Mumps virus is an important human pathogen. In recent years, MuV has reemerged in the United State, with outbreaks occurring in young adults who have been vaccinated. Our work provides insight into a previously unknown mumps virus-host interaction. RPS6KB1 negatively regulates MuV replication, likely through its interaction with the P protein. Understanding virus-host interactions can lead to novel antiviral drugs and enhanced vaccine production.
KEYWORDS: RPS6KB1, kinase, mumps virus, replication
ABSTRACT
Mumps virus (MuV) caused the most viral meningitis before mass immunization. Unfortunately, MuV has reemerged in the United States in the past several years. MuV is a member of the genus Rubulavirus, in the family Paramyxoviridae, and has a nonsegmented negative-strand RNA genome. The viral RNA-dependent RNA polymerase (vRdRp) of MuV consists of the large protein (L) and the phosphoprotein (P), while the nucleocapsid protein (NP) encapsulates the viral RNA genome. These proteins make up the replication and transcription machinery of MuV. The P protein is phosphorylated by host kinases, and its phosphorylation is important for its function. In this study, we performed a large-scale small interfering RNA (siRNA) screen targeting host kinases that regulated MuV replication. The human kinase ribosomal protein S6 kinase beta-1 (RPS6KB1) was shown to play a role in MuV replication and transcription. We have validated the role of RPS6KB1 in regulating MuV using siRNA knockdown, an inhibitor, and RPS6KB1 knockout cells. We found that MuV grows better in cells lacking RPS6KB1, indicating that it downregulates viral growth. Furthermore, we detected an interaction between the MuV P protein and RPS6KB1, suggesting that RPS6KB1 directly regulates MuV replication and transcription.
IMPORTANCE Mumps virus is an important human pathogen. In recent years, MuV has reemerged in the United State, with outbreaks occurring in young adults who have been vaccinated. Our work provides insight into a previously unknown mumps virus-host interaction. RPS6KB1 negatively regulates MuV replication, likely through its interaction with the P protein. Understanding virus-host interactions can lead to novel antiviral drugs and enhanced vaccine production.
INTRODUCTION
Mumps virus (MuV) is a human pathogen that causes an acute infection resulting in symptoms like parotitis, fever, and nausea. Additionally, it is highly neurotropic and has been shown to invade the central nervous system (CNS). Prior to mass vaccination in the late 1960s, MuV was the leading cause of aseptic meningitis, encephalitis, and deafness in children (1). Mass vaccination dramatically reduced MuV infections across the United States. However, there has been a recent resurgence in MuV infections that has occurred mostly in vaccinated populations. In 2006, there was an outbreak on college campuses in Iowa that resulted in over 6,500 diagnosed cases of MuV. Since 2006, there have been several years with over 1,000 diagnosed cases in the United States. In 2016 and 2017, there were over 6,000 cases, rivaling the Iowa outbreak (2, 3). Currently, there are no antiviral drugs approved for MuV infections. Understanding the interactions of host and viral proteins, using the currently circulating 2006 MuV Iowa outbreak strain, will aid in developing new treatments.
MuV is a member of the genus Rubulavirus, in the family Paramyxoviridae, and has a nonsegmented, negative-strand RNA genome of 15,384 nucleotides (1, 4). The genome contains seven genes that are transcribed into nine viral proteins in the order 3′-NP-V/I/P-M-F-SH-HN-L-5′, with RNA synthesis beginning at the 3′ end. The RNA genome serves as a template for replication of the viral RNA (vRNA) genome and mRNA synthesis, and it associates with nucleocapsid proteins (NPs) to form the helical ribonucleoprotein (RNP), which protects the genome from degradation. The RNP associates with the phosphoprotein (P) and the large protein (L) to form the viral RNA-dependent RNA polymerase (vRdRp) complex. The L protein is responsible for initiation, elongation, and termination of RNA synthesis, in addition to adding the 5′ cap and 3′ poly(A) tail to transcribed viral mRNA. The P protein interacts with NP in the RNP, as well as free NP, and is essential for the formation of the vRdRp (1, 4, 5).
The P proteins of paramyxoviruses are highly phosphorylated, and their phosphorylation status impacts vRNA and viral mRNA synthesis. Previous studies, with parainfluenza virus 5 (PIV5), respiratory syncytial virus (RSV), and MuV have shown that mutating residues within P affects RNA synthesis (6–9). It is thought that phosphorylation of the P protein is achieved by host kinases, because paramyxovirus viral proteins lack inherent kinase enzymatic activity (4, 9). Furthermore, several paramyxovirus P proteins have been shown to interact with host kinases, including MuV P, which was shown to interact with the human kinase Polo-like kinase 1 (PLK1) (10–13). Additional host kinases that are important for MuV P phosphorylation and RNA synthesis have yet to be discovered.
In this work, we used a small interfering RNA (siRNA) screen to identify host kinases that are involved in MuV replication and examined the effect of one specific kinase, ribosomal protein S6 kinase beta-1 (RPS6KB1), on MuV replication.
RESULTS
Identification of host kinases that are important for MuV replication.
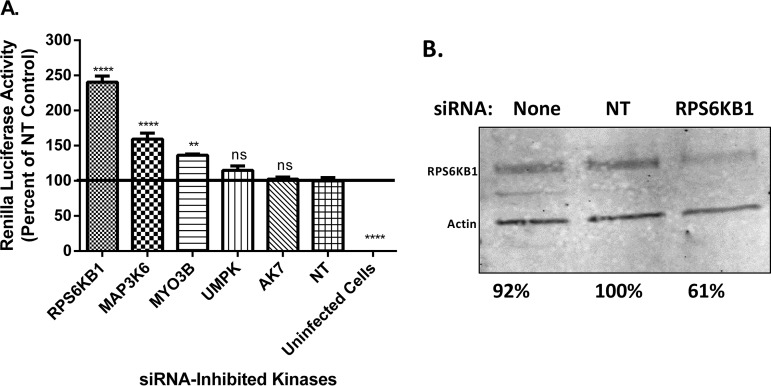
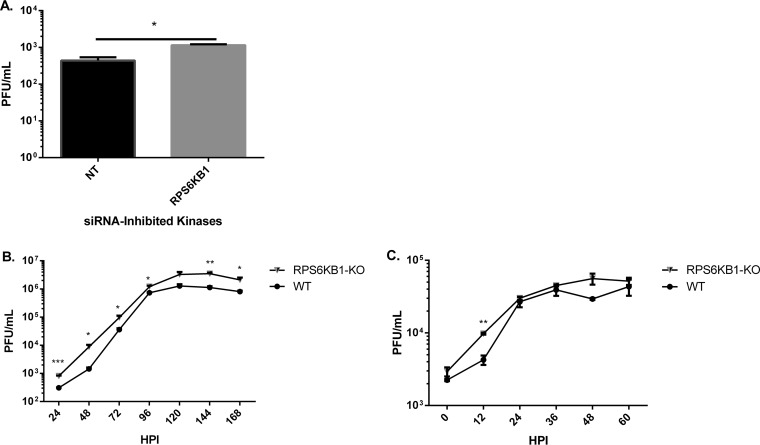
To identify host kinases that are important for MuV replication, an siRNA screen was performed using a library of 720 different siRNAs targeting human protein kinases. Vero-E6 cells were reverse transfected with 4 different siRNAs per kinase in 96-well plates that also contained a positive, a negative, and a TOX control. Using a Z-score analysis, we found several candidates that upregulated or downregulated viral replication. In addition to MuV, a closely related virus in the same genus, PIV5, was also screened. Interestingly, there were many differences in the kinases that play a role in their replication. Of the top 50 repressors, only 5 were similar between PIV5 and MuV, all of which were chosen for further study. Of the top 50 enhancers, 18 were similar between PIV5 and MuV, but only 4 were chosen for further study, and they are not discussed in this work. The candidates were chosen for further study because of their observed roles in MuV, PIV5, or both. Notably, many of these candidates have been previously implicated in interacting with other viruses (10). To validate the siRNA screen, new siRNAs targeting different sequences of the kinase mRNA transcripts from the screen were used. UMPK and AK7, upon further validation, had no significant effect on MuV replication. A few candidates were shown to significantly increase MuV luciferase activity when knocked down, including MYO3B, MAP3K6, and RPS6KB1. siRNA targeting RPS6KB1 (siRPS6KB1) consistently increased MuV activity the most compared to the other siRNA-inhibited kinases (Fig. 1A). A Western blot was performed to examine the amount of RPS6KB1 siRNA knockdown compared to the nontarget control (NT). It was found that the siRNA knockdown was approximately 40% percent that of the control (Fig. 1B). The cellular toxicities of all the siRNA-inhibited kinases were assayed by measuring the amounts of ATP produced. RPS6KB1 reduced cellular viability less than 10% compared to the nontarget control and was not found to be significant (data not shown). siRNA-inhibited RPS6KB1 was not cytotoxic to HeLa cells and increased MuV luciferase activity compared to the nontarget control. Upon further investigation, RPS6KB1 had no significant effect on PIV5 growth, despite the screening results, and no further testing occurred (data not shown).
