Birds are important hosts of viruses that have the potential to cause zoonotic infections in humans. However, only a few antiviral genes (called viral restriction factors) have been described in birds, mostly because birds lack counterparts of highly studied mammalian restriction factors. Tetherin/BST-2 is a restriction factor, originally described in humans, that blocks the release of newly formed virus particles from infected cells. Recent work identified BST-2 in nonmammalian vertebrate species, including birds. Here, we report the BST-2 sequence in domestic chicken and describe its antiviral activity against a prototypical avian retrovirus, avian sarcoma and leukosis virus (ASLV). We also identify BST-2 genes in multiple avian species and show that they evolve rapidly in birds, which is an important indication of their relevance for antiviral defense. Analysis of avian BST-2 genes will shed light on defense mechanisms against avian viral pathogens.
KEYWORDS: avian retrovirus, chicken, restriction factor, tetherin
ABSTRACT
Tetherin/BST-2 is an antiviral protein that blocks the release of enveloped viral particles by linking them to the membrane of producing cells. At first, BST-2 genes were described only in humans and other mammals. Recent work identified BST-2 orthologs in nonmammalian vertebrates, including birds. Here, we identify the BST-2 sequence in domestic chicken (Gallus gallus) for the first time and demonstrate its activity against avian sarcoma and leukosis virus (ASLV). We generated a BST-2 knockout in chicken cells and showed that BST-2 is a major determinant of an interferon-induced block of ASLV release. Ectopic expression of chicken BST-2 blocks the release of ASLV in chicken cells and of human immunodeficiency virus type 1 (HIV-1) in human cells. Using metabolic labeling and pulse-chase analysis of HIV-1 Gag proteins, we verified that chicken BST-2 blocks the virus at the release stage. Furthermore, we describe BST-2 orthologs in multiple avian species from 12 avian orders. Previously, some of these species were reported to lack BST-2, highlighting the difficulty of identifying sequences of this extremely variable gene. We analyzed BST-2 genes in the avian orders Galliformes and Passeriformes and showed that they evolve under positive selection. This indicates that avian BST-2 is involved in host-virus evolutionary arms races and suggests that BST-2 antagonists exist in some avian viruses. In summary, we show that chicken BST-2 has the potential to act as a restriction factor against ASLV. Characterizing the interaction of avian BST-2 with avian viruses is important in understanding innate antiviral defenses in birds.
IMPORTANCE Birds are important hosts of viruses that have the potential to cause zoonotic infections in humans. However, only a few antiviral genes (called viral restriction factors) have been described in birds, mostly because birds lack counterparts of highly studied mammalian restriction factors. Tetherin/BST-2 is a restriction factor, originally described in humans, that blocks the release of newly formed virus particles from infected cells. Recent work identified BST-2 in nonmammalian vertebrate species, including birds. Here, we report the BST-2 sequence in domestic chicken and describe its antiviral activity against a prototypical avian retrovirus, avian sarcoma and leukosis virus (ASLV). We also identify BST-2 genes in multiple avian species and show that they evolve rapidly in birds, which is an important indication of their relevance for antiviral defense. Analysis of avian BST-2 genes will shed light on defense mechanisms against avian viral pathogens.
INTRODUCTION
The genomes of mammals and other vertebrates contain genes whose presumed primary function is to block foreign pathogens (here, we will focus on the blocking of retroviruses). These genes, called viral restriction factors (RFs), target various stages of the retroviral replication cycle, usually at a conserved step, which makes it hard for the virus to circumvent by simple escape mutations (1, 2). In turn, retroviruses often develop elaborate countermeasures to inactivate the RFs or target them for degradation. Repeated cycles of genetic conflict between host RFs and viruses accelerate evolutionary change in RF genes, which manifests as a strong positive selection signature. Further, RFs are often induced by type I interferons (IFNs) and constitute the effector components of innate immune defense.
Many RFs have been identified in mammals, especially in primates, but only a few in birds. Importantly, birds lack orthologs of the two most widely studied mammalian RFs: tripartite motif 5 (TRIM5) and apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3 (APOBEC3) (3). One of the known avian RFs is Daxx in chicken, which restricts the avian sarcoma and leukosis virus (ASLV) by promoting the epigenetic silencing of integrated proviruses (4). Three other chicken proteins—CCCH-type zinc-finger antiviral protein (ZAP), TRIM62, and cholesterol 25-hydroxylase (CH25H)—have been reported to inhibit ASLV, but without detailed characterization of their inhibitory mechanism or the replication stage blocked (5–7). There are other candidates for retroviral RFs in birds, but so far, they have only been shown to block nonretroviral viruses. The following candidates have been found in chicken: viperin, interferon-induced proteins with tetratricopeptide repeats (IFITs), interferon-inducible transmembrane proteins (IFITM), and the Mx protein; however, the antiviral activity of the Mx protein in chicken remains controversial (8–14).
Tetherin, also known as bone marrow stromal antigen 2 (BST-2), was initially identified in human cells as a RF that blocks the release of retroviruses by physically linking (tethering) newly formed virions to the cell membrane (15, 16). The unique topology of the BST-2 protein, with an N-terminal transmembrane (TM) domain and a C-terminal glycosyl-phosphatidylinositol (GPI) anchor, allows it to span both the viral and cellular membranes (17). BST-2 forms homodimers and does not require interaction with specific viral components except for the viral membrane. This nonspecific mode of action allows BST-2 to target not just retroviruses but also many other enveloped viruses (reviewed in reference 18). Several viral BST-2 antagonists have been described, which are at least partially able to block its effects, either by its degradation or by interfering with its transport and recycling from the cellular membrane. These antagonists include the human immunodeficiency virus type 1 (HIV-1) Vpu protein, Nef in simian immunodeficiency virus (SIV), Env in HIV-2, GP in the Ebola virus, K5 in Kaposi’s sarcoma-associated herpesvirus (KSHV), and a growing list of other viral proteins (15, 16, 19–21).
BST-2 was initially described only in primates and several other mammalian species. Recently, however, the deeper evolutionary origins of this gene and its surrounding genetic locus were revealed. BST-2 orthologs capable of antiviral activity were found in the genomes of multiple vertebrates, including birds, alligators, turtles, and various representatives of fish (22, 23). These distant orthologs have low sequence similarity to the mammalian proteins, but all retain the typical structural topology that is considered crucial for their antiviral function. While BST-2 was identified in some bird species, it was reported to be absent in others and has never been described in chicken. Chicken (Gallus gallus) represents an important model organism, and the study of its viral diseases is of high priority due to its importance in human nutrition (24).
In this study, we report for the first time the sequence of chicken BST-2 (chBST-2), and we characterize its inhibitory activity against a prototypic avian retrovirus, ASLV, as well as against HIV-1. We also describe BST-2 orthologs in multiple avian species, including all those species in which the gene was previously reported to be missing. Finally, we have detected positive selection acting on avian BST-2, reflecting its involvement in a host-virus evolutionary arms race.
RESULTS
Identification of chBST-2 in the chicken genome.
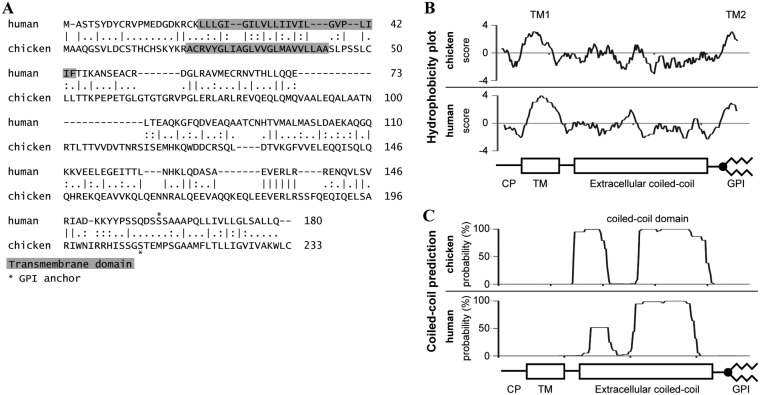
We identified the chBST-2 sequence based on sequence homology with previously reported avian BST-2 genes (22, 23). It resides on chicken chromosome 28 in a locus syntenic to other vertebrate BST-2 genes, between the conserved CILP2 and GTPBP3 genes. There are no confidently annotated genes at this locus in the current chicken genome assembly (galGal6), only various gene predictions. We verified our chBST-2 coding sequence prediction by comparison with publicly available chicken Illumina transcriptome sequencing (RNA-seq) data and by reverse transcription-PCR (RT-PCR) from chicken cDNA (see Materials and Methods) and submitted the sequence to GenBank (accession number MN326300). The predicted chBST-2 amino acid sequence shows a low degree of conservation with the human ortholog, with 23% identity and 36% similarity (Fig. 1A). However, the predicted secondary structure seems to be well conserved: hydrophobicity plots show an N-terminal TM domain and a C-terminal hydrophobic domain, including a GPI prediction, in both proteins (Fig. 1B). Also, structural analysis predicts coiled-coil extracellular domain structures for both chicken and human BST-2 proteins (Fig. 1C). The cytoplasmic domains are similar in size but quite different in primary sequence, including a missing YxY motif in chBST-2, a motif that takes part in the NF-κB signaling pathway in human cells (25). Human BST-2 has three extracellular domain cysteines; chicken BST-2 has only two. However, it was previously shown that a mutated human BST-2 with only a single cysteine has intact antiviral activity (26). Overall, based on the sequence analysis, the chBST-2 is predicted to encode a functional antiviral protein.
