Over 130 modifications have been identified in cellular RNAs, which play critical roles in many cellular processes, from modulating RNA stability to altering translation efficiency. One such modification, 5-methylcytosine, is relatively abundant in mammalian mRNAs, but its precise location and function are not well understood. In this study, we identified unexpectedly high levels of m5C in the murine leukemia virus RNA, precisely mapped its location, and showed that ALYREF, a reader protein that specifically recognizes m5C, regulates viral production. Together, our findings provide a high-resolution atlas of m5C in murine leukemia virus and reveal a functional role of m5C in viral replication.
KEYWORDS: 5-methylcytosine, ALYREF, NSUN2, RNA modification, epitranscriptome, murine leukemia virus, retrovirus
ABSTRACT
RNA modifications play diverse roles in regulating RNA function, and viruses co-opt these pathways for their own benefit. While recent studies have highlighted the importance of N6-methyladenosine (m6A)—the most abundant mRNA modification—in regulating retrovirus replication, the identification and function of other RNA modifications in viral biology have been largely unexplored. Here, we characterized the RNA modifications present in a model retrovirus, murine leukemia virus (MLV), using mass spectrometry and sequencing. We found that 5-methylcytosine (m5C) is highly enriched in viral genomic RNA relative to uninfected cellular mRNAs, and we mapped at single-nucleotide resolution the m5C sites, which are located in multiple clusters throughout the MLV genome. Further, we showed that the m5C reader protein ALYREF plays an important role in regulating MLV replication. Together, our results provide a complete m5C profile in a virus and its function in a eukaryotic mRNA.
IMPORTANCE Over 130 modifications have been identified in cellular RNAs, which play critical roles in many cellular processes, from modulating RNA stability to altering translation efficiency. One such modification, 5-methylcytosine, is relatively abundant in mammalian mRNAs, but its precise location and function are not well understood. In this study, we identified unexpectedly high levels of m5C in the murine leukemia virus RNA, precisely mapped its location, and showed that ALYREF, a reader protein that specifically recognizes m5C, regulates viral production. Together, our findings provide a high-resolution atlas of m5C in murine leukemia virus and reveal a functional role of m5C in viral replication.
INTRODUCTION
Among the over 130 modifications identified in cellular RNAs, >40 are present in mammals (1). The mammalian mRNA transcriptome contains at least 4 major internal modifications, N6-methyladenosine (m6A), pseudouridine (Ψ), N1-methyladenosine (m1A), and 5-methylcytosine (m5C) (2). Of these, m6A is the most abundant and has been the most extensively studied; m6A affects almost all processes in mRNA metabolism, including splicing, stability, localization, and translation. However, the other modifications, including m5C, are much less extensively studied, in part due to the inadequacy of their mapping technologies (3, 4).
Entirely dependent on the host cell machinery for their replication, viruses are known to co-opt RNA modification pathways. Indeed, m6A was first detected in influenza virus and Rous sarcoma virus over 40 years ago (5, 6). However, m6A function and topology in viruses were not determined until recently, when multiple studies determined the m6A landscape in HIV RNA and showed that perturbation of m6A biology can significantly influence retrovirus replication (7–9). Furthermore, an array of other viruses, including hepatitis C virus, Zika virus, and dengue virus, also contain m6A modifications in their RNA genomes that play a functional role (10, 11). Despite a proliferation of studies investigating the role of m6A in the viral life cycle, the identification and function of other RNA modifications have been largely unexplored. m5C modifications were recently discovered in retroviruses, and depletion of the m5C methyltransferase NSUN2 decreased viral replication (12, 13). However, a single-nucleotide profile of m5C is still lacking for any virus. Moreover, a mechanistic understanding of m5C’s role in viral RNA or the proteins viral m5C interacts with has not previously been achieved.