FIG 1.
Examination of kinases in MuV replication. (A) Effects of siRNAs targeting kinases on MuV replication. HeLa cells were transfected with 2 different siRNAs targeting different sequences of each kinase mRNA transcript. The next day, cells were infected with rMuV-rluc at an MOI of 0.1; 24 hpi, the cells were lysed, and luciferase was measured. Values were normalized to those of the NT. The error bars represent standard errors of the mean (SEM) with 3 replicates each; individual experiments were performed independently 3 times. P values were calculated by one-way ANOVA: **, P < 0.01; ****, P < 0 0.0001; ns, not significant. (B) Detection of RPS6KB1 using Western blot analysis. Lysates from the luciferase assay were collected, and replicates were combined and mixed with loading buffer before being resolved by SDS-PAGE and immunoblotted using an anti-RPS6KB1 and an anti-actin antibody. The values were normalized to those of the nontarget control.
MuV grew to a higher titer in cells lacking RPS6KB1.
To examine the growth rates of MuV in cells lacking RPS6KB1, siRNAs against RPS6KB1 were used. MuV grew to a higher titer in RPS6KB1 siRNA-treated cells than the nontarget control at 24 h postinfection (hpi) (Fig. 2A). To examine MuV growth over a longer period, HapI cells lacking RPS6KB1 (RPS6KB1-KO cells) were infected with MuV at a low (0.1) and a high (3) MOI. MuV consistently grew to a higher titer in the RPS6KB1-KO cells than in wild-type (WT) HapI cells at both multiplicities of infection (MOI), suggesting RPS6KB1 has a negative effect on MuV growth (Fig. 2B and C).
FIG 2.
Growth rates of MuV in cells with an RPS6KB1 deficiency. (A) Growth of MuV in siRNA-knocked-down RPS6KB1 HeLa cells. The cells were transfected with RPS6KB1-specific siRNAs, as previously described, and then infected with MuV at an MOI of 0.1. Medium was collected 24 hpi, and plaque assays were performed to determine viral titers. (B) Multistep growth curve of MuV in WT and RPS6KB1-KO HapI cells. The cells were infected at an MOI of 0.1, and medium was collected every 24 h for plaque assay. (C) Single-step growth curve of MuV in WT and RPS6KB1-KO HapI cells. The cells were infected at an MOI of 3, and medium was collected every 12 h for plaque assay. P values were calculated using multiple T tests. *, P < 0.05; **, P < 0.01; ***, P <0.001. The error bars represent SEM of data from 3 replicates. Individual experiments were performed 3 times.
Viral protein expression is increased in cells lacking RPS6KB1.
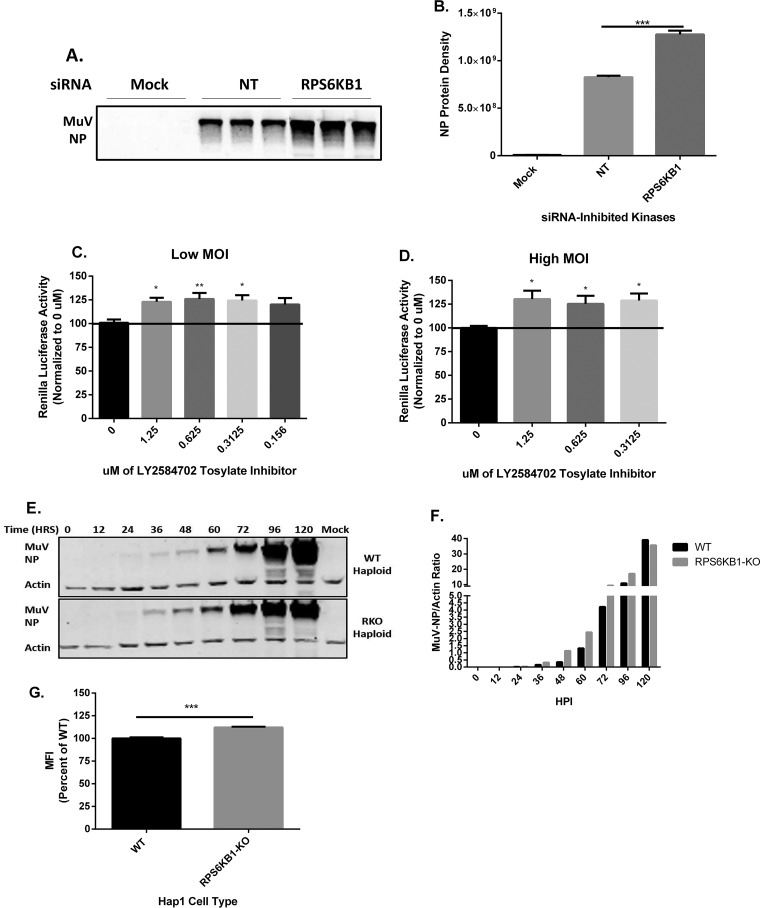
RPS6KB1-specific siRNAs were used to examine MuV protein expression in cells. Lysates were collected and immunoblotted with monoclonal anti-MuV NP (Fig. 3A). At 24 hpi, the NP protein density was greater in the siRPS6KB1-transfected cells than in the nontarget control (Fig. 3B). Cells were subjected to various concentrations of an RPS6KB1-specific inhibitor and then infected with recombinant MuV expressing Renilla luciferase (rMuV-rluc) to measure the luciferase production 24 hpi. Luciferase activity was increased using as little as 0.3 μM the inhibitor at both a low (0.1) and a high (3) MOI (Fig. 3C and D). To examine viral protein expression in cells lacking RPS6KB1, WT and RPS6KB1-KO HapI cells were infected at a low MOI. Lysates from infected cells were taken at various time points and immunoblotted for anti-MuV NP and for anti-actin as a loading control (Fig. 3E). The relative NP densities were calculated, and MuV protein expression was slightly higher in the RPS6KB1-KO HapI cells starting at 24 hpi and gradually increased over time (Fig. 3F). To confirm that the HapI cells lacking RPS6KB1 caused an increase in MuV protein expression at an early time point, flow cytometry was performed. WT and RPS6KB1-KO HapI cells were infected with MuV and then stained for anti-MuV NP. MuV grown in RPS6KB1-KO HapI cells had a greater mean fluorescence intensity (MFI) than MuV grown in WT HapI cells (Fig. 3G). MuV protein expression was increased in cells with inhibited or knocked-out RPS6KB1.
FIG 3.