FIG 1.
Sequence and secondary structure prediction of chicken BST-2. (A) Pairwise alignment of chicken and human BST-2 amino acid sequences. Dashes represent gaps; bars, colons, and dots indicate identical, strongly similar, and weakly similar amino acids, respectively. The predicted positions of transmembrane regions and GPI anchor attachment (omega site) are shown. Hydrophobicity (B) and coiled-coil (C) scores were predicted from primary protein sequences and described in Materials and Methods and plotted for both chicken and human BST-2, in a scale that equalizes their lengths. Schematic drawings describe the approximate positions of predicted cytoplasmic part (CP), transmembrane (TM), extracellular, and GPI domains.
Endogenous chBST-2 is required for an IFN-induced block of ASLV release.
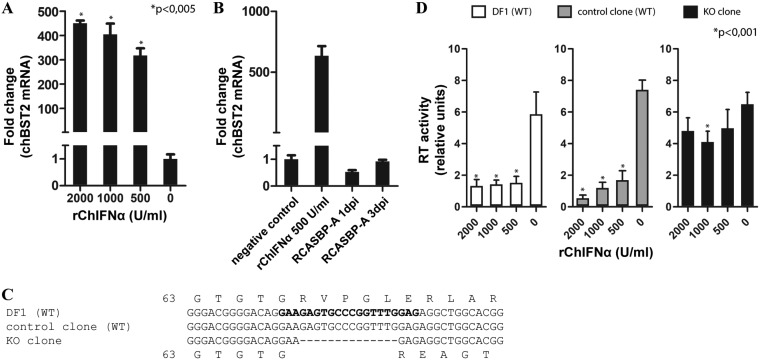
First, we tested whether chBST-2 is IFN inducible (like human BST-2). Recombinant chicken IFN-α (rChIFN-α) treatment of chicken DF-1 cells strongly induced chBST-2 mRNA expression in a dose-dependent manner (Fig. 2A). The baseline level of chBST-2 expression in DF-1 cells—a chicken fibroblast cell line—is very low. Infection of these cells with ASLV-based replication-competent ASLV long-terminal repeat with a Splice acceptor (RCAS) vector (27) does not induce chBST-2 mRNA (Fig. 2B). To evaluate the importance of endogenous chicken BST-2 for retroviral restriction, we generated a chBST-2 knockout in DF-1 cells using the CRISPR/Cas9 technique. The knockout clone contained a homozygous 14-bp deletion, predicted to result in a frameshift after amino acid glycine at position 67 and expression of a nonfunctional protein (Fig. 2C). The homozygous presence of the 14-bp deletion was verified both from genomic DNA and RNA isolated from the knockout DF-1 cell clone. As controls in subsequent infection experiments, we used a normal population of DF-1 cells and a DF-1 cell clone with an intact chBST-2 locus.
FIG 2.
chBST-2 knockout and analysis of IFN-induced block of ASLV release. (A) DF-1 cells were treated with increasing amounts of rChIFN-α. After 24 h, the medium was exchanged and IFN was added at the same concentration as before. Two days after the first IFN treatment, total RNA from the cells was isolated and used to quantify chBST-2 mRNA expression by qRT-PCR. The data were normalized to chicken GAPDH. (B) DF-1 cells were infected with RCASBP(A)GFP at a multiplicity of infection (MOI) of 1 and harvested for RNA isolation on days 1 or 3 postinfection (dpi). The chBST-2 mRNA expression was determined by qRT-PCR. As positive control, cells treated with rChIFN-α were used. (C) Alignment of chBST-2 sequences around the region targeted by the CRISPR/Cas9 construct (sequences used in the targeting oligonucleotide are in bold type). The 14-bp deletion in the knockout (KO) clone is shown. Predicted amino acid translation starting at glycine 63 is shown for both WT and KO variants. (D) Cells (WT DF-1, control clone, and KO clone) were infected with RCASBP(A) vector and passaged several times to achieve complete virus spread. Each of the three cell types was seeded on 12-well plates and, after 1 h, treated with rChIFN-α at the indicated concentration. After 24 h, the medium was exchanged and rChIFN-α was added at the same concentration as before. Forty-eight hours after cell seeding, the virus-containing cell culture media were harvested and used to quantify the amount of reverse transcriptase activity by the product-enhanced reverse transcriptase (PERT) assay as described in Materials and Methods. The means and standard deviations from three (panels A and B) and six (panel D) independent replicates are shown. Statistical significance calculated using Welch’s t test is indicated above the graphs.
The cells were infected with the RCAS vector and passaged several times to allow the virus to spread throughout the culture. In such chronically infected cells, new infection is blocked by receptor interference, and effects of various treatments on virus production can be monitored. These infected cells were then seeded and treated with various doses of rChIFN-α. After 2 days, the release of RCAS particles was quantified by a product-enhanced reverse transcriptase assay (RT assay) using the cell culture medium. The DF-1 cells and the wild-type (WT) clone showed a strong IFN-dependent block of RCAS production (Fig. 2D). In the chBST-2 knockout clone, the block was almost absent, demonstrating a major contribution of chBST-2 to late-phase retroviral restriction in DF-1 cells.
chBST-2 expression blocks ASLV release from chicken DF-1 cells.
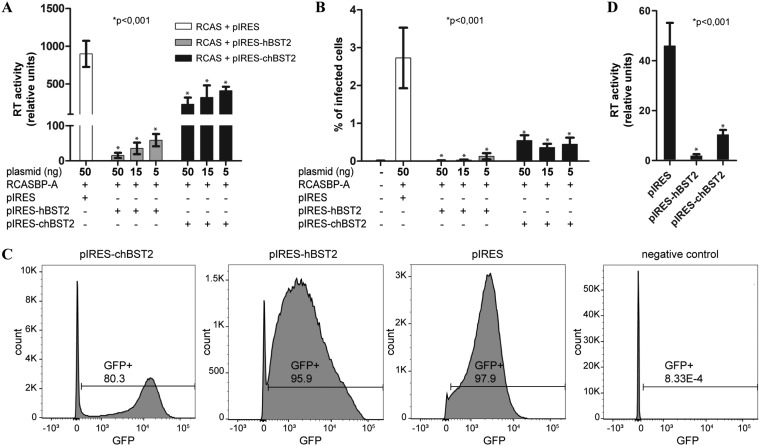
Next, we sought to determine the effect of ectopic expression of chBST-2 on RCAS particle production and infectivity. We constructed plasmids expressing chicken or human BST-2, joined together with a green fluorescent protein (GFP) through an internal ribosomal entry site (IRES). This design enabled indirect monitoring of BST-2 protein expression (with no specific antibody being available) by following the GFP that is cotranslated with BST-2 from the bicistronic vector mRNA. The RCAS vector encoding a GFP reporter [RCASBP(A)GFP] was cotransfected with the chicken (pIRES-chBST2) or human (pIRES-hBST2) tetherin-expressing plasmids into DF-1 cells. The levels of virus release were determined 2 days later by RT assays of the cell medium. Both plasmids significantly inhibited virus release compared to that in mock-transfected control cells (Fig. 3A).
FIG 3.
