Few studies have addressed the mechanisms of immune control in HIV-infected subjects in India, where an estimated 2.7 million people are living with HIV. We focus here on a study cohort in Delhi on one of the most prevalent HLA-B alleles, HLA-B*52:01, present in 22.5% of infected individuals. HLA-B*52:01 has consistently been shown in other cohorts to be associated with protection against HIV disease progression, but studies have been limited by the low prevalence of this allele in North America and Europe. Among the C-clade-infected individuals, we show that HLA-B*52:01 is the most protective of all the HLA-B alleles expressed in the Indian cohort and is associated with the highest absolute CD4 counts. Further, we show that the mechanism by which HLA-B*52:01 mediates immune protection is, at least in part, related to the inability of HIV to evade the HLA-B*52:01-restricted p24 Gag-specific CD8+ T-cell response without incurring a significant loss to viral replicative capacity.
KEYWORDS: C clade, CTL, compensatory mutation, escape mutation, HLA, HLA-B*52:01, Indian, viral replicative capacity, human immunodeficiency virus, p24 Gag
ABSTRACT
HLA-B*52:01 is strongly associated with protection against HIV disease progression. However, the mechanisms of HLA-B*52:01-mediated immune control have not been well studied. We here describe a cohort with a majority of HIV C-clade-infected individuals from Delhi, India, where HLA-B*52:01 is highly prevalent (phenotypic frequency, 22.5%). Consistent with studies of other cohorts, expression of HLA-B*52:01 was associated with high absolute CD4 counts and therefore a lack of HIV disease progression. We here examined the impact of HLA-B*52:01-associated viral polymorphisms within the immunodominant C clade Gag epitope RMTSPVSI (here, RI8; Gag residues 275 to 282) on viral replicative capacity (VRC) since HLA-mediated reduction in VRC is a central mechanism implicated in HLA-associated control of HIV. We observed in HLA-B*52:01-positive individuals a higher frequency of V280T, V280S, and V280A variants within RI8 (P = 0.0001). Each of these variants reduced viral replicative capacity in C clade viruses, particularly the V280A variant (P < 0.0001 in both the C clade consensus and in the Indian study cohort consensus p24 Gag backbone), which was also associated with significantly higher absolute CD4 counts in the donors (median, 941.5 cells/mm3; P = 0.004). A second HLA-B*52:01-associated mutation, K286R, flanking HLA-B*52:01-RI8, was also analyzed. Although selected in HLA-B*52:01-positive subjects often in combination with the V280X variants, this mutation did not act as a compensatory mutant but, indeed, further reduced VRC. These data are therefore consistent with previous work showing that HLA-B molecules that are associated with immune control of HIV principally target conserved epitopes within the capsid protein, escape from which results in a significant reduction in VRC.
IMPORTANCE Few studies have addressed the mechanisms of immune control in HIV-infected subjects in India, where an estimated 2.7 million people are living with HIV. We focus here on a study cohort in Delhi on one of the most prevalent HLA-B alleles, HLA-B*52:01, present in 22.5% of infected individuals. HLA-B*52:01 has consistently been shown in other cohorts to be associated with protection against HIV disease progression, but studies have been limited by the low prevalence of this allele in North America and Europe. Among the C-clade-infected individuals, we show that HLA-B*52:01 is the most protective of all the HLA-B alleles expressed in the Indian cohort and is associated with the highest absolute CD4 counts. Further, we show that the mechanism by which HLA-B*52:01 mediates immune protection is, at least in part, related to the inability of HIV to evade the HLA-B*52:01-restricted p24 Gag-specific CD8+ T-cell response without incurring a significant loss to viral replicative capacity.
INTRODUCTION
HLA class I allele variation has consistently been the strongest genetic factor influencing HIV disease outcome (1), in particular, polymorphism among HLA-B alleles (2, 3). Previous studies have shown that HLA-B*52:01 is strongly protective against HIV disease progression (4–7). However, our understanding of the underlying mechanisms of HLA-B*52:01-mediated immune control of HIV has been limited due to the low prevalence of the allele in the populations most commonly studied. The analyses that have been undertaken to date are based on B clade cohorts in Japan, where the HLA-B*52:01 is relatively prevalent (8–11). In the current study, we focus on a North Indian population in which HLA-B*52:01 is expressed in more than 20% of subjects (http://www.allelefrequencies.net/) and where the C clade is the predominant subtype (12). To date there have been very few studies investigating the impact of HLA on HIV in India, which has the world’s third-highest burden, with 2.7 million infections (12).
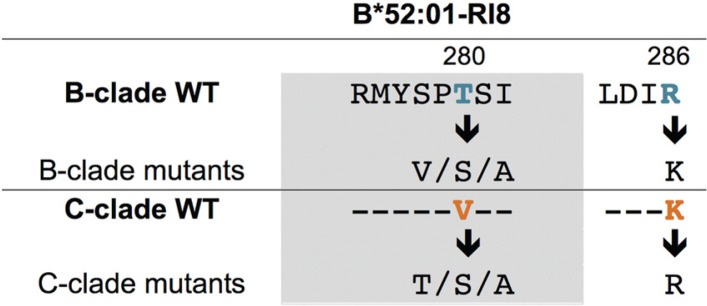
There has been only one optimally defined HLA-B*52:01-restricted HIV-specific epitope. The C clade epitope RMYSPVSI (Gag residues 275 to 282; here, RI8) is in the conserved capsid protein (CA) p24 Gag (9, 13–16). The consensus Gag sequence differs between clade B and clade C at Gag-280 (position 6 within the RI8 epitope), with threonine in B clade and valine in C clade viruses. In a comprehensive analysis identifying associations between HIV B clade amino acid polymorphisms and particular HLA class I allele expression, the T280S mutation is selected in subjects expressing HLA-B*52:01 (n = 1,888 treatment-naive, chronically B-clade-infected individuals; q value, <0.05) (17). Previous studies have shown that successful viral control mediated by protective HLA alleles is typically associated with HIV-specific cytotoxic T lymphocytes (CTL) targeting highly conserved regions of the HIV proteome (16, 18) and driving the selection of escape mutants that reduce viral replicative capacity (VRC) (19–21). Several studies have shown that VRC plays a critical role in disease progression (22, 23). With respect to the particular HLA-B*52:01-associated mutations within the p24 RI8 epitope, a study of B-clade-infected individuals indicated that T280S, T280V, and T280A, are all escape mutations which lower VRC (11).
Of note, six amino acids downstream of the RI8 epitope is an HLA-B*52:01-associated arginine-to-lysine substitution, R286K, in B clade virus and a lysine-to-arginine substitution, K286R, in C clade virus. R286K is observed in linkage disequilibrium with, and dependent on, the T280S escape mutation in B clade infection (17). This occurrence of a mutation that is selected subsequent to an escape mutation is suggestive of a compensatory mutation that can in part restore VRC to wild-type (WT) levels (24, 25). The R286K mutation in B clade virus is therefore potentially a compensatory mutation for T280S.
The focus of this study was, first, to determine whether HLA-B*52:01 is protective against disease progression in C clade infection and, second, to investigate the impact of HLA-B*52:01-mediated selection pressure on viral replicative capacity in C clade infection. The HLA-B*52:01-associated escape mutation in B clade infection involves substitution of Gag-280T, but in C clade virus the consensus amino acid at Gag-280 is valine. Based on data from previous studies (25, 26), we hypothesized that the impact of particular escape mutants on viral replicative capacity might be highly clade specific. To test this hypothesis, we therefore carried out assays of viral replicative capacity (VRC) using a green fluorescent protein (GFP) reporter cell line to assess the impact of these HLA-B*52:01-restricted mutants on VRC in different backbones of p24 Gag.
RESULTS
In a C-clade-infected North Indian cohort, HLA-B*52:01-positive individuals have higher absolute CD4 counts and higher frequencies of the V280X mutation.
The study cohort comprised 169 antiretroviral therapy (ART)-naive individuals attending an outpatient clinic at the Centre for AIDS and Related Diseases (CARD) Division of the National Centre for Disease Control (NCDC), Delhi (Table 1), 87% of whom were C clade infected, as determined via ART drug resistance testing. Absolute CD4 counts but no viral load data were available for the HIV-infected study cohort. To determine the impact of HLA-B*52:01 on these C clade viruses, we sequenced autologous gag in 140 C-clade-infected individuals, among which there were 138 individuals with complete p24 Gag sequences available. The following results and analysis will be based on these 138 C-clade-infected subjects.
TABLE 1.
Characteristics of study participants from New Delhi, India
| Characteristic | Value for the characteristic (n = 169)a |
|---|---|
| Recruitment period (yr) | 2011–2014 |
| No. of subjects of Indian ethnicity | 169 (100) |
| Sex | |
| Male | 92 (54.4) |
| Female | 76 (45.0) |
| Transgender | 1 (0.6) |
| CD4+ T cell count (cells/mm3) | 447.7 (311–584)b |
| HLA-B typing | |
| HLA-B*52:01 positive | 38 (22.5) |
| HLA-B*52:01 negative | 131 (77.5) |
| HIV subtype (n = 161)c | |
| C | 140 (86.9) |
| B | 19 (11.8) |
| A1 | 2 (1.2) |
Data are the number (%) of patients unless otherwise indicated. CD4+ T-cell measurements were performed at enrollment.
