Ebola virus infects human and nonhuman primates, causing severe infections that are often fatal. The epidemics, in West and Central Africa, emphasize the urgent need to develop antiviral therapies. The Ebola virus large protein (L), which is the central protein for viral RNA replication/transcription, harbors a methyltransferase domain followed by a C-terminal domain of unknown function. We show that the C-terminal domain regulates the L protein methyltransferase activities and consequently participates in viral replication and escape of the host innate immunity.
KEYWORDS: capping, Ebola, epitranscriptomic, methyltransferase, RNA
ABSTRACT
The large (L) protein of Ebola virus is a key protein for virus replication. Its N-terminal region harbors the RNA-dependent RNA polymerase activity, and its C terminus contains a cap assembling line composed of a capping domain and a methyltransferase domain (MTase) followed by a C-terminal domain (CTD) of unknown function. The L protein MTase catalyzes methylation at the 2′-O and N-7 positions of the cap structures. In addition, the MTase of Ebola virus can induce cap-independent internal adenosine 2′-O-methylation. In this work, we investigated the CTD role in the regulation of the cap-dependent and cap-independent MTase activities of the L protein. We found that the CTD, which is enriched in basic amino acids, plays a key role in RNA binding and in turn regulates the different MTase activities. We demonstrated that the mutation of CTD residues modulates specifically the different MTase activities. Altogether, our results highlight the pivotal role of the L protein CTD in the control of viral RNA methylation, which is critical for Ebola virus replication and escape from the innate response in infected cells.
IMPORTANCE Ebola virus infects human and nonhuman primates, causing severe infections that are often fatal. The epidemics, in West and Central Africa, emphasize the urgent need to develop antiviral therapies. The Ebola virus large protein (L), which is the central protein for viral RNA replication/transcription, harbors a methyltransferase domain followed by a C-terminal domain of unknown function. We show that the C-terminal domain regulates the L protein methyltransferase activities and consequently participates in viral replication and escape of the host innate immunity.
INTRODUCTION
Ebola virus is an emerging virus that causes severe epidemics, such as the unprecedented devastating outbreak in West Africa of 2015 that caused more than 11,000 deaths (1). Although Ebola has faded from public attention, outbreaks continue to occur, especially in the Democratic Republic of the Congo (DRC), where the current epidemic, which was declared in August 2018, has already killed more than 2,000 people. International efforts have led to control of the epidemic (3). Ebola virus infects human and nonhuman primates, leading to hemorrhagic fever that is fatal in most cases. Although vaccines are currently in development (4, 5) and some therapeutic antibodies have shown antiviral activities (6), therapeutic compounds that limit viral replication are needed to treat infected people or contact patients. However, the fine-tuning of the virus replication process remains poorly understood.
Ebolavirus belongs to the Mononegavirales order, which includes other important human pathogens, such as measles, respiratory syncytial virus, human metapneumovirus (hMPV), and rabies virus. These viruses have similar genetic organizations and common replication strategies. The Ebolavirus genus contains the following six species: Zaire ebolavirus (Ebola virus, EBOV), Sudan ebolavirus (Sudan virus, SUDV), Tai Forest ebolavirus (Tai Forest virus, TAFV), Bundibugyo ebolavirus (Bundibugyo virus, BDBV), Reston ebolavirus (Reston virus, RESTV), and Bombali ebolavirus (Bombali virus, BOMV). These viruses and Marburg marburgvirus (Marburg virus, MARV), which causes acute hemorrhagic fever, Lloviu cuevavirus, and Mengla dianlovirus form the Filoviridae family (7).
The negative-sense RNA genome of filoviruses is about 19 kb long and encodes the following seven major proteins: the nucleoprotein (NP), the viral proteins VP35 and VP40, the glycoprotein (GP), the viral proteins VP30 and VP24, and the “large” protein (L) (8, 9). The viral life cycle is initiated by the interaction of the GP envelope protein with cell surface determinants to allow the virus entry in the cell by macropinocytosis. The NP-encapsidated viral genome is then released in the host cell cytoplasm, where the virus replication cycle is initiated. The L protein, which contains the RNA-dependent RNA polymerase (RdRp) domain, as well as NP, VP35, and VP30, are required for replication. First, viral mRNAs are transcribed using a discontinuous transcription mechanism that generates seven monocistronic capped and poly-adenylated RNA species (10, 11). The polymerase complex initiates mRNA synthesis at a conserved gene-start (GS) sequence and transcribes genes up to the gene-stop sequence (GE) (12). The poly-A tail is then added by a polymerase-mediated slippage mechanism at a conserved poly(U)6 tract in the GE sequence (13). Then, the RdRp scans the intergenic sequences and again starts RNA transcription at the next GS sequence. Mononegavirales mRNAs are cotranscriptionally capped by an unusual sequence of capping reactions. This process, which has been extrapolated from the vesicular stomatitis virus (VSV) model, takes place when the nascent RNA chain length has reached a length of 31 nucleotides (14). The cap synthesis starts with the formation of a covalent adduct between a conserved histidine residue of the polyribonucleotidyl transferase (PRNTase) and the 5′-phosphate of the viral mRNA. In the presence of GDP, the PRNTase transfers a GDP moiety to the 5′-phosphate of the covalently bound RNA, leading to the formation of capped RNA (Gppp-RNA) (15). The 5′ cap structure is subsequently methylated at the 2′-O position of the first nucleotide (N1) and at the N-7 position of the guanosine, yielding cap-1 structures (mGpppNm) (16). This cap-1 structure is undistinguishable from cellular caps and plays a critical role in virus replication because it protects viral mRNAs from the host cell 5′ exonucleases and allows the eukaryotic translation initiation factor 4 (eIF4e)-dependent initiation of mRNA translation (54, 55). Additionally, the cap-dependent 2′-O-methylation is a self-marker that limits the detection of viral RNAs by the innate immunity sensors of the retinoic acid-inducible gene-I (RIG-I)-like family. Thus, viruses that express RNAs without cap 2′-O-methylation (mGpppN-RNA or cap-0) are detected early by RIG-I receptors in infected cells, leading to the secretion type I interferon (IFN α/β) (17, 18). IFNs stimulate the expression of IFN-stimulated genes, such as IFN induced protein with tetratricopeptide repeats (IFIT 1/3), that sequester miss-capped RNAs (i.e., cap-0), leading to reduced virus propagation (19, 20). Therefore, capping enzymes, such as cap-MTases, are considered potent antiviral targets. Indeed, N-7 methylation inhibition is supposed to limit RNA translation into viral proteins, and 2′-O-MTase inhibitors should unmask viral RNA to intracellular innate immunity sensors (21).