Block of ASLV release in chicken cells with ectopic expression of chBST-2. (A) Chicken DF-1 cells (1.2 × 104 per well) were seeded on a 96-well plate. After 24 h, the cells were transfected using 0.3 μl X-tremeGENE reagent (Sigma) with 100 ng of RCASBP(A)GFP and various doses (5 ng, 15 ng, or 50 ng) of pIRES-hBST2, pIRES-chBST2, or pIRES plasmids. Two days later, samples of cell culture media were collected and analyzed for RT activity by the PERT assay. Addition of up to 50 ng of control plasmid DNA (in place of BST-2 constructs) in transfection did not influence the PERT results (data not shown). (B) Virus-containing culture media from the experiment described above were used for infection of naive DF-1 cells. After 2 days, the cells were washed with PBS, trypsinized, and fixed in 1% paraformaldehyde. The percentage of GFP-positive cells was quantified by fluorescence-activated cell sorting using an LSR II analyzer (Becton, Dickinson). (C) Flow cytometry analysis of chicken cells stably expressing human or chicken BST-2. The stably expressing cells were established as described in Materials and Methods. The GFP expression from the pIRES vector is shown in each plot. (D) To quantify the ASLV release block in stable transfectants, 1.5 × 105 cells per well were seeded in 12-well plates and, after 24 h, transfected using 2 μl of Lipofectamine 3000 (Thermo Fisher) with 100 ng RCASBP(A) and 900 ng empty control plasmid. After 2 days, the reverse transcriptase activity in culture media was quantified by the PERT assay. The means and standard deviations from six independent replicates are shown, and statistical significance (Welch’s t test) is indicated above the graphs.
In addition to determining the RT activity, we also quantified the amount of infectious RCAS virus released in this experiment. Equal volumes of virus-containing cell culture media were used to infect naive DF-1 cells. After 2 days, the percentage of infected cells was determined by flow cytometry analysis for GFP, which is carried by the RCAS vector. A similar pattern of inhibition was obtained, with human BST-2 causing a strong decrease of total infectious virus released, and chicken BST-2 having a smaller but statistically significant effect (Fig. 3B).
We also generated stable DF-1 cell lines expressing chBST-2 or hBST-2, or the empty pIRES plasmid. DF-1 cells were first transfected with the respective plasmid, selected for 2 weeks with G418, and then twice sorted for GFP by flow cytometry (both G418 resistance and GFP are present in the pIRES and pIRES-BST2 vectors), until the vast majority of cells were GFP positive (Fig. 3C). All three stable cell lines were then transfected with the RCAS vector, and the virus release was determined 2 days later by the RT assay. Again, both human and chicken BST-2 caused a significant decrease of RCAS virions released (Fig. 3D), without apparent signs of cellular toxicity.
chBST-2 expression blocks HIV-1 release from human cells.
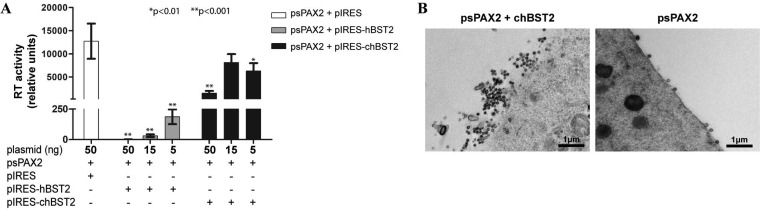
Next, we tested whether chBST-2 can inhibit the release of a heterologous retrovirus, HIV-1. pIRES-chBST2 or pIRES-hBST2 plasmids were cotransfected with an HIV-1-based gag-pol expression plasmid, psPAX2, into human 293T cells, and the release of virus-like particles (VLPs) was determined 2 days later by RT assays of the cell culture medium. Both plasmids inhibited the production of HIV-1 VLPs, with human BST-2 again having a stronger effect than the chicken BST-2 (Fig. 4A). The high level of production of HIV-1 VLPs in 293T cells enabled us to visualize the effect of chBST-2 by electron microscopy. Negative staining revealed clustering of VLPs on the surfaces of chBST-2-transfected 293T cells, consistent with the effect of chBST-2 on the retention of nascent particles (Fig. 4B).
FIG 4.
Block of HIV-1 release in human cells with ectopic expression of chBST-2. (A) Human 293T cells (2 × 104 per well) were seeded on 96-well plates. After 24 h, the cells were transfected using 0.3 μl X-tremeGENE reagent with 100 ng of psPAX2 HIV-1 gag-pol vector and various doses (5 ng, 15 ng, or 50 ng) of pIRES-hBST2, pIRES-chBST2, or pIRES plasmids. Two days later, samples of cell culture media were collected and analyzed for RT activity by the PERT assay. Addition of up to 50 ng of control plasmid DNA (in place of BST-2 constructs) in transfection did not influence the PERT results (data not shown). The means and standard deviations from six independent replicates are shown, and statistical significance (Welch’s t test) is indicated above the graphs. (B) Human 293T cells (6 × 105) were seeded on 6-well plates and, after 24 h, transfected with 2 μg of psPAX2 HIV-1 gag-pol vector and 0.8 μg of pIRES-chBST2 plasmid or with the psPAX2 vector only. Two days later, the cells were processed for transmission electron microscopy as described in Materials and Methods.
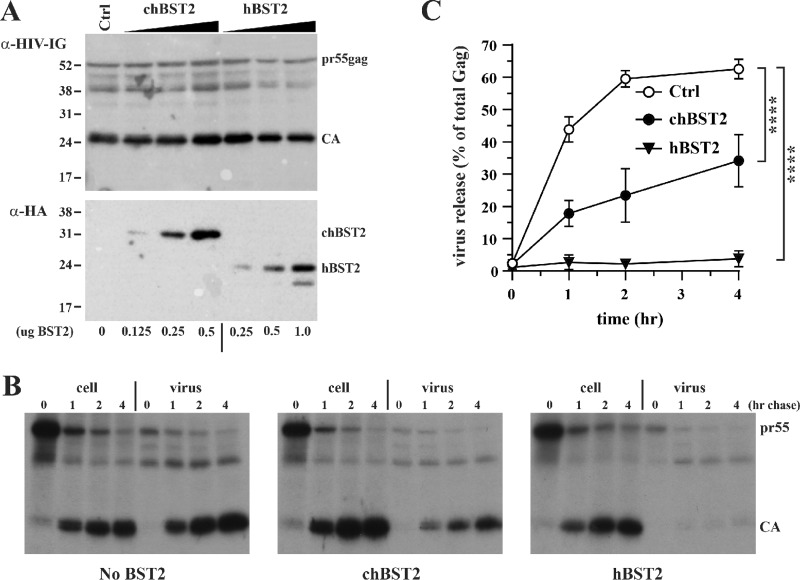
We also took advantage of the HIV-1 heterologous system to confirm that chBST-2 restricts viral particle production at the level of virus release and not at the level of intracellular production of viral proteins. We used an established method, which includes pulse-chase labeling of viral proteins and enables the determination of the fate of the proteins by immunoprecipitation with anti-Gag antibody (28). The proportion of viral proteins that are inside the cell versus that released into the supernatant can then be calculated. In this experiment, a hemagglutinin (HA)-tagged form of chBST-2 and a Vpu-negative HIV-1 NL43 construct (29) were used. The use of tagged constructs allowed us to first estimate by immunoblotting the amounts of constructs needed to achieve similar levels of BST-2 expression (Fig. 5A); these conditions were then used in pulse-chase experiments.
FIG 5.
Characterization of the chBST-2-induced block by pulse-chase labeling of viral proteins. (A) To assess the relative expression of human and chicken BST-2 in the presence of Vpu-defective HIV-1, HEK293T cells were transfected with 4 μg of pNL43Udel in the presence of 1 μg empty vector DNA (Ctrl) or increasing amounts of chBST-2 (0.125, 0.25, or 0.5 μg plasmid DNA) or hBST-2 (0.25, 0.5, or 1 μg plasmid DNA). Total amounts of transfected DNA were adjusted to 5 μg DNA. Cells were harvested 24 h after transfection and processed for immunoblotting against HIV-1-Ig (top) or HA tag-specific antibodies (bottom). Based on these results, we decided to use 0.3 μg of chicken BST-2 vector and 0.75 μg of human BST-2 vector for the subsequent pulse-chase studies. (B) HEK293T cells were transfected with 4.5 μg of pNL43Udel together with 0.75 μg empty vector (No BST-2), 0.3 μg of chicken tetherin vector (chBST2), or 0.75 μg human tetherin vector (hBST2). Total amounts of transfected DNA were adjusted to 5.25 μg using empty vector DNA. After 24 h, cells were metabolically labeled for 25 min with [35S]-Expre35S35S-label and, after removing unincorporated isotope, were chased for up to 4 h in complete DMEM. Cell extracts and virus-containing supernatants were immunoprecipitated with an HIV-positive patient serum and separated by SDS-PAGE. Gels were dried, and labeled proteins were visualized by fluorography. Representative fluorographs from one of three independent experiments are shown. (C) Gag proteins (Pr55 and p24 CA) were quantified by phosphorimage analysis. The proportion of total Gag protein released from the cells was calculated for each time point, and results were blotted as a function of time. Error bars represent standard deviations from three independent experiments. Statistical significance was determined by two-way analysis of variance (ANOVA). ****, P < 0.0001.
