregulation of 1.06-fold), suggesting that EMT primarily mediates translational downregulation of RPs (fig. S13, D to E). Thus, in cancer cells circulating in the blood, persistent epithelial cell fate is associated with higher protein translation and proliferative potential.
Clinical outcome data were available for the first cohort of patients, making it possible to interrogate CTC single-cell RNA-seq data for markers correlated with patient survival (Supplemental Data S2). Using a Cox proportional hazards model, we identified 765 genes associated with worse Overall Survival (OS) (Hazard Ratio (HR): 1.25, FDR: 0.25) (fig. S14A). These genes overlapped significantly with genes associated with adverse outcomes in multiple published datasets (table S1). GSEA of these adverse outcome genes revealed highly significant enrichment of protein synthesis pathways and of the RPs themselves (e.g. FDR = 3.98 x 10−72 for Reactome: Translation) (fig. S14, B to C). High mean expression of all RP genes was associated with significantly worse OS (HR: 3.4, p = 0.0078) (Fig. 4C), as was elevated expression of RPL15 itself (HR: 3.4, p = 0.011) (fig. S15). Consistent with the contribution of RPL15 to cellular heterogeneity in advanced metastatic phenotypes, this correlation was less evident within bulk primary breast cancer tissues, although in one dataset (24), high RP expression was correlated with reduced Progression Free Survival (PFS) (RPL (large subunit): HR: 1.34, p = 0.00023; RPS (small subunit): HR: 1.34, p = 0.00057) (fig. S16).
The enhancement of tumorigenesis by eukaryotic initiation factors (eIFs) and other functional regulators of translation (12, 25-28) has led to the suggestion that targeting the translational machinery may be selectively toxic to malignant cells (29, 30). The FDA-approved translational inhibitor omacetaxine prevents the initial elongation step of protein synthesis by occupying the A-site cleft and preventing proper aminoacyl-tRNA positioning (31, 32). We found that omacetaxine inhibited global translation in breast cancer CTCs (fig. S17A). In combination with the cyclin-dependent kinase (CDK) 4/6 inhibitor palbociclib, which suppresses cell cycle progression and is used in breast cancer treatment (33), omacetaxine showed significantly increased efficacy against RPL15-CTCs compared to parental CTCs in vitro (Fig. 4D and fig. S17B). To extend these results to an in vivo mouse model, we used intracardiac injections to maximize mouse metastatic burden. In this model, omacetaxine-palbociclib treatment showed dramatically increased efficacy against metastases derived from RPL15-CTCs compared to parental CTCs (Fig. 4E). These early results suggest that simultaneous therapeutic targeting of the cell translational machinery and cell proliferation pathways may merit investigation as a method to suppress an aggressive subset of CTCs that are characterized by high expression of RP genes.
Based on two convergent lines of evidence—an in vivo CRISPRa screen for pro-metastatic genes in a mouse model and unsupervised clustering of single-cell RNA-seq from human breast cancer CTCs—we propose a model whereby the epithelial state mediates translational upregulation of RPs and regulators of cellular proliferation, thus contributing to the metastatic propensity of CTCs. The direct role of structural RPs in this phenotype extends the previously established contributions of mTOR and MAPK oncogenic signaling pathways (34, 35), translation initiation factors eIF4A, eIF4E and eIF4G (26, 27, 36, 37), and specific tRNA pools (38). In vivo genome-wide CRISPRa screening for a complex phenotype is unlikely to be saturating for all genes modulating protein translation, but the identification of hits in structural RP genes points to a previously unappreciated regulatory role of these genes.
Enlarged nucleoli, resulting from aberrant ribosomal biosynthesis, were first observed in cancer cells over 100 years ago (39) and germline mutations in RP genes, including RPL15, cause the cancer-associated ribosomopathy Diamond-Blackfan anemia (DBA) (40). Increased expression of some RPs has been reported in tumor specimens (41-43), although its significance has been unclear. In addition to demonstrating the functional consequences of RP expression on metastasis, our observation that RPL15 overexpression increases ribosomal content and global translation has mechanistic implications. Ribosome biogenesis is a highly coordinated process between rRNAs, RPs and assembly factors, with RPs implicated as critical platforms for ribosome assembly (44). In yeast, both Rpl15 and Rpl35 are recruited by ribosome assembly factors, including Nop4, Nop7, and Erb1, to establish a critical large subunit assembly platform, and both also bind rRNA at critical domain interfaces during pre-60S assembly (45, 46), suggesting that Rpl15 and Rpl35 may be rate-limiting for large subunit formation. Importantly, in both yeast and a model of DBA, altered ribosome concentrations lead to transcript-specific differences in translational efficiency (13, 47). While our data point to a RPL15-dependent increase in total ribosomal content, we cannot exclude the possibility of RPL15-specific effects leading to altered ribosome composition or extra-ribosomal RP functions (48, 49).
