The cellular protease furin mediates proteolytic cleavage of many host and pathogen proteins and plays an important role in viral envelope glycoprotein maturation. MuV, an enveloped RNA virus of the Paramyxoviridae family and an important human pathogen, enters the cell through the fusion of its envelope with the plasma membrane of the target cell. Membrane fusion is mediated by the viral attachment protein and the F protein. Cleavage of MuV-F into two subunits by furin is a prerequisite for fusion and virus infectivity. Here, we show that LAMPs support the furin-mediated cleavage of MuV-F. Expression levels of LAMPs affect the processing of MuV-F and MuV-mediated membrane fusion. Among LAMPs, the interferon-stimulated gene product LAMP3 is most critical in certain cells. Our study provides potential targets for anti-MuV therapeutics.
KEYWORDS: mumps virus, furin, LAMP, fusion protein
ABSTRACT
Mumps virus (MuV), an enveloped RNA virus of the Paramyxoviridae family and the causative agent of mumps, affects the salivary glands and other glandular tissues as well as the central nervous system. The virus enters the cell by inducing the fusion of its envelope with the plasma membrane of the target cell. Membrane fusion is mediated by MuV envelope proteins: the hemagglutinin-neuraminidase and fusion (F) protein. Cleavage of the MuV F protein (MuV-F) into two subunits by the cellular protease furin is a prerequisite for fusion and virus infectivity. Here, we show that 293T (a derivative of HEK293) cells do not produce syncytia upon expression of MuV envelope proteins or MuV infection. This failure is caused by the inefficient MuV-F cleavage despite the presence of functional furin in 293T cells. An expression cloning strategy revealed that overexpression of lysosome-associated membrane proteins (LAMPs) confers on 293T cells the ability to produce syncytia upon expression of MuV envelope proteins. The LAMP family comprises the ubiquitously expressed LAMP1 and LAMP2, the interferon-stimulated gene product LAMP3, and the cell type-specific proteins. The expression level of the LAMP3 gene, but not of LAMP1 and LAMP2 genes, differed markedly between 293T and HEK293 cells. Overexpression of LAMP1, LAMP2, or LAMP3 allowed 293T cells to process MuV-F efficiently. Furthermore, these LAMPs were found to interact with both MuV-F and furin. Our results indicate that LAMPs support the furin-mediated cleavage of MuV-F and that, among them, LAMP3 may be critical for the process, at least in certain cells.
IMPORTANCE The cellular protease furin mediates proteolytic cleavage of many host and pathogen proteins and plays an important role in viral envelope glycoprotein maturation. MuV, an enveloped RNA virus of the Paramyxoviridae family and an important human pathogen, enters the cell through the fusion of its envelope with the plasma membrane of the target cell. Membrane fusion is mediated by the viral attachment protein and the F protein. Cleavage of MuV-F into two subunits by furin is a prerequisite for fusion and virus infectivity. Here, we show that LAMPs support the furin-mediated cleavage of MuV-F. Expression levels of LAMPs affect the processing of MuV-F and MuV-mediated membrane fusion. Among LAMPs, the interferon-stimulated gene product LAMP3 is most critical in certain cells. Our study provides potential targets for anti-MuV therapeutics.
INTRODUCTION
Mumps is an acute infectious disease in humans with a range of systemic symptoms. Mumps virus (MuV), the causative agent of the disease, is transmitted by inhalation of respiratory droplets or contact with oral secretions. It affects the salivary glands (typically the parotid grand), pancreas, testis, ovary, mammary glands, and kidney. MuV also infects the central nervous system (CNS), causing meningitis, encephalitis, and sensorineural hearing loss (1, 2). The mechanism underlying its unique tropism for the glandular tissues and CNS remains poorly understood.
MuV, an enveloped nonsegmented negative-strand RNA virus, is a member of the genus Orthorubulavirus in the family Paramyxoviridae. It enters the host cell through membrane fusion mediated by two envelope glycoproteins, a hemagglutinin-neuraminidase (HN) and a fusion (F) protein (1, 2). The MuV HN protein (MuV-HN) binds to the sialoglycan receptor, especially the trisaccharide containing ɑ2,3-linked sialic acid, and then triggers a conformational change of the F protein, leading to the fusion of the viral envelope with the plasma membrane of the host cell (3–5). Later in infection, the two envelope proteins expressed on infected cells also mediate the fusion with neighboring cells, forming syncytia (3). The MuV F protein (MuV-F) is initially synthesized as an inactive precursor F0 in the endoplasmic reticulum, which is then proteolytically activated by the host cell protease furin, a member of the proprotein convertase family (6). Furin is ubiquitously expressed in many tissues and cleaves MuV F0 at the R-R-H-K-R (R, arginine; H, histidine; K, lysine) motif in the trans-Golgi network to produce the disulfide-linked heterodimers F1 and F2. Cleavage of F0 into F1 and F2 is essential for virus-to-cell and cell-to-cell membrane fusion and virus infectivity (1). The F proteins of other paramyxoviruses such as human parainfluenza virus 3, parainfluenza virus 5, Newcastle disease virus virulent strains, measles virus (MeV), and respiratory syncytial virus (RSV) also contain the furin cleavage site (3, 7–11).
The lysosome-associated membrane proteins (LAMPs) are integral membrane glycoproteins present predominantly in lysosomes that are involved in multiple cell functions (12, 13). The LAMP family comprises LAMP1, LAMP2, LAMP3 (also called CD208 or dendritic cell [DC]-LAMP), LAMP4 (also called CD68 or macrosialin), and LAMP5 (also called brain and dendritic cell [BAD]-LAMP) (14–18). The LAMP2 gene produces three splicing variants, LAMP2A, LAMP2B, and LAMP2C, which differ in their sequences of the transmembrane and cytosolic tail (19). LAMP1, LAMP2A, and LAMP2B are ubiquitously expressed in human tissues and cell types, whereas LAMP2C, LAMP3, LAMP4, and LAMP5 are expressed in limited types of cells. LAMP3 is a highly specific marker for mature DCs in humans. It is induced upon DC maturation and likely plays a role in the transfer of the major histocompatibility complex class II molecules loaded with peptides to the cell surface (15). LAMP3 is also induced in various cells by type I interferons (IFNs) (20).
In the present study, we found that an insufficient cleavage of MuV-F is responsible for the lack of syncytium formation in 293T cells expressing MuV envelope proteins and that overexpression of LAMPs confers on 293T cells the ability to form syncytia. Thus, LAMPs appear to play an important role in the efficient cleavage of MuV-F by furin, and their expression levels may contribute to the MuV tropism.
RESULTS
293T cells are deficient in cell-cell fusion mediated by MuV envelope proteins.
