All NNS RNA viruses, including the human pathogens rabies, measles, respiratory syncytial virus, Nipah, and Ebola, possess an essential L-protein cofactor, required to access the N-RNA template and coordinate the various enzymatic activities of L. The polymerase cofactors share a similar modular organization of a soluble N-binding domain and a template-binding domain separated by a central oligomerization domain. Using a prototype of NNS RNA virus gene expression, vesicular stomatitis virus (VSV), we determined the importance of P oligomerization. We find that oligomerization of VSV P is not required for any step of viral mRNA synthesis but is required for efficient RNA replication. We present evidence that this likely occurs through the stage of loading soluble N onto the nascent RNA strand as it exits the polymerase during RNA replication. Interfering with the oligomerization of P may represent a general strategy to interfere with NNS RNA virus replication.
KEYWORDS: genome replication, oligomerization, phosphoprotein, polymerase, vesicular stomatitis virus
ABSTRACT
Nonsegmented negative-strand (NNS) RNA viruses possess a ribonucleoprotein template in which the genomic RNA is sequestered within a homopolymer of nucleocapsid protein (N). The viral RNA-dependent RNA polymerase (RdRP) resides within an approximately 250-kDa large protein (L), along with unconventional mRNA capping enzymes: a GDP:polyribonucleotidyltransferase (PRNT) and a dual-specificity mRNA cap methylase (MT). To gain access to the N-RNA template and orchestrate the LRdRP, LPRNT, and LMT, an oligomeric phosphoprotein (P) is required. Vesicular stomatitis virus (VSV) P is dimeric with an oligomerization domain (OD) separating two largely disordered regions followed by a globular C-terminal domain that binds the template. P is also responsible for bringing new N protomers onto the nascent RNA during genome replication. We show VSV P lacking the OD (PΔOD) is monomeric but is indistinguishable from wild-type P in supporting mRNA transcription in vitro. Recombinant virus VSV-PΔOD exhibits a pronounced kinetic delay in progeny virus production. Fluorescence recovery after photobleaching demonstrates that PΔOD diffuses 6-fold more rapidly than the wild type within viral replication compartments. A well-characterized defective interfering particle of VSV (DI-T) that is only competent for RNA replication requires significantly higher levels of N to drive RNA replication in the presence of PΔOD. We conclude P oligomerization is not required for mRNA synthesis but enhances genome replication by facilitating RNA encapsidation.
IMPORTANCE All NNS RNA viruses, including the human pathogens rabies, measles, respiratory syncytial virus, Nipah, and Ebola, possess an essential L-protein cofactor, required to access the N-RNA template and coordinate the various enzymatic activities of L. The polymerase cofactors share a similar modular organization of a soluble N-binding domain and a template-binding domain separated by a central oligomerization domain. Using a prototype of NNS RNA virus gene expression, vesicular stomatitis virus (VSV), we determined the importance of P oligomerization. We find that oligomerization of VSV P is not required for any step of viral mRNA synthesis but is required for efficient RNA replication. We present evidence that this likely occurs through the stage of loading soluble N onto the nascent RNA strand as it exits the polymerase during RNA replication. Interfering with the oligomerization of P may represent a general strategy to interfere with NNS RNA virus replication.
INTRODUCTION
The 241-kDa large (L) protein of vesicular stomatitis virus (VSV) contains all the enzymatic activities necessary to copy its viral ribonucleoprotein (RNP) template (1–4). That RNP template comprises the genomic RNA completely encased within the viral nucleocapsid (N) protein (5, 6). Responding to specific RNA signals, the RNA-dependent RNA polymerase of L (LRdRP) transcribes the RNP template into a 47-nucleotide (nt) 5′ triphosphate leader RNA and 5 monocistronic mRNAs that are indistinct to host cell mRNA with respect to their 5′ cap and 3′ polyadenylate (7–10). The enzymatic activities necessary for formation of the viral 5′ mRNA cap structure reside within L and differ markedly from those of the host cell and other organisms (11). During viral mRNA cap formation, a GDP:polyribonucleotidyltransferase (LPRNT) transfers the 5′ end of the nascent RNA chain onto a GDP acceptor through a covalent L-pRNA (L-monophosphate RNA) intermediate. The resulting GpppA cap structure is then modified by a dual-specificity L-encoded RNA cap methylase (LMT), which first modifies the ribose-2′-O position associated with first transcribed nucleotide generating GpppAm and subsequently methylate, the guanine ring of the cap at the N7 position, to yield 7mGpppAm (12). The 3′ polyadenylate tail is generated by a chattering mechanism in which the LRdRP reiteratively copies one or more members of an oligo(U) tract resident at the end of each gene (13).
The RNP template is also used to generate full-length genomic RNPs through an antigenomic RNP intermediate. RNA replication requires the LRdRP and a continuous supply of soluble N protein that coats the nascent RNA chain during its synthesis (14). Although the mRNA capping enzymes are not required during replication, as the 5′ end of the replication products are unmodified triphosphate, this process of terminal initiation depends on a priming loop donated from the LPRNT into the LRdRP (4, 15). Despite containing all of the enzymes necessary for RNA synthesis, L cannot copy the N-RNA template without forming a complex with its cofactor, a 29-kDa oligomeric phosphoprotein (P) (1). Complex formation of L with P plays a key role in also organizing the various enzymatic domains of L with respect to one another (16). This defines the minimal viral machinery for RNA synthesis as the N-RNA template and the viral P and L proteins.
Structural studies of the VSV replication machinery highlight the master role of P in orchestrating RNA synthesis. The genomic RNA is completely coated by N, which must be transiently dissociated during copying by L. A globular C-terminal domain of P (PCTD), corresponding to residues 195 to 265, contacts the N-RNA template by binding at a unique interface that forms between adjacent N molecules on the template (17). That PCTD is separated from a largely disordered N-terminal region of P by an oligomerization domain (POD) comprising residues 107 to 177 (18). Although the N-terminal domain of P is largely disordered, complex formation with L defines key contacts between L and two segments of P encompassing residues 49 to 56 and 82 to 105 (19). A short stretch of the N terminus of P (residues 5 to 34) also binds a groove between the N- and C-terminal lobes of monomeric N (N0), occluding binding of RNA (20). This likely reflects how soluble N protein is loaded onto the nascent RNA chain during RNP assembly that is concomitant with viral genome replication.
The central role of P in regulating gene expression prompted us to probe the requirement for its oligomerization in VSV. We deleted the oligomerization domain of P, validated that the resulting protein is monomeric, interrogated the consequences for gene expression in vitro and in cells, and generated a recombinant virus lacking POD. The data demonstrate that viral mRNA synthesis is unaffected by P protein oligomerization, and they identify a pronounced kinetic delay in viral RNA replication for monomeric P. Probing of the underlying mechanism behind this delay demonstrates that the oligomerization of P facilitates genome RNA replication at a lower N protein concentration, likely by directly influencing the loading of N onto the nascent transcript. This work defines a key function for oligomerization of the P protein of VSV that likely extends to other viruses in the order Mononegavirales.
RESULTS
VSV P lacking its oligomerization domain is monomeric in solution.