Finally, while mesenchymal cell states are associated with cancer cell migration, proliferative potential is correlated with epithelial phenotypes and the mesenchymal-to-epithelial transition (MET) (50). Indeed, our work suggests that the increased ribosomal content in epithelial cell fates may contribute to their enhanced metastatic potential. The identification of a well-demarcated subset of breast cancer CTCs defined by high ribosomal content and conferring an adverse prognosis raises the possibility of pharmacologic targeting of metastatic-competent cancer cells.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to all the patients who participated in this study. We thank W. Wu for artistic support in the creation of figures; L. Libby for technical support; and C. Van Rechem for technical assistance.
Funding:
This work was supported by NIH grant 2R01CA129933, the Breast Cancer Research Foundation, the Howard Hughes Medical Institute, and the National Foundation for Cancer Research (D.A.H.), NIH Quantum Grant 2U01EB012493 (M.T., D.A.H.), NIH grant U01CA214297 (M.T., D.A.H., S.M.), ESSCO Breast Cancer Research (S.M.), NCI grant K12CA087723, Susan G. Komen Foundation CCR15224703 (A.B.), T32GM007753 (R.Y.E., M.K.), 1F30CA232407-01 (R.Y.E.), 1F30CA224588-01 (M.K.), NIGMS R01GM100202 (S.V.), T32CA009361, Susan G. Komen Foundation PDF16376429 (N.V.J.), American Cancer Society 132140-PF-18-127-01-CSM, ASCO Young Investigator Award (D.S.M.).
Footnotes
Competing interests:
D.T.T., M.T., D.A.H., and S.M. are founders of and own equity in TellBio, Inc., which is involved with CTC therapeutics and diagnostic. At this time there has been no funding received or license that has been given to TellBio, Inc. for this work. D.T.T. is also a founder and owns equity ROME Therapeutics and PanTher Therapeutics, which is not related to this work. D.T.T. has received consulting fees from Merrimack Pharmaceuticals, Ventana Roche, Foundation Medicine, Inc., and EMD Millipore Sigma, which are not related to this work. D.T.T.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners Healthcare in accordance with their conflict of interest policies. The other authors declare no competing interests.
Data and materials availability:
Raw data from RNA-Seq of primary human breast cancer CTCs, ribosome profiling of RPL15-CTCs and microarray data from polysome profiling of TGF-β treated MCF10A cells were deposited in Gene Expression Omnibus (GEO) database under accession number GSE143626. All other data and materials are available from the corresponding author upon request.
References and Notes
- 1.Gomis RR, Gawrzak S, Tumor cell dormancy. Mol Oncol 11, 62–78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Micalizzi DS, Maheswaran S, Haber DA, A conduit to metastasis: circulating tumor cell biology. Genes Dev 31, 1827–1840 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan H et al. , 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. N Engl J Med 377, 1836–1846 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozkumur E et al. , Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med 5, 179ra147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karabacak NM et al. , Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc 9, 694–710 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu M et al. , Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 345, 216–220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jordan NV et al. , HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature 537, 102–106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konermann S et al. , Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ban N et al. , A new system for naming ribosomal proteins. Curr Opin Struct Biol 24, 165–169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klinge S, Voigts-Hoffmann F, Leibundgut M, Ban N, Atomic structures of the eukaryotic ribosome. Trends Biochem Sci 37, 189–198 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Ben-Shem A et al. , The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334, 1524–1529 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Pelletier J, Thomas G, Volarevic S, Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer 18, 51–63 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Cheng Z et al. , Small and Large Ribosomal Subunit Deficiencies Lead to Distinct Gene Expression Signatures that Reflect Cellular Growth Rate. Mol Cell 73, 36–47 e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS, The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7, 1534–1550 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robledo S et al. , The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA 14, 1918–1929 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamashita R et al. , Comprehensive detection of human terminal oligo-pyrimidine (TOP) genes and analysis of their characteristics. Nucleic Acids Res 36, 3707–3715 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T, Interdependence of cell growth and gene expression: origins and consequences. Science 330, 1099–1102 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Ting DT et al. , Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep 8, 1905–1918 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aceto N et al. , Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyamoto DT et al. , RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 349, 1351–1356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitfield ML et al. , Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell 13, 1977–2000 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu M et al. , Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339, 580–584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arima Y et al. , Rb depletion results in deregulation of E-cadherin and induction of cellular phenotypic changes that are characteristic of the epithelial-to-mesenchymal transition. Cancer Res 68, 5104–5112 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Mihaly Z et al. , A meta-analysis of gene expression-based biomarkers predicting outcome after tamoxifen treatment in breast cancer. Breast Cancer Res Treat 140, 219–232 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Robichaud N, Sonenberg N, Ruggero D, Schneider RJ, Translational Control in