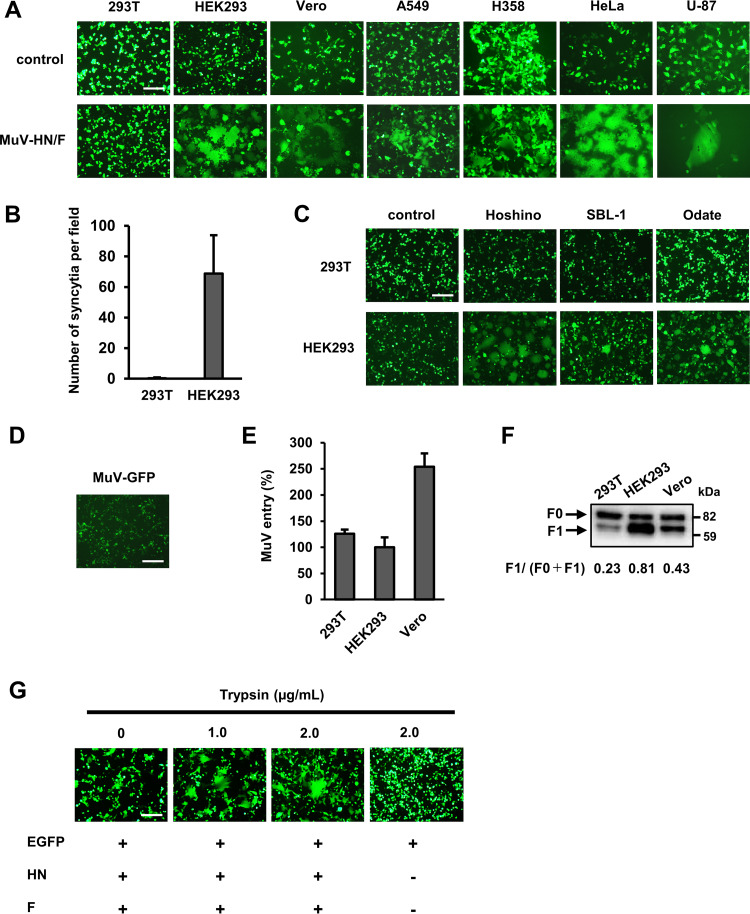
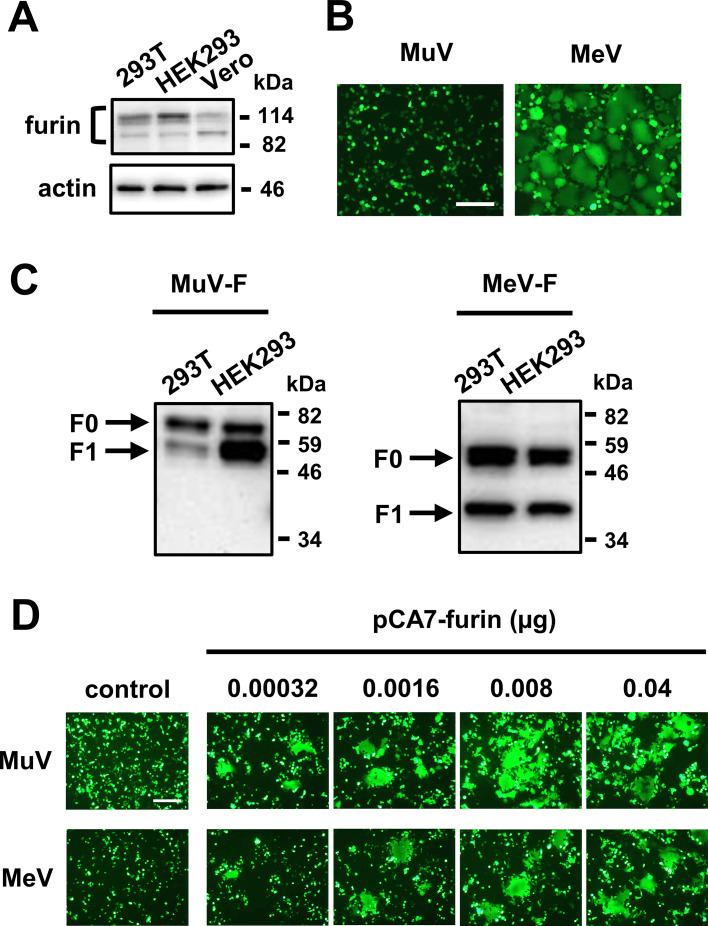
Expression of MuV envelope proteins usually induces cell-cell fusion in receptor-expressing cells, producing syncytia. In our attempt to identify host factors determining cell tropism of MuV other than the sialic acid receptor, we looked for cell lines that failed to develop syncytia after expression of MuV envelope proteins. Human and nonhuman primate cell lines derived from different tissues were transfected with expression plasmids, respectively, encoding the HN and F proteins of the MuV Hoshino strain, together with that encoding the enhanced green fluorescent protein (EGFP). At 24 h after transfection, EGFP-positive syncytia were observed in HEK293 (human embryonic kidney) and Vero (African green monkey kidney) cells, whereas syncytia were hardly detected in 293T cells (a derivative of HEK293 [ATCC]) (Fig. 1A). Other human cell lines, A549 (lung), H358 (lung/bronchiole), HeLa (cervix), and U-87 (brain), also developed apparent syncytia after transfection (Fig. 1A). The difference in syncytium formation between 293T and HEK293 cells was quantitatively apparent (Fig. 1B). Besides the HN and F proteins of the Hoshino strain, those of the MuV SBL-1 and Odate strains also failed to induce syncytia in 293T cells (Fig. 1C). Furthermore, 293T cells from the Riken BioResource Research Center exhibited the same phenotype as those from ATCC (data not shown).
FIG 1.
293T cells are deficient in cell-cell fusion mediated by MuV envelope proteins. (A) 293T, HEK293, Vero, A549, H358, HeLa, and U-87 cells transfected with pCA7-MuV-F (or empty vector for control), -MuV-HN, and -EGFP were observed for syncytium formation using fluorescence microscopy at 24 h posttransfection. Scale bar, 200 μm. (B) Syncytia (>2,000 μm2) in 293T and HEK293 cells transfected with pCA7-MuV-F, -MuV-HN, and -EGFP were counted in 40 fields at 24 h posttransfection, and the average numbers ± standard deviations (SDs) of syncytia per field are shown. (C) 293T and HEK293 cells transfected with plasmids, respectively, encoding the HN and F proteins of the MuV Hoshino, SBL-1, or Odate strains (or empty vector for control) and pCA7-EGFP were observed at 24 h posttransfection. (D) 293T cells infected with the GFP-expressing recombinant MuV were observed using fluorescence microscopy at 48 h postinfection. (E) 293T, HEK293, and Vero cells were infected with the GFP-expressing recombinant MuV. At 24 h postinfection, GFP-positive cells were counted to evaluate the efficiency of virus entry. The value in HEK293 cells was set to 100%, and data indicate the mean ± SDs of triplicate samples. The data are representative of three independently performed experiments. (F) 293T, HEK293, and Vero cells were transfected with pCA7-Flag-MuV-F and pCA7-MuV-HN. Efficiency of the cleavage of MuV-F was examined at 24 h posttransfection by Western blotting with anti-Flag ab. The data shown are representative of three independently performed experiments. Protein band signals were quantified, and the mean ratios and SDs of F1 to (F0 + F1) of the three experiments were 0.23 ± 0.04 for 293T, 0.81 ± 0.04 for HEK293, and 0.43 ± 0.06 for Vero. The mean ratios are shown in the figure. (G) 293T cells cultured in Opti-MEM serum-free medium were transfected with pCA7-MuV-F, -MuV-HN, and -EGFP. At 24 h after transfection, the cells were treated with lysine-acetylated-trypsin at the indicated concentrations or left untreated. 293T cells transfected with pCA7-EGFP only were also treated to examine nonspecific effects of trypsin on the cell monolayer. Syncytium formation was observed using a fluorescence microscope at 24 h after the trypsin treatment.