The pulse-chase experiments showed that chBST-2 decreases the proportion of total Gag that was released (Fig. 5B and C), confirming that the inhibitory effect is at the level of virus release. The inhibitory effect of hBST-2 was again stronger than chBST-2, even when their expression was approximately equal.
BST-2 orthologs are present in the genomes of multiple avian species and evolve under positive selection.
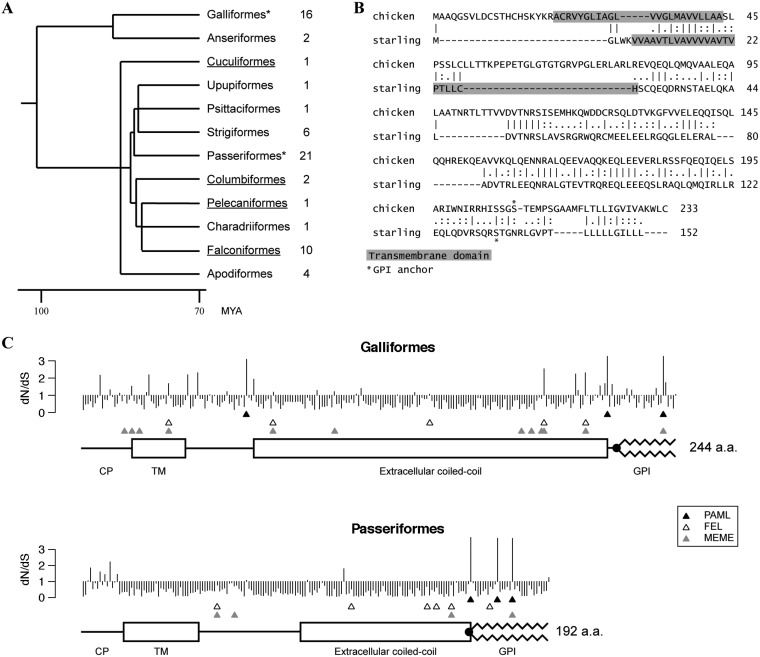
Previous work reported the presence of BST-2 orthologs in the genomes of several avian species (22, 23). It also indicated the absence of BST-2 in a subset of birds. We have systematically screened publicly available avian genomes and “raw” short-read sequences in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) to identify avian BST-2 orthologs. We have successfully identified BST-2 genes in 66 species belonging to 12 avian orders (Fig. 6A; see also Fig. S1 in the supplemental material), including all the species previously reported to lack this gene (e.g., crested ibis [Nipponia nippon], common cuckoo [Cuculus canorus], and rock pigeon [Columba livia]). The main obstacle to finding new BST-2 orthologs, especially in new avian orders, is the very high sequence variability. For example, the pairwise identity of predicted amino acid sequences of chicken (order Galliformes) and common starling (Sturnus vulgaris; order Passeriformes) is only 26% (Fig. 6B). In many avian species, systematic manual analysis of multiple open reading frames (ORFs) in the syntenic genomic region was required, involving a search for the presence of weak sequence similarities and for typical BST-2 secondary structure predictions. All newly identified avian BST-2 orthologs have predicted transmembrane, coiled-coil, and GPI domains in the expected configuration (see Fig. S1 in the supplemental material). Furthermore, in the avian genomic contigs where sufficient sequence context was available, the conserved syntenic position of BST-2 orthologs between the GTPBP3 and CILP2 genes was maintained. In two avian species, we were not able to identify an intact BST-2 gene. In the zebra finch (Taeniopygia guttata), there was no sequence in the syntenic locus with sufficient similarity with orthologs from other passeriforms and with the expected predicted secondary structures. The wild turkey (Meleagris gallopavo) BST-2 gene contains an early stop codon and expresses transcripts that are not predicted to encode a functional protein (our unpublished data).
FIG 6.
Avian BST-2 orthologs and positive selection analysis. (A) A time-calibrated phylogeny of avian orders was generated in TimeTree (64). Only orders from which BST-2 sequences were analyzed are shown (other avian orders were not analyzed or the genomic data were not available); the number of species analyzed in each order is indicated on the right. The two orders we tested for positive selection are marked by asterisks. Underlining highlights avian orders which contain species previously considered to lack BST-2 orthologs. MYA, million years ago. (B) Pairwise sequence alignment of chicken and starling (Sturnus vulgaris) BST-2 amino acid sequences. Dashes represent gaps; bars, colons, and dots indicate identical, strongly similar, and weakly similar amino acids, respectively. The predicted positions of transmembrane regions and GPI anchor attachment (omega site) are shown. (C) The graph shows the estimated ratio of nonsynonymous to synonymous evolutionary changes (dN/dS) (posterior means, PAML’s model 8) at each codon position of BST-2 in aligned Galliformes and Passeriformes sequences. Three rows of triangles indicate sites identified by each algorithm as being under positive selection: the top row (▲) shows sites identified by PAML with BEB probability of ≥0.9, the middle row (△) shows FEL sites, and the third row (gray triangles) shows MEME sites. Protein sequence annotations below each graph were generated based on a single ungapped sequence for each alignment (G. gallus for the Galliformes, and S. vulgaris for Passeriformes), and coordinates were transformed to aligned coordinates.
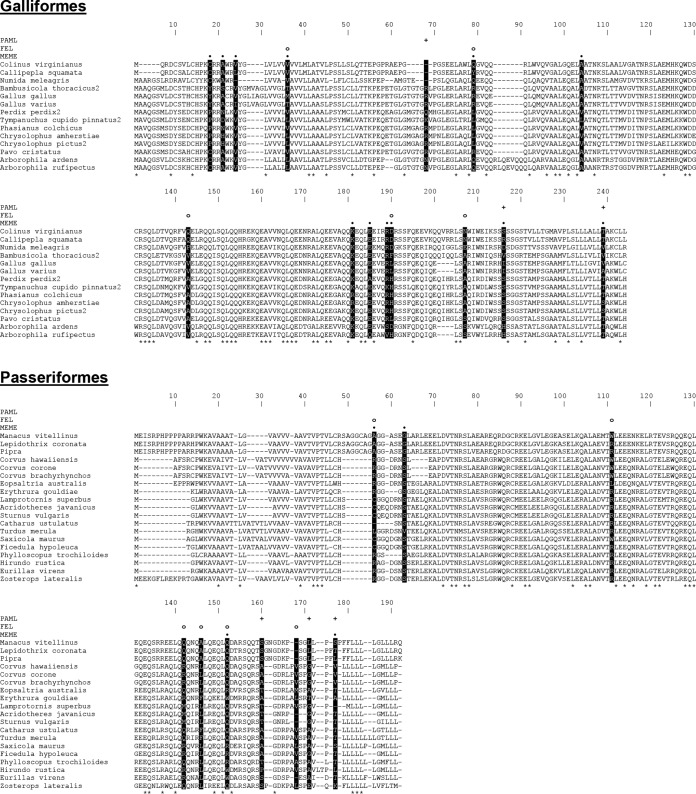
Furthermore, we assessed the evolutionary selective pressures acting on avian BST-2. Two avian orders with the highest number of available BST-2 sequences were selected for evolutionary analyses: Galliformes and Passeriformes. We aligned nucleotide sequences (see Fig. S2 and S3 in the supplemental material) and performed positive selection analysis, using the PAML package (30). The results indicated the presence of significant positive selection in BST-2 genes in both avian orders (Table 1). Additionally, we used three complementary algorithms that predict positively selected codons: PAML BEB (31), FEL (32), and MEME (33). All three algorithms detected multiple positively selected sites across the entire BST-2 protein sequence (Fig. 6C and 7).
TABLE 1.
BST-2 in Galliformes and Passeriformes is evolving under positive selection
| Order | No. of sequences | Alignment length (aa) | Overall dN/dS (PAML model 0) | PAML model 8 vs model:a
|
Model 8 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 8a |
7 |
||||||||||
| 2Δlnλ | dfb | P value | 2Δlnλ | dfb | P value | % sites under positive selection | dN/dS of selected class | ||||
| Galliformes | 14 | 244 | 0.58 | 7.2 | 1 | 0.007 | 15.3 | 2 | 0.0005 | 4.3 | 4.0 |
| Passeriformes | 19 | 192 | 0.32 | 7.7 | 1 | 0.006 | 14.8 | 2 | 0.0006 | 2.7 | 4.1 |
Likelihood ratio tests comparing a model that allows positive selection at a subset of sites (model 8) with each of two alternative models that only allow purifying and neutral selection (models 7 and 8a).
df, degrees of freedom.
FIG 7.
Positively selected sites detected in avian BST-2 orthologs by three different algorithms. Amino acid alignments of Galliformes (top) and Passeriformes (bottom) BST-2 are shown. Residues with 100% conservation in each alignment are marked with asterisks below the sequences. Positively selected sites are highlighted as black columns. Predictions of the individual algorithms are depicted above the alignment as plus signs (PAML), open circles (FEL), and black dots (MEME).
