The RBPs of T4-like phages are gp37 and gp38. The interaction between phage T4 RBP gp37 and its receptors has been clarified by many reports. However, the interaction between gp38 and its receptors during phage adsorption is still not completely understood. Here, we identified phage Bp7, which uses gp38 as an RBP, and provided a good model to study the phage-host interaction mechanisms in an enterobacteriophage. Our study revealed that gp38 of phage Bp7 recognizes the outer membrane proteins (OMPs) LamB and OmpC of E. coli K-12 as specific receptors and binds with them reversibly. HepI of the inner-core oligosaccharide is the second receptor and binds with phage Bp7 irreversibly to begin the infection process. Determining the interaction between the phage and its receptors will help elucidate the mechanisms of phage with a broad host range and help increase understanding of the phage infection mechanism based on gp38.
KEYWORDS: bacteriophage Bp7, receptor binding protein, receptors, E. coli K-12
ABSTRACT
Bp7 is a T-even phage with a broad host range specific to Escherichia coli, including E. coli K-12. The receptor binding protein (RBP) of bacteriophages plays an important role in the phage adsorption process and determines phage host range, but the molecular mechanism involved in host recognition of phage Bp7 remains unknown. In this study, the interaction between phage Bp7 and E. coli K-12 was investigated. Based on homology alignment, amino acid sequence analysis, and a competitive assay, gp38, located at the tip of the long tail fiber, was identified as the RBP of phage Bp7. Using a combination of in vivo and in vitro approaches, including affinity chromatography, gene knockout mutagenesis, a phage plaque assay, and phage adsorption kinetics analysis, we identified the LamB and OmpC proteins on the surface of E. coli K-12 as specific receptors involved in the first step of reversible phage adsorption. Genomic analysis of the phage-resistant mutant strain E. coli K-12-R and complementation tests indicated that HepI of the inner core of polysaccharide acts as the second receptor recognized by phage Bp7 and is essential for successful phage infection. This observation provides an explanation of the broad host range of phage Bp7 and provides insight into phage-host interactions.
IMPORTANCE The RBPs of T4-like phages are gp37 and gp38. The interaction between phage T4 RBP gp37 and its receptors has been clarified by many reports. However, the interaction between gp38 and its receptors during phage adsorption is still not completely understood. Here, we identified phage Bp7, which uses gp38 as an RBP, and provided a good model to study the phage-host interaction mechanisms in an enterobacteriophage. Our study revealed that gp38 of phage Bp7 recognizes the outer membrane proteins (OMPs) LamB and OmpC of E. coli K-12 as specific receptors and binds with them reversibly. HepI of the inner-core oligosaccharide is the second receptor and binds with phage Bp7 irreversibly to begin the infection process. Determining the interaction between the phage and its receptors will help elucidate the mechanisms of phage with a broad host range and help increase understanding of the phage infection mechanism based on gp38.
INTRODUCTION
Bacteriophages (phages), viruses that are widely distributed in nature, can naturally infect and lyse bacteria but generally have a narrow host range. The limited host range is determined by the phage-specific recognition system and the resistance mechanism of host bacteria. On the one hand, phage receptor binding proteins (RBPs) recognize receptors on the bacterial surface, leading to phage adsorption and infection. The presence of suitable receptors is crucial for the phage infection process because it determines the fate of infection; on the other hand, bacteria have a variety of resistance mechanisms against phages, including prevention of phage adsorption and DNA entry, the restriction modification system, an abortive infection system, a CRISPR-Cas system, and so on (1).
The virulent T-even phages (T2, T4, T6, etc.) that infect Escherichia coli are classified as a ubiquitous virus superfamily and share morphology (2). Each of them has two sets of six retractable tail fibers. The long tail fiber (LTF) walks on the host cell surface, and the RBP on the tip of the LTF reversibly anchors the virion to specific receptors on the host surface, allowing the short tail fiber (STF) to bind irreversibly to a second set of receptors and trigger the phage infection process. RBPs play an important role in determining phage host range (3, 4). It has been reported that there are two kinds of RBPs in T-even phages. In the T4 phage, the RBP is gp37, which forms a fibrous parallel homotrimer at the tip of the LTF with the help of chaperones gp57 and gp38, and it determines phage adsorption. In the T2 and T6 phages, gp38 is located at the distal end of the LTF and plays a role in receptor binding (5–8).
The susceptibility of bacteria to phage depends on whether there are specific receptors on the bacterial surface recognized by phage RBPs. The phages that infect Gram-negative bacteria can recognize and bind to a variety of bacterial components, such as lipopolysaccharide (LPS), outer membrane proteins (OMPs), pili, or capsular components. In E. coli, OMP and LPS constitute T-even-type phage receptors; OMP is specific for individual phages, and LPS is common to all phages (9). LPS consists of lipid A, core oligosaccharide, and O antigen (10). There are more than 180 types of O antigen that determine the serotype of E. coli (11). Phages using the O antigen as receptors are more restricted by E. coli serotype. The inner core part of polysaccharide is highly conserved, and the outer core part varies according to the composition and arrangement of polysaccharides. The core polysaccharide of LPS is regulated by a set of biosynthetic genes (12). Using the core polysaccharide as the receptor, phages are capable of infecting various serotypes of E. coli strains. Knockout or site-directed mutagenesis of genes involved in LPS biosynthesis can change the feature of LPS and is commonly used for detecting LPS sites recognized by phages (13).
As mentioned above, the attachment of phages to host cell surface receptors is the first step of infection. Enterobacteriophages recognize one or more receptors, including OMP and LPS (9, 14). We speculated that phages capable of recognizing multiple receptors or receptors present ubiquitously on the bacterial surface have a broad host range because they have a greater chance of finding a corresponding receptor and binding to the surface of bacteria. This kind of phage may have more value in phage therapy than would a phage with a narrow host spectrum.
In our previous work, it was found that enterobacteriophage Bp7, which was isolated from a chicken farm, had a broad host range and was not limited by host serotype. Genomic sequencing and analysis showed that this phage is one of eight Dhakavirus members belonging to the Tevenvirinae subfamily, Myoviridae family, and Caudovirales order (15). The interaction between RBPs and host receptors that determines the host range of phage Bp7 remains unknown. In this study, we identified the RBP of phage Bp7 and analyzed its role in the infection process to explain its broad-host-range feature. We found that LamB and OmpC of E. coli K-12 are the specific receptors of Bp7, and the RBP gp38 attaches and binds reversibly to these proteins, allowing phage Bp7 to irreversibly bind the inner core polysaccharide of E. coli K-12. Characterization of the RBPs and receptors of phage Bp7 will shed light on the interaction between phages and their hosts.
RESULTS
Identification of phage Bp7 receptors.
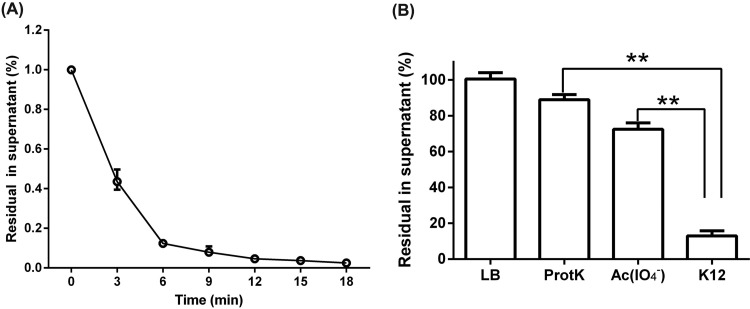
The cell surface of E. coli mainly consists of outer membrane proteins (OMPs) and lipopolysaccharides (LPS), which are commonly exploited as receptors of bacteriophage (9). It is necessary to test whether the OMPs or LPS of E. coli K-12 can be recognized by phage Bp7. E. coli K-12 cells treated with either proteinase K (to destroy OMPs) or sodium acetate containing periodate (IO4−) (to destroy LPS) were incubated with phage Bp7 to determine the possible nature of the receptor. A rapid adsorption effect of phage Bp7 binding to E. coli K-12 was observed. The binding was completed in 6 min, and approximately 88% of phage Bp7 adsorbed to the host cell (Fig. 1A). After the bacteria were treated with proteinase K or sodium acetate, there was a significant increase in free phage particles in the supernatant (P < 0.01), indicating that the phage could not effectively bind to the surface of E. coli K-12 without OMPs or LPS (Fig. 1B). Therefore, Bp7 recognized both OMP and LPS receptors. This result is consistent with the features of T2, T4, and T6 phages (9).
