Papillomaviruses infect stratified squamous epithelia, and the viral life cycle is linked to epithelial differentiation. Additionally, changes occur in viral and host gene expression, and immune cells are activated to modulate the infectious process. In vitro studies with keratinocytes cannot fully model the complex viral and host responses and do not reflect the contribution of local and migrating immune cells. We show that female C57BL/6J mice are susceptible to a transient papillomavirus cervicovaginal infection, and mice deficient in select genes involved in innate immune responses are susceptible to persistent infection with variable manifestations of histopathological abnormalities. The results of our studies support a working model of innate immunity to papillomaviruses, and the model provides a framework for more in-depth studies. A better understanding of mechanisms of early viral clearance and the development of approaches to induce clearance will be important for cancer prevention and the treatment of HPV-related diseases.
KEYWORDS: innate immunity, papillomaviruses
ABSTRACT
We demonstrate that female C57BL/6J mice are susceptible to a transient lower genital tract infection with MmuPV1 mouse papillomavirus and display focal histopathological abnormalities resembling those of human papillomavirus (HPV) infection. We took advantage of strains of genetically deficient mice to study in vivo the role of innate immune signaling in the control of papillomavirus. At 4 months, we sacrificed MmuPV1-infected mice and measured viral 757/3139 spliced transcripts by TaqMan reverse transcription-PCR (RT-PCR), localization of infection by RNAscope in situ hybridization, and histopathological abnormities by hematoxylin and eosin (H&E) staining. Among mice deficient in receptors for pathogen-associated molecular patterns, MyD88−/− and STING−/− mice had 1,350 and 80 copies of spliced transcripts/μg RNA, respectively, while no viral expression was detected in MAVS−/− and Ripk2−/− mice. Mice deficient in an adaptor molecule, STAT1−/−, for interferon signaling had 46,000 copies/μg RNA. Among mice with targeted deficiencies in the inflammatory response, interleukin-1 receptor knockout (IL-1R−/−) and caspase-1−/− mice had 350 and 30 copies/μg RNA, respectively. Among mice deficient in chemokine receptors, CCR6−/− mice had 120 copies/μg RNA, while CXCR2−/− and CXCR3−/− mice were negative. RNAscope confirmed focal infection in MyD88−/−, STAT1−/−, and CCR6−/− mice but was negative for other gene-deficient mice. Histological abnormalities were seen only in the latter mice. Our findings and the literature support a working model of innate immunity to papillomaviruses involving the activation of a MyD88-dependent pathway and IL-1 receptor signaling, control of viral replication by interferon-stimulated genes, and clearance of virus-transformed dysplastic cells by the action of the CCR6/CCL20 axis.
IMPORTANCE Papillomaviruses infect stratified squamous epithelia, and the viral life cycle is linked to epithelial differentiation. Additionally, changes occur in viral and host gene expression, and immune cells are activated to modulate the infectious process. In vitro studies with keratinocytes cannot fully model the complex viral and host responses and do not reflect the contribution of local and migrating immune cells. We show that female C57BL/6J mice are susceptible to a transient papillomavirus cervicovaginal infection, and mice deficient in select genes involved in innate immune responses are susceptible to persistent infection with variable manifestations of histopathological abnormalities. The results of our studies support a working model of innate immunity to papillomaviruses, and the model provides a framework for more in-depth studies. A better understanding of mechanisms of early viral clearance and the development of approaches to induce clearance will be important for cancer prevention and the treatment of HPV-related diseases.
INTRODUCTION
Papillomaviruses preferentially infect stratified squamous epithelia, and the completion of the viral life cycle is tightly linked to epithelial differentiation (1). Basal cells are the site of initial infection and the expression of early viral messages (2). However, when these cells leave the basal layer and differentiate in the superficial layers of the epithelium, a switch to the expression of late viral messages and capsid proteins as well as vegetative genome replication occurs to produce new virions (3). Cellular differentiation is accompanied by changes in viral and host gene expression (4). Additionally, immune cells are activated locally or traffic to the epithelium and modulate the infectious process (5). Although papillomaviruses broadly infect mucosal and cutaneous epithelia, infection is focal within the epithelium and can be anatomically localized, as in the case of human papillomaviruses (HPVs), where metaplastic transformation zones of the cervix, anus, and oropharynx are more susceptible to oncogenesis than other sites (6). Studies of HPV pathogenesis are challenging given the dynamic nature of the infected cell as it differentiates, the relatively long period of time required for papillomaviruses to complete their life cycle, and anatomical differences in tissue-specific responses to infection and disease. Much of this process can be recapitulated by in vitro studies using immortalized keratinocytes harboring persistent viral genomes, cells ectopically infected with viral gene expression vectors, or raft cultures of papillomavirus-infected keratinocytes (7–9). However, in vitro studies with keratinocytes do not reflect the contribution of local and migrating host immune cells to viral pathogenesis. Cervical and oropharyngeal tissue samples from individuals with HPV infection typically provide a single-time-point picture of host and viral gene expression. Primary infections are clinically silent and less commonly sampled. Thus, immunological studies of papillomavirus pathogenesis and potential interventions are hindered by the lack of a tractable small-animal model that manifests the full spectrum of infection and the host response.
Bovine, rabbit, and canine papillomaviruses have been successfully used for several decades to study papillomavirus biology and as surrogates for HPV diseases, diagnosis, treatment, and vaccine assessment, as reviewed by Rector and Van Ranst (10) and Christensen et al. (11). However, a limitation of these animal models is that none allow the study of vaginal and cervical infections. Additionally, these animals do not offer the conveniences of a compact size, full biological characterization, and the availability of reagents that are provided by the mouse.
In 2010, a mouse papillomavirus (now designated MmuPV1) was identified in a colony of nude mice in India (12, 13). Several research groups, including our own, have worked with this mouse model (12, 14–25). Although first thought to be a strictly cutaneous papillomavirus due to isolation from muzzle lesions, MmuPV1 has recently been shown to infect vaginal, anal, and oral mucosae (14–16, 19). MmuPV1 was first isolated from immunodeficient mice (12). However, while infection and disease are more severe in immunodeficient mice, several strains of “immunocompetent” mice have recently been shown to be susceptible to infection (20, 23–26). Nevertheless, most laboratories have reported that MmuPV1 does not produce disease in C57BL/6 mice, suggesting that they may be resistant to infection.
We report here that C57BL/6J mice are susceptible to a transient cervicovaginal infection that clears within several weeks. In these mice, focal histopathological abnormalities progressed from cervical intraepithelial neoplasia grade 1 (CIN1)-like lesions to CIN3-like lesions, before they completely resolved. The observation that C57BL/6J mice are susceptible to transient, but not persistent, infection together with the availability of numerous strains of genetically deficient mice on a C57BL/6 genetic background allowed us to take a novel in vivo approach to the study of innate immunity to papillomaviruses. We reasoned that disruption of key pathways of innate immunity would convert a transient infection into a persistent infection and thus provide insights into how innate immunity controls papillomaviruses. For our initial studies, we targeted key adaptor molecules that play central roles in several signaling pathways involved in the initial sensing of pathogens by innate immunity. We also targeted an effector function of the inflammasome, caspase-1, and several cytokine and chemokine receptors. We found that a number of gene-deficient mice are susceptible to a persistent vaginal infection, defined as the detection of viral gene expression and histological abnormalities at 4 months postinfection. Our findings and the literature support a working model of innate immunity to papillomaviruses involving the activation of a MyD88-dependent pathway and interleukin-1 receptor (IL-1R) signaling, the control of viral replication by interferon (IFN)-stimulated genes (ISGs), and the clearance of virus-transformed dysplastic cells by the action of the CCR6/CCL20 axis.
RESULTS
SCID mice challenged with MmuPV1 develop persistent genital tract lesions with morphology reminiscent of human HSIL/CIN2-3.
To confirm that MmuPV1 can infect the lower female genital tract and to morphologically characterize related lesions, we challenged medroxyprogesterone-pretreated mice intravaginally with a wart extract containing 6 × 108 MmuPV1 genomes following trauma induced by a cervical brush. B6.CB17-Prkdcscid/SzJ (SCID) mice and wild-type C57BL/6J mice were infected and sacrificed at 4 months postinfection. In SCID mice, a mean of 40.3 × 109 copies/μg total RNA (standard deviation [SD], ±56 × 109 copies) of the P533 promoter-initiated transcript with splice junctions at positions 757 and 3139 (757/3139 transcript) were detected in tissue samples by TaqMan reverse transcription-PCR (RT-PCR). Histological examination of tissue showed areas of a high-grade squamous intraepithelial lesion (HSIL)/CIN2 to -3 (CIN2-3) in the vulvar squamous epithelium (Fig. 1a and b), vagina (Fig. 1d and e), and cervix (Fig. 1g and h). Their histological appearance closely resembles that of lesions seen in human tissue samples, illustrated by near-full-thickness cytological atypia, intensely stained nuclei (hyperchromasia), irregular nuclear membranes, increased nuclear size and nuclear-to-cytoplasmic (N:C) ratio, and readily recognized mitotic figures and dyskeratosis. RNAscope in situ hybridization showed that areas of abnormal histology corresponded to sites of viral infection (Fig. 1c, f, and i). Histological abnormalities and viral staining by RNAscope were not observed in endometrial glands or stroma or in uterine tissue.
FIG 1.