DISCUSSION
In this study, we identified chicken tetherin/BST-2 and analyzed its antiviral activity. We found that it is an IFN-inducible protein in chicken, like in mammals. CRISPR/Cas9-mediated knockout in chicken DF-1 cells provides evidence that BST-2 is a major factor required for an IFN-dependent block of ASLV production (Fig. 2D). However, the rescue of ASLV production in BST-2 knockout cells was not complete, which suggests that other, as yet unidentified, IFN-induced RFs are involved. The only other late-phase ASLV replication block reported before was ascribed to interferon-stimulated gene 15 (ISG15) and the ISG15-involved modification (ISGylation) of proteins that participate in retroviral budding (34). However, it has since been found that the ISG15 gene is missing in birds (35). The study by Kuang et al. used not chicken, but human ISG15 and its specific ligases, ectopically expressed in chicken DF-1 cells to characterize the ASLV production block. Interestingly, even though ISG15 itself is missing, the entire ISG15 enzymatic modification machinery is present in chicken cells (35). As some avian genes are extremely difficult to identify by sequencing due to their high GC content (36), it remains possible that avian ISG15 will be found or that another ubiquitin homolog may substitute for its role in birds. In such a case, ISGylation or a similar modification could be responsible for part of the IFN-induced retrovirus restriction in chicken cells.
In addition to determining the role of endogenous chBST-2, we examined the consequences of ectopic expression of the cloned chBST-2 protein on RCAS production. Both transient and stable expression of chBST-2 significantly inhibited the amount of RCAS particles produced from DF-1 cells (Fig. 3). Furthermore, we have shown that chBST-2 can inhibit other retroviruses—HIV-1 VLPs and a Vpu-HIV-1 vector—in a heterologous system of human cells (Fig. 4 and 5). This system also allowed us to show by pulse-chase labeling of viral proteins that the chBST-2-induced block is at the level of virus release. The inhibitory effect of the chicken protein was consistently lower than the inhibition caused by the expression of human BST-2. This difference was observed for ASLV release in chicken cells and also for HIV-1 release in human cells, even under conditions of similar expression of BST-2 proteins. This suggests that chBST-2 has lower inhibitory potency than the human protein and that the difference is not due to an unknown ASLV-encoded BST-2 antagonist.
BST-2 was initially considered to exist exclusively in mammals. Heusinger et al. (22) have shown the presence of BST-2 orthologs in many other vertebrates, including fish, reptiles, and birds. They noted the low primary sequence conservation and the maintenance of structural features only. However, they still proposed the absence of BST-2 in several avian species. A study that closely followed (23) started from the fact that BST-2 is an orphan gene, without detectable homologs outside eutherian mammals. It showed that BST-2 is one of three adjacent genes that share similar architecture but no sequence similarity and proposed a scenario in which these genes arose by segmental duplication in vertebrate ancestors. The study by Blanco-Melo et al. (23) was not focused specifically on birds and therefore did not identify the missing avian BST-2 genes. Neither study reported the sequence of chicken BST-2 (although they mentioned its existence). A single representative of avian BST-2 proteins, from the peregrine falcon (Falco peregrinus, order Falconiformes), was expressed in human cells and shown to have antiviral effect against Vpu-deficient HIV-1 (23). In our present study, we report the sequence of chicken BST-2 and provide the first experimental proof of BST-2 antiviral activity in avian cells.
We obtained a large number of BST-2 orthologs in birds through a systematic search of available sequence data (Fig. 6A; see also Fig. S1 in the supplemental material). The birds included representatives of all the species previously reported to lack this gene. As mentioned above, our search frequently required detailed analysis of multiple ORFs in the target region and consideration of secondary structure prediction rather than primary sequence conservation. These strategies were especially important for searches in avian orders where BST-2 had not yet been identified. Based on the current results, it is likely that almost all avian species, with the possible exceptions of zebra finch and turkey mentioned above, harbor sequences orthologous to BST-2.
Many host proteins that interact with viruses are engaged in evolutionary arms races, whereby host and virus have conflicting interests in establishing or avoiding interaction. Primate BST-2 evolves under positive selection, and at least some of this rapid evolution appears to be driven by the need for BST-2 to escape antagonism by lentiviral Vpu and Nef proteins (37–39). Strong positive selection was detected in BST-2 in both avian orders analyzed—Galliformes and Passeriformes (Fig. 6C; Table 1). Given that BST-2 targets the viral membrane, probably nonspecifically, it is likely that the positive selection we observed on avian BST-2 is not driven by an arms race with its target but rather with as-yet-unidentified viral antagonists of BST-2 (as seen for primate lentiviruses).
Positive selection is often found at sites that represent host-viral interaction interfaces (40). Unlike for primate BST-2, where rapidly evolving sites are mostly in the N-terminal region, positive selection is found throughout avian tetherin, especially in the C-terminal region. This observation could suggest that viral antagonists targeting avian tetherin interact with a different region than antagonists of primate tetherin. In addition to amino acid-altering changes, we note that avian BST-2 sequences have also acquired several length-altering insertion/deletion changes (indels) during their evolution. No methods exist to test whether these indels are adaptive, but it could be interesting to test their functional impact.
We used ASLV/RCAS in this work because it represents a prototypic avian retrovirus and an important avian pathogen (41). The experiments performed in this study used either overexpression of chBST-2 or its induction by IFN to achieve ASLV inhibition. Infection of DF-1 cells by RCAS did not induce the mRNA expression of either chBST-2 or IFN (Fig. 2B and data not shown). However, the published data in this matter are contradictory, with a field ASLV strain showing strong IFN induction (42) and with an RCAS vector showing low induction or even suppression of IFN (43). It is still unclear whether IFN is induced during in vivo ASLV infection of chickens and in which tissues chBST-2 could be induced with subsequent antiviral effects. Alternatively, chBST-2 might be constitutively expressed in some cell types in chicken, as is the case in mammals (44, 45).
Aside from its inhibitory effect on virus release, BST-2 was shown to affect cell signaling. Specifically, it induces NF-κB-dependent proinflammatory responses, and an YxY motif located in the BST-2 cytoplasmic domain was shown to be critical for the signaling function (25, 46, 47). Inspection of the cytoplasmic domain of chicken BST-2 did not reveal a similar YxY motif (Fig. 1A), suggesting that chicken tetherin may not have the signaling function inherent to human tetherin. However, future experiments will have to address this question experimentally.
In summary, we have described a chicken ortholog of BST-2/tetherin and shown its potential to act as a RF against avian retroviruses. The inhibitory effects observed in some experiments presented here are relatively mild, and future investigations should determine the significance of avian BST-2 in viral restriction. RFs are known to be key determinants of cross-species transmission (48). In the future, it would be interesting to test various avian BST-2 genes for their activity against other enveloped avian viruses, for example, avian influenza virus and Marek’s disease virus. These viruses belong to families that are restricted by BST-2 in mammalian cells (49, 50). Our finding of positive selection suggests that antagonists of BST-2 may exist in some avian viruses; identification of these antagonists will be important in our future understanding of BST-2’s role in protecting birds from viral diseases.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic kidney cell line HEK293T was grown in Dulbecco’s modified Eagle’s medium (DMEM; Sigma-Aldrich) supplemented with 7% fetal calf serum (FCS), 100 μg/ml streptomycin, and 100 U/ml penicillin under 5% CO2 atmosphere at 37°C. Permanent chicken fibroblast cell line DF-1 (51) was grown in a mixture of 2 parts DMEM and 1 part F-12 medium (Sigma-Aldrich) supplemented with 5% calf serum, 1% fetal calf serum, 1% chicken serum, streptomycin, and penicillin under 3% CO2 atmosphere at 37°C. RCASBP(A)GFP and RCASBP(A) are replication-competent retroviral vectors based on ASLV subgroup A (27, 52). psPAX2 (Addgene number 12260) is a second-generation lentiviral packaging plasmid harboring HIV-1 gag and pol. For the pulse-chase experiments, a Vpu-negative HIV-1 NL43 construct (29) was used.
Computational identification of avian BST-2 orthologs from RNA-seq and genomic data.