Data in parentheses represent the IQR.
HIV subtype was determined by p24 Gag sequences for n = 161 subjects.
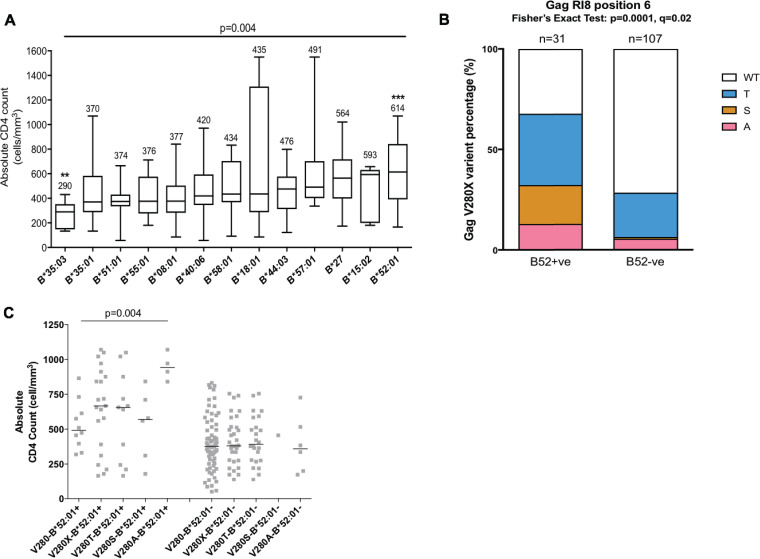
Although the duration of infection is unknown for any of the study subjects, differences among HLA-B alleles were associated with a significant impact on CD4 count (P = 0.004, Kruskal-Wallis test) (Fig. 1A). Previously identified protective alleles such as HLA-B*57:01, -B*58:01, and -B*27 (16) were also associated with somewhat higher CD4 counts in the current cohort, while HLA-B*35 alleles had significantly lower CD4 counts (Fig. 1A). In particular, HLA-B*52:01-positive subjects had significantly higher absolute CD4 counts than HLA-B*52:01-negative subjects (median of 614 cells/mm3 versus a median of 411 cells/mm3; uncorrected P value of <0.0001; the P value threshold following Bonferroni’s correction for 13 HLA-B molecules with more than three individuals expressing each molecule [see Materials and Methods] was 0.0038). HLA-B*35:03 was also significantly correlated with lower absolute CD4 counts (median of 290 cells/mm3 versus median of 432 cells/mm3; P = 0.0008), and this allele has also been previously reported to be associated with worse disease outcomes (27, 28).
FIG 1.
Frequency of B*52:01-restricted mutations and the association between CD4 count and HLA-B alleles (n = 138). (A) Association between HLA-B alleles and CD4 counts. Vertical bar represents the median CD4 counts, boxes indicate the IQR, and brackets are the maximum and minimum values. The median CD4 count is marked on the top of each bar. A Kruskal-Wallis test was applied (P = 0.004). Here, the individual P value was corrected by Bonferroni’s correction (P value threshold of 0.0038). The asterisk represents a statistically significant difference after Bonferroni correction for multiple tests between the absolute CD4 count in individuals expressing a particular HLA allele and in the remaining cohort. (B) Comparison of the percentages of the wild type (WT) and V280X variants (where X is T, S, or A) at Gag-280 in HLA-B*52:01-positive (B52+ve) and -negative (B52−ve) individuals. P < 0.0001 (Fisher’s exact test); q = 0.006. (C) CD4 counts of each V280X mutant in HLA- B*52:01-positive and -negative individuals. Bar represents the median of CD4 counts. A Mann-Whitney U test and Steel-Dwass test were performed for post hoc analysis (P < 0.05 of V280A compared to results with the wild type). **, P < 0.01; ***, P < 0.001.
Despite the difference between B and C clade viruses at Gag-280 in the consensus sequence (valine in C clade and threonine in B clade viruses) (Table 2), the distinctive HLA-B*52:01 footprint in Gag at position 6 in the RI8 epitope (RMTSPVSI) was observed within these C-clade-infected individuals, similar to the findings in B-clade-infected cohorts (Fig. 1B and Table 3) (10, 16). The frequency of V280X T/S/A variants (where X is either serine, threonine, or alanine) was 71% in HLA-B*52:01-positive individuals in contrast to 30% in HLA-B*52:01-negative individuals (Fig. 1B) (P < 0.0001 by Fisher’s exact test; q = 0.02). Of the three variants, the V280A mutant in HLA-B*52:01-positive individuals was the least frequent but nonetheless was associated with a significantly higher absolute CD4 count than the wild-type V280 among HLA-B*52:01-positive subjects (P = 0.004) (Fig. 1C). Any V280X mutant had a higher median CD4 count in HLA-B*52:01-positive individuals than in those whose autologous virus-encoded valine (wild type) at this position. This is unexpected since in the great majority of cases escape mutations are associated with higher viral loads and lower CD4 counts (29). In contrast, the V280X mutations were not associated with a difference in absolute CD4 counts among the HLA-B*52:01-negative subjects. Taking these observations together, HLA-B*52:01 was highly protective in this C-clade-infected cohort, and, among the HLA-B*52:01-positive subjects studied, the V280X escape mutants and in particular the V280A mutant were associated with higher CD4 counts.
TABLE 2.
Amino acids at Gag-280 and Gag-286 of B and C clade viruses
TABLE 3.
Frequency of the Gag V280X variant with or without K286R in HLA-B*52:01-positive and -negative C-clade-infected individuals
| Gag type | B*52:01 Gag275–286 sequencea | Frequency (%) by HLA-B*52:01 status |
P value | |
|---|---|---|---|---|
| Positive (n = 31) | Negative (n = 107) | |||
| Wild type | RMYSPV SILDIK | 22.6 | 63.6 | 0.0001 |
| V280T | -----T------ | 16.1 | 18.7 | 1 |
| V280S | -----S------ | 19.4 | 0 | 0.0001 |
| V280A | -----A------ | 12.9 | 4.7 | 0.1144 |
| K286R | -----------R | 9.7 | 7.5 | 0.7102 |
| V280T/K286R | -----T-----R | 19.4 | 3.7 | 0.0088 |
| V280S/K286R | -----S-----R | 0 | 0.9 | 1 |
| V280A/K286R | -----A-----R | 0 | 0.9 | 1 |
The RI8 sequence is highlighted in the wild-type consensus sequence, and positions 280 and 286, respectively, are shown in boldface.
None of the HLA-B*52:01-positive individuals carried both V280(S/A) and K286R.
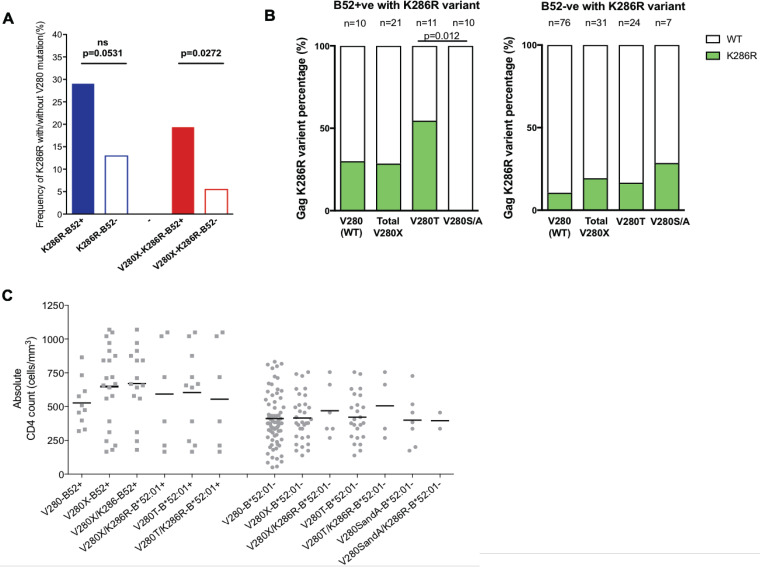
As previously described in B clade infection (17), the HLA-B*52:01-associated variants at Gag-280 were also linked to changes at Gag-286, five amino acids downstream of the carboxy terminus of the RI8 epitope, especially in HLA-B*52:01-positive subjects (Table 3 and Fig. 2A). Note that at this position (Gag-286), there is an arginine-to-lysine substitution (R286K) in B clade virus and a lysine-to-Arginine substitution (K286R) in C clade virus (Table 2). However, K286R was selected only in combination with V280T in our C-clade-infected individuals and not in the subjects carrying V280S and V280A (P = 0.012) (Table 3 and Fig. 2B). We hypothesized that if K286R is a compensatory mutation for V280T, the combination with V280T might be expected to result in decreased absolute CD4 counts among the HLA-B*52:01-positive patients. In fact, no such difference was observed in the CD4 counts of either HLA-B*52:01-positive or -negative individuals, with or without K286R (Fig. 2C). Therefore, on the basis of these CD4 data and on the fact that T280(S/A) variants are found in association with K286R in HLA-B*52:01-negative individuals (Fig. 2B), the K286R mutation is linked to evasion from the HLA-B*52:01-mediated RI8-specific response, as opposed to being a compensatory mutation compensating for a fitness cost to the V280T substitution. In order to address whether K286R had any compensatory effect on VRC in association with V280X, we therefore included these variants in our analyses of the impact of the HLA-B*52:01-RI8 V280X variants on VRC.