Hematoxylin and eosin (H&E) staining and RNAscope in situ hybridization of the lower genital tract of SCID mice 4 months after vaginal challenge with MmuPV1. Formalin-fixed genital tract tissue was harvested 4 months after infection with MmuPV1 and processed for H&E staining. (a, b, d, and e) Images of H&E staining at ×100 and ×400 magnifications showing high-grade squamous intraepithelial lesions of the vulva (HSIL/vulvar intraepithelial lesion grade 3 [VIN3]) (a and b) and vagina (HSIL/vaginal intraepithelial neoplasia [VAIN]) (d and e). (g and h) Images of H&E staining at ×100 and ×400 magnifications showing cervical HSIL/CIN3 arising from the squamocolumnar junction. (c, f, and i) Abundant viral mRNAs are detected in all locations by RNAscope in situ hybridization.
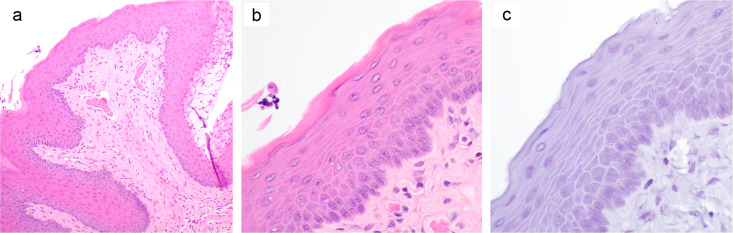
Infected wild-type C57BL/6J mice did not show any signs of MmuPV1 infection when sacrificed at 4 months postchallenge. The mice were negative for viral early transcripts by RT-quantitative PCR (qRT-PCR). Tissue sections showed a normal cervicovaginal squamous epithelium with maturation toward the surface upon histological examination, and RNAscope in situ hybridization showed no detectable viral RNA (Fig. 2).
FIG 2.
H&E staining of the lower genital tract of a C57BL/6J mouse 4 months after vaginal challenge with MmuPV1. Formalin-fixed genital tract tissue was harvested 4 months after infection with MmuPV1 and processed for H&E staining. Representative H&E images at ×100 and ×400 magnifications (a and b) and an RNAscope in situ hybridization image at a ×400 magnification (c) are shown.
Wild-type C57BL/6J mice challenged intravaginally with MmuPV1 develop transient genital tract lesions.
In order to determine whether wild-type C57BL/6J mice are susceptible to transient MmuPV1 infection, we challenged mice intravaginally and sacrificed mice on days 4, 7, 11, and 30 postchallenge. Lower genital tract tissues were processed for the detection of 757/3139 spliced transcripts by TaqMan RT-PCR, RNAscope in situ hybridization, and hematoxylin and eosin (H&E) staining. The mean numbers of viral 757/3139 spliced transcript copies per microgram of total RNA on days 4, 7, and 11 (±SD) were 7.3 × 104 (±4.5 × 104), 259 × 104 (±392 × 104), and 60.2 × 104 (±39.8 × 104), respectively, and by day 30, there were 0 copies, suggesting that the infection peaks after 1 week and is controlled rapidly thereafter. On day 4, H&E staining (Fig. 3a) showed normal basal layer squamous epithelium and overlying endocervical glandular epithelium. Despite the absence of histological abnormalities, focal areas of infection with MmuPV1 virus were detected by RNAscope in situ hybridization in the basal layer of the epithelium (Fig. 3b). By day 7, H&E staining (Fig. 3c) showed an emerging/incipient high-grade squamous intraepithelial lesion (HSIL/CIN2-3) characterized by atypical parabasal cells in the middle and upper thirds of the epithelium with an increased N:C ratio, dyskeratosis, and occasional mitoses. MmuPV1 virus was detected diffusely within the basal epithelium by RNAscope in situ hybridization (Fig. 3d). On day 11, H&E staining (Fig. 3e) showed a classic high-grade squamous intraepithelial lesion (HSIL/CIN2-3) characterized by a larger diffusion of the atypical parabasal cells described above. MmuPV1 virus was detected by RNAscope in situ hybridization, but the signal was weaker than that in day 7 mice and more focal in distribution (Fig. 3f). By day 30, histological abnormalities had completely resolved, and H&E staining was indistinguishable from that of control mock-infected mice. H&E staining (Fig. 3g) showed normal basal layer squamous epithelium and overlying endocervical glandular epithelium. MmuPV1 virus was no longer detected by RNAscope in situ hybridization (Fig. 3h). Mock-infected mice were negative by RT-PCR on days 4, 7, and 11 postchallenge and exhibited no histological abnormalities. Thus, the MmuPV1-infected C57BL/6J mouse provides a murine model of rapid immune control of an active papillomavirus infection within the female genital tract.
FIG 3.
Hematoxylin and eosin staining and RNAscope in situ hybridization of the vaginal epithelium at serial time points after infection of C57BL/6J mice with MmuPV1. Formalin-fixed genital tract tissue was harvested at different time points after infection with MmuPV1 and processed for H&E staining. (a, b, d, e, g, h, j, and k) Representative images of H&E stains from mice on day 4 (a and b), day 7 (d and e), day 11 (g and h), and day 30 (j and k) after MmuPV1 vaginal challenge. (c, f, i, and l) Representative images for RNAscope in situ hybridization for mice on day 4 (c), day 7 (f), day 11 (i), and day 30 (l) after MmuPV1 challenge.
Select strains of immune gene-deficient mice on a C57BL/6J genetic background develop persistent MmuPV1 infection after vaginal challenge.
In order to identify innate immune pathways critical to the clearance of MmuPV1 infection in C57BL/6 mice, we infected mice deficient in select immune genes identified as being relevant in the HPV literature and based upon the availability of mouse strains. In order to determine whether immunodeficient mice would develop a persistent infection, we sacrificed mice at 4 months postchallenge. We chose a time point sufficiently long after immunocompetent mice had cleared infection (1 month) to be certain that we could distinguish persistence from delayed clearance. Cohorts of 3 to 6 gene-deficient mice were challenged intravaginally. After sacrifice, lower genital tract tissues were processed for the detection of viral 757/3139 spliced transcripts by TaqMan RT-PCR, RNAscope in situ hybridization, and H&E staining.
Clearance of MmuPV1 infection in the lower female genital tract is dependent on activation of a MyD88 signaling pathway.
Pathogens are recognized by pattern recognition receptors (PRRs), which then activate intracellular signaling pathways and ultimately lead to the expression of immune response genes. Several studies indicate that HPVs have evolved a number of mechanisms to blunt recognition by receptors for pathogen-associated molecular patterns. HPV16 E6 and E7 and HPV38 downregulate the expression of Toll-like receptor 9 (TLR9) (27). In HPV18-positive cells, TLR3 levels are reduced (28). Overexpression of the nuclear DNA sensor IFI16 impairs HPV18 replication (29). HPV18 E7 binds to the cytoplasmic sensor STING and inhibits its activity (30). HPV E2 has also been reported to suppress STING (31). RIG-I/MDA5 expression is downregulated in HPV-positive cells (28), and a recent study reported that HPV E6 protein inhibits RIG-I-mediated innate immune signaling by impairing RIG-I’s ability to interact with its downstream adaptor protein, MAVS (32). To evaluate signaling pathways activated by the engagement of TLRs, we infected MyD88 knockout (MyD88−/−) mice. We used STING knockout mice to evaluate the cytosolic DNA sensor pathway, MAVS knockout mice for the RIG-I pathway, and Ripk2 knockout mice for the NOD receptor pathway.
MyD88−/− mice (n = 3) had a mean of 1,347 copies/μg total RNA (SD, ±945 copies) of viral 757/3139 spliced transcripts in genital tissue at 4 months postchallenge (Fig. 4), a level significantly higher than that of mice with no detectable viral transcripts (P = 0.03 by a one-tailed t test). STING−/− mice (n = 3) had a mean of 80 copies/μg total RNA (SD, ±70.6 copies) (P = 0.06 by a one-tailed t test versus viral transcript-negative mice), while MAVS−/− (n = 3) and Ripk2−/− (n = 3) mice had no detectable copies of viral transcripts by RT-PCR (Fig. 4). Histological examination of tissue from MyD88−/− mice showed less cytoplasmic differentiation than in low-grade squamous intraepithelial lesions ([LSIL]/CIN1), focal areas of dysplastic cells extending to the middle one-third of the epithelium with an increased N:C ratio, and rare dyskeratotic cells, resembling HSIL/CIN2 (Fig. 5a and b). RNAscope in situ hybridization confirmed the presence of focal areas of infection in MyD88−/− mice, although the signal was very weak compared to that of wild-type mice with a transient infection on day 4 (Fig. 5c). Of note, the mean copy number of viral transcripts in MyD88−/− mice at 4 months postchallenge (1,347 copies/μg RNA) was 2 log10 units lower than that in wild-type C57BL/6 mice at 4 days postchallenge (73,000 copies/μg total RNA). The vaginal and cervical epithelia of STING−/− mice were histologically unremarkable, as were those of MAVS−/− and Ripk2−/− mice. RNAscope in situ hybridization was negative for STING−/−, MAVS−/−, and Ripk2−/− mice.
FIG 4.
MmuPV1 757/3139 spliced transcript levels in genital tract tissue 4 days after infection of wild-type C57BL/6J mice and 4 months after infection of gene-deficient mice. Mice were challenged intravaginally with MmuPV1. After sacrifice, genital tissue was dissected and solubilized in TRIzol. RNA was tested by a TaqMan RT-PCR assay using primers spanning the 757/3139 splice junction. Results are expressed as copy numbers of transcripts per microgram of total RNA and presented in a box plot with a superimposed scatterplot of individual data points. A break was made in the scale between 500 and 1,000 copies/μg total RNA in order to allow for an expanded depiction of data points below 500 copies. No data points are excluded. Group sizes are 3, with the exception of STAT1−/−, CCR6−/−, and IL-1R−/− mice (n = 6).
