Echovirus 30 (E30) is a prevalent enterovirus causing regular outbreaks in both children and adults in different parts of the world. It is therefore surprising that relatively little is known of its infectious entry pathway. We set out to generate a cDNA clone and gradient purified the virus in order to study the early entry events in human cells. We have recently studied other enterovirus B group viruses, like echovirus 1 (EV1) and coxsackievirus A9 (CVA9), and found many similarities between those viruses, allowing us to define a so-called “enterovirus entry pathway.” Here, E30 is reminiscent of these viruses, for example, by not relying on acidification for infectious entry. However, despite not using the clathrin entry pathway, E30 accumulates in classical early endosomes.
KEYWORDS: aseptic meningitis, early entry, echovirus 30, enterovirus
ABSTRACT
Echovirus 30 (E30), a member of the enterovirus B species, is a major cause of viral meningitis, targeting children and adults alike. While it is a frequently isolated enterovirus and the cause of several outbreaks all over the world, surprisingly little is known regarding its entry and replication strategy within cells. In this study, we used E30 strain Bastianni (E30B) generated from an infectious cDNA clone in order to study early entry events during infection in human RD cells. E30B required the newly discovered Fc echovirus receptor (FcRn) for successful infection, but not the coxsackievirus and adenovirus receptor (CAR) or decay-accelerating factor (DAF), although an interaction with DAF was observed. Double-stranded RNA replication intermediate was generated between 2 and 3 h postinfection (p.i.), and viral capsid production was initiated between 4 and 5 h p.i. The drugs affecting Rac1 (NSC 23766) and cholesterol (filipin III) compromised infection, whereas bafilomycin A1, dyngo, U-73122, wortmannin, and nocodazole did not, suggesting the virus follows an enterovirus-triggered macropinocytic pathway rather than the clathrin pathway. Colocalization with early endosomes and increased infection due to constitutively active Rab5 expression suggests some overlap and entry to classical early endosomes. Taken together, these results suggest that E30B induces an enterovirus entry pathway, leading to uncoating in early endosomes.
IMPORTANCE Echovirus 30 (E30) is a prevalent enterovirus causing regular outbreaks in both children and adults in different parts of the world. It is therefore surprising that relatively little is known of its infectious entry pathway. We set out to generate a cDNA clone and gradient purified the virus in order to study the early entry events in human cells. We have recently studied other enterovirus B group viruses, like echovirus 1 (EV1) and coxsackievirus A9 (CVA9), and found many similarities between those viruses, allowing us to define a so-called “enterovirus entry pathway.” Here, E30 is reminiscent of these viruses, for example, by not relying on acidification for infectious entry. However, despite not using the clathrin entry pathway, E30 accumulates in classical early endosomes.
INTRODUCTION
Meningeal inflammation lacking an identifiable bacterial origin is a common neurological syndrome known as aseptic meningitis. Its clinical course, however similar, is generally milder than that of its bacterial counterpart; nonetheless, viral meningitis occurs more frequently and leads to the hospitalization of 26,000 to 42,000 people every year in the United States alone, thus representing a significant economical and societal burden (1–3). Many different viruses can trigger the development of viral meningitis, such as herpesviruses, influenzaviruses, and arboviruses (4; www.cdc.gov/meningitis/viral.html). Since the introduction of the mumps, measles, and rubella (MMR) combination vaccine in 1988, however, nonpolio enteroviruses have taken over as the leading cause of the disease, accounting for over 90% of all cases in which the etiological agent has been identified (1, 4–7). Among these, group B coxsackieviruses and echoviruses are the most commonly isolated types, in particular, echovirus 30 (E30) (7).
E30, a picornavirus belonging to the Enterovirus B genus, is a frequently isolated, positive-sense RNA virus of approximately 7,500 nucleotides enclosed by a nonenveloped protein capsid. Outbreaks of E30-related aseptic meningitis have been recorded every 3 to 5 years in many regions of the world, including Europe, Asia, and the United States (8–14). E30 is the enterovirus type that, over time, has been most frequently reported in humans with aseptic meningitis and has been demonstrated to form a phylogenetic cluster with other notable echoviruses such as echovirus 21 (E21), echovirus 25 (E25), and echovirus 29 (E29) (2, 15). Moreover, the 5′ noncoding region of E30 shows between 68% (coxsackievirus A24 [CVA24]) and 93% (coxsackievirus B3 [CVB3]) homology with other human enteroviruses and appears to contain some coxsackie B-like genomic features (16). Despite often being the subject of medical and epidemiological reports, E30 has been grievously overlooked with regard to its life cycle and infection mechanisms. Using an infectious E30 strain Bastianni (E30B) cDNA clone, this project aimed to study early events in the echovirus life cycle, as well as to pinpoint key cellular components necessary for viral entry into the host cell. We show that E30B represents a typical enterovirus B group virus using an enterovirus-triggered macropinocytic entry pathway leading to rapid replication, which does not require endosomal acidification to facilitate infection. However, it sets itself apart from its closest enterovirus relatives by showing accumulation in classical early endosomes. Being the first report detailing the mechanism of early entry and infection by E30B, this study may open the door to a deeper understanding of the life cycle and infection mechanisms of this pathogen.
RESULTS
E30B displays efficient replication and infection kinetics in human RD cells.
E30B virus stocks were produced from a newly constructed cDNA clone, as described in Materials and Methods. Transmission electron microscopy (TEM) visualization of the negatively stained, gradient-purified E30B revealed typical enteroviral particles, as E30B preparations consisted mainly of intact icosahedral viral particles with a low quantity of empty capsids (Fig. 1A and B). In order to visualize the entry and life cycle of E30B, we performed immunolabeling of E30B-infected RD and A549 cells during various time points postinfection (p.i.). Labeling of the replication intermediate using J2, an antibody specifically geared toward double-stranded RNA (dsRNA), showed that the earliest signs of viral replication appeared between 2 and 3 h p.i. in both RD cells (Fig. 1C) and A549 cells (data not shown). Subsequent quantification of the dsRNA signal using a larger number of cells from confocal images showed that dsRNA was already detectable at 2.5 h p.i., and the intensity of the signal increased exponentially as the infection progressed (Fig. 1D).
FIG 1.
