In the past few decades, respiratory syncytial virus (RSV) has still been a major health concern worldwide. The vaccine-enhance disease (VED) has hindered RSV vaccine development. A truncated hepatitis B virus core protein vaccine containing the conserved region (amino acids 144 to 204) of the RSV G protein (HBc-tG) had previously been shown to induce effective immune responses and confer protection against RSV infection in mice but to also lead to VED. In this study, we investigated the effect of IL-35 on the host response and immunopathology following RSV infection in vaccinated mice. Our results indicate that HBc-tG together with IL-35 elicited a balanced immune response and protected mice against RSV infection without vaccine-enhanced immunopathology. Applying a systems biology method, we identified Il10 to be the key regulator in reducing the excessive lung inflammation. Our study provides new insight into the function of IL-35 and its regulatory mechanism of VED at the network level.
KEYWORDS: Il10, interleukin-35, regulatory T cells, respiratory syncytial virus, transcriptome analysis, vaccine-enhanced disease
ABSTRACT
Respiratory syncytial virus (RSV) is the most important cause of lower respiratory tract infection in infants and young children. The vaccine-enhanced disease (VED) has greatly hindered the development of an RSV vaccine. Currently, there are no licensed vaccines for RSV. In this study, immunization of mice with hepatitis B virus core particles containing a conserved region of the G protein (HBc-tG) combined with interleukin-35 (IL-35) elicited a Th1-biased response and a high frequency of regulatory T (Treg) cells and increased the levels of IL-10, transforming growth factor β, and IL-35 production. Importantly, immunization with HBc-tG together with IL-35 protected mice against RSV infection without vaccine-enhanced immunopathology. To explore the mechanism of how IL-35 reduces lung inflammation at the gene expression level, transcription profiles were obtained from lung tissues of immunized mice after RSV infection by the Illumina sequencing technique and further analyzed by a systems biology method. In total, 2,644 differentially expressed genes (DEGs) were identified. Twelve high-influence modules (HIMs) were selected from these DEGs on the basis of the protein-protein interaction network. A detailed analysis of HIM10, involved in the immune response network, revealed that Il10 plays a key role in regulating the host response. The selected DEGs were consistently confirmed by quantitative real-time PCR (qRT-PCR). Our results demonstrate that IL-35 inhibits vaccine-enhanced immunopathology after RSV infection and has potential for development in novel therapeutic and prophylactic strategies.
IMPORTANCE In the past few decades, respiratory syncytial virus (RSV) has still been a major health concern worldwide. The vaccine-enhance disease (VED) has hindered RSV vaccine development. A truncated hepatitis B virus core protein vaccine containing the conserved region (amino acids 144 to 204) of the RSV G protein (HBc-tG) had previously been shown to induce effective immune responses and confer protection against RSV infection in mice but to also lead to VED. In this study, we investigated the effect of IL-35 on the host response and immunopathology following RSV infection in vaccinated mice. Our results indicate that HBc-tG together with IL-35 elicited a balanced immune response and protected mice against RSV infection without vaccine-enhanced immunopathology. Applying a systems biology method, we identified Il10 to be the key regulator in reducing the excessive lung inflammation. Our study provides new insight into the function of IL-35 and its regulatory mechanism of VED at the network level.
INTRODUCTION
Respiratory syncytial virus (RSV) is the major cause of pneumonia and bronchiolitis in infants and young children worldwide (1–3). Globally, there are an estimated 33 million annual cases of RSV-associated lower respiratory tract infection in children under the age of 5 years (4, 5). RSV infections in older adults are also increasingly recognized as a cause of severe respiratory disease, with complications leading to prolonged hospital stays and high mortality rates (6). Therefore, there is an urgent need to develop an effective vaccine for RSV.
In the 1960s, a formalin-inactivated RSV (FI-RSV) vaccine was used to immunize infants, but subsequent natural RSV infection of the vaccinees resulted in enhanced pulmonary disease (7–9). Previous studies showed that the enhanced pulmonary disease was associated with an exaggerated Th2-type response, which could recruit infiltrating granulocytes, especially eosinophils (10, 11). Depletion of the Th2 cytokine interleukin-4 (IL-4) or IL-13 in mice immunized with FI-RSV inhibited the development of pulmonary disease after RSV challenge (12–14). Thus, induction of a balanced immune response would be critical for a safe RSV vaccine.
Regulatory T (Treg) cells are a subset of CD4+ T cells that specifically express the forkhead box P3 (Foxp3) transcription factor. Previous reports showed that Treg cells can prevent a Th2-type immune response and pulmonary eosinophilia during RSV infection (15), that Treg cells secrete anti-inflammatory cytokines, such as IL-10, IL-35, and transforming growth factor β (TGF-β), and that IL-10 is important in limiting the pathogenic T cell activation and proliferation provoked by RSV infection (16, 17). More remarkably, the defective immune regulation in patients with RSV vaccine-augmented lung disease was restored by the selective chemoattraction of Treg cells (18, 19). These studies revealed that Treg cells modulate the dysregulated immune response to RSV infection.
IL-35 is a novel anti-inflammatory cytokine composed of Epstein-Barr virus-induced gene 3 (EBI3) and p35 subunits (20, 21). IL-35 attenuated established collagen-induced arthritis in mice, with suppression of IL-17 production and promotion of IL-10 expression (22). In an experimental autoimmune uveitis model, IL-35 inhibited autoreactive Th1/Th17 cells and promoted the expansion of Treg cells (23). A recent study demonstrated that IL-35 decelerates the inflammatory process by regulating inflammatory cytokine secretion and the M1/M2 macrophage ratio in patients with psoriasis (24). However, the role of IL-35 in RSV vaccine-enhanced disease (VED) is unclear.
Our previous study showed that mice immunized with hepatitis B virus core particles containing a conserved region of the G protein (HBc-tG) develop enhanced pulmonary inflammation after RSV infection (25). In this study, we investigated the role of the anti-inflammatory cytokine IL-35 in the vaccine-enhanced immunopathology induced by HBc-tG. Combined with transcriptome analysis, we explored the molecular mechanism of RSV vaccine-enhanced immunopathology. Applying an emerging systems biology approach, we quantified the interactions between differentially expressed genes (DEGs) and identified groups of strongly connected genes, allowing us to detect high-influence modules (HIMs) representing host response networks.
RESULTS
Construction and characterization of recombinant IL-35.
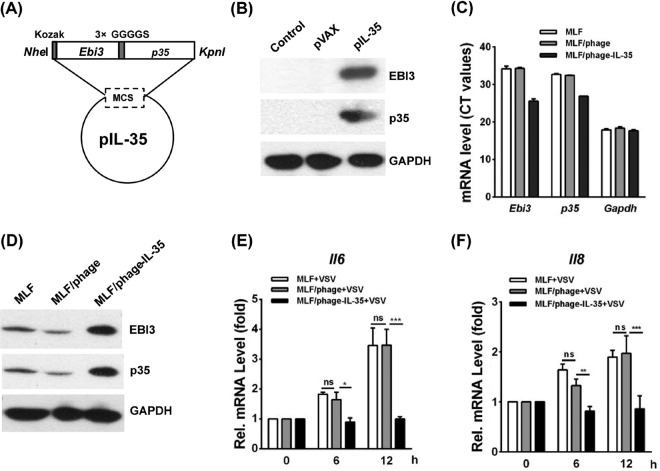
The Ebi3 and p35 genes linked by 3×GGGGS were cloned into the eukaryotic expression vector pVAX to generate recombinant plasmid pVAX-IL-35 (pIL-35) (Fig. 1A). To confirm the expression of the IL-35 protein, pIL-35 was transfected into 293T cells. At 48 h posttransfection (hpt), EBI3 or p35 expression was determined by Western blotting with an anti-EBI3 or anti-p35 antibody, respectively (Fig. 1B).
FIG 1.