Effects of inhibiting RPS6KB1 on MuV protein expression. (A) Detection of MuV NP protein expression in siRNA-transfected HeLa cells at 24 hpi. HeLa cells were transfected with siRNA and then infected at an MOI of 0.1 with MuV 24 h posttransfection in triplicate. Lysates were resolved by SDS-PAGE and immunoblotted using a monoclonal anti-MuV NP antibody. A representative blot is shown. (B) Summary of quantified NP protein densities from Western blotting (WB). (C and D) Luciferase activity of MuV using LY2584702 tosylate inhibitor, an RPS6KB1-specific inhibitor, in HeLa cells at MOI of 0.1 (low) and 3 (high). The cells were infected, and 1 hpi, the medium was replaced with different concentrations of the inhibitor. At 24 hpi, cell lysate luciferase activity was quantified and normalized to the luciferase activity of cells treated with 0 μM inhibitor. The error bars represent SEM of data from 4 replicates. (E) Detection of MuV NP expression in WT and RPS6KB1-KO HapI cells. The cells were infected with MuV at an MOI of 0.1. Lysates were collected at each time point and resolved by SDS-PAGE, and both MuV NP and actin were immunoblotted for. Actin served as a loading control. A representative blot is shown. (F) Summary of quantified NP protein densities from WB. The relative NP protein density was calculated by taking the ratio of NP to actin and then graphed to compare protein expression between WT and RPS6KB1-KO HapI cells. The values represent the results of one of three independent experiments. (G) Flow cytometry analysis of WT and RPS6KB1-KO HapI cells infected with MuV at an MOI of 0.1. At 24 hpi, the cells were fixed and stained with an anti-MuV NP antibody to measure the MFI of infected cells. Values were standardized to the infected WT HapI cell values. The error bars represent SEM of data from 4 replicates. P values were calculated by one-way ANOVA or Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Individual experiments were performed 3 times.
MuV minigenome activity is increased in the absence of RPS6KB1.
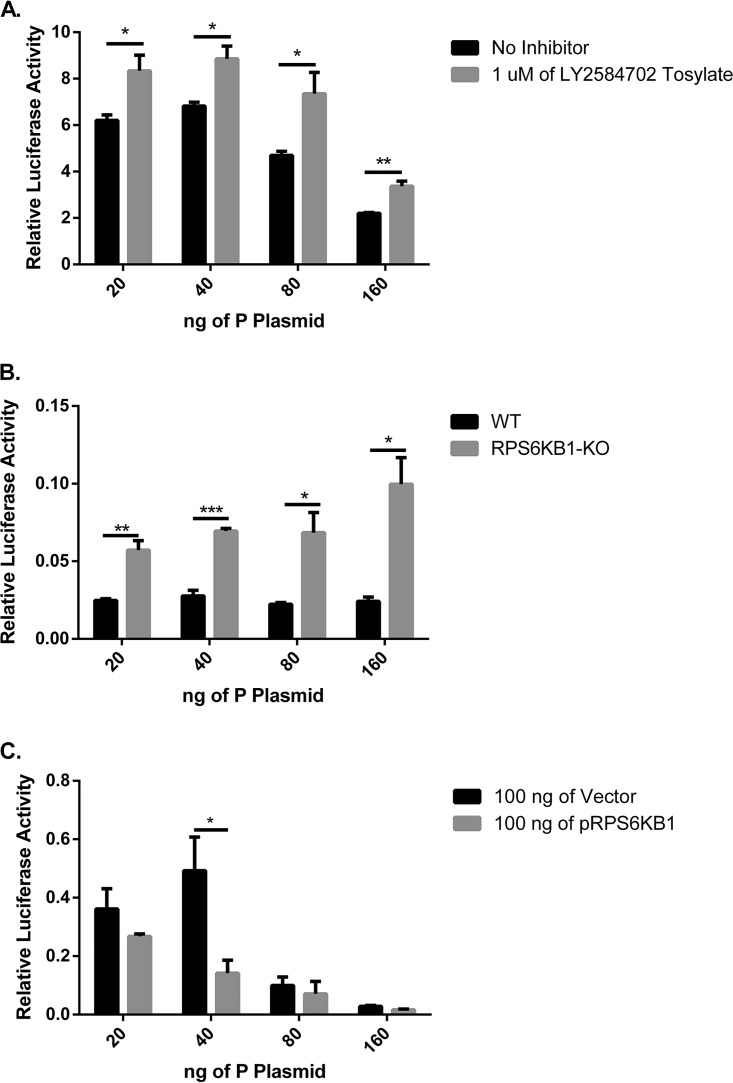
To determine if RPS6KB1 plays a role in viral RNA synthesis, a minigenome system was used. BSR-T7 cells were transfected with the minigenome plasmids with varying concentrations of MuV P plasmid. Multiple concentrations of MuV P plasmid were transfected to find the optimal minigenome activity in the presence of the inhibitor and to examine if different amounts of MuV P plasmid would affect the phenotype. The cells were then either mock treated or treated with the RPS6KB1 inhibitor. MuV minigenome activity was significantly increased at all concentrations of transfected MuV P plasmid in the presence of the RPS6KB1 inhibitor (Fig. 4A). A minigenome assay was also performed in HapI cells. The transfection efficiency of the HapI cells was much lower than that of BSR-T7 cells, causing the overall minigenome activity to be lower. Regardless, the same trend was observed, and MuV minigenome activity was significantly increased in RPS6KB1-KO cells compared to WT HapI cells (Fig. 4B). To determine if overexpression of RPS6KB1 would diminish minigenome activity, varying amounts of MuV P plasmid and a plasmid containing RPS6KB1 (pRPS6KB1) were transfected into BSR-T7 cells. MuV minigenome activity was reduced in the cells overexpressing RPS6KB1, suggesting RPS6KB1 plays a negative role in MuV RNA synthesis (Fig. 4C).
FIG 4.
MuV minigenome activity in the absence of RPS6KB1. (A) MuV minigenome activity in the presence of an RPS6KB1 inhibitor. BSR-T7 cells were either mock treated or treated with 1 μM a RPS6KB1-specific inhibitor, LY2584702 tosylate. (B) MuV minigenome activity in RPS6KB1-KO HapI cells compared to WT HapI cells. (C) MuV minigenome activity with overexpressed RPS6KB1. One hundred nanograms of a vector control or plasmid containing RPS6KB1 was transfected into BSR-T7 cells. The ratio of Renilla luciferase activity to firefly luciferase activity was calculated and graphed. Multiple concentrations of the MuV P plasmid were transfected (20, 40, 80, and 160 ng/well) to obtain optimal minigenome activity. P values were calculated using multiple T tests. *, P < 0.05; **, P < 0.01; ***, P < 0.001. The error bars represent SEM of data from 3 replicates. Individual experiments were performed 3 times.
Viral replication and transcription are increased in cells lacking RPS6KB1.
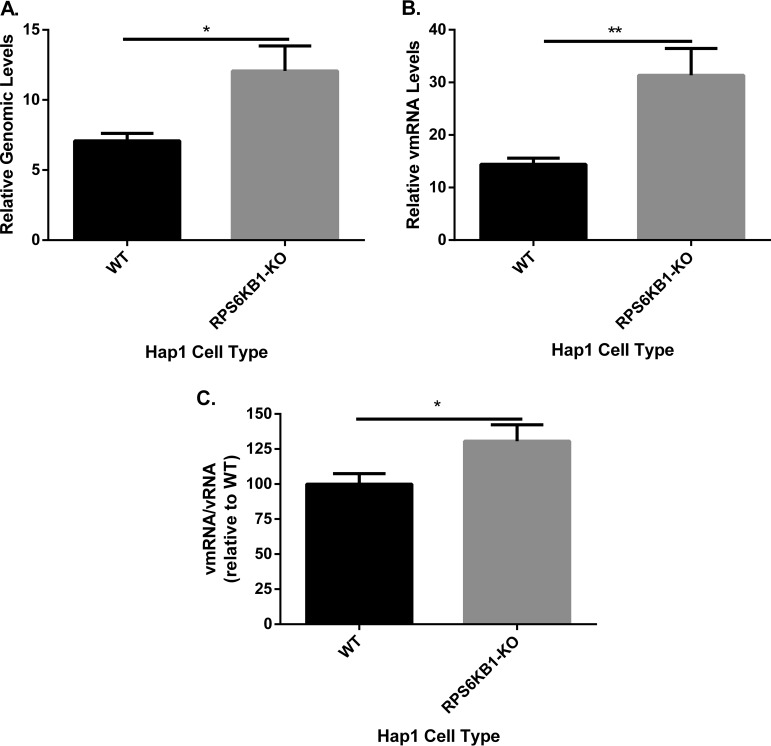
To investigate the role RPS6KB1 has in viral RNA synthesis, real-time PCR was used to compare levels of viral mRNA and genomic RNA (vRNA) in HapI cells. The cells were infected with MuV at high MOI to measure a single round of MuV RNA synthesis. Total RNA was extracted from infected WT and RPS6KB1-KO HapI cells. MuV genomic vRNA was increased in RPS6KB1-KO cells compared with WT HapI cells (Fig. 5A). In addition, MuV mRNA was increased, which is consistent with increased protein expression (Fig. 5B) (3). There was an increased ratio of viral mRNA to genomic vRNA in the RPS6KB1-KO cells compared to WT HapI cells (Fig. 5C). This suggests that RPS6KB1 plays a role in transcription by negatively regulating the viral RNA synthesis machinery.