Data sets of Illumina RNA-seq data from chicken and other birds were downloaded from the NCBI Sequence Read Archive (SRA). Sequence reads originating from chBST-2 were identified using sequences of previously reported avian BST-2 orthologs as blastn queries (53) or by mapping with CLC genomics workbench software (Qiagen Bioinformatics) to the genomic sequences of the syntenic region. Short reads were assembled using the CLC genomics workbench and Lasergene SeqMan Pro (DNASTAR) software. In genomic contigs, positions of BST-2 orthologs were determined either by BLAST (blastn or tblastn) or by systematic analysis of ORFs predicted with ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/). The ORFs were scanned for more subtle homologies to known avian BST-2 genes and for typical BST-2 secondary structures. Transmembrane helices were predicted from primary protein sequences using TMHMM v2.0 (54). Coiled-coil regions were predicted with the algorithm of Lupas et al. (55) at https://npsa-prabi.ibcp.fr/NPSA/npsa_lupas.html. Predicted GPI modification sites were identified using server http://mendel.imp.ac.at/gpi/gpi_server.html (56).
RNA isolation, cDNA synthesis, and cloning of chicken and human BST-2.
Total RNA was isolated from cultured cells by RNAzol RT (Molecular Research Center). Two hundred fifty nanograms of total RNA was reverse transcribed with ProtoScript II reverse transcriptase (NEB) and the SMART RACE (Clontech) protocol. The entire chicken and human BST-2 coding sequences were amplified from the cDNA of each species using the following primer pairs: chicken, 5′-tactcgagccaccATGGCTGCGCAGGGCA and 5′-caggatccTCAGCACAGCCACTTTGCAACG; human, 5′-tactcgagccaccATGGCATCTACTTCGTATGACTATTG and 5′-caggatccTCACTGCAGCAGAGCGCTG. The capital letters in the primer sequences denote regions complementary to BST-2. Lowercase letters in forward primers indicate the Kozak sequence (GCCACC), XhoI restriction site, and a short sequence overhang to allow for efficient cutting with the restriction enzyme; lowercase letters in reverse primers indicate BamHI restriction site and a short sequence overhang. The PCR products were subcloned into pGEM-T Easy vector (Promega) and verified by Sanger sequencing. BST-2 sequences were then transferred to the pIRES2-EGFP vector (BD Biosciences Clontech) using the XhoI and BamHI restriction sites. For the pulse-chase assays in human cells, the chicken BST-2 sequence was subcloned into the pcDNA3.1 vector (Thermo Fisher) with a single N-terminal HA-tag.
Generation of chBST-2 stable transfectants in DF-1 cells.
DF-1 cells were transfected with the respective plasmid vector using X-tremeGENE HP DNA transfection reagent (Sigma) according to the manufacturer’s protocol. After 3 days, selection with G418 (5 ng/ml, later increased to 10 ng/ml) was started and maintained for 2 weeks. Then, cells were sorted for GFP positivity using the Influx cell sorter (Becton, Dickinson) and expanded before further experiments.
Quantitative real-time PCR.
The quantitative real-time PCRs (qRT-PCRs) included cDNA samples (1.5 μl), MESA GREEN qPCR MasterMix Plus (Eurogentec), and primers targeting chicken GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (5′-CATCGTGCACCACCAACTG and 5′-CGCTGGGATGATGTTCTGG) or chBST-2 (5′-CAACAGGGCTCTCCAGGAGG and 5′-GGCATCTCTGTGCTCCCACT). The samples were run on a CFX96TM real-time instrument (Bio-Rad) with a 3-step protocol: 1 cycle of 8 min at 95°C and then 40 cycles of 15 s at 95°C, 25 s at 60°C, and 35 s at 72°C. Cycles of quantification (Cq) values were generated by the CFX Manager software. The specificity of the PCR products was confirmed by melting curve analysis.
Product-enhanced reverse transcriptase assay.
The samples (2 μl of virus-containing culture medium) were lysed in 8 μl of solution containing 1% Triton X-100, 0.4 U/μl RNasin, and 1× M-MLV RT buffer (Promega) at room temperature for 15 to 30 min. Then, 2 master mixes were prepared, with the following amounts per reaction: mix A contained 20 ng of the template phage MS2 RNA (Roche), 0.5 μl of 10 mM MS2 reverse primer (5′-GCCTTAGCAGTGCCCTGTCT), and 12.1 μl water. Mix B contained 3.6 μl of 5× M-MLV RT buffer and 0.8 μl of 10 mM deoxynucleoside triphosphates (dNTPs). Mix A was incubated at 65°C for 5 min and cooled down to room temperature to allow primer annealing. Mixes A and B were pooled and split into 18-μl aliquots, and 2 μl of sample lysate was added to each aliquot. The reaction mixture was incubated at 37°C for 60 min and inactivated at 70°C for 10 min. The newly generated MS2 cDNA was then quantified by real-time PCR, each reaction mixture containing 1.5 μl of cDNA sample with qPCR MasterMix Plus, MS2 primers (forward 5′-AACATGCTCGAGGGCCTTA and reverse primer mentioned above), and MS2 probe (6-carboxyfluorescein [FAM]-TGGGATGCTCCTACATG-6-carboxytetramethylrhodamine [TAMRA]) in a 15-μl reaction mixture. The samples were run on a CFX96 real-time instrument with a 3-step protocol: 1 cycle of 10 min at 95°C and then 40 cycles of 15 s at 95°C, 20 s at 60°C, and 20 s at 72°C. Cycles of quantification (Cq) values were generated by the CFX Manager software.
CRISPR/Cas9-mediated knockout of chBST-2.
We used genome editing tools for knockout of endogenous chicken chBST-2 in DF-1 cells as previously described for other chicken genes (57). Briefly, the CRISPR design tool (58) was used to identify guide RNA (gRNA) sequences in the first exon of chBST-2. The gRNA was synthesized as sense and antisense oligonucleotides (gRNA sequences, 5′-CACCGAAGAGTGCCCGGTTTGGAG and 5′-AAACCTCCAAACCGGGCACTCTTC; purchased from Eurogentec), mixed, phosphorylated, denatured, and annealed. Annealed oligonucleotides were ligated into the pX458 vector (59) to create pX458-tetherin. DF-1 cells were seeded on P60 dishes and, after 24 h, transfected with pX458-tetherin using Lipofectamine 3000 according to the manufacturer’s protocol. Three days posttransfection, the GFP-positive cells were sorted into 96-well plates using the Influx cell sorter (Becton, Dickinson) in single-cell sort mode. A pool of 10,000 cells was used to determine CRISPR/Cas9 efficiency by the T7 endonuclease assay (60). Cell clones were expanded and used for further analysis of CRISPR/Cas9-introduced mutations. From each clone, cell lysate was prepared by washing with phosphate-buffered saline (PBS), resuspending in lysis buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.2 mM CaCl2, 0.001% Triton X-100, 0.001% SDS, 1 mg/ml proteinase K), and then incubating at 58°C for 1 h followed by protease inactivation at 95°C for 10 min. The presence of mutations was determined by PCR amplification of the target region and Sanger sequencing using 2 μl of cell lysates. We chose a clone containing a 14-nucleotide deletion and a clone without any change in the BST-2 target region for further work.
Transmission electron microscopy.
HEK293T cells were quickly washed with Sörensen buffer (SB; 0.1 M sodium/potassium phosphate buffer, pH 7.3) at 37°C, fixed with 2.5% glutaraldehyde in SB for 2 h, washed with SB, and postfixed with 1% OsO4 solution in SB for 2 h. The cells were dehydrated in a series of acetones of increasing concentration, and embedded in Epon-Durcupan resin. Polymerized blocks were cut into 200-nm-thin sections, collected on 200-mesh-size copper grids, and stained with a saturated aqueous solution of uranyl acetate for 4 min. The sections were examined on an FEI Morgagni 268 transmission electron microscope operated at 80 kV. The images were captured using Mega View III charge-coupled-device (CCD) camera (Olympus Soft Imaging Solutions).
Immunoblot analysis.
Cells were washed once with PBS, suspended in PBS (100 μl/106 cells), and mixed with equal volumes of 2× sample buffer. Samples were heated at 95°C, subjected to SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes, and incubated with primary antibodies. Chicken and human tetherin were identified targeting the HA tag using a mouse monoclonal antibody to HA (catalog number [Cat. no.] H3663; Sigma-Aldrich). HIV proteins were detected using human HIV-Ig antibody (Cat. no. 3957; NIH Research and Reference Reagent Program). Membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies (GE Healthcare, Piscataway, NJ, USA), and proteins were visualized by enhanced chemiluminescence (Clarity Western ECL substrate 170–5061; Bio-Rad Laboratories, Hercules, CA, USA).
Metabolic labeling and pulse-chase analysis.
For the pulse-chase analysis, HEK293T cells were grown as described before but with 10% FCS. For transient transfection, 3 × 106 cells were plated in a 25-cm2 flask and grown overnight. The following day, cells were transfected using Lipofectamine Plus (Invitrogen) according to the manufacturer’s instructions. Total amounts of plasmid DNA in all samples were equalized with empty vector DNA as appropriate.