Cancer. Cold Spring Harb Perspect Biol, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robichaud N et al. , Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene 34, 2032–2042 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robichaud N et al. , Translational control in the tumor microenvironment promotes lung metastasis: Phosphorylation of eIF4E in neutrophils. Proc Natl Acad Sci U S A 115, E2202–E2209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petroulakis E et al. , p53-dependent translational control of senescence and transformation via 4E-BPs. Cancer Cell 16, 439–446 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Bhat M et al. , Targeting the translation machinery in cancer. Nat Rev Drug Discov 14, 261–278 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Devlin JR et al. , Combination Therapy Targeting Ribosome Biogenesis and mRNA Translation Synergistically Extends Survival in MYC-Driven Lymphoma. Cancer Discov 6, 59–70 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Rosshandler Y, Shen AQ, Cortes J, Khoury HJ, Omacetaxine Mepesuccinate for Chronic Myeloid Leukemia. Expert Rev Hematol 9, 419–424 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Gurel G, Blaha G, Moore PB, Steitz TA, U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome. J Mol Biol 389, 146–156 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner NC et al. , Palbociclib in Hormone-Receptor-Positive Advanced Breast Cancer. N Engl J Med 373, 209–219 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N, mTOR, translation initiation and cancer. Oncogene 25, 6416–6422 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Topisirovic I, Sonenberg N, mRNA translation and energy metabolism in cancer: the role of the MAPK and mTORC1 pathways. Cold Spring Harb Symp Quant Biol 76, 355–367 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Lazaris-Karatzas A, Montine KS, Sonenberg N, Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5' cap. Nature 345, 544–547 (1990). [DOI] [PubMed] [Google Scholar]
- 37.Avdulov S et al. , Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell 5, 553–563 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Gingold H et al. , A dual program for translation regulation in cellular proliferation and differentiation. Cell 158, 1281–1292 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Pianese G, Beitrag zur histologie und aetiologie der carcinoma. histologische und experimentelle untersuchungen. Beitr. Pathol. Anat. Allgem. Pathol 142, 1–193 (1896). [Google Scholar]
- 40.Mills EW, Green R, Ribosomopathies: There's strength in numbers. Science 358, (2017). [DOI] [PubMed] [Google Scholar]
- 41.de Las Heras-Rubio A, Perucho L, Paciucci R, Vilardell J, ME LL, Ribosomal proteins as novel players in tumorigenesis. Cancer Metastasis Rev 33, 115–141 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Wang H et al. , Overexpression of ribosomal protein L15 is associated with cell proliferation in gastric cancer. BMC Cancer 6, 91 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q et al. , Cloning and characterization of full-length human ribosomal protein L15 cDNA which was overexpressed in esophageal cancer. Gene 263, 205–209 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Pena C, Hurt E, Panse VG, Eukaryotic ribosome assembly, transport and quality control. Nat Struct Mol Biol 24, 689–699 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Granneman S, Petfalski E, Tollervey D, A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J 30, 4006–4019 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanghai ZA et al. , Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature 556, 126–129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khajuria RK et al. , Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis. Cell 173, 90–103 e119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi Z et al. , Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of mRNAs Genome-wide. Mol Cell 67, 71–83 e77 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warner JR, McIntosh KB, How common are extraribosomal functions of ribosomal proteins? Mol Cell 34, 3–11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gunasinghe NP, Wells A, Thompson EW, Hugo HJ, Mesenchymal-epithelial transition (MET) as a mechanism for metastatic colonisation in breast cancer. Cancer Metastasis Rev 31, 469–478 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Debnath J, Muthuswamy SK, Brugge JS, Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Joung J et al. , Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc 12, 828–863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gandin V et al. , Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J Vis Exp, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vasudevan S, Steitz JA, AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 128, 1105–1118 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He SL, Green R, Polysome analysis of mammalian cells. Methods Enzymol 530, 183–192 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Edwards A, Haas W, Multiplexed Quantitative Proteomics for High-Throughput Comprehensive Proteome Comparisons of Human Cell Lines. Methods Mol Biol 1394, 1–13 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Ting L, Rad R, Gygi SP, Haas W, MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods 8, 937–940 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McAlister GC et al. , MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem 86, 7150–7158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lapek JD Jr. et al. , Detection of dysregulated protein-association networks by high-throughput proteomics predicts cancer vulnerabilities. Nat Biotechnol 35, 983–989 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eng JK, McCormack AL, Yates JR, An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5, 976–989 (1994). [DOI] [PubMed] [Google Scholar]
- 61.Huttlin EL et al. , A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elias JE, Gygi SP, Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4, 207–214 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Aceto N et al. , AR Expression in Breast Cancer CTCs Associates with Bone Metastases. Mol Cancer Res 16, 720–727 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.