Here, we applied mass spectrometry and sequencing to evaluate the RNA modifications present in a model retrovirus, the murine leukemia virus (MLV). We found that m5C modifications are highly enriched in MLV RNA and mapped their locations at single-nucleotide resolution. We also found that the m5C reader protein ALYREF, which plays a crucial role in nuclear-cytoplasmic export of m5C-modified mRNA transcripts (14), regulates MLV replication. Our results provide a full viral m5C profile and a mechanistic understanding of how m5C modification exerts its function for viral biology.
RESULTS AND DISCUSSION
m5C is highly enriched in MLV genomic RNA.
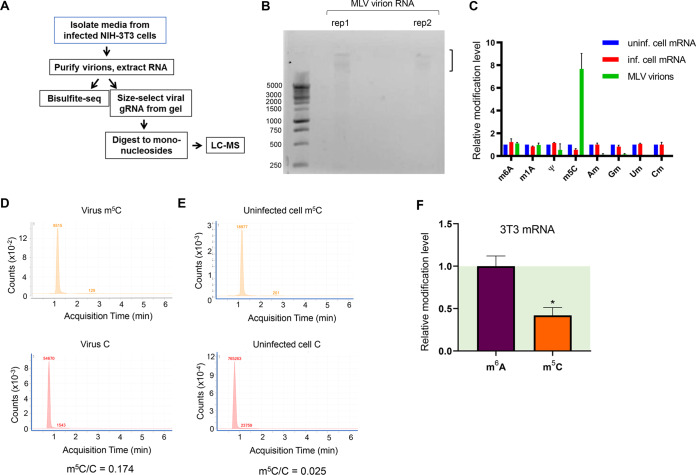
To evaluate which RNA modifications are present in retroviral RNAs, we first purified MLV particles from infected mouse fibroblasts (NIH 3T3 cells) and extracted virion RNA. Because retroviruses selectively incorporate many host cell RNAs, such as tRNAs, snRNAs, and 7SL RNA, that are known to harbor extensive modifications (15, 16), we size-selected MLV genomic RNA (gRNA) on an agarose gel to enable more accurate assessment of viral RNA modification levels (Fig. 1A and B).
FIG 1.
m5C is enriched in MLV genomic RNA. (A) Overview of experimental approach to identify RNA modifications in murine leukemia virus (MLV) by mass spectrometry and bisulfite sequencing. (B) Viral genomic RNA isolated from agarose gels for mass spectrometry analysis of modification contents. (C) QQQ liquid chromatography-mass spectrometry (LC-MS) results for MLV genomic RNA and infected mouse fibroblasts (MLV-3T3 cells) were compared to those for uninfected 3T3 cells. Values for uninfected cells were set to 1. m6A, N6-methyladenosine; m1A, N1-methyladenosine; m5C, 5-methylcytosine; Am, 2′-O-methyladenosine; Gm, 2′-O-methylguanosine; Um, 2′-O-methyluridine; Cm, 2′-O-methylcytodine. (D) LC-MS spectrum counts for m5C and C in the genomic RNA isolated from virions. The y axis shows the signal intensity, and the numerical values show the peak area. (E) LC-MS spectrum counts for m5C and C in the poly(A)-selected, RiboMinus-treated cellular mRNA. The y axis shows the signal intensity, and the numerical values show the peak area. (F) Relative m6A and m5C modification in the cellular transcriptome. After transfection of mouse fibroblasts with a plasmid encoding wild-type MLV (pNCA), RNA was extracted after 48 h and cellular mRNAs were purified. RNA modification levels were evaluated by LC-MS. Values for uninfected cells were set to 1. n = 3 biological replicates; values are means plus standard errors of the means (SEM). *, P ≤ 0.05.
We measured the total modified nucleotide content of the size-selected MLV RNAs by triple-quadrupole (QQQ) liquid chromatography-mass spectrometry (LC-MS). We found that MLV RNA had unexpectedly high enrichment of m5C compared to the mRNA of uninfected host cells (Fig. 1C to E). In contrast, we found m6A levels in MLV gRNA comparable to that in the uninfected host mRNA and only low levels of pseudouridines and 2′-O-methyl residues in MLV gRNA. After transfecting a plasmid encoding wild-type MLV, we found little change in the global cellular m6A level but a 2-fold decrease in the m5C level within host mRNAs (Fig. 1F), indicating that MLV infection also influences m5C modification in the cellular transcriptome. Our results are qualitatively consistent with a recent report on the MLV virus but have significant quantitative differences (12). These discrepancies may be due to the viral strains or cellular conditions used in these studies or to the potential contamination by other packaged nonviral RNAs in the virion.
