Andes orthohantavirus (ANDV) is endemic in Argentina and Chile and is the primary etiological agent of hantavirus cardiopulmonary syndrome (HCPS) in South America. ANDV is distinguished from other hantaviruses by its unique ability to spread from person to person. In a previous report, we identified a novel ANDV protein, ANDV-NSs. Until now, ANDV-NSs had no known function. In this new study, we established that ANDV-NSs acts as an antagonist of cellular innate immunity, the first line of defense against invading pathogens, hindering the cellular antiviral response during infection. This study provides novel insights into the mechanisms used by ANDV to establish its infection.
KEYWORDS: ANDV, IFN-β, MAVS, MDA5, NSs protein, RIG-I
ABSTRACT
The small messenger RNA (SmRNA) of the Andes orthohantavirus (ANDV), a rodent-borne member of the Hantaviridae family of viruses of the Bunyavirales order, encodes a multifunctional nucleocapsid (N) protein and for a nonstructural (NSs) protein of unknown function. We have previously shown the expression of the ANDV-NSs, but only in infected cell cultures. In this study, we extend our early findings by confirming the expression of the ANDV-NSs protein in the lungs of experimentally infected golden Syrian hamsters. Next, we show, using a virus-free system, that the ANDV-NSs protein antagonizes the type I interferon (IFN) induction pathway by suppressing signals downstream of the melanoma differentiation-associated protein 5 (MDA5) and the retinoic acid-inducible gene 1 (RIG-I) and upstream of TBK1. Consistent with this observation, the ANDV-NSs protein antagonized mitochondrial antiviral-signaling protein (MAVS)-induced IFN-β, NF-κB, IFN-regulatory factor 3 (IRF3), and IFN-sensitive response element (ISRE) promoter activity. Results demonstrate that ANDV-NSs binds to MAVS in cells without disrupting the MAVS-TBK-1 interaction. However, in the presence of the ANDV-NSs ubiquitination of MAVS is reduced. In summary, this study provides evidence showing that the ANDV-NSs protein acts as an antagonist of the cellular innate immune system by suppressing MAVS downstream signaling by a yet not fully understand mechanism. Our findings reveal new insights into the molecular regulation of the hosts’ innate immune response by the Andes orthohantavirus.
IMPORTANCE Andes orthohantavirus (ANDV) is endemic in Argentina and Chile and is the primary etiological agent of hantavirus cardiopulmonary syndrome (HCPS) in South America. ANDV is distinguished from other hantaviruses by its unique ability to spread from person to person. In a previous report, we identified a novel ANDV protein, ANDV-NSs. Until now, ANDV-NSs had no known function. In this new study, we established that ANDV-NSs acts as an antagonist of cellular innate immunity, the first line of defense against invading pathogens, hindering the cellular antiviral response during infection. This study provides novel insights into the mechanisms used by ANDV to establish its infection.
INTRODUCTION
Andes orthohantavirus (ANDV), a rodent-borne member of the Hantaviridae family of viruses of the Bunyavirales order, is the primary etiological agent of hantavirus cardiopulmonary syndrome (HCPS), a respiratory disease characterized by the development of vascular leakage syndrome, in South America (1). ANDV is endemic in Argentina and Chile (1), with the long-tailed pygmy rice rat (Oligoryzomys longicaudatus) serving as its main reservoir host (2). ANDV mainly infects humans through aerosolized excreta and secretions from infected rodents. However, unlike with other Hantaviridae viruses, person-to-person transmission of ANDV has been documented (3, 4).
The ANDV genome consists of three negative polarity single-stranded RNA segments designated large (L), medium (M), and small (S), packed into helical nucleocapsids (5, 6). Transcription of the genomic RNA generates the L, M, and S messenger RNAs (mRNAs). The LmRNA encodes a viral RNA-dependent RNA polymerase, which is required for viral RNA transcription and replication (7). The MmRNA encodes a glycoprotein precursor, which is cotranslationally processed to yield two viral envelope glycoproteins (Gc and Gn), which mediate virion assembly and cell entry (8). The SmRNA encodes a nucleocapsid (N) protein and, from an overlapping (+1) open reading frame (ORF), a nonstructural protein (NSs) (9). The ANDV N protein is multifunctional and, among other functions, packs the viral genome and antagonizes the host immune response (10). To date, the role of the ANDV-NSs protein remains unknown.
The NSs proteins of other members of the Bunyavirales order are nonessential for virus replication, but they contribute to viral pathogenesis by acting as interferon (IFN) antagonists (11–21). Therefore, it was conceivable to predict that ANDV-NSs shared a similar function, serving as an antagonist of the cellular type I IFN antiviral response. In general, viral infections trigger the cellular antiviral response, which leads to the activation of type I IFN and proinflammatory cytokines (22, 23). The cytosolic retinoic acid-inducible gene 1 (RIG-I) and melanoma differentiation-associated protein 5 (MDA5) sensors, also referred to as the RIG-I-like receptors (RLRs), detect small amounts of virus-associated double-stranded RNA (dsRNA) and 5′ triphosphorylated RNA (24). RLRs then direct signaling toward the mitochondrial antiviral signaling protein (MAVS), their downstream effector. Upon activation, MAVS forms functional prion-like aggregates and recruits the downstream signaling molecules of the RLR signaling pathway (25), triggering the phosphorylation of IFN-regulatory factor 3 (IRF3) (26, 27). In uninfected cells, IRF3 is predominantly cytoplasmic, yet upon activation, phosphorylated IRF3 dimers translocate into the nucleus, inducing the expression of type I IFN genes (26, 27). Infected cells then secrete IFN, which activates the IFN signaling pathway in neighboring cells (22, 23).
ANDV is known to elicit a poor type I IFN response in infected cells and animals (28–30). ANDV has evolved redundant strategies to delay early IFN induction for efficient viral replication. The ANDV-Gn and ANDV-Gc glycoproteins, as well as the hantavirus N protein, are capable of limiting host cell innate immune responses by directly targeting the IFN induction pathway (28, 29, 31–35). Nonetheless, a close examination of the reported data suggests that the expression of ANDV-N, ANDV-Gn, and ANDV-Gc, alone or in combination, cannot fully explain the overall reduction of IFN-β expression in infected cells (28). To us, this suggested the existence of a yet unknown viral component needed to fully antagonize cellular antiviral defenses. Here, we provide strong evidence indicating that the ANDV-NSs contributes to counteracting the cellular host antiviral defense by antagonizing cellular innate immune responses. We show that the ANDV-NSs inhibits the type I IFN induction pathway by suppressing MAVS downstream signaling.
RESULTS
ANDV infection impairs IRF3 activation in cells.
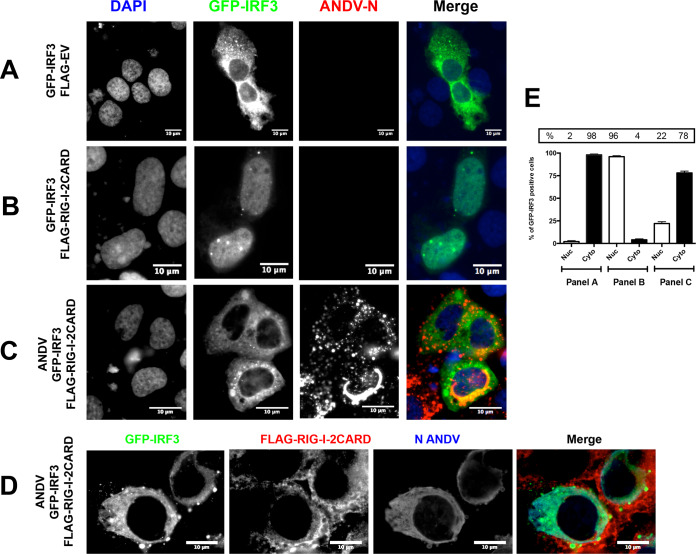
ANDV infection does not elicit IRF3 activation (29). Thus, at early times postinfection with ANDV, human lung endothelial (HMVEC-L) cells (29), human non-small-cell lung adenocarcinoma (A549) cells (28), and a human hepatoma cell line (Huh-7) (28) are unable to induce a type I IFN response. Interested by this observation, we sought to evaluate the intracellular localization of IRF3 during infection with ANDV when an effector of the IFN induction pathway is overexpressed. For this, Huh-7 cells were transfected with the plasmid pEGFP-C1-hIRF3, which encodes the human IRF3 protein fused to the green fluorescent protein (GFP) expressed from a human cytomegalovirus promoter, either with an inducer plasmid encoding the N-terminal 2CARD domain of RIG-I (20, 36, 37) or with the FLAG-empty vector (FLAG-EV), which corresponds to the same DNA plasmid lacking the protein-coding region, as a negative control. In about 98% of the cells transfected with the FLAG-EV plasmid overexpressed GFP-IRF3 fusion protein localized to the cytoplasm (Fig. 1A and E, “panel A”), and the recombinant GFP-IRF3 protein relocalized to the nucleus in about 96% of the cells upon activation of the IFN-production pathway by transfection with the inducer plasmid encoding the N-terminal 2CARD domain of RIG-I (Fig. 1B and E, “panel B”). This result demonstrated the ability of the GFP-IRF3 fusion protein to respond to RIG-I signaling in Huh-7 cells. Infection of Huh-7 cells with ANDV, followed by the detection of the ANDV-N protein, abrogated the ability of RIG-I to induce GFP-IRF3 translocation to the nucleus (Fig. 1C and D); in about 78% of the cells expressing the N-terminal 2CARD domain of RIG-I, the GFP-IRF3 fusion protein remained in the cytoplasm (Fig. 1C, D, and E, “panel C”).
FIG 1.
ANDV infection impairs RIG-I-induced IRF3 activation. Huh-7 cells were mock infected (A and B) or infected with ANDV at a multiplicity of infection (MOI) of 1 (C and D). At 24 h postinfection (h p.i.), cells were transfected with green fluorescent protein (GFP)-IRF3 together with FLAG-EV (A) or a plasmid encoding FLAG-RIG-I-2CARD (B to D). At 24 h posttransfection (h p.t.), the cells were fixed using 4% paraformaldehyde (PFA) and permeabilized with phosphate-buffered saline (PBS)-Triton X-100, and cells were incubated with an anti-ANDV-N antibody (A to C) or with a rabbit anti-ANDV-N antibody plus mouse anti-FLAG antibody (D). (A, B, and C) Alexa Fluor anti-mouse 594 antibody was used as the secondary antibody, and Vectashield with 4′,6-diamidino-2-phenylindole (DAPI) was used as the mounting medium. (D) Alexa Fluor anti-mouse 594 antibody plus an Alexa Fluor anti-rabbit 350 antibody were used as a secondary antibody and Dako without DAPI was used as the mounting medium. The images were obtained by Olympus epifluorescence microscopy and processed with ImageJ. (E) Numbers of cells with a nuclear and cytoplasmic distribution of GFP-IRF3 from panels A (32 cells), B (29 cells), and C (59 cells) were counted by hand using ImageJ and results presented as a percentage. From panel C, only the GFP-IRF3 and ANDV-N protein-positive cells were counted. As a statistical test, a one-way analysis of variance (ANOVA) using the standard error of the mean (SEM) was performed.
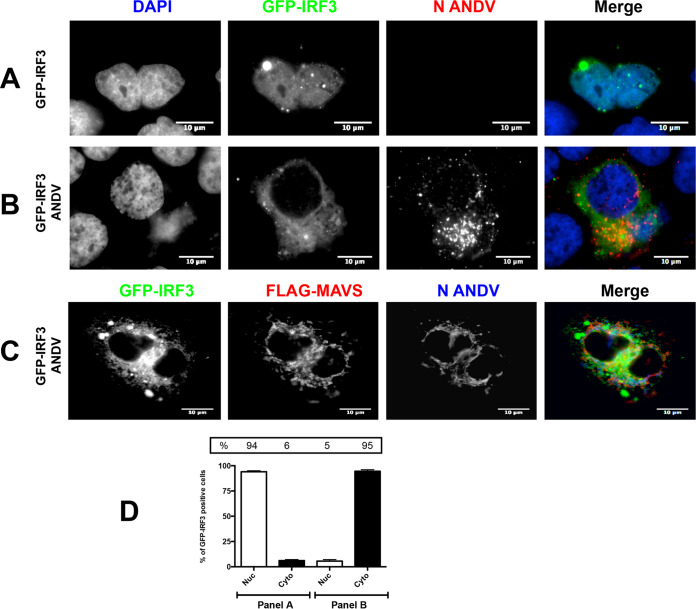
Next, we wondered whether the presence of ANDV could also abrogate IRF-3 activation when a plasmid overexpressing a FLAG-MAVS protein, a common adaptor protein for both MDA5 and RIG-I signaling, was used as an effector of the IFN induction pathway. For this, Huh-7 cells were transfected with the plasmid pEGFP-C1-hIRF3 with an inducer plasmid encoding a FLAG-tagged MAVS protein and infected, or not, with ANDV. In the absence of ANDV, overexpressed GFP-IRF3 fusion protein localized to the nucleus in about 94% of the cells (Fig. 2A and D, “panel A”), suggesting IRF-3 activation when cells are transfected with the plasmid encoding a FLAG-tagged MAVS protein. The infection of Huh-7 cells with ANDV, followed by the detection of the ANDV-N protein, abrogated the ability of FLAG-MAVS to induce GFP-IRF3 translocation to the nucleus (Fig. 2B, C, and D, “panel B”). In 95% of ANDV-infected cells, the GFP-IRF3 fusion protein remained in the cytoplasm (Fig. 2B, C, and D, “panel B”). Therefore, ANDV replication not only avoids eliciting IRF3 activation, as shown by Spiropoulou et al. (29), but the virus also antagonizes IRF3 activation by directly hindering the RLR signaling pathway in Huh-7 cells.