E30B displays efficient replication and infection kinetics. (A, B) Transmission electron microscope imaging of purified virus particles. Scale bars are 500 nm (A) and 100 nm (B). (C) Immunofluorescence staining of RD cells at 2 h, 3 h, and 7 h p.i. with E30B. The presence of dsRNA indicates viral replication. Red, dsRNA (J2); blue, nuclei (4′,6-diamidino-2-phenylindole [DAPI]). Scale bar is 15 μm. (D) Time-resolved quantification of intracellular dsRNA accumulation in RD cells after E30B infection through measurement of the anti-dsRNA (J2) signal intensity. Results are presented as mean values of 12 areas containing 5 to 6 cells each (± standard error of the mean [SEM]). (E) Time-resolved quantitative RT-PCR following the intracellular accumulation of E30B RNA in RD cells. A high CT value corresponds to a small amount of intracellular viral RNA. Results are presented as mean values of 3 replicates (± standard deviation [SD]).
Growth curve analysis through quantitative reverse transcriptase PCR (RT-PCR) monitoring the E30B infection progression in RD cells also showed a low viral load before 3 h p.i., as evidenced by the high qRT-PCR cycle threshold (CT) value, followed by an increase in intracellular E30B RNA starting from 4 h p.i., confirming the dsRNA immunofluorescence (IF) labeling (Fig. 1E). These results indicated that E30B adapted extremely well to RD cell culture and reached a high viral titer.
E30B infection was also followed using confocal microscopy by labeling the capsid with antibodies against VP1. E30B capsid protein could be visualized using the monoclonal rhesus monkey antiserum from ATCC originally prepared against human E30 virus (Fig. 2). The vesicular label was scattered and mostly peripheral for the first 4 h p.i., after which the cytoplasmic, more widespread signal increased. The labeling was also performed with the monoclonal mouse antiserum (clone 5-D8/1; Dako) reactive against several members of the enterovirus B group virus VP1 capsid proteins, which showed a similar distribution in infected cells (data not shown). Together, these data support the notion that E30B appears to exhibit similar efficient replication and infection kinetics as other members of the enterovirus B genus (17, 18).
FIG 2.

Immunofluorescence staining of the E30B capsid in infected RD cells. Infected cells are marked with an asterisk (*). Purple, VP1 capsid protein (antibody made in rhesus monkeys); blue, nuclei (DAPI). Scale bar is 10 μm.
E30B uses DAF as its attachment receptor.
Members of the CVB cluster within the enterovirus B genus utilize the coxsackievirus and adenovirus receptor (CAR) to attach to and enter their respective host cells (19–21). While some enteroviruses also interact with the decay-accelerating factor (DAF) receptor, this interaction in itself is often insufficient for virus entry into the cell (22–24). We performed a radioactive E30B receptor binding assay to assess the propensity for attachment of E30B to CAR and DAF. Based on this experiment, the virus does not bind to CAR but preferentially attaches to the DAF receptor, a feature that is in line with previous findings (Fig. 3) (22, 25, 26). CHO-cells stably transfected with CAR (CHO-CAR) or DAF (CHO-DAF) were confirmed for strong DAF or CAR expression, respectively, using immunofluorescence and fluorescence-activated cell sorter (FACS) (data not shown).
FIG 3.

E30B binding assay. Fifty thousand counts per minute of metabolically labeled E30B were bound on ice to 150,000 CHO cells for 1 h and washed. Results are presented as mean values of 3 replicates (± SEM).CHO cells stably transfected with CAR (CHO-CAR) or DAF (CHO-DAF) were tested for strong DAF or CAR expression by immunofluorescence and FACS (data not shown).
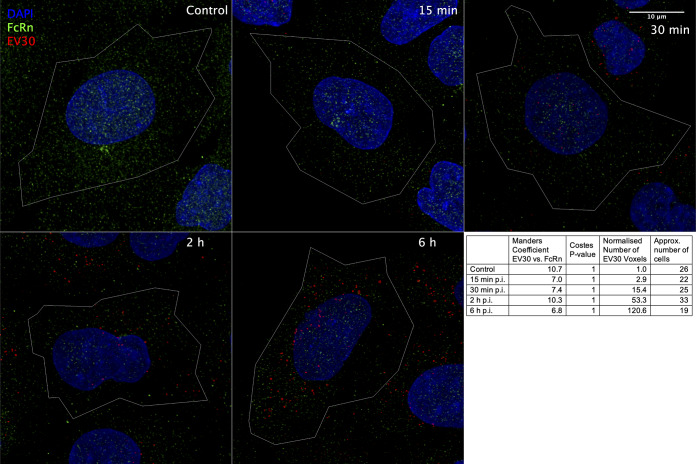
It has recently been shown that CD64, the Fc receptor (FcRn), acts as a pan-receptor for all echoviruses, including E30 (27, 28). We therefore performed double labeling using antibodies against FcRn and capsid antibodies during E30B infection. Despite the presence of FcRn in A549 cells, we found no difference in the distribution of the receptor in infected cells compared to noninfected cells. The receptor showed a small vesicular appearance in all studied time points. In addition, we found no apparent colocalization of E30B with FcRn at any infected time point (Fig. 4). This was confirmed by careful quantification of colocalization using automatic thresholding for colocalization. The Mander’s coefficient was kept at a maximum of 10% in all studied time points, including in the noninfected control cells, suggesting that the colocalization was caused by the background noise.
FIG 4.
Colocalization analysis of E30B and FcRn in A549 cells. Example of the localization of E30B and FcRn signals from representative cells for each time point shown as a maximum intensity projection. The Mander’s coefficient represents the percentage of E30B voxels colocalizing with the FcRn voxels.
As this was quite unexpected due to recent results on the importance of FcRn as an echoviral receptor also for E30B, we decided to perform small interfering RNA (siRNA) knockdown of the receptors studied here: FcRn, DAF, and CAR. Despite having no effect on the cellular distribution of FcRn during E30B infection, the siRNA treatment of FcRn completely abolished E30B infection in A549 cells (Fig. 5). In addition, siRNA knockdown of CAR or DAF, respectively, did not affect the infection of E30B. Furthermore, we used another enterovirus, namely CVB5, as a control, as it has been shown to use the CAR and DAF receptors, but not FcRn, during infection (20, 23, 28). Indeed, our results showed that, in contrast to E30B, CVB5 infection was not affected by FcRn knockdown, but instead, the infection was clearly decreased by CAR or DAF siRNA treatment (Fig. 5). Taken together, these results show that E30B requires FcRn for successful infection, although DAF, unlike CAR, may function as a coreceptor for attachment.
FIG 5.

The effect of CAR, DAF, and FcRn siRNA knockdown on E30B infection. A549 cells were transfected for 48 h with pooled siRNAs against CAR, DAF, or FcRn or with negative control siRNA (CTRLsi) and infected with E30B or CVB5 for 6 h. Representative image of Western blotting where the infection was detected by immunolabeling of VP1 and γ-tubulin as a loading control (top). Quantification of the infection from Western blots by normalizing the VP1 signal to γ-tubulin (bottom). Results are presented as mean values of 3 replicates (± SEM). Statistical significance was determined using an unpaired t-test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Effects of pharmacological inhibitors on early E30B entry.