To understand why 293T cells do not develop syncytia upon expression of MuV envelope proteins, they were infected with the GFP-expressing recombinant MuV (produced in Vero cells). The 293T cells were infectible with the virus, indicating that they express MuV receptors on the surface, but still did not develop syncytia (Fig. 1D). Then, virus entry in different cells was evaluated by counting the number of GFP-expressing cells at 24 h after MuV infection (before syncytia started to emerge). Unlike the results with the fusion assay above, the efficiency of MuV entry into 293T cells was comparable with that into HEK293 cells (Fig. 1E). We next examined the structure of MuV-F in 293T cells since it must be proteolytically cleaved to be fusogenic. The expression plasmid encoding the Flag-tagged MuV-F was transfected into cells together with that encoding MuV-HN. The cleavage of the F protein was not as efficient in 293T cells as that in HEK293 or Vero cells (Fig. 1F). To determine whether this inefficient cleavage of the F protein was responsible for the failure of 293T cells to develop syncytia, we exogenously added trypsin to cleave the F protein. The cells cultured in serum-free medium were transfected with expression plasmids, respectively, encoding the HN and F proteins and then (24 h later) treated with trypsin or control medium. Syncytia were clearly detected in 293T cells treated with trypsin but not in those treated with control medium (Fig. 1G). The results indicate that cell-cell fusion was not observed in 293T cells because the processing of MuV-F was inefficient in them.
Furin alone is not sufficient for the efficient cleavage of MuV-F.
MuV-F is cleaved by the host cell protease furin (1). Thus, it is possible that furin is missing or defective in 293T cells. Western blot analysis showed that 293T cells express comparable amounts of furin with HEK293 and Vero cells (Fig. 2A). We then examined whether the F protein of MeV, another human paramyxovirus, is properly processed in 293T cells. Like MuV-F, the MeV F protein (MeV-F) is cleaved by furin (21), and expression of the MeV attachment protein and F protein induces cell-cell fusion in receptor-expressing cells. Upon expression of the MeV attachment protein of the Edmonston strain that uses the ubiquitously expressed CD46 as a receptor (22) and F protein, 293T cells developed syncytia, in contrast to those after expression of MuV-HN and -F (Fig. 2B). Furthermore, MeV-F was cleaved to the same extent in 293T and HEK293 cells, unlike MuV-F (Fig. 2C). We also performed the fusion assay using LoVo cells lacking furin (23). As expected, no syncytium was detected in LoVo cells upon expression of MuV-HN and -F or that of MeV envelope proteins (Fig. 2D). However, expression of furin by transfection restored the ability of LoVo cells to develop syncytia mediated by MuV and MeV envelope proteins in a dose-dependent manner. These results indicate that furin is necessary but not sufficient for the efficient cleavage of MuV-F and that 293T cells are deficient in some host factors present in HEK293 and LoVo cells, which supports the activity of furin to cleave MuV-F.
FIG 2.
Furin alone is not sufficient for the efficient cleavage of MuV-F. (A) Expression levels of furin in 293T, HEK293, and Vero cells were examined by Western blotting. (B) 293T cells transfected with pCA7-MuV-F, -MuV-HN, and -EGFP or pCA7-MeV-F, pCAGGS-MeV-H, and pCA7-EGFP were observed for syncytium formation using fluorescence microscopy at 24 h posttransfection. Scale bar, 200 μm. (C) 293T and HEK293 cells were transfected with pCA7-Flag-MuV-F and pCA7-MuV-HN, or pCA7-Flag-MeV-F and pCAGGS-MeV-H. The cleavage of MuV-F or MeV-F was examined at 24 h posttransfection by Western blotting with anti-Flag ab. (D) LoVo cells were transfected with pCA7-MuV-F, -MuV-HN, and -EGFP or pCA7-MeV-F, pCAGGS-MeV-H, and pCA7-EGFP, together with indicated amounts of pCA7-furin. The cells were observed under fluorescence microscopy at 36 h posttransfection.
Transfection with LAMP family genes supports cell-cell fusion mediated by MuV envelope proteins in 293T cells.
To identify the host factor required for the cleavage of MuV-F, we performed an expression cloning using a cDNA library generated from Vero cells. Sibling selection of cDNA pools capable of causing cell-cell fusion in 293T cells upon expression of MuV envelope proteins identified a cDNA clone 40-25-11 (Fig. 3A). DNA sequencing revealed that this cDNA clone had a high level of similarity to the human LAMP1 gene (96.4% identity at the nucleotide level in the coding region). Expression of the human LAMP1 cDNA also supported syncytium formation in 293T cells (Fig. 3B). Surprisingly, no mutation was found in the LAMP1 gene of 293T cells, and there was no significant difference in the expression level of LAMP1 between 293T and HEK293 cells, as detected by Western blotting (Fig. 3C).
FIG 3.
Transfection with LAMP family genes supports the efficient cleavage of MuV-F and cell-cell fusion mediated by MuV envelope proteins in 293T cells. (A) Expression cloning by using a cDNA library generated from Vero cells. 293T cells were transfected with the plasmid DNA (clone 40-25-11) from the library (or pCA7 for negative control), pCA7-MuV-F, -MuV-HN, and -EGFP. At 48 to 72 h after transfection, the cells were observed under a fluorescence microscope. The cDNA clone (no. 40-5-26) is a negative clone from the library. Scale bar, 200 μm. (B) 293T cells were transfected with pCA7-MuV-F, -MuV-HN, and -EGFP together with pCA7-LAMP1, -LAMP2A, -LAMP2B, or -LAMP3 and observed for syncytium formation using fluorescence microscopy at 48 h posttransfection. Scale bar, 200 μm. (C) Expression levels of LAMP1 were examined by Western blotting in 293T and HEK293 cells. Actin was used as a loading control. The data shown are representative of three independently performed experiments. Protein band signals were quantified, and relative expression levels of LAMP1 were normalized to the level of actin. The mean value in HEK293 was set to 1. The mean values and SDs of the three experiments were 0.92 ± 0.11 for 293T and 1 ± 0.24 for HEK293. The mean values are shown in the figure. (D) 293T and HEK293 cells were transfected with pCA7-MuV-F, -MuV-HN, and -EGFP, infected with MuV at a multiplicity of infection (MOI) of 10, or left untreated (control). At 48 h after transfection or infection, relative gene expression levels of LAMP1, LAMP2, and LAMP3 in those cells were quantified by RT-qPCR. Data were normalized to those of GAPDH, and the average value for untreated 293T cells was set to 1. Data indicate the means ± SDs of three independent experiments. *, P < 0.05; **, P < 0.01; two-tailed Student's t test. (E) 293T and HEK293 cells were transfected with pCA7-LAMP1, -LAMP2A, -LAMP2B, and -LAMP3 or empty vector, together with pCA7-Flag-MuV-F and pCA7-MuV-HN. Expression levels of MuV-F were examined at 48 h posttransfection by Western blotting with anti-Flag ab. The data shown are representative of three independently performed experiments. Protein band signals were quantified, and the mean ratios and SDs of F1 to (F0 + F1) of the three experiments were 0.17 ± 0.01, 0.30 ± 0.01, 0.24 ± 0.01, 0.27 ± 0.01, and 0.43 ± 0.03 for 293T cells transfected with empty vector (pCA7), pCA7-LAMP1, -LAMP2A, -LAMP2B, and -LAMP3, respectively, and 0.86 ± 0.03 for HEK293 cells transfected with the empty vector. The mean ratios are shown in the figure.