FIG 2.
ANDV infection impairs MAVS-induced IRF3 activation. Huh-7 cells were mock infected (A) or infected with ANDV at an MOI of 1 (B and C). At 24 h p.i., cells were transfected with GFP-IRF3 together with a plasmid encoding FLAG-MAVS (A to C). At 24 h p.t., the cells were fixed using 4% PFA and permeabilized with PBS-Triton, and covers were incubated with a mouse anti-ANDV-N antibody (A and B). Alexa Fluor anti-mouse 594 antibodies were used as a secondary antibody, and Vectashield with DAPI was used as the mounting medium. (C) Covers were incubated with a mouse anti-FLAG together with a rabbit anti-ANDV-N antibody. Alexa Fluor anti-mouse 594 antibodies and Alexa Fluor anti-rabbit 350 antibodies were used as a secondary antibody, while Dako without DAPI was used as the mounting medium. The images were obtained by Olympus epifluorescence microscopy and processed with ImageJ. (D) The numbers of cells with a nuclear and cytoplasmic distribution of GFP-IRF3 from panels A (25 cells) and B (75 cells) were counted and results presented as a percentage. From panel B, only the GFP-IRF3 and ANDV-N protein-positive cells were counted. As a statistical test, a one-way ANOVA using the standard error of the mean (SEM) was performed.
The NSs protein is detected during ANDV infection.
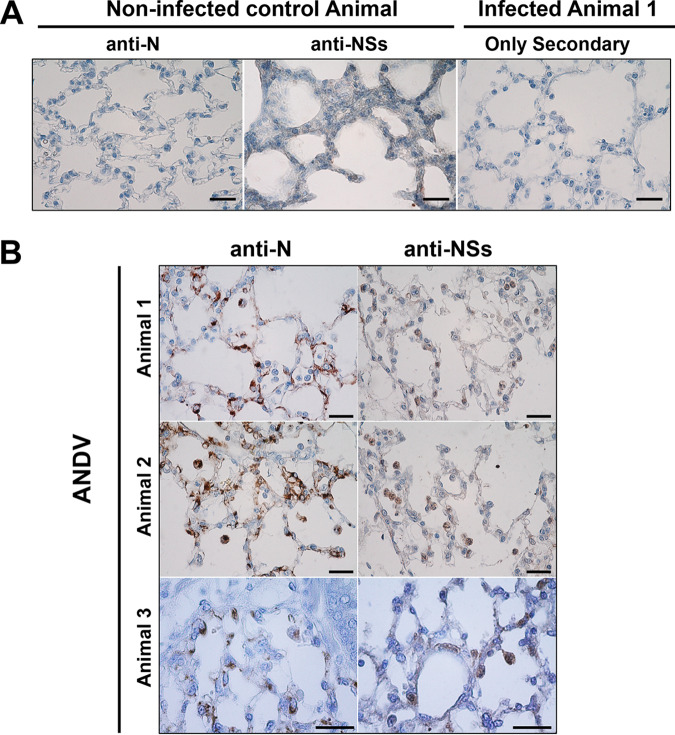
In a previous report, we described the expression of the ANDV-NSs protein in ANDV-infected Vero E6 cells (9). We wondered if, similarly to the NSs proteins of other members of the Bunyavirales order (11–19), the ANDV-NSs protein could play a part in the inhibition of innate immunity observed during ANDV infection (Fig. 1 and 2). However, to date, the expression of the ANDV-NSs protein has only been demonstrated in cell culture-infected cells (9). So, before addressing its putative function, we decided to evaluate the expression of the ANDV-NSs protein in lung samples obtained from experimentally ANDV-infected animals. For this, lung tissue samples of three golden Syrian hamsters inoculated with Andes/ARG, a strain isolated from the lung of an Oligoryzomys longicaudatus specimen captured in the Patagonian forest ecoregion (38, 39), were analyzed. Previous studies using the same ANDV/hamster lethal HCPS model showed that infectious virus was detected in serum at day 8 postinoculation (p.i.) and that animals died at around day 10 p.i. (39–41). Based on these reports and due to the limited access to animal samples, we arbitrarily decided to evaluate the possible expression of the ANDV-NSs by immunohistochemistry in lung samples harvested at 8 days p.i. The expression of the ANDV-N protein was followed as a positive indicator of viral infection. As expected, no staining was observed when the anti-N or anti-NSs antibodies were used in tissue obtained from noninfected animals or when only the secondary antibody was used in tissue obtained from infected animals (Fig. 3A). However, both the ANDV-N and ANDV-NSs proteins were detected in the lung samples of infected animals (Fig. 3B, brown staining). Thus, we confirmed our previous findings (9), and we conclude that the ANDV-NSs protein is expressed, at least at day 8, in ANDV-infected Syrian hamsters.
FIG 3.
ANDV-NSs is detected in the lungs of experimentally inoculated golden Syrian hamsters. Male golden Syrian hamsters were inoculated with a mock control or with the Andes/ARG strain of ANDV as described in the Materials and Methods. Shown are lung samples collected from representative animals in each group, necropsied at day 8 postinoculation with a mock sample or with the virus and tested for the presence of the N and NSs viral proteins by immunohistochemistry. (A) Lung tissue sample obtained from mock-inoculated animals was processed for the ANDV-N (far left) or the ANDV-NSs (middle) proteins using an anti-N (94) or an anti-NSs (9) antibody, respectively. Lungs obtained from an animal infected with the Andes/ARG strain of ANDV were stained using the same procedure but omitting the addition of the primary anti-ANDV protein antibody and using only the secondary antibody (far right). (B) Lung tissue samples obtained from three independent animals infected with the Andes/ARG strain of ANDV were tested for the presence of the N and NSs viral protein by immunohistochemistry using an anti-N (94) or an anti-NSs (9) antibody. The cellular nuclei of the lung tissues were stained with hematoxylin-eosin. Bars, 50 μm.
The ANDV-NSs protein suppresses the type I IFN induction pathway downstream of MDA5 and RIG-I and upstream of TBK1.
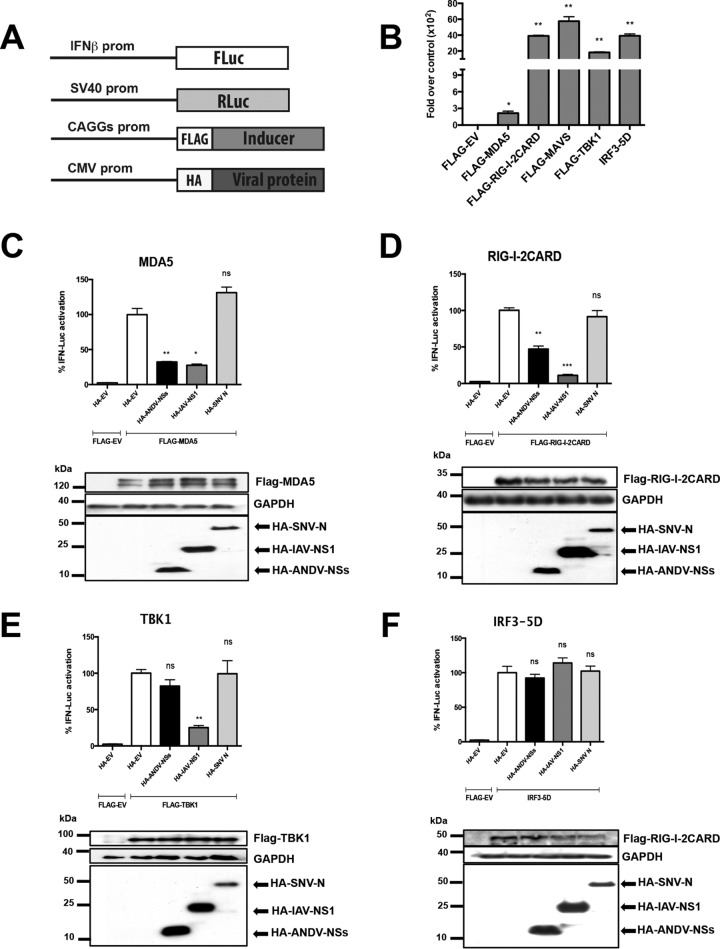
As the ANDV-NSs is indeed expressed during ANDV infection in cells (9) and in animals (Fig. 3), we next sought to address its possible function as an antagonist of the type I IFN response. It was noteworthy that the ANDV-Gn, -Gc, and -N proteins, when expressed individually or in combination, inhibit the IFN induction pathway (28, 29, 31–35). In consequence, the inhibition of IRF3 activation observed during ANDV infection (Fig. 1 and 2) could be due to the individual or combined expression of the ANDV-Gn, -Gc, or -N proteins (28, 29, 31–35). Due to the lack of a reverse genetics system for ANDV, and to the known functions of the ANDV-Gn, -Gc, and -N proteins, we decided to explore the potential role of the ANDV-NSs protein as an antagonist of the type I IFN response in a virus-free system. This experimental approach allowed us to avoid interference from other ANDV proteins (28, 29, 31–35). HEK 293T cells were selected for the experiments because they can be efficiently transfected and have proved useful in characterization of the inhibition of innate immunity exerted by the other ANDV proteins (33, 42), as well as by proteins from other members of the Bunyavirales order (12, 20, 43, 44). So, a plasmid expressing a hemagglutinin (HA)-tagged ANDV-NSs protein was cotransfected into HEK 293T cells along with plasmids expressing the FLAG-tagged version of MDA5, RIG-I, TBK1, or IRF3-5D and the Firefly (FLuc) (IFN-β–FLuc) or the Renilla (RLuc) (SV40-RLuc) luciferase reporters (Fig. 4A). Luciferase mRNA expression from the IFN-β–FLuc plasmid is driven by the IFN-β promoter (45), while expression from the RLuc reporter, which served as a control for transfection efficiency between samples, is driven by the SV40 promoter (Fig. 4A). The FLAG-EV plasmid was used as a negative control because it does not induce the RLR signaling pathway (Fig. 1). An empty plasmid corresponding to the backbone used to construct the viral protein-expressing vectors (HA-EV), a plasmid driving expression on the influenza A virus (IAV) HA-NS1 protein, which is known to block the type I IFN induction pathway (46–48), and a plasmid encoding the HA-Sin Nombre virus (SNV) N protein, which does not suppress the RLR pathway (28, 33), were also used as controls. The type I IFN induction pathway was activated by individually overexpressing the FLAG-tagged versions of the various effectors of the pathway (20, 33) (Fig. 4B). At 18 h posttransfection, cell lysates were subjected to a dual-luciferase reporter assay using the FLuc/RLuc ratio as a readout for gene expression. The expression of MDA5 (Fig. 4C), RIG-I-2CARD (Fig. 4D), TBK1 (Fig. 4E), or IRF3-5D (Fig. 4F), as well as that of the viral proteins ANDV-NSs, IAV-NS1, and SNV-N, was confirmed by Western blot analysis (Fig. 4, lower panels). The results showed that neither the HA-EV- nor the SNV-N-expressing plasmid hinders the activation of the type I IFN induction pathway by the different effector proteins (Fig. 4) (28, 33). As expected (46–48), IAV-NS1 significantly suppressed the activity of the IFN-β promoter by 73, 89, and 75% when the pathway was induced by the overexpression of MDA5 (Fig. 4C), RIG-I (Fig. 4D), and TBK1 (Fig. 4E), respectively. No effect on the activity of the IFN-β promoter was observed when cells were transfected with IAV-NS1 in combination with IRF3-5D (Fig. 4F), which is a constitutively active form of IRF3 (37). The ANDV-NSs protein significantly suppressed transactivation of the IFN-β promoter by 68 and 53% when induced by MDA5 (Fig. 4C) and RIG-I (Fig. 4D), respectively. The ANDV-NSs protein had no impact on the activity of the IFN-β promoter when TBK1 (Fig. 4E) or IRF3-5D (Fig. 4F) induced the pathway. These findings indicate that the ANDV-NSs protein antagonizes the type I IFN induction pathway by suppressing signals downstream of MDA5 and RIG-I and upstream of TBK1.
FIG 4.