After 24 h, the transfected HEK293T cells were harvested by scraping, washed with PBS, and suspended in 7 ml of labeling medium (methionine- and cysteine-free RPMI [MP Biomedical] containing 5% fetal calf serum [FCS]). Samples were incubated for 20 min at 37°C to deplete the intracellular methionine/cysteine pool. Cells were then labeled for 30 min at 37°C in 200 μl of labeling medium supplemented with 30 μl (300 μCi) of [35S]-Expre35S35S-label (NEG072; PerkinElmer). After the labeling period, unincorporated isotope was removed, and equal aliquots of cells were added to 1 ml of prewarmed complete DMEM and chased for the selected times. Cells and virus-containing supernatants were harvested separately at each time point and stored on dry ice until all samples had been collected. For immunoprecipitation of intracellular and virus-associated Gag proteins, cells and virus-containing supernatants were lysed in 200 μl of Triton X-100-based lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 10% glycerol) and incubated on ice for 5 min. After lysis, the cell extracts were pelleted at 10,000 rpm for 10 min, and clarified supernatants were added to 1.0 ml of 0.1% bovine serum albumin (BSA)-PBS at 4°C. Cell and virus lysates were immunoprecipitated (1 h, 4°C on a rotator) with human HIV-Ig antibody (Cat. no. 3957; NIH Research and Reference Reagent Program) immobilized to protein A Sepharose beads (P-3391; Sigma-Aldrich). Precipitated proteins were solubilized by boiling in sample buffer and separated by SDS-PAGE. Gels were fixed and dried. Gels were exposed to Kodak XMR film, and proteins were visualized by fluorography. For protein quantitation, gels were exposed to imaging plates, and analysis of the relevant bands was performed using a Fujifilm FLA-7000 phosphorimager.
Statistical analysis.
For the evaluation of statistical significance of the differences in chBST-2 expression, RT activity, and percentage of infected cells, we performed Welch’s t tests. This test is robust to unequal standard deviations of the data sets tested.
Analysis of positive selection.
Starting with 16 Galliformes and 21 Passeriformes tetherin sequences, two Galliformes sequences from quail species were removed because they have large in-frame deletions. Two Passeriformes sequences (Ficedula albicollis and Corvus cornix) were removed, as each is identical to BST-2 from a related species and therefore adds no information for evolutionary analyses. In-frame nucleotide alignments of the remaining 14 Galliformes and 19 Passeriformes sequences were generated using MACSE (61) with default settings except for an increased frameshift penalty (-fs 300) (see Fig. S3 and S4 in the supplemental material). Phylogenies were generated from each alignment using PHYML (62) (parameters: -m GTR –pinv e –alpha e -f e). There was no evidence of gene conversion in either alignment according to the GARD algorithm (63) (run via http://www.datamonkey.org/), using the general discrete model of site-to-site rate variation and 3 rate classes (gene conversion should be excluded before running positive selection analyses, as it can cause false signatures of positive selection).
The alignments and phylogenies were used as input for PAML (Phylogenetic Analysis by Maximum Likelihood) (30), where model 8 (allows positive selection at a subset of codons) was compared with model 7 or model 8a (all codons are under neutral or purifying selection). P values were generated by comparing twice the difference in log-likelihoods to the chi-squared distribution (30). In order to check for robustness, analyses were run using two different values for starting omega (0.4 and 3), with two different codon frequency choices (F3 × 4 and “codon table”) and with and without using the “cleandata” option to remove codons that contain gaps in one or more aligned sequences. Results were consistent using all eight combinations of these parameters. The results shown in Table 1 and Fig. 5C use starting omega 0.4, the F3 × 4 codon model, and cleandata of 0 (i.e., retaining all codons). FEL (32) and MEME (33) analyses were performed using the http://www.datamonkey.org/ website (choosing the option to allow synonymous site variation in FEL), and selected sites were identified using the website’s default statistical thresholds (P < 0.1).
Data availability.
The chBST-2 coding sequence is available in GenBank under accession number MN326300.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bernd Kaspers for the gift of recombinant chicken interferon.
This work was funded by grant 17-23675S from the Czech Science Foundation (to D.E.). We also acknowledge institutional support from project RVO 68378050. K.S. and H.F. were supported by the Intramural Research Program of the NIH, NIAID (1 Z01 AI000669). J.M.Y. was funded by a grant from the Mathers Foundation (PI, Harmit S. Malik) and NIH grant P50GM082250 (PI, Nevan Krogan; subaward to Michael Emerman and Harmit S. Malik). We acknowledge use of the Microscopy Centre–Electron Microscopy CF, IMG AS CR, supported by the MEYS CR (LM2018129 Czech-BioImaging).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Chemudupati M, Kenney AD, Bonifati S, Zani A, McMichael TM, Wu L, Yount JS. 2019. From APOBEC to ZAP: diverse mechanisms used by cellular restriction factors to inhibit virus infections. Biochim Biophys Acta Mol Cell Res 1866:382–394. doi: 10.1016/j.bbamcr.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neil S, Bieniasz P. 2009. Human immunodeficiency virus, restriction factors, and interferon. J Interferon Cytokine Res 29:569–580. doi: 10.1089/jir.2009.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krishnan A, Iyer LM, Holland SJ, Boehm T, Aravind L. 2018. Diversification of AID/APOBEC-like deaminases in metazoa: multiplicity of clades and widespread roles in immunity. Proc Natl Acad Sci U S A 115:E3201–E3210. doi: 10.1073/pnas.1720897115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haugh KA, Shalginskikh N, Nogusa S, Skalka A, Katz RA, Balachandran S. 2014. The interferon-inducible antiviral protein Daxx is not essential for interferon-mediated protection against avian sarcoma virus. Virol J 11:100. doi: 10.1186/1743-422X-11-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu M, Ma X, Cui X, Zhou J, Li C, Huang L, Shang Y, Cheng Z. 2017. Inhibition of avian tumor virus replication by CCCH-type zinc finger antiviral protein. Oncotarget 8:58865–58871. doi: 10.18632/oncotarget.19378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li L, Feng W, Cheng Z, Yang J, Bi J, Wang X, Wang G. 2019. TRIM62-mediated restriction of avian leukosis virus subgroup J replication is dependent on the SPRY domain. Poultry Science 98:6019–6025. doi: 10.3382/ps/pez408. [DOI] [PubMed] [Google Scholar]
- 7.Xie T, Feng M, Dai M, Mo G, Ruan Z, Wang G, Shi M, Zhang X. 2019. Cholesterol-25-hydroxylase is a chicken ISG that restricts ALV-J infection by producing 25-hydroxycholesterol. Viruses 11:498. doi: 10.3390/v11060498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schusser B, Reuter A, von der Malsburg A, Penski N, Weigend S, Kaspers B, Staeheli P, Hartle S. 2011. Mx is dispensable for interferon-mediated resistance of chicken cells against influenza A virus. J Virol 85:8307–8315. doi: 10.1128/JVI.00535-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ko J-H, Jin H-K, Asano A, Takada A, Ninomiya A, Kida H, Hokiyama H, Ohara M, Tsuzuki M, Nishibori M, Mizutani M, Watanabe T. 2002. Polymorphisms and the differential antiviral activity of the chicken Mx gene. Genome Res 12:595–601. doi: 10.1101/gr.210702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goossens KE, Karpala AJ, Rohringer A, Ward A, Bean A. 2015. Characterisation of chicken viperin. Mol Immunol 63:373–380. doi: 10.1016/j.molimm.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Shah M, Bharadwaj MSK, Gupta A, Kumar R, Kumar S. 2019. Chicken viperin inhibits Newcastle disease virus infection in vitro: a possible interaction with the viral matrix protein. Cytokine 120:28–40. doi: 10.1016/j.cyto.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Santhakumar D, Rohaim M, Hussein HA, Hawes P, Ferreira HL, Behboudi S, Iqbal M, Nair V, Arns CW, Munir M. 2018. Chicken interferon-induced protein with tetratricopeptide repeats 5 antagonizes replication of RNA viruses. Sci Rep 8:6794. doi: 10.1038/s41598-018-24905-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith SE, Gibson MS, Wash RS, Ferrara F, Wright E, Temperton N, Kellam P, Fife M. 2013. Chicken interferon-inducible transmembrane protein 3 restricts influenza viruses and lyssaviruses in vitro. J Virol 87:12957–12966. doi: 10.1128/JVI.01443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S, Wang L, Chen J, Zhang L, Wang S, Goraya MU, Chi X, Na Y, Shao W, Yang Z, Zeng X, Chen S, Chen J-L. 2017. Avian interferon-inducible transmembrane protein family effectively restricts avian Tembusu virus infection. Front Microbiol 8:672. doi: 10.3389/fmicb.2017.00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neil SJD, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, Bieniasz PD. 2009. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauter D. 2014. Counteraction of the multifunctional restriction factor tetherin. Front Microbiol 5:163. doi: 10.3389/fmicb.2014.