FIG 1.
Identification of phage Bp7 receptors. (A) Adsorption kinetics of Bp7 to E. coli K-12. The adsorption rate of Bp7 to E. coli K-12 at different times was tested by the plaque assay and is shown as residual PFU percentages in the supernatant. (B) Effects of the different treatments of E. coli K-12 on phage Bp7 adsorption. E. coli K-12 was treated by ProtK or sodium acetate containing IO4−, and the adsorption effects of phage Bp7 on E. coli K-12 are shown as residual PFU percentages in the supernatant. All assays were performed in triplicate. **, P < 0.01.
Identification of receptor binding proteins of phage Bp7.
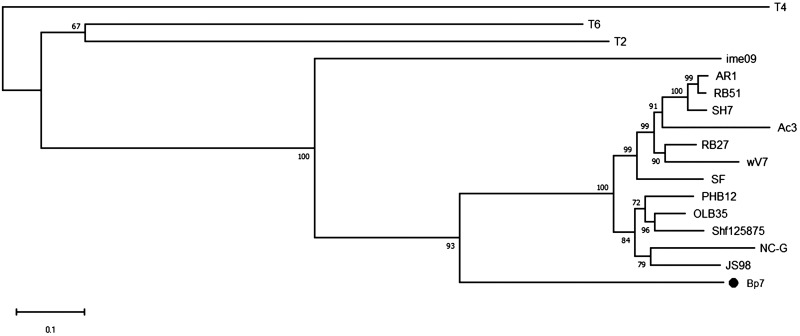
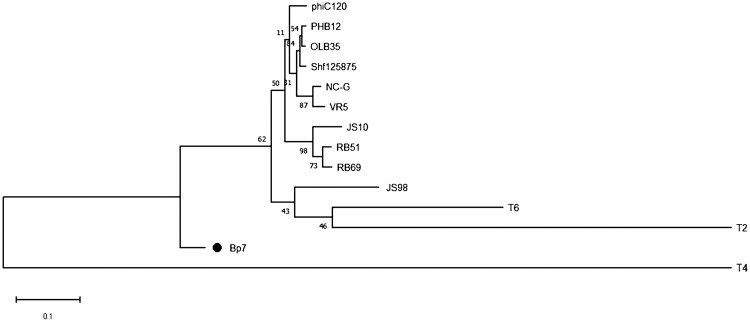
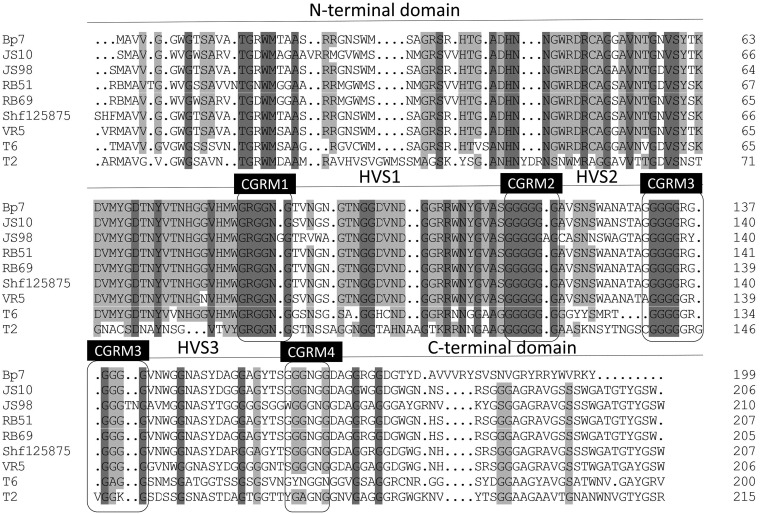
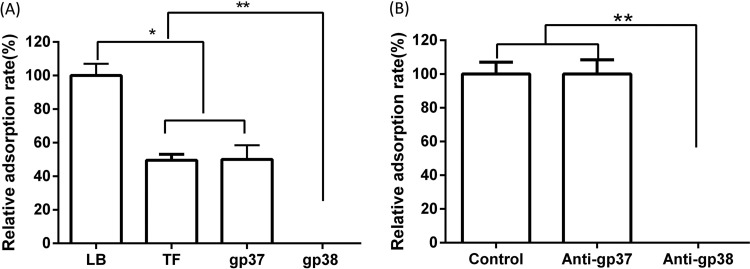
For T-even phages, the RBP at the tip of long tail fibers plays an important role in the process of phage adsorption. RBP gp37 in T4 phage and gp38 in T2 and T6 phages are responsible for the initial specific and reversible binding to receptors on the host cell surface (3, 9). To determine the RBP of Bp7, the amino acid sequences of gp37 and gp38 were subjected to a BLAST search in the nonredundant protein sequence database, and the homology was analyzed. A relationship with the RBP of phage T2 and that of phage T6 was observed, but these RBPs were distantly related to that of T4 (Fig. 2 and 3). It is generally believed that phage RBPs in the same species have similar amino acid sequences. The RBP of phage T2 and phage T6 is gp38, so we predicted that gp38 is the RBP of phage Bp7. It has been reported that the C-terminal specificity domain of RBP gp38 determines the host receptor affinity, and there are a series of hypervariable segments (HVSs) separated by a set of highly conserved glycine-rich motifs (CGRMs) in this specificity domain (9). This pattern of sequence conservation is also observed in the alignment of gp38 sequences of E. coli phages (JS98, JS10, RB51, RB69, Shf125875, VR5, T6, and T2) (Fig. 4). The alignment showed that there were four CGRMs separated by three HVSs, suggesting that gp38 in these phages used the same mechanism of attaching to the receptors on the host cell surface. To further verify the RBP of Bp7, the protein competitive effects of gp37 and gp38 and their antibody-blocking effect on phage Bp7 adsorption were tested. The gp38 protein or its antibody completely inhibited Bp7 adsorption (Fig. 5A and B), indicating that gp38 is the key factor for Bp7 adsorption to the surface of E. coli K-12. The gp37 protein showed a nonspecific competition effect on Bp7, although it caused an ∼55% decrease in the Bp7 adsorption rate compared with that of the LB control group (P < 0.05); there was no significant difference with the trigger factor (TF) control group (P > 0.05), which is the expressed TF protein of the pCold-TF vector (Fig. 5A). Additionally, the gp37 antibody had no inhibitory effect on Bp7 binding to E. coli K-12 (Fig. 5B). Therefore, gp38 was identified as the RBP of phage Bp7.
FIG 2.
Evolutionary relationships of phage Bp7 based on gp37 phylogenetic analysis. A total of 17 orthologous groups were selected for the phylogenetic analysis. The evolutionary tree was constructed by the neighbor-joining (NJ) method with the default parameter. The numbers at the nodes indicate the bootstrap probabilities.
FIG 3.
Evolutionary relationships of phage Bp7 based on gp38 phylogenetic analysis. A total of 14 orthologous groups were selected for the phylogenetic analysis. The evolutionary tree was constructed by NJ method with the default parameter. The numbers at the nodes indicate the bootstrap probabilities.
FIG 4.
gp38 amino acid sequence pattern comparison of phage Bp7 with those of 8 other phages. Residues that are conserved in all aligned sequences are shown on a gray background (the darker gray indicates higher sequence homology), and nonconserved residues are shown on a white background. A mosaic character is shown in all of the sequences. Boxes show the highly conserved glycine-rich motifs (CGRMs); hypervariable segments (HVSs) are separated by a set of CGRMs.
FIG 5.
Identification of receptor binding protein of phage Bp7. The tail fiber protein gp38 was identified as a receptor binding protein. (A) Recombined proteins gp37 and gp38 were competed in a phage Bp7 adsorption assay. LB and TF proteins were used as controls, and the adsorption rate was set as 100%. (B) Anti-gp37 and anti-gp38 antibody serum-blocking phage Bp7 adsorption assay. Serum collected before immunization was used as a control, and the adsorption rate was set as 100%. All assays were performed in triplicate, and significance was determined by Student's t test for comparison between the control group and the treated group. *, P < 0.05; **, P < 0.01.