FIG 5.
Hematoxylin and eosin staining and RNAscope in situ hybridization of vaginal epithelia from MmuPV1-infected MyD88, STAT1, and CCR6 knockout mice at 4 months postchallenge. Formalin-fixed genital tract tissue was harvested 4 months after infection with MmuPV1 and processed for H&E staining. (a, b, d, e, g, and h) Representative images of H&E staining at ×100 and ×400 magnifications for MyD88−/− mice (a and b), STAT1−/− mice (d and e), and CCR6−/− mice (g and h). (c, f, and i) Representative images at a ×400 magnification for RNAscope in situ hybridization and an insert of a magnified image of a localized area for MyD88−/−, STAT1−/−, and CCR6−/− mice.
Clearance of MmuPV1 infection in the lower female genital tract is dependent on STAT1, which mediates interferon signaling.
The activation of pathogen recognition receptors leads to the expression of soluble mediators of innate immunity, principally interferons. Most viruses need to overcome the potent antiviral activity of IFNs, and HPV is no exception. The HPV E7 protein interacts with and inactivates interferon regulatory factor 1 (IRF-1) (33). HPV E7 protein also binds to p48, a component of the STAT-containing ISGF3 complex, blocking the translocation of the complex to the nucleus (34). HPV16 E7 can also repress IFN signaling by binding to IRF-9 (35). HPV18 E6 can inhibit IFN signaling by physically associating with TYK2, impairing the phosphorylation of STAT1 and STAT2 (36). HPV E6 binds to IRF-3 and inhibits its transcriptional activity (37). Microarray analysis of keratinocytes harboring HPV16 and -31 showed that STAT1 is repressed (38), and the E6 and E7 proteins of these HPVs inhibit STAT1 in vitro (39). We evaluated the role of IFN using mice deficient in an adaptor protein in the signaling pathway, STAT1.
STAT1−/− mice (n = 6) had a mean of 46,154 copies/μg total RNA (SD, ±94,327 copies) of viral 757/3139 spliced transcripts in genital tissue at 4 months postchallenge (Fig. 4), a level significantly higher than that of mice negative for viral transcripts (P = 0.024 by a Mann-Whitney rank sum test). Histological examination showed a proliferation of basal/parabasal cells that extend no further than the lower layer of the epithelium. Despite occasional cytologic atypia, the upper layer exhibited differentiation; thus, the findings are consistent with LSIL/CIN1 (Fig. 5d and e). RNAscope in situ hybridization confirmed the presence of focal areas of infection in STAT1−/− mice, although the signal was very weak (Fig. 5f).
STAT1−/− mice mount an antigen-specific CD8+ T cell response.
In order to determine whether viral replication would trigger an adaptive cellular immune response, we measured the frequency of antigen-specific interferon gamma (IFN-γ)-secreting CD8+ T cells responding to an E6 epitope (KNIVFVTVR) that we previously mapped (20). In the spleen, three STAT1−/− mice had frequencies of antigen-specific CD8+ T cells of 0.7%, 0.4%, and 0.1%, respectively (Fig. 6). The frequency of antigen-specific cells in control cultures that were not stimulated with peptide was <0.01% for all mice.
FIG 6.
E6 antigen-specific IFN-γ-secreting CD8+ T cells in STAT1−/− mice 4 months after MmuPV1 infection. Splenocytes from STAT1−/− mice 4 months after MmuPV1 infection were stimulated overnight with or without peptide, and the frequency of antigen-specific IFN-γ-secreting CD8+ T cells was determined by an intracellular cytokine flow assay. Data are presented as CD8/IFN-γ dot plots generated with DIVA software. The frequency of antigen-specific cells is indicated to the right of the dot plot for peptide-stimulated cultures. Background stimulation in the absence of peptide was <0.01% for all cultures.
Clearance of MmuPV1 infection in the lower female genital tract is dependent on activation of IL-1 and signaling through the IL-1 receptor.
A hallmark of HPV infection is the absence of an inflammatory response. The key cytokine that orchestrates inflammation is the IL-1 family of cytokines (40). HPV16 E6 and E7 oncoproteins have been shown to enhance the release of IL-1α from cultures of normal cervical keratinocytes (41). Infection of human keratinocytes with HPV16 also induces the secretion of IL-1β. Conversely, a recent study showed that IL-1β production can be blocked by E6 through inhibition of IRF-6 transcription (42). Genome-wide expression profiling of human keratinocytes harboring episomal copies of HPV16 and -18 showed that the proinflammatory cytokines IL-1α and IL-1β are downregulated (43), and in a separate study, expression profiling showed that a network of genes strongly interconnected by IL-1β is also downregulated (44). Thus, HPV may interfere with IL-1 production as a means of immune evasion. The conflicting data for the induction and inhibition of IL-1 may be attributable to differences in experimental systems.
The inflammasome is a multimeric protein complex that activates proinflammatory IL-1 family cytokines through the proteolytic activity of caspase-1. To evaluate the role of the inflammasome, we challenged caspase-1 knockout mice, and as a complementary approach, we challenged IL-1 receptor (IL-1R) knockout mice.
IL-1R−/− mice (n = 6) had a mean of 349 copies/μg total RNA (SD, ±385 copies) of viral 757/3139 spliced transcripts in genital tissue at 4 months postchallenge (P = 0.024 by a Mann-Whitney rank sum test versus mice negative for viral transcripts), and caspase-1−/− mice (n = 3) had a mean of 30 copies/μg total RNA (SD, ±15 copies) (P = 0.014 by a one-tailed t test versus mice negative for viral transcripts) (Fig. 4). The vaginal and endocervical epithelia of IL-1R−/− and caspase-1−/− mice were unremarkable, and RNAscope in situ hybridization was negative.
Clearance of MmuPV1 infection in the lower female genital tract is dependent on activation of the CCR6/CCL20 axis.
Chemokines are chemotactic cytokines that control the migratory patterns and positioning of immune cells. Keratinocytes readily upregulate the production and secretion of chemokines in response to danger signals (35, 44–46). HPV may evade innate immunity by interfering with chemokine expression. HPV16 E6 and E7 oncoproteins reduce the basal and PRR- or cytokine-stimulated expression of chemokines by keratinocytes (44, 47, 48), and conditioned medium from HPV E6/E7-positive keratinocytes has a reduced ability to support the chemotaxis of immune cells (49). HPV8 E7 represses CCL20 transcription by binding to the transcription factor C/EBPβ and preventing binding to the CCL20 promoter (50). Silencing of HPV16 E6 and E7 oncogenes by RNA interference (small interfering RNA [siRNA]) in HPV-transformed keratinocytes has been shown to increase CCL20 secretion and induce a greater migratory capacity of Langerhans cells (LCs) in vitro (51). In cervical lesions of patients, E6 and E7 expression is negatively correlated with CCL20-positive cells and LCs. In one study, greater numbers of CD1a+ Langerhans cells and CCL20+ cells were detected by immunohistochemistry in HPV-negative than in HPV E6- and E7-positive lesions, suggesting that the downregulation of CCL20 by E6 and E7 leads to LC decline and is a key factor in the development of cervical lesions (52). Chemokines have been implicated in other genital tract infections or are known to play a role in keratinocyte wound healing. For example, CXCL10−/− and CXCL9−/− mice as well as mice deficient in the receptor for these chemokines, CXCR3−/− mice, showed a heightened sensitivity to infection with genital herpes simplex virus (53, 54). The poor response of these mice to viral infection was associated with reduced cytotoxic T lymphocyte (CTL) activity and a reduction in the recruitment of plasmacytoid dendritic cells (DCs) and CD80-positive dendritic cells to draining lymph nodes. CXCL2 plays an important role in wound healing, which may be important for papillomaviruses since microtrauma is thought to be important for infection (55). The availability of knockout mice and the redundancy in ligand signaling dictated our choice to challenge mice lacking the receptors for these ligands, CCL20/CCR6, CXCL10/CXCR3, and CXCL2/CXCR2, rather than the ligands themselves.
CCR6−/− mice (n = 6) had a mean of 117 copies/μg total RNA (SD, ±77 copies) of viral 757/3139 spliced transcripts in genital tissue at 4 months postchallenge (Fig. 4), a level significantly higher than that of mice with no detectable viral transcripts (P = 0.019 by a one-tailed t test). There were no detectable copies of viral transcripts by RT-PCR in CXCR2−/− (n = 3) and CXCR3−/− (n = 3) mice (Fig. 4). Histological examination of tissue from CCR6−/− mice showed changes similar to those in MyD88−/− mice, characterized by hyperchromatic dysplastic cells in the middle and upper thirds of the epithelium with features consistent with HSIL/CIN2 (Fig. 5g and h). Despite a lower copy number of viral transcripts (117 copies/μg total RNA) than in MyD88−/− mice (1,347 copies/μg total RNA) and STAT1−/− mice (46,000 copies/μg total RNA), CCR6−/− mice had histological abnormalities comparable to those of MyD88−/− mice and more severe lesions than those of STAT1−/− mice. RNAscope in situ hybridization confirmed the presence of focal areas of infection in CCR6−/− mice (Fig. 5i). The vaginal and cervical epithelia of CXCR2−/− and CXCR3−/− mice were unremarkable, and RNAscope in situ hybridization was negative.
Wild-type and gene-deficient mice mount an anticapsid antibody response.