The ANDV-NSs protein suppresses the IFN induction pathway downstream of MDA5 and RIG-I and upstream of TBK1. (A) Schematic representation of IFN-β–Fluc, SV40-Rluc, pHA-EV, pCAGGs-FLAG-EV, or pCAGGs-FLAG encoding the FLAG versions of MDA5, RIG-I-2CARD, MAVS, TBK1, or IRF3-5D plasmids used in the assays. (B) HEK-293T cells were transfected with IFN-β–Fluc, SV40-Rluc, pHA-EV, pCAGGs-FLAG-EV, or pCAGGs-FLAG encoding the FLAG versions of MDA5, RIG-I-2CARD, MAVS, TBK1, or IRF3-5D, all inducers of the IFN induction pathway by polyethyleneimine (PEI). At 18 h p.t., the cells were lysed, and FLuc and RLuc luciferase activities were determined using a commercial dual-luciferase assay (Promega). Values represent the relative luciferase activity (FLuc/RLuc) obtained for the IFN-β–FLuc reporter plasmids when transfected with the inducer plasmids over the FLuc/RLuc activity obtained when transfecting the FLAG-EV plasmid, which was arbitrary set as 1.Values shown are the mean (±SEM) of at least three independent experiments, each conducted in triplicate. Statistical analysis was performed with a Student’s t test (*, P < 0.05). HEK 293T cells were transfected with plasmids expressing the HA-tagged ANDV-NSs, IAV-NS1, and SNV-N proteins or with HA-EV along with plasmids expressing the FLAG-tagged version of MDA5 (C) RIG-I (D), TBK1 (E), or IRF-5D (F) and the plasmids IFN-β–FLuc and SV40-RLuc. At 18 h p.t., the cells were lysed with passive lysis buffer (Promega), and FLuc and RLuc activities were determined using a dual-luciferase assay (Promega). The relative luciferase activity (FLuc/RLuc) obtained for IFN-β–FLuc when transfected with the plasmid pHA-EV together with the inducer of the IFN production pathway was set to 100%. The values shown are the mean (±SEM) of at least three independent experiments, each conducted in triplicate. Statistical analysis was performed by ANOVA followed by Dunnett’s multiple-comparison test (*, P < 0.05). A representative Western blot for each set of experiments is shown below its corresponding bar chart.
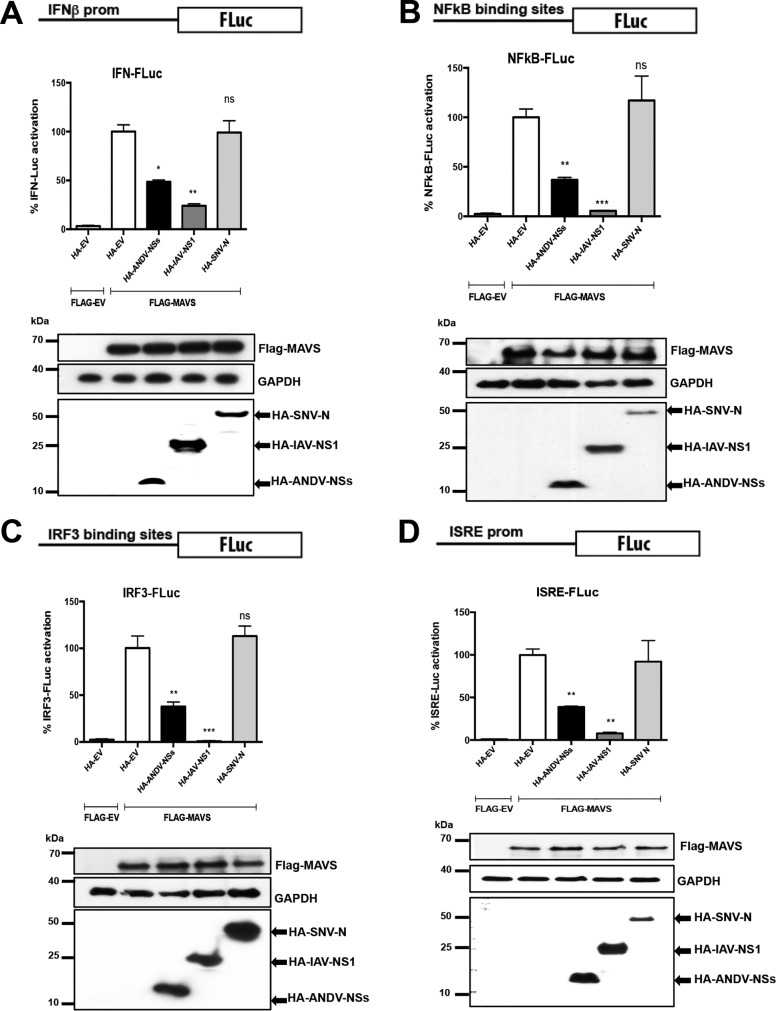
The ANDV-NSs protein targets the adaptor protein MAVS.
In the IFN induction pathway, MAVS are downstream of MDA5 and RIG-I and upstream of TBK1 (49–52). Thus, we next wondered whether MAVS were the target of the ANDV-NSs protein. To evaluate this hypothesis, HEK 293T cells were cotransfected with FLAG-EV or a plasmid encoding FLAG-MAVS along with the HA-EV plasmid, or with plasmids encoding either ANDV-NSs, IAV-NS1, or SNV-N, together with different luciferase reporters. As reporters, we used the plasmids IFN-β–FLuc, NF-κB-FLuc, IRF3-FLuc, and ISRE-FLuc, for which transcription is driven by the IFN-β promoter (45), an NF-κB responsive promoter with two NF-κB binding sites (53), a promoter with three IRF3 binding sites (45), or the interferon-stimulated response element (ISRE) promoter (53), respectively. Cells were also cotransfected with the SV40-RLuc plasmid as a control for transfection efficiency between samples. At 18 h posttransfection, cell lysates were subjected to a dual-luciferase reporter assay using the FLuc/RLuc ratio as a readout for gene expression. The expression of MAVS and the viral proteins ANDV-NSs, IAV-NS1, and SNV-N was confirmed by Western blot (Fig. 5, lower panels). Neither the HA-EV- nor the SNV-N-expressing plasmid hindered MAVS-induced transcriptional activation of the IFN-β (Fig. 5A), NF-κB (Fig. 5B), IRF3 (Fig. 5C) or ISRE (Fig. 5D) promoters. IAV-NS1 reduced MAVS-induced FLuc activity by 76, 95, 98, and 92% from the IFN-β–FLuc, NF-κB-FLuc, IRF3-FLuc, and ISRE-FLuc plasmids, respectively (Fig. 5). ANDV-NSs antagonized MAVS-induced IFN-β promoter activity by 52% (Fig. 5A), NF-κB promoter activity by 74% (Fig. 5B), IRF3 promoter activity by 63% (Fig. 5C), and ISRE promoter activity by 61% (Fig. 5D). From these results, we conclude that the ANDV-NSs protein targets MAVS signaling in HEK 293T cells.
FIG 5.
The ANDV-NSs protein targets the adaptor protein MAVS. HEK 293T cells were cotransfected with FLAG-EV or with a plasmid encoding FLAG-MAVS along with HA-EV or with plasmids encoding either HA-ANDV-NSs, HA-IAV-NS1, or HA-SNV-N, together with the IFN-β–FLuc (A), NF-κB–FLuc (B), IRF3-FLuc (C), or ISRE-FLuc (D) reporter plasmids. The cells were also cotransfected with the SV40-RLuc plasmid as a control for transfection efficiency between samples. At 18 h p.t., the cells were lysed using passive lysis buffer (Promega), and FLuc and RLuc activities were measured using a dual-luciferase assay (Promega). The relative luciferase activity (FLuc/RLuc) obtained for the IFN-β–FLuc, NF-κB–FLuc, IRF3-FLuc, and ISRE-FLuc reporter plasmids when transfected with plasmid pHA-EV together with FLAG-MAVS was set to 100%. The values shown are the mean (±SEM) of at least three independent experiments, each conducted in triplicate. Statistical analysis was performed by ANOVA followed by Dunnett’s multiple-comparison test (*, P < 0.05). A representative Western blot for each set of experiments is shown below its corresponding bar chart.
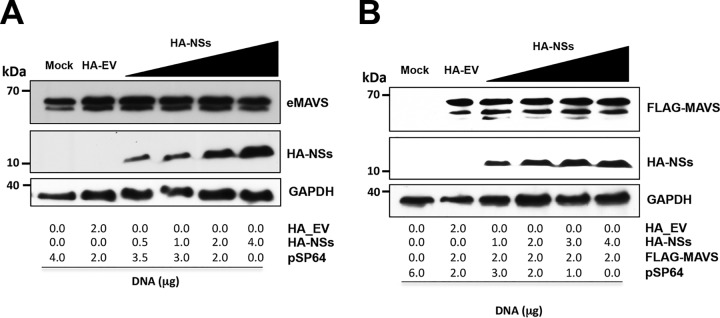
The suppression of MAVS-induced TBK1 activation by the ANDV-NSs protein could be due to a reduction in MAVS content, which could ultimately impact its signaling. To test this possibility, we evaluated if the expression of the ANDV-NSs protein altered the levels of the endogenous or the overexpressed MAVS. For this, HEK 293T cells were transfected, or not, with increasing amounts of the HA-NSs plasmid or with the HA-EV control plasmid (Fig. 6). Alternatively, cells were transfected, or not, with the FLAG–MAVS-expressing plasmid. The levels of endogenous MAVS (eMAVS) (Fig. 6A), FLAG-MAVS (Fig. 6B), and the HA-NSs proteins were followed by Western blot analysis using GAPDH as a loading control. The overexpression of the ANDV-NSs protein did not affect eMAVS (Fig. 6A) or FLAG-MAVS (Fig. 6B) protein levels, suggesting that ANDV-NSs protein is not involved in the regulation of MAVS stability.
FIG 6.
The HA-NSs protein expression does not impact on the levels of endogenous or recombinant MAVS. HEK 293T cells were transfected with different amounts of HA-NSs expression plasmid (A and B) or together with a constant amount of FLAG-MAVS (B). At 24 h p.t., cells were lysed and Western blotting was performed using mouse anti-MAVS, mouse anti-HA, and mouse anti-GAPDH primary antibodies (A) or mouse anti-FLAG, mouse anti-HA and mouse anti-GAPDH primary antibodies (B).
The ANDV-NSs protein suppresses MAVS-induced type I IFN expression in cells.
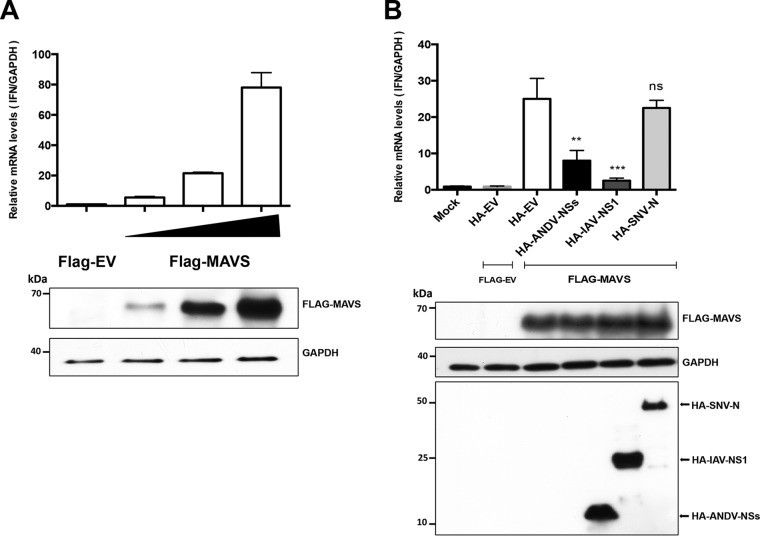
The above data indicate that the ANDV-NSs protein antagonizes the type I IFN induction pathway by suppressing MAVS signaling. If so, overexpression of ANDV-NSs should antagonize the expression of endogenous IFN-β induction by MAVS overexpression. To investigate this, we first determined if the overexpression of the FLAG-MAVS protein induced the expression of endogenous IFN-β. For this, HEK-293T cells were transfected with the FLAG-EV or different concentrations of the FLAG-MAVS plasmid (6.25, 12.5, or 25 ng). Total RNA from cells was extracted and used as the template in a reverse transcription-quantitative PCR (RT-qPCR) designed to detect the endogenous IFN-β transcript (54). As an internal normalizer, the same template was used in an RT-qPCR designed to detect endogenous GAPDH mRNA (9). The level of IFN-β mRNA was expressed relative to the level of mRNA of GAPDH, with the IFN-β mRNA/GAPDH mRNA ratio obtained in the presence of the FLAG-EV control vector set to 1. As expected, the overexpression of FLAG-MAVS induced the expression of endogenous IFN-β in a concentration-dependent manner (Fig. 7A). Next, a similar experiment was conducted, but cells were cotransfected with the plasmid overexpressing FLAG-MAVS (12.5 ng) together with the HA-EV, the ANDV-NSs, the HA-IAV-NS1, or the HA-SNV-N. As a negative control, cells were transfected with the HA-EV together with the FLAG-EV. Untransfected cells (mock) were used to determine the basal levels of endogenous IFN-β mRNA. Results showed that the transfection of the HA-EV plasmid in combination with FLAG-EV did not alter the levels of endogenous IFN-β mRNA (compare to mock in Fig. 7B). Transfection of the FLAG-MAVS plasmid in the presence of the HA-EV or HA-SNV-N induced the expression of endogenous IFN-β expression by 25- and 23-fold, respectively (Fig. 7B), while the expression of HA-IAV-NS1 antagonized the induction of endogenous IFN-β (only a 3-fold induction) by the overexpressed FLAG-MAVS protein. As predicted based on results shown in Fig. 5, the expression of the ANDV-NSs protein also antagonized, without fully abolishing, the induction of endogenous IFN-β (only an 8-fold induction) by the FLAG-MAVS protein. This result confirms our previous conclusions, indicating that the ANDV-NSs protein antagonizes the MAVS-induced type I IFN signaling pathway.