Identification of host OMP receptor targets of gp38.
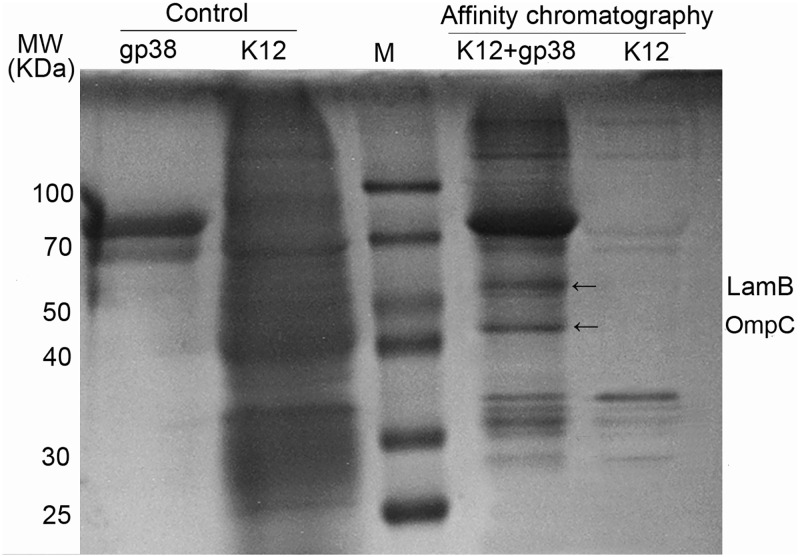
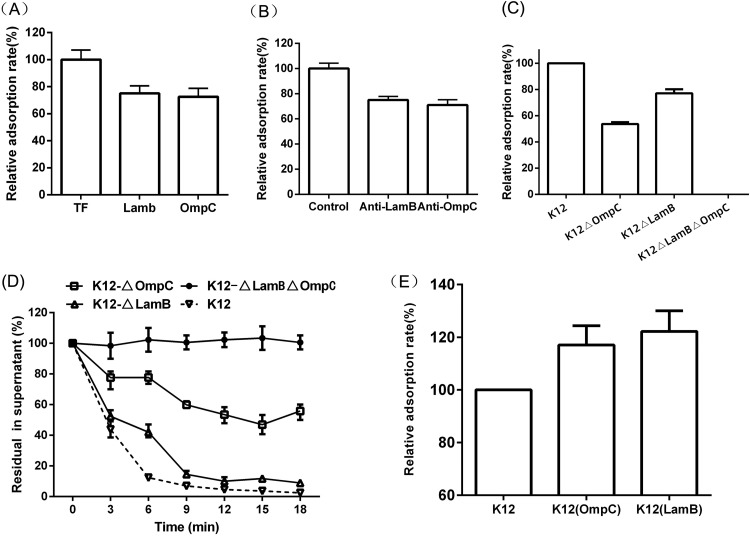
It is considered that the first step of phage binding to bacteria is mediated by the interaction of its RBP with host cell surface receptors (16). The His6 fusion protein gp38 was coupled to Ni-agarose resin and incubated with E. coli K-12 lysates. The OMP receptors of E. coli K-12 recognized by gp38 were pulled down by affinity chromatography. Two protein bands recovered specifically from E. coli K-12 lysates were analyzed by mass spectrometry, and the captured polypeptides were LamB and OmpC (Fig. 6). To further verify the OMP receptors of Bp7, the LamB or OmpC adsorption-blocking effect and the competitive adsorption effect of their antibodies were tested. The soluble fusions and the polyclonal antibodies caused an ∼25% reduction in phage adsorption (Fig. 7A and B), indicating that LamB and OmpC could block phage Bp7 adsorption to the surface of E. coli K-12. To further test the roles of LamB and OmpC in phage Bp7 infection, the mutant strains E. coli K-12 ΔlamB, K-12 ΔompC, and K-12 ΔlamB ΔompC were constructed using the scarless Cas9 assisted recombineering (no-SCAR) system, and the phage adsorption rates were assayed. The results showed that E. coli K-12 ΔompC and E. coli K-12 ΔlamB caused a 50% and 20% decrease in phage binding activity based on the adsorption assay, respectively, and the E. coli K-12 ΔlamB ΔompC double-mutant strain was completely resistant to phage Bp7 (Fig. 7C). Phage Bp7 was incubated with the mutant strains, and virions in the supernatant showed a similar trend over time (Fig. 7D). To avoid the phenotype caused by a spontaneous mutation elsewhere in the genome, each single-mutant strain was complemented by the corresponding wild-type gene, and the phage adsorption rate increased to the same level as the wild-type strain (data not shown). Furthermore, LamB or OmpC overexpression in E. coli K-12 led to an ∼20% increase in phage adsorption efficiency (Fig. 7E). The results confirmed that LamB and OmpC of E. coli K-12 were the receptors of phage Bp7 and played a role in the phage adsorption process.
FIG 6.
Identification of OMP receptors on the surface of E. coli K-12 recognized by phage Bp7. The OMPs on the E. coli K-12 surface targeted by phage Bp7 were identified by affinity chromatography. Recombined protein gp38 and E. coli K-12 were used as a control. The protein bands which could bind with gp38 protein specifically are shown by black arrows. The common bands in all lanes were nonspecifically bound to the nickel column. MW, molecular weight (marked in lane M).
FIG 7.
LamB and OmpC acted as the receptors of phage Bp7. (A) Recombined proteins LamB and OmpC competed in a phage Bp7 adsorption assay. The TF protein was used as a control, and the adsorption rate was set as 100%. (B) LamB and OmpC antibody serum-blocking phage Bp7 adsorption assay. Naive serum was used as a control, and the adsorption rate was set as 100%. (C) The mutant strains E. coli K-12 ΔOmpC, E. coli K-12 ΔLamB, and E. coli K-12 ΔOmpC ΔLamB were constructed by the no-SCAR system, the adsorption rates of Bp7 were tested by a plaque assay, E. coli K-12 was used as a control, and the adsorption rate was set as 100%. (D) The adsorption kinetics of Bp7 to E. coli K-12 ΔOmpC, E. coli K-12 ΔLamB, and E. coli K-12 ΔOmpC ΔLamB were tested by a plaque assay and are shown as residual PFU percentages in the supernatant, and E. coli K-12 was used as a control. (E) Plasmid pCold-OmpC or pCold-LamB was transferred into E. coli K-12 and induced by IPTG to acquire the OMP-overexpressing strain of E. coli K-12. The adsorption effects of Bp7 were tested by a plaque assay. The adsorption rate of phage Bp7 to the E. coli K-12 wild-type strain was set as 100%. All assays were performed in triplicate.
As outer membrane porins, LamB and OmpC are abundant in Gram-negative bacteria and form pores that allow the passive diffusion of small molecules across the outer membrane; they also serve as receptors of bacteriophages (2, 14, 17). The results of the present study are consistent with the notion that OmpC serves as a receptor for many T-even phages infecting Shigella flexneri and E. coli (8, 18). LamB is seldom reported as the phage receptor, except for phages K10, TP1, and λ (14, 19). Phages K10 and λ belong to the Siphoviridae family, and phage TP1, a mutant strain of phage TuIa with gp37 as the RBP, belongs to the Myoviridae family. In this work, the RBP of phage Bp7, gp38, was also specifically recognized and bound with LamB. This result indicated that although phages have different RBPs and adsorption mechanisms, their receptors tend to be components widely present on the host cell surface, increasing the chance of infection. Because LamB and OmpC are widely distributed on the surface of E. coli, phage Bp7 could recognize them at the same time, so Bp7 had a greater chance of binding with different E. coli strains and showed a broad host range.
Identification of LPS as a receptor of phage Bp7.