FIG 5.
Viral RNA synthesis in cells with RPS6KB1 deficiency. Cells were infected at an MOI of 5, and total RNA was collected from 4 independent samples at 2 and 24 hpi. (A) Genome replication of MuV (vRNA) in WT and RPS6KB1-KO cells. Genome-specific primers were used to detect genomic RNA. (B) MuV mRNA. Oligo(dT) primers were used to generate the mRNAs. (C) Viral mRNA relative to genomic RNA. The ratio of vmRNA to genomic vRNA was calculated and then normalized to the WT HapI control. A MuV F-specific probe was used for real-time PCR after cDNA synthesis. The values were calculated after normalization to genomic-RNA levels at 2 hpi. P values were calculated using multiple T tests. *, P < 0.5; **, P < 0.01. The error bars represent SEM of data from 4 replicates of 3 individual experiments.
MuV has reduced P phosphorylation in cells lacking RPS6KB1.
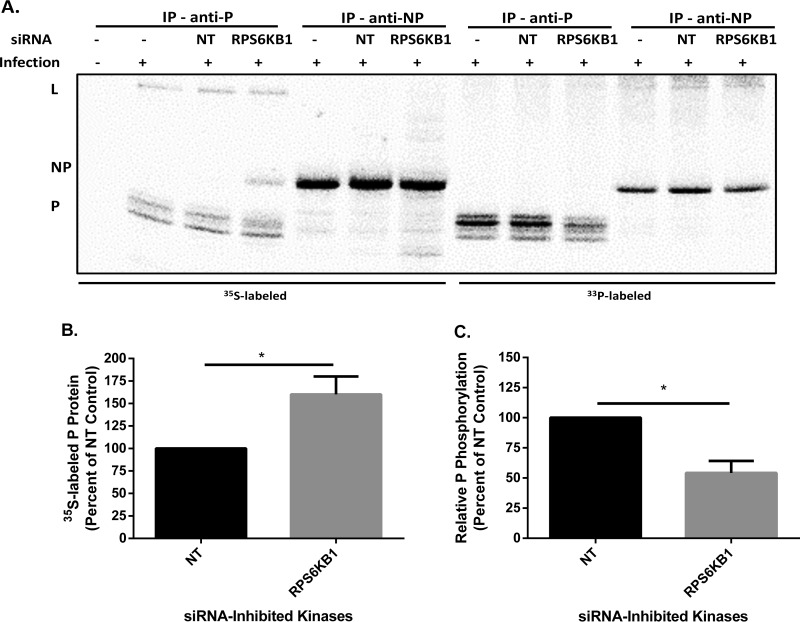
To determine if RPS6KB1 plays a role in MuV P phosphorylation, cells were transfected with siRNA and then infected with MuV. Cells were labeled with 35S or 33P and then immunoprecipitated with monoclonal anti-MuV P or monoclonal anti-MuV NP. 35S-labeled P protein was increased in the RPS6KB1 siRNA-transfected cells compared to the nontarget control. Interestingly, MuV NP was coimmunoprecipitated with MuV P in the RPS6KB1 siRNA-transfected cells, which was not observed in the nontarget control. Additionally, more MuV P was coimmunoprecipitated in the RPS6KB1 siRNA-treated cells with the anti-MuV NP antibody (Fig. 6A). These results are consistent with the previous findings that MuV protein expression is increased in cells lacking RPS6KB1 (Fig. 6B). MuV P phosphorylation was significantly decreased in the RPS6KB1 siRNA-transfected cells, which suggests RPS6KB1 phosphorylates MuV P (Fig. 6C). The slight reduction in phosphorylation is feasible because MuV P has several phosphorylation sites that all undergo phosphorylation changes during an infection, and proteins were radiolabeled for only 6 to 8 h (7). The results indicate that RPS6KB1 phosphorylates MuV P, which interferes with transcription and replication, causing a negative effect on MuV growth.
FIG 6.
MuV P and NP phosphorylation in cells with RPS6KB1 deficiency. (A) Representative radio blot. HeLa cells were transfected with siRNA, infected with MuV at an MOI of 0.1, and then labeled with 35S or 33P for 6 to 8 h. Cell lysates were immunoprecipitated with monoclonal anti-MuV P or anti-MuV NP antibody and resolved on an SDS-PAGE gel. (B) Total P protein quantification. Shown is 35S-labeled P protein from infected HeLa cells. Values were standardized to that of the nontarget control. (C) Relative phosphorylation levels of P in infected cells. The relative level was calculated as a ratio of phosphorylated protein (33P) to total protein (35S) and standardized to the nontarget control. P values were calculated using Student's t test. *, P < 0.5. The error bars represent SEM of data from 3 individual experiments.
RPS6KB1 interacts with MuV P.
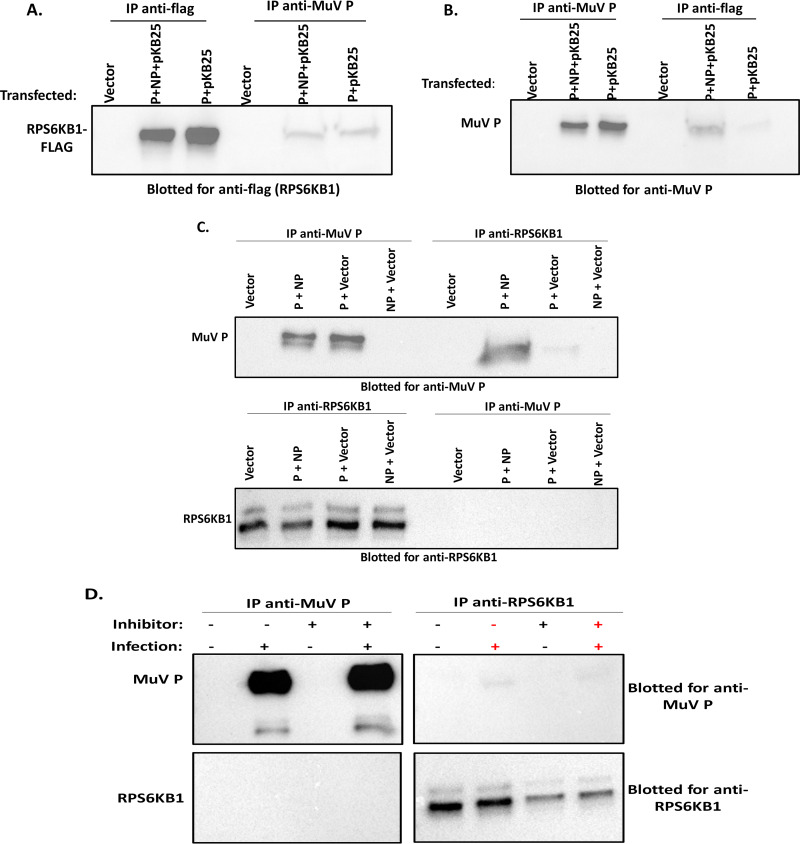
To examine if RPS6KB1 has a direct effect on MuV P, cells were transfected with different combinations of MuV P, MuV NP, pCAGGS-GFP, and a plasmid encoding RPS6KB1 with a Flag tag (pKB25). Lysates were immunoprecipitated with either a monoclonal anti-MuV P or monoclonal anti-Flag antibody and then immunoblotted. The cells that were transfected with MuV P plus MuV NP plus pKB25 and immunoprecipitated/immunoblotted with their reciprocal antibody each showed a band of the corresponding size (Fig. 7A and B). For example, when an anti-MuV P antibody was used for immunoprecipitation (IP), the RPS6KB1-Flag protein was detected with an anti-Flag antibody in the immunoblot (Fig. 7A). The cells transfected with MuV P plus pKB25 alone also showed the correct corresponding bands, suggesting that MuV P and RPS6KB1 interact independently of MuV NP (Fig. 7A and B). Interestingly, compared to lysate from cells transfected with P plus pKB25 plus NP, lysate from cells transfected with P plus pKB25 had less MuV P when immunoprecipitated with anti-Flag (Fig. 7B). However, this was not seen when anti-MuV P antibody was used for the immunoprecipitation and anti-Flag was immunoblotted for (Fig. 7A).