FIG 2.
Frequency of Gag K286R and the CD4 count of each mutant group (n = 138). (A) Frequency of Gag V280X, K286R, and V280X/K286R variants in HLA-B*52:01-positive and -negative individuals. (B) Comparison of the percentages of K286R in wild-type (WT) and V280X viruses in HLA-B*52:01-positive and -negative individuals. (C) CD4 count in wild-type and V280X viruses with/without K286R in HLA-B*52:01-positive and -negative individuals. The bar represents the median of the CD4 count. Fisher’s exact test for frequency difference and a Mann-Whitney U test and Steel-Dwass test post hoc analysis for CD4 counts were applied. ns, not significant.
V280X variants in C clade viruses significantly reduce VRC, and K286R is not a compensatory mutation.
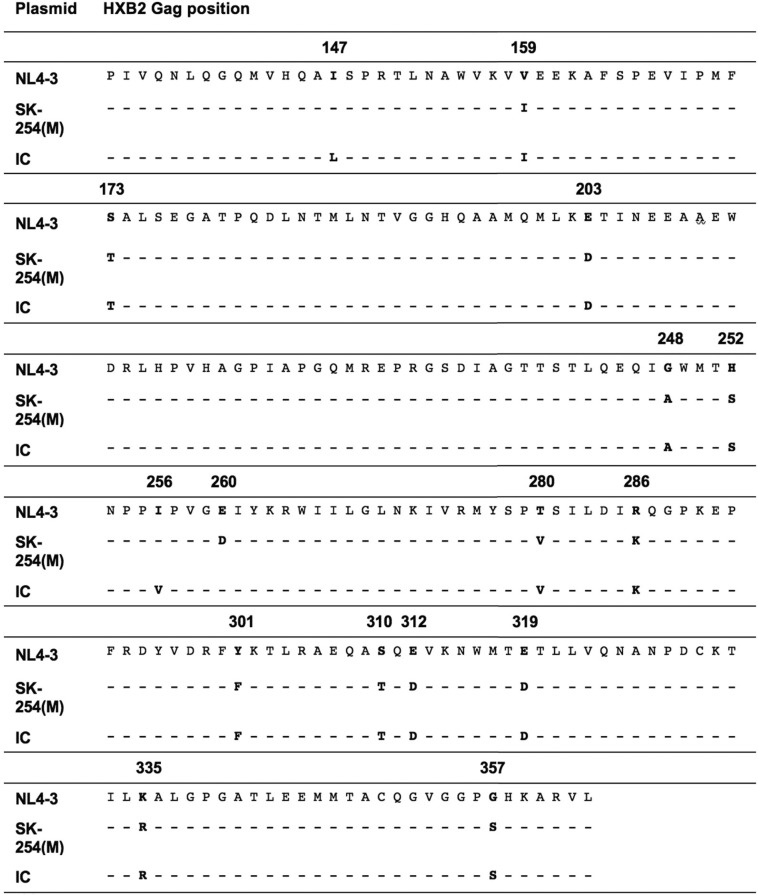
To examine whether the mutations at Gag-280 and Gag-286 affect VRC, viral fitness assays were performed using three virus backbones: the Indian cohort C clade (IC) consensus, the C clade consensus, and the reference B clade strain NL4-3. An Indian C clade consensus sequence was generated from gag proviral sequences determined from 138 subjects of the study cohort (Fig. 3). The C clade consensus sequence was determined from the Los Alamos HIV database (http://www.hiv.lanl.gov/). The IC sequence differs by 3 amino acids in p24 Gag from the C clade consensus sequence at I147L, I256V, and D260E (the C clade consensus residues are shown first in Table 4). These sequences were the three backbones of the chimeric viruses used in these fitness assays. The T280(V/S/A) HLA-B*52:01-associated mutations in B clade infection, with and without R286K, were introduced into the B clade backbone (NL4-3), and mutations of V280X with or without K286R were introduced into the C clade backbones of the C clade consensus and the Indian C clade (IC) consensus (Table 2).
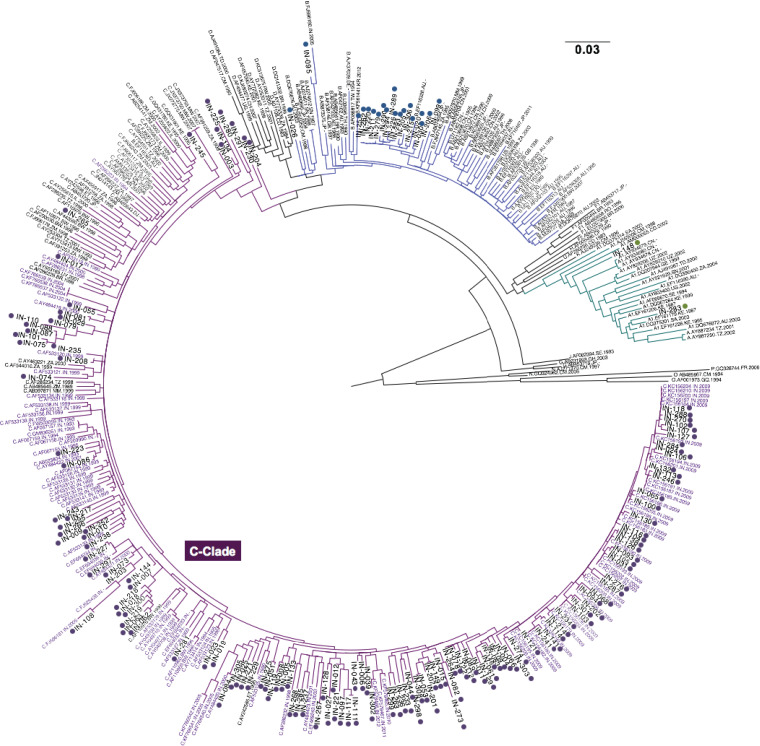
FIG 3.
Maximum likelihood phylogenetic tree of p24 Gag proviral sequences of HIV-1 group M, N, P, and O. The C clade sequences are shown in purple (n = 138), B clade sequences are in blue, and A clade sequences are in green. Subjects recruited in our cohort are numbered with the prefix “IN” and highlighted with dots. Reference Indian sequences randomly selected from 1993 to 2002 are highlighted in purple. Other reference sequences are shown in black. All of the reference sequences are from the Los Alamos database (http://www.hiv.lanl.gov/).
TABLE 4.
p24 Gag sequences of NL4-3, SK254(M), and IC
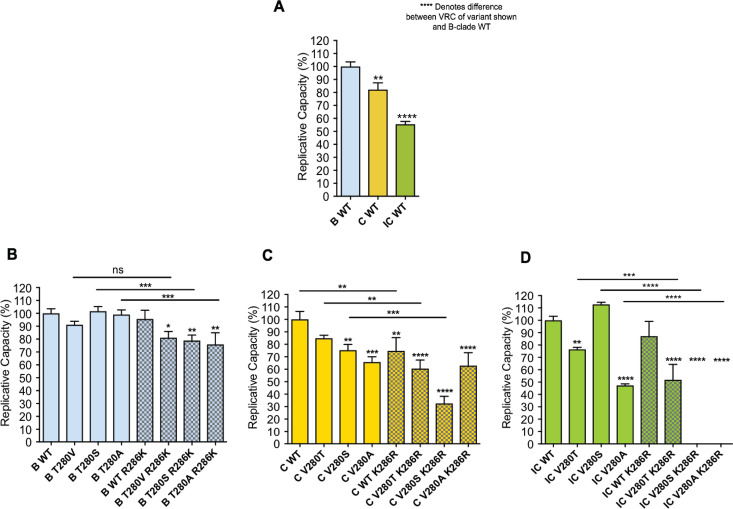
First, as expected, VRCs of C clade viruses were lower than those of the B clade (30–32) (Fig. 4A). The IC viruses were the least fit of all, indicating that I147L, I256V, and D260E (the residues unique to the IC virus sequence compared to the C clade consensus) contributed to lowering VRC in C clade viruses. Second, among B clade viruses, changes at position 280 had very little impact on VRC (Fig. 4B). Furthermore, the putative R286K mutation in combination with T280X reduced VRC, significantly so in the cases of T280(S/A), and therefore did not serve as a compensatory mutation in this setting. In the C clade consensus virus, all V280X mutants, in particular V280A, lowered the VRC significantly (Fig. 4C), and, as in the B clade backbone, the K286R variant significantly lowered the VRC. In the Indian consensus virus, the V280(S/A) variants but not the V280T variant reduced VRC (Fig. 4D), and again there was no evidence that K286R acted as a compensatory mutant. Strikingly, in the Indian consensus sequences, the combination of V280(S/A) and K286R that was not observed in any HLA-B*52:01-positive subjects completely abrogated the ability of the virus to replicate.
FIG 4.