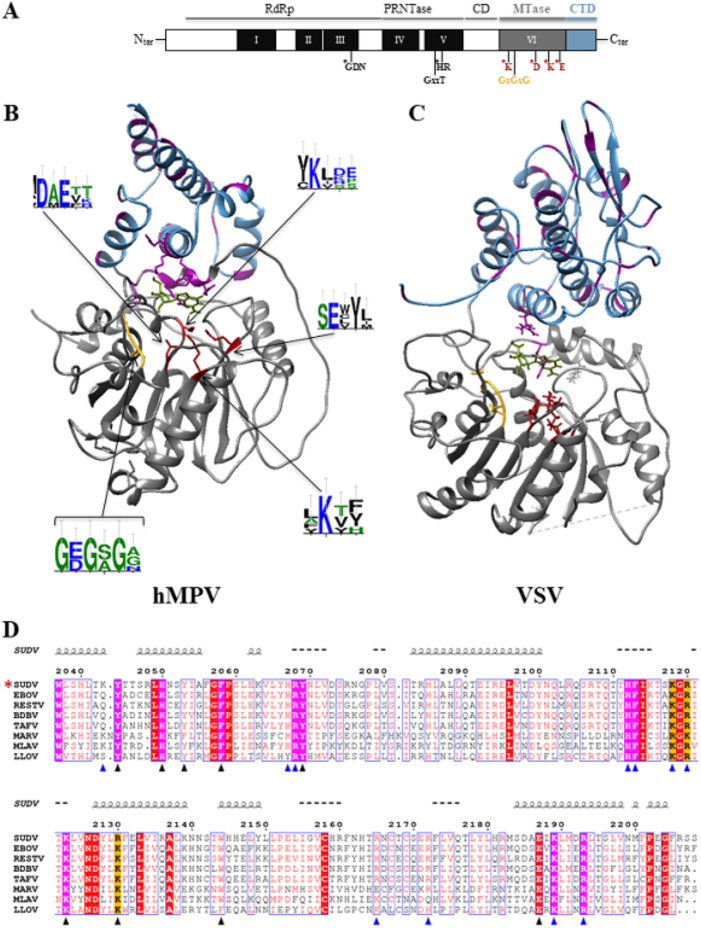
Several cryo-electron microscopy (cryo-EM) structures of the full-length mononegavirus L protein were recently described (22–25). While L protein C-terminal domains were not visible in most structures, the VSV L protein was fully unraveled (22). This study revealed an organization with five different topological domains (Fig. 1A). The RdRp contains three conserved regions (CRI, CRII, and CRIII) and is intimately associated with the PRNTase domain (CRIV), forming the “donut-like” structure observed with negative staining (26). The RdRp domain is organized in a right-hand “fingers-palm-thumb” structure, typical of polymerases. The cap domain (PRNTase), which has an original fold, projects a loop near the catalytic site of the polymerase. This structural interplay suggests that the PRNTase domain participates in polymerization initiation, similarly to polymerases with a priming loop (27). The structure corroborates biochemical analysis results showing the interplay between RdRp and capping activities (14). The donut-like cluster (RdRp and PRNTase domains) is followed by three globular structures that correspond to the connector domain (CD), the MTase domain, and a small C-terminal domain (CTD). The CD separates the PRNTase domain from the MTase domain, which is composed of a bundle of eight helices (22). The MTase domain contains a K-D-K-E catalytic tetrad, characteristic of 2′-O-MTases. The VSV MTase structure is organized in a typical Rossman fold of S-adenosyl methionine (SAM)-dependent methyltransferases. The X-ray structure of the hMPV MTase+CTD segment confirmed the presence of the methyltransferase fold with a SAM-binding site (GxGxG) that positions the released methyl group in front of the catalytic tetrad (28) (Fig. 1B and C). However, unlike other viral MTases, the hMPV MTase+CTD lacks a canonical cap-binding site (29–31). Thus, it was proposed that the RNA substrate is accommodated in an unusual RNA-binding groove formed by the MTase domain overlaid by the CTD enriched in basic and aromatic amino acids (in purple in Fig. 1B and C), known to interact with the nucleotide bases by stacking interactions (28).
FIG 1.
Bioinformatic analysis of the C-terminal domains (MTase+CTD) of mononegavirus L protein based on hMPV and VSV structures and alignment of the filovirus CTD sequences. (A) Domain organization of the mononegavirus L protein with the six conserved regions (CRI to CRV, black boxes; CRVI, gray box) that contain motifs important for the different activities of the L protein (motifs mapped with asterisks). The Sudan ebolavirus (SUDV) methyltransferase domain (MTase) encompasses amino acids 1693 to 2036, based on the alignment with the vesicular stomatitis virus (VSV) L protein. The SAM-binding site (GxGxG) and the 2′-O catalytic tetrad K-D-K-E are also shown (asterisks). The C-terminal domain (CTD) of SUDV L follows the MTase domain (amino acids 2037 to 2210). (B) X-ray structure of human metapneumovirus (hMPV) MTase (gray) and CTD (blue) domains at a resolution of 2.2 Å (26) (PDB ID, 4UCZ). The residues of the catalytic tetrad and of the SAM-binding site are in red and yellow, respectively. Aromatic and basic residues are in purple. WebLogo (47) was used to illustrate the degree of amino acid conservation based on previously published alignments (34). (C) Structure of VSV MTase (gray) (PDB ID, 5A22) and CTD (blue) (53). The residues involved in the catalytic tetrad and SAM-binding site are in red and yellow, respectively. Aromatic and basic residues are in purple. (D) CTD domains in the following L protein sequences selected from GenBank: EBOV (Zaire ebolavirus, AAG40171.1), SUDV (Sudan ebolavirus, YP_138527.1), TAFV (Tai Forest ebolavirus, ALT19766.1), BDBV (Bundibugyo ebolavirus, AKB09568.1), RESTV (Reston ebolavirus, APA16576.1), MARV (Marburg virus, CAA82542.1), LLOV (Lloviu cuevavirus, YP_004928143.1), and MLAV (Mengla dianlovirus, AZL87829.1). An asterisk indicates the strain under study. The alignment was generated with Seaview and analyzed with ESPript. The numbers on top of the alignment indicate the amino acid positions in the SUDV sequence. Strictly conserved aromatic and basic amino acids are highlighted in purple, and arginine or lysine basic residues are highlighted in orange. Mutations leading to soluble proteins are indicated by blue triangles, and mutations leading to insoluble proteins are indicated by black triangles. Spirals just above the alignment indicate the positions of α helices, based on the secondary structure prediction server (NPS@: PHD secondary structure prediction).
The MTase activities of the VSV L protein were first demonstrated in vitro. This protein sequentially catalyzes the RNA cap structure methylation at the ribose 2′-O and guanosine N-7 positions (32). The MTase catalytic activity was confirmed using recombinant hMPV and SUDV MTase+CTD proteins (28, 33). Indeed, it was reported that these MTase+CTDs methylate the 2′-O position of the first 5′-RNA nucleotide (N1) of capped and triphosphate RNAs. The subsequent methylation of the N-7 guanosine of the cap structure was demonstrated by incubating the SUDV MTase+CTD protein with longer RNA substrates. We also demonstrated that the SUDV MTase catalyzes cap-independent methylation, leading to internal adenosine 2′-O-methylation (33), an activity that is not obviously detected using the hMPV MTase (28). The role of these epitranscriptomic RNA modifications is not yet fully understood. However, it was recently reported that similar internal 2′-O-methylation on the HIV RNA genome protects viral RNAs from detection by the RIG-like receptor melanoma differentiation-associated protein 5 (MDA5) (34). Therefore, the filovirus MTase might have evolved toward a dual activity, targeting the cap structure present at the 5′ end of mRNA and adenosine residues inside the RNA.
In this work, we investigated the role of the SUDV CTD domain of the L protein in the MTase activities by mutational analysis. We found that the CTD plays a key role in RNA substrate binding and thereby regulates the different MTase activities. In addition, the mutational analysis allowed the identification of key CTD residues that regulate the different MTase activities of the SUDV L protein. These results suggest that SUDV CTD is involved in the RNA methylation process and in the control of internal methylation versus cap methylation.
RESULTS
The CTD of the SUDV L protein contains basic and aromatic amino acids that could be involved in RNA recognition.