FIG 7.
MuV P and RPS6KB1 interact. (A and B) 293T cells were transfected with plasmids containing MuV P, MuV NP, and RPS6KB1-Flag (pKB25). The cells were lysed and then immunoprecipitated with either monoclonal anti-MuV P or monoclonal anti-Flag. (A) Lysates blotted for anti-Flag. (B) Lysates blotted for anti-MuV P. (C) 293T cells transfected with plasmids containing MuV P and/or MuV NP. The cells were lysed and then immunoprecipitated and immunoblotted with either monoclonal anti-MuV P or monoclonal anti-RPS6KB1. (D) Vero cells were infected with MuV at an MOI of 0.1. After 1 h, the inoculum was replaced with medium containing 1 μM LY2584702 tosylate inhibitor or fresh medium; 24 hpi, the cells were lysed, immunoprecipitated, and immunoblotted with either monoclonal anti-MuV P or monoclonal anti-RPS6KB1. Each experiment was performed independently 3 times, and a representative blot is shown for each.
To confirm that the interaction was not due to the presence of the recombinant protein with a Flag tag, another immunoprecipitation was performed using cellular RPS6KB1. Cells were transfected with MuV P and MuV NP and immunoprecipitated/immunoblotted for MuV P or RPS6KB1. When anti-RPS6KB1 was immunoprecipitated, MuV P was coimmunoprecipitated in samples containing MuV P plus MuV NP and MuV P plus vector, suggesting the proteins were interacting, while MuV NP plus vector did not show the interaction. The immunoprecipitation of RPS6KB1 was able to pull down MuV P; however, the reciprocal immunoprecipitation (IP, MuV P; immunoblotting [IB], RPS6KB1) did not pull down RPS6KB1 (Fig. 7C). Lastly, we confirmed that the interaction occurs during a MuV infection. Vero cells were infected with MuV, and after 1 h, the medium was replaced with medium containing 1 μM LY2584702 (tosylate) inhibitor or fresh medium. The cells were lysed and immunoprecipitated/immunoblotted for either MuV P or RPS6KB1. MuV P was coimmunoprecipitated when RPS6KB1 was immunoprecipitated, while the reciprocal did not appear on the blot (Fig. 7D). The inhibitor did enhance the amount of MuV P and decrease the amount of RPS6KB1 on the immunoblots but did not enhance the interaction as predicted. These results show that RPS6KB1 and MuV P interact during transfection and infection, suggesting that RPS6KB1 negatively regulates MuV P through a direct interaction.
DISCUSSION
In this study, we performed a large-scale siRNA screen to identify host kinases that affect MuV growth. We found several candidates that enhanced the growth of MuV using a recombinant MuV expressing Renilla luciferase. After assessing the top 50 candidates from the screen, some candidates were chosen for further analysis. Validation of the screen with some of the candidates showed that, while the results were not consistent between different cell types or pools of siRNA, RPS6KB1 was consistent throughout these validation steps. Interestingly, in the initial siRNA screen, siRPS6KB1 had a much larger effect on PIV5 than it did on MuV. Upon further investigation with new siRNA against RPS6KB1 and HapI cells lacking RPS6KB1, the impact of RPS6KB1 on PIV5 was not consistent. This indicates the importance of validating initial screening results. Although PIV5 and MuV are closely related viruses, the host kinase mechanisms they use to replicate within a cell differ somewhat. This was different from two other kinases, PLK1 and AKT, which were found to regulate both PIV5 and MuV through similar mechanisms (10–12).
RPS6KB1 (also referred to as S6K1 and p70S6K1) is a member of the AGC subfamily of serine/threonine protein kinases and is encoded by the human gene Rps6kb1. The gene mainly encodes two isoforms of the protein, p70 and p85. p70, which localizes to the cytoplasm, is considered the major isoform and is essential for cell cycle progression. RPS6KB1 promotes protein synthesis, cell survival, and growth and regulates the DNA damage response (14–17). Recently, it has been observed that hyperphosphorylation and/or overexpression of RPS6KB1 is common in many types of cancer, and therefore, it has become a target for anticancer treatments (18, 19). RPS6KB1 was shown to be essential for optimal viral infection in CD4+ T cells by CCR5-tropic human immunodeficiency virus type 1 (HIV-1), but direct phosphorylation of an HIV-1 protein was not tested (20). Interestingly, Kaposi’s sarcoma-associated herpesvirus (KSHV) encodes a viral protein kinase that is homologous to cellular RPS6KB1, and it was shown that deleting the kinase inhibited virus lytic replication (21). Lastly, one study found that using rapamycin, an inhibitor of the mammalian target of rapamycin (mTOR) kinase, which has been shown to phosphorylate RPS6KB1, reduced Rift Valley fever virus (RVFV) titers in vitro and increased survivability of mice in vivo (22). To our knowledge, this is the first time that RPS6KB1 has been shown to interact with MuV P protein and to act as a negative regulator of viral replication and transcription.
We examined the effect of RPS6KB1 on MuV growth rates and protein expression using siRNA and an inhibitor. We found that using either mechanism of inhibition was sufficient to increase protein expression and growth rates in a variety of cell lines. Confirmation of this phenotype was performed using a RPS6KB1 knockout HapI cell line. While siRNA and inhibitors lose their efficiency over a few days, the cell line has the advantage of allowing us to examine the growth effect for up to 7 days postinfection. Furthermore, the modest effect of inhibiting or knocking out RPS6KB1 was likely due to phosphorylation of P by additional kinases.
The role of RPS6KB1 in replication and transcription was confirmed using a minigenome system, in which viral RNA synthesis could be dissected without involving viral entry and egress. Minigenome activity was increased in the presence of the RPS6KB1-specific inhibitor and in cells lacking RPS6KB1, and viral synthesis of both vRNA and mRNA was increased in cells lacking RPS6KB1. Our findings suggest RPS6KB1 negatively regulates the production of new genomic RNA and viral mRNAs, causing a decrease in titer and protein expression.
Next, we examined the role of phosphorylation in MuV and found that RPS6KB1 phosphorylates MuV P. Cells transfected with RPS6KB1-specific siRNA showed a significant decrease in MuV P phosphorylation, while MuV NP phosphorylation seemed unaffected. Similar results were observed when an RPS6KB1-specific inhibitor was used (data not shown). Radiolabeling experiments were also performed in HapI cells, but not enough viral protein was produced in the labeling period to produce quantifiable data (data not shown). However, we expect a greater effect would have been seen in the RPS6KB1 knockout cells. Interestingly, cells with reduced RPS6KB1 seemed to have increased MuV P and MuV NP interactions, suggesting that RPS6KB1 may interfere with MuV viral RNA polymerase complex formation or binding of MuV P to MuV NP, although this needs to be further explored.
Lastly, we examined the interaction between RPS6KB1 and MuV and found that it binds to the MuV P protein. The P-RPS6KB1 interaction was examined in three different systems to ensure the validity of the interaction. First, a recombinant RPS6KB1 protein containing a Flag tag was transfected into cells, along with MuV P and NP, and the interaction was observed using either anti-MuV P or anti-Flag. To confirm that MuV P was binding to RPS6KB1 and not the Flag tag, we performed another immunoprecipitation using cellular RPS6KB1. Since most cell culture lines are cancer cell lines, RPS6KB1 is naturally upregulated and highly expressed in many lines, making it easy to detect by Western blotting (14). One pitfall of using cellular RPS6KB1 is that the anti-MuV P antibody does not coimmunoprecipitate the cellular RPS6KB1form, but the anti-RPS6KB1 antibody does coimmunoprecipitate MuV P. Although MuV NP is not required for the P-RPS6KB1 interaction, it appears to enhance the interaction. This also occurs with another kinase that negatively regulates MuV growth, PLK1 (11). Lastly, we confirmed that the interaction occurs during a viral infection in cell culture. In our previous work, it was noted that the addition of a kinase inhibitor may help prolong the interaction with its counterpart, so the RPS6KB1-specific inhibitor was added postinfection to determine whether it enhanced the interaction (11). The presence of the inhibitor, however, did not seem to have any effect on the RPS6KB1-P interaction. Taking the data together, we speculate that RPS6KB1 negatively regulates MuV growth by binding to MuV P and effecting subsequent phosphorylation.