Other members of the enterovirus B subgroup have been previously shown to prefer the nonclathrin pathway to enter their respective host cells (21, 29–31). Through the use of chemical inhibitors known to affect the action of several key elements of cell entry, we attempted to assess which cellular components or pathways are indispensable for virus infection and replication to occur. Subconfluent monolayers of RD cells were treated with the different compounds for 30 min prior to virus addition, after which the infection was allowed to proceed for 6 h at 37°C. After incubation, the cells were fixed and labeled with a pan-enteroviral VP1 capsid protein antibody. Immunofluorescent labeling of the viral capsid allowed visual distinction between infected and uninfected cells using confocal microscopy. The entry inhibitor drug NSC 23766 (inhibiting Rac1) drastically reduced the pathogen’s capacity for infection, suggesting that this cellular component is essential for virus entry. In contrast, phosphoinositide 3-kinases (PI3K), endosomal acidification, phospholipase C activation, dynamin, and microtubule (de)polymerization appeared not to influence E30B infection, as evidenced by the use of wortmannin, bafilomycin A1, U-73122, Dyngo 4a, and nocodazole, respectively (Fig. 6).
FIG 6.
Pharmacological inhibition of early E30B entry. Immunofluorescence staining of RD cells 6 h p.i. with E30B. Cells were pretreated with inhibitory chemicals 30 min before addition of the virus. The presence of capsid indicates viral replication. Red, viral capsid (Dako); blue, nuclei (DAPI). Scale bar is 20 μm.
None of the compounds significantly affected cell survival in the used concentration compared to a control group, as evidenced by the evaluation of the cell toxicity assay (Fig. 7). This indicated the observed cell deaths were natural rather than a toxic chemical effect.
FIG 7.
Cell viability assay of RD cells treated with pharmacological compounds. Cell viability measurement of RD cells 6.5 h after treatment with indicated inhibitory chemicals. Results are presented as mean values of 6 replicates (± SD). CTRL, untreated control cells.
To confirm the confocal microscopy data, the capacity of these drugs to interfere with viral replication was further evaluated by quantification of the amount of intracellular viral RNA using qRT-PCR (Fig. 8). As previously shown, NSC 23766 affected RNA replication, confirming our previous microscopy results. The treatment with NSC 23766 prevented RNA replication, resulting in an amount of viral RNA that was below the detectable level.
FIG 8.
Pharmacological inhibition of early E30B entry. Quantitative RT-PCR measuring the intracellular accumulation of E30B RNA in RD cells treated with pharmacological inhibitors, 6 h p.i. Cells were pretreated with inhibitory chemicals 30 min before addition of the virus. A high CT value corresponds to a small amount of intracellular viral RNA. Results are presented as mean values of 3 replicates (± SD). POS, positive control for infection without the presence of the vehicle; NEG, negative control for infection; DMSO, dimethyl sulfoxide, positive control for infection in the presence of the vehicle.
In addition, the effect of the cholesterol-modifying drug filipin III on E30B infection was studied using qRT-PCR (Fig. 9A). The results showed that filipin treatment decreased replication, as the CT value increased from 20 for the control infection to 25 for the filipin treatment, corresponding to a 30-fold decrease in viral RNA amount. The results also showed that the lowest effective concentration of filipin was not cytotoxic to RD cells (Fig. 9B).
FIG 9.
The effect of the cholesterol-modifying drug filipin on E30B infection. (A) Quantitative RT-PCR measuring the intracellular accumulation of E30B RNA in RD cells treated with different concentrations of the caveolae pathway inhibitor filipin, 6 h p.i. Results are presented as mean values of 3 replicates (± SD). CTRL E30B, control infection without filipin. Statistical significance was determined using an unpaired t-test. ****, P < 0.0001. (B) Cell viability assay of RD cells after treatment with different filipin concentrations for 6 h. Results are presented as mean values of 3 replicates (± SD). CTRL, untreated control cells.
E30B colocalizes with early endosomes during early entry.
The early endosomes are a preferred sorting station for several incoming vesicles, regardless of the origin of the plasma membrane-derived vesicle. In our earlier studies with echovirus 1 (EV1) and coxsackievirus A9 (CVA9), we found negligible colocalization of the viruses with early endosomes (17, 32). Our results here with bafilomycin A1 and nocodazole showed that acidification of the endosomal structures and microtubule-dependent targeting of E30B to the perinuclear region was not necessary for infection. However, we were curious to find out if E30B would still enter the early sorting or early recycling endosomes; therefore, we infected RD cells with E30B and used confocal immunofluorescence microscopy to visualize possible colocalization of the virus with early endosomal antigen (EEA1) for early-sorting endosomes and the transferrin receptor for recycling early endosomes (Fig. 10). After an incubation period of 5 min, colocalization of E30B with the endosomal markers was rather low; however, this colocalization increased dramatically after 30 min, suggesting that E30B does indeed invade the early endosomal compartments but with delayed kinetics in comparison to cargo relying on clathrin-dependent entry. After 5 min, E30B colocalized to some extent both with internalized transferrin as well as with EEA1. Also, transferrin and EEA1 showed good colocalization of their signals, which is expected given that transferrin passes the sorting early endosomes on its way to recycling early endosomes. In contrast, as previously described, CVA9 did not colocalize with either of the endosomal markers, indicating that CVA9, unlike E30B, does not enter the early endosomal compartments at any time (17). Interestingly, after 30 min of E30B entry, there was much higher colocalization between transferrin and EEA1, and the volume of the colocalized structures had increased due to virus infection. This suggests that transferrin recycling and the overall dynamics of early endosomes were affected by the E30B infection.
FIG 10.
E30B colocalizes with early endosomes during early entry. E30, or CVA9 as a control, was bound to RD cells on ice and washed, and incubation was continued at 37°C with transferrin-Alexa Fluor 488 for 5 or 30 min, after which cells were fixed and labeled also for EEA1. Green, transferrin receptor (transferrin-Alexa Fluor 488); red, early endosomal antigen (EEA1); purple, VP1 capsid protein (antibody made in rhesus monkeys). Scale bar is 10 μm.