Profiling the MLV m5C landscape.
We next determined the locations of the m5C modifications in MLV RNA. Currently, RNA bisulfite sequencing (RNA-BisSeq) is the only m5C mapping method at single-base resolution for RNA by sequencing (4). RNA-BisSeq converts all unmethylated cytosines to uridine, but the 5-methylated cytosines are protected from conversion. When this method is applied to the mammalian mRNA transcriptome, however, the signal-to-noise ratio is drastically reduced, in part due to the low coverage obtained for each mRNA position. We reasoned that m5C mapping would be more precise using a retroviral model system where high coverage can be obtained.
We performed RNA-BisSeq for both the viral RNA from purified cell-free virions and the mRNA transcriptome from host cells that were infected with MLV. As a positive control for our procedure, we found that the two known m5C sites in the 28S rRNA from the cellular samples were modified at 88 to 98% levels, whereas the surrounding unmodified C residues were quantitatively converted to uridine and exhibited low levels of BisSeq noise (Fig. 2A). In both MLV RNA from purified virions and MLV RNA transcripts in total cellular mRNA, we found multiple high-confidence m5C sites, defined as the sites with >20% C reads remaining after bisulfite treatment in both biological replicates (Fig. 2B to E). We validated the mapping of the m5C sites using reverse transcription-PCR (RT-PCR) followed by Sanger sequencing of the bisulfite-treated samples (Fig. 2F). We found six of the eight high-confidence MLV gRNA m5C sites clustered within two regions: one in the untranslated region upstream of gag and the other within the env gene (Fig. 2D and E). We found another site located near the gag-pol boundary.
FIG 2.
m5C sites are mapped in MLV RNA at single-nucleotide resolution. (A) Mouse 28S rRNA m5C site identification by bisulfite sequencing. IGV tracks display reads aligned to the 3′ end of the mouse 28S rRNA sequence with two known m5C sites. The m5C modification percentage at each site is indicated. (B) IGV alignments of RNA-BisSeq data from two independent replicates of MLV virion RNA showing the clustered sites in the upstream gag region and the env region. Reads were aligned to the bisulfite-converted MLV genome sequence in which all C residues were changed to T. Sites with high levels of unconverted C (in blue) are indicative of m5C sites, with the modification fraction shown on the right. The y axis indicates read depth. (C) IGV alignments of RNA-BisSeq data from two independent replicates of cellular mRNA showing the clustered sites in the upstream gag region and the env region of MLV gRNA transcripts. (D) Distribution and stoichiometry of m5C sites on the MLV genome from virion samples. The y axis shows the m5C fractions. The x axis indicates the genomic location of the m5C sites, with a diagram of the viral genome shown at the top. (E) Distribution and stoichiometry of m5C sites on the MLV genome from cellular mRNA samples. (F) Validation of the three high-confidence m5C sites located upstream of gag carried out by RT-PCR followed by Sanger sequencing of bisulfite-treated samples obtained from virions or infected cells. Blue arrows show the locations of the m5C sites.
Five of the eight high-confidence sites showed strong conservation among common MLV isolates (Fig. 3) and were often present in related strains, such as Friend leukemia virus. Of the two m5C sites in the env region, 100% of examined isolates shared one site and >75% harbored the other site (Fig. 3C). Given the high mutational rate of these viral species, this level of conservation supports a potential functional relevance for these sites.
FIG 3.
Conservation of m5C sites in the retroviral genomic RNA. (A) Conservation of upstream gag region m5C sites among 17 MLV isolates. Viral genomic sequences were aligned via ClustalW on the Jalview multiple-alignment editor. The locations of candidate m5C sites from BisSeq data are indicated by arrows. (B) Conservation of the env region m5C sites among MLV isolates. (C) Neighbor-joining tree of MLV isolates constructed using Jalview.