In order to determine whether viral replication would trigger an adaptive humoral immune response, we measured anticapsid antibody responses at 4 months postchallenge by a pseudovirion enzyme-linked immunosorbent assay (ELISA) (Fig. 7). At a 1:400 serum dilution, the mean absorbance value for wild-type mice (0.987 optical density at 450 nm [OD450] units [SD, 0.486 units]) was significantly higher than that for mock-infected control mice (0.035 OD450 units [SD, 0.022 units]) (P = 0.01 by a t test). The mean absorbance values among gene-deficient mice were 0.286 OD450 units (SD, 0.176 units), 0.962 OD450 units (SD, 0.447 units), 0.780 OD450 units (SD, 0.344 units), and 0.132 OD450 units (SD, 0.026 units) for STAT1−/−, IL-1R−/−, CCR6−/−, and MyD88−/− mice, respectively. The responses of gene-deficient mice were significantly higher than that of mock-infected mice (P < 0.05 by a t test). However, while IL-1R−/− and CCR6−/− mice had responses comparable to those of wild-type mice, STAT1−/− and MyD88−/− mice had significantly lower responses than those of wild-type mice (P < 0.05 by a t test). As expected, the mean absorbance value for SCID mice at a 1:400 dilution (0.043 OD450 units [SD, 0.001 units]) did not differ significantly from that for mock-infected mice (0.035 OD450 units [SD, 0.022 units]).
FIG 7.
Anti-MmuPV1 capsid antibody response 4 months after infection of wild-type and gene-deficient mice. Serial 2-fold dilutions from 1:50 to 1:6,400 of sera collected at 4 months postchallenge were tested in a pseudovirion ELISA for anticapsid antibodies. The absorbance at 450 nm is plotted against the dilution on a log10 scale. Panels show responses of 6 wild-type (WT) mice and a representative mock-infected control mouse (A), responses of 6 STAT1−/− mice and a representative SCID mouse (B), responses of 6 IL-1R−/− mice and a representative SCID mouse (C), and responses of 3 MyD88−/− mice, 3 CCR6−/− mice, and a representative SCID mouse (D).
DISCUSSION
The introduction of molecular screening with licensed tests has demonstrated that genital high-risk HPV infection is extremely common, and the vast majority of cases resolve spontaneously (56–58). Since it is only the persistent infections that can progress to cancer, a better understanding of the mechanisms of early viral clearance is critical. Likewise, the development of approaches to induce such viral clearance will be informed by such studies. Innate immunity is an important component of the host defense against infection and against damaged or infected host cells. All cells can contribute to innate immunity by producing secreted factors involved in the inflammatory response, including interferons, cytokines, and chemokines (59, 60). Innate immunity also involves a cellular component, including macrophages, dendritic cells, neutrophils, and natural killer (NK) cells, which contribute directly to host defense and support an adaptive immune response (61, 62). These cells can be prepositioned in tissues or recruited in response to inflammation. This coordinated response of tissue-specific soluble mediators and immune cells is difficult to study without an in vivo model of infection. Here, we describe a murine model with which to study the role of key pathways in the rapid immune control of an active papillomavirus infection of the female genital tract.
We first demonstrated the susceptibility of C57BL/6J mice to a transient infection with MmuPV1 and showed that MmuPV1 infection results in characteristic papillomavirus-induced precancerous lesions, with focal histopathological abnormalities indistinguishable from the histopathology of HPV infection in women. The abnormalities progressed from CIN1-like lesions to CIN3-like lesions, before clearing by day 30 postinfection. We then took advantage of numerous gene-deficient mice on a C57BL/6 genetic background to investigate in vivo mechanisms of innate immunity to papillomaviruses.
Our studies revealed several important negative findings. The absence of infection or histopathological abnormalities 4 months after challenge of Ripk2−/− and MAVS−/− mice suggests that the NOD and RIG-I receptor pathways do not play an important role in the control of MmuPV1 infection in the lower genital tract. CXCR3−/− mice are highly susceptible to genital herpes simplex virus 2 (53, 54), but this was not the case for MmuPV1 infection. CXCR3 is an inflammatory chemokine receptor whose expression is associated with CD4+ type 1 helper (Th1) cells and CD8+ cytotoxic T lymphocytes (CTLs) as well as innate lymphocytes such as NK cells, NKT cells, plasmacytoid DCs, and subsets of B cells (63). CXCR3 is thought to participate in the localization of these first‐line defenders at sites of infection and inflammation (64). Our data suggest that the subset of cells expressing CXCR3 does not play a role in the early clearance of MmuPV1 infection in the genital tract. CXCR2−/− mice are highly susceptible to Chlamydia muridarum infection of the genital tract (65), but the loss of this receptor had no effect on susceptibility to papillomavirus infection. Microtrauma is thought to be important for papillomavirus infection, and this might be expected to initiate a wound-healing response. CXCR2+ cells, which include neutrophils and macrophages, play an important role in the inflammatory response to wound healing (66, 67). If such a response occurs following a papillomavirus infection, it does not appear to contribute to viral clearance. CXCR2 is a permissive receptor that can bind CXCL1-CXCL3 and CXCL5-CXCL8; thus, it is unlikely that this family of chemokines plays an important role in the clearance of MmuPV1 infection.
The other gene-deficient mice that we studied showed some evidence of infection, with or without signs of histopathology. However, the level of viral replication, measured by TaqMan RT-PCR for viral transcripts, was several log10 units lower than that detected in wild-type mice with a transient infection and many orders of magnitude lower than that seen in immunodeficient SCID mice. This finding is consistent with the known redundancy in innate immune pathways and the expectation that knocking out a single pathway will not confer a phenotype like that of SCID mice. In addition, gene-deficient mice may retain the ability to mount an adaptive immune response. In fact, STAT1−/− mice generated a detectable E6 antigen-specific IFN-γ-secreting CD8+ T cell response. Thus, the low level of viral transcripts in gene-deficient mice could be attributed to an intact adaptive immune response that partially controlled viral replication but could not completely clear infection. The highest level of viral gene expression determined by RT-PCR at 4 months postinfection was seen in STAT1−/− mice. These findings are strong evidence for an important role for interferons in the control of papillomavirus infection and are consistent with the known ability of HPV oncoproteins to inhibit interferon expression, as discussed above. In addition, interferons were used in the past to treat HPV infections (68). Moderately high levels of viral gene expression were observed in MyD88−/− mice, and somewhat lower levels were observed in IL-1R−/− mice, while STING−/− and caspase-1−/− mice had very low levels. Interferon is induced through several PRR signaling pathways, and MyD88 is a key adaptor molecule for all the TLRs, except TLR3, and also for the IL-1 receptor (69). We hypothesize that that expression of interferon is primarily the result of a MyD88-dependent pathway that involves signaling through the IL-1 receptor and probably one or more TLRs. Further support for the involvement of IL-1 comes from the presence of a persistent infection in mice lacking caspase-1, which plays a critical role in the proteolytic cleavage of IL-1 family cytokine precursor proteins. However, the lower level of viral gene expression in caspase-1−/− mice than in IL-1R−/− mice suggests that the activation of the IL-1 signaling pathway is largely caspase-1 independent. The most likely explanation for this independence is that the pathway is activated by IL-1α since pro-IL-1α is biologically active and can be converted to IL-1α by proteases other than caspase-1 (70). In addition, keratinocytes, which are the primary target of papillomavirus infection, are rich in preformed IL-1α (40, 71). The higher level of viral gene expression in MyD88−/− mice than in IL-1R−/− and caspase-1−/− mice suggests that signaling through the IL-1 receptor is not the only mechanism for the induction of antiviral responses to papillomavirus. It is likely that one or more TLRs are activated by papillomaviruses, but which TLR recognizes papillomaviruses is still unknown. The importance of TLR activation during papillomavirus infection is supported by the known therapeutic effect of imiquimod, a TLR7/8 agonist (72). We previously reported that HPV16 L1 virus-like particles activate bone marrow-derived dendritic cells to produce type I interferon and Th1 immune signals via MyD88, and we reported the potential contribution of TLR4 to the IgG response (73, 74). Future studies could address this question in vivo by infecting specific TLR gene-deficient mice.
STING−/− mice also had evidence of a persistent infection, which is consistent with the reported ability of HPV E7 and E2 to suppress STING, as discussed above. While our studies support a role for STING in the control of papillomavirus infection, the low viral copy number (80 copies/μg RNA), the undetectable staining by RNAscope in situ hybridization, and the absence of histological abnormalities indicate that the STING signaling pathway does not play as significant a role as MyD88-dependent signaling pathways. Among the chemokine receptor gene-deficient mice, only CCR6−/− mice showed evidence of infection at 4 months postchallenge. This finding is consistent with the reported role of the CCR6/CCL20 axis in human papillomavirus infection, as discussed above.