Viral replicative capacity (VRC) of T280X/R286K B clade and V280X/K286R C clade mutant viruses. (A) Relative VRC of the consensus sequence normalized to that of the B clade wild type (WT) (NL4-3). (B) Relative VRC of B clade viruses normalized to that of the B clade wild type (NL4-3). (C) Relative VRC of C clade viruses normalized to that of the C clade wild type [SK-254(M)]. (D) Relative VRC of IC viruses normalized to that of the IC wild type. All bars are shown as means with standard deviations. Patterned bars represent the R286K/K286R mutant. Clades are indicated according to the color legend [blue, NL4-3; yellow, SK-254(M); green, Indian C clade consensus (IC)]. One-way ANOVA multiple comparison and Dunnett’s correction were used for calculating P values. Asterisks indicate P values for comparison of the results with the mutant virus to those with the wild type (ns, not significant [P ≥ 0.05]; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Emergence of compensatory mutations L135F and I223T for V280S/K286R and V280A/K286R during viral inoculation in vitro.
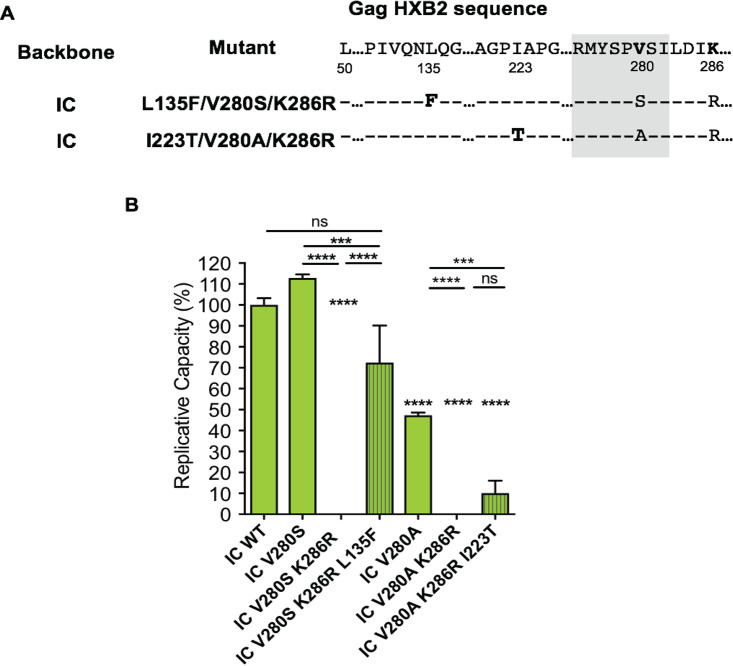
For some less fit viruses, the inoculation time in vitro for the viral growth could take up to 60 days. As described above, the IC viruses carrying V280(S/A) and K286R variants failed to grow to GFP detectability in three independent experimental repeats continued for ≥60 days. However, viral sequences could be determined by RNA extraction from the harvested supernatant after prolonged culture.
Viral sequencing showed that variants were selected in vitro. For example, L135F and I223T, which were harvested on days 73 and 71 of cell culture, respectively, emerged once out of three independent experiments in Indian C clade variants V280S/K286R and V280A/K286R, respectively (Fig. 5A). Sanger sequencing showed a clear substitution by the harvest point (frequency of GFP-positive cells of ≥30%). L135F in IC virus effectively restored the VRC of the V280S/K286R variant to 75% of that of the wild-type level, and I223T improved the VRC of the V280A/K286R variant to a modest degree (Fig. 5B).
FIG 5.
Identification and VRC of the developed variants during viral inoculation. (A) Positions of the developed variants in Gag HXB2. (B) VRC of L135F/V280S/K286R and I223T/V280A/K286R variants in IC. All bars are shown as means with standard deviations. Striped bars represent developed mutants. One-way ANOVA multiple comparison and Dunnett’s correction were used for calculating P values. Asterisks indicate P values for results with the mutants compared to those with the wild type. (ns, not significant [P ≥ 0.05]; ***, P < 0.001; ****, P < 0.0001).
DISCUSSION
This study is the first to evaluate HLA-B*52:01-associated immune control of HIV in C-clade infection. As with previous studies in the B clade, these C clade data demonstrate that HLA-B*52:01 is strongly associated with a better disease outcome, as reflected by a higher absolute CD4 count. Also, in parallel with the findings in B clade infection (11), we observed that the HLA-B*52:01-driven escape mutations at Gag-280 resulted in a cost to viral fitness. In particular, the V280A mutant was associated with a substantial reduction in viral replicative capacity (VRC), consistent with the observation of higher absolute CD4 counts in HLA-B*52:01-positive subjects whose autologous virus encoded this variant. Finally, the K286R mutant flanking the RI8 epitope did not appear to have a compensatory effect in restoring VRC to the level of the wild type, suggesting that its selection in combination with certain Gag-280 variants is driven by an alternative mechanism.
It was unexpected that the V280X mutants observed in association with HLA-B*52:01 in this Indian cohort are all linked with higher CD4 counts than the wild-type V280. In particular, the V280A variant in the Gag RI8 epitope was associated with an especially high CD4 count. Analysis was limited by the absence of viral load data from this cohort, but the HLA-B associations observed here between HLA-B*57/58:01 and -B*52:01 with high CD4 counts, and therefore protection against disease progression, and between HLA-B*35 and in particular HLA-B*35:03 with low CD4 counts, and therefore more rapid disease progression, are remarkably consistent with other studies showing HLA associations with low and high viral loads, respectively (17). It is difficult to envisage how an escape mutant would be selected were it not to benefit the virus, and this is indeed the norm (31). One may speculate that strong selection pressure may select for a variant that confers low VRC when the alternative is elimination. It is possible also that alternative, less costly variants than V280A are selected over the course of time by the virus, as shown in longitudinal studies (33). Previous studies have shown that a low VRC contributes to suppression of viremia (33–35). However, a low VRC alone does not prevent HIV disease progression. Apart from the single RI8-specific HLA-B*52:01-restricted response and Gag-280 variant being considered here, the contributions of all components of the immune system to the control of HIV are critically important to disease outcome.
In addition to the immune responses made by different subjects, the viral backbone as a whole has a major influence on the impact of Gag-280 variants on VRC, as illustrated clearly by use of three backbones in the current experiments. In contrast to previous studies (11), in our hands the Gag-280 variants had a more substantial impact in the C clade backbones tested and did not lower VRC significantly in the B clade backbone. Also, as shown previously (25, 26), the impact of a particular mutation is strikingly backbone dependent even when backbones of the same clade are compared. In the current study, for example, the V280A/K286R combination reduced VRC somewhat in the C clade consensus SK-254(M) backbone but abrogated replication altogether when it was introduced into the IC backbone.
The putative compensatory mutation at Gag-286 is a single residue that we have found here that alters the VRC. Intriguingly, we observed that R286K in B clade and K286R in C clade viruses not only failed to rescue the VRC of T280A and V280(T/S/A) but also contributed to further lowering of the VRC. In the absence of additional samples to explain this finding, one may speculate that the mechanism of selection of Gag-286 variants in HLA-B*52:01 may be to reduce CTL recognition further, potentially via an effect on processing (36, 37). If the Gag-280 variant merely reduced recognition without abrogating it altogether, there might still be selection pressure for an additional escape mutation to decrease epitope presentation. The Gag-280 variants might be selected first because the cost to VRC is typically less than the cost resulting from the Gag-286 variant (Fig. 4).
To track the escape pathway by which Gag-V280 achieves the preferred viral solution of V280T or V280S would require longitudinal studies. However, within this cross-sectional analysis, it is noteworthy that the C clade consensus codon for V280 is GTC. The virus therefore needs to transition through the low-fitness V280A (GCC) before reaching the higher fitness position V280T (ACC) or V280S (TCC) in the landscape. This pathway explains why V280A is selected at all when other, higher fitness escape options are available. However, this does not take into account qualitative differences between these variants in terms of the extent to which they mediate escape from CTL recognition.
Of note, we observed two compensatory mutations, L135F for the V280S/K286R and I223V for the V280A/K286R variants, during the long viral inoculation (∼100 days). This again highlights the importance of the viral backbone and the limitations of considering individual variants introduced into a reference strain of virus. The accumulation of compensatory mutations in vitro has been previously described after several passages of virus through the host cell (38). In vivo, one might anticipate that HIV is similarly capable of selecting similar compensatory mutations that restore VRC following the selection of the escape mutant that incurred a fitness cost to the virus. The mutation to phenylalanine at position 135 (CA position 3) might alter the stability of the capsid as this residue is at a monomer-monomer interface. Residue 223 (CA position 91) is in the capsid N-terminal domain, on the face of the capsid that would be exposed to the cytoplasm upon entry. However, these two mutations are distant from residues 280 and 286. It is unlikely that the compensatory effects of L135F and I223V are via direct interaction. The mechanism of how these two mutations located in the β-hairpin (L135F) and cyclophilin A loop (I223V) compensate the V280S/K286R and V280A/K286R variants, respectively, remains unclear.