In addition to these high-confidence sites, we identified multiple additional m5C candidate sites that harbor lower stoichiometry (Fig. 2D and E). Adding together the m5C modification fraction at all detected sites, we obtained a total of 3.5 to 4 m5C in MLV RNA. In comparison, cellular mRNAs were estimated to contain <0.5 m5C per mRNA; thus, MLV exhibits a striking enrichment of m5C in its RNA, as already indicated by our mass spectrometry results.
Consistent with our findings of m5C enrichment, Courtney et al. (12, 13) recently applied an antibody immunoprecipitation-based approach to identify m5C peaks in retroviral genomes. While our results provide a more conservative estimate (8 high-confidence sites), their data suggest as many as 40 m5C sites in MLV. However, since antibody immunoprecipitation-based m5C detection methods cannot determine m5C at single-nucleotide resolution or its stoichiometry, it is unclear how many of those sites have a >20% m5C fraction.
The presence of >10 m5C sites in the MLV gRNA makes the assessment of the biological function of these sites through site mutations difficult, as mutations can introduce non-m5C-related effects. We therefore proceeded with our biological study with perturbations of potential writer and reader proteins for the m5C modification.
The m5C reader protein ALYREF modulates MLV replication.
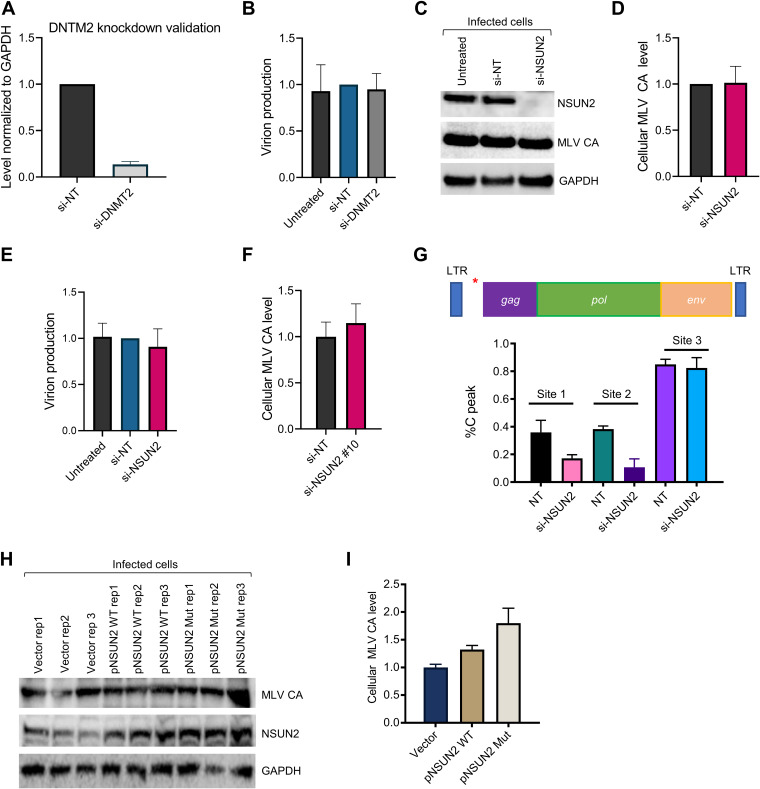
Cellular RNA modifications are installed by writer enzymes, may be removed by erasers, and are recognized by reader proteins. At least eight m5C methyltransferases are known to methylate cellular RNA, seven from the NSUN family and DNMT2 (3, 17). Unlike the m6A writer complex METTL3/METTL14, which installs m6A only in mRNA, all known m5C writers also methylate tRNA or rRNA. Among these, NSUN2, also known as a tRNA m5C methyltransferase, is the best candidate for installing m5C in mRNA (18). No m5C eraser has been identified. To identify a potential function for m5C modification in the retroviral life cycle, we knocked down NSUN2 and DNMT2, two potential m5C writer proteins for the viral transcripts. To our surprise, knocking down either protein did not significantly affect viral protein production or viral release (Fig. 4A to F). In contrast to our findings, prior studies reported a role for NSUN2 (13) or DNMT2 (19) in the HIV life cycle. To further test whether this lack of effect for NSUN2 might be due to the small interfering RNA (siRNA) pool used in our experiment, we tested an individual siRNA targeting NSUN2, but again, no difference in viral protein levels was detected (Fig. 4F). This discrepancy may be due to viral or cell condition differences. Moreover, it is also possible that more than one m5C methyltransferase targets MLV RNA and that downregulation of a single writer does not produce a distinctive phenotype under our conditions. This possibility is supported by our observation that NSUN2 knockdown affected