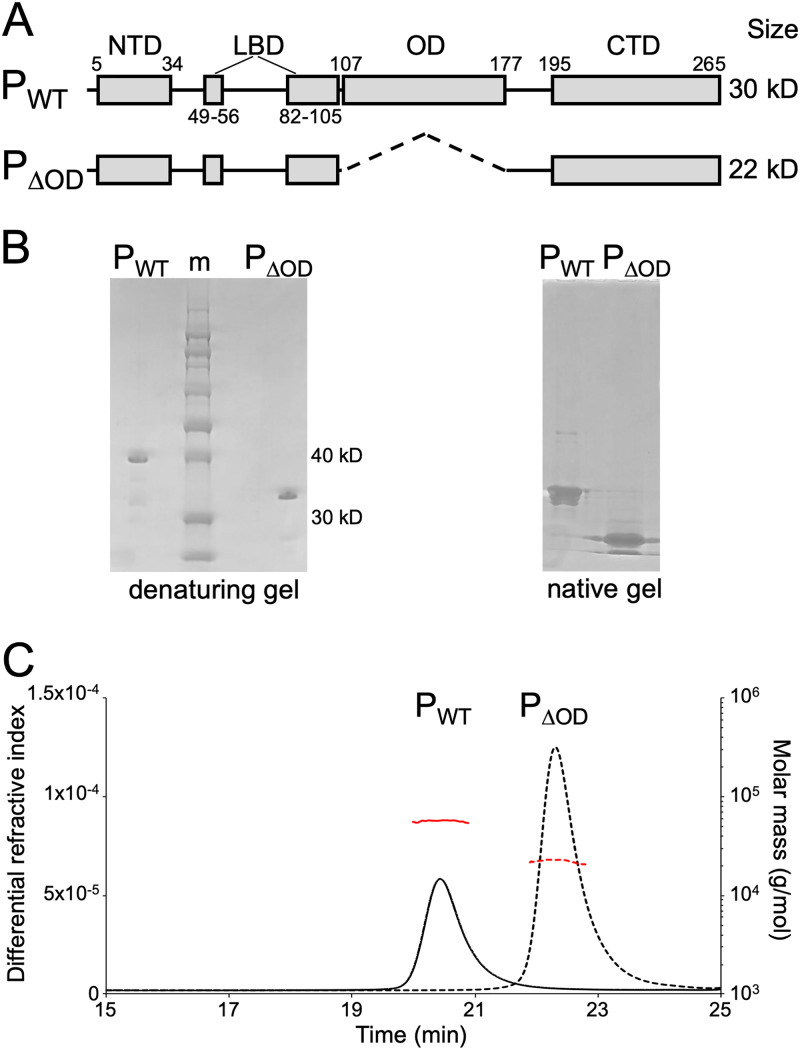
To probe the function of P protein oligomerization in assembly and function of the VSV replication machinery, we expressed and purified from bacteria a variant of P lacking residues 107 to 177, termed PΔOD (Fig. 1A and B). Comparison of identically prepared wild-type P (PWT) and PΔOD on denaturing and native protein gels reveals that deletion of the oligomerization domain yields a P variant whose mobility is altered, consistent with the loss of the 8-kDa OD (Fig. 1B). To provide evidence that PΔOD is present as a monomer in solution, we used size exclusion chromatography and multiple-angle laser light scattering (SEC-MALS). Both PWT and PΔOD elute as single monodisperse peaks (Fig. 1C) and allowed us to calculate the molar mass of the corresponding protein. For PWT, the measured weight-averaged molar mass of 56.2 kDa was within the range of the expected mass of a full-length P dimer (60.6 kDa). The weight-average molar mass (22.6 kDa) for PΔOD is in good agreement with the expected molar mass for a monomer (22.4 kDa). Thus, deletion of the oligomerization domain of VSV P renders the protein monomeric in solution.
FIG 1.
Characterization of PΔOD. (A) Schematic of VSV PWT and PΔOD showing their modular organization with the N-terminal domain (NTD), L-binding domains (LBD), oligomerization domain (OD), and C-terminal domain (CTD) represented as rectangles. (B) Analysis of purified PWT and PΔOD proteins by polyacrylamide gel electrophoresis on a denaturing gel (left) and a native gel (right). Sizes of two bands of the protein ladder (m) are indicated. (C) SEC-MALS analysis of purified PWT (solid lines) and PΔOD (dashed lines) proteins. The horizontal red traces show the inferred molecular mass. Predicted molecular masses for monomeric and dimeric PWT are 30.3 kDa and 60.6 kDa, respectively. For monomeric and dimeric PΔOD, predicted molecular masses are 22.4 kDa and 44.8 kDa, respectively. Observed experimental molecular masses are 56.2 kDa and 22.6 kDa for PWT and PΔOD, respectively.
PΔOD is functional for RNA synthesis in vitro.
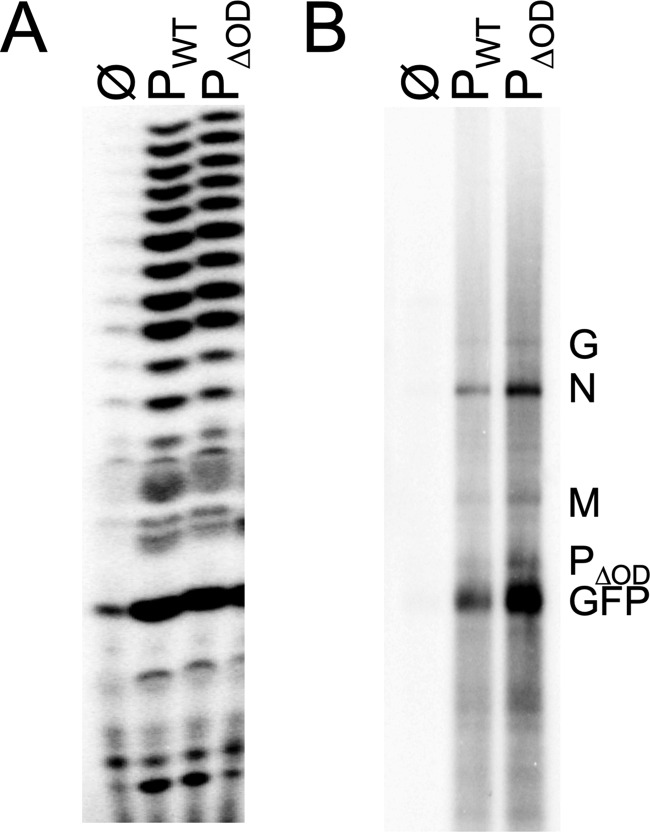
We next compared the ability of PWT and PΔOD to stimulate LRdRP activity using an in vitro assay with chemically synthesized RNA as the template (21). At equimolar amounts, PΔOD and PWT stimulate the LRdRP indistinguishably (Fig. 2A). The natural template for RNA synthesis is, however, the genomic RNA completely encased in a nucleocapsid protein sheath. To compare the ability of PWT and PΔOD to facilitate L-mediated copying of the natural template, we isolated N-RNA from virions and reconstituted RNA synthesis in vitro. Briefly, N-RNA is incubated with purified L and equimolar amounts of PWT or PΔOD, and RNA synthesis assessed by incorporation of [32P]GTP with subsequent analysis of the products on acid-agarose gels (Fig. 2B). The results of this analysis demonstrate that both PΔOD and PWT have the ability to produce the 5 viral mRNAs, confirming that the oligomerization domain of P is not required for mRNA synthesis. We observed, however, an apparent increase in mRNAs produced by PΔOD, and this was seen consistently among experiments.