We speculate that RPS6KB1 phosphorylates MuV by binding to the P protein. However, the site(s) of the interaction has not yet been identified. Since RPS6KB1 is an AGC kinase, it has a conserved binding motif, R/L-X-R-X-X-S/T, which is found in MuV P in two locations in the MuV P N-terminal domain (NTD) and partially found (R-X-X-S/T) once in the C-terminal domain (CTD) (23). Future studies will focus on elucidating interactions between RPS6KB1 and P, as well as the role of NP in this interaction.
Understanding the mechanism by which a host kinase can regulate viral replication is a useful tool and has translational applications. Kinases that positively regulate viruses can be used as potential antiviral drugs, many of which are already FDA approved and used as cancer treatments, while kinases that negatively regulate viral replication can be used to enhance viral vaccine yields in cell culture. To our knowledge, this is the first time RPS6KB1 has been shown to negatively regulate a virus. We speculate that MuV uses RPS6KB1 to downregulate its viral transcription and replication to prevent overproduction of viral proteins and progeny virions within a host, which would activate the innate immune system and eventually clear the infection. MuV uses the MuV V protein in tandem with host kinase phosphorylation to balance its replication within a host. The fact that RPS6KB1 could be knocked out in a HapI cell line suggests that other cell lines could also have RPS6KB1 knocked out. As MuV growth is highly regulated by type I interferons (IFNs), developing an RPS6KB1 knockout in an IFN-deficient cell line, such as Vero cells, would be beneficial (24). A cell line lacking RPS6KB1 may enhance the growth of MuV for vaccine production.
MATERIALS AND METHODS
Cells.
293T cells were maintained in Dulbecco’s modified Eagle medium (DMEM) with 5% fetal bovine serum (FBS) and 1% penicillin-streptomycin (P/S) (Mediatech Inc., Manassas, VA). Vero and HeLa cells were maintained in DMEM supplemented with 5% FBS and 1% P/S. BSR-T7 cells were maintained in DMEM supplemented with 5% FBS, 1% P/S, 10% tryptose phosphate broth (TPB), and 400 μg/ml G418 sulfate antibiotic (Mediatech Inc.). HapI cells (Horizon Discovery, Cambridge, United Kingdom) were maintained in Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 10% FBS and 1% P/S. All the cell lines were incubated at 37°C with 5% CO2. The cells were passed at an appropriate dilution 1 or 2 days prior to use in order to achieve 80% to 90% confluence upon infection or 60% to 80% confluence upon transfection. 293T and BSR-T7 cells were used for transfection experiments, HeLa cells were used for transfection and then infection experiments, and HapI and Vero cells were used for infection experiments.
Plasmids and transfections.
Plasmids were constructed using standard molecular-cloning techniques and are available upon request (25). A Flag tag was added to the 3′ end of recombinant RPS6KB1, which was cloned into the pCAGGS expression vector, along with MuV NP, P, and L genes. Plasmids containing firefly luciferase (pFF-luc) and the MuV minigenome (pBH526/pMG-Rluc) containing Renilla luciferase under the T7 promoter were used in the MuV minigenome assays (7).
JetPrime was used to transfect cells with plasmids, and Interferin was used to transfect cells with siRNA according to the manufacturer’s protocols (Polyplus Transfection Inc., New York, NY).
Viruses and infections.
As previously described, WT MuV and rMuV-rLuc were propagated from an early passage and sequenced to confirm that no mutations occurred (25, 26). In brief, a T150 flask of 90 to 100% confluent Vero cells was infected at an MOI of 0.01 for 1 h. The inocula were then replaced with fresh DMEM supplemented with 2% FBS and 1% P/S, and virus was propagated for 2 or 3 days. The viruses were collected after syncytia formed, and plaque assays were performed to determine the titer (7, 26). Viral infections were performed at an MOI of 0.01, 0.1, 1, or 3. Inocula were prepared in DMEM supplemented with 2% FBS and 1% P/S and incubated on cells for 1 or 2 h. The inocula were then replaced with fresh DMEM supplemented with 2% FBS and 1% P/S. HapI cells were inoculated with IMDM supplemented with 10% FBS and 1% P/S.
siRNA screen and cytotoxicity assay.
A library of 720 siRNA pools targeting protein kinase families (Dharmacon siArray siRNA library and human genome, G-003505 [Thermo Fisher Scientific, Lafayette, CO]) was utilized. Using DharmaFect 1 transfection reagent (Dharmacon) and following the manufacturer’s instructions, Vero-E6 cells (96-well format) were reverse transfected with siRNAs at a 50 nM final concentration. Each 96-well plate had additional control wells with a nontarget control siRNA (siControl nontarget siRNA; Dharmacon), siRNA targeting AKT1, and TOX control siRNA and wells without any siRNA. At 2 days posttransfection, cells were infected with rMuV-rluc or rPIV5-rLuc virus at an MOI of 1 or 2, respectively. The transfected/infected cells were lysed for Renilla luciferase assay at 1 day postinfection.
Cytotoxic effects of siRNAs on cells were determined using a ToxiLight bioassay kit (Lonza). Supernatants of gene-silenced cells were harvested at 2 days posttransfection and before infection and assayed for adenylate kinase (AK) release according to the manufacturer’s protocol.
Renilla luciferase and cytotoxicity assays.
Two different Mission predesigned siRNAs (Sigma-Aldrich, St. Louis, MO) were transfected into 60 to 80% confluent HeLa cells using Interferin (Polyplus Transfection Inc.) to a final concentration of 5 nM. NT, also purchased from Sigma-Aldrich, was used as the control. The catalog numbers for the predesigned siRNAs are as follows: RPS6KB1 (NM_003161), MAP3K6 (NM_004672), MYO3B (NM_001083615), UMPK (NM_016308), and AK7 (NM_152327). Individual sequences are available upon request. Twenty-four hours later, the cells were infected with a recombinant MuV expressing Renilla luciferase protein (rMuV-rluc) at an MOI of 0.1. After another 24 h, the cells were lysed in 100 μl of 5× Renilla luciferase lysis buffer (Promega, Madison, WI) by vigorous shaking; 40 μl of lysate from each well was used to run the Renilla luciferase assay according to the manufacturer’s protocol, and the light intensity was detected using a GloMax 96 microplate luminometer (Promega). Three replicates of each sample were compared to the nontarget control, which was used to normalize the activities. The lysates of each replicate were combined and collected for Western blot analysis.
LY2584702 tosylate inhibitor (Selleck Chemicals, Houston, TX), an RPS6KB1 inhibitor, was dissolved in DMSO (dimethyl sulfoxide) to make a 10 mM stock solution. HeLa cells were infected in triplicate with rMuV-rluc for 1 h in a 12-well plate. During this time, the inhibitor was diluted 2-fold starting at 10 μM. After infection, the diluted inhibitor, plus medium, was added back to the cells for 24 h. The Renilla luciferase assay was performed as described above.
To determine if the siRNAs and RPS6KB1 inhibitor had any negative effects on HeLa cells, the CellTiter-Glo luminescent cell viability assay (Promega) was used. siRNAs were transfected, and the inhibitor was added as described above. After 24 or 48 h, the cells were lysed with 100 μl of CellTiter-Glo reagent (Promega) according to the manufacturer’s protocol. Sample lysates were run straight and diluted 10-fold in triplicate. Either the nontarget control or 0.1% DMSO was used as the control, and values were normalized to these values.
Multistep and single-step growth curves.