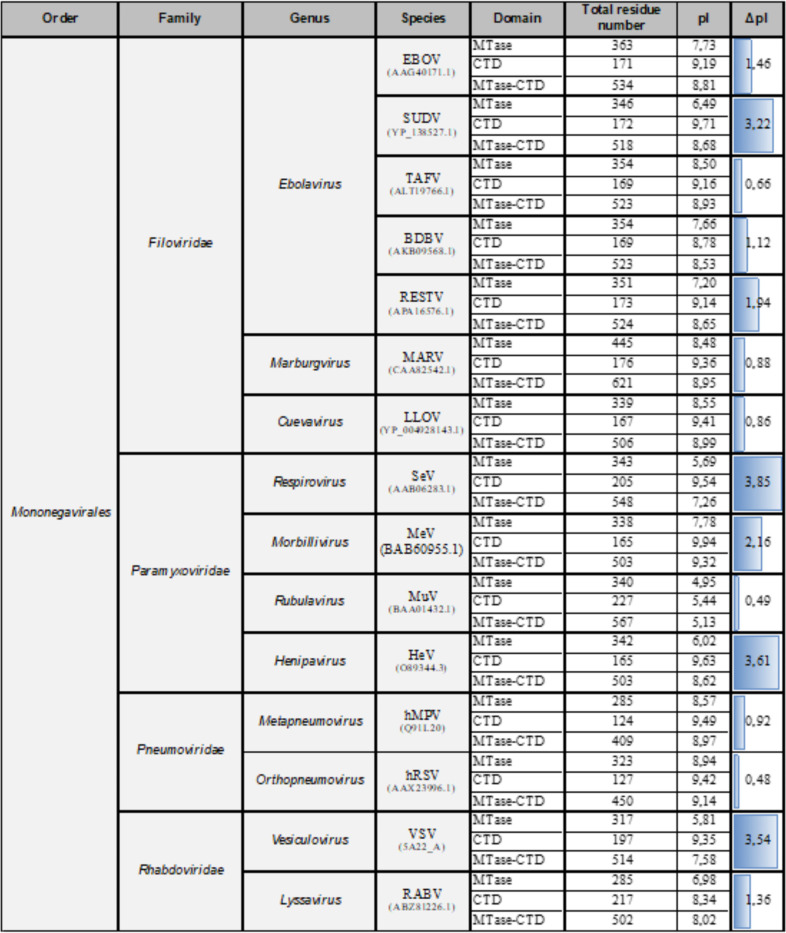
The C terminus of the Mononegavirales L protein contains a conserved MTase domain followed by a less conserved CTD domain with unknown function(s) (Fig. 1). Unfortunately, due to their important differences in length (from ∼120 residues in Pneumoviridae to ∼240 residues in Rhabdoviridae) and sequence, we could not align the CTD of Mononegavirales (Table 1). Nevertheless, comparison of the isoelectric points (pI) of the different CTDs showed that they were all quite basic and varied between 8.33 and 9.94, except for Rubulavirus (Table 1). To identify conserved features among filovirus CTDs (residues 2037 to 2210, numbering corresponding to the SUDV sequence), we aligned their sequences (Fig. 1D). The alignment revealed the presence of 10 conserved aromatic (n = 5) and basic (n = 5) amino acids (highlighted in magenta). In addition, we also noted the presence of three positions with either arginine or lysine basic residues (highlighted in orange) (Fig. 1D).
TABLE 1.
Isoelectric point analysis of mononegavirus MTase and CTD domainsa
Based on the alignment of different viruses belonging to the Mononegavirales order, the residue content of the methyltransferase (MTase) and C-terminal domains (CTD) of their L protein was analyzed. For each virus, the MTase, CTD, and MTase+CTD total number of residues was determined. The isoelectric points (pI) were estimated with ProtParam. The difference between the MTase and CTD pIs were calculated and modeled with a blue gauge normalized to the greatest difference.
The L protein CTD is essential for SUDV MTase activities.
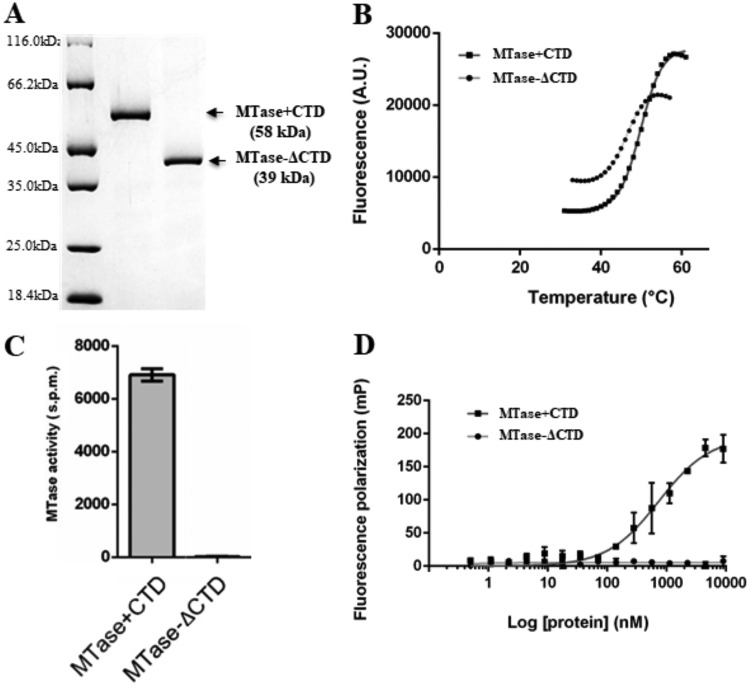
We produced the C-terminal region of the SUDV L protein (MTase+CTD) and the MTase domain alone (MTase-ΔCTD) in bacteria. SDS-PAGE followed by Coomassie blue staining showed that recombinant MTase+CTD and MTase-ΔCTD had the expected apparent molecular weight of 58 kDa and 39 kDa, respectively (Fig. 2A). After confirming their identity with matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF), we analyzed their folding and stability with a thermal shift assay (TSA) (Fig. 2B). The analysis revealed the typical folded-to-unfolded transition curves of globular proteins (35), with a melting temperature (Tm) value of 54.3°C for MTase+CTD and 46.0°C for MTase-ΔCTD. Although the Tm difference between MTase+CTD and MTase-ΔCTD was significant (∼8°C), the two proteins presented the classical native to unfolded phases, suggesting that both adopt a folded conformation and that the CTD contributes to MTase thermal resistance, potentially indicative of a close structural relation. Altogether, these results confirmed that both proteins are folded. We next investigated CTD involvement in MTase activity by comparing the enzymatic activity of both proteins using a 13-nucleotide-long capped RNA (GpppG-SUDV12) substrate, which mimics the conserved 5′ sequence of SUDV 5′ transcripts, in the presence of a radiolabeled methyl donor (3H-SAM). Measurement of the radioactivity transferred to the RNA substrate (Fig. 2C) showed that MTase-ΔCTD was not active, whereas MTase+CTD induced the methylation of the capped-RNA substrate. We previously confirmed the specificity of the observed methylation by showing that mutation of each residue of the conserved K-D-K-E catalytic tetrad in the MTase domain leads to a strong decrease of MTase+CTD MTase activities (36).
FIG 2.
Production and purification of recombinant SUDV MTase+CTD and MTase-ΔCTD and CTD effect on MTase activity and RNA binding. (A) Recombinant Sudan ebolavirus (SUDV) MTase+CTD (58 kDa) and MTase-ΔCTD (39 kDa) were purified using affinity chromatography and then separated using SDS-PAGE before Coomassie blue staining. (B) Thermal shift assay of recombinant SUDV MTase+CTD (dark gray) and MTase-ΔCTD (light gray). The melting temperatures (Tm) were calculated using the Boltzmann sigmoidal regression and were evaluated at 54.6°C and 46°C, respectively. Raw data are shown. (C) MTase activity evaluation of SUDV MTase+CTD (gray) and MTase-ΔCTD (dark) incubated with a 13-mer capped RNA substrate that mimics the capped 5′ extremity of viral mRNA (GpppG-SUDV12). MTase activity was determined with a filter binding assay. Data are the mean ± standard deviation (n = 6). (D) RNA-binding analysis by fluorescence polarization using a 13-mer capped SUDV RNA substrate labeled at the 3′ end pCp-Cy5 and SUDV MTase-ΔCTD (light gray) and MTase+CTD (dark gray). The dissociation coefficient (Kd) was calculated with site-specific binding regression analysis with Hill slope. MTase+CTD affinity for GpppG-SUDV12 was 740 ± 108 nM. Data are the mean ± standard deviation (n = 3).