only 2 of 3 examined m5C sites (Fig. 4G). Thus, our findings suggest that an additional methyltransferase may be involved beyond NSUN2. We also found that overexpression of wild-type NSUN2 slightly increased viral protein levels; however, enhanced cellular levels of MLV capsid were also detected upon overexpression of a catalytically dead version of NSUN2 (Fig. 4H and I). Thus, this effect may be independent of NSUN2’s methyltransferase role. As it is a major tRNA m5C modification enzyme, it should be noted that any experiment involving NSUN2 can generate pleotropic effects on cellular translation and viral biology.
FIG 4.
The putative m5C writer NSUN2 does not have a strong effect on viral replication. (A) Quantitative RT-PCR confirming depletion of DNMT2 mRNA following siRNA knockdown. n = 3 biological replicates; values are means plus SEM. GAPDH mRNA levels were used as a control to normalize expression. (B) Viral release was determined after DNMT2 knockdown. 3T3 cells were transfected with nontargeting (NT) or DNMT2 siRNAs and infected with MLV 24 h later. After 48 h of incubation, medium was harvested and MLV capsid (CA) levels were determined. n = 3 biological replicates; values are means plus SEM. (C) Viral production was determined after NSUN2 depletion. 3T3 cells were transfected with NT or NSUN2 siRNAs (Smartpool) and infected with MLV 24 h later. Following 48 h of incubation, medium was collected and CA levels determined by immunoblotting. A representative Western blot demonstrating the effect of NSUN2 depletion is shown, and successful NSUN2 knockdown was confirmed. GAPDH was detected as a loading control. (D) Quantification of data from panel C. n = 5 biological replicates; values are means plus SEM. (E) Viral release, as determined by MLV CA levels from medium samples, was monitored after NSUN2 depletion (Smartpool siRNAs). n = 7 biological replicates; values are means plus SEM. (F) Viral release was determined after transfection of an individual siRNA targeting the 3′ UTR of NSUN2 (si-NSUN2 10). n = 3 biological replicates; values are means plus SEM. (G) Effect of NSUN2 knockdown on viral m5C levels. Quantification of Sanger sequencing results from bisulfite-treated samples obtained from 3T3 cells transfected with the indicated siRNAs and infected with MLV. Results for two independent replicates are shown. Sites 1, 2, and 3 are the m5C sites located at positions 817, 818, and 819 of the MLV genome (pNCA); the approximate location of this m5C cluster is indicated by a red asterisk on the schematic at the top. (H) Effect of NSUN2 overexpression on cellular MLV CA levels. 3T3 cells were transfected with a control vector, wild-type (WT) NSUN2, or a catalytically dead NSUN2 mutant (Mut) plasmid and infected with MLV. After 48 h of incubation, CA levels were determined by immunoblotting. (I) Quantification of data from panel H. n = 3 biological replicates; values are means plus SEM.