Due to the involvement of early endosomes in E30B infection, we investigated the role of Rab5 in E30B infection by transfecting RD cells with different Rab5 constructs (Fig. 11). This small GTPase is important for the dynamics of early endosomes, particularly for their homotypic fusion (33). In addition, Rab5 and some of its effectors have been shown to regulate macropinosome dynamics (34, 35). Our experiment showed that E30B infection was approximately 40% lower (P < 0.01) in cells transfected with a dominant negative Rab5 construct (pEGFP-Rab5-S34N) compared to the wild-type Rab5 control. In addition, overexpression of the constitutively active Rab5 gene markedly improved E30B infection, as transfection with the constitutively active Rab5 (pEYFP-Rab5-Q79L) resulted in a circa 70% increase of infection compared to the wild-type Rab5 (P < 0.05). In contrast, the infection of CVA9 was less affected by the dominant negative Rab5 (P < 0.05). and in comparison to E30B, constitutively active Rab5 decreased the infection of CVA9, which has also been previously shown (17). Altogether, these results further suggest that, in contrast to CVA9, E30B uses early endosomes as a route of entry.
FIG 11.
The effect of Rab5 on E30B infection. RD cells were transfected with a wild-type (pEGFP-Rab5-WT), a dominant negative (pEGFP-Rab5-S34N), or a constitutively active (pEGFP-Rab5-Q79L) Rab5 construct. Cells and plasmids were incubated for 48 h to allow for stable expression, followed by infection for 6 h with E30B or CVA9, after which the infection was detected using immunofluorescence microscopy. The infection percentage of transfected cells was quantified based on VP1 signal, and 350 to 550 transfected cells were calculated per sample in total. Results are normalized to wild-type control and presented as mean values of 3 replicates (± SEM). Statistical significance was determined using an arcsine square root transformation followed by an unpaired t-test. *, P < 0.05; ** P < 0.01.
DISCUSSION
Despite its role as a principal cause of viral meningitis and the consequent extensive epidemiological and diagnostic attention it has received, the life cycle and replication mechanics of E30 have long been overlooked. Here, we report the development of a novel infectious cDNA clone—the first of its kind, to the best of our knowledge—which actively replicates and infects in cell culture, allowing us to study and investigate early entry events in E30 infection using immunofluorescence and confocal microscopy.
Time-resolved analysis of the early infection progression suggested that E30B shows very similar infection and replication kinetics to other enterovirus B members, a notion that is supported by our findings regarding dsRNA production and initiation of viral replication (Fig. 1) (17, 18).
Members of the CVB cluster within the enterovirus B genus tend to favor the CAR receptor to facilitate attachment and entry into their respective host cells (19–21). In some cases, the DAF receptor may function as a coreceptor for attachment, but this interaction in itself is often insufficient to establish infection (22–24). We found that E30B does not bind to CAR and appears to attach to DAF on the plasma membrane, which is in accordance with previous findings (Fig. 3) (22, 25, 26). To our surprise, however, siRNA knockdown of DAF did not prevent the infection of E30B, indicating that although DAF promotes E30B binding to cells, it is not needed for infection. In contrast, the siRNA knockdown of FcRn prevented E30B infection despite the fact that the distribution of the receptor in the cytoplasm did not appear to be affected by E30B infection, nor did it colocalize with the virus after entry. Taken together, these results suggest that while DAF may facilitate the binding of E30B on cells and may function as a coreceptor, the FcRn receptor is absolutely required for successful infection, which is in accordance with previous studies (27, 28).
Early endosomes function as cellular sorting stations that accumulate various uptake vesicles from the plasma membrane. Delivery of these vesicles can occur via different routes, some of which can be hijacked by viruses to facilitate their entry into the host cell. The increased colocalization of E30B with both EEA1 and transferrin suggests that E30B does indeed accumulate in early endosomes. However, there were several lines of evidence to suggest that E30B does not use clathrin-dependent entry to early endosomes. First of all, inhibition of dynamin had no effect on infectivity. Second, the entry to the early endosomes took longer than the typical clathrin cargos, which accumulate in early endosomes within minutes. In addition, expression of the dominant negative small GTPase Rab5 construct decreased E30B infection. Furthermore, transfection of a constitutively active Rab5 construct increased E30B infection, suggesting that an increased amount of homotypic fusion of early endosomes, and supposedly with other incoming vesicles, promoted E30B infection (Fig. 11). This led us to believe that E30B can make use of early endosomes but does not rely on clathrin-dependent entry to facilitate its entry into these organelles.
In addition to entry, the results suggested that the progression of infection was not dependent on acidification, which is typical for the clathrin-dependent pathway. This was proven by the lack of an inhibitory effect of bafilomycin A1. Furthermore, the absence of an effect with nocodazole suggests there is no explicit need for endosomal acidification, microtubule transport to perinuclear regions and late endosomes, or recycling of early endosomes to establish infection. The results thus altogether indicate that entry into early endosomal structures occurs not via clathrin-coated pits but rather through cholesterol-containing raft domains, following a longer route to reach its destination.
In conclusion, E30B proved to be a typical enterovirus by not relying on acidification to ensure infection; E30B showed a preference for DAF over CAR for cellular attachment but demonstrated the Fc receptor to be an absolute requirement for infection. In contrast to EV1 and CVA9, E30B depends on sorting to early endosomes for efficient uncoating and infection. This study represents, to the best of our knowledge, the first in-depth examination of E30B early entry and virus-host cell interaction mechanics. The development of a viable, efficiently replicating E30B clone enables examination of the virus’s life cycle and its behavior in vitro, opening the door to the development of better treatment strategies and care.
MATERIALS AND METHODS
Cells and viruses.
Human RD and A549 cell lines, as well as CHO and GMK cells, were purchased from the American Tissue Culture Collection (ATCC). Additionally, two distinct lines of recombinant CHO cells (stably expressing human coxsackie and adenovirus receptor [CHO-CAR] and human decay-accelerating factor [CHO-DAF]) were previously constructed by H.C. Selinka (36). Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich), 1% penicillin-streptomycin, and 2 mM l-glutamine (Sigma-Aldrich) at 37°C and 5.0% CO2.
An E30B infectious clone was designed using the prototype E30 clone Bastianni sequence (GenBank accession no. AF311938.1) and subsequently produced and cloned into a pUC57 cloning vector (GenScript). A previously described hammerhead ribozyme structure containing an inactivated AscI restriction enzyme site (G39C and C48G) was added at the 5′ untranscribed region (UTR), as well as a 28-adenine nucleotide poly(A) tail at the 3′ UTR (37). The plasmid was introduced into NovaBlue competent cells, which were incubated in LB medium (1% tryptone, 0.5% yeast extract, and 0.5% NaCl) and plated on LB/Amp (100 μg/ml ampicillin) plates. Plasmid DNA was isolated and purified using the GeneJet plasmid miniprep kit (Life Sciences).