FIG 7.
The ANDV-NSs inhibits MAVS induced transcriptional activation of endogenous IFN-β. (A) HEK 293T cells were transfected with FLAG-EV vector or with increasing amounts of FLAG-MAVS, and (B) HEK 293T cells were mock transfected or transfected with FLAG-EV or FLAG-MAVS plus HA-EV, HA-ANDV-NSs, HA-IAV-NS1, or HA-SNV-N. At 18 h posttransfection, the cells were collected, and total RNA was extracted from transfected cells and used as a template in a reverse transcription-quantitative PCR (RT-qPCR) assay designed to detects human IFN-β mRNA and GAPDH mRNA (9, 54). Values are the mean (± standard deviation [SD]) of results from three independent experiments, each conducted in duplicate. Statistical analysis was performed with ANOVA followed by Dunnett’s multiple-comparison test; *, P < 0.05 versus Flag-EV plus HA-EV. A representative Western blot for each set of experiments is shown below its corresponding bar chart.
The ANDV-NSs protein and MAVS interact in HEK 293T cells.
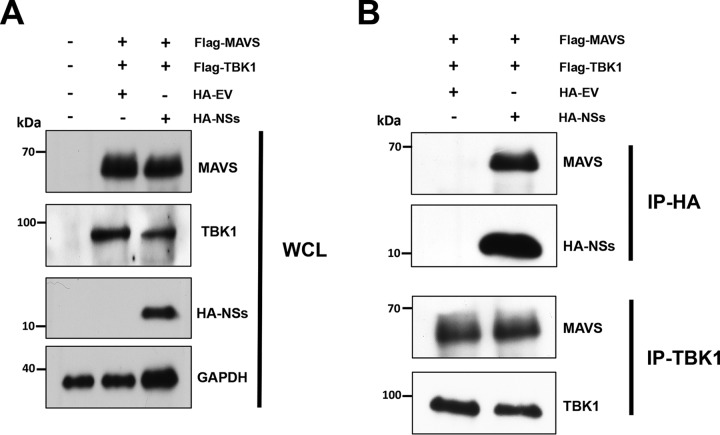
ANDV-NSs blocks type I IFN induction to a lesser extent than the IAV-NS1 protein does (Fig. 4 and 5). The Uukuniemi virus (UUKV)-NSs protein, which is also a weak IFN antagonist, acts by binding to MAVS and blocking its downstream signaling (20). Based on these observations (20), we sought to determine if the ANDV-NSs protein bound with MAVS and, if so, to establish if the MAVS-TBK-1 interaction was abolished in the presence of the ANDV-NSs. For this, HEK293T cells were cotransfected with the FLAG-MAVS and FLAG-TBK1 plasmids together with the HA-EV or HA-NSs plasmids. Then, 24 h later, cells were lysed (whole-cell lysate [WCL]), and the expression of MAVS, TBK-1, and the HA-NSs was confirmed by Western blotting using GAPDH as a loading control (Fig. 8A). WCL were then subjected to immunoprecipitation using an anti-HA or anti-TBK1 antibody. Interestingly, FLAG-MAVS coimmunoprecipitated with the HA-NSs protein, suggesting that both proteins interact in cells (Fig. 8B). However, and in contrast to the UUKV-NSs protein (20), the interaction between MAVS and TBK-1 was not disrupted by the presence of the ANDV-NSs protein (Fig. 8B).
FIG 8.
The HA-NSs protein and Flag-MAVS bind in HEK 293T cells. HEK293T cells were cotransfected with Flag-MAVS, Flag-TBK1, and HA-EV or HA-NSs plasmids. At 24 h p.t., cells were lysed, and immunoprecipitation (IP) assays were performed using protein A/G agarose coated with the corresponding antibody. The beads were washed extensively 4 times and incubated with loading buffer at 95°C to rescue the supernatant, which was used for Western blotting. (A) Whole-cell lysate (WCL) of each sample and (B) IP fractions with the corresponding primary antibody highlighted on the left side of the panels. Western blotting was performed using mouse anti-TBK1, mouse anti-MAVS, mouse anti-HA, and mouse anti-GAPDH. horseradish peroxidase (HRP)-conjugated protein A/G was used to detect the primary antibodies.
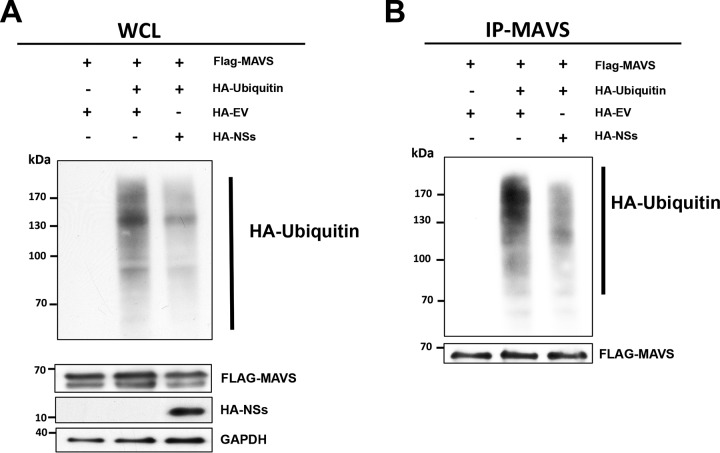
Intrigued by the above results, we decided to further explore the mechanism by which the ANDV-NSs protein reduced MAVS signaling. MAVS aggregation, the hallmark of its activation and antiviral function, requires MAVS polyubiquitination (55, 56). Virally induced MAVS aggregation is needed for IRF3 phosphorylation and subsequent IFN-β production (25). To evaluate the possible impact of the ANDV-NSs protein on MAVS ubiquitination, HEK293T were transfected with a plasmid expressing an HA-tagged human ubiquitin (HA-Ub) together with the FLAG-MAVS plasmid, as well as the HA-EV or HA-NSs plasmids, and cells were treated with the proteasome inhibitor MG-132. Results showed that ubiquitination was reduced in cells expressing the FLAG-MAVS together with the ANDV-NSs protein compared to that in cells transfected with the FLAG-MAVS and HA-EV plasmid (Fig. 9A). To determine if the degree of ubiquitination of MAVS was directly altered in the presence of the viral protein, total MAVS were immunoprecipitated using an anti-MAVS antibody, and upon resolution, the Western blot was developed either using a mouse anti-HA-antibody to evaluate the relative level of ubiquitination of the immunoprecipitated MAVS (57, 58) or with an anti-MAVS antibody. Results showed a reduction in HA-Ub in the immunoprecipitated MAVS in cells expressing the ANDV-NSs protein (Fig. 9B), suggesting that the inhibition of MAVS signaling by the ANDV-NSs protein could be due to a reduction of MAVS ubiquitination.
FIG 9.
The ANDV-NSs protein reduced MAVS polyubiquitination in cells. HEK 293T cells were cotransfected with HA-EV or HA-NSs plus Flag-MAVS and HA-ubiquitin vectors. At 24 h posttransfection, cells were treated with MG-132 (5 μM) for 6 h and lysed. (A) Whole-cell lysate (WCL) was used to evaluate total ubiquitinated proteins as well as the overexpression of MAVS and the ANDV-NSs proteins. GAPDH was detected as a loading control. (B) MAVS IP fractions were generated from the WCL by immunoprecipitation assay using an anti-MAVS antibody. Western blotting was performed using a mouse anti-HA to visualize ubiquitinated MAVS and an anti-MAVS to confirm MAVS immunoprecipitation. Horseradish peroxidase (HRP)-conjugated protein A/G was used to detect the primary antibodies.
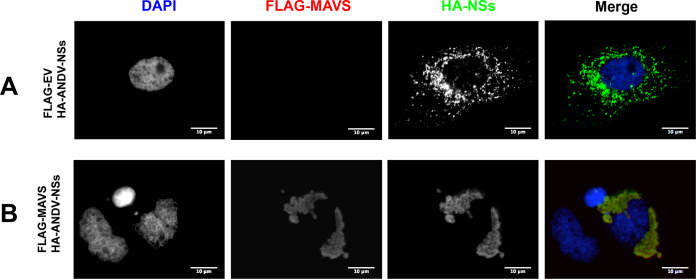
The ANDV-NSs protein interacts with MAVS and blocks its downstream signaling in Huh-7 cells.
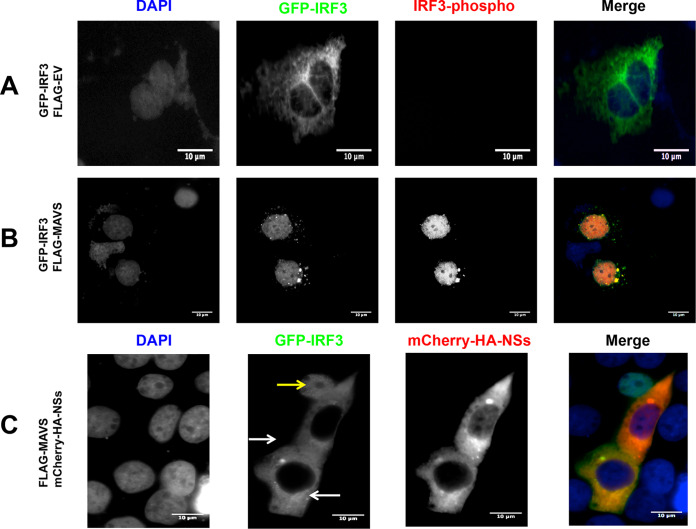
The initial experiments of this study were conducted using Huh-7 cells. Thus, we wondered if MAVS signaling was also blocked by the ANDV-NSs protein in Huh-7 cells or if this observation was only valid for HEK293T. For this, Huh-7 cells were transfected with the plasmid pEGFP-C1-hIRF3 together with the FLAG-EV plasmid or the FLAG-MAVS-encoding plasmid. The overexpressed IRF3-GFP fusion protein was localized to the cytoplasm in cells transfected with the FLAG-EV plasmid (Fig. 10A). The recombinant IRF3-GFP protein was phosphorylated and relocalized to the cell nucleus upon the transfection of cells with the FLAG-MAVS plasmid (Fig. 10B). Thus, as expected, IRF3 is activated when cells are transfected with the FLAG-MAVS plasmid (Fig. 10B). When the FLAG-MAVS plasmid was transfected together with the mCherry-HA-NSs plasmid, the IRF3-GFP protein remained in the cytoplasm of cells expressing the ANDV-NSs protein (Fig. 10C).
FIG 10.
The ANDV-NSs protein inhibits MAVS-induced IRF3 activation in Huh-7 cells. Huh-7 cells were transfected with GFP-IRF3 together with the plasmid FLAG-EV (A) or FLAG-MAVS (B). Huh-7 cells were transfected with GFP-IRF3 together with the plasmids FLAG-MAVS and mCherry-HA-NSs (C). The latter expresses an mCherry-tagged version of the ANDV-NSs protein. At 24 h p.t., the cells were fixed with 4% PFA and permeabilized with PBS-Triton X-100. The cells, shown in panels A and B were incubated with a rabbit anti-phospho-IRF3 primary antibody and an Alexa Fluor 594 donkey anti-rabbit secondary antibody. Vectashield with DAPI was used as a mounting medium in panels A, B, and C. (C) GFP fluorescence served as a marker for IRF3, DAPI marked the cell nuclei, and mCherry fluorescence-labeled ANDV-NSs. The white arrow highlights an NSs-positive cell, while the yellow arrow points to an NSs-negative cell. The images were obtained on an Olympus epifluorescence microscope and processed with ImageJ.
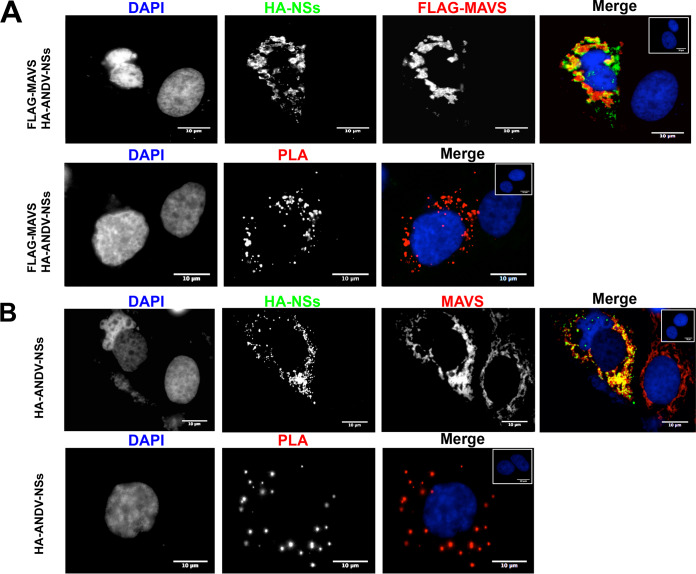
Next, we sought to determine if the cellular localization of the overexpressed ANDV-NSs was altered when MAVS are also overexpressed. For this, HEK293 cells were transfected with the FLAG-MAVS or the FLAG-EV plasmid in combination with the plasmid encoding ANDV-NSs. The expression and localization of the overexpressed proteins were followed by immunofluorescence (IF) (Fig. 11). Interestingly, the cellular distribution of the ANDV-NSs protein varied when in the presence of the FLAG-MAVS proteins (compare Fig. 11A and B). Consistent with the coimmunoprecipitation data (Fig. 8), the analysis of different cells coexpressing both the ANDV-NSs and the FLAG-MAVS protein suggested that these proteins colocalized (Fig. 11). To confirm the protein-protein interaction between MAVS and ANDV-NSs directly within Huh-7 cells, we performed an in situ proximity ligation assay (PLA) (59, 60) in cells expressing both proteins. Note that mock-transfected cells used as negative controls are shown in the upper corner of the panels (Fig. 12). Expression of the ANDV-NSs and FLAG-MAVS in Huh-7 cells transfected with the HA-NSs of FLAG-MAVS plasmid was assessed by immunofluorescence (Fig. 12A, upper). The PLA was conducted using a rabbit anti-HA and a mouse anti-FLAG as primary antibodies (Fig. 12A, lower). The PLA signal indicated that overexpressed ANDV-NSs and FLAG-MAVS interact within cells (Fig. 12A, lower), confirming the coimmunoprecipitation results obtained in HEK 293T cells (Fig. 8).