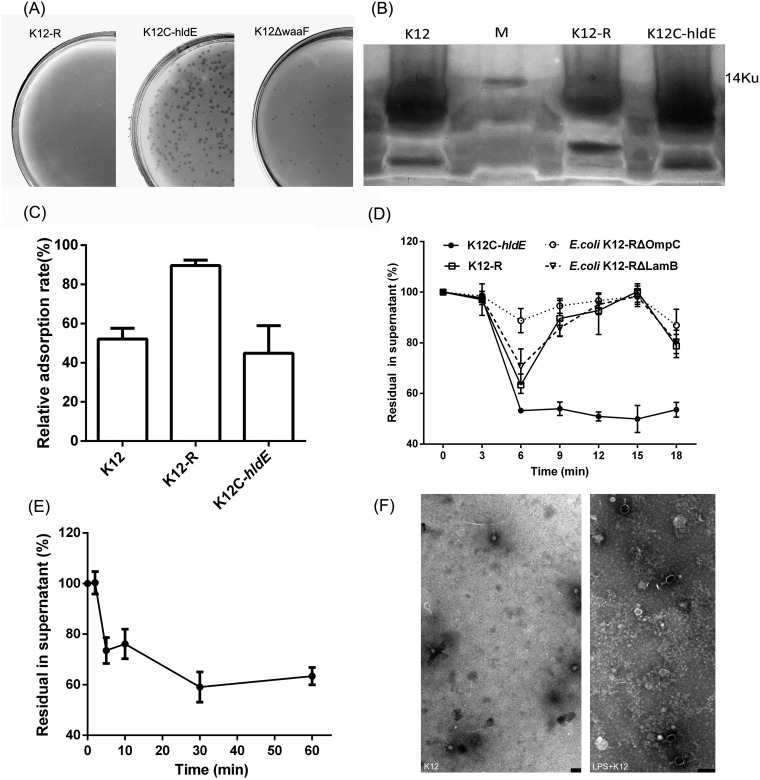
It has been reported that phage T2 recognizes LPS as a receptor to which STF irreversibly binds (20). LPS is a component of the outer leaflet of the outer membrane of Gram-negative bacteria and is composed of three regions, lipid A, a core oligosaccharide, and O antigen. The LPS of E. coli usually acts as the receptor of many T-even phages and is crucial for the phage STF to attach and inject its genome into the host cell (19). Bp7 was cultured with E. coli K-12, and a naturally mutant strain, E. coli K-12-R, was obtained; this mutant strain showed complete resistance to phage Bp7 (Fig. 8A). To find the mutation site, the genomes of E. coli K-12-R and E. coli K-12 were sequenced and aligned. There was a two-base deletion in the hldE gene (positions 241 and 242) of K-12-R that caused early transcription termination (data not shown). hldE (formerly known as waaE or rfaE) encodes HepI transferase, which has bifunctional enzyme activity of heptose 7-phosphate kinase and heptose 1-phosphate adenylyltransferase; HepI transferase is responsible for inner core LPS biosynthesis and regulates the production of heptose-less LPS (21). To verify the tolerance of E. coli K-12-R caused by the hldE gene mutation, the hldE-T plasmid was constructed and transformed into E. coli K-12-R to acquire the complement strain E. coli K-12C-hldE. After induction by isopropyl-β-d-thiogalactopyranoside (IPTG), E. coli K-12C-hldE carrying the hldE-T plasmid formed plaques after incubation with phage Bp7 (Fig. 8A). To eliminate other possible LPS adsorption sites and confirm the receptor role of HepI, waaF, which encodes HepII of E. coli K-12, was knocked out by the no-SCAR system to acquire the E. coli K-12 ΔwaaF mutant strain. The results showed that phage Bp7 could form plaques on the lawn of E. coli K-12 ΔwaaF (Fig. 8A), thereby confirming that HepI of inner core LPS was a critical part of the receptor of Bp7. This is consistent with previous reports that the inner core oligosaccharide in E. coli is necessary for the efficient binding of many T-even phages (19).
FIG 8.
LPS acted as the second receptor of phage Bp7. (A) The plasmid hldE-T was transferred into the mutant strain E. coli K-12-R to acquire the complement strain K-12C-hldE, the waaF gene of E. coli K-12 was knocked out by the no-SCAR system to construct the mutant strain E. coli K-12 ΔwaaF, and the susceptibility of E. coli K-12 ΔwaaF to phage Bp7 was tested by the plaque assay to identify the role of the hldE gene. Left, E. coli K-12-R was tolerant to phage Bp7; middle, the K-12C-hldE complemented strain carrying the hldE-T plasmid became susceptible to phage Bp7 after induced by IPTG; and right, the E. coli K-12 ΔwaaF was susceptible to phage Bp7. (B) LPS was purified from the E. coli K-12, E. coli K-12-R, and E. coli K-12C-hldE strains and detected by silver-staining SDS-PAGE. (C) Phage Bp7 was incubated with LPS of E. coli K-12, E. coli K-12-R, or E. coli K-12C-hldE, and free virions in the supernatant were tested by the plaque assay to identify the LPS effects on Bp7 adsorption in vitro. The adsorption rate of phage Bp7 was set as 100%. All assays were performed in triplicate. (D) The mutant strains of E. coli K-12-R ΔLamB and E. coli K-12-R ΔOmpC were constructed by the no-SCAR system, and the adsorption kinetics of Bp7 to E. coli K-12-R, E. coli K-12-R ΔLamB, E. coli K-12-R ΔOmpC, and E. coli K-12-R/hldE were tested by the plaque assay to identify the second receptor role of LPS. The phage Bp7 adsorption rates are shown as residual PFU percentages in supernatant. All assays were performed in triplicate. (E) The adsorption effects of phage Bp7 preincubated with LPS at different times was tested by a plaque assay. Phage Bp7 without LPS treatment was used as a control, and the adsorption rate was set as 100%. All assays were performed in triplicate. (F) LPS-triggered DNA release from phage Bp7 in vitro was tested by TEM. Phage Bp7 was used as a control (left); after incubation with LPS, phage Bp7 released its DNA, and the head of phage particles changed from white to black (right). Scale bar = 200 nm.
Analysis of the role of LPS in phage Bp7 adsorption.
To verify the role of LPS in the Bp7 attachment process, LPS was extracted and purified from E. coli K-12, E. coli K-12-R, and E. coli K-12C-hldE and detected by SDS-PAGE. The results showed that LPS of E. coli K-12-R was truncated down to the inner core oligosaccharide (Fig. 8B). Furthermore, the effects of purified LPS on Bp7 in vitro were tested. LPS of E. coli K-12 was incubated with Bp7 for 10 min, and approximately 48% virions could not bind host cells, indicating that LPS could inhibit Bp7 from attaching to host cell in vitro (Fig. 8C). LPS of K-12-R showed almost no inhibitory effect on Bp7 adsorption, and the binding rate of phage Bp7 increased again to 45% after incubating with the LPS of K-12C-hldE (Fig. 8C), indicating that hldE gene regulation of the inner core LPS was essential for phage Bp7 adsorption. There are many reports describing that LPS can act as the second receptor and irreversibly bind with the STF of T-even phages (20). Phage Bp7 also belongs to T-even phages; therefore, LPS may play a second receptor role. To confirm this hypothesis, the adsorption kinetics of phage Bp7 on K-12-R and K-12C-hldE were determined. Approximately 50% of virions could rapidly adsorb to K-12C-hldE in 6 min (Fig. 8D). Although no plaque on E. coli K-12-R incubated with Bp7 was formed, a periodic phage binding-release cycle was observed. The binding was completed in 6 min and released again at 15 min (Fig. 8D). Furthermore, the E. coli K-12-R ΔLamB and E. coli K-12-R ΔOmpC mutant strains were constructed. These strains showed a periodic phage binding-release cycle similar to that of E. coli K-12-R, and OmpC showed a higher affinity to phage Bp7 than did LamB (Fig. 8D). The results suggested that phage Bp7 could reversibly bind with LamB and OmpC of E. coli K-12-R but could not infect cells, indicating that the LPS of E. coli K-12 acted as the second receptor of Bp7, binding with STF of phage Bp7 irreversibly and promoting the injection of genetic material into host cells. To further confirm the role of LPS, the DNA release of phage Bp7 induced by LPS of E. coli K-12 in vitro was tested by plaque assay and transmission electron microscopy (TEM) observation. After incubation with LPS, the adsorption of phage Bp7 showed a significant decrease in 5 min (Fig. 8E), and the head of phage particles changed from white to black in TEM, indicating that LPS could induce DNA release from phage Bp7 in vitro (Fig. 8F). All of the above-described results suggested that the LPS of E. coli K-12 was the second receptor of phage Bp7 and could induce the phage injection process.