Transfection and virus production and purification.
RD cells were grown in a 6-well plate to 70 to 90% confluence, transfected with E30B-pUC57 using Lipofectamine 2000 transfection reagent (Invitrogen), and incubated at 37°C until complete cytopathic effect (CPE) was observed. Following transfection, cells were subjected to three rounds of freeze-thawing to ensure maximal virus yield, and generated viruses were further propagated through five serial passages to ensure adequate adaptation to the cell line. For each passage, 1.0 ml of lysate from either the transfection or the previous passage was added to subconfluent RD cells grown in a T25 flask, which was subsequently incubated for 1 h at room temperature. After incubation, the inoculum was removed, a fresh cell medium was applied, and cells were further cultured at 37°C until CPE was visible, or for a maximum of 5 days. To obtain the purified virus, E30B was propagated in 5-layer flasks containing RD cells and subsequently purified using sucrose gradients as previously described (38). The cell culture medium for virus propagation and purification consisted of serum-free DMEM (Sigma-Aldrich) supplemented with 1% penicillin-streptomycin and 2 mM l-glutamine (Sigma-Aldrich).
Transmission electron microscopy.
Transmission electron microscopy (TEM) imaging was performed as previously described (39). Briefly, Butwar-coated copper grids were hydrophilized through glow discharging with an EMS/SC7620 Mini sputter coater (Quorum Technologies) as per the manufacturer’s instructions before incubation with E30B for 15 s. Excess virus was removed, after which the remaining virions were negatively stained by incubating the grid with 1% phosphotungstic acid in water (pH 7.4) for 1 min. After incubation, excess stain was removed, and E30B (subjected to 5 min of heat treatment at 50°C prior to application) was added. Samples were dried overnight and subsequently evaluated using a JEM-1400 transmission electron microscope (JEOL).
Confocal immunofluorescence imaging.
RD and A549 cells were cultured on coverslips to subconfluency, washed once with phosphate-buffered saline (PBS), and subsequently infected with E30B. After incubation at 37°C, the cells were fixed at selected time points using 4% paraformaldehyde (PFA) for 20 min, permeabilized using 0.2% Triton X-100 for 5 min, and antibody labeled. The samples were mounted using Mowiol (Sigma-Aldrich) containing DABCO (1,4-diazabicyclo[2,2,2]octane, Sigma-Aldrich) and evaluated using an Olympus FluoView FV1000 laser scanning confocal microscope or Leica SP8 with Leica’s lighting-optimized settings using a voxel size of 35 nm in the x-y plane and 245 nm in the z-axis.
The following antibodies were used: mouse monoclonal antisera against enterovirus VP1 capsid protein (catalog no. M7064; Dako), mouse monoclonal antisera against human EEA1 (catalog no. 610457; BD Biosciences), and mouse monoclonal antisera against dsRNA (J2) (catalog no. 10010200; Scicons); rabbit monoclonal antisera against Fc receptor (CD64) (catalog no. ab193148; Abcam) and rabbit polyclonal antisera against EEA1 (catalog no. ab2900; Abcam); rhesus monkey monoclonal antiserum against human echovirus 30 (catalog no. VR-1072 AS/MK; ATCC); Alexa Fluor 488 goat polyclonal IgG against mouse (catalog no. A-11029; Thermo Fisher Scientific); Alexa Fluor 555 goat polyclonal IgG against mouse (catalog no. A-21422; Thermo Fisher Scientific); Alexa Fluor 488 goat polyclonal IgG against rabbit (catalog no. A-11008; Thermo Fisher Scientific); Alexa Fluor 555 goat polyclonal IgG against rabbit (catalog no. A-21428; Thermo Fisher Scientific); and Alexa Fluor 647 goat polyclonal IgG against rhesus monkey (catalog no. 6200-31; SouthernBiotech). Antibody dilutions were prepared in 3% bovine serum albumin (BSA) in PBS.
Receptor binding assay.
Radioactive 35S-labeled E30B was produced as previously described (39). Briefly, RD monolayers were grown to subconfluency, washed once with PBS, and infected for 3 h at 37°C with E30B diluted in low-methionine-cysteine medium supplemented with 1% FBS (Sigma-Aldrich). After incubation, the medium was replaced with low-methionine-cysteine medium supplemented with 1% FBS and 50 μCi/ml of [35S]methionine-cysteine (EasyTag Express 35S protein labeling mix [35S]; PerkinElmer), and infection was allowed to continue for 9 h at 37°C. Cell lysates were collected after repeated freeze-thaw cycles, after which cell debris was pelleted through centrifugation at 4°C using an SL 16R rotor (2,500 × g for 10 min) (Thermo Fisher Scientific). The supernatant was incubated with 0.3% (wt/vol) sodium deoxycholate (DOC) and 0.6% (vol/vol) NP-40 substitute for 30 min on ice. Membrane structures were pelleted through centrifugation at 4°C using an SL 16R rotor (4,000 × g for 10 min), and the supernatant was applied to 40% sucrose cushions. Samples were ultracentrifuged at 4°C using an SW 41 rotor (35,000 rpm for 2.5 h) (Beckman Coulter). The liquid above each cushion, as well as one 500-μl fraction, was discarded, while three subsequent 500-μl fractions were collected and applied to 5 to 20% continuous sucrose gradients. Gradients were subjected to centrifugation at 4°C using an SW 41 rotor (35,000 rpm for 2 h) and fractioned into 500-μl aliquots starting from the top, which were consequently analyzed through the addition of 4 ml of Ultima Gold MV scintillation cocktail (PerkinElmer) and application of the Tri-Carb 2910TR liquid scintillation analyzer scintillation counting method (PerkinElmer).
CHO cells stably transfected with CAR (CHO-CAR) or DAF (CHO-DAF) were tested for strong DAF or CAR expression by immunofluorescence and FACS. Each adherent cell culture was individually detached using trypsin (Sigma-Aldrich) before 150,000 cells per replicate were washed, resuspended in 2 mM MgCl-PBS, and subsequently incubated at 4°C with 50,000 CPM of 35S-labeled E30B (corresponding to a multiplicity of infection [MOI] of 850). After 1 h, cells were washed to remove unbound virions, resuspended in 4 ml of Ultima Gold MV scintillation cocktail, and analyzed using the Tri-Carb 2910TR liquid scintillation analyzer scintillation counting method.
siRNA transfections.