A limitation of our cohort study is the lack of assessment of the CD8+ T-cell responses to confirm the RI8-specific CTL pressure on the virus due to the unavailability of patient samples. Since the Gag V280 variants are located at the nonanchoring position (position 6) in the RI8 epitope, it is likely that peptides with variant sequences can still be presented by the B*52:01 major histocompatibility (MHC) molecule. The cytotoxic T-cell response is therefore dependent on the recognition by the individual’s T cell receptors (TCRs). Previous studies have shown various degrees of RI8 recognition without cross-reactivity in mainly B-clade-infected Japanese (11, 39, 40) and Caucasian populations (41). In this paper, our data strongly suggest that a robust CD8+ T-cell response might be present in the B*52:01-positive Indian patients. Further immunological study will be required to verify the CTL response, the rate of CTL response in relation to the putative escape mutations, and potentially the diversity of TCRs in the B*52:01 Indian population against HIV-1 C clade infection. A longitudinal follow-up in these individuals will also help to reveal if the RI8 epitope is dominant with or without the variants in the CTL hierarchy.
One factor that may contribute to the apparent impact of HLA-B molecules on HIV disease outcome is the presence of other HLA alleles. In this study, only HLA-B typing was undertaken as the size of the cohort was small (n = 138) and as the focus of the study was HLA-B*52:01. However, HLA-C molecules have been shown to play an important role also in immune control of HIV (42), and they may especially play a role in immune control in HLA-B*52:01-positive individuals. The reason for this is that HLA-B*52:01 is in tight linkage disequilibrium with HLA-C*12:02 (11, 43). The HLA-B*52:01- and HLA-C*12:02-haplotypes were the most prevalent and protective in the Japanese cohorts. The HLA-C-restricted CTL responses and the escape mutations were associated with a lower plasma viral load (11, 44–47). Two recent studies in Japan further revealed that epitopes in polymerase (Pol) were selected by HLA-B*52:01 and -C*12:02 in HIV B clade infection (45, 48). Mutations in these epitopes reduced the viral fitness and contributed to better disease progression. In fact, HLA-C*12 is one of the most highly expressed HLA-C molecules and, similar to other highly expressed HLA-C molecules, is therefore associated with improved control of HIV (42). Similar to HLA-B*52:01, which is a Bw4-80I molecule and therefore a ligand for KIR3DL1 (44), HLA-C alleles are also the ligands for killer cell immunoglobulin-like receptors (KIR). By reducing the binding affinity of KIR2DL2 to the respective HLA complexes, HLA-C*12:02 induces KIR2DL2+ NK cell activity to achieve effective viral control (45). The relationship between certain HLA class I molecules such as HLA-B*52:01 and immune control of HIV is therefore a complex one, requiring more than the interaction between the dominant Gag CD8+ T-cell response and viral replicative capacity to explain it.
In summary, HLA-B*52:01 is a protective allele in not only HIV B clade but also C clade infection. The fitness cost of the mutants at Gag-280, the same P6 position in the targeted epitope, might vary due to different structural changes, but the low VRC is likely to be one major factor that contributes to benefit the host, resulting in relatively high CD4 counts and protection against HIV disease progression. Of note, in other autoimmune and infectious diseases, HLA-B*52 (HLA-B*52:01) has been associated with worse prognosis and outcomes. It is highly susceptible to Takayasu arteritis (49–51), cutaneous Leishmaniasis lesions (52), Henoch-Schönlein purpura (53), Behçet’s disease (54), Crohn’s disease (55), and dengue fever (56). Evidently, the underlying mechanism for these diverse influences of HLA-B*52:01 in various diseases is yet to be defined.
MATERIALS AND METHODS
Study subjects.
In total, 169 chronically HIV-infected individuals were recruited in a North India cohort. The characteristics of the studied participants are shown in Table 1. Informed consent was provided for participation of the subjects in the study. Ethics approval was given by National Centre for Disease Control (NCDC), Delhi, India, and permission for carrying the proviral DNA samples required in this study to Oxford, United Kingdom, was obtained from Indian Council for Medical Research (ICMR), India.
Four-digit HLA-B class I typing was performed from genomic DNA by sequence-based typing at the Clinical Laboratory Improvement Amendments/American Society for Histocompatibility and Immunogenetics (CLIA/ASHI)-accredited laboratory of William Hildebrand at the University of Oklahoma Health Sciences Center, Oklahoma, using locus-specific PCR amplification of class I exons 2 and 3 and heterozygous DNA sequencing. Resolution of ambiguities was undertaken according to the ASHI committee recommendations (57). In total, 42 HLA-B alleles were present in this cohort. CD4 counts of only 13 alleles were analyzed when more than three individuals carried each allele.
CD4+ T cell count data were determined by flow cytometry using a standard clinical protocol and obtained from the AIDS Division of the National Centre for Disease Control (NCDC), Delhi. The mean CD4+ T cell count was 448 cell/mm3 (interquartile range [IQR], 311 to 584 cell/mm3). Viral load data were unavailable in this cohort. Sequences that belonged to clades other than C were excluded from further analysis, including CD4 data analysis and identification of HLA-associated polymorphisms.
Amplification and sequencing of proviral DNA.
Genomic DNA was extracted during the separation of peripheral blood mononuclear cells (PBMCs), and the Gag sequences were then amplified by nested PCR as previously described (58, 59). The primers for the PCRs were 5′-GACTAGCGGAGGCTAGAAG-3′ (G00) and 5′-AGGGGTCGTTGCCAAAGA-3′ (G01) for the first round and 5′-CAGCCAAAATTACCCTATAGTGCAG-3′ (G60) and 5′-ATTGCTTCAGCCAAAACTCTTGC-3′ (G25) for the second round. The PCR products were purified and subjected to automated nucleotide sequencing using dideoxy terminator sequencing chemistry on an automated DNA sequencer (ABI Genetic Analyzer 3730xl). As previously described (27), sequences were analyzed by using Sequencher, version 5.0.1 (Gene Codes Corporation). HXB sequence was used as a reference for all residue numbers.
Site-directed mutagenesis of NL4-3, SK-254(M), and IC.
All the mutations were introduced by using a QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies). The fitness assay performed in this study was based on ΔGag-protease pNL4-3 with three different insertions of Gag-protease sequences as the backbones: (i) pNL4-3, an HIV-1 subtype B NL4-3; (ii) SK-254(M), a modified version of patient-derived HIV-1 Gag-protease sequence (SK-254; GenBank accession number HM593258) with p24 Gag identical to consensus C, as previously described (24); (iii) the Indian consensus (IC) sequence, determined based on the cohort subjects (n = 138) by using Geneious, version 7.0.4 (Table 4). Custom-designed mutagenesis forward and reverse primers for I147L, I256V, D260E, 280X, and 286X are listed in Table 5. Note that due to the proximity of I256V and D260E in the IC sequence, the mutagenesis was performed twice, with one mutation each time. All the mutations were confirmed by sequencing.
TABLE 5.
HLA-B*52:01 custom-designed mutagenesis forward and reverse primers
| HXB2 position in Gag | Clade | Mutant | Primer direction | Primer sequence (5′→3′)a |
|---|---|---|---|---|
| 147 | C | I→L | Forward | CAA ATG GTA CAC CAA GCC TTA TCA CCT AGA ACT TTG AA |
| C | Reverse | TT CAA AGT TCT AGG TGA TAA GGC TTG GTG TAC CAT TTG C | ||
| 256 | C | I→V | Forward | G ATG ACT AGT AAC CCA CCT GTC CCA GTG GGA G |
| C | Reverse | C TCC CAC TGG GAC AGG TGG GTT ACT AGT CAT C | ||
| 260 | C | D→E | Forward | CA CCT ATC CCA GTG GGA GAG ATC TAT AAA AGA TGG ATA AT |
| C | Reverse | AT TAT CCA TCT TTT ATA GAT CTC TCC CAC TGG GAT AGG TG | ||
| 280 | B | T→V | Forward | AAA ATA GTA AGA ATG TAT AGC CCT GTC AGC ATT CTG GAC ATA AGA CAA GG |
| B | Reverse | CC TTG TCT TAT GTC CAG AAT GCT GAC AGG GCT ATA CAT TCT TAC TAT TTT | ||
| B | T→S | Forward | A GTA AGA ATG TAT AGC CCT AGC AGC ATT CTG GAC ATA AG | |
| B | Reverse | CT TAT GTC CAG AAT GCT GCT AGG GCT ATA CAT TCT TAC T | ||
| B | T→A | Forward | ATA GTA AGA ATG TAT AGC CCT GCC AGC ATT CTG GAC ATA AG | |
| B | Reverse | CT TAT GTC CAG AAT GCT GGC AGG GCT ATA CAT TCT TAC TAT | ||
| C | V→T | Forward | T AAA ATA GTA AGA ATG TAT AGC CCT ACC AGC ATC TTG GAC ATA AAA CAA GGG | |
| C | Reverse | CCC TTG TTT TAT GTC CAA GAT GCT GGT AGG GCT ATA CAT TCT TAC TAT TTT A | ||
| C | V→S | Forward | AAA ATA GTA AGA ATG TAT AGC CCT AGC AGC ATC TTG GAC ATA AAA CAA GGG | |
| C | Reverse | CCC TTG TTT TAT GTC CAA GAT GCT GCT AGG GCT ATA CAT TCT TAC TAT TTT A | ||
| C | V→A | Forward | A GTA AGA ATG TAT AGC CCT GCC AGC ATC TTG GAC ATA AAA C | |
| C | Reverse | G TTT TAT GTC CAA GAT GCT GGC AGG GCT ATA CAT TCT TAC T | ||
| 286 | B | R→K | Forward | C AGC ATT CTG GAC ATA AAA CAA GGA CCA AAG GAA CCC |
| B | Reverse | GGG TTC CTT TGG TCC TTG TTT TAT GTC CAG AAT GCT G | ||
| C | K→R | Forward | C AGC ATC TTG GAC ATA AGA CAA GGG CCA AAG GAA CC | |
| C | Reverse | GG TTC CTT TGG CCC TTG TCT TAT GTC CAA GAT GCT G |
Mutant amino acids are shown in boldface.