Expression of constructed IL-35 in 293T cells and the mRNA levels of Il6 and Il8 in MLFs, MLF/phage, and MLF/phage-IL-35 stimulated with VSV. (A) The sequence of Ebi3 and p35, linked by 3×GGGGS, was inserted into the NheI/KpnI sites of pVAX to generate pIL-35. (B) Western blots were performed to measure IL-35 levels in 293T cells that had been transfected with pVAX or pIL-35 for 48 h. IL-35 was detected with anti-mouse EBI3 or anti-mouse p35 antibody (the molecular mass of IL-35 is ∼50 kDa). (C) Ebi3 and p35 mRNA levels in MLFs, MLF/phage, and MLF/phage-IL-35. The mRNA level was determined from the threshold cycle (CT) values of the PCR amplification. Low threshold cycle values indicate high mRNA levels, while high threshold cycle values represent low mRNA levels. (D) IL-35 expression in MLFs was confirmed by Western blotting with anti-mouse EBI3 antibody and anti-mouse p35 antibody (the molecular mass of IL-35 is ∼50 kDa). (E and F) qRT-PCR analysis of Il6 (E) and Il8 (F) mRNA levels in MLFs, MLF/phage, and MLF/phage-IL-35 after VSV stimulation for 6 h and 12 h. Pairwise comparisons of the values from 3 experiments were performed using a t test. ***, P < 0.01; **, P < 0.01; *, P < 0.05; ns, not significant. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Rel., relative.
To test the activity of IL-35 in vitro under a stable condition, we generated mouse lung fibroblasts (MLFs) that stably overexpressed IL-35 (MLF/phage-IL-35). The generated MLF/phage was used as a negative control. The mRNA and protein levels of IL-35 were verified by quantitative real-time PCR (qRT-PCR) and Western blotting, respectively (Fig. 1C and D). To investigate the effect of IL-35 on IL-6 and IL-8 expression, MLF, MLF/phage, or MLF/phage-IL-35 was stimulated with vesicular stomatitis virus (VSV) treatment. Then, the mRNA levels of Il6 and Il8 in the cell lysates were tested by qRT-PCR. The data showed that the mRNA levels of Il6 and Il8 observed in MLF/phage-IL-35 were significantly decreased compared with those observed in MLF or MLF/phage at 0, 6, and 12 h after VSV stimulation (P < 0.05) (Fig. 1E and F), suggesting that IL-35 significantly inhibited IL-6 and IL-8 expression in stimulated MLFs.
RSV-specific immune response induced by HBc-tG+pIL-35.
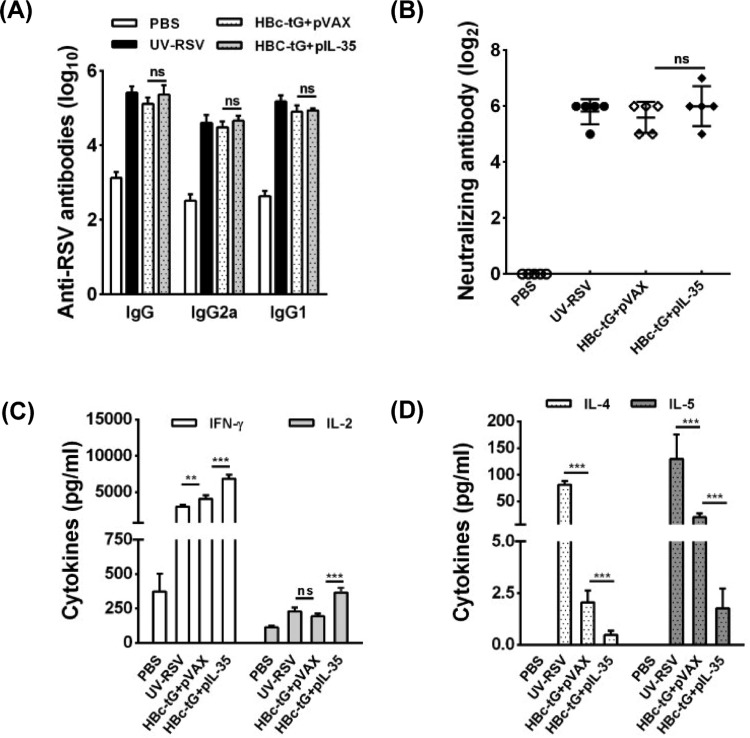
To evaluate the specific antibody response induced by HBc-tG combined with the plasmid pIL-35 (HBc-tG+pIL-35), we measured the RSV-specific IgG, IgG1, and IgG2a concentrations in the sera of immunized mice by enzyme-linked immunosorbent assay (ELISA). The data demonstrated that vaccination with HBc-tG+pIL-35 generated IgG, IgG1, and IgG2a antibody levels similar to those generated by HBc-tG combined with the plasmid pVAX (HBc-tG+pVAX) in mice (P > 0.05) (Fig. 2A). The neutralizing antibody titers in the sera of immunized mice were measured using an in vitro plaque reduction assay. Both HBc-tG+pVAX and HBc-tG+pIL-35 induced similar RSV-neutralizing antibody levels (P > 0.05) (Fig. 2B). These data indicate that IL-35 had no impact on the specific antibody response induced by HBc-tG.
FIG 2.
RSV-specific humoral and cellular responses induced by HBc-tG+pIL-35. Mice were immunized i.p. three times with PBS, UV-RSV, or HBc-tG+pIL-35. Serum samples and spleen cells were collected at 2 weeks after the final immunization. Cells were stimulated with heat-inactivated RSV A2, and the supernatants were collected after 72 h of incubation. (A) RSV-specific IgG, IgG1, and IgG2a antibodies were determined by ELISA. (B) RSV-neutralizing antibody titers were determined by a plaque reduction assay. (C and D) Th1 cytokine (IFN-γ, IL-2) (C) and Th2 cytokine (IL-4 and IL-5) (D) concentrations were measured by ELISA. Data are presented as the mean values ± SD for five mice in each group. Pairwise comparisons of values were performed using a t test or one-way ANOVA. ***, P < 0.01; **, P < 0.01; ns, not significant.
Next, we determined the concentrations of Th1-type (interferon gamma [IFN-γ] and IL-2) and Th2-type (IL-4 and IL-5) cytokines in the culture supernatants of splenocytes from immunized mice. Mice vaccinated with HBc-tG+pVAX, HBc-tG+pIL-35, or UV-inactivated RSV (UV-RSV) induced both Th1 and Th2 cytokine production (Fig. 2C and D). Vaccination with HBc-tG+pIL-35 resulted in significantly increased IFN-γ and IL-2 levels (P < 0.01) (Fig. 2C) and decreased IL-4 and IL-5 production (P < 0.01) compared with the findings obtained by vaccination with HBc-tG+pVAX (Fig. 2D). These results reveal that HBc-tG+pVAX induced a mixed Th1/Th2 response and that the Th2-type response induced by HBc-tG was significantly suppressed by IL-35.
Pulmonary viral load and pathology in mice immunized with HBc-tG+pIL-35 following RSV infection.
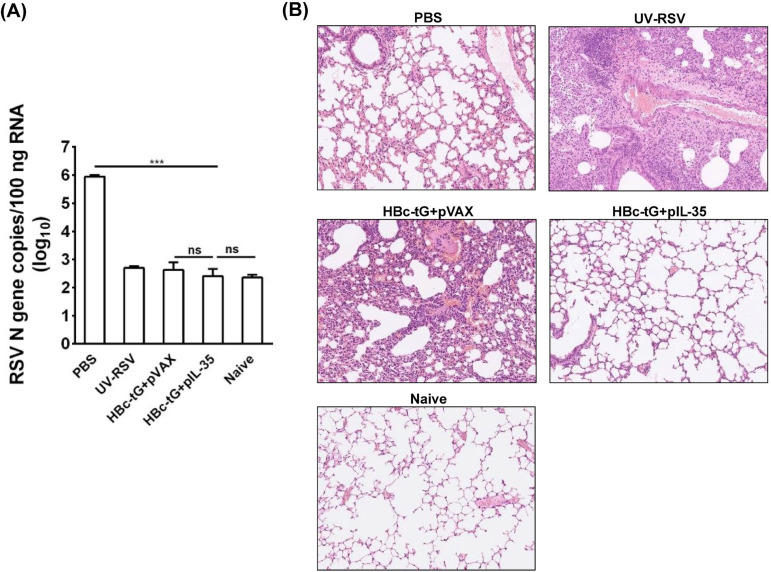
To investigate the effect of HBc-tG+pIL-35 vaccination on lung viral clearance, immunized mice were infected intranasally (i.n.) with RSV at 2 weeks after the final immunization. The RSV load in the lungs was measured on day 4 postchallenge by qRT-PCR. Naive mice that received no treatment were used as a negative control. The phosphate-buffered saline (PBS)-treated mice had high RSV N gene copy numbers in the lungs (∼106 copies/100 ng total RNA), whereas low RSV N gene copy numbers (<103 copies) were observed in the lungs of mice vaccinated with HBc-tG+pVAX and HBc-tG+pIL-35, similar to the findings for naive mice (P > 0.05) (Fig. 3A), demonstrating that vaccination with HBc-tG+pVAX or HBc-tG+pIL-35 effectively inhibited RSV replication in mice.