A549 cells were reverse transfected using DharmaFect transfection reagent (Horizon Discovery) according to the manufacturer’s instructions. The pool of three target-specific siRNAs against CAR, DAF, or FcRn (Santa Cruz Biotechnology) or AllStars negative-control siRNA (kindly gifted by the Johanna Ivaska Laboratory, University of Turku, Turku, Finland) were added in a final concentration of 11.4 nM, and the transfection was allowed to proceed for 48 h at 37°C in DMEM supplemented with 10% FBS and 1% GlutaMax. Next, 200 PFU/cell of E30B or coxsackievirus B5 (CVB5) were added in DMEM supplemented with 1% FBS and 1% GlutaMax and bound on ice for 1 h, after which the excess virus was washed away (40). The infection was then allowed to proceed at 37°C in DMEM supplemented with 10% FBS and 1% GlutaMax for 6 h, after which the cells were collected into 2× Laemmli buffer containing β-mercaptoethanol.
SDS-PAGE and Western blotting.
The samples were boiled and separated in a 4 to 20% Mini-Protean TGX stain-free gel (Bio-Rad). Next, the proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore) and blocked overnight with 5% BSA and 0.05% Tween in Tris-buffered saline (TBS). Blots were immunolabeled with mouse monoclonal antisera against enterovirus VP1 capsid protein, and rat monoclonal antisera against γ-tubulin (Abcam) was used as a loading control. The primary antibodies were detected using corresponding horseradish peroxidase-conjugated secondary antibodies (Cell Signaling). Finally, the chemiluminescent substrate SuperSignal West Pico Plus (Thermo Fisher Scientific) was incubated for 5 min, and chemiluminescence was detected using the ChemiDoc MP (Bio-Rad).
Pharmacological inhibitor assay.
RD cells were cultured on coverslips to subconfluency. The cells were washed once with PBS and subsequently incubated at 37.0°C in DMEM supplemented with 50.0 nM bafilomycin A1 (targeting vacuolar type H+-ATPase; catalog no. 196000; Calbiochem), 33.0 μM nocodazole (affecting microtubule assembly/disassembly; catalog no. 487928; Calbiochem), 100.0 μM wortmannin (inhibiting phosphoinositide 3-kinase; catalog no. 681675; Calbiochem), 1.0 mM NSC 23766 (targeting Rac1; catalog no. 2161; Tocris BioScience), 10.0 μM U-73122 (affecting phospholipase C; catalog no. 662035; Calbiochem), or 12.5 μM Dyngo 4a (inhibiting dynamin; catalog no. 120689; Abcam) for 30 min prior to E30B addition (17). The infection assay was carried out for 6 h at 37°C, after which the cells were fixed using 4% PFA for 20 min and antibody labeled, and the presence of virus capsid protein was determined.
The cellular toxicity of the pharmacological inhibitors was evaluated using the CellTiter-Glo luminescent cell viability assay (Promega) according to the manufacturer’s protocol.
Filipin assay.
RD cells were cultured until subconfluency and incubated for 30 min with a 1-, 2-, or 3-μg/ml concentration of filipin III (catalog no. F4767; Sigma-Aldrich). Next, E30B was added onto the cells (200 PFU/cell) and bound on ice for 1 h, after which excess virus was washed away. The infection was allowed to proceed for 6 h in DMEM supplemented with 10% FBS, 1% penicillin-streptomycin, and 1% GlutaMax, also, including 1, 2, or 3 μg/ml of filipin. Finally, the medium was removed, and viral RNA was isolated from lysed cells using the QIAamp viral RNA extraction kit (Qiagen) according to the manufacturer’s instructions. Reverse transcription was carried out for positive-sense RNA using 1.2 μM antisense primer (5′-GAAACACGGACACCCAAAGTA-3′), 20 U Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega), deoxynucleoside triphosphates (dNTPs) (Promega), and 4 U RNasin RNase inhibitor (Promega). Five microliters from the 40-μl reaction mixture were subsequently used in a PCR, which also included SYBR green Supermix (Bio-Rad) and 600 nM of both antisense primer (5′-GAAACACGGACACCCAAAGTA-3′) and sense primer (5′-CGGCCCCTGAATGCGGCTAA-3′). The amplification was carried out on the C1000 Touch thermal cycler with CFX96 Touch real-time PCR detection system (Bio-Rad) using the following protocol: 95°C for 10 min, 40 cycles of 95°C for 15 s to 60°C for 1 min, and a final melting step at 72 to 95°C, 1°C/5 s. The assay also contained negative controls to confirm the specificity of the products.
The cytotoxicity of filipin was evaluated using the CellTiter-Glo luminescent cell viability assay (Promega) according to the manufacturer’s protocol.
Quantification of viral infection.
Viral RNA was extracted from infected RD cell cultures using the QIAamp viral RNA extraction kit according to the manufacturer’s protocol and subsequently copied to cDNA using the TaqMan reverse transcription reagents kit (Applied Biosystems). Two-step qRT-PCRs were carried out on the 7500 real-time PCR system (Applied Biosystems) with 7500 SDS analysis software. Each reaction was prepared using 1 μl cDNA, 1× Power SYBR green master mix (Applied Biosystems), and 200 nM (each) of primers 5UTR-F (5′-CGTTGCGGAGTGTTTCGTTC-3′) and 5UTR-R (5′-TCCGCAGTTAGGATTAGCCG-3′) directed against the 5′ UTR of the genome in a final reaction volume of 20 μl. The following thermocycling program was applied: reverse transcription at 50°C for 2 min, TaqMan DNA polymerase activation, and simultaneous reverse transcriptase inactivation at 95°C for 10 min, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. Each reaction was run in triplicate. Standard curves were generated by running the aforementioned protocol using the E30B cDNA template in triplicate.
Transferrin-EEA1 assay.
RD cells were cultured on coverslips to subconfluency, washed once with PBS, and incubated with purified E30B or CVA9 (Griggs strain) for 1 h on ice. After virus binding, the medium was removed, cells were gently washed with 0.5% BSA-PBS, and 50 μg/ml of transferrin-Alexa Fluor 488 conjugate (transferrin from human serum, Alexa Fluor 488 conjugate; Invitrogen) in DMEM supplemented with 0.2% BSA was added. Infection was carried out at 37°C and terminated at 5-min and 30-min time points by fixing the cells with 4% PFA for 20 min, after which the cells were antibody labeled and imaged.
Plasmid transfections.