DISCUSSION
There are many reports about the Myoviridae phage infection mechanism. The RBP, located at the tip of the LTF, recognizes specific primary receptors initially; after at least three LTFs have bound, an irreversible second interaction is formed between extended STFs and a second set of receptors, followed by phage DNA injection (22–24). The primary receptors on the host cell surface are specific for different phages and determine their host range. OMP and LPS are currently recognized as the receptors for T-even phages (20). In T-even phages, the interaction mechanism between the RBP gp37 and recognized receptors, represented by phage T4, has been clearly reported, but in phages represented by the T2 and T6 phages, the molecular interactions between RBP gp38 and host receptors are not fully understood. Marti et al. (8) reported that the T4-like Salmonella phage S16 could recognize and bind with OmpC and LPS core, and its long tail fiber proteins gp37 and gp38 played roles as RBPs. Phage Bp7 belongs to T-even phages, one of eight members of the Dhakavirus genus. To date, there is no report about the interaction mechanism between Dhakavirus phage members and their host bacteria. In the current study, phage Bp7 and host bacteria of E. coli K-12 were used to identify the characteristics of the Dhakavirus phage adsorption process.
In this study, the RBP of phage Bp7 was identified as gp38. This protein showed homology and a similar conserved amino acid sequence pattern with phage T2 and T6, which used gp38 as an RBP. To further confirm the RBP role of gp38, we analyzed the effect of gp38 fusion and its antibody on Bp7 adsorption, and as expected, they completely inhibited on Bp7 attachment to host cells in vitro.
E. coli K-12 was used in this study to identify the host components that interact with phage Bp7. The heptose of the core oligosaccharide and two OMPs (OmpC and LamB) were identified as receptors of phage Bp7. This conclusion was supported by the following experiments. First, both sodium acetate- and proteinase K-treated E. coli K-12 had a decreased phage Bp7 adsorption rate. Then, two OMPs (OmpC and LamB) of E. coli K-12 were found to have a binding relationship with the RBP gp38 in an affinity chromatography assay. Furthermore, OMP fusions and purified LPS of E. coli K-12 decreased phage Bp7 binding to host cells in vitro. Finally, OMP single mutants of E. coli K-12 showed a decrease in phage adsorption efficiency. The LPS mutant strain and OMP double-mutant strain were completely resistant to phage Bp7, and OMP-overexpressing strains increased phage binding efficiency.
In addition, OMPs were identified as the primary receptors, and HepI acted as the second receptor of phage Bp7. The LPS mutant strain that was completely tolerant to Bp7 still had a periodic binding-release cycle with phage Bp7, and purified LPS could trigger Bp7 DNA release in vitro.
The data in this study, together with those from previous reports, revealed the phage-host interaction mechanisms in phages with gp38 as the RBP. Phages walk on the bacterial surface, and gp38, located at the tip of six LTFs, recognizes suitable OMPs on the bacterial surface and binds with them reversibly, bringing phages close to the host cell surface. STFs then adsorb to LPS irreversibly, attaching the phage to the bacterial surface, and the phage injects its genetic material into the host cell. OmpC and LamB, as OMP components, are widely spread on the bacterial surface, and heptose in the LPS inner core region is conserved in E. coli, increasing the chance that Bp7 will find a suitable receptor on the bacterial surface; therefore, Bp7 is not limited by serotypes and has a broad host range.
The LPS of Gram-negative bacteria is important to the structural integrity and distribution of OMPs (25–27). The relationship of the LPS-OMP complex affects phage adsorption efficiency. For example, two OMPs were found to be Bp7 receptors in this study; OMP single-mutant strains caused a 50% to ∼78% decrease in adsorption of Bp7, and the double-mutant strain was completely resistant to Bp7, but OMP fusion proteins could only slightly decrease the binding efficiency in vitro. Additionally, the LPS mutant could tolerate phage Bp7 completely, but purified LPS inhibited ∼50% phage adsorption. This might be because there was cooperation between LPS and OMP on the bacterial surface in the process of phage adsorption. OMPs can move in the bacterial membrane, and when multiple phage LTFs attach to host OMPs, OMPs gather at the phage to perform their LTF binding function, which will increase the chance of phage STF adsorbing to LPS irreversibly; however, there is no LPS-OMP cooperation in vitro, so weak inhibition was observed in the phage Bp7 adsorption process. However, whether there is LPS-OMP cooperation remains unknown. Another possible explanation is that the receptor conformation is important for the recognition and binding of phages, and the conformations of fusion OMPs or purified LPS may be altered, leading to a low adsorption rate (18).
In conclusion, the RBP of the isolated enterobacteriophage Bp7 is gp38, which reversibly bound with primary receptors OmpC and LamB and recognized HepI of inner core LPS as the second receptor, providing Bp7 a broad host range. Our results elucidated the interaction mechanism between the E. coli host and phages with the gp38 RBP. This information may also contribute to the identification of the phage infection processes of other members of the Dhakavirus genus.
MATERIALS AND METHODS
Bacteria and phages.
E. coli K-12 (GenBank accession no. CP040663) and its mutant strain E. coli K-12-R (accession no. CP040664) were cultured in LB at 37°C and stored in 30% glycerol at –80°C until use. Bacteriophage Bp7 (accession no. NC_019500.1) was isolated from a chicken farm (15). The characteristics of the strains and plasmids used in this study are presented in Table 1. All primers used in this study are listed in Table 2.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains or plasmid | Relevant characteristics or descriptiona |
|---|---|
| E. coli strains | |
| BL21 | E. coli engineering bacterium used as competent cells |
| K-12 | Wild-type strain, host bacterium of phage Bp7 |
| K-12-R | ΔhldE mutant naturally derived from strain K-12 and resistant to phage Bp7, heptose-less LPS |
| K-12C-hldE | Complemented ΔhldE strain, transformed with a pMD19-T vector with cloned hldE |
| K-12 ΔlamB | ΔlamB mutant derived from strain K-12, CRISPR-Cas9 technology knockout of lamB gene |
| K-12 ΔompC | ΔompC mutant derived from strain K-12, CRISPR-Cas9 technology knockout of ompC gene |
| K-12 ΔwaaF | ΔwaaF mutant derived from strain K-12, CRISPR-Cas9 technology knockout of waaF gene |
| K-12 ΔompC ΔlamB | ΔompC ΔlamB mutant derived from strain K-12, CRISPR-Cas9 technology knockout of ompC and lamB genes |
| K-12-R ΔompC | ΔompC mutant derived from strain K-12-R, CRISPR-Cas9 technology knockout of ompC gene |
| K-12-R ΔlamB | ΔlamB mutant derived from strain K-12-R, CRISPR-Cas9 technology knockout of lamB gene |
| Plasmids | |
| pKDsg-ack | sgRNA plasmid, Specr |
| pKDsg-LamB | Derived from sgRNA plasmid, retargeting the lamB gene, Specr |
| pKDsg-OmpC | Derived from sgRNA plasmid, retargeting the ompC gene, Specr |
| pKDsg-WaaF | Derived from sgRNA plasmid, retargeting the waaF gene, Specr |
| pCas9 | CRISPR Cas9 plasmid, expression of Cas9 protein used as cleaving gene, Ampr |
| pCold-TF-LamB | Fusion cold shock expression plasmid with a TF chaperone fusion tag, expresses LamB protein, Ampr |
| pCold-TF-OmpC | Fusion cold shock expression plasmid with a TF chaperone fusion tag, expresses LamB protein, Ampr |
| pCold-TF-gp37 | Fusion cold shock expression plasmid with a TF chaperone fusion tag, expresses gp37 protein, Ampr |
| pCold-TF-gp38 | Fusion cold shock expression plasmid with a TF chaperone fusion tag, expresses gp38 protein, Ampr |
| pMD19-T-hldE | Cloning plasmid, expression of hldE protein, Ampr |
sgRNA, single guide RNA; Specr, spectinomycin resistance; Ampr, ampicillin resistance.
TABLE 2.