FIG 2.
Functional analysis of PΔOD in vitro. Reactions were performed in the absence (Ø) or presence of equimolar amounts of PWT or PΔOD using purified L and either a synthetic, naked 19-nt RNA template corresponding to the 3′ leader sequence of the VSV genome (A) or a purified, encapsidated N-RNA template (B). Radioactive products were analyzed by gel electrophoresis on a 20% acrylamide gel (A) or a 1.75% acid-agarose gel containing 6 M urea (B). (A) n = 1 replicate; (B) representative experiment (n = 3 replicates).
A recombinant VSV lacking the oligomerization domain of P is defective in viral multiplication.
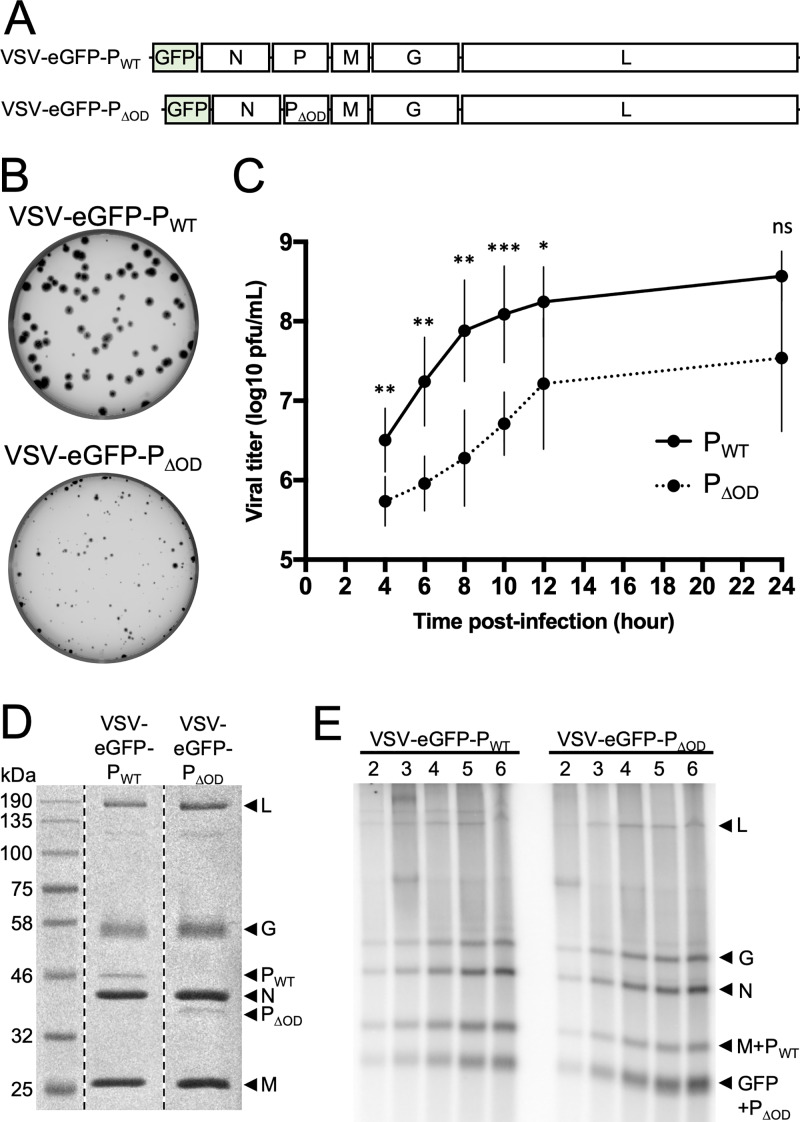
To examine whether oligomerization of P is required for viral amplification in cell culture, we deleted residues 107 to 177 of P in an infectious molecular clone of the virus that expresses enhanced green fluorescent protein (eGFP) as a marker of infection (Fig. 3A). The resulting virus, VSV-eGFP-PΔOD, exhibits a growth defect in cell culture, as is evident by both its plaque size (Fig. 3B) and a 4-h delay before the onset of the exponential phase of virion production in a single-step growth assay (Fig. 3C). Molecular characterization of the virus revealed that the genomic sequence retained the desired deletion and that the composition of purified VSV-eGFP-PΔOD virions is indistinct to those of VSV-eGFP-PWT by SDS-PAGE with respect to the relative amounts of the 5 viral proteins and the particle-to-PFU ratio (Fig. 3D). This result indicates that ablation of the oligomerization domain of P has little effect on protein incorporation into particles but attenuates viral growth in cell culture.
FIG 3.
Characterization of a recombinant VSV expressing PΔOD. (A) Schematic representation of the recombinant virus genomes. (B) Viral spreading as seen by plaque assay on Vero cells infected with VSV-eGFP-PWT or VSV-eGFP-PΔOD. (C) Viral growth kinetic on Vero cells infected with VSV-eGFP-PWT (PWT, black line) or VSV-eGFP-PΔOD (PΔOD, dotted line) at an MOI of 3. Supernatants were harvested and titers determined at 4, 6, 8, 10, 12, and 24 h postinfection. Statistical analysis was performed by a paired t test. * P, < 0.05; ** P, < 0.005; *** P, < 0.0005; ns, nonsignificant. (D) Analysis of virion protein content. Gradient-purified virions (108 PFU) were denaturated by SDS and heat and analyzed by SDS-PAGE and Coomassie staining. (E) In phosphate-free media supplemented with radioactive [32P]orthophosphate, BSR-T7 cells were treated with 10 μg/ml actinomycin D and 100 μg/ml cycloheximide and infected at an MOI of 100 with VSV-eGFP-PWT or VSV-eGFP-PΔOD. RNA was extracted at 2, 3, 4, 5, and 6 h postinfection and analyzed on a 1.75% acid-agarose gel containing 6 M urea. Representative experiment (n = 4).
To begin to understand the mechanism underlying the kinetic delay in viral multiplication, we compared the ability of VSV-eGFP-PWT and VSV-eGFP-PΔOD to undertake primary transcription in cells. Cells were treated with cycloheximide and actinomycin D to block protein and cellular RNA synthesis, respectively, and infected with the indicated virus at a multiplicity of infection (MOI) of 100, and viral RNA synthesis was monitored by metabolic incorporation of [32P]orthophosphate. At various times postinfection, total cellular RNA was extracted and analyzed by acid-agarose gel electrophoresis, and the products of primary transcription were detected by phosphor image analysis. The abundance of each viral mRNA was similar for VSV-eGFP-PWT and VSV-eGFP-PΔOD (Fig. 3E), demonstrating that loss of the oligomerization domain of P does not compromise transcription. Consistent with the experiments of Fig. 2B, VSV-eGFP-PΔOD primary transcription appears to generate more mRNA than VSV-eGFP-PWT. Quantitation of the N mRNA signal coupled with statistical analysis yields a P value of 0.0562 (Fig. 3E). This result demonstrates that the polymerase packaged into particles remains associated with the N-RNA template in the absence of the POD and that there is no apparent consequence for the stability of the polymerase complex. Furthermore, because the relative proportions of the viral mRNAs are indistinct, POD is not required to retain the L protein on the template as it navigates gene junctions. These data imply that a step downstream of transcription, likely RNA replication, is responsible for the observed kinetic delay in viral growth.