To explain this unexpected finding, we hypothesized that other members of the LAMP family might also support the cleavage of MuV-F and syncytium formation in 293T cells. To test this idea, we expressed LAMP2A, LAMP2B, and LAMP3 in 293T cells, together with MuV envelope proteins and EGFP, by transfection. LAMP2A and LAMP2B are ubiquitously expressed among different types of cells like LAMP1, while LAMP3 is the product of an IFN-stimulated gene (ISG). These three LAMPs were also found to support syncytium formation in 293T cells (Fig. 3B). We then examined expression levels of the genes encoding LAMP1, LAMP2 (LAMP2A and 2B combined), and LAMP3 in 293T and HEK293 cells by reverse transcription-quantitative PCR (RT-qPCR) (Fig. 3D). The expression level of the LAMP3 gene was apparently different between the two cell lines. Its level in 293T cells was ∼10-fold lower than that in HEK293 cells. Even after transfection or MuV infection, its expression level in 293T cells was still lower than the level in untreated HEK293 cells. In contrast, no remarkable difference was detected in the expression level of the LAMP1 gene between the two cell lines, and the expression level of the LAMP2 gene was higher in 293T cells than that in HEK293 cells. These results suggest that the expression level of LAMP3 accounts for the different phenotypes (the MuV-F cleavage and syncytium formation) observed in 293T and HEK293 cells. Although the processing of MuV-F was enhanced in 293T cells upon transfection with the LAMP1, LAMP2A, LAMP2B, or LAMP3 gene, that with the LAMP3 gene exhibited the strongest effect on the processing (Fig. 3E).
LAMPs interact with both MuV-F and furin.
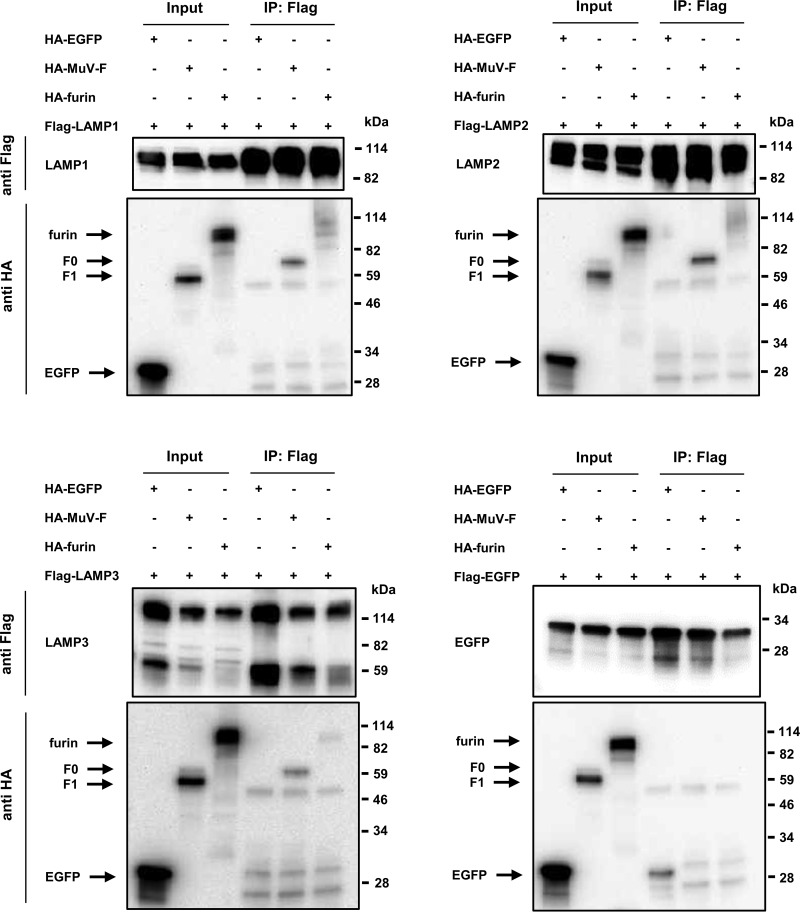
To gain insight into the mechanism by which LAMPs support the cleavage of MuV-F, coimmunoprecipitation (IP) experiments were performed using Flag- and influenza virus hemagglutinin (HA)-tagged proteins in HEK293 cells. Flag- and HA-tagged EGFP were used as controls. The HA-tagged MuV-F (F0, but not F1) and HA-tagged furin coprecipitated with the Flag-tagged LAMP1, LAMP2A, and LAMP3, but not with the Flag-tagged EGFP (Fig. 4). The results indicate that these LAMPs interact with both the inactive precursor F0 of MuV-F and furin. It should be noted that the input expression level of LAMP3 was lower when coexpressed with MuV-F or furin than the expression level with the EGFP control. The mechanism for this is unknown at present, but LAMP3 might become structurally unstable when complexed with MuV-F and/or furin.
FIG 4.
LAMPs interact with both MuV-F and furin. HEK293 cells were transfected with pCA7-Flag-LAMP1, -LAMP2A, -LAMP3, or -EGFP and one of pCA7-HA-MuV-F, -furin, and -EGFP. pCA7-Flag-EGFP and pCA7-HA-EGFP were used as negative controls. Proteins were immunoprecipitated 24 h later with anti-Flag ab, followed by Western blotting of total lysates (Input) and immunoprecipitates (IP) with anti-Flag (upper) or anti-HA ab (lower).
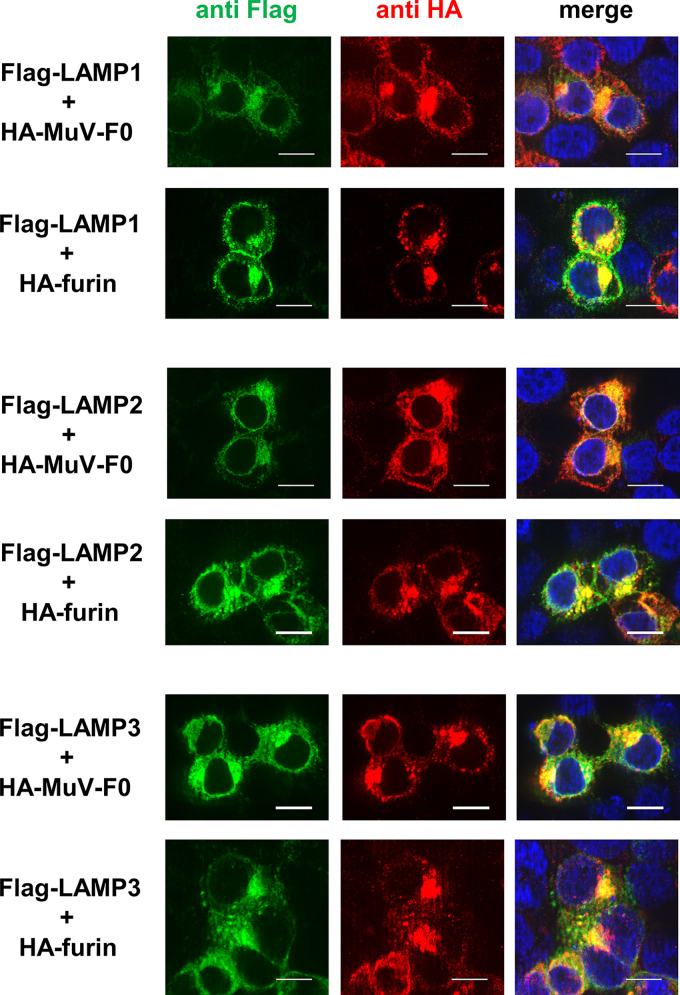
To further examine the interaction of LAMPs with MuV-F and furin in the cell, 293T cells were transfected with the expression plasmid encoding the Flag-tagged LAMP1, LAMP2A, or LAMP3, together with that encoding the HA-tagged MuV-F or furin, and observed using an immunofluorescent microscope. For this experiment, the uncleavable MuV-F was used because the co-IP experiments above indicated that LAMPs interact with F0 but not F1. All of these LAMPs were found to colocalize with both the F0 form of MuV-F and furin in the cytoplasm (Fig. 5).