FIG 3.
RSV load and histopathological analysis of lung tissues from vaccinated mice upon RSV challenge. Mice were immunized i.p. three times and challenged i.n. with RSV 3 weeks after the final immunization. Lungs were harvested at 4 days postchallenge. (A) RSV N gene copy numbers in lung tissues were measured by qRT-PCR. Data are presented as the mean values ± SD for five mice in each group. (B) Hematoxylin-eosin (H&E) staining of lung tissues from immunized mice at day 4 after RSV challenge. An image for a representative section is shown for each experimental group or for a naive mouse.
To evaluate the effect of HBc-tG+pIL-35 vaccination on lung injury upon RSV infection, vaccinated mice were challenged with RSV and lung tissues were assessed for inflammatory pathology. The data showed that mice immunized with HBc-tG+pVAX exhibited severe lung pathology, including extensive lymphocyte infiltration around the blood vessels and alveolar hemorrhage. In contrast, mice vaccinated with HBc-tG+pIL-35 had no signs of inflammation in the lung tissues, which was similar to the findings for naive mice (Fig. 3B). The average inflammation severity scores of vaccinated mice were in the following order: UV-RSV > HBc-tG+pVAX > PBS > HBc-tG+pIL-35 (Table 1). These results demonstrate that IL-35 eliminated the pulmonary damage induced by HBc-tG upon RSV infection.
TABLE 1.
Histopathology scores of lungs in vaccinated mice at day 4 after RSV challenge
| Inoculum | No. of mice with the following lung lesion severity scorea
: |
Mean severity scoreb | ||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| Naive | 0 | 1 | 1 | 0.7 |
| PBS | 2 | 3 | 2 | 2.3 |
| UV-RSV | 4 | 4 | 3 | 3.7 |
| HBc-tG+pVAX | 3 | 4 | 3 | 3.3 |
| HBc-tG+pIL-35 | 1 | 1 | 2 | 1.3 |
The lung inflammation severity scores were defined on a scale of from 0 to 4, as described in Materials and Methods, where 0 indicates that inflammation is not present, 1 indicates minimal inflammation, 2 indicates mild inflammation, 3 indicates moderate inflammation, and 4 indicates marked inflammation.
The mean severity score for mice (n = 3).
Cellular immune response in lungs of mice induced by HBc-tG+pIL-35 following RSV infection.
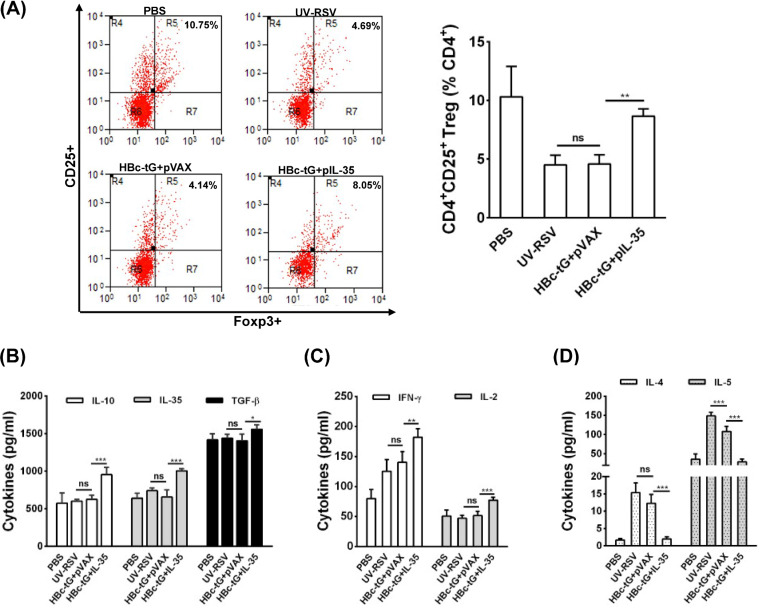
Next, the proportion of CD25+ Foxp3+ Treg cells among the CD4+ T cells and the levels of cytokines in the lungs of vaccinated mice upon RSV challenge were measured. The data showed that the percentage of CD25+ Foxp3+ Treg cells among CD4+ T cells in the lungs of mice immunized with HBc-tG+pIL-35 was significantly increased compared to that in the lungs of mice immunized with HBc-tG+pVAX (P < 0.01) (Fig. 4A). As expected, significantly increased levels of production of pulmonary Treg cell-related cytokines (IL-10, TGF-β, and IL-35) (Fig. 4B) and Th1-type cytokines (IFN-γ and IL-2) (Fig. 4C) and decreased levels of production of Th2-type cytokines (IL-4 and IL-5) (Fig. 4D) were induced by HBc-tG+pIL-35 compared to the levels induced by HBc-tG+pVAX (P < 0.05). These results indicate that HBc-tG+pIL-35 elicited a robust Treg cell response and a weakened inflammatory response in the lungs of vaccinated mice upon subsequent RSV infection.
FIG 4.
Immune response in lungs induced by HBc-tG+pIL-35 after RSV infection. Mice were immunized i.p. three times and challenged i.n. with RSV 3 weeks after the final immunization. Lungs were harvested at day 4 postchallenge. (A) The percentage of CD25+ Foxp3+ Treg cells in CD4+ T cells from lungs was measured by flow cytometry with specific antibody staining. (B to D) Treg cell-related cytokine (IL-35, TGF-β, IL-10) (B), Th1 cytokine (IFN-γ, IL-2) (C), and Th2 cytokine (IL-4 and IL-5) (D) concentrations were measured by ELISA. Data are presented as the mean values ± SD for five mice in each group. Pairwise comparisons of values were performed using a t test or one-way ANOVA. ***, P < 0.01; **, P < 0.01; *, P < 0.05; ns, not significant.
Gene expression profile in lungs of immunized mice after RSV infection.
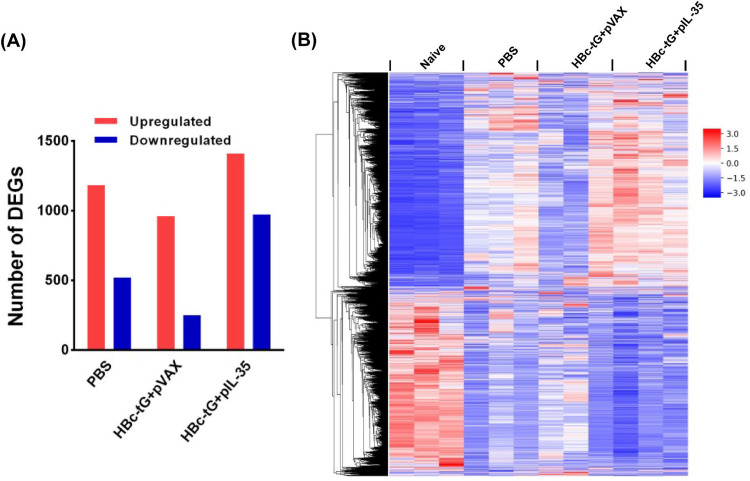
To further investigate the molecular mechanism of how IL-35 inhibits lung inflammation at the gene expression level, we performed transcriptome sequencing of lung tissues from PBS-treated, HBc-tG+pVAX-immunized, and HBc-tG+pIL-35-immunized mice on day 4 after RSV challenge. Naive mice that received no treatment were used as a control. A stronger host transcriptional response in treated mice than in naive mice was observed. In total, 2,644 DEGs were identified from treated mice (see Table S1 in the supplemental material). As shown in Fig. 5A, 1,703, 1,210, and 2,382 DEGs were obtained in the PBS-, HBc-tG+pVAX-, and HBc-tG+pIL-35-treated groups, respectively, compared to their expression in the naive group, and of these, 1,182, 959, and 1,410 DEGs, respectively, were upregulated and 521, 251, and 972 DEGs, respectively, were downregulated in the three groups. The clustering of the DEGs is shown as a heat map (Fig. 5B). The data showed that RSV infection induced the maximum changes in the differentially expressed gene profile in HBc-tG+pIL-35-vaccinated mice, indicating that IL-35 enhanced the host transcriptional response.