We also compared the RNA-binding properties of MTase+CTD, MTase-ΔCTD, and the MTase+CTD catalytic mutants by fluorescence polarization (FP) assay. To this aim, we labeled the capped RNA substrate at its 3′ end by ligation of a fluorescent pCp-Cy5 residue (GpppG-SUDV12-pCp-Cy5), incubated it with increasing concentrations of MTase+CTD, MTase-ΔCTD, or catalytic mutants, and measured the RNA-protein interaction using FP, except for the K1959A mutant, which was not stable (Fig. 2D; Table 2). MTase-ΔCTD barely interacted with the capped RNA substrate. Conversely, MTase+CTD induced an FP signal increase, confirming its interaction with the synthetic capped RNA substrate (apparent dissociation constant [Kd] value of 740 nM). We also observed that the MTase+CTD catalytic mutants without MTase activity (36) maintained most of their RNA-binding properties. However, the K1813A mutation led to a loss of the protein-RNA interaction (Kd ≈ 2.7 μM) (Table 2). This indicates that the catalytic site could also contribute to RNA interaction. Altogether, these results suggest that the SUDV CTD plays a key role in RNA binding and promotes MTase activity.
TABLE 2.
RNA binding assay of catalytic mutantsa
| Strain or catalytic mutant | Fluorescence polarization GpppG-SUDV12 (Kd) (μM) |
|---|---|
| WT | 0.740 (±0.108) |
| K1813A | 4.6 (±0.943) |
| D1924A | 0.581 (±0.05) |
| K1959A | ND |
| E1994A | 1.3 (±0.206) |
RNA-binding assays were performed with fluorescence polarization using a 13-mer SUDV-specific capped RNA (GpppG-SUDV12) labeled at the 3′ end with pCp-Cy5 and wild-type (WT) SUDV MTase+CTD and the indicated catalytic mutant MTase+CTD proteins. The dissociation coefficient (Kd) was calculated by site-specific binding regression with Hill slope. Data are the mean (± standard deviation) (n = 3). ND, not determined.
Effect on MTase activities of single mutations in the conserved residues of SUDV CTD.
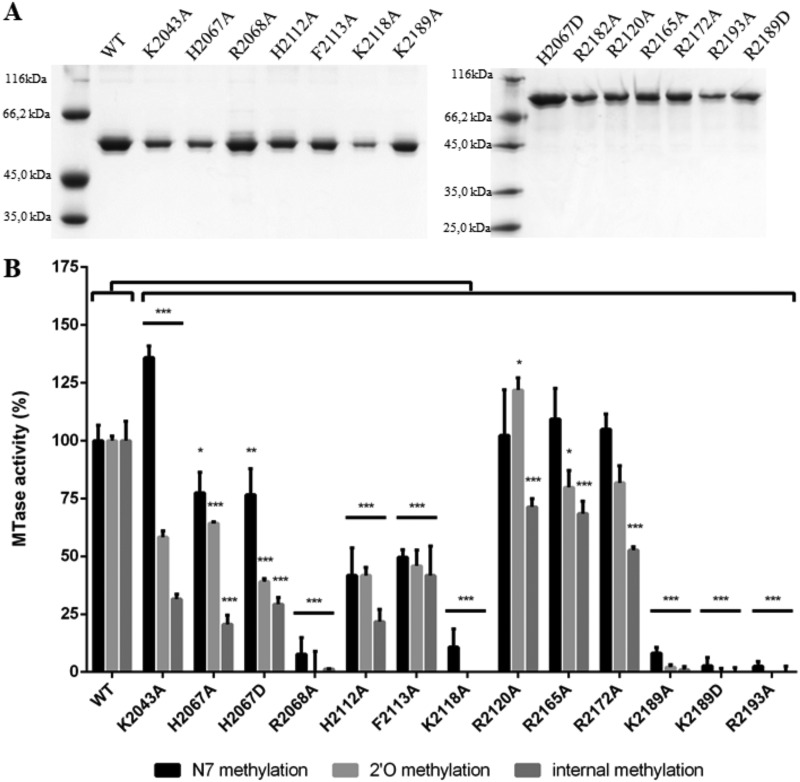
We previously demonstrated that in addition to cap N-7 and 2′-O-methylation, the SUDV MTase+CTD induces internal adenosine 2′-O-methylation (36). To further characterize CTD’s role in RNA recruitment and in the regulation of the different MTase activities catalyzed by MTase+CTD, we mutated the conserved residues in CTD into alanine or aspartic acid (Fig. 1C; Table 3). Among the 25 generated mutants, 13 led to the production of soluble proteins with a molecular weight similar to that of wild-type MTase+CTD (Fig. 3A) and with the typical denaturation curve of folded proteins (Tm value from 50.6°C to 55.7°C) (Table 3). The Tm values of most mutants were close to that of wild-type MTase+CTD, except the Tms of the R2068A and R2172A mutants, which were slightly lower (ΔTm = −3.7°C and ΔTm = –3.1°C, respectively, compared with wild-type MTase+CTD). This suggests that these mutations slightly affected the protein stability, but to a lesser extent than with CTD deletion (ΔTm = –8.3°C). We next tested the mutation effect on RNA binding with the FP assay using GpppG-RNA-Cy5 and mGpppGm-RNA-Cy5 SUDV RNA as canonical substrates for all MTase activities and for internal methylation, respectively (Table 3). All single mutations affected RNA binding, with Kd values ranging from 1 to >9 μM; however, the binding decrease was less important than that observed with MTase-ΔCTD. Moreover, most mutations affected the binding of GpppG-SUDV12 and mGpppGm-SUDV12 in a similar manner (Table 3). Conversely, the K2043A and K2189A mutations reduced the interaction with GpppG-SUDV12 (by about 3 times), particularly with mGpppGm-SUDV12 (Kd > 9 μM and Kd = 8.6 μM, respectively). Then, by incubating wild-type and mutated MTase+CTD proteins with GpppGm(Am)-SUDV12, mGpppA(Am)-SUDV12, or mGpppGm-SUDV12 in the presence of a radiolabeled methyl donor (3H-SAM), we determined the mutation effects on the different MTase activities (i.e., cap N-7 MTase, cap 2′-O-MTase, and internal 2′-O-MTase activities, respectively) (Fig. 3B). The results showed that some mutations affected almost all MTase activities, whereas others impaired only some MTase activities. The mutations R2068A, K2118A, K2189A/D, and R2193A led to a reduction by 90% or more of N-7- and 2′-O-MTase and also internal 2′-O-MTase activities compared with wild-type MTase+CTD. In these mutants, RNA binding was also strongly reduced, except for the K2189A/D mutant, which still partially recognized the RNA substrate (Table 3). This suggested that this mutant protein accommodates the RNA in an inappropriate way for the transfer of the methyl group to the RNA substrate. In addition, residues K2189 and R2194 correspond to a conserved K-K-G motif in Paramyxovirus CTD that was previously proposed to play a key role in RNA binding and N-7 and 2′-O-MTase activities (32). In the second group of mutants, RNA binding and MTase activities were less affected. In this group, we identified two mutations (K2043A, H2067D) that impaired more strongly the 2′-O-MTase activities (cap 2′-O-MTase and internal 2′-O-A MTase) than the N-7 MTase activity. As expected, binding of these two mutants to mGpppGm-SUDV12 was more affected than that to GpppG-SUDV12. In two other mutants (H2112A and F2113A) of this second group, internal methylation was more strongly limited than cap-dependent MTase activities. This suggests that these mutants can accommodate the RNA substrate in a nonoptimal manner for internal 2′-O-methylation. Altogether, these results confirmed that the CTD plays an important role in the RNA substrate binding and that some conserved basic residues (R2068A, K2118A, K2189A, R2193A, and K2189) are implicated in RNA binding and MTase activities. We also identified basic and aromatic residues that impair specifically the 2′-O-MTase activities more strongly than the N-7 MTase activity. These observations suggest that the CTD of SUDV L protein selectively contributes to the right positioning of RNA in the MTase catalytic site, thereby regulating the different MTase activities of the protein.
TABLE 3.