WT or RPS6KB1-KO HapI cells were seeded in a 6-well plate in triplicate. Upon confluence, cells were infected at either an MOI of 0.01 (multistep) or an MOI of 5 (single step). Medium was collected from each well at 0, 24, 48, 72, 96, 120, 144, and 168 hpi for the multistep growth curve and at 0, 12, 24, 36, 48, and 60 hpi for the single-step growth curve. Virus titers were determined in Vero cells by plaque assays as previously described (27).
Immunoblotting.
Cell lysates were mixed with 2× Laemmli sample buffer with β-mercaptoethanol (Bio-Rad Laboratories, Inc., Hercules, CA) and heated at 95°C for 5 min. Samples were resolved in 10% SDS-PAGE and transferred to a polyvinylidene difluoride membrane (GE Healthcare, Piscataway, NJ). Immunoblotting, using monoclonal MuV protein-specific antibodies, was performed as previously described (25). The membrane was incubated with mouse anti-MuV NP antibody (1:2,500 dilution) and mouse anti-MuV P antibody (1:2,000 dilution), followed by incubation with Cy3-conjugated goat anti-mouse IgG secondary antibody (1:2,500 dilution; Jackson ImmunoResearch, West Grove, PA) and imaged using a Typhoon 9700 imager (GE Healthcare Life Sciences, Piscataway, NJ).
To compare the viral protein expression levels in infected cells, HapI cells were mock infected or infected with WT MuV (Iowa 06) at an MOI of 0.01. At 0, 12, 24, 36, 48, 60, 72, and 96 hpi, cells were prepared and resolved as described above. Immunoblotting was performed with the primary antibodies mouse anti-MuV NP and rabbit anti-actin antibody at a 1:2,500 dilution (Sigma-Aldrich, St. Louis, MO), followed by Cy3-conjugated goat anti-mouse IgG secondary antibody and Cy3-conjugated goat anti-rabbit IgG secondary antibody at 1:2,500 dilutions (Jackson Immuno-Research).
Minigenome and dual-luciferase assays.
BSR-T7 and HapI cells in 24-well plates were transfected with pCAGGS-NP (25 ng), pCAGGS-L (500 ng), pMG-RLuc (100 ng), pFF-Luc (1 ng), and various amounts (20, 40, 80, or 160 ng) of pCAGGS-P. For inhibition of RPS6KB1, BSR-T7 cells were treated with either 1 μM RPS6KB1 inhibitor (Selleck Chemicals) or 0.1% DMSO. For overexpression of RPS6KB1, BSR-T7 cells were transfected with a vector control or a plasmid containing RPS6KB1 at 100 ng per well. pCAGGS vector with green fluorescent protein (GFP) was used to confirm the transfection. After 24 h, cells were lysed in 100 μl passive lysis buffer (Promega, Madison, WI) and vigorously shaken for 20 min to permit full lysis. Clear lysate (40 μl) from each well was used to carry out the dual-luciferase assay according to the manufacturer’s protocol (Promega), and the light intensity was detected using a GloMax 96 microplate luminometer (Promega). Relative luciferase activity was defined as the ratio of Renilla luciferase (R-Luc) activity to firefly luciferase (FF-Luc) activity. Three replicates of each sample were used to compare the peak activity of each P concentration with or without RPS6KB1.
Flow cytometry.
WT and RPS6KB1-KO HapI cells in 10-cm dishes were infected with WT MuV at an MOI of 0.1 in quadruplicate. At 24 hpi, the cells were trypsinized and resuspended in 1% formaldehyde in PBS for 1 h at 4°C. They were then washed with PBS, resuspended in 70% ethanol (EtOH), and incubated overnight at 4°C. The cells were stained with anti-mouse MuV NP antibody at 1:100 dilution in 1% bovine serum albumin (BSA)-PBS for 1 h at 4°C. The cells were then washed 3 times with PBS and resuspended in allophycocyanin (APC)-goat anti-mouse IgG secondary antibody (BioLegend, San Diego, CA) at 1:100. The cells were washed with PBS another 3 times and resuspended in 1 ml PBS. Flow cytometry was performed using an LSRII flow cytometer (BD Biosciences, San Jose, CA) and analyzed using BD FACSDiva software version 8.0.1 (BD Biosciences). The mean fluorescence intensity of MuV NP was calculated for each replicate of infected and uninfected cells.
qRT-PCR.
WT and RPS6KB1-KO HapI cells were infected with WT MuV at an MOI of 5 in triplicate in a 12-well plate. Total RNA was collected at 2 and 24 hpi using an RNeasy minikit (Qiagen, Waltham, MA). cDNA was generated via reverse transcription with Super Script III reverse transcriptase (Invitrogen, Frederick, MD). mRNA was generated with Oligo(dT)15 (Invitrogen), and genomic RNA was generated with a vRNA-specific primer flanking the MuV F gene. Synthesized cDNAs were used as templates for quantitative real-time PCR (qRT-PCR). A MuV HN-specific 6-carboxyfluorescein (FAM)-tagged probe (Applied Biosystems, South San Francisco, CA) and TaqMan gene expression master mix (Applied Biosystems) were used according to the manufacturer’s protocol. Real-time PCR was completed using a StepOnePlus RT-PCR system (AB Biosciences, Foster City, CA). Threshold cycle (CT) values were normalized to the genomic RNA values when the inoculum was removed at 2 hpi.
Radiolabeled IP.
To examine the phosphorylation status of MuV P, HeLa and Hap1cells were used. HeLa cells were transfected with siRNA and then infected as described above. HapI cells were infected or mock infected when confluent at MOI of 0.1 and 3. At 24 hpi, the cells were starved in DMEM lacking cysteine-methionine and then labeled with 72.7 μCi/ml 35S-EasyTag Express 35S protein label (PerkinElmer, Waltham, MA) for 6 to 8 h or starved with DMEM lacking sodium phosphate and then labeled with 100 μCi 33P radionuclide orthophosphoric acid (PerkinElmer, Waltham, MA) for 6 to 8 h. The cells were then lysed with whole-cell extraction buffer (WCEB) (50 mM Tris-HCl [pH 8.0], 280 mM NaCl, 0.5% NP-40, 0.2 mM EDTA, 2 mM EGTA, and 10% glycerol) with a mixture of protease inhibitors as previously described (28). The lysate was split into two tubes and immunoprecipitated using recombinant protein G-Sepharose 4B-conjugated beads and either a mouse monoclonal anti-MuV P or anti-MuV NP antibody. After washing 3 times with WCEB, the beads were mixed with 2× Laemmli sample buffer with β-mercaptoethanol (Bio-Rad Laboratories, Inc., Hercules, CA) and heated at 95°C for 5 min. Samples were resolved with 10% SDS-PAGE. Phosphorylation of the P protein was calculated by densitometry analysis of the 33P/35S ratio using ImageQuant TL software (GE Healthcare Life Sciences).
IP.
To detect the interaction between MuV P and RPS6KB1, 293T cells were transfected with 4 μg of pCAGGS-P, 4 μg of pCAGGS-RPS6KB1-Flag (pKB25), and 2 μg of pCAGGS-NP. pCAGGS-GFP was used to normalize the amount of DNA per 10-cm dish. Twenty-four hours posttransfection, the cells were lysed in 1 ml WCEB with a mixture of protease inhibitors. The lysates were immunoprecipitated overnight at 4°C using recombinant protein G-Sepharose 4B-conjugated beads and either mouse monoclonal anti-MuV P or anti-Flag (M2 clone; Sigma-Aldrich, St. Louis, MO) antibodies. After washing 3 times with WCEB, samples were resuspended in 2× Laemmli sample buffer with β-mercaptoethanol (Bio-Rad Laboratories, Inc., Hercules, CA) and heated at 95°C for 5 min. Immunoblotting was performed as described above. The membrane was blotted with anti-MuV P or anti-Flag, followed by incubation with a horseradish peroxidase (HRP)-conjugated rat anti-mouse IgG antibody (Abcam, Cambridge, MA). After extensive washing, the blot was incubated with SuperSignal West Pico Plus chemiluminescent substrate (ThermoFisher Scientific, Waltham, MA) for 5 min and then exposed on a Gel Doc RX+ imaging system with Image Lab for analysis (Bio-Rad Laboratories, Inc.).