The oligomerization domain of P affects protein diffusion in viral replication compartments.
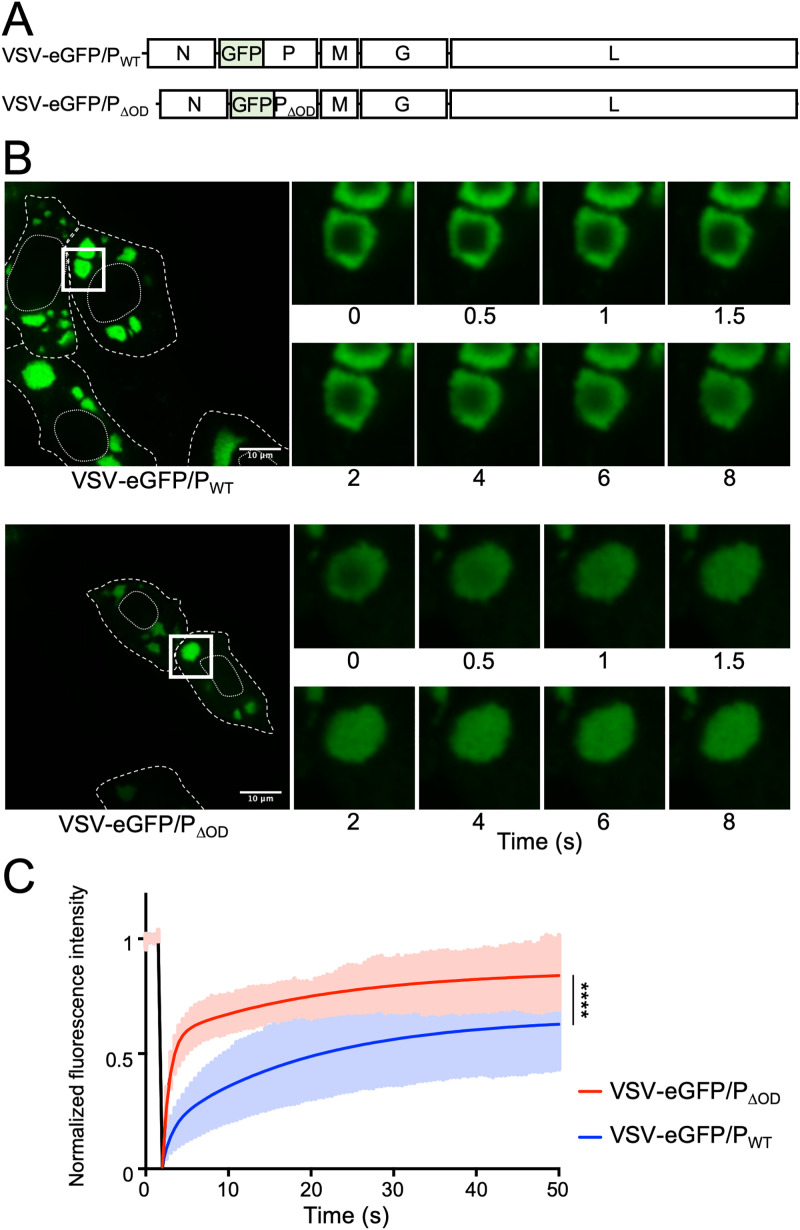
Following infection of cells, the viral replication machinery is found in the cytoplasm in a compartment that is not bounded by a membrane (22, 23). Work with rabies virus implicates the oligomerization domain of its phosphoprotein as required for the reconstitution of such compartments (24). We therefore generated a recombinant virus expressing PΔOD fused to eGFP at its N terminus (termed VSV-eGFP/PΔOD), which allows visualization of the replication compartment (Fig. 4A). The eGFP/P viruses differ from eGFP-P, which encodes an eGFP reporter from a separate transcription unit inserted between the leader and N genes. As seen for wild-type P, we found the eGFP/PΔOD concentrates in structures that look morphologically similar (Fig. 4B). We next compared the properties of the compartments by measuring fluorescence recovery after photobleaching (FRAP) of eGFP/P. To take into account the growth delay of VSV-eGFP-PΔOD, experiments were performed at 6 and 10 hpi for VSV-eGFP/PWT and VSV-eGFP/PΔOD, respectively. Previous work demonstrated that the size of viral replication compartments impacts the diffusion properties of the viral proteins (25). We therefore selected similar-sized structures for this analysis which were present in cells infected with VSV-eGFP/PWT at 6 hpi and cells infected with VSV-eGFP/PΔOD at 10 hpi. We measured a half-maximal recovery time of 12.3 s for eGFP/PWT containing compartments compared with 2.1 s for compartments containing eGFP/PΔOD (Fig. 4B and C). This result demonstrates that PΔOD recovers more rapidly from photobleaching, which means that the protein must undergo a more rapid diffusion and exchange of PΔOD than wild-type P. This is consistent with the expected reduction in intermolecular interactions following ablation of the oligomerization domain.
FIG 4.
FRAP analysis of replication compartments. (A) Schematic representation of the recombinant virus genomes. (B, C) Vero cells were infected at an MOI of 3 with VSV-eGFP/PWT or VSV-eGFP/PΔOD, and eGFP was visualized with a spinning disk confocal microscope at 6 and 10 h postinfection for VSV-eGFP/PWT and VSV-eGFP/PΔOD, respectively. Fluorescence recovery after photobleaching (FRAP) experiments were performed on areas of 4 μm2 located inside compartments. Recovery fluorescence was measured every 500 ms for 50 s. (B) Infected cells before photobleaching (left), and zoomed-in pictures taken at indicated times after photobleaching (right). Dashed and dotted lines delimit the cells and the nucleus, respectively. Squares represent the zoomed-in sections. (C) FRAP data were corrected for background, normalized to the minimum and maximum intensities. The mean is shown on the black line, with the gray zone representing the SD. Mean experimental curves were fitted with double-exponential models (red line; VSV-eGFP/PWT, R2 = 0.997; VSV-eGFP/PΔOD, R2 = 0.996). Statistical comparison of the two data sets was performed using the Kolmogorov-Smirnov test. P < 0.0001.
RNA replication requires higher N protein concentrations for monomeric P.