FIG 11.
The recombinant HA-NSs redistributes in the presence of FLAG-MAVS. Huh-7 cells were cotransfected with HA-NSs and FLAG-EV (A) or FLAG-MAVS (B). At 24 h p.t., cells were fixed and permeabilized with 4% PFA and PBS-Triton. HA-NSs proteins and FLAG-MAVS were detected by immunofluorescence (IF) using a mouse anti-HA and a rabbit anti-FLAG as primary antibodies. Alexa 488 donkey anti-mouse and an Alexa 594 donkey anti-rabbit were used as secondaries antibodies. Vectashield with DAPI was used as the mounting medium. The images were obtained by Olympus epifluorescence microscopy and processed with ImageJ.
FIG 12.
The ANDV-NSs protein and MAVS interact in Huh-7 cells. (A) Huh-7 cells were transfected with plasmids encoding HA-ANDV-NSs and FLAG-MAVS. At 24 h p.t., cells were fixed with 4% PFA and permeabilized using PBS-Triton X-100. ANDV-NSs and MAVS were detected by immunofluorescence (IF) using rabbit anti-HA and mouse anti-FLAG primary antibodies in combination with Alexa Fluor 488 donkey anti-rabbit and Alexa Fluor 594 donkey anti-mouse secondary antibodies (upper panels). The inset in the upper right corner of the upper right panel shows mock-transfected cells as a negative control for IF. PLA secondary antibodies were used following the manufacturer’s instructions (bottom panels). The inset in the upper left corner of the bottom right panel shows transfected cells without primary antibodies but with the PLA secondary antibodies added as a negative control of the PLA reaction. Vectashield with DAPI was used as a mounting medium. The images were obtained on an Olympus epifluorescence microscope and processed with ImageJ. (B) Huh-7 cells were transfected with a plasmid encoding HA-ANDV-NSs in combination with FLAG-EV. At 24 h p.t., cells were fixed with 4% PFA and permeabilized using PBS-Triton X-100. The ANDV-NSs protein and endogenous MAVS were detected by IF using rabbit anti-HA and mouse anti-MAVS primary antibodies and Alexa Fluor 488 donkey anti-rabbit and Alexa Fluor 594 donkey-anti-mouse as secondary antibodies. The inset in the upper right corner of the upper right panel shows mock-transfected cells as a negative control for IF. PLA secondary antibodies were used as indicated above. The inset in the upper right corner of the bottom right panel shows transfected cells without primary antibodies but with the PLA secondary antibodies added as a negative control of the PLA reaction. Vectashield with DAPI was used as a mounting medium. The images were obtained on an Olympus epifluorescence microscope and processed with ImageJ.
Next, we sought to determine if ANDV-NSs could also interact with the endogenous MAVS. For this, Huh-7 cells were transfected with the HA-EV plasmid or the plasmid encoding ANDV-NSs. The endogenous MAVS, as well as the overexpressed ANDV-NSs, were detected by immunofluorescence (Fig. 12B, upper). The PLA was conducted using a rabbit anti-HA and a mouse anti-MAVS as primary antibodies. The PLA signal confirmed the interaction between the ANDV-NSs and eMAVS (Fig. 12B, lower). We conclude that ANDV-NSs and MAVS interact with each other in Huh-7 cells.
DISCUSSION
The IFN response system is one of the earliest defense mechanisms against viral infections (22, 61). Consistent with its antiviral role, exogenously administered IFNs protect against viruses of the Bunyavirales order both in vitro and in vivo (32, 62–67). During infection, type I IFN production is triggered via the recognition of pathogen-associated molecular patterns (PAMPs) by the cell’s pathogen recognition receptors (PRRs) (23, 24, 68). PRRs recognize viral components or by-products of viral origin generated during virus replication. The sensing of PAMPs by PRRs ultimately leads to the expression of numerous cytokines, including IFNs. IFNs, in turn, induce the expression of a broad array of IFN-stimulated genes (ISGs) to establish an antiviral state. Not unexpectedly, pathogenic hantaviruses, such as ANDV, have developed diverse and redundant mechanisms to antagonize IFN-mediated defense triggered by the RLR signaling pathway. The ANDV-N, ANDV-Gn, and ANDV-Gc proteins, alone or in combination, are capable of antagonizing the type I IFN induction pathway in virus-free model systems (28, 29, 31–35). These findings are consistent with data showing a poor IFN response elicited during the early stages of ANDV replication (29). The results presented here extend the initial findings (29), showing that during an active infection ANDV not only evades but also directly antagonizes the RLR signaling pathway, suppressing IRF3 activation (Fig. 1 and 2). In these experiments, we used the intracellular localization of IRF3, a key player in the induction of type I IFNs following viral infection (26), as a readout of activation of the RLR signaling pathway. During infection, the ANDV-N protein was readily detected, indicating virus replication (Fig. 1 and 2). Interestingly, in the presence of ANDV, IRF3 nuclear translocation was abrogated even when RIG-I or MAVS proteins were overexpressed (Fig. 1 and 2). As mentioned previously, either ANDV-N, ANDV-Gn, or ANDV-Gc alone could be responsible for this observation. Nonetheless in this study, we show that the ANDV-NSs, a viral protein expressed during infection (9) (Fig. 3), when expressed alone, inhibits the RLR signaling pathway at the level of MAVS, blocking MDA5 and RIG-I but not TBK1 signaling (Fig. 4 and 5). ANDV-NSs blocked IRF3 activation (Fig. 4) and RLR-induced IFN-β, NF-κB, IRF3, and ISRE promoter activity when evaluated using luciferase reporter plasmids (Fig. 5). Consistent with this observation, the presence of the ANDV-NSs blocked MAVS-induced expression of endogenous IFN-β mRNA (Fig. 7).
In cells, MAVS is regulated at multiple stages, including at its synthesis, activation, and protein stability stages (24, 69–72). Accordingly, viruses have evolved different strategies to inhibit MAVS (68). For example, the VP3 protein of rotavirus drives MAVS degradation (73). The cytomegalovirus (CMV) protein vMIA inhibits MAVS signaling by promoting mitochondrial fission (74). The viral proteases of hepatitis A virus (HAV), hepatitis C virus (HCV), GB virus B, enterovirus 71, poliovirus, rhinovirus, and coxsackievirus cleave MAVS (52, 75–79). HCV NS5A in complex with the mitochondrial protein LRPPRC inhibits MAVS activity (80). The influenza virus protein PB1-F2 binds to MAVS and decreases mitochondrial membrane potential, which is essential for MAVS signaling (81). Results showed that the ANDV-NSs protein binds with MAVS in cells (Fig. 8 and 12), without reducing MAVS levels (Fig. 6) or interfering with the ability of MAVS to interact with TBK-1 (Fig. 8). Thus, we cannot discard the possibility that the ANDV-NSs may act like the influenza virus protein PB1-F2 (81). Even so, in this study, we evaluated MAVS ubiquitination and showed that it is reduced in the presence of the ANDV-NSs protein (Fig. 9). These results suggest that, most probably, one mechanism of action of the ANDV-NSs protein it to interfere with MAVS aggregation, a critical step in MAVS downstream signaling (25, 56). It is therefore tempting to speculate that the reduction of MAVS polyubiquitination is directly linked with the capacity of the ANDV-NSs protein to block MAVS-downstream signaling. However, further experiments, not included in this study, are required to fully dissect the molecular mechanisms by which the ANDV-NSs protein block MAVS polyubiquitination and function. It should be noted that overall cellular ubiquitination is reduced in cells expressing the ANDV-NSs proteins (Fig. 9A). Thus, we cannot discard the possibility that the ubiquitination of other proteins required for the activation of the type I IFN pathway is also targeted by the ANDV-NSs protein. So, despite our efforts to determine how ANDV-NSs impacts RLR signaling, the precise molecular mechanism involved in the inhibition of MAVS signaling by the ANDV-NSs protein remains unknown. A series of experiments are under way to further unveil the molecular mechanism by which ANDV-NSs inhibits the RLR signaling pathway.
A previous report by Cimica et al. argued that the ANDV-NSs protein was dispensable for innate immune antagonism mediated by the ANDV-S gene segment (33). This statement seems to conflict with our findings. However, this report did not address, nor did it properly demonstrate, the role of the ANDV-NSs protein. Using a ΔNSs-ANDV-SmRNA, Cimica et al. showed that the ANDV-N protein selectively inhibits helicase-directed IFN signaling pathway responses mediated by the activation of TBK1 or IKKε and that the ANDV-N protein is not a ubiquitous inhibitor of κB transcription. So, the study of Cimica et al. established that the ANDV-N protein does not require the ANDV-NSs protein to selectively inhibit helicase-directed IFN signaling pathway responses. In consequence, our findings complement those of Cimica et al. (33) by unveiling an additional function of the ANDV-S gene segment encoded proteins.
In summary, in this study, we report that the recently identified ANDV-NSs protein plays a role in inhibiting the induction of type I interferon at the level of the MAVS adaptor protein.
MATERIALS AND METHODS
Cells, viruses, and animal experiments.
Vero E6 (Vero C1008; ATCC CRL 1586), HEK 293T (ATCC CRL-3216), and Huh-7 cells provided by R. Bartenschlager (University of Heidelberg, Heidelberg, Germany) and previously used in the laboratory (82) were grown in Dulbecco’s modified Eagle’s medium (DMEM; HyClone, Logan, UT) containing 10% fetal bovine serum (HyClone, Logan, UT, USA), 1% amphotericin B, 1% penicillin-streptomycin, and 1% nonessential amino acids (Gibco BRL, Life Technologies Corporation, Carlsbad, CA) at 37°C in a 5% CO2 atmosphere. ANDV strain CHI-7913 or strain Andes/ARG were propagated in Vero E6 cells as previously described (9). ANDV infection of Vero E6 or Huh-7 cells was performed using a multiplicity of infection (MOI) of 1, as previously described (9). Viral infection was monitored by detection of the ANDV-N protein by immunofluorescence using monoclonal antibodies as described by Vera-Otarola et al. (9). The work with ANDV was performed in a biosafety level 3 (BSL3) facility at Escuela de Medicina, Pontificia Universidad Católica de Chile. The infection of animals was conducted as described by Martinez and Padula (39). The stock of Andes/ARG used in the infection assay corresponded to the 9th viral passage in Vero E6 cells, and its infectious titer was 2 × 105 PFU/ml (39). Undiluted supernatants were utilized as inocula in experiments. Two- to 4-week-old male golden Syrian hamsters (Mesocricetus auratus), provided by the National Institute of Biologic Production from the National Administration of Health Institutes and Laboratories “Dr. C. G. Malbrán” (Argentina), were infected. Animals were anesthetized by an intramuscular injection of ketamine (approximately 3 mg/100 g of body weight) prior to intramuscular inoculation with 0.4 ml of virus using a 1-ml syringe. Control animals were inoculated with the supernatants of uninfected cells. Upon infection, animals were placed in a cage inside within an aseptic air negative-pressure environmental cabinet (A130SN-Flufrance; France) located in an animal facility specially equipped for that purpose and of exclusive use for these experiments. The development of illness was evaluated, considering the lack of movement, reduced interest in food, and dyspnea as illness markers (39). After 8 days of infection (39), hamsters were vascularly perfused with Bouin fixative for 30 min; then, the tissue samples were dissected out and immersed in Bouin fixative for 2 to 3 days. Samples from the lung were dehydrated in a series of alcohols and embedded in paraffin and sent to the Universidad Austral de Chile, where they were stored. Samples were serially sectioned (4 to 6 μm thick), and adjacent sections were mounted on separate silanized (3-aminopropyltriethoxysilane; Polysciences Inc., Warrington, PA) slides and used for immunohistochemistry. The described animal studies were approved by the Animal Care and Experimentation Committee of the Universidad Austral de Chile (reference no. FONDEF D 99I 1105). Standards were according to the 2008 guidelines of the Chilean Comisión Nacional de Investigación Científica y Tecnológica (CONICYT) for the performance of animal studies and for biosafety requirements. All procedures were conducted in agreement with the Laboratory Animals Limited organization recommendations (www.lal.org.uk).