FIG 5.
Transcriptional profiles in lungs of vaccinated mice upon RSV challenge. (A) The number of DEGs in the lungs of vaccinated mice (n = 3) on day 4 after RSV infection. The upregulated and downregulated genes are indicated by the red and blue bars, respectively. The criteria used for determination of DEGs were a P value of <0.05 after an adjusted false discovery rate (FDR) and a |log2 fold change in expression value| of ≥1. (B) Heat map depicting the expression values for 2,644 DEGs from immunized mice upon RSV challenge. Each column is for a sample from a mouse immunized or treated with the inoculum labeled at the top, and each row represents a gene whose expression value was normalized across that for the naive mice by the Z-score method.
GO functional enrichment analysis of HIMs.
Twelve high-influence modules from 2,644 DEGs (Table S2) were selected on the basis of the initial protein-protein interaction (PPI) network. Gene Ontology (GO) functional enrichment of the 12 HIMs identified 1,068 significant categories (Table S3). Representative GO categories from each HIM are shown in Table 2, and all GO functional enrichments from each HIM are listed in Table S3. HIM1 contained the largest number of DEGs, and the functional enrichment is involved in cell cycle cell division, mitotic nuclear division, and DNA replication. HIM10 was strongly enriched for biological processes involved in the immune response, the positive regulation of T cell proliferation, the immune system process, the inflammatory response, and the regulation of interleukin-10 production. Importantly, our experimental data showed that the level of IL-10 expression observed in mice immunized with HBc-tG+pIL-35 was significantly increased compared to that observed in mice immunized with HBc-tG+pVAX (Fig. 4B). These findings suggest that HIM10 may play a key role in regulating host responses. Immune system process enrichment was also observed in HIM2, HIM3, and HIM6. Other HIMs were enriched in processes involved in signal transduction, lipid transport, and protein ubiquitination.
TABLE 2.
Analysis of 12 high-influence modules
| HIM | No. of genes | Representative GO functional enrichmenta |
|---|---|---|
| HIM1 | 248 | Cell cycle, cell division, mitotic nuclear division, DNA replication |
| HIM2 | 72 | Chemotaxis, G-protein-coupled receptor signaling pathway, chemokine-mediated signaling pathway, immune response |
| HIM3 | 67 | Response to virus, defense response to virus, cellular response to interferon beta, innate immune response, immune system process |
| HIM4 | 31 | Adenylate cyclase-activating G-protein-coupled receptor signaling pathway, signal transduction |
| HIM5 | 37 | Lipoprotein metabolic process, lipid transport |
| HIM6 | 56 | Antigen processing and presentation, immune system process, adaptive immune response, positive regulation of T cell-mediated cytotoxicity |
| HIM7 | 22 | Collagen fibril organization, skeletal system development |
| HIM8 | 37 | Protein ubiquitination, intracellular signal transduction |
| HIM9 | 39 | G-protein-coupled receptor signaling pathway |
| HIM10 | 96 | Immune response, positive regulation of T cell proliferation, immune system process, inflammatory response, regulation of interleukin-10 production |
| HIM11 | 34 | Positive regulation of phagocytosis, respiratory burst |
| HIM12 | 33 | Defense response to Gram-positive bacteria |
Representative Gene Ontology (GO) categories are hypergeometrically enriched (FDR-adjusted P value, <0.05). See Table S3 in the supplemental material for all enriched GO categories and their corresponding P values.
Topological analysis of HIM10 and verification of selected genes.
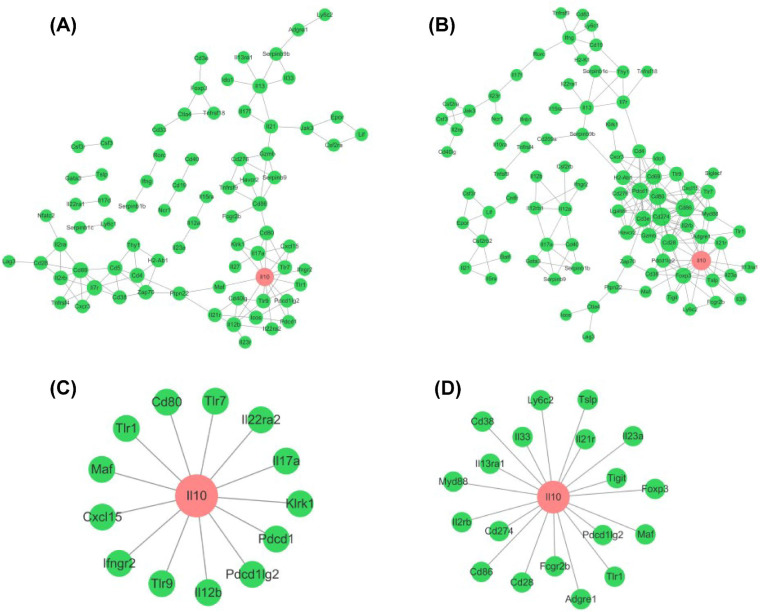
The differential network characteristics of HIM10 were observed between the HBc-tG+pVAX-vaccinated group and the HBc-tG+pIL-35-vaccinated group (Fig. 6A and B). Topological analysis showed that the node with the highest degree in both the HBc-tG+pVAX- and HBc-tG+pIL-35-vaccinated groups (the red nodes in Fig. 6A and B) was for the Il10 gene. The degree of Il10 was 13 in the HBc-tG+pVAX-vaccinated group and 19 in the HBc-tG+pIL-35-vaccinated group. The nodes connected with Il10 in the HBc-tG+pVAX-vaccinated group and the HBc-tG+pIL-35-vaccinated group are represented in Fig. 6C and D, respectively. Among these nodes, only Pdcd1lg2, Maf, and Tlr1 appeared in both vaccinated groups. These data demonstrate that IL-35 altered the relationships between Il10 and its interacting genes, thus regulating the IL-10 signaling pathway and lightening the immunopathology induced by HBc-tG.
FIG 6.
Topological analysis of HIM10. (A and B) The interactions of each gene product from HIM10 in the HBc-tG+pVAX-immunized (A) and HBc-tG+pIL-35-immunized (B) groups. The node with the highest degree is labeled in red. (C and D) The connections between Il10 and other nodes in the HBc-tG+pVAX-immunized (C) and HBc-tG+pIL-35-immunized (D) groups. The STRING database (http://string-db.org/) was used to analyze the interaction network of gene products.
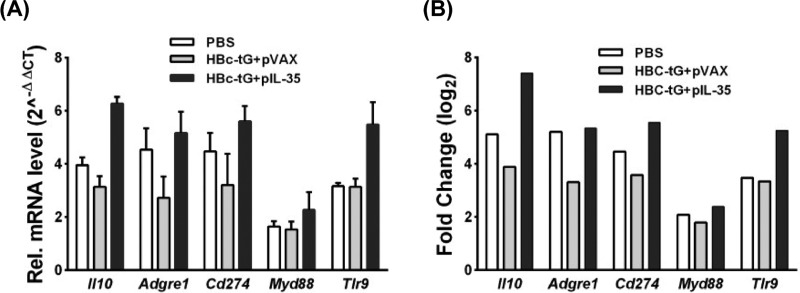
To verify the RNA sequencing data, the several selected genes (Il10, Tlr9, Adgre1, Cd274, and Myd88) were confirmed by qRT-PCR. The mRNA expression levels of these genes (Fig. 7A) were consistent with those determined from the transcriptome sequencing data (Fig. 7B), suggesting that the DEG database obtained from transcriptional sequencing is reliable and may be further studied.