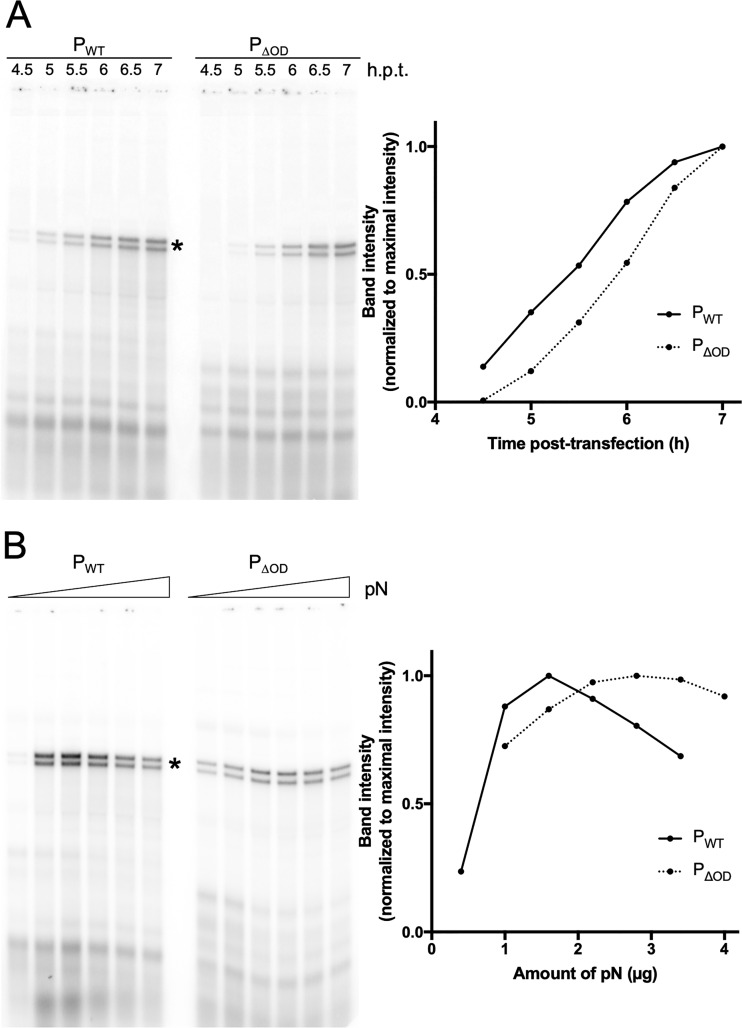
Viral genome replication is obligatorily coupled to encapsidation of the nascent RNA chain by soluble nucleocapsid proteins, and available evidence underscores the central role of P in that assembly process (26, 27). To directly measure the influence of the oligomerization domain of P on RNA replication, we employed a cell-based assay of RNA replication for a well-characterized defective interfering (DI) particle of VSV, DI-T (28). The genome of DI-T contains a 5′ copyback arrangement in which the antigenomic promoter, which exclusively supports replication, drives synthesis from both the genomic and antigenomic strands. In this assay, BSR-T7 cells are infected with DI-T particles and transfected with different amounts of plasmids expressing L, N, and PWT or PΔOD. The products of RNA replication are then directly detected by metabolic incorporation of radioactive orthophosphate in the presence of actinomycin D, purified, and analyzed by acid-agarose electrophoresis (Fig. 5A). The acid-agarose gels facilitate the separation of the positive and negative strands of the DI genome RNA, allowing us to measure both genome and antigenome synthesis. Quantitation of the DI-T genome and antigenome bands demonstrates that they increase over time at a similar rate and reach the same levels for PWT and PΔOD, but it highlights a delay in accumulation of replication products for PΔOD (Fig. 5A).
FIG 5.
Effect of PΔOD on viral RNA synthesis. (A) BSR-T7 cells were infected with DI-T for 1 h and transfected with plasmids coding for L, N, and PWT or PΔOD. At 1.5, 2, 2.5, 3, 3.5, or 4 h posttransfection, cells were incubated for 3 h in phosphate-free media supplemented with radioactive [32P]orthophosphate and 10 μg/ml actinomycin D. RNA was harvested and analyzed on a 1.75% agarose gel containing 6 M urea (left). DI-T band intensities were quantified and plotted as percentage of maximal intensity (right). Times of harvest posttransfection are indicated. DI-T bands are marked with an asterisk. Representative experiment (n = 4). (B) BSR-T7 cells were infected with DI-T for 1 h and transfected with plasmids coding for L, N, and PWT or PΔOD. Increasing amounts of plasmid coding for N were transfected with 0.4, 1, 1.6, 2.2, 2.8, and 3.4 μg for PWT and 1, 1.6, 2.2, 2.8, 3.4, and 4 μg for PΔOD. Five hours posttransfection, cells were incubated for 3 h in phosphate-free media supplemented with radioactive [32P]orthophosphate and 10 μg/ml actinomycin D. RNA was harvested and analyzed on a 1.75% agarose gel containing 6 M urea (left). DI-T band intensities were quantified and plotted as percentage of maximal intensity (right). Representative experiment (n = 2).
We next hypothesized that the oligomerization domain of P could serve to concentrate N during the process of nascent strand encapsidation, which would suggest that a higher concentration of N would further stimulate replication for PΔOD. It was previously established that altering the concentration of N plasmid transfected into cells alters the concentration of N protein and that there is an optimal N-P ratio to support maximal replication (29). The optimal N plasmid concentration to support maximal replication is higher for PΔOD than PWT, consistent with higher concentrations of N protein being required for replication (Fig. 5B). Together with the time course experiment, these results suggest that P oligomerization plays a role in genome replication by lowering the N concentration threshold required to initiate the replication phase of the cycle.
DISCUSSION
The major conclusions of this study are (i) the oligomerization domain of VSV P results in its dimerization, (ii) P dimerization is dispensable for all aspects of viral mRNA transcription, and (iii) P dimerization stimulates RNA replication. We also present evidence in support of a mechanism where the dimerization-mediated stimulation of replication is likely accomplished by facilitating encapsidation of the nascent replicative product at an optimal N protein concentration that is lowered by the presence of the oligomerization domain. We suggest that this is a central function of the P proteins of all Mononegavirales and that interfering with the oligomeric state of P may serve as a general mechanism to inhibit replication of these viruses.
VSV P protein dimerization.
The oligomeric status of VSV P has been variably reported as trimer (30), tetramer (31), and, most recently, following structural studies of the oligomerization domain, as a dimer (18, 32). The SEC-MALS data presented here provide support that P exists as a homodimer and that homodimerization is mediated by the oligomerization domain. During the preparation of the manuscript, Gérard and colleagues published structural analyzes of purified PΔOD confirming PΔOD is monomeric in solution and showing its global architecture is not affected by the loss of the OD (33). In addition to forming a homodimer, P has additional viral binding partners, including N-RNA, N0, and L. Using electron cryomicroscopy, we recently defined key contacts between L and two regions of the P N-terminal domain (PNTD) (19). In that structure, however, P is noncontiguous; thus, we cannot exclude the possibility that 2 separate molecules of P can bind L. Crystallographic studies showed the PCTD binds the N-RNA at an interface only formed by adjacent N protomers (17). The engagement of the N-RNA by the PCTD raises the possibility that even in the absence of an oligomerization domain, P may be able to effectively function as an oligomer with respect to L through its template-binding properties. Additional structural studies are likely to help unravel this question.
Virion production.
Since the production of infectious viral particles is delayed for a virus expressing PΔOD, one or several of the viral cycle steps, including viral entry, primary transcription, genome replication, or particle assembly, must be affected. Using a similar recombinant virus, Gérard and colleagues did not see any strong effect of the loss of the OD on virion production (33). We cannot explain the different conclusion of this earlier study, but this may reflect strain differences or the use of different assays between our studies. Our conclusion that the loss of the OD affects viral replication by influencing a key step involved in viral RNA replication and genome encapsidation comes from biochemical analysis of the RNA products synthesized from a naturally occurring defective interfering particle of VSV. This assay differs from the use of synthetic minigenome in the earlier work. Additional work will be required to understand the basis for this distinction.
Viral replication compartments.