Plasmids.
The plasmids IFN-β–FLuc (p125-Luc) (45), IRF3-FLuc (p55-CIB-Luc) (45), NF-κB–FLuc (53), ISRE-FLuc (53), pCAGGs-FLAG-EV, pCAGGs-FLAG-2CARD-RIG-I (83), pCAGGs-FLAG-MDA5 (84), pCAGGs-FLAG-MAVS (85), pCAGGs-FLAG-TBK1 (86), pEGFP-N1-IRF3, and pCAGGs-IRF3-5D (36) were kindly provided by A. García Sastre (Icahn School of Medicine at Mount Sinai, New York, NY). HA-NS1 from IAV (87) was provided by R. Medina (Escuela de Medicina, Pontificia Universidad Catolica de Chile, Santiago, Chile). The HA-ubiquitin plasmid (57, 58) was kindly provided by G. Gori Savellini (Università degli Studi di Siena, Italy). To generate the plasmid HA-SNV-N, the SNV-N ORF was recovered by PCR from the plasmid pSNV-N TriEx 1.1 (88), which was kindly provided by A. Panganiban (Division of Microbiology, Tulane University, New Orleans, USA), using forward primer 5′-TACGTCTCGAGATGAGCACCCTCAAAGAAGTGCAAG-3′ and reverse primer 5′-TACGTTCTAGATTAAAGTTTAAGTGGTTCTTGGTTAGAGATTTCC-3′. The PCR product was digested with XhoI and XbaI restriction enzymes and cloned into pCI-Neo-HA (89), which was generously provided by R. Soto-Rifo (ICBM, Universidad de Chile, Santiago, Chile). To generate plasmid HA-NSs, the ANDV-NSs ORF was recovered from the plasmid Topo-His-NSs (9) by PCR using forward primer 5′-ACGTGAATTCACCATGCCGAGAAGGCAGTGG-3′ and reverse primer 5′-TACGTGCGGCCGCTTAGATGATCATCAGGCTCAAG-3′. The PCR product was digested with EcoRI and NotI restriction enzymes and cloned into the pCI-Neo-HA vector. To generate the mCherry-HA-NSs plasmid, the mCherry ORF was recovered from the mCherry plasmid (90), which was kindly provided by F. Valiente-Echeverría (ICBM, Universidad de Chile, Santiago, Chile), by PCR using forward and reverse primers 5′-AATCGGAATTCGGATCCATGGTGAGCAAGGGCGAG-3′ and 5′-AATCGGAATTCGGATCCGGACTTGTACAGCTCGTC-3′, respectively. The PCR product was digested with EcoRI and cloned into HA-NSs previously dephosphorylated with Antarctic Phosphatase (New England Biolabs, Ipswich, MA). To generate the SV40-RLuc plasmid, the dl HCV1b plasmid (82) was digested with XhoI and XbaI, refilled using the Klenow fragment (Thermo Fisher Scientific, MA), and ligated. The HA-EV plasmid (empty vector) was constructed using HA-SNV-N as a template, digested by EcoRI and NotI, refilled using the Klenow fragment, and ligated. All PCR assays were performed using Pfu DNA polymerase (Thermo Fisher Scientific). Constructs were verified by sequencing (Psomagen, Rockville, MD).
DNA transfection.
For the IFN-β–FLuc, NF-κB–FLuc, ISRE-FLuc, and IRF3-FLuc experiments, 24 h before transfection, HEK 293T cells were seeded (60,000 cells) in 48-well plates. HEK 293T cells were transfected with IFN-β–FLuc (150 ng) and SV40-RLuc (1.5 ng) together with an inducer plasmid (FLAG-MDA5, FLAG-2CARD-RIG-I, FLAG-MAVS, FLAG-TBK1, or IRF3-5D) (6.25 ng) and the HA-plasmid (30 ng) using polyethyleneimine (PEI) (ratio of DNA:PEI, 1:5; Gibco, Thermo Fisher Scientific). At 18 h posttransfection (h p.t.), the cells were lysed with passive lysis buffer (50 μl; Promega Corporation, WI). Luciferase activity in the lysate (5 μl) was determined using luciferase assay reaction I buffer (LAR I; 10 μl) and Stop and Glo reaction buffer (10 μl) from the Dual-Luciferase assay kit (Promega Corporation) using a Sirius Lumat 9507 luminometer (Berthold Detection Systems GmbH, Pforzheim, Germany). The remaining cellular lysates were pooled for each condition and used in Western blotting assays. Experiments were conducted at least 3 times each in triplicate. For GFP-IRF3 translocation experiments, pEGFP-C1-IRF3 (500 ng) together with FLAG-EV (25 ng) or FLAG-MAVS (25 ng), with or without pmCherry-HA-NSs (500 ng), were transfected in Huh-7 cells seeded (50,000 cells) 24 h before transfection in 24-well plates with glass covers using JetPEI (Polyplus Transfection Company, Illkirch, France). For GFP-IRF3 translocation experiments in ANDV-infected cells, Huh-7 cells were infected with ANDV as described above, and at 24 h postinfection (h p.i.) cells were transfected with pEGFP-C1-IRF3 together with pCAGGs-FLAG-2CARD-RIG-I, as described above. At 24 h p.t., the cells were fixed with 4% paraformaldehyde (PFA) for 20 min and subjected to an immunofluorescence assay.
For the immunoprecipitation assays, 3.6 × 106 HEK 293T cells were seeded in a 10-cm plate format and 24 h later were transfected using PEI with HA-EV (8 μg) or HA-ANDV-NSs (8 μg) together with FLAG-MAVS (8 μg) and HA-ubiquitin (5 μg) plasmids. At 24 h posttransfection, the cells were treated with 5 μM of MG-132 (Sigma) for 6 h before the collection of cells in lysis buffer (NaCl 100 mM, EDTA 2 mM, Tris-HCl [pH 7.5] 50 mM, NaF 50 mM, sodium orthovanadate 1 mM, Triton X-100 1%, and protease inhibitors; Roche Diagnostics, Indianapolis, IN). For the coimmunoprecipitation assays, HEK 293T cells were transfected with the HA-EV (7 μg) or the HA-ANDV-NSs (7 μg) plasmid together with the FLAG-MAVS (7 μg) and FLAG-TBK1 (7 μg) plasmids. At 24 h posttransfection, cells were collected in lysis buffer as described above. For the RT-qPCR experiments, 2.2 × 105 HEK 293T cells were seeded in a 24-well format; 24 h later, cells were transfected with FLAG-EV (25 ng) or increasing amounts of FLAG-MAVS (6.25, 12.5, or 25 ng) using Lipofectamine 2000. The total amount of DNA transfected per well was kept constant at 400 ng by using, when required, the pSP64 poly(A) vector (Promega Corporation) as a filler DNA. In these assays, when used, the plasmids encoding the viral proteins were transfected in HEK 293T cells or cells were mock-transfected or transfected with FLAG-EV or FLAG-MAVS plus HA-EV, HA-ANDV-NSs, HA-IAV-NS1, or HA-SNV-N plasmids using Lipofectamine 2000. At 18 h posttransfection, the cells were washed with phosphate-buffered saline (PBS) and lysed with RLNa buffer (10 mM Tris-HCl [pH 8], 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, and 1 mM dithiothreitol [DTT]) containing 10 U of RiboLock RNase inhibitor (catalog no. EO0381; Thermo Fisher Scientific, Inc.). After incubation for 5 min on ice, 500 μl of TRIzol reagent (catalog no. 15596018; Thermo Fisher Scientific, Inc.) was added following the manufacturer’s instructions. The RNA concentration was quantified by nanospectrophotometry (N60 NanoPhotometer; Implen, Westlake Village, CA). Total RNA (4 μg) was treated with Turbo DNase (catalog no. AM2238; Thermo Fisher Scientific, Inc.), and 200 ng per-sample was used for the RT-qPCR assay. The real-time RT-qPCR experiments were carried out using the Brilliant II SYBR green RT-qPCR one-step mastermix (catalog no. 600835; Agilent Technologies, Santa Clara, CA). Human IFN-β mRNA was amplified using the primers hIFNβ sense (5′-AAACTCATGAGCAGTCTGCA-3′) and hIFNβ antisense (5′-AGGAGATCTTCAGTTTCGGAGG-3′) as previously described (54). A qPCR with no previous RT was carried out to control for contaminant DNA. GAPDH mRNA was detected with the primers GAPDH sense (5′-TCCACCACCCTGTTGCTGTAG-3′) and GAPDH antisense (5′-ACCCACTCCTCCACCTTTGAC-3′) as previously described (9). Data analysis was performed by the previously described threshold cycle (ΔΔCT) method (91, 92).
Western blotting.
The protein concentration in cellular lysates was determined by Bradford assay (Bio-Rad Laboratories, Inc., CA). Proteins (40 to 60 μg) were resolved on a 15% tricine-SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride (PVDF) membrane (Thermo Fisher Scientific, MA, USA). Membranes were blocked (5% skim milk in PBS-Tween 0.1% [PBS-T]) and incubated with an anti-HA mouse monoclonal antibody (H9658; Sigma-Aldrich, St. Louis, MO) at a 1:5,000 dilution, an anti-FLAG mouse monoclonal antibody (F1804; Sigma-Aldrich) at a 1:500 dilution, an anti-IRF3 rabbit polyclonal antibody (sc-9082; Santa Cruz Biotechnology, TX, USA) at a 1:1,000 dilution, or an anti-GAPDH mouse monoclonal antibody (MA5-15738; Thermo) at a 1:5,000 dilution in PBS-T. As secondary antibodies, we used a goat anti-mouse horseradish peroxidase (HRP)-conjugated antibody or a goat anti-rabbit HRP-conjugated antibody (Merck-Millipore, Darmstadt, Germany) at a 1:10,000 dilution (5% skim milk in PBS-T). Proteins were detected using the SuperSignal West Pico or Femto chemiluminescence kits (Thermo Fisher Scientific). Total protein (1 mg) was used for immunoprecipitation assays together with protein A/G Plus agarose (sc-2003; Santa Cruz Biotechnology) loaded with anti-HA mouse monoclonal antibody (H9658; Sigma-Aldrich), anti-MAVS mouse monoclonal antibody (sc-166583; Santa Cruz Biotechnology), or anti-TBK1 mouse monoclonal antibody (sc-73115; Santa Cruz Biotechnology). After 16 h at 4°C with rotation, the beads were washed three times with lysis buffer before directly mixing with tricine loading buffer (2×) and heated (95°C for 5 min). The supernatant was recovered and subjected to Western blot assay as described above but using recombinant protein A/G conjugated with HRP as the secondary antibody (Thermo Fisher Scientific).
Immunofluorescence and proximity ligation assay.
Cells were washed with PBS and fixed with 4% paraformaldehyde (Merck Millipore) in PBS for 10 min at room temperature. Cells were permeabilized with PBS-Tr (PBS-Triton X-100 [0.03%]) for 10 min at room temperature. Coverslips were blocked with 10% bovine serum albumin (BSA) in PBS-Tr for 1 h at room temperature and incubated with the different primary antibodies, namely, anti-FLAG mouse monoclonal antibody (F1804; Sigma-Aldrich), anti-HA rabbit polyclonal antibody (H6908; Sigma-Aldrich), anti-MAVS mouse monoclonal antibody (clone E-3, sc-166583; Santa Cruz Biotechnology), anti-phospho-IRF3 (Ser396) rabbit monoclonal antibody (clone 4D4G, 4947S; Cell Signaling Technology, Inc., MA, USAA), or anti-IRF3 rabbit polyclonal antibody (sc-9082; Santa Cruz Biotechnology) in 5% BSA in PBS-Tr overnight at 4°C. Coverslips were washed 3 times with PBS-Tr and incubated with Alexa Fluor secondary antibodies (Invitrogen) for 2 h at room temperature. Coverslips were washed 3 times with PBS-Tr, once with PBS, and once with ultrapure water. Vectashield H1200 (Vector Laboratories, Inc., Burlingame, CA) with 4′,6-diamidino-2-phenylindole (DAPI) was used as a mounting medium, sealed with nail polish, and stored at 4°C. For the proximity ligation assay (PLA), the Duolink in situ PLA kit was used (Duo 92004 and Duo 92002; Sigma-Aldrich). The steps of fixation and permeabilization were performed as described above. Coverslips were incubated with blocking reagent (PLA reagent; Sigma-Aldrich) for 1 h at 37°C, incubated with primary antibodies in antibody diluent (PLA reagent; Sigma-Aldrich) for 1 h at 37°C, washed three times with PBS, and incubated with the PLA secondary antibodies anti-rabbit Plus and anti-mouse Minus for 1 h at 37°C. Coverslips were washed 3 times with PBS and once with ultrapure water; the ligation (30 min at 37°C) and PCR assay (100 min at 37°C) were performed following the manufacturer’s instructions. Finally, coverslips were washed with PBS and ultrapure water before adding Vectashield with DAPI as a mounting medium. Images were captured using an Olympus model BX51 microscope (60× objective) with MBF Stereo Investigator software version 11. ImageJ version 1.52c (National Institutes of Health, USA) was used to split channels from raw images, to count cells using its edit function and to prepare images for publication.