Virus production and replication kinetics.
All of the plasmids were maxi-prepped according to the manufacturer’s instructions (HiSpeed Plasmid Maxi kit; Qiagen, Hilden, Germany) beforehand. To generate the mutant viruses, the mutated NL4-3, SK-254(M), and IC Gag-protease PCR-amplified and cleaned up products along with the BstEII (New England Biolabs, Ipswich, MA) linearized pNL4-3ΔGag-protease were transfected into GFP reporter GXR cells via electroporation in a Bio-Rad GenePulsar II using 0.4-cm cuvettes at 300 V, 500 μF, and infinite resistance as previously described (18). Virus propagation was then monitored by flow cytometry to detect GFP-expressing infected cells after approximately 2 weeks in culture with GXR cells. Virus culture supernatants were harvested mostly when 30% of cells were GFP positive. Viruses were aliquoted and stored at –80°C until use.
All of the mutations, including the compensatory mutations from long-term culture, were confirmed again by extracting viral RNA from the harvest supernatant using a QIAmp Viral RNA minikit (Qiagen) and Sanger sequencing. Briefly, the extracted RNA was reverse transcribed and amplified for the Gag-protease fragment by using a SuperScript III one-step RT-PCR system with Platinum Taq High Fidelity (Invitrogen) and cDNA amplification (Bioline) as previously described (58, 59). The Sanger sequencing and analysis were the performed as described in the proviral DNA sequencing section. The nucleotide identity between pre- and postinoculation was 99.98%.
Along with the wild type as positive controls and two negative controls without viruses, NL4-3, SK-254, and IC mutant viruses were incubated with GXR cells in a 24-well plate for determination of viral titers, as previously described (58, 59). The percentages of GXR-positive cells were measured by flow cytometry after 48 h. A low multiplicity of infection (MOI) (0.03%) was set as the lowest threshold for determining the amount of virus required for inoculation. The GFP expression was measured from days 2 to 7 (or 8) before it reached saturation at 30% to 40%. The viral replication capacity was defined by the natural semi-log calculation of the mean slope of exponential growth in Excel. This was further calibrated to the normalized value relative to that of the wild-type NL4-3, SK-254(M), and IC. All of the assays were done at least in triplicate.
Phylogenetic analysis.
A BLAST search was carried out to confirm the identity of strains. DNA and protein alignments were created using Clustal X program. Phylogenetic analysis was performed on these sequences to confirm the clade of our sequences. A recombinant identification tool (RIP) was also used to check for occurrence of any recombinant sequences. A maximum likelihood phylogenetic tree using the general time-reversible model of nucleotide substitution was constructed with 1,000 bootstrap replicates using Mega, version 7.0.14, software and viewed using FigTree, version 1.4.0, software. The IC clade consensus sequence was generated using the Gag-protease sequence and the Simple Consensus Maker tool available from the Los Alamos HIV database (http://www.hiv.lanl.gov/). Gag-protease C clade and A, B, F, and D clade reference sequences from the Los Alamos HIV database were included as reference sequences.
Statistical analysis.
All analyses were performed in Prism (version 6.0c; GraphPad). Data were first run to determine whether the distribution was parametric or nonparametric (D’Agostino and Pearson’s omnibus normality test). For two-group analyses, Student's t test (parametric) or Mann-Whitney U test (nonparametric) was performed; for analyses of ≥3 groups, one-way analysis of variance (ANOVA; parametric) or Kruskal-Wallis (nonparametric) followed by post hoc testing was performed. Nominal data categorized in two distinct ways were compared by Fisher’s exact test. P values of <0.05 were considered significant. Post hoc analysis, including Bonferroni’s correction and Steel test for corrected P value was performed, depending on the sample characteristics, sample size, and P value distribution. The viral replication capacity was defined by the log calculation of the mean slope of exponential growth in Microsoft Excel (Mac version, 2011).
Data availability.
All analyzed viral sequences in the cohort are available under GenBank accession numbers MN989432 to MN989571.
ACKNOWLEDGMENTS
P.G. is a Wellcome Trust Investigator (WT104748MA) and is also supported by the NIH (RO1 AI133673). S.S. acknowledges financial support received from the Commonwealth Commission, United Kingdom.
We thank Elizabeth R. Morris, Melvyn W. J. Yap, and Anabel Guedan for their comments on the manuscript. S.S. sincerely thanks Arvind Rai and Shashi Khare for guidance. S.S. also thanks the Centre for AIDS & Related Diseases (CARD), National Centre for Disease Control, Delhi, for providing samples for this study. We acknowledge the Indian Council of Medical Research for permission for the transfer of samples.
REFERENCE
- 1.Carrington M, Walker B. 2012. Immunogenetics of spontaneous control of HIV. Annu Rev Med 63:131–145. doi: 10.1146/annurev-med-062909-130018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiepiela P, Leslie A, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott K, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo M, Altfeld M, James I, Mallal S, Bunce M, Barber L, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia H, Walker B, Goulder P. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 3.International HIV Controllers Study, Pereyra F, Jia X, McLaren PJ, Telenti A, deBakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O’Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, Alvino DM, Vine S, Addo MM, Allen TM, Altfeld M, et al. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fabio G, Scorza R, Lazzarin A, Marchini M, Zarantonello M, D'Arminio A, Marchisio P, Plebani A, Luzzati R, Costigliola P. 1992. HLA-associated susceptibility to HIV-1 infection. Clin Exp Immunol 87:20–23. doi: 10.1111/j.1365-2249.1992.tb06407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antoni G, Guergnon J, Meaudre C, Samri A, Boufassa F, Goujard C, Lambotte O, Autran B, Rouzioux C, Costagliola D, Meyer L, Theodorou I. 2013. MHC-driven HIV-1 control on the long run is not systematically determined at early times post-HIV-1 infection. AIDS 27:1707–1716. doi: 10.1097/QAD.0b013e328360a4bd. [DOI] [PubMed] [Google Scholar]
- 6.Vijaya Lakshmi V, Rakh S, Anu Radha B, Hari Sai Priya V, Pantula V, Jasti S, Suman Latha G, Murthy K. 2006. Role of HLA-B51 and HLA-B52 in susceptibility to pulmonary tuberculosis. Infect Genet Evol 6:436–439. doi: 10.1016/j.meegid.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Teixeira S, de Sá N, Campos D, Coelho A, Guimarães M, Leite T, Veloso V, Morgado M. 2014. Association of the HLA-B*52 allele with non-progression to AIDS in Brazilian HIV-1-infected individuals. Genes Immun 15:256–262. doi: 10.1038/gene.2014.14. [DOI] [PubMed] [Google Scholar]
- 8.Yagita Y, Kuse N, Kuroki K, Gatanaga H, Carlson J, Chikata T, Brumme Z, Murakoshi H, Akahoshi T, Pfeifer N, Mallal S, John M, Ose T, Matsubara H, Kanda R, Fukunaga Y, Honda K, Kawashima Y, Ariumi Y, Oka S, Maenaka K, Takiguchi M. 2013. Distinct HIV-1 escape patterns selected by cytotoxic T cells with identical epitope specificity. J Virol 87:2253–2263. doi: 10.1128/JVI.02572-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chikata T, Carlson J, Tamura Y, Borghan M, Naruto T, Hashimoto M, Murakoshi H, Le A, Mallal S, John M, Gatanaga H, Oka S, Brumme Z, Takiguchi M. 2014. Host-specific adaptation of HIV-1 subtype B in the Japanese population. J Virol 88:4764–4775. doi: 10.1128/JVI.00147-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakai K, Chikata T, Brumme ZL, Brumme CJ, Gatanaga H, Gatanag H, Oka S, Takiguchi M. 2015. Lack of a significant impact of Gag-Protease-mediated HIV-1 replication capacity on clinical parameters in treatment-naive Japanese individuals. Retrovirology 12:98–100. doi: 10.1186/s12977-015-0223-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murakoshi H, Akahoshi T, Koyanagi M, Chikata T, Naruto T, Maruyama R, Tamura Y, Ishizuka N, Gatanaga H, Oka S, Takiguchi M. 2015. Clinical control of HIV-1 by cytotoxic T cells specific for multiple conserved epitopes. J Virol 89:5330–5339. doi: 10.1128/JVI.00020-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seth P. 2010. Evolution of HIV-1 in India. Indian J Virol 21:3–7. doi: 10.1007/s13337-010-0001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hadida F, Haas G, Zimmermann N, Hosmalin A, Spohn R, Samri A, Jung G, Debre P, Autran B. 1995. CTLs from lymphoid organs recognize an optimal HLA-A2-restricted and HLA-B52-restricted nonapeptide and several epitopes in the C-terminal region of HIV-1 Nef. J Immunol 154:4174–4186. [PubMed] [Google Scholar]