Statistical analysis.
Statistical analyses were performed using GraphPad Prism version 6.00 for Windows (GraphPad Software, San Diego, CA). Student's t test and one-way analysis of variance (ANOVA) were used to calculate P values when comparing two groups. When performing multiple comparisons, the Holm-Šidák method, with an alpha of 5%, was used to determine statistical significance.
ACKNOWLEDGMENTS
We appreciate the helpful discussions and technical assistance from members of Biao He’s laboratory and thank Ashley Beavis for her insightful discussions and critical reading of the manuscript.
This work was supported by a grant from the National Institutes of Health (R01AI 097368) and by an endowment of the Fred C. Davison Distinguished University Professor in Veterinary Medicine to B.H.
REFERENCES
- 1.Lamb RA, Parks GD. 2013. Mumps virus, p 957–995. In Knipe DM, Howley PM (ed), Fields virology, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.National Center for Immunization and Respiratory Diseases. 26 April 2019. Mumps cases and outbreaks. https://www.cdc.gov/mumps/outbreaks.html.
- 3.Marin M, Quinlisk P, Shimabukuro T, Sawhney C, Brown C, Lebaron CW. 2008. Mumps vaccination coverage and vaccine effectiveness in a large outbreak among college students—Iowa, 2006. Vaccine 26:3601–3607. doi: 10.1016/j.vaccine.2008.04.075. [DOI] [PubMed] [Google Scholar]
- 4.Lamb RA, Parks GD. 2013. Paramyxoviridae, p 957–995. In Knipe DM, Howley PM (ed), Fields Virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 5.Cox R, Green TJ, Purushotham S, Deivanayagam C, Bedwell GJ, Prevelige PE, Luo M. 2013. Structural and functional characterization of the mumps virus phosphoprotein. J Virol 87:7558–7568. doi: 10.1128/JVI.00653-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asenjo A, Calvo E, Villanueva N. 2006. Phosphorylation of human respiratory syncytial virus P protein at threonine 108 controls its interaction with the M2-1 protein in the viral RNA polymerase complex. J Gen Virol 87:3637–3642. doi: 10.1099/vir.0.82165-0. [DOI] [PubMed] [Google Scholar]
- 7.Pickar A, Xu P, Elson A, Li Z, Zengel J, He B. 2014. Roles of serine and threonine residues of mumps virus P protein in viral transcription and replication. J Virol 88:4414–4422. doi: 10.1128/JVI.03673-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Timani KA, Sun D, Sun M, Keim C, Lin Y, Schmitt PT, Schmitt AP, He B. 2008. A single amino acid residue change in the P protein of parainfluenza virus 5 elevates viral gene expression. J Virol 82:9123–9133. doi: 10.1128/JVI.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lenard J. 1999. Host cell protein kinases in nonsegmented negative-strand virus (Mononegavirales) infection. Pharmacol Ther 83:39–48. doi: 10.1016/s0163-7258(99)00016-9. [DOI] [PubMed] [Google Scholar]
- 10.Sun M, Fuentes SM, Timani K, Sun D, Murphy C, Lin Y, August A, Teng MN, He B. 2008. Akt plays a critical role in replication of nonsegmented negative-stranded RNA viruses. J Virol 82:105–114. doi: 10.1128/JVI.01520-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickar A, Zengel J, Xu P, Li Z, He B. 2016. Mumps virus nucleoprotein enhances phosphorylation of the phosphoprotein by Polo-like kinase 1. J Virol 90:1588–1598. doi: 10.1128/JVI.02160-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun D, Luthra P, Li Z, He B. 2009. PLK1 down-regulates parainfluenza virus 5 gene expression. PLoS Pathog 5:e1000525. doi: 10.1371/journal.ppat.1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villanueva N, Navarro J, Mendez E, Garcia-Albert I. 1994. Identification of a protein kinase involved in the phosphorylation of the C-terminal region of human respiratory syncytial virus P protein. J Gen Virol 75:555–565. doi: 10.1099/0022-1317-75-3-555. [DOI] [PubMed] [Google Scholar]
- 14.Bahrami BF, Ataie-Kachoie P, Pourgholami MH, Morris DL. 2014. p70 ribosomal protein S6 kinase (Rps6kb1): an update. J Clin Pathol 67:1019–1025. doi: 10.1136/jclinpath-2014-202560. [DOI] [PubMed] [Google Scholar]
- 15.Hanks SK, Hunter T. 1995. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9:576–596. doi: 10.1096/fasebj.9.8.7768349. [DOI] [PubMed] [Google Scholar]
- 16.Jeno P, Ballou LM, Novak-Hofer I, Thomas G. 1988. Identification and characterization of a mitogen-activated S6 kinase. Proc Natl Acad Sci U S A 85:406–410. doi: 10.1073/pnas.85.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma SC. 1998. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J 17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen B, Yang L, Zhang R, Gan Y, Zhang W, Liu D, Chen H, Tang H. 2017. Hyperphosphorylation of RPS6KB1, rather than overexpression, predicts worse prognosis in non-small cell lung cancer patients. PLoS One 12:e0182891. doi: 10.1371/journal.pone.0182891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinonen H, Nieminen A, Saarela M, Kallioniemi A, Klefstrom J, Hautaniemi S, Monni O. 2008. Deciphering downstream gene targets of PI3K/mTOR/p70S6K pathway in breast cancer. BMC Genomics 9:348. doi: 10.1186/1471-2164-9-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiredja DD, Tabler CO, Schlatzer DM, Li M, Chance MR, Tilton JC. 2018. Global phosphoproteomics of CCR5-tropic HIV-1 signaling reveals reprogramming of cellular protein production pathways and identifies p70-S6K1 and MK2 as HIV-responsive kinases required for optimal infection of CD4+ T cells. Retrovirology 15:44. doi: 10.1186/s12977-018-0423-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhatt AP, Wong JP, Weinberg MS, Host KM, Giffin LC, Buijnink J, van Dijk E, Izumiya Y, Kung H, Temple BRS, Damania B. 2016. A viral kinase mimics S6 kinase to enhance cell proliferation. Proc Natl Acad Sci U S A 113:7876–7881. doi: 10.1073/pnas.1600587113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell TM, Espina V, Senina S, Woodson C, Brahms A, Carey B, Lin SC, Lundberg L, Pinkham C, Baer A, Mueller C, Chlipala EA, Sharman F, de la Fuente C, Liotta L, Kehn-Hall K. 2017. Rapamycin modulation of p70 S6 kinase signaling inhibits Rift Valley fever virus pathogenesis. Antiviral Res 143:162–175. doi: 10.1016/j.antiviral.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearce LR, Komander D, Alessi DR. 2010. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol 11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 24.Young DF, Galiano MC, Lemon K, Chen YH, Andrejeva J, Duprex WP, Rima BK, Randall RE. 2009. Mumps virus Enders strain is sensitive to interferon (IFN) despite encoding a functional IFN antagonist. J Gen Virol 90:2731–2738. doi: 10.1099/vir.0.013722-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu P, Li Z, Sun D, Lin Y, Wu J, Rota PA, He B. 2011. Rescue of wild-type mumps virus from a strain associated with recent outbreaks helps to define the role of the SH ORF in the pathogenesis of mumps virus. Virology 417:126–136. doi: 10.1016/j.virol.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zengel J, Pickar A, Xu P, Lin A, He B. 2015. Roles of phosphorylation of the nucleocapsid protein of mumps virus in regulating viral RNA transcription and replication. J Virol 89:7338–7347. doi: 10.1128/JVI.00686-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu P, Luthra P, Li Z, Fuentes S, D’Andrea JA, Wu J, Rubin S, Rota PA, He B. 2012. The V protein of mumps virus plays a critical role in pathogenesis. J Virol 86:1768–1776. doi: 10.1128/JVI.06019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luthra P, Sun D, Wolfgang M, He B. 2008. AKT1-dependent activation of NF-κB by the L protein of parainfluenza virus 5. J Virol 82:10887–10895. doi: 10.1128/JVI.00806-08. [DOI] [PMC free article] [PubMed] [Google Scholar]