RD cells were grown on coverslips to subconfluency. Plasmid transfections were carried out for 48 h at 37°C using Lipofectamine 3000 transfection reagent (Invitrogen) according to the manufacturer’s instructions. The cells were infected with 200 PFU/cell of E30B or CVA9 (Griggs strain) by binding the virus on ice for 1 h in DMEM supplemented with 1% FBS and 1% GlutaMax. After excess virus was washed away, the infection was allowed to proceed for 6 h at 37°C in DMEM supplemented with 10% FBS, 1% GlutaMax, and 1% penicillin-streptomycin. Finally, the cells were fixed using 4% PFA for 20 min and labeled with mouse monoclonal antisera against enterovirus VP1 capsid protein. The infection percentage of transfected cells was quantified by evaluating the presence of viral capsid protein.
Plasmid constructs were obtained from the following sources: the dominant negative (pEGFP-Rab5-S34N) and dominant positive (pEYFP-Rab5-Q79L) Rab5 constructs were procured from Lucas Pelkmans (Department of Molecular Life Sciences, University of Zurich, Zurich, Switzerland), and the wild-type Rab5 (pEGFP-Rab5) was acquired from Miguel Seabra (Faculty of Medicine, National Heart and Lung Institute, Imperial College, London, United Kingdom).
Microscopy data analysis.
Microscope settings were optimized for each channel prior to imaging. Confocal immunofluorescence image analysis was executed using the Fiji free open-source software package (41). For the colocalization analysis, the coloc2 plugin was used to measure the Mander’s correlation and Costes significance with a point spread function (PSF) estimation of 8 pixels and 20 iterations (42, 43). Image analysis was executed using the Fiji free open-source software package (41). To visualize colocalizing pixels between transferrin, EEA1, and E30B or CVA9 (Griggs strain), the open-source software BioImageXD (www.bioimagexd.net/) was used. Thresholding for E30B was set with the help of uninfected controls, and for transferrin and EEA1, they were set manually to not contain background signal.
Statistical analysis.
Statistical sample comparison of proportions and ratios was performed using an arcsine square root transformation to convert the data to be more normally distributed, followed by a paired or unpaired t-test. A P-value of < 0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank Kjell Edman (Linnaeus University, Kalmar, Sweden) for valuable discussions and technical assistance. We also acknowledge Sami Salmikangas (University of Jyväskylä, Jyväskylä, Finland) for technical assistance.
We declare no conflicts of interest.
REFERENCES
- 1.Irani DN. 2008. Aseptic meningitis and viral myelitis. Neurol Clin 26:635–655. doi: 10.1016/j.ncl.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khetsuriani N, Quiroz ES, Holman RC, Anderson LJ. 2003. Viral meningitis-associated hospitalizations in the United States, 1988–1999. Neuroepidemiology 22:345–352. doi: 10.1159/000072924. [DOI] [PubMed] [Google Scholar]
- 3.Pallansch MA, Roos RP. 2007. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p 839–893. In Knipe DM, Howley PM (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 4.Connolly KJ, Hammer SM. 1990. The acute aseptic meningitis syndrome. Infect Dis Clin North Am 4:599–622. [PubMed] [Google Scholar]
- 5.Rotbart HA. 2000. Viral meningitis. Semin Neurol 20:277–292. doi: 10.1055/s-2000-9427. [DOI] [PubMed] [Google Scholar]
- 6.Davidson KL, Ramsay ME. 2003. The epidemiology of acute meningitis in children in England and Wales. Arch Dis Child 88:662–664. doi: 10.1136/adc.88.8.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Logan SA, MacMahon E. 2008. Viral meningitis. BMJ 336:36–40. doi: 10.1136/bmj.39409.673657.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamashita K, Miyamura K, Yamadera S, Kato N, Akatsuka M, Hashido M, Inouye S, Yamazaki S. 1994. Epidemics of aseptic meningitis due to echovirus 30 in Japan. A report of the National Epidemiological Surveillance of Infectious Agents in Japan. Jpn J Med Sci Biol 47:221–239. doi: 10.7883/yoken1952.47.221. [DOI] [PubMed] [Google Scholar]
- 9.Oberste MS, Maher K, Kennett ML, Campbell JJ, Carpenter MS, Schnurr D, Pallansch MA. 1999. Molecular epidemiology and genetic diversity of echovirus type 30 (E30): genotypes correlate with temporal dynamics of E30 isolation. J Clin Microbiol 37:3928–3933. doi: 10.1128/JCM.37.12.3928-3933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thoelen I, Lemey P, Van Der Donck I, Beuselinck K, Lindberg AM, Van Ranst M. 2003. Molecular typing and epidemiology of enteroviruses identified from an outbreak of aseptic meningitis in Belgium during the summer of 2000. J Med Virol 70:420–429. doi: 10.1002/jmv.10412. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2003. Outbreaks of aseptic meningitis associated with echoviruses 9 and 30 and preliminary surveillance reports on enterovirus activity—United States, 2003. MMWR Morb Mortal Wkly Rep 52:761–764. [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention (CDC). 2006. Enterovirus surveillance—United States, 2002–2004. MMWR Morb Mortal Wkly Rep 55:153–156. [PubMed] [Google Scholar]
- 13.McWilliam Leitch EC, Bendig J, Cabrerizo M, Cardosa J, Hyypiä T, Ivanova OE, Kelly A, Kroes AC, Lukashev A, MacAdam A, McMinn P, Roivainen M, Trallero G, Evans DJ, Simmonds P. 2009. Transmission networks and population turnover of echovirus 30. J Virol 83:2109–2118. doi: 10.1128/JVI.02109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broberg EK, Simone B, Jansa J, EU/EEA Member State Contributors. 2018. Upsurge in echovirus 30 detections in five EU/EEA countries, April to September, 2018. Euro Surveill 23:1800537. doi: 10.2807/1560-7917.ES.2018.23.44.1800537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oberste MS, Maher K, Kilpatrick DR, Pallansch MA. 1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol 73:1941–1948. doi: 10.1128/JVI.73.3.1941-1948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diedrich S, Driesel G, Schreier E. 1995. Sequence comparison of echovirus type 30 isolates to other enteroviruses in the 5′ noncoding region. J Med Virol 46:148–152. doi: 10.1002/jmv.1890460212. [DOI] [PubMed] [Google Scholar]