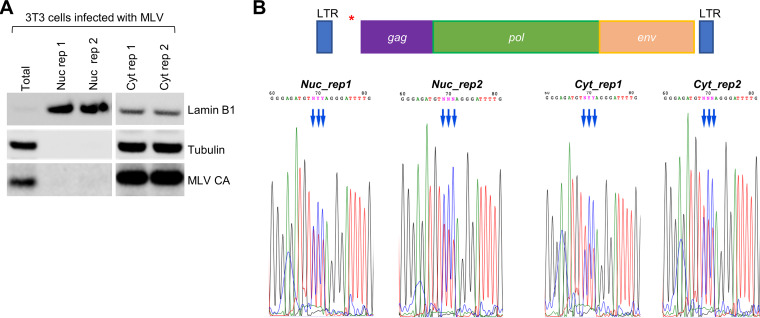
Only two m5C reader proteins have been reported: ALYREF (Aly/REF export factor) is an mRNA nuclear-cytoplasmic export factor (14), and YBX1 modulates mRNA stability (20, 21). ALYREF works in the nucleus, while YBX1 works in the cytoplasm. To determine whether m5C installation of the viral transcript occurs in the nucleus or cytoplasm, we isolated RNAs from the nuclear and cytoplasmic fractions and performed bisulfite treatment followed by Sanger sequencing (Fig. 5A). We found that for at least the m5C cluster between the long terminal repeat (LTR) and gag region, the m5C modifications were fully present in the nuclear fraction (Fig. 5B). We therefore focused our functional investigations on the ALYREF reader protein, although we do not exclude the possibility that the YBX1 protein may also play a role in MLV replication.
FIG 5.
Viral m5C modifications are installed in the nucleus. (A) Western blot of cellular fractionation. Nuclear and cytoplasmic RNAs and proteins were isolated in parallel from 3T3 cells infected with MLV. Antibodies against tubulin and lamin B1 were used to assess the purity of cytoplasmic and nuclear isolations, respectively. (B) Sanger sequencing analysis of bisulfite-treated nuclear and cytoplasmic fractions isolated from infected 3T3 cells. Blue arrows indicate m5C site location. Results for two independent replicates are shown for each fraction. (Top) Schematic of the MLV genome with a red asterisk indicating the location of the examined m5C sites. The m5C sites are located at 817, 818, and 819 of the MLV genome (pNCA).
We found that knocking down the m5C reader protein ALYREF produced a striking effect on viral protein production and viral replication (Fig. 6A to C). Depletion of ALYREF resulted in a 5-fold reduction in the cellular levels of MLV Gag proteins and viral release (Fig. 6B and C). Conversely, overexpression of ALYREF resulted in an increase in cellular Gag protein levels (Fig. 6D). We also examined whether 3T3 cells depleted of endogenous ALYREF could be rescued using a plasmid encoding wild-type ALYREF or an ALYREF mutant (K171A) known to have impaired m5C recognition (14). Supporting the critical importance of ALYREF’s m5C reader function, we found that although the wild-type ALYREF construct could rescue the phenotypic effect, the mutant version could not (Fig. 6E).
FIG 6.
The m5C reader ALYREF regulates MLV production. (A) Effect of ALYREF knockdown on cellular MLV protein levels. After transfection of 3T3 cells with siRNAs targeting ALYREF or nontargeting (NT) siRNAs, cells were infected with MLV for 24 h; after an additional 48 h of incubation, cellular proteins and media were collected. A representative Western blot of MLV capsid protein (CA) levels from infected 3T3 cells following siRNA-mediated knockdown of ALYREF is shown, with GAPDH as a loading control. (B) Quantitative analysis of MLV CA levels in cellular lysates upon ALYREF depletion. Data are means plus SEM for 3 biological replicates; **, P ≤ 0.01. (C) Quantitative analysis of MLV release upon ALYREF knockdown, as determined by MLV capsid levels from harvested medium samples. Data are means plus SEM for 6 biological replicates; **, P ≤ 0.01. (D) Quantitative analysis of MLV CA levels upon ALYREF overexpression as determined by MLV capsid levels in cellular lysates, normalized to GAPDH. Data are means plus SEM for 3 biological replicates. (E) Rescue of ALYREF depletion. 3T3 cells were transfected with an siRNA targeting the 3′ UTR of ALYREF. After 24 h, cells were cotransfected with a vector for wild-type MLV (pNCA) and a plasmid encoding either wild-type ALYREF (pALYREF) or an ALYREF mutant with defective m5C binding (pALYREF-K171A) or a control vector. MLV CA levels were quantified and normalized to the GAPDH loading control. Data are means plus SEM for 2 biological replicates.