RNAscope in situ hybridization confirmed infection in SCID mice and transiently infected wild-type C57BL/6J mice, and the intensity of staining was qualitatively correlated with levels of viral transcripts detected by RT-PCR. RNAscope can detect viral DNA and mRNA; however, the pattern of staining differs. DNA is detected in the upper layers of the epithelium, and mRNA is detected in the lower layers. In wild-type mice on days 4 and 7, staining in the lower levels of the epithelium is evident, while on day 11, staining is more diffuse, likely reflecting both DNA from viral genome amplification and mRNA. For gene-deficient mice at 4 months postchallenge, staining is very weak, and precise localization is more difficult to interpret. There were notable discrepancies between infections detected by RT-PCR and RNAscope in situ hybridization for some strains of gene-deficient mice. As expected, no mouse that was negative for viral transcripts by RT-PCR had a detectable signal by RNAscope in situ hybridization. Several mice that were positive by RT-PCR, including STING−/−, caspase-1−/−, and IL-1R−/− mice, had no detectable evidence of infection by RNAscope in situ hybridization. The transcript levels determined by RT-PCR in these mice (range, 29 to 350 copies/μg RNA) were ∼3 log10 copies lower than those in wild-type mice at 4 days postinfection (73,000 copies/μg RNA) and thus could be attributable to differences in assay sensitivities. Although RNAscope in situ hybridization signals were faint, they were clearly detectable in 3 strains of gene-deficient mice, STAT1−/−, MyD88−/−, and CCR6−/− mice. However, RNAscope positivity did not correlate closely with RT-PCR because viral transcript copy numbers in these three strains of mice ranged from 117 to 46,000 copies/μg RNA. There are several possible explanations for the discordance between RT-PCR and RNAscope in situ hybridization in the detection of infection. In situ hybridization is expected to be less sensitive than RT-PCR. Second, because infection is focal, tissue sections may have missed lesions. A third possibility is differences in the transcripts detected by RNAscope in situ hybridization and the TaqMan RT-PCR assay. The recently reported transcriptome map of MmuPV1 showed that the most abundant viral transcripts are those initiated from the P533 promoter for late viral transcripts, and >90% of all viral transcripts use the 757/3139 splice site (75). The primers and probe for RT-PCR are located downstream of the nucleotide (nt) 533 transcription start site. The probes for the RNAscope in situ hybridization assay span the coding region for the E6/E7 genes from nt 1 to nt 755, which is largely upstream of the nt 533 transcription start site. Thus, while the RNAscope assay is expected to be specific for early transcripts, it may be less sensitive for P533-initiated late transcripts because the probes target only ∼200 bp of the 5′ end of these transcripts.
Histological abnormalities tracked more closely with the RNAscope in situ hybridization signal than the level of viral transcripts determined by RT-PCR. CCR6−/− and MyD88−/− mice, which exhibited the strongest staining in the RNAscope assay among the gene-deficient mice, showed high-grade intraepithelial lesions characterized by near-full-thickness dysplasia with increased nuclear to cytoplasmic (N:C) ratios and dyskeratotic cells. STAT1−/− mice, despite having the highest level of viral transcripts by RT-PCR, showed very weak staining by RNAscope in situ hybridization and low-grade intraepithelial lesions. Caspase-1−/−, STING−/−, and IL-1R−/− mice, with levels of viral transcripts comparable to those of CCR6−/− mice, were negative by the RNAscope assay and had a normal-appearing epithelium. Since RNAscope in situ hybridization primarily detects E6 and E7 transcripts, one explanation for the greater concordance of RNAscope with histological abnormalities is that dysplastic cells preferentially express early transcripts.
The development of an anticapsid antibody response and an E6-specific cellular immune response suggests that infection in the gene-deficient mice, similar to that in wild-type mice, was productive, and the virus completed its life cycle. One caveat is the possibility that the antibody response resulted from an immunizing dose of virus rather than active replication. This possibility is unlikely because a single administration of protein in the genital tract is not a potent route of immunization and would not explain the E6 antigen-specific CD8+ T cell response, which requires the expression of antigen in the cytoplasm of antigen-presenting cells and infected cells. Since mice were sampled only at 4 months postinfection, we do not know the kinetics of the immune response. Because a productive infection in the first week or two after challenge may explain the immune response, whether persistent infection in gene-deficient mice is productive or abortive is unknown and will require further investigation.
Taken together, our findings support a working model of innate immune control of a papillomavirus infection (Fig. 8). When papillomaviruses infect keratinocytes, they engage a TLR that has yet to be definitively identified. Infection also causes the release of stored, preformed pro-IL-1α from keratinocytes. Signaling through TLR and IL-1 receptors is MyD88 dependent and results in the expression of interferon and the CCL20 chemokine. In an autocrine fashion, IFN engages its receptor on keratinocytes and via a STAT1-dependent pathway induces the expression of IFN-stimulated genes (ISGs), a subset of which is known to have antiviral effects (76). One or more of these ISGs block the expression of viral late genes at the transcriptional or posttranscriptional level. The expression of CCL20 recruits CCR6-positive cells of a myeloid or lymphoid lineage, which we hypothesize are principally responsible for the clearance of dysplastic epithelial cells transformed by E6 and E7 early genes. Papillomaviruses may also directly activate dendritic cells to secrete CCL20. According to this model, STAT1−/− mice will have an intact MyD88 signaling pathway and normal production of CCL20. CCR6+ cells will be recruited to the site of infection and clear dysplastic cells, and the mice will exhibit minimal histological abnormities. In contrast, CCR6−/− mice will have a normal IFN response and, consequently, low levels of late viral gene transcripts, but consistent with our hypothesis, an impaired recruitment of CCR6+ cells will prevent the clearance of E6- and E7-expressing dysplastic cells, resulting in histological abnormalities. MyD88−/− mice will have impaired expressions of both IFN and CCL20 and exhibit a dual phenotype of modestly high levels of late viral gene transcript expression and histological abnormalities. While more work remains to be done to confirm the model and fill in the missing pieces, in broad terms, this model is supported by our studies with gene-deficient mice and consistent with what is currently known about innate immune responses to human papillomaviruses.
FIG 8.
Working model of the innate immune response to papillomavirus infection of the female lower genital tract. (A) Papillomaviruses (PV) infect keratinocytes and activate innate immunity via a yet-to-be-identified TLR and the release of preformed pro-IL-1α. Signaling through the MyD88-dependent pathways of the TLR and IL-1R leads to the expression of interferon (IFN) and the chemokine CCL20. IFN binds to the IFN receptor and signals through the STAT1-dependent IFN pathway to induce the expression of interferon-stimulated genes (ISGs) that block late viral gene expression at the transcriptional or posttranscriptional level. (B) CCL20 recruits CCR6-expressing tissue-resident or recruited myeloid or lymphoid cells that clear papillomavirus-infected dysplastic epithelial cells. Papillomaviruses may also directly activate dendritic cells to secrete CCL20.
Whether our findings in the MmuPV1 mouse model of genital infection are generalizable to human papillomavirus infection of women remains to be determined. MmuPV1 is phylogenetically more closely related to cutaneous beta-HPVs than mucosal alpha-HPVs, and it shares a number of genomic and molecular features with high-risk cutaneous beta-HPVs. These include the observation that MmuPV1 lacks an E5 open reading frame (ORF) (13) and contains separate promoters for E6 and E7 (75). Furthermore, MmuPV1 encodes an E6 protein that shares with HPV8 E6 the ability to bind MAML1 and SMAD2/SMAD3 but not E6AP and p53, which are bound by high-risk HPV E6 (21). Whether these molecular differences are important for innate immune responses is unknown and needs to be the subject of future investigations. In support of the relevance of the murine model, MmuPV1-infected mice recapitulate the characteristic preneoplastic histopathological lesions of an HPV infection. Additionally, this model is in agreement with the literature on innate immune responses to HPV. Our proposed model should be a rich source of hypotheses to guide future studies of innate immunity to human papillomaviruses.
MATERIALS AND METHODS
Mice.
Four- to six-week-old female mice of the following strains (with the abbreviated designation in parentheses) were obtained from The Jackson Laboratory (Bar Harbor, ME): C57BL/6J, B6.CB17-Prkdcscid/SzJ (SCID), C57BL/6J-Tmem173gt/J (STING), B6N.129S2-Casp1tm1Flv/J (caspase-1), B6.129S(Cg)-Stat1tm1Dlv/J (STAT1), B6.129P2(SJL)-Myd88tm1.1Defr/J (MyD88), B6;129-Mavstm1Zjc/J (MAVS), B6.129S7-Il1r1tm1Imx/J (IL-1R), B6.129P2-Cxcr3tm1Dgen/J (CXCR3), B6.129P2-Ccr6tm1Dgen/J (CCR6), B6.129S1-Ripk2tm1Flv/J (Ripk2), and B6.129S2(C)-Cxcr2tm1Mwm/J (CXCR2). All animal procedures were performed according to NIH guidelines under protocols approved by the Johns Hopkins University Animal Care and Use Committee.
Cervicovaginal challenge and tissue sampling.
Athymic nude mice were used to propagate MmuPV1 viruses as previously described (20). To prepare a viral stock, tail warts were harvested and then homogenized with glass beads (catalog number G4649; Sigma) in phosphate-buffered saline (PBS) with 0.1 M NaCl by using a mini-bead beater (BioSpec Products). The wart extract was centrifuged at room temperature for 10 min at 10,000 rpm in a microcentrifuge to remove tissue, and the supernatant was collected and stored at −20°C for future infectious challenge. To quantify genome copy numbers, DNA was extracted with a Quick-DNA miniprep plus kit (catalog number D4068; Zymo Research) and amplified via reverse transcription-quantitative PCR (qRT-PCR) using primers 5′-AGAGTGCATGGCTGGCAAGA-3′ and 5′-CATGTGGCGCACCAAGTGAA-3′ and probe 5′-FAM (6-carboxyfluorescein)-TGGCAAGCCGCACGCTTTGGCATCA-TAMRA (6-carboxytetramethylrhodamine)-3′, as previously described (20). To produce a standard curve, religated MmuPV1 genomic DNA was serially diluted in nuclease-free reagent-grade water. Threshold cycle (CT) values were plotted against the copy number to generate a standard curve.