Primers used in this study
| Primer name (target) by use | Oligonucleotide sequence (5′–3′)a |
|---|---|
| CRISPR Cas9 test | |
| Prospacer-F (lamB) | ACGGTATTTACTTTGAGCACgttttagagctagaaatagcaag |
| Prospacer-R (lamB) | GTGCTCAAAGTAAATACCGTgtgctcagtatctctatcactGA |
| Prospacer-F (ompC) | GCTGAAGTTTACAACAAAGAgttttagagctagaaatagcaag |
| Prospacer-R (ompC) | TCTTTGTTGTAAACTTCAGCgtgctcagtatctctatcactGA |
| Prospacer-F (waaF) | TCATGTCGCCAACCCAAGACgttttagagctagaaatagcaag |
| Prospacer-R (waaF) | GTCTTGGGTTGGCGACATGAgtgctcagtatctctatcactGA |
| pKDseq1-F | ccaattgtccatattgcatca |
| gamR | tttataacctccttagagctcga |
| Cas9-F | ATGCTCATCCGGAATTTCAAATATGTATCCGCTCAT |
| Cas9-R | TCATTGCCATACGAATTACCAATGCTTAATCAGTG |
| pBAD-F | ATGCCATAGCATTTTTATCC |
| pBAD-R | GATTTAATCTGTATCAGGC |
| pKD-LamB-F | GGTCTGCTATGTGGTGCT |
| pKD-LamB-R | GCCAAGCTGGAGACCGTT |
| K-12△lamB-F | GAAAAGCAATGACTCAGGAGATAG |
| K-12△lamB-R | CCTTATCCGGCCCAGGTTTTGCT |
| K-12△ompC-F | GGGAGAATGGACTTGCCGACTG |
| K-12△ompC-R | TGATTATCCTCATGCGAACGGTC |
| K-12△waaF-F | CCGCTAATGGTGGAACGCTA |
| K-12△waaF-R | CGGGCTACTCGGACCATACA |
| Chromosomal mutations | |
| LamB | C*A*C*CCCACAAAACACACAAAGCCTGTCACAGGTGATGTGAAAAAAGAAAAGCAATGACTCAGGAGATAGATAGCAAAACCTGGGCCGGATAAGGCGTTTACGCCGCATTCGGCAACCAACGCCTGATGCGACGCTTGCGC |
| OmpC | T*T*A*ATGAGGGTTAATCAGTATGCAGTGGCATAAAAAAGCAAATAAAGGCATATAACAGAGGGTTAATAACTCTCGATTGATATCGAACAAAGGGCCTGCGGGCCCTTTTTTCATTGTTTTCAGCGTACAAACTCAGTTT |
| WaaF | C*A*A*GTTTTGGCAACGACTTCCACATTGCCTTATGTACCGGACCGACTTAGCGCTGCGTATTCTCGAGACGGCCTACGCCCAAAACTAGCAATTTTGTAGCAGCTACCCGCTACAAGAGGTATGCAACGGGCGTGAGTGAC |
| Front-F (lamB) | C*A*C*CCCACAAAACACACAAAGCCTGTCACA |
| Front-R (lamB) | TCCGGCCCAGGTTTTGCTATCTATCTCCTGAGTCATTGCT |
| Front-F (lamB) | C*A*C*CCCACAAAACACACAAAGCCTGTCACA |
| Rear-F (lamB) | AGCAATGACTCAGGAGATAGATAGCAAAACCTGGGCCGGA |
| Rear-R (lamB) | GCGCAAGCGTCGCATCAGGCGTTGGTTGCC |
| Front-F (ompC) | T*T*A*ATGAGGGTTAATCAGTATGCAGTGGC |
| Front-R (ompC) | TGTTCGATATCAATCGAGAGTTATTAACCCTCTGTTATAT |
| Rear-F (ompC) | ATATAACAGAGGGTTAATAACTCTCGATTGATATCGAACA |
| Rear-R (ompC) | AAAACTGAGTTTGTACGCTGAAAACAATG |
| Front-F (waaF) | C*A*A*GTTTTGGCAACGACTTCCACAT |
| Front-R (waaF) | GCTAGTTTTGGGCGTAGGCCGTCTCGAGAATACGCAGCGC |
| Rear-F (waaF) | GCGCTGCGTATTCTCGAGACGGCCTACGCCCAAAACTAGC |
| Rear-R (waaF) | GTCACTCACGCCCGTTGCATACCTC |
| Overexpression test | |
| LamB-F | GATGAGCTCATGATGATTACTCTGCGC (SacI) |
| LamB-R | GGCAAGCTTTTACCACCAGATTTCCATCT (HindIII) |
| OmpC-F | GGCGAGCTCATGAAAGTTAAAGTACTGTC (SacI) |
| OmpC-R | GGCAAGCTTTTAGAACTGGTAAACCAGAC (HindIII) |
*, phosphorothioate bond. The lowercase letters in the primers show plasmid genes, and the uppercase letters indicate the sgRNA sequences of lamB, ompC, and waaF, respectively. The plasmids and maps have been deposited to Addgene (Cambridge, MA), with pCas9cr4 as plasmid no. 62655 and pKDsg-ack as plasmid no. 62654. Restriction sites are in parentheses.
Phage adsorption assay.
The adsorption kinetics of phage Bp7 to E. coli K-12 were assayed as previously described (28). Briefly, 4 ml of Bp7 (1 × 104 PFU/ml) was mixed with an equal volume of E. coli K-12 (1 × 109 CFU/ml) and incubated at room temperature for 18 min. Then, 500 μl of the mixed suspension was collected every 3 min. The sample was centrifuged at 13,000 × g for 1 min, and the phage titer remaining in the supernatant was determined by the double-layer agar method. LB medium only was used as a control in each assay, and the phage titer of the LB control group was set to 100%. Each assay was performed in duplicate and repeated three times.
Identification of the targets of phage Bp7 on the host bacterial surface.
To determine the targets of Bp7 on the host cell surface, E. coli K-12 cells were treated with periodate and proteinase K, as described previously (28). A total of 1.5 ml of E. coli K-12 culture was treated with proteinase K (200 μg/ml) or 50 mM sodium acetate (pH 5.2) containing 100 mM IO4− at 37°C for 2 h and then washed with 1.5 ml LB three times. A total of 500 μl of treated E. coli K-12 (109 CFU/ml) sample was incubated with 500 μl phage Bp7 (105 PFU/ml) for 5 min and then centrifuged at 16,000 × g for 1 min; the phage titer in the supernatant was determined by the double-layer agar method to test the adsorption rate of Bp7. The untreated E. coli K-12 sample was used as a control. Each assay was performed in duplicate and repeated three times.
Identification of the RBP of phage Bp7.
To identify the RBP of Bp7, the amino acid sequences of long tail fiber proteins gp37 (NCBI RefSeq accession no. YP_007004199.1) and gp38 (accession no. YP_007004198.1) were compared with those in the NCBI nonredundant sequence database, and homology analysis was performed. Then, the gp37 and gp38 genes were amplified by PCR and inserted into the pCold-TF vector to construct the recombinant plasmids pCold-gp37 and pCold-gp38, respectively. Proteins were expressed in E. coli BL21(λDE3) as His6 fusions and purified by affinity chromatography. For preparation of the polyclonal antibody, New Zealand White rabbits were subcutaneously immunized with 1 mg of protein three times. For the first immunization, proteins were emulsified with complete Freund’s adjuvant (CFA). For the second immunization, proteins were injected with incomplete Freund’s adjuvant, and for the last immunization, no adjuvant was used. Three days after the final immunization, serum samples were collected and stored at –80°C until use. The expressed putative RBPs’ competitive effects and their antibody-blocking effects on phage Bp7 adsorption were tested. Briefly, to test the competitive effect of gp37 or gp38, 200 μl purified gp37 or gp38 protein (1 mg/ml) was incubated with 200 μl E. coli K-12 (109 CFU/ml), respectively, for 10 min; an equal volume of LB was used as a control, and the expressed trigger factor (TF) protein of the pCold-TF vector (1 mg/ml) was used as a control. A total of 100 μl Bp7 (106 PFU/ml) was added, and the mixture was incubated for 10 min. Then, the phage adsorption rates were tested by the double-layer agar method. To test the antibody-blocking effect of gp37 or gp38, 100 μl serum antibody against gp37 or gp38 was mixed with an equal volume of phage Bp7 (106 PFU/ml) for 10 min. Serum collected before immunization was used as a control. The mixture was incubated with 200 μl E. coli K-12 (109 CFU/ml) for 5 min, and phage adsorption rates were tested by the double-layer agar method. Each experiment was performed in triplicate.