FIG 7.
Quantitative RT-PCR analysis of selected genes. (A) Gene expression levels were measured by qRT-PCR. The gene for glyceraldehyde-3-phosphate dehydrogenase (Gapdh) was used as an internal control, and the relative expression level of each gene was calculated by the comparative 2−ΔΔCT method. All data are presented as the mean values ± SD. (B) Gene expression levels from RNA sequencing data. The x axis shows the validated genes. The y axis is the normalized fold change in expression values for each gene.
DISCUSSION
Currently, there is no licensed RSV vaccine for clinical use (26). The major obstacle in RSV vaccine development is vaccine-enhanced disease. The excessive lung inflammation is caused by an unbalanced immune response, including a predominant Th2-type response and distinct CD4 T cell subsets (27, 28). Therefore, balanced immune responses should be considered for the design of a safe and effective RSV vaccine. In this study, we evaluated the effect of IL-35 on the immune response, antiviral protection, immunopathology, and lung inflammation induced by the RSV subunit vaccine candidate HBc-tG, and we identified some potential regulators associated with VED by a systems biology method.
Our results demonstrated that immunization with both HBc-tG+pVAX and HBc-tG+pIL-35 induced RSV-specific humoral and cellular immune responses and effectively cleared the virus in the lungs of vaccinated mice. However, immunization of mice with HBc-tG+pIL-35 elicited am increased Th1-type response and a decreased Th2-type response and resulted in reduced lung pathological damage upon RSV infection. Importantly, compared with the findings obtained with HBc-tG+pVAX vaccination, HBc-tG+pIL-35 vaccination elicited a significantly increased percentage of CD25+ Foxp3+ Treg cells among CD4+ T cells and significantly increased Treg-related cytokine (IL-10, IL-35, and TGF-β) levels in the lungs of vaccinated mice upon RSV infection. Previous reports showed that Treg cells play a crucial role in controlling the pathology of VED in RSV infection (15, 18, 29). Our results suggest that IL-35 enhances the Treg cell response induced by HBc-tG in mice and elicits balanced immunity, thus attenuating pathological damage upon RSV infection.
In recent years, an emerging systems biology method had been used to study the host response in different cases and to identify key regulators associated with diseases (30–32). This method detects gene expression patterns based on the underlying PPI network without interference with existing knowledge. We explored the molecular mechanism of VED at the gene expression level and identified 2,664 DEGs in the lungs of PBS-treated, HBc-tG+pVAX-immunized, and HBc-tG+pIL-35-immunized mice on day 4 after RSV infection. Twelve HIMs were selected from the PPI network on the basis of these DEGs, and GO functional enrichment was further annotated. Among the 12 HIMs, HIM10 was mainly involved in immune responses, and Il10 was a key regulator in the network of HIM10. IL-10 is a major immune-regulatory cytokine that acts on many immune cells. It has been confirmed that IL-10 plays an important role in preventing RSV-induced disease (18, 33, 34). Differential regulatory networks were observed between mice immunized with HBc-tG+pVAX and mice immunized with HBc-tG+pIL-35. Il10 interacted with the Cd80, Cxcl15, Ifngr2, Il12b, Il17a, Il22ra2, Klrk1, Maf, Pdcd1, Pdcd1lg2, Tlr1, Tlr7, and Tlr9 genes in the HBc-tG+pVAX-vaccinated group, but the genes that interacted with Il10 in the HBc-tG+pIL-35-vaccinated group included Adgre1, Cd28, Cd38, Cd86, Cd274, Fcgr2b, Foxp3, Il2rb, Il13ra1, Il21r, Il23a, Il33, Ly6c2, Maf, Myd88, Pdcd1lg2, Tlr1, Tigit, and Tslp. We also identified that IL-35 upregulated Foxp3 expression. As a transcriptional regulator, Foxp3 plays a crucial role in the development and inhibitory function of Treg cells (35–37), suggesting that IL-35 activated Foxp3 expression and thus enhanced Treg cell inhibition.
Taken together, our results demonstrate that IL-35 can regulate the host immune response and attenuate the RSV vaccine-enhanced inflammation. Thus, these findings of the study provide new insights into the molecular mechanism of vaccine-enhanced immunopathology and will contribute to the development of novel therapeutic and prophylactic strategies for disease caused by RSV infection.
MATERIALS AND METHODS
Cells, virus, and preparation of UV-inactivated virus.
African green monkey kidney cells (Vero cells), human laryngeal epidermoid carcinoma cells (HEp-2 cells), and human embryonic kidney cells (293T cells) were obtained from the China Center for Type Culture Collection (CCTCC; Wuhan, China) and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, New Zealand), 1% penicillin, and 100-U/ml streptomycin. Primary mouse lung fibroblasts (MLFs) were isolated from 8- to 10-week-old mice. Lungs were minced and digested in calcium- and magnesium-free Hanks balanced salt solution (HBSS) buffer supplemented with 10-mg/ml type I collagenase (Sigma-Aldrich) and 20-μg/ml DNase I (Sigma-Aldrich) for 3 h at 37°C with shaking. Cell suspensions were filtered through sterile mesh, and filtered cells were cultured in DMEM containing 10% FBS, 1% penicillin, and 100-U/ml streptomycin. Two days later, adherent fibroblasts were rinsed with HBSS and cultured for subsequent experiments. The respiratory syncytial virus (RSV) A2 strain was obtained from CCTCC, propagated in HEp-2 cells, and quantified in Vero cells. RSV A2 inactivation by UV light was performed as described previously (38). Briefly, 0.5 ml of a purified virus suspension in a 35-mm petri dish was irradiated with UV light for 40 min, and the efficacy of virus inactivation by UV radiation was examined using a plaque assay in Vero cells.
Preparation of protein and plasmids.
Purification by affinity chromatography, renaturation by dialysis, and analysis of HBc-tG were performed as previously described (25).
The single-chain open reading frame of mouse IL-35 cDNA encoding Ebi3 and p35 was optimized and synthesized by Sangon Biotech (Shanghai, China). The Ebi3 and p35 genes were linked by three repeats of GGGGS. The IL-35 fragment was amplified by PCR with specific primers, using the plasmid pUC57-IL-35 as the template (Table 3). The PCR-amplified fragments were then cloned into eukaryotic expression plasmids to generate phage-IL-35 and pVAX-IL-35 (pIL-35), respectively. The recombinant single-chain IL-35 vectors were confirmed by DNA sequencing. Plasmid DNA was purified from Escherichia coli DH5α using an EndoFree plasmid midi kit (Omega Bio-Tek Inc., Norcross, GA, USA) and dissolved in sterile distilled water at 1.0 mg/ml for DNA immunization.
TABLE 3.
Primers used in the experiment
| Name | Sequence (5′–3′) |
|
|---|---|---|
| Forward primer | Reverse primer | |
| phage-IL-35 | GAGCTAGCGCCACCATGGGGTCCAAG | GATCTAGATCAGGCGGAGCTGAGATAGCC |
| pVAX-IL-35 | GAGCTAGCGCCACCATGGGGTCCAAG | GAGGTACCTCAGGCGGAGCTGAGATAGCC |
| Ebi3 | GAAACCCCATGCCAAGTATTG | CAGTATTCAGACTCAGGACAGG |
| p35 | AGACATCACACGGGACCAAAC | CAAGGCACAGGGTCATCATCA |
| RSV N | AGATCAACTTCTGTCATCCAGCAA | TTCTGCACATCATAATTAGGAGTATCAAT |
| Gapdh | CATGGCCTTCCGTGTTCCTA | ATGCCTGCTTCACCACCTTCT |
| Adgre1 | CGTGTTGTTGGTGGCACTGTGA | CCACATCAGTGTTCCAGGAGAC |
| Tlr9 | TGAAAGCATCACCCACACCAA | TGTTGAGCAAGCGGAAGAAGA |
| Myd88 | CAAAGGAACTGGGAGGCATCA | CGGTCGGACACACACAACTTA |
| Cd274 | TGCGGACTACAAGCGAATCACG | CTCAGCTTCTGGATAACCCTCG |
| Il10 | TTTGAATTCCCTGGGTGAGAA | GCTCCACTGCCTTGCTCTTATT |
Transfection and lentivirus-mediated gene transfer.