Immunohistochemistry.
For immunohistochemistry, the immunoperoxidase method described by Sternberger et al. was used (93). Cells were rehydrated and treated with 3% H2O2 for 15 min, washed with distilled water for 5 min, and treated with 10 mM citric acid solution. Cells were incubated with a mouse-made anti-N ANDV (7B3/F7; 1:2,000 dilution) or with the anti-NSs ANDV (NS2-5E7/D9; 1:50 dilution) primary antibodies (described in references 9 and 94), diluted in Tris buffer [pH 7.8] containing 0.7% nongelling seaweed gelatin lambda carrageenan (Sigma-Aldrich) and 0.5% Triton X-100 (Sigma-Aldrich), in a humidified chamber for 16 h at room temperature. Samples were washed and then the biotin-streptavidin-peroxidase method was used (LSAB+ System-HRP, catalog no. K0679; Dako). Biotin-conjugated secondary antibody was detected with streptavidin-horseradish peroxidase (HRP) conjugate and was visualized using 3,3′-diaminobenzidine solution. Nuclear staining was done by the hematoxylin-eosin method. As controls, lung sections from infected hamsters were equally processed but the primary antibody was omitted; negative controls were done from uninfected hamster lungs. Samples were observed on an Axoskope A1 microscope (Carl Zeiss, Gottingen, Germany) coupled to a DXM1200 digital video camera (Nikon).
ACKNOWLEDGMENTS
We thank Adolfo García-Sastre, Gianni Gori Savellini, Rafael Medina, Antonito Panganiban, Ralf Bartenschlager, Fernando Valiente-Echeverria, and Ricardo Soto-Rifo for kindly providing the plasmids used in this study. We are grateful to Benjamin Brennan and Veronica Rezelj for sharing their protocols used for the IFN-Luc reporter assays. We thank the Microscopy Advanced Unit (UMA-MED) and biosafety level 3 facility at the Escuela de Medicina, Pontificia Universidad Católica de Chile.
The work was supported by the Comision Nacional de Investigación Cientifica y Tecnologica (CONICYT), Gobierno de Chile, through grant FONDECYT 11150611 from the Fondo Nacional de Ciencia y Tecnología del Gobierno de Chile to J.V.-O., by CONICYT-Programa de Investigación Asociativa (PIA) ACT1408 to M.L.-L. and J.V.-O., and by the Proyecto P09/016-F de la Iniciativa Científica Milenio (ICM) del Ministerio de Economía, Fomento y Turismo to M.L.-L. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
L.S. and F.L. conducted this work as part of their Ph.D. theses in the Programa de Doctorado en Ciencias Biológicas mención Microbiología, Facultad de Ciencias, Universidad de Chile, and in the Programa de Doctorado en Ciencias Biológicas, mención Genética Molecular y Microbiología, Facultad de Ciencias Biológicas, Pontificia Universidad Católica de Chile, respectively. At the time, L.S. and F.L. were recipients of a Ph.D. fellowship and Beca de Extensión from CONICYT, Chile.
J.V.-O. and M.L.-L. designed the study. J.V.-O. and V.O. performed (when required) the cloning and plasmid construction. J.V.-O., L.S., F.L., K.P., and J.A. performed the experiments in cells. N.D.T. provided the anti-N and anti-NSs antibodies used in the study. P.P. designed and conducted the Syrian hamster infection experiments. F.L., L.S., and C.O. performed the immunohistochemistry analysis in lung tissues of experimentally inoculated Syrian hamsters. M.L.-L. wrote the manuscript. All authors read and approved the final manuscript.
We declare that no conflicts of interest exist.
REFERENCES
- 1.Figueiredo LT, Souza WM, Ferres M, Enria DA. 2014. Hantaviruses and cardiopulmonary syndrome in South America. Virus Res 187:43–54. doi: 10.1016/j.virusres.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Palma RE, Polop JJ, Owen RD, Mills JN. 2012. Ecology of rodent-associated hantaviruses in the Southern Cone of South America: Argentina, Chile, Paraguay, and Uruguay. J Wildl Dis 48:267–281. doi: 10.7589/0090-3558-48.2.267. [DOI] [PubMed] [Google Scholar]
- 3.Padula PJ, Edelstein A, Miguel SD, Lopez NM, Rossi CM, Rabinovich RD. 1998. Hantavirus pulmonary syndrome outbreak in Argentina: molecular evidence for person-to-person transmission of Andes virus. Virology 241:323–330. doi: 10.1006/viro.1997.8976. [DOI] [PubMed] [Google Scholar]
- 4.Martinez VP, Bellomo C, San Juan J, Pinna D, Forlenza R, Elder M, Padula PJ. 2005. Person-to-person transmission of Andes virus. Emerg Infect Dis 11:1848–1853. doi: 10.3201/eid1112.050501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plyusnin A, Vapalahti O, Vaheri A. 1996. Hantaviruses: genome structure, expression and evolution. J Gen Virol 77:2677–2687. doi: 10.1099/0022-1317-77-11-2677. [DOI] [PubMed] [Google Scholar]
- 6.Khaiboullina SF, Morzunov SP, St Jeor SC. 2005. Hantaviruses: molecular biology, evolution and pathogenesis. Curr Mol Med 5:773–790. doi: 10.2174/156652405774962317. [DOI] [PubMed] [Google Scholar]
- 7.Kukkonen SK, Vaheri A, Plyusnin A. 2005. L protein, the RNA-dependent RNA polymerase of hantaviruses. Arch Virol 150:533–556. doi: 10.1007/s00705-004-0414-8. [DOI] [PubMed] [Google Scholar]
- 8.Cifuentes-Munoz N, Salazar-Quiroz N, Tischler ND. 2014. Hantavirus Gn and Gc envelope glycoproteins: key structural units for virus cell entry and virus assembly. Viruses 6:1801–1822. doi: 10.3390/v6041801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vera-Otarola J, Solis L, Soto-Rifo R, Ricci EP, Pino K, Tischler ND, Ohlmann T, Darlix J-L, López-Lastra M. 2012. The Andes hantavirus NSs protein is expressed from the viral small mRNA by a leaky scanning mechanism. J Virol 86:2176–2187. doi: 10.1128/JVI.06223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reuter M, Kruger DH. 2018. The nucleocapsid protein of hantaviruses: much more than a genome-wrapping protein. Virus Genes 54:5–16. doi: 10.1007/s11262-017-1522-3. [DOI] [PubMed] [Google Scholar]
- 11.Bridgen A, Weber F, Fazakerley JK, Elliott RM. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc Natl Acad Sci U S A 98:664–669. doi: 10.1073/pnas.98.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber F, Bridgen A, Fazakerley JK, Streitenfeld H, Kessler N, Randall RE, Elliott RM. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J Virol 76:7949–7955. doi: 10.1128/jvi.76.16.7949-7955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas D, Blakqori G, Wagner V, Banholzer M, Kessler N, Elliott RM, Haller O, Weber F. 2004. Inhibition of RNA polymerase II phosphorylation by a viral interferon antagonist. J Biol Chem 279:31471–31477. doi: 10.1074/jbc.M400938200. [DOI] [PubMed] [Google Scholar]
- 14.Billecocq A, Spiegel M, Vialat P, Kohl A, Weber F, Bouloy M, Haller O. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J Virol 78:9798–9806. doi: 10.1128/JVI.78.18.9798-9806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leonard VH, Kohl A, Hart TJ, Elliott RM. 2006. Interaction of Bunyamwera orthobunyavirus NSs protein with mediator protein MED8: a mechanism for inhibiting the interferon response. J Virol 80:9667–9675. doi: 10.1128/JVI.00822-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaaskelainen KM, Kaukinen P, Minskaya ES, Plyusnina A, Vapalahti O, Elliott RM, Weber F, Vaheri A, Plyusnin A. 2007. Tula and Puumala hantavirus NSs ORFs are functional and the products inhibit activation of the interferon-beta promoter. J Med Virol 79:1527–1536. doi: 10.1002/jmv.20948. [DOI] [PubMed] [Google Scholar]
- 17.Le May N, Mansuroglu Z, Leger P, Josse T, Blot G, Billecocq A, Flick R, Jacob Y, Bonnefoy E, Bouloy M. 2008. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog 4:e13. doi: 10.1371/journal.ppat.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaaskelainen KM, Plyusnina A, Lundkvist A, Vaheri A, Plyusnin A. 2008. Tula hantavirus isolate with the full-length ORF for nonstructural protein NSs survives for more consequent passages in interferon-competent cells than the isolate having truncated NSs ORF. Virol J 5:3. doi: 10.1186/1743-422X-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott RM, Weber F. 2009. Bunyaviruses and the type I interferon system. Viruses 1:1003–1021. doi: 10.3390/v1031003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rezelj VV, Li P, Chaudhary V, Elliott RM, Jin DY, Brennan B. 2017. Differential antagonism of human innate immune responses by tick-borne phlebovirus nonstructural proteins. mSphere 2:e00234-17. doi: 10.1128/mSphere.00234-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rezelj VV, Overby AK, Elliott RM. 2015. Generation of mutant Uukuniemi viruses lacking the nonstructural protein NSs by reverse genetics indicates that NSs is a weak interferon antagonist. J Virol 89:4849–4856. doi: 10.1128/JVI.03511-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonjardim CA, Ferreira PC, Kroon EG. 2009. Interferons: signaling, antiviral and viral evasion. Immunol Lett 122:1–11. doi: 10.1016/j.imlet.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar H, Kawai T, Akira S. 2011. Pathogen recognition by the innate immune system. Int Rev Immunol 30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 24.Reikine S, Nguyen JB, Modis Y. 2014. Pattern recognition and signaling mechanisms of RIG-I and MDA5. Front Immunol 5:342. doi: 10.3389/fimmu.2014.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. 2011. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiscott J. 2007. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem 282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- 27.Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, Chen ZJ. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 28.Levine JR, Prescott J, Brown KS, Best SM, Ebihara H, Feldmann H. 2010. Antagonism of type I interferon responses by new world hantaviruses. J Virol 84:11790–11801. doi: 10.1128/JVI.00916-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spiropoulou CF, Albarino CG, Ksiazek TG, Rollin PE. 2007. Andes and Prospect Hill hantaviruses differ in early induction of interferon although both can downregulate interferon signaling. J Virol 81:2769–2776. doi: 10.1128/JVI.02402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Safronetz D, Zivcec M, Lacasse R, Feldmann F, Rosenke R, Long D, Haddock E, Brining D, Gardner D, Feldmann H, Ebihara H. 2011. Pathogenesis and host response in Syrian hamsters following intranasal infection with Andes virus. PLoS Pathog 7:e1002426. doi: 10.1371/journal.ppat.1002426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alff PJ, Sen N, Gorbunova E, Gavrilovskaya IN, Mackow ER. 2008. The NY-1 hantavirus Gn cytoplasmic tail coprecipitates TRAF3 and inhibits cellular interferon responses by disrupting TBK1-TRAF3 complex formation. J Virol 82:9115–9122. doi: 10.1128/JVI.00290-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alff PJ, Gavrilovskaya IN, Gorbunova E, Endriss K, Chong Y, Geimonen E, Sen N, Reich NC, Mackow ER. 2006. The pathogenic NY-1 hantavirus G1 cytoplasmic tail inhibits RIG-I- and TBK-1-directed interferon responses. J Virol 80:9676–9686. doi: 10.1128/JVI.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cimica V, Dalrymple NA, Roth E, Nasonov A, Mackow ER. 2014. An innate immunity-regulating virulence determinant is uniquely encoded by the Andes virus nucleocapsid protein gene. mBio 5:e01088-13. doi: 10.1128/mBio.01088-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthys V, Mackow ER. 2012. Hantavirus regulation of type I interferon responses. Adv Virol 2012:524024. doi: 10.1155/2012/524024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matthys VS, Cimica V, Dalrymple NA, Glennon NB, Bianco C, Mackow ER. 2014. Hantavirus GnT elements mediate TRAF3 binding and inhibit RIG-I/TBK1-directed beta interferon transcription by blocking IRF3 phosphorylation. J Virol 88:2246–2259. doi: 10.1128/JVI.02647-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basler CF, Mikulasova A, Martinez-Sobrido L, Paragas J, Mühlberger E, Bray M, Klenk H-D, Palese P, García-Sastre A. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J Virol 77:7945–7956. doi: 10.1128/jvi.77.14.7945-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin R, Heylbroeck C, Pitha PM, Hiscott J. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol 18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Padula PJ, Sanchez AJ, Edelstein A, Nichol ST. 2002. Complete nucleotide sequence of the M RNA segment of Andes virus and analysis of the variability of the termini of the virus S, M and L RNA segments. J Gen Virol 83:2117–2122. doi: 10.1099/0022-1317-83-9-2117. [DOI] [PubMed] [Google Scholar]
- 39.Martinez VP, Padula PJ. 2012. Induction of protective immunity in a Syrian hamster model against a cytopathogenic strain of Andes virus. J Med Virol 84:87–95. doi: 10.1002/jmv.22228. [DOI] [PubMed] [Google Scholar]
- 40.Hooper JW, Larsen T, Custer DM, Schmaljohn CS. 2001. A lethal disease model for hantavirus pulmonary syndrome. Virology 289:6–14. doi: 10.1006/viro.2001.1133. [DOI] [PubMed] [Google Scholar]