FIG 5.
LAMPs colocalize with both MuV-F and furin in the cytoplasm. 293T cells were transfected with pCA7-Flag-LAMP1, -LAMP2A, or -LAMP3, together with pCA7-HA-MuV-F0, pCA7-MuV-HN, and pCA7-furin (upper) or together with pCA7-HA-furin, pCA7-MuV-HN, and pCA7-MuV-F0 (lower). They were observed at 24 h posttransfection using a fluorescence microscope. Scale bar, 10 μm.
DISCUSSION
In our attempt to identify host factors determining the MuV tropism for glandular tissues and the CNS, we first found that 293T cells do not develop syncytia upon MuV infection or expression of MuV envelope proteins. We found that 293T cells express receptors for MuV, but they exhibited a defect in processing MuV-F. Since the addition of trypsin to the culture medium allowed 293T cells to develop syncytia, we concluded that the inefficient cleavage of MuV-F is responsible for the failure of 293T cells to develop syncytia. However, furin responsible for the processing of MuV-F was functionally intact in 293T cells (because MeV-F was properly processed), suggesting that some host factor required for the furin-mediated cleavage of MuV-F (but not MeV-F) is defective in 293T cells.
It was then found that overexpression of LAMPs confers on 293T cells the ability to develop syncytia. The LAMP family comprises the ubiquitously expressed LAMP1 and LAMP2 (LAMP2A and 2B), the IFN-inducible LAMP3, and the cell type-specific proteins. Since overexpression of any of LAMP1, LAMP2A, LAMP2B, and LAMP3 allowed 293T cells to develop syncytia upon expression of MuV envelope proteins, these ubiquitously expressed and IFN-inducible LAMPs are all likely involved in the cleavage of MuV-F. Importantly, there were little differences in expression levels of the LAMP1 and LAMP2 genes between 293T cells and parental HEK293 cells that produce syncytia, but the expression level of the LAMP3 gene was much lower in 293T cells than that in HEK293 cells. Even after transfection or MuV infection, it was still lower in 293T cells than that in untreated HEK293 cells.
Thus, although steadily expressed levels of LAMP1 and LAMP2 may support the furin-mediated cleavage of MuV-F (they are presumably responsible for the low level of cleavage observed in 293T cells), they may not be sufficient to cleave MuV-F efficiently and to cause cell-cell fusion in 293T cells, and increased levels of LAMP3 may be required. Indeed, when exogenously expressed, LAMP3 enhanced the processing of MuV-F in 293T cells more strongly than LAMP1 or LAMP2 (Fig. 3E). The reason why 293T cells produce low, steady, and induced levels of LAMP3 remains to be determined, but it may be partly attributed to the defects in the DNA sensor cyclic GMP-AMP synthase (cGAS) and the adaptor protein STING in them (24). Nevertheless, expression of the LAMP3 gene was increased after transfection in 293T cells, indicating that its induction can occur independently of the cGAS-STING pathway.
How do LAMPs support the furin-mediated cleavage of MuV-F? Interestingly, LAMP1 has been shown to play an important role in Lassa virus (LASV) entry. LASV first binds to its cell surface receptor ɑ-dystroglycan, and, after being endocytosed, it then interacts with its intracellular receptor LAMP1 within the endosomal compartment (25). A recent study showed that LAMP1 elevates the pH threshold of the LASV envelope glycoprotein complex for fusion, thereby increasing the overall efficiency of LASV entry and infection (26). Similarly, LAMPs may somehow modify the structure of MuV-F such that it could serve as a good substrate for furin. Alternatively, LAMPs may act as scaffolds to concentrate MuV-F and furin, as LAMPs interact with both of the molecules (Fig. 4). The finding that LAMPs interact with the unprocessed form F0 (but not the processed form F1) of MuV-F is consistent with these two possibilities. Lysosomal proteins, including LAMPs, are synthesized in the endoplasmic reticulum and transported through the Golgi complex to the trans-Golgi network. Then, they can follow the secretory pathway to the plasma membrane and subsequently reach lysosomes by endocytosis. Alternatively, lysosomal proteins can follow a direct intracellular pathway to the endolysosomal system (13). Thus, LAMPs may interact with MuV-F and furin in the trans-Golgi network or other as yet undefined compartments within these pathways to support the processing of MuV-F.
The activity of furin may be finely controlled during virus infections. It has been shown that protease-activated receptor 1, which is induced under proinflammatory conditions, interacts with furin in the trans-Golgi network and interferes with the furin-mediated processing of the human metapneumovirus F protein and human immunodeficiency virus 1 (HIV-1) envelope protein (27, 28). A similar inhibitory activity was also described for two IFN-inducible GTPases, termed guanylate-binding proteins 2 and 5 (29). These proteins decrease the activity of furin and inhibit HIV-1, Zika virus, MeV, and avian influenza A virus replication by affecting the proteolytic cleavage of their envelope proteins. In contrast, LAMPs supported the furin-mediated cleavage of MuV-F. Although the processing of MeV-F appears to occur efficiently in LAMP3-deficient 293T cells (Fig. 2), it is presently unknown whether LAMPs are also required for the furin-mediated processing of other viral and host proteins. Since LAMP3 is the ISG product, it is tempting to speculate that one of its functions is to help furin to cleave and functionally activate host proteins involved in innate immunity and that MuV exploits such LAMP3-furin interaction for its own growth. Interestingly, a study reported that LAMP3 (but not LAMP1 and LAMP2) is induced in human lung A549 cells upon influenza A virus infection and plays an important role in its postentry replication steps (30). However, it should be noted that Vero cells incapable of producing IFNs still developed syncytia (Fig. 1), suggesting that some cells can use steadily expressed LAMPs to process MuV-F efficiently without IFN-induced LAMP3.
We have previously shown that α2,3-sialylated glycans such as α2,3-sialyllactose/sialyllactosamine, sialyl Lewis x, and GM2 ganglioside serve as receptors for MuV, and proposed that the distribution of these glycan motifs may partly account for the MuV tropism (4, 5). The present study suggests that the overall expression levels of LAMPs, including IFN-inducible LAMP3, may also contribute to the MuV tropism.
MATERIALS AND METHODS
Cells and virus.
293T, HEK293, HeLa, U-87, and Vero cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Wako) supplemented with 10% (vol/vol) fetal bovine serum (FBS; Sigma-Aldrich) and penicillin/streptomycin (Gibco). 293T cells were obtained from ATCC as well as from Riken BioResource Research Center. A549 and H358 cells were maintained in RPMI medium (Wako) supplemented with 10% (vol/vol) FBS and penicillin/streptomycin. LoVo cells, obtained from the Japanese Collection of Research Bioresources (JCRB) Cell Bank, National Institutes of Biomedical Innovation, Health and Nutrition, were maintained in Ham’s F-12 with l-glutamine (Nacalai) supplemented with 20% (vol/vol) FBS and penicillin/streptomycin. Cells were transfected with indicated amounts of plasmid DNAs using Polyethylenimine Max (Polysciences) (293T, HEK293, Vero, and HeLa cells), Lipofectamine LTX (Invitrogen) (A549, H358, and U-87 cells), or Lipofectamine 2000 (Invitrogen) (LoVo cells). The GFP-expressing recombinant MuV of the Hoshino strain was recovered from the full-length cDNA plasmid as described previously (31).