DNA transfection was performed according to the manufacturer’s procedure. Briefly, 5 μl of the Lipofectamine 3000 reagent (Invitrogen) and 2.5 μg DNA were diluted into 100 μl of Opti-MEM medium. The diluted DNA was added to each tube with diluted Lipofectamine 3000 reagent, followed by incubation at room temperature for 15 min. 293T cell monolayers, grown on 6-well cell culture plates, were washed twice with Opti-MEM medium and incubated with the DNA transfection mixture for subsequent assays.
293T cells were transfected with phage-IL-35 or the empty vector phage along with the packaging vectors pSPAX2 and pMD2G. The medium was exchanged for fresh medium (10% FBS, 1% streptomycin-penicillin) after 8 h. Forty hours later, the supernatants were harvested to infect MLFs to construct MLFs stably expressing IL-35 (MLF/phage-IL-35) or the negative control (MLF/phage).
Western blotting.
The expressed protein samples were subjected to 12% SDS-PAGE and subsequently characterized by Western blotting as described previously (38). Briefly, the expressed protein samples were separated on 12% polyacrylamide gels, which were transferred onto a polyvinylidene difluoride membrane for Western blot analysis using a mouse anti-EBI3 monoclonal antibody (Rockland Inc., Limerick, PA, USA) and a mouse anti-p35 monoclonal antibody (R&D Systems Inc., Minneapolis, MN, USA).
Immunization and challenge.
All animal studies were approved by the Institutional Animal Care and Use Committee of Wuhan University.
Six- to 8-week-old specific-pathogen-free (SPF) female BALB/c mice (Wuhan University Center for Animal Experiments) were randomly divided into groups with 18 mice in each group. For the HBc-tG+pVAX- and HBc-tG+pIL-35-vaccinated groups, the mice were immunized intraperitoneally (i.p.) with 10 μg HBc-tG in 100 μl at day 2 after intramuscular (i.m.) injection of 100 μg pVAX or pIL-35 adjuvant with 100 μl 25% sucrose. For the UV-RSV-immunized group, mice were immunized i.p. with 1 × 105 PFU of UV-RSV in 100 μl. For the PBS-treated group, mice were immunized i.p. with 100 μl PBS. All mice received a booster administration with the same dose at 2-week intervals. Blood samples were collected by tail vein puncture during preimmunization and at 2 weeks after the final immunization for antibody detection, and splenocytes were isolated for cytokine detection.
Mice were infected intranasally (i.n.) with 3 × 106 PFU of RSV in 100 μl at 2 weeks after the final immunization and sacrificed at day 4 postchallenge for assessment of pulmonary pathology. The lung inflammation severity scores were defined as previously described (25, 38).
ELISA.
RSV-specific antibodies (IgG, IgG1, and IgG2a) in the sera of mice were determined by enzyme-linked immunosorbent assay (ELISA) using purified RSV as the coating antigen (38, 39). Briefly, 96-well microtiter plates (Corning, Corning, NY, USA) were coated with 100 μl of inactivated RSV (1 × 105 PFU/well) in coating buffer at 4°C overnight. Serial dilutions of mouse sera in PBS-Tween 20 containing 1% bovine serum albumin were added to the wells, and then the plates were incubated for 1 h at 37°C. Horseradish peroxidase-conjugated goat anti-mouse IgG (Thermo Fisher Scientific, Inc., Waltham, MA, USA) or IgG1 or IgG2a (Friendbio Science & Technology Co., Ltd., Wuhan, Hubei, China) was added to the plates, and the plates were incubated at 37°C for 1 h. The 3,3′,5,5′-tetramethylbenzidine (TMB; Sigma) substrate in a 100-μl volume was added to each well, and the plates were incubated for 15 to 20 min at room temperature. The reaction was stopped with 100 μl of 2 M H2SO4, and the optical density at 450 nm was measured using an ELISA reader (Multidkan MK3; Thermo Fisher Scientific).
The cytokines in the splenocytes or lung homogenates were quantitatively measured by ELISA as previously described (38, 40). Th1 (IFN-γ, IL-2), Th2 (IL-4, IL-10), and Treg cell-related (IL-10, TGF-β) cytokines in the culture supernatants were quantitatively determined using commercially available ELISA kits (BioLegend, San Diego, CA).
Neutralization assay.
RSV-specific serum neutralizing antibody titers were determined by a plaque reduction assay as described previously (41, 42). Mouse sera were inactivated in 56°C for 30 min to inactivate the complement and then 2-fold serially diluted in DMEM. Purified RSV was diluted to approximately 100 PFU in 100 μl and added to the diluted sera in 100-μl aliquots. The virus-serum mixture or a virus-DMEM control was incubated at 37°C for 1 h. Then, the mixture was added to prewashed confluent monolayers of Vero cells in 24-well plates. After 2 h of incubation, the mixture was removed and 1 ml of methylcellulose overlay (1 volume of 2× DMEM containing 4% FBS, 2% penicillin-streptomycin, and 1 volume of 2% methylcellulose) was added to each well. The plates were incubated for 3 to 4 days at 37°C, and the plaques were stained. The neutralization titer was defined as the log2 of the reciprocal of the highest dilution of serum that reduced the virus titer by 50% (38).
Quantitative real-time PCR (qRT-PCR).
Total RNA was extracted from lung tissues using the RNA Pure reagent (Aidlab Biotechnologies Co., Ltd., Beijing, China) and reverse transcribed into cDNA using Moloney murine leukemia virus reverse transcriptase according to the manufacturer’s recommendations. Real-time PCR was performed using 2× SYBR green master mix (Biotool) in a 7500 real-time system (Applied Biosystems, USA). The primer sequences are listed in Table 3.
Flow cytometry assay.
Cytokine staining was performed according to the manufacturer’s procedure. Lung cells were stained with surface antibodies labeled with CD4-peridinin chlorophyll protein and CD25-phycoerythrin (BioLegend, San Diego, CA), and after fixation and permeabilization, the cells were stained with the intracellular antibody Foxp3-fluorescein isothiocyanate (BioLegend, San Diego, CA). After washing and filtering, the stained cells were analyzed by flow cytometry (Beckman, USA). The data were analyzed with Summit software (Beckman, USA), and the results are presented as the percentage of CD25+ Foxp3+-producing cells among CD4+ T cells.
cDNA library preparation.
RNA purification, reverse transcription, library construction, and sequencing were performed at WuXi Next Code in Shanghai, China, according to the manufacturer’s instructions. The mRNA-focused sequencing libraries from total RNA were prepared using the WuXi in-house protocol. Poly(A) mRNA was purified from total RNA using magnetic beads to which oligo(dT) was attached and then fragmented by the use of fragmentation buffer. Taking these short fragments as templates, the first-strand cDNA was synthesized using reverse transcriptase and random primers, followed by second-strand cDNA synthesis. The synthesized cDNA was subjected to end repair, phosphorylation, and A base addition according to the library construction protocol. Then, sequencing adapters were added to both ends of the cDNA fragments. After PCR amplification of the cDNA fragments, the targets of 250 to 350 bp were cleaned up. After library construction, a double-stranded DNA HiSeq assay with a Qubit (version 2.0) fluorometer (Thermo Fisher Scientific) was used to quantify the concentration of the resulting sequencing libraries, while the size distribution was analyzed using an Agilent 2100 Bioanalyzer system (Agilent). After library validation, an Illumina cBot cluster generation system with HiSeq PE cluster kits (Illumina) was used to generate clusters. Paired-end sequencing was performed using an Illumina HiSeq system following Illumina-provided protocols for 2 × 150 paired-end sequencing at WuXi Next Code in Shanghai, China.
Identification of differentially expressed genes (DEGs).