Effects of single mutations of CTD conserved residues on SUDV MTase+CTD stability and RNA bindinga
| Strain or mutantb | Thermal shift assay |
Fluorescence polarization |
||
|---|---|---|---|---|
| Tm (°C) | ΔTm | GpppG-SUDV12 (Kd) (μM) | mGpppGm-SUDV12 (Kd) (μM) | |
| WT | 54.3 (±0.13) | 0.740 (±0.108) | 0.570 (±0.110) | |
| K2043A | 55.7 (±0.03) | 1.4 | 2 (±0.280) | >9 |
| H2067A | 54.7 (±0.58) | 0.4 | 5 (±1.5) | 4.4 (±1.5) |
| H2067D | 53.1 (±0.70) | –0.7 | 3.5 (±1) | 6.5(±2.3) |
| R2068A | 50.6 (±0.01) | –3.7 | >9 | >9 |
| H2112A | 54.0 (±0.39) | –0.3 | 4.5 (±1.1) | 5.4 (±2.1) |
| F2113A | 53.5 (±0.33) | –0.8 | 6.6 (±1.6) | 3.1 (±0.567) |
| K2118A | 53.7 (±0.24) | –0.6 | >9 | >9 |
| R2120A | 51.3 (±0.16) | –3 | 2.8 (±0.649) | 2.2 (±0.705) |
| R2165A | 53.05 (±0.63) | –1.2 | 1 (±0.238) | 1.4 (±0.254) |
| R2172A | 51.2 (±0.41) | –3.1 | 3.1 (±0.988) | 1.7 (±0.835) |
| K2189A | 55.0 (±0.83) | 0.7 | 1.9 (±0,507) | 8.6 (±2.8) |
| K2189D | 53.85 (±0.28) | –0.45 | 2.4 (±0.509) | 1.3 (±0.478) |
| R2193A | 51.9 (±0.70) | –2.4 | >9 | 7.7 (±2.7) |
Thermostability analysis by thermal shift assay of wild-type (WT) Sudan ebolavirus (SUDV) MTase+CTD and the indicated mutated proteins. Melting temperatures (Tm) were calculated using Boltzmann sigmoidal regression. Tm data are the mean (± standard deviation) of n = 2. ΔTm corresponds to the difference between the Tm of the mutant and WT proteins. RNA binding was tested with fluorescence polarization using 13-mer SUDV-specific capped RNAs (GpppG-SUDV12 and mGpppGm-SUDV12) labeled at the 3′ end with pCp-Cy5 and SUDV MTase+CTD and mutated MTase+CTD proteins. The dissociation coefficient (Kd) was calculated with site-specific binding regression with Hill slope. Data are the mean (± standard deviation) of n = 3.
Mutations that led to insoluble proteins: F2113D, K2043D, W2144A, R2130A, Y2053A, Y2044A, Δ-CTD(2105), H2050A, F2058A, Y2069A, K2123A, and E2187A.
FIG 3.
Effect of single mutations of CTD conserved residues on MTase activities of Sudan ebolavirus MTase+CTD. (A) Wild-type (WT) and mutated SUDV MTase+CTD (58 kDa) were purified using affinity chromatography and then separated using SDS-PAGE analysis before Coomassie blue staining. (B) MTase activities of SUDV WT and mutated MTase+CTD incubated with GpppGm(Am)-SUDV12, mGpppA(Am)SUDV12, or mGpppGm-SUDV12 and a radiolabeled methyl donor (3H-SAM) to assess the cap N-7 MTase (black), cap 2′-O-MTase (light gray), and internal 2′-O-A MTase (dark gray) activities, respectively, by filter binding assay. Data are the mean ± standard deviation (n = 3); *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-way analysis of variance [ANOVA] and multiple comparison Tukey test, WT versus mutation).
DISCUSSION
The L protein of mononegavirus plays a key role in replication/transcription and RNA capping. Structural and biochemical studies have highlighted its organization in five main topological domains (22) that include the RdRp, PRNTase, and MTase activities. However, little is known about the roles of the CD and CTD. Here, we show that in SUDV, the CTD regulates the MTase activities.
Here, we experimentally demonstrated that in SUDV, the CTD is essential for RNA binding, because binding of capped RNA is impaired in its absence (Fig. 3D), and single mutations of conserved basic residues within the CTD affect RNA binding to different extents. We also evaluated the effect of CTD deletion and of single mutations within the CTD on the different MTase activities of the SUDV L protein. All MTase activities are abrogated by CTD deletion as well as RNA binding. In contrast, single mutations in the CTD had different effects on the SUDV MTase activities. Specifically, mutations that strongly impaired RNA binding also reduced all MTase activities (R2193A, R2068A, and K2189A/D). Two other mutations (K2043A, H2067D) affected mainly the 2′-O-MTase activities, and three mutations (H2067A, H2112A, and R2172A) strongly reduced particularly the internal methylation. The different effects on MTase activities are consistent with the RNA-binding properties of the involved mutants, suggesting that the RNA substrate must be differently accommodated in order to present the target nucleotide position to the catalytic site for the specific MTase activity. In other words, in some mutants, RNA binding might occur in a position that does not allow just a specific MTase activity (e.g., internal 2′-O). The high mobility of CD, MTase, and CTD observed using cryo-EM analysis of the VSV L protein also suggests that different conformations might correspond to different activation states of the MTase (22). Altogether, these results suggest that in SUDV, the CTD regulates RNA substrate binding and, in turn, participates in the fine-tuning of the MTase activities.
Although CTD’s role in such regulation is unknown, it is noteworthy that some viral MTases involved in cap-methylation require protein partners for their activities. For instance, interaction of the N-7 MTase of vaccinia virus (VV), which is carried by the D1 subunit, with the stimulatory subunit D12 promotes its MTase activity. In this system, D12 is an allosteric activator of the N-7 MTase. Indeed, structural analysis of the VV D1/D12 complex revealed that the D12 protein is on the opposite side of the MTase RNA-binding site and thus is not directly involved in RNA recognition. Nevertheless, the D12 protein subunit stabilizes the MTase domain and increases substrate recognition (37–39). More recently, it was reported that nsp16, the 2′-O-MTase of coronaviruses (severe acute respiratory syndrome [SARS] and Middle East respiratory syndrome coronavirus [MERS-CoV]), interacts with nsp10, a cofactor essential for its 2′-O-MTase enzymatic activity. In this case, the mechanism of MTase activation is quite different. Indeed, in the absence of nsp10, nsp16 recognizes the RNA substrate with low affinity (40, 41). In the presence of nsp10, nsp16 is stabilized and its catalytic and RNA-binding properties are increased. By interacting with nsp16, nsp10 aligns one of the catalytic lysines (K46 in SARS-CoV nsp16) in helix αZ and extends the RNA-binding groove, resulting in an overall stimulation of nsp16 MTase activity (40). The SUDV MTase activation mechanism might be similar to that of CoV nsp16, because our results suggest a direct participation of CTD in RNA binding. Moreover, our observation extends the list of viral MTases that are regulated by an additional domain or protein, suggesting that such a mechanism plays an important role during the life cycle of several viruses.
Although the CTD domain of the mononegavirus L proteins shows a large degree of diversity in size and sequence, its function as a regulator of the MTase activity might be conserved. Indeed, N-7 methylation is essential for RNA translation into viral proteins, and cap 2′-O-methylation avoids detection by RIG-I sensors (18). Besides the canonical cap-dependent MTase activities, the SUDV MTase shows also an unexpected high level of internal adenosine 2′-O-MTase activity in vitro (36). Conversely, the hMPV MTase induces mainly N-7 and 2′-O-methylation of the cap structure (28). These different MTase specificities might be related to the huge CTD sequence variability in Mononegavirales.