Plasmid constructions.
To construct expression plasmids, the cDNAs encoding human LAMP1, LAMP2A, LAMP2B, LAMP3, and furin were obtained by reverse transcription (RT) of total RNA from HEK293 cells, followed by PCR using specific primers. The DNA fragments encoding the MuV-F and -HN proteins of the SBL-1 strain were amplified by PCR from the template plasmids (4). These cDNAs were cloned into the expression plasmid pCA7 (32), generating pCA7-LAMP1, pCA7-LAMP2A, pCA7-LAMP2B, pCA7-LAMP3, pCA7-furin, pCA7-MuV-F (SBL-1 strain), and pCA7-MuV-HN (SBL-1 strain), respectively. pCAGGS-MuV-F and -HN (Odate strain) were kindly provided by Hiroshi Katoh (Department of Virology III, National Institute of Infectious Diseases, Tokyo, Japan) (33). pCA7-MuV-F (Hoshino strain), pCA7-MuV-HN (Hoshino strain), pCA7-MeV-F (IC-B strain), pCA7-EGFP, and pCAGGS-MeV-H (Edmonston strain) were described previously (4, 34–36). The sequence (amino acid positions 111 to 116, GVPQSR) of the Sendai virus F protein (Fushimi strain) was substituted for the furin cleavage site sequence of MuV-F (amino acid positions 97 to 102, SRRHKR) by site-directed mutagenesis, generating pCA7-MuV-F0 encoding the uncleaved MuV-F0 protein. The DNA fragments encoding Flag- or HA-tagged MuV-F (pCA7-Flag-MuV-F or pCA7-HA-MuV-F), Flag-tagged MeV-F (pCA7-Flag-MeV-F), HA-tagged MuV-F0 (pCA7-HA-MuV-F0), Flag- or HA-tagged EGFP (pCA7-Flag-EGFP or pCA7-HA-EGFP), Flag-tagged LAMP1 (pCA7-Flag-LAMP1), Flag-tagged LAMP2A (pCA7-Flag-LAMP2A), Flag-tagged LAMP3 (pCA7-Flag-LAMP3), and HA-tagged furin (pCA7-HA-furin) were cloned into pCA7.
Fusion assay.
Various cells on 12-well plates were transfected with 1 μg of pCA7-MuV-F, 0.4 μg of pCA7-MuV-HN, and 0.5 μg of pCA7-EGFP (Fig. 1A and C and Fig. 2B) or with 1 μg of pCA7-MeV-F, 0.4 μg of pCAGGS-MeV-H, and 0.5 μg of pCA7-EGFP (Fig. 2B) when they were ∼80 to 90% confluent. LoVo and 293T cells were transfected with pCA7-MuV-F, -MuV-HN and -EGFP, or pCA7-MeV-F, pCAGGS-MeV-H, and pCA7-EGFP as described above, together with different amounts of pCA7-furin and with 1 μg of pCA7-LAMP1, -LAMP2A, -LAMP2B, or -LAMP3, respectively (Fig. 2D and Fig. 3B). For Fig. 1G, culture media of 293T cells in a 12-well dish (Iwaki) were replaced with Opti-MEM serum-free medium (Gibco). The cells were then transfected with 1 μg of pCA7-MuV-F, 0.4 μg of pCA7-MuV-HN, and 0.5 μg of pCA7-EGFP or with pCA7-EGFP only. At 24 h after transfection, transfected cells were treated with lysine-acetylated trypsin (Merck) for 24 h or left untreated. The cell-cell fusion was evaluated under fluorescence microscopy at 24 h after trypsin treatment. For some experiments, syncytium formation was quantitated by counting the number of syncytia (>2,000 μm2) per field using a BZ-X710 microscope with the BZ-X analyzer (Keyence).
Virus entry assay.
Cells were infected with GFP-expressing recombinant MuV for 1 h at 37°C. The infected cells were then washed, and the fresh medium was added. At 24 h after infection, the number of GFP-expressing cells was counted under a fluorescence microscope.
Expression cloning.
A cDNA library was synthesized by using poly(A)-positive (poly[A]+) RNA from Vero cells and the cDNA library construction kit (TaKaRa). cDNA molecules were ligated unidirectionally to pCA7 digested with EcoRI and NotI (pCA7 has been modified to contain the multicloning site). The transformed E. coli was divided into 100 pools, and individual pools were cultured at 37°C overnight. The total number of independent clones (3.9 × 105) was determined by plating a portion of the transformed E. coli. Thus, each pool contained about 4,000 independent clones. Plasmid DNA was extracted from each pool. 293T cells were plated in 12-well plates, and they were transiently transfected with 1 μg of plasmid DNA from each pool, 0.05 μg of pCA7-EGFP, 0.4 μg of pCA7-MuV-HN, and 1.5 μg of pCA7-MuV-F. At 48 to 72 h after transfection, fusion was evaluated by observing EGFP-positive syncytia under a fluorescence microscope. Among 40 pools of cDNA clones tested (no. 1 to no. 40), a single pool (no. 40) produced apparent syncytia. This positive pool was further subdivided (no. 40-1 to no. 40-100), and the screening was repeated until a single clone (no. 40-25-11) was obtained that could allow transfected 293T cells to form syncytia.
Co-IP and Western blot analysis.
Subconfluent monolayers of HEK293 cells on 6-well plates were transfected with 2 μg of pCA7-Flag-LAMP1, -LAMP2A, LAMP3, or -EGFP together with 2 μg of pCA7-HA-MuV-F, pCA7-HA-furin, or pCA7-HA-EGFP. At 24 h after transfection, the cells were washed with phosphate-buffered saline (PBS) and lysed in 800 μl of IP lysis buffer (Thermo Fisher) containing the protease inhibitor cocktail (Sigma). After incubation for 30 min at 4°C, the lysates were centrifuged at 17,360 × g for 30 min at 4°C. A small amount (50 μl) of each supernatant was mixed with an equal volume of the 2× SDS loading buffer (125 mM Tris-HCl, pH 6.8, 10% 2-mercaptoethanol, 4% SDS, 0.1% bromophenol blue, and 20% glycerol) and kept as the whole-cell lysate sample. For preclearing the lysate, the rest of the supernatant was incubated for 30 min at 4°C with protein A-Sepharose (GE Healthcare AB). The precleared cell lysate was collected by centrifugation, and the bead pellet was discarded. The cell lysate was treated with an anti-Flag (catalog no. F1804; Sigma) antibody (ab) for 2 h at 4°C. Protein A-Sepharose was then added to the complexes and incubated for 1 h at 4°C. Complexes with the Sepharose were obtained by centrifugation and washed four times with IP lysis buffer. The polypeptides in the precipitated complexes were fractionated by SDS-PAGE and electroblotted onto polyvinylidene difluoride membranes (Immobilon-P; Millipore). The membranes were incubated with anti-Flag (catalog no. F7425; Sigma) or anti-HA (Y-11; Santa Cruz Biotechnology) ab, followed by incubation with horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (Invitrogen) for detection of the Flag-tagged or HA-tagged proteins. Chemiluminescent signals (Chemi-Lumi One Super; Nacalai Tesque) were detected and visualized using a VersaDoc 5000 imager (Bio-Rad). To detect the cleavage of MuV-F and MeV-F, cells on 12-well plates were transfected with 1 μg of pCA7-Flag-MuV-F and 0.4 μg of pCA7-MuV-HN (Fig. 1F and Fig. 2C), with 1 μg of pCA7-Flag-MeV-F and 0.4 μg of pCAGGS-MeV-H (Fig. 2C) or with 1 μg of pCA7-Flag-MuV-F, 0.4 μg of pCA7-MuV-HN, and 1 μg of pCA7-LAMP1, -LAMP2A, -LAMP2B, -LAMP3, or pCA7 (Fig. 3E). At 24 or 48 h posttransfection, cells were lysed in IP lysis buffer. To detect human furin, LAMP1, and β-actin, cells were lysed in IP lysis buffer. The lysates were fractionated by SDS-PAGE and blotted onto membranes as described above. Anti-Flag (to detect MuV-F or MeV-F), anti-furin (catalog no. ab3467; Abcam), anti-LAMP1 (catalog no. ab25630; Abcam), anti-β-actin (clone BA3R; BioVision), HRP-conjugated anti-rabbit IgG, and HRP-conjugated anti-mouse IgG (catalog no. sc-2005; Santa Cruz Biotechnology) were used for immunoblotting.