The transcriptional level of each expressed gene was calculated and normalized to the number of fragments per kilobase of exon per million fragments mapped (FPKM). DESeq software was applied to analyze differential gene expression in biological samples, which was used to calculate and to compare the differences in the gene expression profiles between the PBS-treated, HBc-tG+pVAX-immunized, or HBc-tG+pIL-35-immunized groups and the naive group. In this study, we had three biological repeats for each group, and the correlation of the number of counts detected between parallel libraries was assessed statistically by calculating the Pearson correlation. The P value threshold in multiple tests and analyses was determined by the false discovery rate (FDR). Genes with an FDR of <0.05 and a |log2 fold change in expression value| of ≥1 were considered DEGs.
Construction of protein-protein interaction (PPI) network.
The Search Tool for the Retrieval of InteractiNg Genes (STRING) database (43) and Biological General Repository for Interaction Data Sets (BioGRID) (44) were used to build the PPI networks for the identified DEGs to predict their interactions. A combination score of >0.5 was used as the threshold. The topological properties of the PPI network were visualized and analyzed with Cytoscape software (45). We used the ClusterONE algorithm to further identify the high-influence modules (HIMs) in the network, as previously described (30, 46).
Gene Ontology (GO) enrichment analysis of HIMs.
The Database for Annotation, Visualization, and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/) provides a comprehensive set of functional annotation tools for biological interpretation of large gene lists (47). We used DAVID to group the functions of the DEGs and identified enriched biological processes associated with the DEGs in the HIMs from the GO database.
Statistics.
Statistical analyses of the data were performed using Student’s t test or one-way analysis of variance (ANOVA). A P value of less than 0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Key R&D Program of China (grant 2017YFA0505801), the National Natural Science Foundation of China (grant 11831015), and the Natural Science Foundation of Hubei Province Innovation Group (grant 2017CFA022).
Jie Yang established the methodology and performed the investigation, data analysis, and writing; Chen Ma established the methodology and performed data analysis; Yu Zhao established the methodology; Anjing Fan performed data analysis; Xiufen Zou supervised the study and reviewed the findings; Zishu Pan performed conceptualization and supervision and reviewed the findings.
We declare no conflicts of interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Shi T, McAllister DA, O'Brien KL, Simoes EAF, Madhi SA, Gessner BD, Polack FP, Balsells E, Acacio S, Aguayo C, Alassani I, Ali A, Antonio M, Awasthi S, Awori JO, Azziz-Baumgartner E, Baggett HC, Baillie VL, Balmaseda A, Barahona A, Basnet S, Bassat Q, Basualdo W, Bigogo G, Bont L, Breiman RF, Brooks WA, Broor S, Bruce N, Bruden D, Buchy P, Campbell S, Carosone-Link P, Chadha M, Chipeta J, Chou M, Clara W, Cohen C, de Cuellar E, Dang D-A, Dash-Yandag B, Deloria-Knoll M, Dherani M, Eap T, Ebruke BE, Echavarria M, de Freitas Lázaro Emediato CC, Fasce RA, Feikin DR, Feng L, et al. 2017. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 390:946–958. doi: 10.1016/S0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall CB, Weinberg GA, Blumkin AK, Edwards KM, Staat MA, Schultz AF, Poehling KA, Szilagyi PG, Griffin MR, Williams JV, Zhu Y, Grijalva CG, Prill MM, Iwane MK. 2013. Respiratory syncytial virus-associated hospitalizations among children less than 24 months of age. Pediatrics 132:e341–e348. doi: 10.1542/peds.2013-0303. [DOI] [PubMed] [Google Scholar]
- 3.Resch B. 2012. Burden of respiratory syncytial virus infection in young children. World J Clin Pediatr 1:8–12. doi: 10.5409/wjcp.v1.i3.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bont L, Checchia PA, Fauroux B, Figueras-Aloy J, Manzoni P, Paes B, Simões EAF, Carbonell-Estrany X. 2016. Defining the epidemiology and burden of severe respiratory syncytial virus infection among infants and children in Western countries. Infect Dis Ther 5:271–298. doi: 10.1007/s40121-016-0123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higgins D, Trujillo C, Keech C. 2016. Advances in RSV vaccine research and development—a global agenda. Vaccine 34:2870–2875. doi: 10.1016/j.vaccine.2016.03.109. [DOI] [PubMed] [Google Scholar]
- 6.Pangesti KNA, Abd El Ghany M, Walsh MG, Kesson AM, Hill-Cawthorne GA. 2018. Molecular epidemiology of respiratory syncytial virus. Rev Med Virol 28:e1968. doi: 10.1002/rmv.1968. [DOI] [PubMed] [Google Scholar]
- 7.Kapikian AZ, Mitchell RH, Chanock RM, Shvedoff RA, Stewart CE. 1969. An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am J Epidemiol 89:405–421. doi: 10.1093/oxfordjournals.aje.a120954. [DOI] [PubMed] [Google Scholar]
- 8.Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol 89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 9.Fulginiti VA, Eller JJ, Sieber OF, Joyner JW, Minamitani M, Meiklejohn G. 1969. Respiratory virus immunization. I. A field trial of two inactivated respiratory virus vaccines; an aqueous trivalent parainfluenza virus vaccine and an alum-precipitated respiratory syncytial virus vaccine. Am J Epidemiol 89:435–448. doi: 10.1093/oxfordjournals.aje.a120956. [DOI] [PubMed] [Google Scholar]
- 10.Waris ME, Tsou C, Erdman DD, Zaki SR, Anderson LJ. 1996. Respiratory syncytial virus infection in BALB/c mice previously immunized with formalin-inactivated virus induces enhanced pulmonary inflammatory response with a predominant Th2-like cytokine pattern. J Virol 70:2852–2860. doi: 10.1128/JVI.70.5.2852-2860.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becker Y. 2006. Respiratory syncytial virus (RSV) evades the human adaptive immune system by skewing the Th1/Th2 cytokine balance toward increased levels of Th2 cytokines and IgE, markers of allergy—a review. Virus Genes 33:235–252. doi: 10.1007/s11262-006-0064-x. [DOI] [PubMed] [Google Scholar]
- 12.Connors M, Kulkarni AB, Firestone CY, Holmes KL, Morse HC III, Sotnikov AV, Murphy BR. 1992. Pulmonary histopathology induced by respiratory syncytial virus (RSV) challenge of formalin-inactivated RSV-immunized BALB/c mice is abrogated by depletion of CD4+ T cells. J Virol 66:7444–7451. doi: 10.1128/JVI.66.12.7444-7451.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connors M, Giese NA, Kulkarni AB, Firestone CY, Morse HC III, Murphy BR. 1994. Enhanced pulmonary histopathology induced by respiratory syncytial virus (RSV) challenge of formalin-inactivated RSV-immunized BALB/c mice is abrogated by depletion of interleukin-4 (IL-4) and IL-10. J Virol 68:5321–5325. doi: 10.1128/JVI.68.8.5321-5325.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson TR, Parker RA, Johnson JE, Graham BS. 2003. IL-13 is sufficient for respiratory syncytial virus G glycoprotein-induced eosinophilia after respiratory syncytial virus challenge. J Immunol 170:2037–2045. doi: 10.4049/jimmunol.170.4.2037. [DOI] [PubMed] [Google Scholar]
- 15.Durant LR, Makris S, Voorburg CM, Loebbermann J, Johansson C, Openshaw PJ. 2013. Regulatory T cells prevent Th2 immune responses and pulmonary eosinophilia during respiratory syncytial virus infection in mice. J Virol 87:10946–10954. doi: 10.1128/JVI.01295-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Annacker O, Asseman C, Read S, Powrie F. 2003. Interleukin-10 in the regulation of T cell-induced colitis. J Autoimmun 20:277–279. doi: 10.1016/S0896-8411(03)00045-3. [DOI] [PubMed] [Google Scholar]
- 17.Beiting DP, Gagliardo LF, Hesse M, Bliss SK, Meskill D, Appleton JA. 2007. Coordinated control of immunity to muscle stage Trichinella spiralis by IL-10, regulatory T cells, and TGF-beta. J Immunol 178:1039–1047. doi: 10.4049/jimmunol.178.2.1039. [DOI] [PubMed] [Google Scholar]