Identification of OMP receptors on host bacteria recognized by phage Bp7.
Host bacterial OMP receptors recognized by the RBP of phage Bp7 were identified by affinity chromatography, as previously described (28). The protein gp38 fused to a His6 tag was covalently coupled to Ni-agarose resin (CWBiotech Co., Ltd.) in Tris-HCl buffer (pH 7.9) for 2 h and incubated with 10 mM ethanolamine at 4°C for 30 min to block unreacted sites. A total of 1 ml lysate from exponential-phase E. coli K-12 cells was mixed with the resin and incubated at 4°C overnight to pull down the receptor proteins. The resin was washed with 20 mM imidazole buffer three times to remove nonspecifically bound proteins. Then, bound proteins were eluted with 500 mM imidazole buffer and identified by SDS-PAGE. Specific bands were purified from the gel and sequenced by Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China).
Effect of OMP receptors on phage Bp7 adsorption analysis.
The genes of two identified OMPs were amplified by PCR and inserted into the pCold-TF vector to construct the recombinant plasmids, from which soluble His6 fusion proteins were expressed in E. coli BL21(λDE3) and purified by affinity chromatography. New Zealand White rabbits were immunized with 1 mg recombinant protein to obtain polyclonal antibody products, as described above. The effects of OMPs and their antibodies on phage adsorption were tested. In brief, to test the blocking effect of OMP, 100 μl purified OMP (1 mg/ml) was incubated with 100 μl Bp7 (106 PFU/ml) at 37°C for 10 min, and an equal volume of TF protein (1 mg/ml) was used as a control; 100 μl E. coli K-12 (109 CFU/ml) was added, and the mixture was incubated for 10 min. The phage adsorption rates were tested by the double-layer agar method. Each experiment was performed in triplicate. To test the competitive adsorption effect of OMP antibody, 100 μl serum of OMP antibody was mixed with an equal volume of E. coli K-12 (109 CFU/ml) at 37°C for 10 min, and naive serum was used as a control. The mixture was incubated with 100 μl phage Bp7 (106 PFU/ml) at 37°C for 5 min, and the phage adsorption rates were tested by the double-layer agar method. Each experiment was performed in triplicate.
Construction and verification of OMP mutants.
The genes encoding the possible receptors of E. coli K-12 were subjected to deletion by the no-SCAR system, as described previously (29). Briefly, plasmid pCas9cr4 was transferred into E. coli K-12, which is sensitive to phage Bp7, and screened by 100 μg/ml ampicillin (Amp); the pKDsg-LamB or pKDsg-OmpC plasmid was transferred into the resulting strain and screened by 50 μg/ml spectinomycin (Spec). The positive clone possessing both the pCas9cr4 plasmid and pKDsg-LamB or pKDsg-OmpC plasmid was cultivated and induced by 50 mM l-arabinose to express the λ red genes. A 140-bp DNA fragment (600 ng) containing both sides of the target OMP gene-coding region (70 bp upstream of the coding sequence and 70 bp downstream of the transcriptional terminator) with 3 phosphorothioate bonds at the 5′ end was electrically transferred into the induced positive clone and screened with 100 μg/ml Amp, 50 μg/ml Spec, and 100 ng/ml anhydrotetracycline (aTc). The target OMP knockout strains were confirmed by PCR. The constructed mutant strains were incubated at 37°C to eliminate the temperature-sensitive plasmid pKDsg-OmpC or pKDsg-LamB. Based on the mutant strain E. coli K-12 ΔompC, the lamB gene was deleted by the no-SCAR system, as described above, to construct the E. coli K-12 ΔompC ΔlamB double-mutant strain. The adsorption rates and adsorption kinetics of phage Bp7 on the OMP mutant strains were assayed using the double-layer agar method, as described above.
To further confirm the role of OmpC and LamB, the lamB and ompC genes of E. coli K-12 were amplified by PCR and inserted into the pCold-TF vector to construct the recombinant plasmids pCold-LamB and pCold-OmpC, respectively. Plasmids were transferred into E. coli K-12 and induced by 1 mM IPTG at 16°C for 8 h to overexpress the OmpC and LamB proteins. The adsorption rates of phage Bp7 were tested by the double-layer agar method. The E. coli K-12 wild-type strain was used as a control, and each assay was performed in triplicate.
Selection of phage-resistant mutants and genome analysis.
Host strains resistant to phage Bp7 were isolated during the phage cultivation process. Then, 100 μl E. coli K-12 (109 CFU/ml) and 100 μl Bp7 (109 PFU/ml) were mixed in 5 ml LB and cultured at 37°C overnight. After the culture turbidity changed from turbid to clear and became turbid again, 100 μl resistant strain culture was spread onto double-layer phage plates, and the lawn on the agar layer contained 109 PFU/ml phage Bp7. The plates were incubated overnight, and colonies of phage-resistant mutants were purified by repeated single-colony isolation. A resistant strain named E. coli K-12-R was selected and sequenced by Biozeron Biotech Co., Ltd. (Shanghai, China). The genome sequence of the resistant strain was compared with that of the wild-type strain and analyzed to identify mutations that corresponded to phage resistance. The adsorption kinetics of phage Bp7 to E. coli K-12-R were assayed by the double-layer agar method, as described above.
Complementation test with plasmid hldE-T.
The hldE gene of E. coli K-12 was amplified by PCR to construct the recombinant plasmid hldE-T. Plasmid hldE-T was transferred into E. coli K-12-R, and the complement strain E. coli K-12C-hldE was acquired. The complementation effect with plasmid hldE-T was tested by the double-layer agar method. A total of 100 μl E. coli K-12C-hldE carrying hldE-T plasmid culture was mixed with 1 μl IPTG (100 mM), and the mixture was spread onto double-layer phage plates. The lawn that formed on the agar layer contained 109 PFU/ml phage Bp7. The E. coli K-12C-hldE carrying the hldE-T plasmid without IPTG was used as a control. The plates were incubated overnight to test the phage adsorption rates. Each experiment was performed in triplicate. The adsorption kinetics of phage Bp7 were assayed as described above.
Effect of LPS on phage Bp7 adsorption analysis.
LPS was extracted from fresh overnight cultures of E. coli K-12, the K-12-R mutant strain, and the K-12C-hldE complemented strain using an LPS extraction kit (iNTRON Biotechnology, South Korea). Purified LPS was determined using a Tris-Tricine 15% SDS-PAGE gel and subjected to silver staining. The in vitro effects of purified LPS from E. coli K-12, K-12-R, and K-12C-hldE in the phage adsorption process were determined by a plaque-forming assay. Phage Bp7 (104 PFU/ml) was incubated with LPS (2.5 μg/ml, solubilized in 10 mM Tris-HCl [pH 8.0]) at 37°C for 10 min, and phage Bp7 with LB was used as a control. The mixture was incubated with E. coli K-12 (109 CFU/ml) at 37°C for 5 min, and the remaining free virions were tested by the double-layer agar method, as described above. Samples were analyzed in triplicate. The in vitro phage DNA ejection induced by LPS was monitored. Phage Bp7 (109 PFU/ml) was incubated with LPS (2.5 μg/ml) at 37°C for 1 h, and the adsorption rates were tested at 3 min, 5 min, 10 min, 30 min, and 60 min by a plaque assay. Each experiment was performed in triplicate. Additionally, preincubated phage Bp7 was negatively stained with 2% (wt/vol) aqueous uranyl acetate on a carbon-coated grid and observed by TEM (JEM-1200EX). Phage Bp7 was used as a control. To confirm the receptor role of HepI (hldE), the waaF gene, which is related to the synthesis of HepII of inner-core LPS of E. coli K-12, was knocked out as described above, and the effect of phage adsorption on the mutant strain E. coli K-12 ΔwaaF was tested using a plaque assay, as described above. Moreover, based on E. coli K-12-R, the mutant strains E. coli K-12-R ΔOmpC and E. coli K-12-R ΔLamB were constructed as described above, and the adsorption kinetics of phage Bp7 to E. coli K-12-R ΔOmpC and E. coli K-12-R ΔLamB were assayed as described above.