RT-qPCR.
For RT-qPCR, total RNA was extracted from transfected, MuV-infected, and untreated cells with the TRIzol reagent (Invitrogen), treated with DNase I (Promega), and reverse transcribed into cDNAs using SuperScript III (Invitrogen) and oligo(dT) primer. PCR (45 cycles of 95°C for 15 s, 55°C for 10 s, and 72°C for 30 s) was then performed with Thunderbird SYBR qPCR Mix (ToYoBo) using a CFX96 real-time system (Bio-Rad). Primers used for this assay were 5′- TCTCAGTGAACTACGACACCA-3′ and 5′-AGTGTATGTCCTCTTCCAAAAGC-3′ for human LAMP1 mRNA, 5′-TGGCAATGATACTTGTCTGCTG-3′ and 5′-ACGGAGCCATTAACCAAATACAT-3′ for LAMP2 mRNA, 5′-GCGTCCCTGGCCGTAATTT-3′ and 5′-TGCTTGCTTAGCTGGTTGCT-3′ for human LAMP3 mRNA, and 5′-CTGCACCACCAACTGCTTAG-3′ and 5′-AGGTCCACCACTGACACGTT-3′ for human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA, respectively. The amounts of the individual mRNAs were normalized to that of GAPDH mRNA and expressed as relative values. Data were analyzed with the CFX Maestro software (Bio-Rad).
Immunofluorescence staining.
293T cells seeded on coverslips, which had been precoated with 10 μg of poly-d-lysine hydrobromide (Sigma) per milliliter in a 24-well plate, were transfected with 0.2 μg of pCA7-Flag-LAMP1, -LAMP2A, or -LAMP3; 0.2 μg of pCA7-HA-MuV-F0; 0.08 μg of pCA7-MuV-HN; and 0.2 μg of pCA7-furin or with 0.2 μg of pCA7-Flag-LAMP1, -LAMP2A, or LAMP3; 0.2 μg of pCA7-HA-furin; 0.08 μg of pCA7-MuV-HN; and 0.2 μg of pCA7-MuV-F0. At 24 h posttransfection, the cells were fixed and permeabilized with PBS containing 3.6% formaldehyde and 0.5% Triton X-100. The cells were treated with 10% normal donkey serum and then incubated with appropriate combinations of primary and secondary antibodies. The following abs were used: mouse anti-Flag ab, rabbit anti-HA ab (catalog no. 561; MBL), Alexa Fluor 488-conjugated donkey anti-mouse IgG (H+L) (Molecular Probes), and Alexa Fluor 594-conjugated donkey anti-rabbit IgG (H+L) (Molecular Probes). The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Lonza). The stained cells were observed using a BZ-X710 fluorescence microscope (Keyence) equipped with an optical sectioning algorithm system (to obtain clear images without fluorescence blurring).
ACKNOWLEDGMENTS
We thank Takashi Nakagawa for support and advice and Hiroshi Katoh for plasmids.
This work was supported by JSPS KAKENHI grant number JP17K19563 (to Y.Y.), the Naito Foundation (to T.H.), and the Takeda Science Foundation (to T.H.). Part of this work was performed in the Cooperative Research Project Program of the Medical Institute of Bioregulation, Kyushu University.
Y.S. and Y.Y. designed research; A.U. and M.K. performed research; A.U., M.K., Y.S., S.O., T.H., and Y.Y. analyzed data; and U.A., Y.S., and Y.Y. wrote the paper. All authors edited the manuscript.
REFERENCES
- 1.Rubin SA, Sauder CJ, Carbone KM. 2013. Mumps virus, p 1024–1041. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Rubin S, Eckhaus M, Rennick LJ, Bamford CG, Duprex WP. 2015. Molecular biology, pathogenesis and pathology of mumps virus. J Pathol 235:242–252. doi: 10.1002/path.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamb RA, Parks GD. 2013. Paramyxoviridae, p 957–995. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 4.Kubota M, Takeuchi K, Watanabe S, Ohno S, Matsuoka R, Kohda D, Nakakita SI, Hiramatsu H, Suzuki Y, Nakayama T, Terada T, Shimizu K, Shimizu N, Shiroishi M, Yanagi Y, Hashiguchi T. 2016. Trisaccharide containing alpha2,3-linked sialic acid is a receptor for mumps virus. Proc Natl Acad Sci U S A 113:11579–11584. doi: 10.1073/pnas.1608383113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kubota M, Matsuoka R, Suzuki T, Yonekura K, Yanagi Y, Hashiguchi T. 2019. Molecular mechanism of the flexible glycan receptor recognition by mumps virus. J Virol 93:e00344-19. doi: 10.1128/JVI.00344-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas G. 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol 3:753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spriggs MK, Collins PL. 1986. Human parainfluenza virus type 3: messenger RNAs, polypeptide coding assignments, intergenic sequences, and genetic map. J Virol 59:646–654. doi: 10.1128/JVI.59.3.646-654.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paterson RG, Harris TJ, Lamb RA. 1984. Fusion protein of the paramyxovirus simian virus 5: nucleotide sequence of mRNA predicts a highly hydrophobic glycoprotein. Proc Natl Acad Sci U S A 81:6706–6710. doi: 10.1073/pnas.81.21.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toyoda T, Sakaguchi T, Imai K, Inocencio NM, Gotoh B, Hamaguchi M, Nagai Y. 1987. Structural comparison of the cleavage-activation site of the fusion glycoprotein between virulent and avirulent strains of Newcastle disease virus. Virology 158:242–247. doi: 10.1016/0042-6822(87)90261-3. [DOI] [PubMed] [Google Scholar]
- 10.Richardson C, Hull D, Greer P, Hasel K, Berkovich A, Englund G, Bellini W, Rima B, Lazzarini R. 1986. The nucleotide sequence of the mRNA encoding the fusion protein of measles virus (Edmonston strain): a comparison of fusion proteins from several different paramyxoviruses. Virology 155:508–523. doi: 10.1016/0042-6822(86)90212-6. [DOI] [PubMed] [Google Scholar]
- 11.Collins PL, Huang YT, Wertz GW. 1984. Nucleotide sequence of the gene encoding the fusion (F) glycoprotein of human respiratory syncytial virus. Proc Natl Acad Sci U S A 81:7683–7687. doi: 10.1073/pnas.81.24.7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eskelinen EL. 2006. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol Aspects Med 27:495–502. doi: 10.1016/j.mam.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Saftig P, Klumperman J. 2009. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol 10:623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 14.Fukuda M. 1991. Lysosomal membrane glycoproteins. Structure, biosynthesis, and intracellular trafficking. J Biol Chem 266:21327–21330. [PubMed] [Google Scholar]