- 41.Wahl-Jensen V, Chapman J, Asher L, Fisher R, Zimmerman M, Larsen T, Hooper JW. 2007. Temporal analysis of Andes virus and Sin Nombre virus infections of Syrian hamsters. J Virol 81:7449–7462. doi: 10.1128/JVI.00238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z, Mir MA. 2015. Andes virus nucleocapsid protein interrupts protein kinase R dimerization to counteract host interference in viral protein synthesis. J Virol 89:1628–1639. doi: 10.1128/JVI.02347-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kohl A, Clayton RF, Weber F, Bridgen A, Randall RE, Elliott RM. 2003. Bunyamwera virus nonstructural protein NSs counteracts interferon regulatory factor 3-mediated induction of early cell death. J Virol 77:7999–8008. doi: 10.1128/jvi.77.14.7999-8008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blakqori G, Delhaye S, Habjan M, Blair CD, Sanchez-Vargas I, Olson KE, Attarzadeh-Yazdi G, Fragkoudis R, Kohl A, Kalinke U, Weiss S, Michiels T, Staeheli P, Weber F. 2007. La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J Virol 81:4991–4999. doi: 10.1128/JVI.01933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoneyama M, Suhara W, Fukuhara Y, Sato M, Ozato K, Fujita T. 1996. Autocrine amplification of type I interferon gene expression mediated by interferon stimulated gene factor 3 (ISGF3). J Biochem 120:160–169. doi: 10.1093/oxfordjournals.jbchem.a021379. [DOI] [PubMed] [Google Scholar]
- 46.Haye K, Burmakina S, Moran T, Garcia-Sastre A, Fernandez-Sesma A. 2009. The NS1 protein of a human influenza virus inhibits type I interferon production and the induction of antiviral responses in primary human dendritic and respiratory epithelial cells. J Virol 83:6849–6862. doi: 10.1128/JVI.02323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Talon J, Horvath CM, Polley R, Basler CF, Muster T, Palese P, Garcia-Sastre A. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J Virol 74:7989–7996. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, Li M, Zheng H, Muster T, Palese P, Beg AA, Garcia-Sastre A. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J Virol 74:11566–11573. doi: 10.1128/jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 50.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 51.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 52.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 53.Versteeg GA, Rajsbaum R, Sánchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, Inn K-S, Fernandez-Sesma A, Jung J, García-Sastre A. 2013. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity 38:384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richtsteiger R, Henke-Gendo C, Schmidtke M, Harste G, Heim A. 2003. Quantitative multiplex real-time PCR for the sensitive detection of interferon beta gene induction and viral suppression of interferon beta expression. Cytokine 24:190–200. doi: 10.1016/j.cyto.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 55.Paz S, Vilasco M, Arguello M, Sun Q, Lacoste J, Nguyen TL, Zhao T, Shestakova EA, Zaari S, Bibeau-Poirier A, Servant MJ, Lin R, Meurs EF, Hiscott J. 2009. Ubiquitin-regulated recruitment of IκB kinase epsilon to the MAVS interferon signaling adapter. Mol Cell Biol 29:3401–3412. doi: 10.1128/MCB.00880-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu B, Zhang M, Chu H, Zhang H, Wu H, Song G, Wang P, Zhao K, Hou J, Wang X, Zhang L, Gao C. 2017. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat Immunol 18:214–224. doi: 10.1038/ni.3641. [DOI] [PubMed] [Google Scholar]
- 57.Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villán E, García-Sastre A, Gack MU. 2012. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog 8:e1003059. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosebeck S, Rehman AO, Apel IJ, Kohrt D, Appert A, O’Donnell MA, Ting AT, Du M-Q, Baens M, Lucas PC, McAllister-Lucas LM. 2014. The API2-MALT1 fusion exploits TNFR pathway-associated RIP1 ubiquitination to promote oncogenic NF-κB signaling. Oncogene 33:2520–2530. doi: 10.1038/onc.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 60.Soderberg O, Leuchowius KJ, Gullberg M, Jarvius M, Weibrecht I, Larsson LG, Landegren U. 2008. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods 45:227–232. doi: 10.1016/j.ymeth.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 61.Rajsbaum R, Garcia-Sastre A. 2013. Viral evasion mechanisms of early antiviral responses involving regulation of ubiquitin pathways. Trends Microbiol 21:421–429. doi: 10.1016/j.tim.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morrill JC, Jennings GB, Cosgriff TM, Gibbs PH, Peters CJ. 1989. Prevention of Rift Valley fever in rhesus monkeys with interferon-alpha. Rev Infect Dis 11(Suppl 4):S815–S825. doi: 10.1093/clinids/11.supplement_4.s815. [DOI] [PubMed] [Google Scholar]
- 63.Andersson I, Lundkvist A, Haller O, Mirazimi A. 2006. Type I interferon inhibits Crimean-Congo hemorrhagic fever virus in human target cells. J Med Virol 78:216–222. doi: 10.1002/jmv.20530. [DOI] [PubMed] [Google Scholar]
- 64.Livonesi MC, de Sousa RL, Badra SJ, Figueiredo LT. 2007. In vitro and in vivo studies of the onterferon-alpha action on distinct Orthobunyavirus. Antiviral Res 75:121–128. doi: 10.1016/j.antiviral.2007.01.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tamura M, Asada H, Kondo K, Takahashi M, Yamanishi K. 1987. Effects of human and murine interferons against hemorrhagic fever with renal syndrome (HFRS) virus (Hantaan virus). Antiviral Res 8:171–178. doi: 10.1016/0166-3542(87)90071-4. [DOI] [PubMed] [Google Scholar]
- 66.Temonen M, Lankinen H, Vapalahti O, Ronni T, Julkunen I, Vaheri A. 1995. Effect of interferon-alpha and cell differentiation on Puumala virus infection in human monocyte/macrophages. Virology 206:8–15. doi: 10.1016/s0042-6822(95)80014-x. [DOI] [PubMed] [Google Scholar]
- 67.Oelschlegel R, Kruger DH, Rang A. 2007. MxA-independent inhibition of Hantaan virus replication induced by type I and type II interferon in vitro. Virus Res 127:100–105. doi: 10.1016/j.virusres.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 68.Zinzula L, Tramontano E. 2013. Strategies of highly pathogenic RNA viruses to block dsRNA detection by RIG-I-like receptors: hide, mask, hit. Antiviral Res 100:615–635. doi: 10.1016/j.antiviral.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pan Y, Li R, Meng JL, Mao HT, Zhang Y, Zhang J. 2014. Smurf2 negatively modulates RIG-I-dependent antiviral response by targeting VISA/MAVS for ubiquitination and degradation. J Immunol 192:4758–4764. doi: 10.4049/jimmunol.1302632. [DOI] [PubMed] [Google Scholar]
- 70.Liu B, Gao C. 2018. Regulation of MAVS activation through post-translational modifications. Curr Opin Immunol 50:75–81. doi: 10.1016/j.coi.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 71.Brubaker SW, Gauthier AE, Mills EW, Ingolia NT, Kagan JC. 2014. A bicistronic MAVS transcript highlights a class of truncated variants in antiviral immunity. Cell 156:800–811. doi: 10.1016/j.cell.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu L, Peng L, Gu T, Yu D, Yao YG. 2019. The 3′UTR of human MAVS mRNA contains multiple regulatory elements for the control of protein expression and subcellular localization. Biochim Biophys Acta Gene Regul Mech 1862:47–57. doi: 10.1016/j.bbagrm.2018.10.017. [DOI] [PubMed] [Google Scholar]
- 73.Ding S, Zhu S, Ren L, Feng N, Song Y, Ge X, Li B, Flavell RA, Greenberg HB. 2018. Rotavirus VP3 targets MAVS for degradation to inhibit type III interferon expression in intestinal epithelial cells. Elife 7:e39494. doi: 10.7554/eLife.39494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Castanier C, Garcin D, Vazquez A, Arnoult D. 2010. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep 11:133–138. doi: 10.1038/embor.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A 102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang Y, Liang Y, Qu L, Chen Z, Yi M, Li K, Lemon SM. 2007. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc Natl Acad Sci U S A 104:7253–7258. doi: 10.1073/pnas.0611506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang B, Xi X, Lei X, Zhang X, Cui S, Wang J, Jin Q, Zhao Z. 2013. Enterovirus 71 protease 2Apro targets MAVS to inhibit anti-viral type I interferon responses. PLoS Pathog 9:e1003231. doi: 10.1371/journal.ppat.1003231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen Z, Benureau Y, Rijnbrand R, Yi J, Wang T, Warter L, Lanford RE, Weinman SA, Lemon SM, Martin A, Li K. 2007. GB virus B disrupts RIG-I signaling by NS3/4A-mediated cleavage of the adaptor protein MAVS. J Virol 81:964–976. doi: 10.1128/JVI.02076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mukherjee A, Morosky SA, Delorme-Axford E, Dybdahl-Sissoko N, Oberste MS, Wang T, Coyne CB. 2011. The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog 7:e1001311. doi: 10.1371/journal.ppat.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Refolo G, Ciccosanti F, Di Rienzo M, Perdomo AB, Romani M, Alonzi T, Tripodi M, Ippolito G, Piacentini M, Fimia GM. 2019. Negative regulation of MAVS-mediated antiviral signaling by the mitochondrial protein LRPPRC during HCV infection. Hepatology 69:34–50. doi: 10.1002/hep.30149. [DOI] [PubMed] [Google Scholar]
- 81.Varga ZT, Grant A, Manicassamy B, Palese P. 2012. Influenza virus protein PB1-F2 inhibits the induction of type I interferon by binding to MAVS and decreasing mitochondrial membrane potential. J Virol 86:8359–8366. doi: 10.1128/JVI.01122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barría MI, González A, Vera-Otarola J, León U, Vollrath V, Marsac D, Monasterio O, Pérez-Acle T, Soza A, López-Lastra M. 2009. Analysis of natural variants of the hepatitis C virus internal ribosome entry site reveals that primary sequence plays a key role in cap-independent translation. Nucleic Acids Res 37:957–971. doi: 10.1093/nar/gkn1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nistal-Villán E, Gack MU, Martínez-Delgado G, Maharaj NP, Inn K-S, Yang H, Wang R, Aggarwal AK, Jung JU, García-Sastre A. 2010. Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. J Biol Chem 285:20252–20261. doi: 10.1074/jbc.M109.089912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sánchez-Aparicio MT, Feinman LJ, García-Sastre A, Shaw ML. 2018. Paramyxovirus V proteins interact with the RIG-I/TRIM25 regulatory complex and inhibit RIG-I signaling. J Virol 92. doi: 10.1128/JVI.01960-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nistal-Villán E, Rodríguez-García E, Di Scala M, Ferrero-Laborda R, Olagüe C, Vales Á, Carte-Abad B, Crespo I, García-Sastre A, Prieto J, Larrea E, González-Aseguinolaza G. 2015. A RIG-I 2CARD-MAVS200 chimeric protein reconstitutes IFN-β induction and antiviral response in models deficient in type I IFN response. J Innate Immun 7:466–481. doi: 10.1159/000375262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Varga ZT, Ramos I, Hai R, Schmolke M, Garcia-Sastre A, Fernandez-Sesma A, Palese P. 2011. The influenza virus protein PB1-F2 inhibits the induction of type I interferon at the level of the MAVS adaptor protein. PLoS Pathog 7:e1002067. doi: 10.1371/journal.ppat.1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kochs G, García-Sastre A, Martínez-Sobrido L. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol 81:7011–7021. doi: 10.1128/JVI.02581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mir MA, Panganiban AT. 2008. A protein that replaces the entire cellular eIF4F complex. EMBO J 27:3129–3139. doi: 10.1038/emboj.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Soto-Rifo R, Rubilar PS, Limousin T, de Breyne S, Décimo D, Ohlmann T. 2012. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J 31:3745–3756. doi: 10.1038/emboj.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Valiente-Echeverria F, Melnychuk L, Vyboh K, Ajamian L, Gallouzi IE, Bernard N, Mouland AJ. 2014. eEF2 and Ras-GAP SH3 domain-binding protein (G3BP1) modulate stress granule assembly during HIV-1 infection. Nat Commun 5:4819. doi: 10.1038/ncomms5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 92.Yuan JS, Reed A, Chen F, Stewart CN Jr. 2006. Statistical analysis of real-time PCR data. BMC Bioinformatics 7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sternberger LA, Hardy PH Jr, Cuculis JJ, Meyer HG. 1970. The unlabeled antibody enzyme method of immunohistochemistry: preparation and properties of soluble antigen-antibody complex (horseradish peroxidase-antihorseradish peroxidase) and its use in identification of spirochetes. J Histochem Cytochem 18:315–333. doi: 10.1177/18.5.315. [DOI] [PubMed] [Google Scholar]
- 94.Tischler ND, Rosemblatt M, Valenzuela PD. 2008. Characterization of cross-reactive and serotype-specific epitopes on the nucleocapsid proteins of hantaviruses. Virus Res 135:1–9. doi: 10.1016/j.virusres.2008.01.013. [DOI] [PubMed] [Google Scholar]