We do not know whether SUDV L induces the methylation of adenosines on its own genomic RNA and/or on viral or cellular mRNA in infected cells. The role of such methylation in the viral life cycle is elusive, and several hypotheses, not necessarily exclusive, can be proposed. It was recently reported that other viruses, such as human immunodeficiency viruses (HIV), methylate their own RNA. In this case, the virus recruits the host cell MTase FTSJ3 that catalyzes the genomic RNA methylation to escape detection by MDA5 (34). It might be hypothesized that SUDV and filoviruses, in general, have evolved to induce epitranscriptomic RNA modifications, instead of hijacking a cellular enzyme. In both cases, the resulting methylation might limit detection of viral mRNA by cellular sensors at the early stage of viral infection. Internal 2′-O-methylation of RNA might also have proviral effects by inducing resistance toward cellular 3′-exonuleases involved in RNA decay. Indeed, in plants, it has been shown that microRNAs and small interfering RNAs are 2′-O-methylated at their 3′ end, and such methylation induces resistance against 3′-5′ exonucleases (42, 43) or stabilizes RNAs, thus limiting their degradation (44). Such mechanisms might be used also by viruses to limit exonuclease-mediated RNA turnover induced by IFN (45). Internal 2′-O-methylation of viral RNA might also mediate an antiviral effect by interfering with the RNA replication process. Indeed, the reverse transcriptase of retroviruses is impaired by 2′-O-methylation occurring inside the RNA template at low deoxynucleoside triphosphate (dNTP) concentrations (46). Similarly, VSV and Dengue virus polymerases are sensitive to 2′-O-methyl presence inside their RNA template (47, 48). Thus, the regulation of the MTase activities of L is probably essential for the virus life cycle, and the CTD might be involved in such processes in order to control the temporality and the nature of RNA methylation (i.e., mRNA, poly-A tail, RNA). Studies using cryo-EM of C-terminal domains of mononegavirus L proteins show that the MTase and CTD domains (22, 25) harbor different positions, which might reflect the switching on/off of specific MTase activities regulated by the CTD domain. Additional work is required to address these hypotheses. Finally, 2′-O-methylation may regulate viral protein expression during the different replication steps. For instance, it was reported that internal 2′-O-methylation in cellular mRNA inhibits protein translation (49). The identification of mutations in the SUDV CTD that uncouple the different MTase activities of the L protein might help to elucidate the role of epitranscriptomic RNA modifications, such as internal methylation, in filovirus infection. In addition, this work suggests that the regulation of viral MTase is probably a key factor for efficient virus replication. Designing specific inhibitors of these activities might contribute to the development of combined antiviral strategies that simultaneously lead to the activation of the innate immunity system, especially through the RIG-I/MDA5 pathway, and to inhibition of viral replication.
MATERIALS AND METHODS
Cloning and recombinant protein production.
Codon-optimized SUDV MTase+CTD synthetic genes (Biomers) were cloned in the pET14b vector for bacterial expression (36). The MTase-ΔCTD construct was derived from MTase+CTD with PCR (Pfu Turbo DNA polymerase; Agilent technologies) using primers (forward MTase-ΔCTD: 5′GGCTGAGCCACCTGACCAAGTATACCACCTGATGACTGCACAACAGCTATATCGCGTTCGGCTTTCCG-3′ and reverse MTase-ΔCTD: 5′CGGAAAGCCGAACGCGATATAGCTGTTGTGCAGTCATCAGGTGGTATACTTGGTCAGGTGGCTCAGCC-3′) that flank the sequence of interest and was expressed in bacteria. Mutations were introduced with PCR amplification of the wild-type sequence using mutation-carrying primers (designed using QuikChange primer design) and the Pfu Turbo DNA polymerase. After introduction of the constructs in the Escherichia coli T7 strain (New England Biolabs [NEB]) by transformation, bacteria were cultured at 30°C until the optical density at 600 nm (OD600nm) reached 0.6, after which the temperature was reduced to 17°C and 20 μM IPTG (isopropyl-β-d-thiogalactopyranoside) (Sigma) was added. After 16 h of induction, bacteria were centrifuged (8,000 × g for 20 min at 4°C) in a Sorvall Lynx 6000 centrifuge (Thermo), and pellets were stored at –80°C.
Recombinant protein purification.
All recombinant proteins were purified using a similar protocol, except for the elution step. Bacterial pellets were thawed on ice and lysed in lysis buffer (50 mM Tris pH 8, 150 mM NaCl, 5% glycerol, 30 mM imidazole, 1 mM phenylmethylsulfonyl fluoride [PMSF], 100 μg/ml lysozyme, 1 μg/ml DNase, 0.1% Triton X-100) supplemented with the detergent mix BugBuster (Merck Millipore) only for MTase+CTD purification. The volume of lysis buffer corresponded to 10 × the (final OD600nm value × culture volume [ml]) value. After clarification (18,000 × g, 30 min, 4°C), lysates were incubated with the CoNTA resin (Thermo Fisher; 0.5 ml/liter culture) at 4°C for 1 h, with gentle shaking. Beads were transferred to 25-ml cartridges and washed with 2 × 20 ml of 50 mM Tris pH 8, 1 M NaCl, 5% glycerol, 30 mM imidazole, and 2 × 10 ml of buffer 50 mM Tris pH 8, 150 mM NaCl, and 5% glycerol. Proteins were eluted using two different buffers—50 mM Tris pH 8, 150 mM NaCl, 5% glycerol 1 M arginine for MTase+CTD and 50 mM Tris pH 8, 150 mM NaCl, 5% glycerol, and 150 mM imidazole for MTase-ΔCTD. In this case, imidazole was removed by size exclusion chromatography using Superdex S75 16/60 filtration columns (GE Healthcare). Finally, proteins were concentrated on Amicon ultrafilters (EMD Millipore), and the final concentration was determined with the OD280nm. Proteins were stored in 50% glycerol at –20°C.
Synthesis of RNA substrates.
RNA sequences were chemically synthesized on solid support using an ABI 394 oligonucleotide synthesizer. After RNA elongation with 2′-O-pivaloyloxymethyl phosphoramidite ribonucleotides and 2′-O-methyl phosphoramidite ribonucleotides (ChemGenes, USA), the 5′-hydroxyl group was phosphorylated, and the resulting H-phosphonate derivative was oxidized and activated into a phosphoroimidazolidate derivative to react with GDP (GpppRNA). After deprotection and release from the solid support, GpppRNAs were purified using ion exchange-high-performance liquid chromatography (IEX-HPLC), and their purity (>95%) was confirmed using MALDI-TOF spectrometry. N-7 methylation of the purified GpppRNAs was performed using incubation with N-7-hMTase (50, 51).
MTase activity assay (filter binding assay).
To evaluate methyltransferase activities, a radioactive test was set up by mixing 4 μM recombinant proteins with 1 μM purified synthetic RNAs (RNA sequences in Table 4), 10 μM SAM and 0.5 μM 3H-SAM (Perkin Elmer) in an optimized MTase assay buffer (50 mM Tris-HCl), supplemented with 10 mM arginine for MTase-ΔCTD, and optimum pH for the different MTase activities as follows: pH 7 for the N-7-cap MTase activity, pH 8 for the 2′-O-cap MTase, and pH 8.5 for internal methylation (33). Reactions were stopped by 10-fold dilution in water after 3 h at 30°C. Samples were transferred to DEAE filtermats (Perkin Elmer) using a Filtermat Harvester (Packard Instruments). Methyl transfer was then evaluated as described before (52). Briefly, the RNA-retaining filtermats were washed twice with 10 mM ammonium formate pH 8, twice with water, and once with ethanol. Then, they were soaked with liquid scintillation fluid to measure the 3H-methyl transfer to the RNA substrates using a Wallac MicroBeta TriLux liquid scintillation counter 13. For comparison, it was assumed that the different experimental groups were independent, and data followed a Gaussian distribution with the same variance. Two-way ANOVA and multiple comparison Tukey tests were used (Prism) to evaluate differences between groups. The level of significance for α = 0.05 is indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
TABLE 4.
List of synthetic RNAsa
| RNA | Sequence | Origin |
|---|---|---|
| GpppG-SUDV12 | GpppGAUGAAGAUUAAG | Chemical synthesis |
| GpppGm(Am)-SUDV12 | GpppGmAmUGAmAmGAmUUAmAmG | Chemical synthesis |
| mGpppGm-SUDV12 | mGpppGmAUGAAGAUUAAG | Chemical synthesis |
| mGpppA(Am)-SUDV12 | mGpppAUGAmUGAmAmGAmUUAm | Chemical synthesis |
Xm, nucleotide methylated on position 2′-O of the ribose; mX, nucleotide methylated on position N7 of the base.
Fluorescence polarization (FP).