For rabies virus, the POD is reported to be essential for the formation of the viral replication compartment in infected cells (24). This is not the case for VSV-eGFP/PΔOD, which still forms compartments. While such compartments form in cells infected with VSV-eGFP/PΔOD, the properties of the compartments differ with respect to the diffusion of eGFP/PWT, as evidenced by a 6-fold faster recovery rate following photobleaching. We interpret that increased recovery rate as reflective of the loss of the oligomerization domain, effectively decreasing the number of partners for P in its monomeric versus dimeric states, and that those interactions contribute to the slower recovery observed in wild-type infected cells. As the properties of inclusions alter with their size, which itself is related to the stage of the infectious cycle, we cannot exclude that the difference in P diffusion might also be linked to the acquisition of the data at 6 versus 10 hours postinfection (hpi) rather than intrinsic differences between PWT and PΔOD.
Viral transcription.
Reconstitution of transcription in vitro on naked RNA or on purified N-RNA templates was unaffected by the loss of the OD. Consistent with this observation, measures of primary mRNA transcription by the polymerase molecules brought into cells with the infecting virus were also unaffected. Collectively, those data provide compelling evidence that no step of transcription, including initiation, cap formation, elongation, polyadenylation, termination, and reinitiation, require the presence of the OD. We consistently observed an increase in levels of transcription with PΔOD, although quantitation of the N mRNA indicated that this difference was not significant. Additional work will be required to understand the basis of this observation. An earlier model of migration of the polymerase complex of Sendai virus, a related virus in the family Paramyxoviridae, posited that the polymerase cartwheels along the N-RNA template via the interaction between the PCTD of its tetrameric P and the nucleocapsid (34). Such a cartwheeling model suggests that dissociation of the polymerase at gene junctions would be favored when interactions between P and the N-RNA template are compromised. Studies with measles virus found that modulation of the interaction between its PCTD and the N-RNA alters the relative abundance of viral mRNAs (35, 36). In contrast, deletion of the oligomerization domain of VSV P does not alter the gradient of transcripts produced from the template, suggesting that the interaction between the L protein and template is not compromised. This suggests that either modifying the strength of the VSV PCTD-N-RNA interaction by tuning its affinity or its avidity has different outcomes, or the interaction of VSV PCTD with the N-RNA is not a key determinant of transcription reinitiation at gene junctions.
RNA replication.
During RNA replication, in addition to the requirements for P as a cofactor for the polymerase and its role in binding the N-RNA template, P loads soluble N protomers onto the nascent RNA (26, 27). The availability of a pool of N0-P for loading onto that nascent strand is thought to regulate polymerase activity during leader synthesis, coupling the nascent strand encapsidation to genome replication (14, 37). Absent robust assays for the in vitro reconstitution of RNA encapsidation, direct tests of the role of the POD in encapsidation are not possible, and therefore, in this study, we used a cell-based assay that reports on encapsidation indirectly by measuring the accumulation of the products of RNA replication. Using that assay, we attained evidence that when equal amounts of N and L are available, the kinetics with which RNA replication is established with PΔOD are slower. We also found that replication is further stimulated by increasing the amount of N in cells through increasing levels of an N expression plasmid. The structure of the VSV L-P complex reveals that the region of P that binds N0 is positioned by the RNA exit channel from the polymerase poised to encapsidate the nascent RNA strand (19). We cannot exclude the possibility that the OD itself plays a role during the replication, nor that changing the spatial arrangement of the N-terminal and C-terminal domains impairs proper function. However, as both PNTD of a PWT dimer can bind simultaneously an N monomer, perhaps dimerization of P facilitates a local increase in the concentration of N0 at the site of encapsidation favoring RNA replication (32). As PWT is a dimer, its interaction with N0 is likely enhanced by avidity, thus potentially further increasing the recruitment of N0 at the encapsidation site. Robust in vitro encapsidation assays and structural studies of polymerase complexes during RNA replication will likely prove informative.
MATERIALS AND METHODS
Cells.
BSR-T7 cells (a kind gift from K. Conzelmann) (38) and African green monkey kidney Vero cells (ATCC CCL-81) were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Corning Inc.; product no. 10-013-CV) containing 10% fetal bovine serum (FBS; Tissue Culture Biologicals; catalog no. 101) at 37°C and 5% CO2.
Plasmids.
For bacterial expression and purification, a gBlocks gene fragment (Integrated DNA Technologies, Inc.) coding for 6×His/PΔOD was cloned in a pET16b vector (16). To rescue recombinant VSV, pVSV1+-eGFP-PΔOD was derived from pVSV1+-eGFP (39). pVSV1+-eGFP and a gBlocks gene fragment containing the sequence of the end of the N gene, the N-P intergenic region, and the PΔOD gene were digested with AloI and BstZI17I restriction enzymes and ligated together.
pVSV1+-eGFP/PΔOD was derived from pVSV1+-eGFP/P (40) after amplification of the PΔOD gene from pVSV1+-eGFP-PΔOD and insertion by ligation.
Viruses.
VSV-eGFP-PWT and VSV-eGFP/PWT were described previously (39, 40). VSV-eGFP-PΔOD and VSV-eGFP/PΔOD were rescued following previously described protocol (41) using pVSV1+-eGFP-PΔOD and pVSV1+-eGFP/PΔOD plasmids, respectively. Virus stocks were grown on BSR-T7 cells, and we determined the titer by plaque assay on BSR-T7 or Vero cells. Briefly, cells were seeded in DMEM-10% FBS and infected 1 day later for 1 h with viruses at a multiplicity of infection (MOI) of 0.01. Virus suspensions were replaced by DMEM-2% FBS, and cell supernatants were harvested when 95% of the cells were infected and ready to detach (about 24 h for VSV-eGFP-PWT). For gradient-purified viruses, infected cell supernatant was first concentrated through a 15% sucrose cushion in NTE (10 mM Tris-HCl pH 7.4, 100 mM NaCl, and 1 mM EDTA) at 110,000 × g for 2 h at 4°C. Pellets were resuspended overnight at 4°C in NTE, put on top of a linear 15 to 45% sucrose gradient in NTE, and centrifuged at 200,000 × g for 3 h at 4°C. Bands corresponding to virus were collected in 0.5-ml tubes by side puncture of the tube and diluted 10 times in NTE.
Proteins and nucleocapsid purification.
P and PΔOD were purified from BL21(DE3) Escherichia coli cells and the L protein from Spodoptera frugiperda 21 (Sf21) cells as previously described (16). Briefly, after cell lysis, proteins were affinity purified with HisTrap HP (Ge Healthcare) followed by gel filtration (Superdex 200 HR 10/30; GE Healthcare). P proteins were stored in 20 mM Tris (pH 7.4), 150 mM NaCl, and 1 mM dithiothreitol (DTT) buffer, and L proteins were stored in 50 mM Tris-HCl (pH 7.4), 200 mM NaCl, 15% glycerol, and 1 mM DTT buffer. N-RNA templates were purified from gradient-purified VSV-eGFP-PΔOD virions as previously described (42).
Size exclusion chromatography with multiangle light scattering.
SEC-MALS analysis was performed on an Agilent 1260 Infinity liquid chromatography system in phosphate-buffered saline (PBS) by use of a Wyatt Dawn Heleos II multiangle light scattering detector and Optilab T-rEX refractive index detector at the Center for Macromolecular Interactions, Harvard Medical School. Data were processed using Astra 7, and weight-averaged molar mass was fit using the Zimm method and a protein refractive index increment of 0.185. Fitting errors of 0.7% and 1.4% were achieved for PWT and PΔOD, respectively.