- 17.Huttunen M, Waris M, Kajander R, Hyypiä T, Marjomäki V. 2014. Coxsackievirus A9 infects cells via nonacidic multivesicular bodies. J Virol 88:5138–5151. doi: 10.1128/JVI.03275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pietiäinen V, Marjomäki V, Upla P, Pelkmans L, Helenius A, Hyypiä T. 2004. Echovirus 1 endocytosis into caveosomes requires lipid rafts, dynamin II, and signaling events. Mol Biol Cell 15:4911–4925. doi: 10.1091/mbc.e04-01-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, Horwitz MS, Crowell RL, Finberg RW. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 20.Martino TA, Petric M, Weingartl H, Bergelson JM, Opavsky MA, Richardson CD, Modlin JF, Finberg RW, Kain KC, Willis N, Gauntt CJ, Liu PP. 2000. The coxsackie-adenovirus receptor (CAR) is used by reference strains and clinical isolates representing all six serotypes of coxsackievirus group B and by swine vesicular disease virus. Virology 271:99–108. doi: 10.1006/viro.2000.0324. [DOI] [PubMed] [Google Scholar]
- 21.Marjomäki V, Turkki P, Huttunen M. 2015. Infectious entry pathway of enterovirus B species. Viruses 7:6387–6399. doi: 10.3390/v7122945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergelson JM, Chan M, Solomon KR, St John NF, Lin H, Finberg RW. 1994. Decay-accelerating factor (CD55), a glycosylphosphatidylinositol-anchored complement regulatory protein, is a receptor for several echoviruses. Proc Natl Acad Sci U S A 91:6245–6248. doi: 10.1073/pnas.91.13.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shafren DR, Bates RC, Agrez MV, Herd RL, Burns GF, Barry RD. 1995. Coxsackieviruses B1, B3, and B5 use decay accelerating factor as a receptor for cell attachment. J Virol 69:3873–3877. doi: 10.1128/JVI.69.6.3873-3877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milstone AM, Petrella J, Sanchez MD, Mahmud M, Whitbeck JC, Bergelson JM. 2005. Interaction with coxsackievirus and adenovirus receptor, but not with decay-accelerating factor (DAF), induces A-particle formation in a DAF-binding coxsackievirus B3 isolate. J Virol 79:655–660. doi: 10.1128/JVI.79.1.655-660.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powell RM, Schmitt V, Ward T, Goodfellow I, Evans DJ, Almond JW. 1998. Characterization of echoviruses that bind decay accelerating factor (CD55): evidence that some haemagglutinating strains use more than one cellular receptor. J Gen Virol 79:1707–1713. doi: 10.1099/0022-1317-79-7-1707. [DOI] [PubMed] [Google Scholar]
- 26.Rothe D, Werk D, Niedrig S, Horbelt D, Grunert HP, Zeichhardt H, Erdmann VA, Kurreck J. 2009. Antiviral activity of highly potent siRNAs against echovirus 30 and its receptor. J Virol Methods 157:211–218. doi: 10.1016/j.jviromet.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Morosky S, Wells AI, Lemon K, Evans AS, Schamus S, Bakkenist CJ, Coyne CB. 2019. The neonatal Fc receptor is a pan-echovirus receptor. Proc Natl Acad Sci U S A 116:3758–3763. doi: 10.1073/pnas.1817341116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao X, Zhang G, Liu S, Chen X, Peng R, Dai L, Qu X, Li S, Song H, Gao Z, Yuan P, Liu Z, Li C, Shang Z, Li Y, Zhang M, Qi J, Wang H, Du N, Wu Y, Bi Y, Gao S, Shi Y, Yan J, Zhang Y, Xie Z, Wei W, Gao GF. 2019. Human neonatal Fc receptor is the cellular uncoating receptor for enterovirus B. Cell 177:1553–1565. doi: 10.1016/j.cell.2019.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Triantafilou K, Triantafilou M. 2004. Lipid-raft-dependent coxsackievirus B4 internalization and rapid targeting to the Golgi. Virology 326:6–19. doi: 10.1016/j.virol.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 30.Patel KP, Coyne CB, Bergelson JM. 2009. Dynamin- and lipid raft-dependent entry of decay-accelerating factor (DAF)-binding and non-DAF-binding coxsackieviruses into nonpolarized cells. J Virol 83:11064–11077. doi: 10.1128/JVI.01016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krieger SE, Kim C, Zhang L, Marjomaki V, Bergelson JM. 2013. Echovirus 1 entry into polarized Caco-2 cells depends on dynamin, cholesterol, and cellular factors associated with macropinocytosis. J Virol 87:8884–8895. doi: 10.1128/JVI.03415-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marjomäki V, Pietiäinen V, Matilainen H, Upla P, Ivaska J, Nissinen L, Reunanen H, Huttunen P, Hyypiä T, Heino J. 2002. Internalization of echovirus 1 in caveolae. J Virol 76:1856–1865. doi: 10.1128/jvi.76.4.1856-1865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M. 1992. The small GTPase Rab5 functions as a regulatory factor in the early endocytic pathway. Cell 70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- 34.Feliciano WD, Yoshida S, Straight SW, Swanson JA. 2011. Coordination of the Rab5 cycle on macropinosomes. Traffic 12:1911–1922. doi: 10.1111/j.1600-0854.2011.01280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Egami Y, Taguchi T, Maekawa M, Arai H, Araki N. 2014. Small GTPases and phosphoinositides in the regulatory mechanisms of macropinosome formation and maturation. Front Physiol 5:374. doi: 10.3389/fphys.2014.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selinka HC, Wolde A, Pasch A, Klingel K, Schnorr JJ, Küpper JH, Lindberg AM, Kandolf R. 2002. Comparative analysis of two coxsackievirus B3 strains: putative influence of virus-receptor interactions on pathogenesis. J Med Virol 67:224–233. doi: 10.1002/jmv.2211. [DOI] [PubMed] [Google Scholar]
- 37.Israelsson S, Sävneby A, Ekström JO, Jonsson N, Edman K, Lindberg AM. 2014. Improved replication efficiency of echovirus 5 after transfection of colon cancer cells using an authentic 5′ RNA genome end methodology. Invest New Drugs 32:1063–1070. doi: 10.1007/s10637-014-0136-z. [DOI] [PubMed] [Google Scholar]
- 38.Abraham G, Colonno RJ. 1984. Many rhinovirus serotypes share the same cellular receptor. J Virol 51:340–345. doi: 10.1128/JVI.51.2.340-345.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myllynen M, Kazmertsuk A, Marjomäki V. 2016. A novel open and infectious form of echovirus 1. J Virol 90:6759–6770. doi: 10.1128/JVI.00342-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turkki P, Laajala M, Stark M, Vandesande H, Sallinen-Dal Maso H, Shroff S, Sävneby A, Galitska G, Lindberg AM, Marjomäki V. 2019. Slow infection due to lowering the amount of intact versus empty particles is a characteristic feature of coxsackievirus B5 dictated by the structural proteins. J Virol 93:e01130-19. doi: 10.1128/JVI.01130-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manders EMM, Verbeek FJ, Aten JA. 1993. Measurement of co‐localization of objects in dual‐colour confocal images. J Microsc 169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 43.Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. 2004. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J 86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]