Using T4 RNA ligase 1 (20 units; New England Biolabs), cyanine 5-cytidine-5-phosphate-3-(6-aminohexyl)phosphate (12.5 μM; Jena Bioscience) was ligated to the 3′ ends of the RNA substrates (10 μM) in T4 RNA ligase 1 buffer (NEB) and 1 mM ATP (16°C, overnight). The ligase was removed using RNA precipitation in 3 M sodium acetate supplemented with 1 μg/μl of glycogen (Thermo Scientific). Fluorescent RNA was incubated (at room temperature for 5 min) with increasing protein concentrations in reaction buffer (20 mM Tris pH 8 or 8.5, 150 mM NaCl, and 5% glycerol) supplemented with 50 mM arginine for MTase-ΔCTD. This corresponded to the final concentration of arginine in the fluorescence polarization (FP) assay. FP measurements were performed in a microplate reader (PHERAstar FS; BMG Labtech) with an optical module equipped with polarizers and using excitation and emission wavelengths of 590 and 675 nm, respectively. Dissociation constants (Kd) were determined using Hill slope curve fitting (Prism).
Thermal shift assay (TSA).
A mix of 40 μM recombinant proteins diluted (1/5) in 50 mM Tris pH 8, 150 mM NaCl, 5% glycerol, and 0.02% (vol/vol) SYPRO orange dye (Thermo Scientific) was prepared to a total volume of 25 μl. To have the same arginine concentration as for MTase+CTD, the buffer reaction for MTase-ΔCTD was supplemented with 100 mM arginine, which corresponds to the final concentration of arginine in the assay. Samples were placed in semiskirted 96-well PCR plates (Bio-Rad), sealed, and heated in a CFX96 Touch quantitative PCR (qPCR) machine (Bio-Rad) from 25 to 95°C at a rate of 1°C min−1. Fluorescence changes were monitored at 492 and 610 nm (excitation and emission wavelengths), respectively. Melting temperatures (Tm) were calculated using a Boltzmann sigmoidal regression for fitting (Prism).
ACKNOWLEDGMENTS
We thank Xavier De Lamballerie and Yannick Boehmann for their help in the initiation of this study.
The program was initiated in a research program funded by the Délégation Générale pour l’Armement (DGA) (grant 2009.34.0038)/Aix-Marseille Université Ph.D. fellowship for B. Martin. C. Valle was funded by the National Research Agency ANR under the program ANR Rab-cap (ANR-16_CE11_0031_01).
C. Valle, B. Martin, B. Coutard, and E. Decroly conceived and designed the study. C. Valle, B. Martin, B. Coutard, and E. Decroly performed the experiments, using material prepared by F. Debart and J.-J. Vasseur. C. Valle, B. Martin, B. Coutard, and E. Decroly analyzed the data and wrote the paper. B. Canard contributed to the writing of the paper. I. Imbert participated in the review of the paper.
REFERENCES
- 1.Shultz JM, Espinel Z, Espinola M, Rechkemmer A. 2016. Distinguishing epidemiological features of the 2013–2016 West Africa Ebola virus disease outbreak. Disaster Heal 3:78–88. doi: 10.1080/21665044.2016.1228326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reference deleted.
- 3.WHO. 3 June 2019. Ebola DRC KIVU 2018 dashboard. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 4.Henao-Restrepo AM, Longini IM, Egger M, Dean NE, Edmunds WJ, Camacho A, Carroll MW, Doumbia M, Draguez B, Duraffour S, Enwere G, Grais R, Gunther S, Hossmann S, Kondé MK, Kone S, Kuisma E, Levine MM, Mandal S, Norheim G, Riveros X, Soumah A, Trelle S, Vicari AS, Watson CH, Kéïta S, Kieny MP, Røttingen J-A. 2015. Efficacy and effectiveness of an rVSV-vectored vaccine expressing Ebola surface glycoprotein: interim results from the Guinea ring vaccination cluster-randomised trial. Lancet 386:857–866. doi: 10.1016/S0140-6736(15)61117-5. [DOI] [PubMed] [Google Scholar]
- 5.Wells CR, Pandey A, Parpia AS, Fitzpatrick MC, Meyers LA, Singer BH, Galvani AP. 2019. Ebola vaccination in the Democratic Republic of the Congo. Proc Natl Acad Sci U S A 116:10178–10183. doi: 10.1073/pnas.1817329116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saphire EO, Schendel SL, Gunn BM, Milligan JC, Alter G. 2018. Antibody-mediated protection against Ebola virus. Nat Immunol 19:1169–1178. doi: 10.1038/s41590-018-0233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhn JH, Becker S, Ebihara H, Geisbert TW, Johnson KM, Kawaoka Y, Lipkin WI, Negredo AI, Netesov SV, Nichol ST, Palacios G, Peters CJ, Tenorio A, Volchkov VE, Jahrling PB. 2010. Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch Virol 155:2083–2103. doi: 10.1007/s00705-010-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez A, Kiley MP, Holloway BP, Auperin DD. 1993. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res 29:215–240. doi: 10.1016/0168-1702(93)90063-S. [DOI] [PubMed] [Google Scholar]
- 9.Elliott LH, Sanchez A, Holloway BP, Kiley MP, McCormick JB. 1993. Ebola protein analyses for the determination of genetic organization. Arch Virol 133:423–436. doi: 10.1007/BF01313780. [DOI] [PubMed] [Google Scholar]
- 10.Mühlberger E, Trommer S, Funke C, Volchkov V, Klenk HD, Becker S. 1996. Termini of all mRNA species of Marburg virus: sequence and secondary structure. Virology 223:376–380. doi: 10.1006/viro.1996.0490. [DOI] [PubMed] [Google Scholar]
- 11.Weik M, Modrof J, Klenk H-D, Becker S, Mühlberger E. 2002. Ebola virus VP30-mediated transcription is regulated by RNA secondary structure formation. J Virol 76:8532–8539. doi: 10.1128/JVI.76.17.8532-8539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whelan SPJ, Barr JN, Wertz GW. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol 283:61–119. doi: 10.1007/978-3-662-06099-5_3. [DOI] [PubMed] [Google Scholar]
- 13.Jordan PC, Liu C, Raynaud P, Lo MK, Spiropoulou CF, Symons JA, Beigelman L, Deval J. 2018. Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLoS Pathog 14:e1006889. doi: 10.1371/journal.ppat.1006889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tekes G, Rahmeh AA, Whelan SPJ. 2011. A freeze frame view of vesicular stomatitis virus transcription defines a minimal length of RNA for 5′ processing. PLoS Pathog 7:e1002073. doi: 10.1371/journal.ppat.1002073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogino T, Banerjee AK. 2007. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol Cell 25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 16.Shatkin AJ. 1976. Capping of eucaryotic mRNAs. Cell 9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 17.Schuberth-Wagner C, Ludwig J, Bruder AK, Herzner A-M, Zillinger T, Goldeck M, Schmidt T, Schmid-Burgk JL, Kerber R, Wolter S, Stümpel J-P, Roth A, Bartok E, Drosten C, Coch C, Hornung V, Barchet W, Kümmerer BM, Hartmann G, Schlee M. 2015. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2′O-methylated self RNA. Immunity 43:41–51. doi: 10.1016/j.immuni.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devarkar SC, Wang C, Miller MT, Ramanathan A, Jiang F, Khan AG, Patel SS, Marcotrigiano J. 2016. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc Natl Acad Sci U S A 113:596–601. doi: 10.1073/pnas.1515152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson B, VanBlargan LA, Xu W, White JP, Shan C, Shi P-Y, Zhang R, Adhikari J, Gross ML, Leung DW, Diamond MS, Amarasinghe GK. 2018. Human IFIT3 modulates IFIT1 RNA binding specificity and protein stability. Immunity 48:487–499.e5. doi: 10.1016/j.immuni.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi YJ, Bowman JW, Jung JU. 2018. A talented duo: IFIT1 and IFIT3 patrol viral RNA caps. Immunity 48:474–476. doi: 10.1016/j.immuni.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shuman S. 2002. What messenger RNA capping tells us about eukaryotic evolution. Nat Rev Mol Cell Biol 3:619–625. doi: 10.1038/nrm880. [DOI] [PubMed] [Google Scholar]