In vitro RNA synthesis assays.
In vitro RNA synthesis assays on naked RNA were performed as previously described (21) using purified L protein and either PWT or PΔOD. Transcription assays on encapsidated RNA were performed using N-RNA extracted from VSV-eGFP-PΔOD virions and purified PWT, PΔOD, and L proteins (42).
Analysis of primary transcripts.
BSR-T7 cells were seeded in 6-well plates and incubated 1 day later in phosphate-free DMEM (Gibco; catalog no. 11971-025) for 30 min followed by a 30-min incubation in phosphate-free DMEM containing 10 μg/ml actinomycin D (Sigma; catalog no. A5156) and 100 μg/ml cycloheximide (VWR; catalog no. 94271). Cells were then infected for 30 min with sucrose cushion-purified virus at an MOI of 100. Virus solutions were replaced by 1 ml phosphate-free DMEM containing 10 μg/ml actinomycin D, 100 μg/ml cycloheximide, and 10 μl of phosphorus-32 radionuclide (PerkinElmer; catalog no. NEX053H005MC). At 2, 3, 4, 5, and 6 h postinfection, RNA was extracted using TRIzol reagent (Invitrogen; catalog no. 15596018) following the manufacturer’s protocol. RNA was boiled at 100°C for 1 min, incubated on ice for 2 min, mixed with a 1.33× loading buffer (33.3 mM citrate pH 3, 8 M urea, 20% sucrose, and 0.001% bromophenol blue), and analyzed on a 25 mM citrate, pH 3, 1.75% agarose, 6 M urea gel run for 18 h at 4°C and 180 V. Gels were fixed (in 30% methanol and 10% acetic acid), dried, and exposed overnight to a phosphor screen (GE Healthcare), and the radiolabeled RNA products were visualized using a Typhoon FLA 9500 scanner (GE Healthcare).
Analysis of DI-T replication.
BSR-T7 cells were seeded in 6-well plates and infected 1 day later with a recombinant vaccinia virus expressing the T7 polymerase (vTF7-3) and DI-T particles for 1 h at 37°C in Dulbecco’s PBS (DPBS; Sigma; catalog no. 59300C). Cells were then transfected using Lipofectamine 2000 with plasmids expressing L (0.24 μg), N (2.9 μg or indicated amounts), and P or PΔOD (0.8 μg). Five hours later or at indicated hours, the medium was removed, and cells were incubated in 1 ml phosphate-free DMEM containing 10 μg/ml actinomycin D for 30 min before the addition of 10 μl of phosphorus-32 radionuclide. Cells were incubated for 3 h at 37°C before RNA was harvested by the use of TRIzol reagent following the manufacturer’s protocol. RNA was boiled at 100°C for 1 min, incubated on ice for 2 min, mixed with a 1.33× loading buffer (33.3 mM citrate pH 3, 8 M urea, 20% sucrose, and 0.001% bromophenol blue), and analyzed on a 25 mM citrate, pH 3, 1.75% agarose, and 6 M urea gel run for 18 h at 4°C and 180 V. Gels were fixed (in 30% methanol and 10% acetic acid), dried, and exposed overnight to a phosphor screen (GE Healthcare), and the radiolabeled RNA products were visualized using a Typhoon FLA 9500 scanner (GE Healthcare). DI-T band intensities were quantified with ImageJ software.
FRAP of viral replication compartments.
Vero cells were plated at 3 × 105 cells per chamber in a 8-well chambered cover glass (Cellvis) and infected 20 and 24 h later at an MOI of 3 by VSV-eGFP/PΔOD and VSV-eGFP/PWT, respectively. Thirty hours after seeding, the photobleaching was performed using the Vector photomanipulation module attached to a Marianas spinning disk confocal platform (3i, Denver, Colorado). The images were acquired using a Plan-Apochromat 100×/1.4 oil lens (Carl Zeiss, Jena, Germany). The incubation system (Okolab, Naples, Italy) was set at 5% CO2 and 37°C. Four images were acquired prior to bleaching and imaging with the 488-nm laser.
For data analysis, mean background fluorescence was measured from an area outside the cells and subtracted from other measurements. Mean fluorescence intensities of each photobleached area were also corrected for the photobleaching that occurred during image acquisition postbleach and normalized by the average fluorescence prebleach. Photobleaching postbleach was measured on nonbleached compartments. After normalization, mean recovery was calculated using EasyFRAP (43) and fit with a double-exponential model: Y(t) = Y0 + Afast(1 − e−Kfast*t) + Aslow(1 − e−Kslow*t).
ACKNOWLEDGMENTS
We thank Kelly Arnett and the Center for Macromolecular Interactions at the Harvard Medical School Department of Biological Chemistry and Molecular Pharmacology for assistance with SEC-MALS measurements and helpful discussions.
This study was supported by NIH grant AI059371 to S.P.J.W.
REFERENCES
- 1.Emerson SU, Yu Y. 1975. Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J Virol 15:1348–1356. doi: 10.1128/JVI.15.6.1348-1356.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hercyk N, Horikami SM, Moyer SA. 1988. The vesicular stomatitis virus L protein possesses the mRNA methyltransferase activities. Virology 163:222–225. doi: 10.1016/0042-6822(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 3.Hunt DM, Mehta R, Hutchinson KL. 1988. The L protein of vesicular stomatitis virus modulates the response of the polyadenylic acid polymerase to S-adenosylhomocysteine. J Gen Virol 69:2555–2561. doi: 10.1099/0022-1317-69-10-2555. [DOI] [PubMed] [Google Scholar]
- 4.Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC, Whelan S. 2015. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emerson SU, Wagner RR. 1972. Dissociation and reconstitution of the transcriptase and template activities of vesicular stomatitis B and T virions. J Virol 10:297–309. doi: 10.1128/JVI.10.2.297-309.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green TJ, Zhang X, Wertz GW, Luo M. 2006. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313:357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- 7.Abraham G, Banerjee AK. 1976. Sequential transcription of the genes of vesicular stomatitis virus. Proc Natl Acad Sci U S A 73:1504–1508. doi: 10.1073/pnas.73.5.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ball LA, White CN. 1976. Order of transcription of genes of vesicular stomatitis virus. Proc Natl Acad Sci U S A 73:442–446. doi: 10.1073/pnas.73.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colonno RJ, Banerjee AK. 1978. Complete nucleotide sequence of the leader RNA synthesized in vitro by vesicular stomatitis virus. Cell 15:93–101. doi: 10.1016/0092-8674(78)90085-5. [DOI] [PubMed] [Google Scholar]