- 15.de Saint-Vis B, Vincent J, Vandenabeele S, Vanbervliet B, Pin JJ, Ait-Yahia S, Patel S, Mattei MG, Banchereau J, Zurawski S, Davoust J, Caux C, Lebecque S. 1998. A novel lysosome-associated membrane glycoprotein, DC-LAMP, induced upon DC maturation, is transiently expressed in MHC class II compartment. Immunity 9:325–336. doi: 10.1016/s1074-7613(00)80615-9. [DOI] [PubMed] [Google Scholar]
- 16.Holness CL, Simmons DL. 1993. Molecular cloning of CD68, a human macrophage marker related to lysosomal glycoproteins. Blood 81:1607–1613. doi: 10.1182/blood.V81.6.1607.1607. [DOI] [PubMed] [Google Scholar]
- 17.Gottfried E, Kunz-Schughart LA, Weber A, Rehli M, Peuker A, Muller A, Kastenberger M, Brockhoff G, Andreesen R, Kreutz M. 2008. Expression of CD68 in non-myeloid cell types. Scand J Immunol 67:453–463. doi: 10.1111/j.1365-3083.2008.02091.x. [DOI] [PubMed] [Google Scholar]
- 18.Defays A, David A, de Gassart A, De Angelis Rigotti F, Wenger T, Camossetto V, Brousset P, Petrella T, Dalod M, Gatti E, Pierre P. 2011. BAD-LAMP is a novel biomarker of nonactivated human plasmacytoid dendritic cells. Blood 118:609–617. doi: 10.1182/blood-2010-11-319699. [DOI] [PubMed] [Google Scholar]
- 19.Perez L, McLetchie S, Gardiner GJ, Deffit SN, Zhou D, Blum JS. 2016. LAMP-2C inhibits MHC class II presentation of cytoplasmic antigens by disrupting chaperone-mediated autophagy. J Immunol 196:2457–2465. doi: 10.4049/jimmunol.1501476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ. 2013. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res 41:D1040–1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffin DE. 2013. Measles Virus, p 1042–1069. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 22.Yanagi Y, Takeda M, Ohno S, Hashiguchi T. 2009. Measles virus receptors. Curr Top Microbiol Immunol 329:13–30. doi: 10.1007/978-3-540-70523-9_2. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi S, Nakagawa T, Kasai K, Banno T, Duguay SJ, Van de Ven WJ, Murakami K, Nakayama K. 1995. A second mutant allele of furin in the processing-incompetent cell line, LoVo. Evidence for involvement of the homo B domain in autocatalytic activation. J Biol Chem 270:26565–26569. doi: 10.1074/jbc.270.44.26565. [DOI] [PubMed] [Google Scholar]
- 24.Sun LJ, Wu JX, Du FH, Chen X, Chen Z. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jae LT, Raaben M, Herbert AS, Kuehne AI, Wirchnianski AS, Soh TK, Stubbs SH, Janssen H, Damme M, Saftig P, Whelan SP, Dye JM, Brummelkamp TR. 2014. Lassa virus entry requires a trigger-induced receptor switch. Science 344:1506–1510. doi: 10.1126/science.1252480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hulseberg CE, Fénéant L, Szymańska KM, White JM. 2018. Lamp1 increases the efficiency of Lassa virus infection by promoting fusion in less acidic endosomal compartments. mBio 9:e01818-17. doi: 10.1128/mBio.01818-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aerts L, Hamelin ME, Rheaume C, Lavigne S, Couture C, Kim W, Susan-Resiga D, Prat A, Seidah NG, Vergnolle N, Riteau B, Boivin G. 2013. Modulation of protease activated receptor 1 influences human metapneumovirus disease severity in a mouse model. PLoS One 8:e72529. doi: 10.1371/journal.pone.0072529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim W, Zekas E, Lodge R, Susan-Resiga D, Marcinkiewicz E, Essalmani R, Mihara K, Ramachandran R, Asahchop E, Gelman B, Cohen EA, Power C, Hollenberg MD, Seidah NG. 2015. Neuroinflammation-induced interactions between protease-activated receptor 1 and proprotein convertases in HIV-associated neurocognitive disorder. Mol Cell Biol 35:3684–3700. doi: 10.1128/MCB.00764-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braun E, Hotter D, Koepke L, Zech F, Groß R, Sparrer KMJ, Müller JA, Pfaller CK, Heusinger E, Wombacher R, Sutter K, Dittmer U, Winkler M, Simmons G, Jakobsen MR, Conzelmann K-K, Pöhlmann S, Münch J, Fackler OT, Kirchhoff F, Sauter D. 2019. Guanylate-binding proteins 2 and 5 exert broad antiviral activity by inhibiting furin-mediated processing of viral envelope proteins. Cell Rep 27:2092–2104.e10. doi: 10.1016/j.celrep.2019.04.063. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Z, Xue QH, Wan YL, Yang YW, Wang JW, Hung T. 2011. Lysosome-associated membrane glycoprotein 3 is involved in influenza A virus replication in human lung epithelial (A549) cells. Virol J 8:384. doi: 10.1186/1743-422X-8-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ninomiya K, Kanayama T, Fujieda N, Nakayama T, Komase K, Nagata K, Takeuchi K. 2009. Amino acid substitution at position 464 in the haemagglutinin-neuraminidase protein of a mumps virus Urabe strain enhanced the virus growth in neuroblastoma SH-SY5Y cells. Vaccine 27:6160–6165. doi: 10.1016/j.vaccine.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 32.Takeda M, Ohno S, Seki F, Nakatsu Y, Tahara M, Yanagi Y. 2005. Long untranslated regions of the measles virus M and F genes control virus replication and cytopathogenicity. J Virol 79:14346–14354. doi: 10.1128/JVI.79.22.14346-14354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh H, Kubota T, Ihara T, Maeda K, Takeda M, Kidokoro M. 2016. Cross-neutralization between human and African bat mumps viruses. Emerg Infect Dis 22:703–706. doi: 10.3201/eid2204.151116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tahara M, Takeda M, Seki F, Hashiguchi T, Yanagi Y. 2007. Multiple amino acid substitutions in hemagglutinin are necessary for wild-type measles virus to acquire the ability to use receptor CD46 efficiently. J Virol 81:2564–2572. doi: 10.1128/JVI.02449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komune N, Ichinohe T, Ito M, Yanagi Y. 2011. Measles virus V protein inhibits NLRP3 inflammasome-mediated interleukin-1beta secretion. J Virol 85:13019–13026. doi: 10.1128/JVI.05942-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeuchi K, Takeda M, Miyajima N, Kobune F, Tanabayashi K, Tashiro M. 2002. Recombinant wild-type and edmonston strain measles viruses bearing heterologous H proteins: role of H protein in cell fusion and host cell specificity. J Virol 76:4891–4900. doi: 10.1128/jvi.76.10.4891-4900.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]