- 22.Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC, Whelan S. 2015. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilman MSA, Liu C, Fung A, Behera I, Jordan P, Rigaux P, Ysebaert N, Tcherniuk S, Sourimant J, Eléouët J-F, Sutto-Ortiz P, Decroly E, Roymans D, Jin Z, McLellan JS. 2019. Structure of the respiratory syncytial virus polymerase complex. Cell 179:193–204.e14. doi: 10.1016/j.cell.2019.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan J, Qian X, Lattmann S, El Sahili A, Yeo TH, Jia H, Cressey T, Ludeke B, Noton S, Kalocsay M, Fearns R, Lescar J. 2020. Structure of the human metapneumovirus polymerase phosphoprotein complex. Nature 577:275–279. doi: 10.1038/s41586-019-1759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdella R, Aggarwal M, Okura T, Lamb RA, He Y. 2020. Structure of a paramyxovirus polymerase complex reveals a unique methyltransferase-CTD conformation. Proc Natl Acad Sci U S A 117:4931–4941. doi: 10.1073/pnas.1919837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahmeh AA, Schenk AD, Danek EI, Kranzusch PJ, Liang B, Walz T, Whelan SPJ. 2010. Molecular architecture of the vesicular stomatitis virus RNA polymerase. Proc Natl Acad Sci U S A 107:20075–20080. doi: 10.1073/pnas.1013559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selisko B, Potisopon S, Agred R, Priet S, Varlet I, Thillier Y, Sallamand C, Debart F, Vasseur J-J, Canard B. 2012. Molecular basis for nucleotide conservation at the ends of the dengue virus genome. PLoS Pathog 8:e1002912. doi: 10.1371/journal.ppat.1002912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paesen GC, Collet A, Sallamand C, Debart F, Vasseur J-J, Canard B, Decroly E, Grimes JM. 2015. X-ray structure and activities of an essential Mononegavirales L-protein domain. Nat Commun 6:8749. doi: 10.1038/ncomms9749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egloff M-P, Benarroch D, Selisko B, Romette J-L, Canard B. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J 21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi P-Y. 2006. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol 80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi P-Y, Li H. 2007. Structure and function of flavivirus NS5 methyltransferase. J Virol 81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahmeh AA, Li J, Kranzusch PJ, Whelan S. 2009. Ribose 2′-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J Virol 83:11043–11050. doi: 10.1128/JVI.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin B, Canard B, Decroly E. 2017. Filovirus proteins for antiviral drug discovery: structure/function bases of the replication cycle. Antiviral Res 141:48–61. doi: 10.1016/j.antiviral.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Ringeard M, Marchand V, Decroly E, Motorin Y, Bennasser Y. 2019. FTSJ3 is an RNA 2′-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature 565:500–504. doi: 10.1038/s41586-018-0841-4. [DOI] [PubMed] [Google Scholar]
- 35.Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, Carver T, Asel E, Springer BA, Lane P, Salemme FR. 2001. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen 6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 36.Martin B, Coutard B, Guez T, Paesen GC, Canard B, Debart F, Vasseur J-J, Grimes JM, Decroly E. 2018. The methyltransferase domain of the Sudan ebolavirus L protein specifically targets internal adenosines of RNA substrates, in addition to the cap structure. Nucleic Acids Res 46:7902–7912. doi: 10.1093/nar/gky637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao X, Shuman S. 1994. Intrinsic RNA (guanine-7) methyltransferase activity of the vaccinia virus capping enzyme D1 subunit is stimulated by the D12 subunit. Identification of amino acid residues in the D1 protein required for subunit association and methyl group transfer. J Biol Chem 269:24472–24479. [PubMed] [Google Scholar]
- 38.De la Peña M, Kyrieleis OJP, Cusack S. 2007. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. EMBO J 26:4913–4925. doi: 10.1038/sj.emboj.7601912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higman MA, Christen LA, Niles EG. 1994. The mRNA (guanine-7-)methyltransferase domain of the vaccinia virus mRNA capping enzyme. Expression in Escherichia coli and structural and kinetic comparison to the intact capping enzyme. J Biol Chem 269:14974–14981. [PubMed] [Google Scholar]
- 40.Chen Y, Su C, Ke M, Jin X, Xu L, Zhang Z, Wu A, Sun Y, Yang Z, Tien P, Ahola T, Liang Y, Liu X, Guo D. 2011. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog 7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aouadi W, Blanjoie A, Vasseur J-J, Debart F, Canard B, Decroly E. 2017. Binding of the methyl donor S-adenosyl-l-methionine to Middle East respiratory syndrome coronavirus 2′-O-methyltransferase nsp16 promotes recruitment of the allosteric activator nsp10. J Virol 91:e02217-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramachandran V, Chen X. 2008. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirino Y, Mourelatos Z. 2007. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol 14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- 44.Prusiner P, Yathindra N, Sundaralingam M. 1974. Effect of ribose O(2′)-methylation on the conformation of nucleosides and nucleotides. Biochim Biophys Acta Nucleic Acids Protein Synth 366:115–123. doi: 10.1016/0005-2787(74)90325-6. [DOI] [PubMed] [Google Scholar]
- 45.Cooper DA, Jha BK, Silverman RH, Hesselberth JR, Barton DJ. 2014. Ribonuclease L and metal-ion-independent endoribonuclease cleavage sites in host and viral RNAs. Nucleic Acids Res 42:5202–5216. doi: 10.1093/nar/gku118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Motorin Y, Muller S, Behm-Ansmant I, Branlant C. 2007. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol 425:21–53. doi: 10.1016/S0076-6879(07)25002-5. [DOI] [PubMed] [Google Scholar]
- 47.Morin B, Whelan S. 2014. Sensitivity of the polymerase of vesicular stomatitis virus to 2′ substitutions in the template and nucleotide triphosphate during initiation and elongation. J Biol Chem 289:9961–9969. doi: 10.1074/jbc.M113.542761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong H, Chang DC, Hua MHC, Lim SP, Chionh YH, Hia F, Lee YH, Kukkaro P, Lok S-M, Dedon PC, Shi P-Y. 2012. 2′-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog 8:e1002642. doi: 10.1371/journal.ppat.1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elliott BA, Ho H-T, Ranganathan SV, Vangaveti S, Ilkayeva O, Abou Assi H, Choi AK, Agris PF, Holley CL. 2019. Modification of messenger RNA by 2′-O-methylation regulates gene expression in vivo. Nat Commun 10:3401. doi: 10.1038/s41467-019-11375-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barral K, Sallamand C, Petzold C, Coutard B, Collet A, Thillier Y, Zimmermann J, Vasseur J-J, Canard B, Rohayem J, Debart F, Decroly E. 2013. Development of specific dengue virus 2′-O- and N7-methyltransferase assays for antiviral drug screening. Antiviral Res 99:292–300. doi: 10.1016/j.antiviral.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Thillier Y, Decroly E, Morvan F, Canard B, Vasseur J-J, Debart F. 2012. Synthesis of 5′ cap-0 and cap-1 RNAs using solid-phase chemistry coupled with enzymatic methylation by human (guanine-N7)-methyl transferase. RNA 18:856–868. doi: 10.1261/rna.030932.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bollati M, Milani M, Mastrangelo E, Ricagno S, Tedeschi G, Nonnis S, Decroly E, Selisko B, de Lamballerie X, Coutard B, Canard B, Bolognesi M. 2009. Recognition of RNA cap in the Wesselsbron virus NS5 methyltransferase domain: implications for RNA-capping mechanisms in Flavivirus. J Mol Biol 385:140–152. doi: 10.1016/j.jmb.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 53.Züst R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beelman CA, Parker R. 1995. Degradation of mRNA in eukaryotes. Cell 81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 55.Fischer PM. 2009. Cap in hand: targeting eIF4E. Cell Cycle 8:2535–2541. doi: 10.4161/cc.8.16.9301. [DOI] [PubMed] [Google Scholar]