- 10.Colonno RJ, Banerjee AK. 1978. In vitro RNA transcription by the New Jersey serotype of vesicular stomatitis virus. II. Characterization of the leader RNA. J Virol 26:188–194. doi: 10.1128/JVI.26.1.188-194.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogino T, Banerjee AK. 2007. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol Cell 25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Rahmeh AA, Li J, Kranzusch PJ, Whelan SP. 2009. Ribose 2’-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J Virol 83:11043–11050. doi: 10.1128/JVI.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barr JN, Wertz GW. 2001. Polymerase slippage at vesicular stomatitis virus gene junctions to generate poly(A) is regulated by the upstream 3’-AUAC-5’ tetranucleotide: implications for the mechanism of transcription termination. J Virol 75:6901–6913. doi: 10.1128/JVI.75.15.6901-6913.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patton JT, Davis NL, Wertz GW. 1984. N protein alone satisfies the requirement for protein synthesis during RNA replication of vesicular stomatitis virus. J Virol 49:303–309. doi: 10.1128/JVI.49.2.303-309.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogino M, Gupta N, Green TJ, Ogino T. 2019. A dual-functional priming-capping loop of rhabdoviral RNA polymerases directs terminal de novo initiation and capping intermediate formation. Nucleic Acids Res 47:299–309. doi: 10.1093/nar/gky1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rahmeh AA, Schenk AD, Danek EI, Kranzusch PJ, Liang B, Walz T, Whelan SP. 2010. Molecular architecture of the vesicular stomatitis virus RNA polymerase. Proc Natl Acad Sci U S A 107:20075–20080. doi: 10.1073/pnas.1013559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green TJ, Luo M. 2009. Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P Proc Natl Acad Sci U S A 106:11713–11718. doi: 10.1073/pnas.0903228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding H, Green TJ, Lu S, Luo M. 2006. Crystal structure of the oligomerization domain of the phosphoprotein of vesicular stomatitis virus. J Virol 80:2808–2814. doi: 10.1128/JVI.80.6.2808-2814.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenni S, Bloyet LM, Diaz-Avalos R, Liang B, Whelan SPJ, Grigorieff N, Harrison SC. 2020. Structure of the vesicular stomatitis virus l protein in complex with its phosphoprotein cofactor. Cell Rep 30:53–60.e5. doi: 10.1016/j.celrep.2019.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leyrat C, Yabukarski F, Tarbouriech N, Ribeiro EA, Jensen MR, Blackledge M, Ruigrok RWH, Jamin M. 2011. Structure of the vesicular stomatitis virus N0-P complex. PLoS Pathog 7:e1002248. doi: 10.1371/journal.ppat.1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morin B, Rahmeh AA, Whelan SP. 2012. Mechanism of RNA synthesis initiation by the vesicular stomatitis virus polymerase. EMBO J 31:1320–1329. doi: 10.1038/emboj.2011.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinrich BS, Cureton DK, Rahmeh AA, Whelan SP. 2010. Protein expression redirects vesicular stomatitis virus RNA synthesis to cytoplasmic inclusions. PLoS Pathog 6:e1000958. doi: 10.1371/journal.ppat.1000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinrich BS, Maliga Z, Stein DA, Hyman AA, Whelan S. 2018. Phase transitions drive the formation of vesicular stomatitis virus replication compartments. mBio 9:e02290-17. doi: 10.1128/mBio.02290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikolic J, Le Bars R, Lama Z, Scrima N, Lagaudriere-Gesbert C, Gaudin Y, Blondel D. 2017. Negri bodies are viral factories with properties of liquid organelles. Nat Commun 8:58. doi: 10.1038/s41467-017-00102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y, Su JM, Samuel CE, Ma D. 2019. Measles virus forms inclusion bodies with properties of liquid organelles. J Virol 93. doi: 10.1128/JVI.00948-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masters PS, Banerjee AK. 1988. Complex formation with vesicular stomatitis virus phosphoprotein NS prevents binding of nucleocapsid protein N to nonspecific RNA. J Virol 62:2658–2664. doi: 10.1128/JVI.62.8.2658-2664.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peluso RW, Moyer SA. 1988. Viral proteins required for the in vitro replication of vesicular stomatitis virus defective interfering particle genome RNA. Virology 162:369–376. doi: 10.1016/0042-6822(88)90477-1. [DOI] [PubMed] [Google Scholar]
- 28.Pattnaik AK, Ball LA, LeGrone A, Wertz GW. 1995. The termini of VSV DI particle RNAs are sufficient to signal RNA encapsidation, replication, and budding to generate infectious particles. Virology 206:760–764. doi: 10.1016/s0042-6822(95)80005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pattnaik AK, Wertz GW. 1990. Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J Virol 64:2948–2957. doi: 10.1128/JVI.64.6.2948-2957.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y, Greenfield NJ, Cleverley DZ, Lenard J. 1996. The transcriptional form of the phosphoprotein of vesicular stomatitis virus is a trimer: structure and stability. Biochemistry 35:14569–14573. doi: 10.1021/bi9613133. [DOI] [PubMed] [Google Scholar]
- 31.Gao Y, Lenard J. 1995. Multimerization and transcriptional activation of the phosphoprotein (P) of vesicular stomatitis virus by casein kinase-II. EMBO J 14:1240–1247. doi: 10.1002/j.1460-2075.1995.tb07107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yabukarski F, Leyrat C, Martinez N, Communie G, Ivanov I, Ribeiro EA Jr, Buisson M, Gerard FC, Bourhis JM, Jensen MR, Bernado P, Blackledge M, Jamin M. 2016. Ensemble structure of the highly flexible complex formed between vesicular stomatitis virus unassembled nucleoprotein and its phosphoprotein chaperone. J Mol Biol 428:2671–2694. doi: 10.1016/j.jmb.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 33.Gerard FC, Jamin M, Blackledge M, Blondel D, Bourhis JM. 2019. Vesicular stomatitis virus phosphoprotein dimerization domain is dispensable for virus growth. J Virol 94:e01789-19. doi: 10.1128/JVI.01789-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Curran J. 1998. A role for the Sendai virus P protein trimer in RNA synthesis. J Virol 72:4274–4280. doi: 10.1128/JVI.72.5.4274-4280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bloyet LM, Brunel J, Dosnon M, Hamon V, Erales J, Gruet A, Lazert C, Bignon C, Roche P, Longhi S, Gerlier D. 2016. Modulation of re-initiation of measles virus transcription at intergenic regions by PXD to NTAIL binding strength. PLoS Pathog 12:e1006058. doi: 10.1371/journal.ppat.1006058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox RM, Krumm SA, Thakkar VD, Sohn M, Plemper RK. 2017. The structurally disordered paramyxovirus nucleocapsid protein tail domain is a regulator of the mRNA transcription gradient. Sci Adv 3:e1602350. doi: 10.1126/sciadv.1602350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta AK, Banerjee AK. 1997. Expression and purification of vesicular stomatitis virus N-P complex from Escherichia coli: role in genome RNA transcription and replication in vitro. J Virol 71:4264–4271. doi: 10.1128/JVI.71.6.4264-4271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchholz UJ, Finke S, Conzelmann KK. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol 73:251–259. doi: 10.1128/JVI.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, Perrimon N. 2005. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev 19:445–452. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schott DH, Cureton DK, Whelan SP, Hunter CP. 2005. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc Natl Acad Sci U S A 102:18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whelan SP, Ball LA, Barr JN, Wertz GT. 1995. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci U S A 92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Rahmeh A, Morelli M, Whelan SP. 2008. A conserved motif in region V of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J Virol 82:775–784. doi: 10.1128/JVI.02107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koulouras G, Panagopoulos A, Rapsomaniki MA, Giakoumakis NN, Taraviras S, Lygerou Z. 2018. EasyFRAP-web: a web-based tool for the analysis of fluorescence recovery after photobleaching data. Nucleic Acids Res 46:W467–W472. doi: 10.1093/nar/gky508. [DOI] [PMC free article] [PubMed] [Google Scholar]