- 14.Brander C, Yang O, Jones N, Lee Y, Goulder P, Johnson R, Trocha A, Colbert D, Hay C, Buchbinder S, Bergmann C, Zweerink H, Wolinsky S, Blattner W, Kalams S, Walker B. 1999. Efficient processing of the immunodominant, HLA-A*0201-restricted human immunodeficiency virus type 1 cytotoxic T-lymphocyte epitope despite multiple variations in the epitope flanking sequences. J Virol 73:10191–10198. doi: 10.1128/JVI.73.12.10191-10198.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brumme Z, Brumme C, Carlson J, Streeck H, John M, Eichbaum Q, Block B, Baker B, Kadie C, Markowitz M, Jessen H, Kelleher A, Rosenberg E, Kaldor J, Yuki Y, Carrington M, Allen T, Mallal S, Altfeld M, Heckerman D, Walker B. 2008. Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J Virol 82:9216–9227. doi: 10.1128/JVI.01041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goulder P, Walker B. 2012. HIV and HLA class I: an evolving relationship. Immunity 37:426–440. doi: 10.1016/j.immuni.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlson J, International HIV Adaptation Collaborative, Brumme C, Martin E, Listgarten J, Brockman M, Le A, Chui C, Cotton L, Knapp D, Riddler S, Haubrich R, Nelson G, Pfeifer N, DeZiel C, Heckerman D, Apps R, Carrington M, Mallal S, Harrigan P, John M, Brumme Z. 2012. Correlates of protective cellular immunity revealed by analysis of population-level immune escape pathways in HIV-1. J Virol 86:13202–13216. doi: 10.1128/JVI.01998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miura T, Brockman M, Schneidewind A, Lobritz M, Pereyra F, Rathod A, Block B, Brumme Z, Brumme C, Baker B, Rothchild A, Li B, Trocha A, Cutrell E, Frahm N, Brander C, Toth I, Arts E, Allen T, Walker B. 2009. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare Gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte recognition. J Virol 83:5961–5961. doi: 10.1128/JVI.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawashima Y, Pfafferott K, Frater J, Matthews P, Payne R, Addo M, Gatanaga H, Fujiwara M, Hachiya A, Koizumi H, Kuse N, Oka S, Duda A, Prendergast A, Crawford H, Leslie A, Brumme Z, Brumme C, Allen T, Brander C, Kaslow R, Tang J, Hunter E, Allen S, Mulenga J, Branch S, Roach T, John M, Mallal S, Ogwu A, Shapiro R, Prado JG, Fidler S, Weber J, Pybus OG, Klenerman P, Ndung’u T, Phillips R, Heckerman D, Harrigan PR, Walker BD, Takiguchi M, Goulder P. 2009. Adaptation of HIV-1 to human leukocyte antigen class I. Nature 458:641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Picado J, Prado J, Fry E, Pfafferott K, Leslie A, Chetty S, Thobakgale C, Honeyborne I, Crawford H, Matthews P, Pillay T, Rousseau C, Mullins J, Brander C, Walker B, Stuart D, Kiepiela P, Goulder P. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol 80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kløverpris H, Leslie A, Goulder P. 2015. Role of HLA adaptation in HIV evolution. Front Immunol 6:665. doi: 10.3389/fimmu.2015.00665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goepfert P, Lumm W, Farmer P, Matthews P, Prendergast A, Carlson J, Derdeyn C, Tang J, Kaslow R, Bansal A, Yusim K, Heckerman D, Mulenga J, Allen S, Goulder P, Hunter E. 2008. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J Exp Med 205:1009–1017. doi: 10.1084/jem.20072457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prince J, Claiborne D, Carlson J, Schaefer M, Yu T, Lahki S, Prentice H, Yue L, Vishwanathan S, Kilembe W, Goepfert P, Price M, Gilmour J, Mulenga J, Farmer P, Derdeyn C, Tang J, Heckerman D, Kaslow R, Allen S, Hunter E. 2012. Role of transmitted Gag CTL polymorphisms in defining replicative capacity and early HIV-1 pathogenesis. PLoS Pathog 8:e1003041. doi: 10.1371/journal.ppat.1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crawford H, Prado J, Leslie A, Hue S, Honeyborne I, Reddy S, van der Stok M, Mncube Z, Brander C, Rousseau C, Mullins J, Kaslow R, Goepfert P, Allen S, Hunter E, Mulenga J, Kiepiela P, Walker B, Goulder P. 2007. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J Virol 81:8346–8351. doi: 10.1128/JVI.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright JK, Naidoo VL, Brumme ZL, Prince JL, Claiborne DT, Goulder PJR, Brockman MA, Hunter E, Ndung'u T. 2012. Impact of HLA-B*81-associated mutations in HIV-1 Gag on viral replication capacity. J Virol 86:3193–3199. doi: 10.1128/JVI.06682-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai M, Muenchhoff M, Adland E, Carlqvist A, Roider J, Cole D, Sewell A, Carlson J, Ndung’u T, Goulder P. 2016. Paediatric non-progression following grandmother-to-child HIV transmission. Retrovirology 13:1. doi: 10.1186/s12977-016-0300-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matthews PC, Adland E, Listgarten J, Leslie A, Mkhwanazi N, Carlson JM, Harndahl M, Stryhn A, Payne RP, Ogwu A, Huang K-HG, Frater J, Paioni P, Kloverpris H, Jooste P, Goedhals D, van Vuuren C, Steyn D, Riddell L, Chen F, Luzzi G, Balachandran T, Ndung'u T, Buus S, Carrington M, Shapiro R, Heckerman D, Goulder PJR. 2011. HLA-A*7401-mediated control of HIV viremia is independent of its linkage disequilibrium with HLA-B*5703. J Immunol 186:5675–5686. doi: 10.4049/jimmunol.1003711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ragonnet-Cronin M, Shilaih M, Günthard H, Hodcroft E, Böni J, Fearnhill E, Dunn D, Yerly S, Klimkait T, Aubert V, Yang W, Brown A, Lycett S, Kouyos R, Brown A. 2016. A direct comparison of two densely sampled HIV epidemics: the UK and Switzerland. Sci Rep 6:32251. doi: 10.1038/srep32251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao X, Nelson GW, Karacki P, Martin MP, Phair J, Kaslow R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, O'Brien SJ, Carrington M. 2001. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N Engl J Med 344:1668–1675. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
- 30.Juarez-Molina C, Valenzuela-Ponce H, Avila-Rios S, Garrido-Rodriguez D, Garcia-Tellez T, Soto-Nava M, Garcia-Morales C, Goulder P, Reyes-Teran G. 2014. Impact of HLA-B*35 subtype differences on HIV disease outcome in Mexico. AIDS 28:1687–1690. doi: 10.1097/QAD.0000000000000322. [DOI] [PubMed] [Google Scholar]
- 31.Carlson JM, Du VY, Pfeifer N, Bansal A, Tan VYF, Power K, Brumme CJ, Kreimer A, DeZiel CE, Fusi N, Schaefer M, Brockman MA, Gilmour J, Price MA, Kilembe W, Haubrich R, John M, Mallal S, Shapiro R, Frater J, Harrigan PR, Ndung'u T, Allen S, Heckerman D, Sidney J, Allen TM, Goulder PJR, Brumme ZL, Hunter E, Goepfert PA. 2016. Impact of pre-adapted HIV transmission. Nat Med 22:606–613. doi: 10.1038/nm.4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adland E, Paioni P, Thobakgale C, Laker L, Mori L, Muenchhoff M, Csala A, Clapson M, Flynn J, Novelli V, Hurst J, Naidoo V, Shapiro R, Huang K, Frater J, Prendergast A, Prado J, Ndung’u T, Walker B, Carrington M, Jooste P, Goulder P. 2015. Discordant impact of HLA on viral replicative capacity and disease progression in pediatric and adult HIV infection. PLoS Pathog 11:e1004954. doi: 10.1371/journal.ppat.1004954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedrich TC, Dodds EJ, Yant LJ, Vojnov L, Rudersdorf R, Cullen C, Evans DT, Desrosiers RC, Mothé BR, Sidney J, Sette A, Kunstman K, Wolinsky S, Piatak M, Lifson J, Hughes AL, Wilson N, O'Connor DH, Watkins DI. 2004. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat Med 10:275–281. doi: 10.1038/nm998. [DOI] [PubMed] [Google Scholar]