- 18.Li C, Zhou X, Zhong Y, Li C, Dong A, He Z, Zhang S, Wang B. 2016. A recombinant G protein plus cyclosporine A-based respiratory syncytial virus vaccine elicits humoral and regulatory T cell responses against infection without vaccine-enhanced disease. J Immunol 196:1721–1731. doi: 10.4049/jimmunol.1502103. [DOI] [PubMed] [Google Scholar]
- 19.Loebbermann J, Durant L, Thornton H, Johansson C, Openshaw PJ. 2013. Defective immunoregulation in RSV vaccine-augmented viral lung disease restored by selective chemoattraction of regulatory T cells. Proc Natl Acad Sci U S A 110:2987–2992. doi: 10.1073/pnas.1217580110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collison LW, Workman CJ, Kuo TT, Boyd K, Wang Y, Vignali KM, Cross R, Sehy D, Blumberg RS, Vignali DA. 2007. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature 450:566–569. doi: 10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- 21.Collison LW, Vignali DA. 2008. Interleukin-35: odd one out or part of the family? Immunol Rev 226:248–262. doi: 10.1111/j.1600-065X.2008.00704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niedbala W, Wei XQ, Cai B, Hueber AJ, Leung BP, McInnes IB, Liew FY. 2007. IL-35 is a novel cytokine with therapeutic effects against collagen-induced arthritis through the expansion of regulatory T cells and suppression of Th17 cells. Eur J Immunol 37:3021–3029. doi: 10.1002/eji.200737810. [DOI] [PubMed] [Google Scholar]
- 23.Wang RX, Yu CR, Mahdi RM, Egwuagu CE. 2012. Novel IL27p28/IL12p40 cytokine suppressed experimental autoimmune uveitis by inhibiting autoreactive Th1/Th17 cells and promoting expansion of regulatory T cells. J Biol Chem 287:36012–36021. doi: 10.1074/jbc.M112.390625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Lin Y, Li C, Zhang X, Cheng L, Dai L, Wang Y, Wang F, Shi G, Li Y, Yang Q, Cui X, Liu Y, Wang H, Zhang S, Yang Y, Xiang R, Li J, Yu D, Wei Y, Deng H. 2016. IL-35 decelerates the inflammatory process by regulating inflammatory cytokine secretion and M1/M2 macrophage ratio in psoriasis. J Immunol 197:2131–2144. doi: 10.4049/jimmunol.1600446. [DOI] [PubMed] [Google Scholar]
- 25.Qiao L, Zhang Y, Chai F, Tan Y, Huo C, Pan Z. 2016. Chimeric virus-like particles containing a conserved region of the G protein in combination with a single peptide of the M2 protein confer protection against respiratory syncytial virus infection. Antiviral Res 131:131–140. doi: 10.1016/j.antiviral.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Boyoglu-Barnum S, Chirkova T, Anderson LJ. 2019. Biology of infection and disease pathogenesis to guide RSV vaccine development. Front Immunol 10:1675. doi: 10.3389/fimmu.2019.01675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roman M, Calhoun WJ, Hinton KL, Avendano LF, Simon V, Escobar AM, Gaggero A, Diaz PV. 1997. Respiratory syncytial virus infection in infants is associated with predominant Th-2-like response. Am J Respir Crit Care Med 156:190–195. doi: 10.1164/ajrccm.156.1.9611050. [DOI] [PubMed] [Google Scholar]
- 28.Knudson CJ, Hartwig SM, Meyerholz DK, Varga SM. 2015. RSV vaccine-enhanced disease is orchestrated by the combined actions of distinct CD4 T cell subsets. PLoS Pathog 11:e1004757. doi: 10.1371/journal.ppat.1004757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruckwardt TJ, Bonaparte KL, Nason MC, Graham BS. 2009. Regulatory T cells promote early influx of CD8+ T cells in the lungs of respiratory syncytial virus-infected mice and diminish immunodominance disparities. J Virol 83:3019–3028. doi: 10.1128/JVI.00036-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Jin S, Lei L, Pan Z, Zou X. 2015. Deciphering deterioration mechanisms of complex diseases based on the construction of dynamic networks and systems analysis. Sci Rep 5:9283. doi: 10.1038/srep09283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Cheng X, Liu H, Zheng C, Rao K, Fang Y, Zhou H, Xiong S. 2014. Identification of key genes and crucial modules associated with coronary artery disease by bioinformatics analysis. Int J Mol Med 34:863–869. doi: 10.3892/ijmm.2014.1817. [DOI] [PubMed] [Google Scholar]
- 32.Li C, Bankhead A III, Eisfeld AJ, Hatta Y, Jeng S, Chang JH, Aicher LD, Proll S, Ellis AL, Law GL, Waters KM, Neumann G, Katze MG, McWeeney S, Kawaoka Y. 2011. Host regulatory network response to infection with highly pathogenic H5N1 avian influenza virus. J Virol 85:10955–10967. doi: 10.1128/JVI.05792-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christiaansen AF, Knudson CJ, Weiss KA, Varga SM. 2014. The CD4 T cell response to respiratory syncytial virus infection. Immunol Res 59:109–117. doi: 10.1007/s12026-014-8540-1. [DOI] [PubMed] [Google Scholar]
- 34.Lotz MT, Peebles RS Jr. 2012. Mechanisms of respiratory syncytial virus modulation of airway immune responses. Curr Allergy Asthma Rep 12:380–387. doi: 10.1007/s11882-012-0278-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu L, Barbi J, Pan F. 2017. The regulation of immune tolerance by FOXP3. Nat Rev Immunol 17:703–717. doi: 10.1038/nri.2017.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Devaud C, Darcy PK, Kershaw MH. 2014. Foxp3 expression in T regulatory cells and other cell lineages. Cancer Immunol Immunother 63:869–876. doi: 10.1007/s00262-014-1581-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rudensky AY. 2011. Regulatory T cells and Foxp3. Immunol Rev 241:260–268. doi: 10.1111/j.1600-065X.2011.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Qiao L, Hu X, Zhao K, Zhang Y, Chai F, Pan Z. 2016. Baculovirus vectors expressing F proteins in combination with virus-induced signaling adaptor (VISA) molecules confer protection against respiratory syncytial virus infection. Vaccine 34:252–260. doi: 10.1016/j.vaccine.2015.11.027. [DOI] [PubMed] [Google Scholar]
- 39.Quan FS, Kim Y, Lee S, Yi H, Kang SM, Bozja J, Moore ML, Compans RW. 2011. Viruslike particle vaccine induces protection against respiratory syncytial virus infection in mice. J Infect Dis 204:987–995. doi: 10.1093/infdis/jir474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao HY, Yu SL, Sia C, Chen Y, Chitra E, Chen IH, Venkatesan N, Leng CH, Chong P, Chow YH. 2009. Immunogenic properties of RSV-B1 fusion (F) protein gene-encoding recombinant adenoviruses. Vaccine 27:5460–5471. doi: 10.1016/j.vaccine.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 41.McGinnes LW, Gravel KA, Finberg RW, Kurt-Jones EA, Massare MJ, Smith G, Schmidt MR, Morrison TG. 2011. Assembly and immunological properties of Newcastle disease virus-like particles containing the respiratory syncytial virus F and G proteins. J Virol 85:366–377. doi: 10.1128/JVI.01861-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murawski MR, McGinnes LW, Finberg RW, Kurt-Jones EA, Massare MJ, Smith G, Heaton PM, Fraire AE, Morrison TG. 2010. Newcastle disease virus-like particles containing respiratory syncytial virus G protein induced protection in BALB/c mice, with no evidence of immunopathology. J Virol 84:1110–1123. doi: 10.1128/JVI.01709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. 2009. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oughtred R, Stark C, Breitkreutz B-J, Rust J, Boucher L, Chang C, Kolas N, O'Donnell L, Leung G, McAdam R, Zhang F, Dolma S, Willems A, Coulombe-Huntington J, Chatr-Aryamontri A, Dolinski K, Tyers M. 2019. The BioGRID interaction database: 2019 update. Nucleic Acids Res 47:D529–D541. doi: 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang W, Xu J, Li Y, Zou X. 2019. Integrating network topology, gene expression data and GO annotation information for protein complex prediction. J Bioinform Comput Biol 17:1950001. doi: 10.1142/S021972001950001X. [DOI] [PubMed] [Google Scholar]
- 47.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.