Mice were inoculated subcutaneously with 3 mg progesterone in the form of medroxyprogesterone (Depo-Provera; Pfizer) in 100 μl PBS. Three days following injection, the mice were sedated intraperitoneally (i.p.) with 0.1 ml/10 g of body weight of a ketamine-xylazine mixture (100 mg/10 mg in 10 ml PBS). A cervical brush was inserted into the vaginal canal and rotated gently 15 times to create minor abrasions, and 25 μl of the viral supernatant was then pipetted into the vaginal canal. After gently abrading the site a second time, a second aliquot of 25 μl of virus was added. The total virus delivered was calculated to be 6 × 108 viral genome equivalents per mouse. At designated times postchallenge, mice were sacrificed, and the lower genital tract from the vaginal introitus to the cervix was dissected. One-third of the tissue, spanning the length of the genital tract, was placed into TRIzol for RNA extraction, and the remaining tissue was placed into formalin. Formalin-fixed tissue was embedded in paraffin, and 4-μm sections were prepared for RNAscope chromogenic in situ hybridization and hematoxylin and eosin staining.
Spliced transcript TaqMan reverse transcription-PCR assay.
We designed a TaqMan assay with an upstream primer in the C terminus of the E7 open reading frame downstream of the P533 promoter (nt 695 to nt 714, 5′-TAGCTTTGTCTGCCCGCACT-3′) and a reverse primer downstream of the 757/3139 splice junction site (nt 3213 to nt 3232, 5′-GTCAGTGGTGTCGGTGGGAA-3′). The probe was also located downstream of the splice junction site at nt 3179 to nt 3202 (5′-FAM-CGGCCCGAAGACAACACCGCCACG-3′-TAMRA). Nucleotide numbering is based on the reference sequence for Mus musculus papillomavirus type 1 isolate MusPV (GenBank accession number NC_014326). The amplicon was sequenced, and a search alignment was done to confirm the specificity of the primers and probe in the 159-nucleotide fragment.
Total cellular RNA was extracted using Direct-zol RNA miniprep plus (Zymo Research) according to the manufacturer’s instructions. RT was performed using the high-capacity cDNA RT kit (Invitrogen Corporation, San Diego, CA, USA). The primers and probe were added to universal PCR master mix (Applied Biosystems, Life Technologies, Foster City, CA, USA) at 500 and 250 nM, respectively, in a final volume of 20 μl using an ABI 7000 sequence detector (Applied Biosystems, Life Technologies). In addition to the MmuPV1 TaqMan assay, all samples were tested for transcripts of the endogenous control, the beta-actin gene (TaqMan assay identification Mm00607939_s1; Applied Biosystems, Life Technologies), to assess the efficiency of RNA extraction. Reactions were performed in triplicate for each primer-probe set. A standard for the 757/3139 spliced transcript fragment was obtained by cloning a 229-base pair fragment containing the 159-bp amplicon into a pUC57 plasmid using the Plasmid DNA Preparation Services of GenScript (Piscataway, NJ, USA). A linear relationship between the input target DNA copy numbers and CT values was obtained from 10-fold serial dilutions of the recombinant plasmid DNA. The absolute copy numbers of the 757/3139 spliced transcripts were inferred from the regression line of plasmid DNA standards and were normalized by micrograms of total RNA from samples.
Based on the published transcriptome of MmuPV1 (75), the assay is expected to detect most viral transcripts since the majority are initiated from the P533 promoter and/or utilize the 757/3139 splice junction. The assay will also detect transcripts that are initiated at 4 upstream promoters and use the 757/3139 splice junction site, including the major transcripts for the E6 and E7 genes. Importantly, the assay will detect both early and late transcripts. Late transcripts initiated from the P7107 promoter that utilize the 7243/3139 splice junction and singly spliced late transcripts that use the splice junction at position 5243 (∼35% of L1 transcripts) will not be detected. The assay will not detect the major E2 transcript that utilizes the 757/2493 splice junction or transcripts encoding E1, which are not spliced.
RNAscope in situ hybridization.
Custom RNA in situ hybridization probes to detect the full-length E6/E7 mRNA coding region sequence (nt 1 to nt 755) of MmuPV1 were designed and produced by Advanced Cell Diagnostics for use with the RNAscope 2.0 assay kit. To ensure RNA integrity and assay procedures, adjacent sections were also hybridized with a probe for the endogenous housekeeping gene ubiquitin (positive control) and the bacterial gene dapB (negative control). The assay was performed as previously described (20).
Intracellular cytokine staining.
Splenocytes were stimulated with 1 μg/ml of MmuPV1 E6 peptide (KNIVFVTVR) in the presence of brefeldin A (10 μg/ml) overnight at 37°C with 5% CO2. Cells were permeabilized with permeabilization buffer and stained with Pacific Blue-conjugated anti-mouse CD3, clone 17A2; allophycocyanin (APC)-conjugated anti-mouse CD8α, clone 53-6.7; and phycoerythrin (PE)-conjugated anti-mouse interferon gamma (IFN-γ), clone XMG1.2. Reagents were purchased from BioLegend. Flow cytometry was performed on an LSR-II flow cytometer, and data were analyzed using FACSDiva software. Gating was done on forward- and side-scatter parameters to select for lymphocytes and singlets. CD8+ T lymphocytes were identified on a CD3/CD8 dot plot of gated lymphocytes, and IFN-γ-secreting cells were identified on a CD8/IFN-γ dot plot of gated CD8+ T cells. A minimum of 22,000 CD8+ T cells were analyzed.
Enzyme-linked immunosorbent assay for anti-MmuPV1 capsid antibody.
An enzyme-linked immunosorbent assay (ELISA) was used to measure serum levels of antibody against MmuPV1 L1L2 pseudovirus (PsV). PsVs were produced in the human embryonic kidney cell line 293TT, which expresses high levels of the simian virus 40 (SV40) large T antigen. 293TT cells were cotransfected with a plasmid driving the expression of MmuPV1 L1 and L2 (catalog number 89926; Addgene) and a pcDNA3.1 plasmid, according to the protocol described previously by Buck et al. (77). For antigen binding, 100 ng/well of Optiprep gradient-purified PsV in Dulbecco’s phosphate-buffered saline (DPBS) was used to coat ELISA plates overnight at 4°C. The next day, plates were blocked with DPBS containing 4% skim milk and 0.2% Tween 20 for 1.5 h at room temperature and subsequently incubated with diluted sera (serial dilution starting at 1:50) in blocking buffer for 1 h at room temperature, followed by incubation with a horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG secondary antibody (catalog number A4416; Sigma-Aldrich) at a 1:10,000 dilution in blocking buffer for 1 h at room temperature. An HRP substrate (3,3′,5,5′-tetramethylbenzidine) was added, and the reaction was stopped with 0.36 N H2SO4 after a 25-min incubation in the dark at room temperature. The absorbance was read at dual wavelengths of 450 nm and 620 nm, and the background absorbance at 620 nm was subtracted from that at 450 nm. Data were displayed graphically as the absorbance at 450 nm on the y axis and as the reciprocal dilution on a log10 scale on the x axis.
ACKNOWLEDGMENTS
This work was supported in part by the following grants from the National Institutes of Health: R01CA233486, R01CA237067, P50CA098252, and P30CA06973. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was funded in part by the Sapienza University of Rome (Finanziamenti di Ateneo per la Ricerca Scientifica, 2019, RP11916B6EC60AF3) and by the MIUR Programmi di Ricerca Scientifica di Rilevante Interesse Nazionale (PRIN 2017, 20179JHAMZ_002). Fabiana Cannella was supported by a fellowship from the Institute Pasteur-Cenci Bolognetti Foundation, Sapienza University, Rome, Italy.