- 34.Leslie A, Pfafferott K, Chetty P, Draenert R, Addo M, Feeney M, Tang Y, Holmes E, Allen T, Prado J, Altfeld M, Brander C, Dixon C, Ramduth D, Jeena P, Thomas S, John A, Roach T, Kupfer B, Luzzi G, Edwards A, Taylor G, Lyall H, Tudor-Williams G, Novelli V, Martinez-Picado J, Kiepiela P, Walker B, Goulder P. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med 10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi M, Igarashi H, Takeda A, Kato M, Matano T. 2005. Reversion in vivo after inoculation of a molecular proviral DNA clone of simian immunodeficiency virus with a cytotoxic-T-lymphocyte escape mutation. J Virol 79:11529–11532. doi: 10.1128/JVI.79.17.11529-11532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimbwa P, Milicic A, Frater J, Scriba T, Willis A, Goulder P, Pillay T, Gunthard H, Weber J, Zhang H, Phillips R. 2007. Precise identification of a human immunodeficiency virus type 1 antigen processing mutant. J Virol 81:2031–2038. doi: 10.1128/JVI.00968-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Draenert R, Le Gall S, Pfafferott K, Leslie A, Chetty P, Brander C, Holmes E, Chang S, Feeney M, Addo M, Ruiz L, Ramduth D, Jeena P, Altfeld M, Thomas S, Tang Y, Verrill C, Dixon C, Prado J, Kiepiela P, Martinez-Picado J, Walker B, Goulder P. 2004. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J Exp Med 199:905–915. doi: 10.1084/jem.20031982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.González-Ortega E, Ballana E, Badia R, Clotet B, Esté J. 2011. Compensatory mutations rescue the virus replicative capacity of VIRIP-resistant HIV-1. Antiviral Res 92:479–483. doi: 10.1016/j.antiviral.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 39.Koibuchi T, Allen TM, Lichterfeld M, Mui SK, O'Sullivan KM, Trocha A, Kalams SA, Johnson RP, Walker BD. 2005. Limited sequence evolution within persistently targeted CD8 epitopes in chronic human immunodeficiency virus type 1 infection. J Virol 79:8171–8181. doi: 10.1128/JVI.79.13.8171-8181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe K, Murakoshi H, Tamura Y, Koyanagi M, Chikata T, Gatanaga H, Oka S, Takiguchi M. 2013. Identification of cross-clade CTL epitopes in HIV-1 clade A/E-infected individuals by using the clade B overlapping peptides. Microbes Infect 15:874–886. doi: 10.1016/j.micinf.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Pereyra F, Heckerman D, Carlson J, Kadie C, Soghoian D, Karel D, Goldenthal A, Davis O, DeZiel C, Lin T, Peng J, Piechocka A, Carrington M, Walker B. 2014. HIV control is mediated in part by CD8+ T-cell targeting of specific epitopes. J Virol 88:12937–12948. doi: 10.1128/JVI.01004-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Apps R, Qi Y, Carlson JM, Chen H, Gao X, Thomas R, Yuki Y, Del Prete GQ, Goulder P, Brumme ZL, Brumme CJ, John M, Mallal S, Nelson G, Bosch R, Heckerman D, Stein JL, Soderberg KA, Moody MA, Denny TN, Zeng X, Fang J, Moffett A, Lifson JD, Goedert JJ, Buchbinder S, Kirk GD, Fellay J, McLaren P, Deeks SG, Pereyra F, Walker B, Michael NL, Weintrob A, Wolinsky S, Liao W, Carrington M. 2013. Influence of HLA-C expression level on HIV control. Science 340:87–91. doi: 10.1126/science.1232685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naruto T, Gatanaga H, Nelson G, Sakai K, Carrington M, Oka S, Takiguchi M. 2012. HLA class I-mediated control of HIV-1 in the Japanese population, in which the protective HLA-B*57 and HLA-B*27 alleles are absent. J Virol 86:10870–10872. doi: 10.1128/JVI.00689-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O'Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. 2007. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet 39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Z, Kuroki K, Kuse N, Sun X, Akahoshi T, Qi Y, Chikata T, Naruto T, Koyanagi M, Murakoshi H, Gatanaga H, Oka S, Carrington M, Maenaka K, Takiguchi M. 2016. HIV-1 control by NK cells via reduced interaction between KIR2DL2 and HLA-C*12:02/C*14:03. Cell Rep 17:2210–2220. doi: 10.1016/j.celrep.2016.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chikata T, Murakoshi H, Koyanagi M, Honda K, Gatanaga H, Oka S, Takiguchi M. 2017. Control of HIV-1 by an HLA-B*52:01-C*12:02 protective haplotype. J Infect Dis 216:1415–1424. doi: 10.1093/infdis/jix483. [DOI] [PubMed] [Google Scholar]
- 47.Chikata T, Tran G, Murakoshi H, Akahoshi T, Qi Y, Naranbhai V, Kuse N, Tamura Y, Koyanagi M, Sakai S, Nguyen D, Nguyen D, Nguyen H, Nguyen T, Oka S, Martin M, Carrington M, Sakai K, Nguyen K, Takiguchi M. 2017. HLA class I-mediated HIV-1 control in Vietnamese infected with HIV-1 subtype A/E. J Virol 92:e01749-17. doi: 10.1128/JVI.01749-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zou C, Murakoshi H, Kuse N, Akahoshi T, Chikata T, Gatanaga H, Oka S, Hanke T, Takiguchi M. 2019. Effective suppression of HIV-1 replication by cytotoxic T lymphocytes specific for Pol epitopes in conserved mosaic vaccine immunogens. J Virol 93:e02142-18. doi: 10.1128/JVI.02142-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Terao C, Yoshifuji H, Mimori T. 2014. Recent advances in Takayasu arteritis. Int J Rheum Dis 17:238–247. doi: 10.1111/1756-185X.12309. [DOI] [PubMed] [Google Scholar]
- 50.Terao C, Matsumura T, Yoshifuji H, Kirino Y, Maejima Y, Nakaoka Y, Takahashi M, Amiya E, Tamura N, Nakajima T, Origuchi T, Horita T, Matsukura M, Kochi Y, Ogimoto A, Yamamoto M, Takahashi H, Nakayamada S, Saito K, Wada Y, Narita I, Kawaguchi Y, Yamanaka H, Ohmura K, Atsumi T, Tanemoto K, Miyata T, Kuwana M, Komuro I, Tabara Y, Ueda A, Isobe M, Mimori T, Matsuda F. 2015. Takayasu arteritis and ulcerative colitis: high rate of co-occurrence and genetic overlap. Arthritis Rheumatol 67:2226–2232. doi: 10.1002/art.39157. [DOI] [PubMed] [Google Scholar]
- 51.Chen S, Luan H, Li L, Zeng X, Wang T, Li Y, Yuan H. 2017. Relationship of HLA-B*51 and HLA-B*52 alleles and TNF-α-308A/G polymorphism with susceptibility to Takayasu arteritis: a meta-analysis. Clin Rheumatol 36:173–181. doi: 10.1007/s10067-016-3445-0. [DOI] [PubMed] [Google Scholar]
- 52.Ribas-Silva R, Ribas A, Ferreira E, Silveira T, Borelli S. 2015. Association between HLA-C*04 and American cutaneous leishmaniasis in endemic region of southern Brazil. Genet Mol Res 14:14929–14935. doi: 10.4238/2015.November.18.58. [DOI] [PubMed] [Google Scholar]
- 53.Ren S, Yang G, Liu C, Zhang C, Shou Q, Yu S, Li W, Su X. 2012. Association between HLA-A and -B polymorphisms and susceptibility to Henoch-Schönlein purpura in Han and Mongolian children from Inner Mongolia. Genet Mol Res 11:221–228. doi: 10.4238/2012.February.3.2. [DOI] [PubMed] [Google Scholar]
- 54.Soto-Vega E, García-Muñoz R, Richaud-Patin Y, Zúñiga-Ramos J, Crispín JC, Díaz-Jouanen E, Flores-Suárez LF, Llorente L, Granados J. 2004. Class I and class II MHC polymorphisms in Mexican patients with Behçet’s disease. Immunol Lett 93:211–215. doi: 10.1016/j.imlet.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 55.Kinouchi Y, Matsumoto K, Negoro K, Takagi S, Takahashi S, Hiwatashi N, Shimosegawa T. 2003. HLA-B genotype in Japanese patients with Crohn’s disease. Dis Colon Rectum 46(10 Suppl):S10–S14. [DOI] [PubMed] [Google Scholar]
- 56.Stephens H, Klaythong R, Sirikong M, Vaughn D, Green S, Kalayanarooj S, Endy T, Libraty D, Nisalak A, Innis B, Rothman A, Ennis F, Chandanayingyong D. 2002. HLA-A and -B allele associations with secondary dengue virus infections correlate with disease severity and the infecting viral serotype in ethnic Thais. Tissue Antigens 60:309–318. doi: 10.1034/j.1399-0039.2002.600405.x. [DOI] [PubMed] [Google Scholar]
- 57.Cano P, Klitz W, Mack SJ, Maiers M, Marsh SG, Noreen H, Reed EF, Senitzer D, Setterholm M, Smith A, Fernández-Viña M. 2007. Common and well-documented HLA alleles: report of the ad-hoc committee of the American Society for Histocompatiblity and immunogenetics. Hum Immunol 68:392–417. doi: 10.1016/j.humimm.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 58.Brockman M, Schneidewind A, Lahaie M, Schmidt A, Miura T, DeSouza I, Ryvkin F, Derdeyn C, Allen S, Hunter E, Mulenga J, Goepfert P, Walker B, Allen T. 2007. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J Virol 81:12608–12618. doi: 10.1128/JVI.01369-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wright JK, Brumme ZL, Carlson JM, Heckerman D, Kadie CM, Brumme CJ, Wang B, Losina E, Miura T, Chonco F, van der Stok M, Mncube Z, Bishop K, Goulder PJR, Walker BD, Brockman MA, Ndung'u T. 2010. Gag-protease-mediated replication capacity in HIV-1 subtype C chronic infection: associations with HLA type and clinical parameters. J Virol 84:10820–10831. doi: 10.1128/JVI.01084-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All analyzed viral sequences in the cohort are available under GenBank accession numbers MN989432 to MN989571.