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Tortorec A, Neil S. 2009. Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J Virol 83:11966–11978. doi: 10.1128/JVI.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez LA, Yang SJ, Hauser H, Exline CM, Haworth KG, Oldenburg J, Cannon PM. 2010. Ebola virus glycoprotein counteracts BST-2/tetherin restriction in a sequence-independent manner that does not require tetherin surface removal. J Virol 84:7243–7255. doi: 10.1128/JVI.02636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mansouri M, Viswanathan K, Douglas JL, Hines J, Gustin J, Moses AV, Fruh K. 2009. Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi’s sarcoma-associated herpesvirus. J Virol 83:9672–9681. doi: 10.1128/JVI.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heusinger E, Kluge SF, Kirchhoff F, Sauter D. 2015. Early vertebrate evolution of the host restriction factor tetherin. J Virol 89:12154–12165. doi: 10.1128/JVI.02149-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanco-Melo D, Venkatesh S, Bieniasz PD. 2016. Origins and evolution of tetherin, an orphan antiviral gene. Cell Host Microbe 20:189–201. doi: 10.1016/j.chom.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burt DW. 2007. Emergence of the chicken as a model organism: implications for agriculture and biology. Poult Sci 86:1460–1471. doi: 10.1093/ps/86.7.1460. [DOI] [PubMed] [Google Scholar]
- 25.Galão RP, Le Tortorec A, Pickering S, Kueck T, Neil S. 2012. Innate sensing of HIV-1 assembly by tetherin induces NFκB-dependent proinflammatory responses. Cell Host Microbe 12:633–644. doi: 10.1016/j.chom.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrew AJ, Miyagi E, Kao S, Strebel K. 2009. The formation of cysteine-linked dimers of BST-2/tetherin is important for inhibition of HIV-1 virus release but not for sensitivity to Vpu. Retrovirology 6:80. doi: 10.1186/1742-4690-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes SH. 2004. The RCAS vector system. Folia Biol (Praha) 50:107–119. [DOI] [PubMed] [Google Scholar]
- 28.Sukegawa S, Miyagi E, Bouamr F, Farkašová H, Strebel K. 2018. Mannose receptor 1 restricts HIV particle release from infected macrophages. Cell Rep 22:786–795. doi: 10.1016/j.celrep.2017.12.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klimkait T, Strebel K, Hoggan MD, Martin MA, Orenstein JM. 1990. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J Virol 64:621–629. doi: 10.1128/JVI.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 31.Yang Z, Wong WSW, Nielsen R. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol 22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 32.Kosakovsky Pond SL, Frost S. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol 22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 33.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet 8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuang Z, Seo EJ, Leis J. 2011. Mechanism of inhibition of retrovirus release from cells by interferon-induced gene ISG15. J Virol 85:7153–7161. doi: 10.1128/JVI.02610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magor KE, Miranzo Navarro D, Barber MRW, Petkau K, Fleming-Canepa X, Blyth GAD, Blaine AH. 2013. Defense genes missing from the flight division. Dev Comp Immunol 41:377–388. doi: 10.1016/j.dci.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hron T, Pajer P, Pačes J, Bartůněk P, Elleder D. 2015. Hidden genes in birds. Genome Biol 16:164. doi: 10.1186/s13059-015-0724-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNatt MW, Zang T, Hatziioannou T, Bartlett M, Fofana IB, Johnson WE, Neil SJD, Bieniasz PD. 2009. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog 5:e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta RK, Hué S, Schaller T, Verschoor E, Pillay D, Towers GJ. 2009. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog 5:e1000443. doi: 10.1371/journal.ppat.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim ES, Malik HS, Emerman M. 2010. Ancient adaptive evolution of tetherin shaped the functions of Vpu and Nef in human immunodeficiency virus and primate lentiviruses. J Virol 84:7124–7134. doi: 10.1128/JVI.00468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daugherty MD, Malik HS. 2012. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet 46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 41.Payne LN, Nair V. 2012. The long view: 40 years of avian leukosis research. Avian Pathol 41:11–19. doi: 10.1080/03079457.2011.646237. [DOI] [PubMed] [Google Scholar]
- 42.Gao Y, Liu Y, Guan X, Li X, Yun B, Qi X, Wang Y, Gao H, Cui H, Liu C, Zhang Y, Wang X, Gao Y. 2015. Differential expression of immune-related cytokine genes in response to J group avian leukosis virus infection in vivo. Mol Immunol 64:106–111. doi: 10.1016/j.molimm.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 43.Lin W, Xu Z, Yan Y, Zhang H, Li H, Chen W, Chen F, Xie Q. 2018. Avian leukosis virus subgroup J Attenuates type I interferon production through blocking IκB phosphorylation. Front Microbiol 9:1089. doi: 10.3389/fmicb.2018.01089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blasius AL, Giurisato E, Cella M, Schreiber RD, Shaw AS, Colonna M. 2006. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J Immunol 177:3260–3265. doi: 10.4049/jimmunol.177.5.3260. [DOI] [PubMed] [Google Scholar]
- 45.Homann S, Smith D, Little S, Richman D, Guatelli J. 2011. Upregulation of BST-2/tetherin by HIV infection in vivo. J Virol 85:10659–10668. doi: 10.1128/JVI.05524-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sauter D, Hotter D, Engelhart S, Giehler F, Kieser A, Kubisch C, Kirchhoff F. 2013. A rare missense variant abrogates the signaling activity of tetherin/BST-2 without affecting its effect on virus release. Retrovirology 10:85. doi: 10.1186/1742-4690-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tokarev A, Suarez M, Kwan W, Fitzpatrick K, Singh R, Guatelli J. 2013. Stimulation of NF-κB activity by the HIV restriction factor BST2. J Virol 87:2046–2057. doi: 10.1128/JVI.02272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warren CJ, Sawyer SL. 2019. How host genetics dictates successful viral zoonosis. PLoS Biol 17:e3000217. doi: 10.1371/journal.pbio.3000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blondeau C, Pelchen-Matthews A, Mlcochova P, Marsh M, Milne RSB, Towers GJ. 2013. Tetherin restricts herpes simplex virus 1 and is antagonized by glycoprotein M. J Virol 87:13124–13133. doi: 10.1128/JVI.02250-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gnirß K, Zmora P, Blazejewska P, Winkler M, Lins A, Nehlmeier I, Gärtner S, Moldenhauer A-S, Hofmann-Winkler H, Wolff T, Schindler M, Pöhlmann S. 2015. Tetherin sensitivity of influenza A viruses is strain specific: role of hemagglutinin and neuraminidase. J Virol 89:9178–9188. doi: 10.1128/JVI.00615-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Himly M, Foster DN, Bottoli I, Iacovoni JS, Vogt PK. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248:295–304. doi: 10.1006/viro.1998.9290. [DOI] [PubMed] [Google Scholar]
- 52.Federspiel MJ, Hughes SH. 1997. Retroviral gene delivery. Methods Cell Biol 52:179–214. doi: 10.1016/S0091-679X(08)60379-9. [DOI] [PubMed] [Google Scholar]
- 53.Wheeler DL, Church DM, Edgar R, Federhen S, Helmberg W, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, Suzek TO, Tatusova TA, Wagner L. 2004. Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res 32:D35–D40. doi: 10.1093/nar/gkh073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 55.Lupas A, Van Dyke M, Stock J. 1991. Predicting coiled coils from protein sequences. Science 252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 56.Eisenhaber B, Bork P, Eisenhaber F. 1999. Prediction of potential GPI-modification sites in proprotein sequences. J Mol Biol 292:741–758. doi: 10.1006/jmbi.1999.3069. [DOI] [PubMed] [Google Scholar]
- 57.Koslová A, Kučerová D, Reinišová M, Geryk J, Trefil P, Hejnar J. 2018. Genetic resistance to avian leukosis viruses induced by CRISPR/Cas9 editing of specific receptor genes in chicken cells. Viruses 10:605. doi: 10.3390/v10110605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud J-B, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, Joly J-S, Concordet J-P. 2016. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol 17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. 2010. A Rapid and General Assay for Monitoring Endogenous Gene Modification. Methods Mol Biol 649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 61.Ranwez V, Douzery EJP, Cambon C, Chantret N, Delsuc F. 2018. MACSE v2: toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol Biol Evol 35:2582–2584. doi: 10.1093/molbev/msy159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 63.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost S. 2006. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol 23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- 64.Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol 34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The chBST-2 coding sequence is available in GenBank under accession number MN326300.