Data availability.
The genome sequences of E. coli K-12 and E. coli K-12-R are available in the GenBank database under accession numbers CP040663 and CP040664, respectively.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant 31600149).
We thank Yigang Tong (Beijing University of Chemical Technology) for providing plasmids and technical support.
REFERENCES
- 1.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 2.Grose JH, Casjens SR. 2014. Understanding the enormous diversity of bacteriophages: the tailed phages that infect the bacterial family Enterobacteriaceae. Virology 468–470:421–443. doi: 10.1016/j.virol.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leiman PG, Arisaka F, van Raaij MJ, Kostyuchenko VA, Aksyuk AA, Kanamaru S, Rossmann MG. 2010. Morphogenesis of the T4 tail and tail fibers. Virol J 7:355. doi: 10.1186/1743-422X-7-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu B, Margolin W, Molineux IJ, Liu J. 2013. The bacteriophage T7 virion undergoes extensive structural remodeling during infection. Science 339:576–579. doi: 10.1126/science.1231887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montag D, Riede I, Eschbach ML, Degen M, Henning U. 1987. Receptor-recognizing proteins of T-even type bacteriophages. Constant and hypervariable regions and an unusual case of evolution. J Mol Biol 196:165–174. doi: 10.1016/0022-2836(87)90519-5. [DOI] [PubMed] [Google Scholar]
- 6.Yap ML, Rossmann MG. 2014. Structure and function of bacteriophage T4. Future Microbiol 9:1319–1327. doi: 10.2217/fmb.14.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riede I, Drexler K, Schwarz H, Henning U. 1987. T-even-type bacteriophages use an adhesin for recognition of cellular receptors. J Mol Biol 194:23–30. doi: 10.1016/0022-2836(87)90712-1. [DOI] [PubMed] [Google Scholar]
- 8.Marti R, Zurfluh K, Hagens S, Pianezzi J, Klumpp J, Loessner MJ. 2013. Long tail fibres of the novel broad-host-range T-even bacteriophage S16 specifically recognize Salmonella OmpC. Mol Microbiol 87:818–834. doi: 10.1111/mmi.12134. [DOI] [PubMed] [Google Scholar]
- 9.Trojet SN, Caumont-Sarcos A, Perrody E, Comeau AM, Krisch HM. 2011. The gp38 adhesins of the T4 superfamily: a complex modular determinant of the phage’s host specificity. Genome Biol Evol 3:674–686. doi: 10.1093/gbe/evr059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knirel YA, Prokhorov NS, Shashkov AS, Ovchinnikova OG, Zdorovenko EL, Liu B, Kostryukova ES, Larin AK, Golomidova AK, Letarov AV. 2015. Variations in O-antigen biosynthesis and O-acetylation associated with altered phage sensitivity in Escherichia coli 4s. J Bacteriol 197:905–912. doi: 10.1128/JB.02398-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rojas-Macias MA, Stahle J, Lutteke T, Widmalm G. 2015. Development of the ECODAB into a relational database for Escherichia coli O-antigens and other bacterial polysaccharides. Glycobiology 25:341–347. doi: 10.1093/glycob/cwu116. [DOI] [PubMed] [Google Scholar]
- 12.Heinrichs DE, Yethon JA, Whitfield C. 1998. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol Microbiol 30:221–232. doi: 10.1046/j.1365-2958.1998.01063.x. [DOI] [PubMed] [Google Scholar]
- 13.Šimoliūnas E, Vilkaitytė M, Kaliniene L, Zajančkauskaitė A, Kaupinis A, Staniulis J, Valius M, Meškys R, Truncaitė L. 2015. Incomplete LPS core-specific Felix01-like virus vB_EcoM_VpaE1. Viruses 7:6163–6181. doi: 10.3390/v7122932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatterjee S, Rothenberg E. 2012. Interaction of bacteriophage l with its E. coli receptor, LamB. Viruses 4:3162–3178. doi: 10.3390/v4113162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C, Li W, Liu W, Zou L, Yan C, Lu K, Ren H. 2013. T4-like phage Bp7, a potential antimicrobial agent for controlling drug-resistant Escherichia coli in chickens. Appl Environ Microbiol 79:5559–5565. doi: 10.1128/AEM.01505-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kutter E, Bryan D, Ray G, Brewster E, Blasdel B, Guttman B. 2018. From host to phage metabolism: hot tales of phage T4’s takeover of E. coli. Viruses 10:387. doi: 10.3390/v10070387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Washizaki A, Yonesaki T, Otsuka Y. 2016. Characterization of the interactions between Escherichia coli receptors, LPS and OmpC, and bacteriophage T4 long tail fibers. Microbiologyopen 5:1003–1015. doi: 10.1002/mbo3.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Islam MZ, Fokine A, Mahalingam M, Zhang Z, Garcia-Doval C, van Raaij MJ, Rossmann MG, Rao VB. 2019. Molecular anatomy of the receptor binding module of a bacteriophage long tail fiber. PLoS Pathog 15:e1008193. doi: 10.1371/journal.ppat.1008193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertozzi Silva J, Storms Z, Sauvageau D. 2016. Host receptors for bacteriophage adsorption. FEMS Microbiol Lett 363:fnw002. doi: 10.1093/femsle/fnw002. [DOI] [PubMed] [Google Scholar]
- 20.Yoichi M, Abe M, Miyanaga K, Unno H, Tanji Y. 2005. Alteration of tail fiber protein gp38 enables T2 phage to infect Escherichia coli O157:H7. J Biotechnol 115:101–107. doi: 10.1016/j.jbiotec.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Valvano MA, Marolda CL, Glaskin-Clay M, Bittner M, Simon TL, Klena JD. 2000. The rfaE gene from Escherichia coli encodes a bifunctional protein involved in biosynthesis of the lipopolysaccharide core precursor ADP-l-glycero-d-manno-heptose. J Bacteriol 182:488–497. doi: 10.1128/JB.182.2.488-497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu F, Mizushima S. 1982. Roles of lipopolysaccharide and outer membrane protein OmpC of Escherichia coli K-12 in the receptor function for bacteriophage T4. J Bacteriol 151:718–722. doi: 10.1128/JB.151.2.718-722.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu B, Margolin W, Molineux IJ, Liu J. 2015. Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proc Natl Acad Sci U S A 112:E4919–E4928. doi: 10.1073/pnas.1501064112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossmann MG, Mesyanzhinov VV, Arisaka F, Leiman PG. 2004. The bacteriophage T4 DNA injection machine. Curr Opin Struct Biol 14:171–180. doi: 10.1016/j.sbi.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Botos I, Noinaj N, Buchanan SK. 2017. Insertion of proteins and lipopolysaccharide into the bacterial outer membrane. Philos Trans R Soc Lond B Biol Sci 372:20160224. doi: 10.1098/rstb.2016.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sperandeo P, Martorana AM, Polissi A. 2017. Lipopolysaccharide biogenesis and transport at the outer membrane of Gram-negative bacteria. Biochim Biophys Acta Mol Cell Biol Lipids 1862:1451–1460. doi: 10.1016/j.bbalip.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Simpson BW, Trent MS. 2019. Pushing the envelope: LPS modifications and their consequences. Nat Rev Microbiol 17:403–416. doi: 10.1038/s41579-019-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao X, Cui Y, Yan Y, Du Z, Tan Y, Yang H, Bi Y, Zhang P, Zhou L, Zhou D, Han Y, Song Y, Wang X, Yang R. 2013. Outer membrane proteins Ail and OmpF of Yersinia pestis are involved in the adsorption of T7-related bacteriophage Yep-phi. J Virol 87:12260–12269. doi: 10.1128/JVI.01948-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reisch CR, Prather KL. 2015. The no-SCAR (Scarless Cas9 Assisted Recombineering) system for genome editing in Escherichia coli. Sci Rep 5:15096. doi: 10.1038/srep15096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequences of E. coli K-12 and E. coli K-12-R are available in the GenBank database under accession numbers CP040663 and CP040664, respectively.