REFERENCES
- 1.Doorbar J. 2005. The papillomavirus life cycle. J Clin Virol 32(Suppl 1):S7–S15. doi: 10.1016/j.jcv.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 2.McBride AA. 2017. Mechanisms and strategies of papillomavirus replication. Biol Chem 398:919–927. doi: 10.1515/hsz-2017-0113. [DOI] [PubMed] [Google Scholar]
- 3.Longworth MS, Laimins LA. 2004. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol Mol Biol Rev 68:362–372. doi: 10.1128/MMBR.68.2.362-372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porter SS, Stepp WH, Stamos JD, McBride AA. 2017. Host cell restriction factors that limit transcription and replication of human papillomavirus. Virus Res 231:10–20. doi: 10.1016/j.virusres.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song D, Li H, Li H, Dai J. 2015. Effect of human papillomavirus infection on the immune system and its role in the course of cervical cancer. Oncol Lett 10:600–606. doi: 10.3892/ol.2015.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. 2007. Human papillomavirus and cervical cancer. Lancet 370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 7.Bienkowska-Haba M, Luszczek W, Myers JE, Keiffer TR, DiGiuseppe S, Polk P, Bodily JM, Scott RS, Sapp M. 2018. A new cell culture model to genetically dissect the complete human papillomavirus life cycle. PLoS Pathog 14:e1006846. doi: 10.1371/journal.ppat.1006846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biryukov J, Meyers C. 2015. Papillomavirus infectious pathways: a comparison of systems. Viruses 7:4303–4325. doi: 10.3390/v7082823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow LT, Duffy AA, Wang HK, Broker TR. 2009. A highly efficient system to produce infectious human papillomavirus: elucidation of natural virus-host interactions. Cell Cycle 8:1319–1323. doi: 10.4161/cc.8.9.8242. [DOI] [PubMed] [Google Scholar]
- 10.Rector A, Van Ranst M. 2013. Animal papillomaviruses. Virology 445:213–223. doi: 10.1016/j.virol.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Christensen ND, Budgeon LR, Cladel NM, Hu J. 2017. Recent advances in preclinical model systems for papillomaviruses. Virus Res 231:108–118. doi: 10.1016/j.virusres.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingle A, Ghim S, Joh J, Chepkoech I, Bennett JA, Sundberg JP. 2011. Novel laboratory mouse papillomavirus (MusPV) infection. Vet Pathol 48:500–505. doi: 10.1177/0300985810377186. [DOI] [PubMed] [Google Scholar]
- 13.Joh J, Jenson AB, King W, Proctor M, Ingle A, Sundberg JP, Ghim SJ. 2011. Genomic analysis of the first laboratory-mouse papillomavirus. J Gen Virol 92:692–698. doi: 10.1099/vir.0.026138-0. [DOI] [PubMed] [Google Scholar]
- 14.Cladel NM, Budgeon LR, Balogh KK, Cooper TK, Hu J, Christensen ND. 2015. A novel pre-clinical murine model to study the life cycle and progression of cervical and anal papillomavirus infections. PLoS One 10:e0120128. doi: 10.1371/journal.pone.0120128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cladel NM, Budgeon LR, Balogh KK, Cooper TK, Hu J, Christensen ND. 2016. Mouse papillomavirus MmuPV1 infects oral mucosa and preferentially targets the base of the tongue. Virology 488:73–80. doi: 10.1016/j.virol.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cladel NM, Budgeon LR, Cooper TK, Balogh KK, Hu J, Christensen ND. 2013. Secondary infections, expanded tissue tropism, and evidence for malignant potential in immunocompromised mice infected with Mus musculus papillomavirus 1 DNA and virus. J Virol 87:9391–9395. doi: 10.1128/JVI.00777-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Handisurya A, Day PM, Thompson CD, Bonelli M, Lowy DR, Schiller JT. 2014. Strain-specific properties and T cells regulate the susceptibility to papilloma induction by Mus musculus papillomavirus 1. PLoS Pathog 10:e1004314. doi: 10.1371/journal.ppat.1004314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Handisurya A, Day PM, Thompson CD, Buck CB, Pang YY, Lowy DR, Schiller JT. 2013. Characterization of Mus musculus papillomavirus 1 infection in situ reveals an unusual pattern of late gene expression and capsid protein localization. J Virol 87:13214–13225. doi: 10.1128/JVI.02162-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu J, Budgeon LR, Cladel NM, Balogh K, Myers R, Cooper TK, Christensen ND. 2015. Tracking vaginal, anal and oral infection in a mouse papillomavirus infection model. J Gen Virol 96:3554–3565. doi: 10.1099/jgv.0.000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang RT, Wang JW, Peng S, Huang TC, Wang C, Cannella F, Chang YN, Viscidi RP, Best SRA, Hung CF, Roden RBS. 2017. Spontaneous and vaccine-induced clearance of Mus musculus papillomavirus 1 infection. J Virol 91:e00699-17. doi: 10.1128/JVI.00699-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyers JM, Uberoi A, Grace M, Lambert PF, Munger K. 2017. Cutaneous HPV8 and MmuPV1 E6 proteins target the NOTCH and TGF-beta tumor suppressors to inhibit differentiation and sustain keratinocyte proliferation. PLoS Pathog 13:e1006171. doi: 10.1371/journal.ppat.1006171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulz E, Gottschling M, Ulrich RG, Richter D, Stockfleth E, Nindl I. 2012. Isolation of three novel rat and mouse papillomaviruses and their genomic characterization. PLoS One 7:e47164. doi: 10.1371/journal.pone.0047164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sundberg JP, Stearns TM, Joh J, Proctor M, Ingle A, Silva KA, Dadras SS, Jenson AB, Ghim SJ. 2014. Immune status, strain background, and anatomic site of inoculation affect mouse papillomavirus (MmuPV1) induction of exophytic papillomas or endophytic trichoblastomas. PLoS One 9:e113582. doi: 10.1371/journal.pone.0113582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uberoi A, Yoshida S, Frazer IH, Pitot HC, Lambert PF. 2016. Role of ultraviolet radiation in papillomavirus-induced disease. PLoS Pathog 12:e1005664. doi: 10.1371/journal.ppat.1005664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang JW, Jiang R, Peng S, Chang YN, Hung CF, Roden RB. 2015. Immunologic control of Mus musculus papillomavirus type 1. PLoS Pathog 11:e1005243. doi: 10.1371/journal.ppat.1005243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cladel NM, Budgeon LR, Balogh KK, Cooper TK, Brendle SA, Christensen ND, Schell TD, Hu J. 2017. Mouse papillomavirus infection persists in mucosal tissues of an immunocompetent mouse strain and progresses to cancer. Sci Rep 7:16932. doi: 10.1038/s41598-017-17089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasan UA, Zannetti C, Parroche P, Goutagny N, Malfroy M, Roblot G, Carreira C, Hussain I, Muller M, Taylor-Papadimitriou J, Picard D, Sylla BS, Trinchieri G, Medzhitov R, Tommasino M. 2013. The human papillomavirus type 16 E7 oncoprotein induces a transcriptional repressor complex on the Toll-like receptor 9 promoter. J Exp Med 210:1369–1387. doi: 10.1084/jem.20122394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reiser J, Hurst J, Voges M, Krauss P, Munch P, Iftner T, Stubenrauch F. 2011. High-risk human papillomaviruses repress constitutive kappa interferon transcription via E6 to prevent pathogen recognition receptor and antiviral-gene expression. J Virol 85:11372–11380. doi: 10.1128/JVI.05279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lo Cigno I, De Andrea M, Borgogna C, Albertini S, Landini MM, Peretti A, Johnson KE, Chandran B, Landolfo S, Gariglio M. 2015. The nuclear DNA sensor IFI16 acts as a restriction factor for human papillomavirus replication through epigenetic modifications of the viral promoters. J Virol 89:7506–7520. doi: 10.1128/JVI.00013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lau L, Gray EE, Brunette RL, Stetson DB. 2015. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science 350:568–571. doi: 10.1126/science.aab3291. [DOI] [PubMed] [Google Scholar]
- 31.Sunthamala N, Thierry F, Teissier S, Pientong C, Kongyingyoes B, Tangsiriwatthana T, Sangkomkamhang U, Ekalaksananan T. 2014. E2 proteins of high risk human papillomaviruses down-modulate STING and IFN-kappa transcription in keratinocytes. PLoS One 9:e91473. doi: 10.1371/journal.pone.0091473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiang C, Pauli EK, Biryukov J, Feister KF, Meng M, White EA, Munger K, Howley PM, Meyers C, Gack MU. 2018. The human papillomavirus E6 oncoprotein targets USP15 and TRIM25 to suppress RIG-I-mediated innate immune signaling. J Virol 92:e01737-17. doi: 10.1128/JVI.01737-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perea SE, Massimi P, Banks L. 2000. Human papillomavirus type 16 E7 impairs the activation of the interferon regulatory factor-1. Int J Mol Med 5:661–666. doi: 10.3892/ijmm.5.6.661. [DOI] [PubMed] [Google Scholar]
- 34.Barnard P, McMillan NA. 1999. The human papillomavirus E7 oncoprotein abrogates signaling mediated by interferon-alpha. Virology 259:305–313. doi: 10.1006/viro.1999.9771. [DOI] [PubMed] [Google Scholar]
- 35.Andersen JM, Al-Khairy D, Ingalls RR. 2006. Innate immunity at the mucosal surface: role of Toll-like receptor 3 and Toll-like receptor 9 in cervical epithelial cell responses to microbial pathogens. Biol Reprod 74:824–831. doi: 10.1095/biolreprod.105.048629. [DOI] [PubMed] [Google Scholar]
- 36.Li S, Labrecque S, Gauzzi MC, Cuddihy AR, Wong AH, Pellegrini S, Matlashewski GJ, Koromilas AE. 1999. The human papilloma virus (HPV)-18 E6 oncoprotein physically associates with Tyk2 and impairs Jak-STAT activation by interferon-alpha. Oncogene 18:5727–5737. doi: 10.1038/sj.onc.1202960. [DOI] [PubMed] [Google Scholar]
- 37.Ronco LV, Karpova AY, Vidal M, Howley PM. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev 12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nees M, Geoghegan JM, Hyman T, Frank S, Miller L, Woodworth CD. 2001. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-kappaB-responsive genes in cervical keratinocytes. J Virol 75:4283–4296. doi: 10.1128/JVI.75.9.4283-4296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong S, Mehta KP, Laimins LA. 2011. Suppression of STAT-1 expression by human papillomaviruses is necessary for differentiation-dependent genome amplification and plasmid maintenance. J Virol 85:9486–9494. doi: 10.1128/JVI.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dinarello CA. 2009. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol 27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 41.Iglesias M, Yen K, Gaiotti D, Hildesheim A, Stoler MH, Woodworth CD. 1998. Human papillomavirus type 16 E7 protein sensitizes cervical keratinocytes to apoptosis and release of interleukin-1alpha. Oncogene 17:1195–1205. doi: 10.1038/sj.onc.1202054. [DOI] [PubMed] [Google Scholar]
- 42.Ainouze M, Rochefort P, Parroche P, Roblot G, Tout I, Briat F, Zannetti C, Marotel M, Goutagny N, Auron P, Traverse-Glehen A, Lunel-Potencier A, Golfier F, Masson M, Robitaille A, Tommasino M, Carreira C, Walzer T, Henry T, Zanier K, Trave G, Hasan UA. 2018. Human papillomavirus type 16 antagonizes IRF6 regulation of IL-1beta. PLoS Pathog 14:e1007158. doi: 10.1371/journal.ppat.1007158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klymenko T, Gu Q, Herbert I, Stevenson A, Iliev V, Watkins G, Pollock C, Bhatia R, Cuschieri K, Herzyk P, Gatherer D, Graham SV. 2017. RNA-Seq analysis of differentiated keratinocytes reveals a massive response to late events during human papillomavirus 16 infection, including loss of epithelial barrier function. J Virol 91:e01001-17. doi: 10.1128/JVI.01001-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karim R, Meyers C, Backendorf C, Ludigs K, Offringa R, van Ommen GJ, Melief CJ, van der Burg SH, Boer JM. 2011. Human papillomavirus deregulates the response of a cellular network comprising of chemotactic and proinflammatory genes. PLoS One 6:e17848. doi: 10.1371/journal.pone.0017848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karim R, Tummers B, Meyers C, Biryukov JL, Alam S, Backendorf C, Jha V, Offringa R, van Ommen GJ, Melief CJ, Guardavaccaro D, Boer JM, van der Burg SH. 2013. Human papillomavirus (HPV) upregulates the cellular deubiquitinase UCHL1 to suppress the keratinocyte’s innate immune response. PLoS Pathog 9:e1003384. doi: 10.1371/journal.ppat.1003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lebre MC, van der Aar AM, van Baarsen L, van Capel TM, Schuitemaker JH, Kapsenberg ML, de Jong EC. 2007. Human keratinocytes express functional Toll-like receptor 3, 4, 5, and 9. J Invest Dermatol 127:331–341. doi: 10.1038/sj.jid.5700530. [DOI] [PubMed] [Google Scholar]
- 47.Huang SM, McCance DJ. 2002. Down regulation of the interleukin-8 promoter by human papillomavirus type 16 E6 and E7 through effects on CREB binding protein/p300 and P/CAF. J Virol 76:8710–8721. doi: 10.1128/jvi.76.17.8710-8721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kleine-Lowinski K, Rheinwald JG, Fichorova RN, Anderson DJ, Basile J, Münger K, Daly CM, Rösl F, Rollins BJ. 2003. Selective suppression of monocyte chemoattractant protein-1 expression by human papillomavirus E6 and E7 oncoproteins in human cervical epithelial and epidermal cells. Int J Cancer 107:407–415. doi: 10.1002/ijc.11411. [DOI] [PubMed] [Google Scholar]
- 49.Guess JC, McCance DJ. 2005. Decreased migration of Langerhans precursor-like cells in response to human keratinocytes expressing human papillomavirus type 16 E6/E7 is related to reduced macrophage inflammatory protein-3alpha production. J Virol 79:14852–14862. doi: 10.1128/JVI.79.23.14852-14862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sperling T, Ołdak M, Walch-Rückheim B, Wickenhauser C, Doorbar J, Pfister H, Malejczyk M, Majewski S, Keates AC, Smola S. 2012. Human papillomavirus type 8 interferes with a novel C/EBPbeta-mediated mechanism of keratinocyte CCL20 chemokine expression and Langerhans cell migration. PLoS Pathog 8:e1002833. doi: 10.1371/journal.ppat.1002833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caberg JH, Hubert P, Herman L, Herfs M, Roncarati P, Boniver J, Delvenne P. 2009. Increased migration of Langerhans cells in response to HPV16 E6 and E7 oncogene silencing: role of CCL20. Cancer Immunol Immunother 58:39–47. doi: 10.1007/s00262-008-0522-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang B, Xue M. 2015. Correlation of E6 and E7 levels in high-risk HPV16 type cervical lesions with CCL20 and Langerhans cells. Genet Mol Res 14:10473–10481. doi: 10.4238/2015.September.8.8. [DOI] [PubMed] [Google Scholar]
- 53.Thapa M, Carr DJ. 2009. CXCR3 deficiency increases susceptibility to genital herpes simplex virus type 2 infection: uncoupling of CD8+ T-cell effector function but not migration. J Virol 83:9486–9501. doi: 10.1128/JVI.00854-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thapa M, Welner RS, Pelayo R, Carr DJ. 2008. CXCL9 and CXCL10 expression are critical for control of genital herpes simplex virus type 2 infection through mobilization of HSV-specific CTL and NK cells to the nervous system. J Immunol 180:1098–1106. doi: 10.4049/jimmunol.180.2.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ridiandries A, Tan JTM, Bursill CA. 2018. The role of chemokines in wound healing. Int J Mol Sci 19:3217. doi: 10.3390/ijms19103217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ho GY, Studentsov YY, Bierman R, Burk RD. 2004. Natural history of human papillomavirus type 16 virus-like particle antibodies in young women. Cancer Epidemiol Biomarkers Prev 13:110–116. doi: 10.1158/1055-9965.epi-03-0191. [DOI] [PubMed] [Google Scholar]
- 57.Huh WK, Ault KA, Chelmow D, Davey DD, Goulart RA, Garcia FA, Kinney WK, Massad LS, Mayeaux EJ, Saslow D, Schiffman M, Wentzensen N, Lawson HW, Einstein MH. 2015. Use of primary high-risk human papillomavirus testing for cervical cancer screening: interim clinical guidance. Gynecol Oncol 136:178–182. doi: 10.1016/j.ygyno.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 58.Ronco G, Dillner J, Elfström KM, Tunesi S, Snijders PJ, Arbyn M, Kitchener H, Segnan N, Gilham C, Giorgi-Rossi P, Berkhof J, Peto J, Meijer CJ, International HPV Screening Working Group. 2014. Efficacy of HPV-based screening for prevention of invasive cervical cancer: follow-up of four European randomised controlled trials. Lancet 383:524–532. doi: 10.1016/S0140-6736(13)62218-7. [DOI] [PubMed] [Google Scholar]
- 59.Chen K, Liu J, Cao X. 2017. Regulation of type I interferon signaling in immunity and inflammation: a comprehensive review. J Autoimmun 83:1–11. doi: 10.1016/j.jaut.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 60.Turner MD, Nedjai B, Hurst T, Pennington DJ. 2014. Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochim Biophys Acta 1843:2563–2582. doi: 10.1016/j.bbamcr.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 61.Colonna M. 2018. Innate lymphoid cells: diversity, plasticity, and unique functions in immunity. Immunity 48:1104–1117. doi: 10.1016/j.immuni.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rivera A, Siracusa MC, Yap GS, Gause WC. 2016. Innate cell communication kick-starts pathogen-specific immunity. Nat Immunol 17:356–363. doi: 10.1038/ni.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Groom JR, Luster AD. 2011. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol 89:207–215. doi: 10.1038/icb.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuo PT, Zeng Z, Salim N, Mattarollo S, Wells JW, Leggatt GR. 2018. The role of CXCR3 and its chemokine ligands in skin disease and cancer. Front Med (Lausanne) 5:271. doi: 10.3389/fmed.2018.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee HY, Schripsema JH, Sigar IM, Lacy SR, Kasimos JN, Murray CM, Ramsey KH. 2010. A role for CXC chemokine receptor-2 in the pathogenesis of urogenital Chlamydia muridarum infection in mice. FEMS Immunol Med Microbiol 60:49–56. doi: 10.1111/j.1574-695X.2010.00715.x. [DOI] [PubMed] [Google Scholar]
- 66.Kobayashi Y. 2008. The role of chemokines in neutrophil biology. Front Biosci 13:2400–2407. doi: 10.2741/2853. [DOI] [PubMed] [Google Scholar]
- 67.Koh TJ, DiPietro LA. 2011. Inflammation and wound healing: the role of the macrophage. Expert Rev Mol Med 13:e23. doi: 10.1017/S1462399411001943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stentella P, Frega A, Di Renzi F, Palazzetti PL, Pachi A. 1996. Topic and systemic administration of natural alfa interferon in the treatment of female and male HPV genital infections. Clin Exp Obstet Gynecol 23:29–36. [PubMed] [Google Scholar]
- 69.Kawai T, Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 70.Kim B, Lee Y, Kim E, Kwak A, Ryoo S, Bae SH, Azam T, Kim S, Dinarello CA. 2013. The interleukin-1alpha precursor is biologically active and is likely a key alarmin in the IL-1 family of cytokines. Front Immunol 4:391. doi: 10.3389/fimmu.2013.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Di Paolo NC, Shayakhmetov DM. 2016. Interleukin 1alpha and the inflammatory process. Nat Immunol 17:906–913. doi: 10.1038/ni.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Witte CJ, van de Sande AJ, van Beekhuizen HJ, Koeneman MM, Kruse AJ, Gerestein CG. 2015. Imiquimod in cervical, vaginal and vulvar intraepithelial neoplasia: a review. Gynecol Oncol 139:377–384. doi: 10.1016/j.ygyno.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 73.Yang R, Murillo FM, Cui H, Blosser R, Uematsu S, Takeda K, Akira S, Viscidi RP, Roden RB. 2004. Papillomavirus-like particles stimulate murine bone marrow-derived dendritic cells to produce alpha interferon and Th1 immune responses via MyD88. J Virol 78:11152–11160. doi: 10.1128/JVI.78.20.11152-11160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang R, Murillo FM, Delannoy MJ, Blosser RL, Yutzy WH, Uematsu S, Takeda K, Akira S, Viscidi RP, Roden RB. 2005. B lymphocyte activation by human papillomavirus-like particles directly induces Ig class switch recombination via TLR4-MyD88. J Immunol 174:7912–7919. doi: 10.4049/jimmunol.174.12.7912. [DOI] [PubMed] [Google Scholar]
- 75.Xue XY, Majerciak V, Uberoi A, Kim BH, Gotte D, Chen X, Cam M, Lambert PF, Zheng ZM. 2017. The full transcription map of mouse papillomavirus type 1 (MmuPV1) in mouse wart tissues. PLoS Pathog 13:e1006715. doi: 10.1371/journal.ppat.1006715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schoggins JW, Rice CM. 2011. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol 1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Buck CB, Pastrana DV, Lowy DR, Schiller JT. 2004. Efficient intracellular assembly of papillomaviral vectors. J Virol 78:751–757. doi: 10.1128/jvi.78.2.751-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]