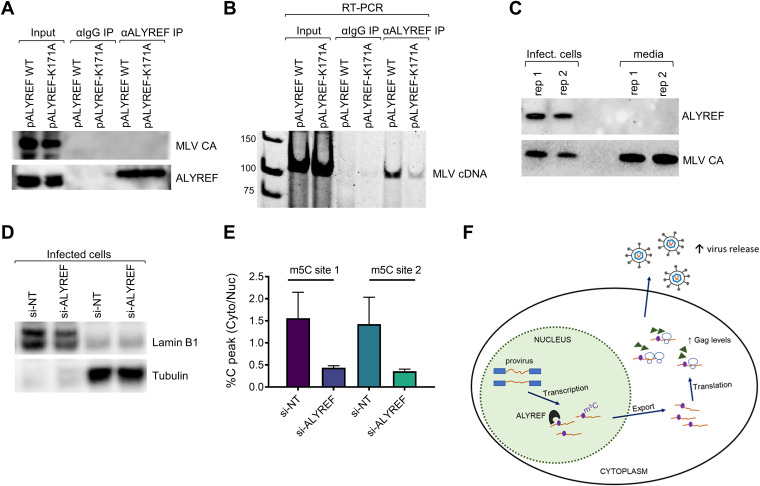
We next asked whether ALYREF directly interacts with MLV gRNA. After carrying out endogenous ALYREF RNA immunoprecipitation (RIP), we could amplify RT-PCR products using primers targeting MLV regions containing m5C from the ALYREF RIP samples but not from the corresponding IgG negative control (Fig. 7A and B). Notably, the ALYREF-K171A mutant showed reduced binding to MLV RNA relative to wild-type ALYREF, suggesting an important role for m5C in bridging this interaction. Despite its large influence on MLV production, we did not find ALYREF proteins in the viral particles (Fig. 7C), consistent with ALYREF playing a role mostly in the nuclear-cytoplasmic export of MLV transcripts. Further supporting this model, we demonstrated that ALYREF knockdown resulted in a decrease in the cytoplasmic-to-nuclear ratio of m5C in viral RNA (Fig. 7D and E), as would be expected if fewer m5C-containg viral transcripts were being exported to the cytoplasm.
FIG 7.
ALYREF interaction with viral RNA and a functional model of viral m5C modifications. (A and B) ALYREF interaction with viral RNA was evaluated by pulldown and RT-PCR. 3T3 cells were transfected with wild-type (WT) ALYREF or the ALYREF-K171A mutant with impaired m5C binding and then infected with MLV. (A) A representative Western blot is shown, confirming immunoprecipitation of endogenous ALYREF. IgG antibody (αIgG) immunoprecipitation was performed as a negative control. Capsid levels were detected to confirm cells were infected by virus. (B) Endogenous ALYREF RNA pulldown was followed by reverse transcription of extracted RNA and PCR. RT-PCR products, with primers amplifying the region upstream of gag containing three high-confidence m5C sites, were visualized by gel electrophoresis. (C) Viral packaging of ALYREF evaluated by Western blotting. MLV CA protein was used as a control. (D) Cellular fractionation of ALYREF-depleted cells. 3T3 cells were transfected with NT or ALYREF siRNAs and infected with MLV 24 h later. Nuclear and cytoplasmic RNAs and proteins were isolated in parallel from cells. A representative Western blot is shown; antibodies against tubulin and lamin B1 were used to assess fractionation purity. (E) Quantification of Sanger sequencing results for bisulfite-treated nuclear and cytoplasmic fractions isolated from si-NT or si-ALYREF cells infected with MLV. The cytoplasmic-to-nuclear ratio of unconverted C sites, which is indicative of m5C, is shown. The two examined m5C locations, sites 1 and 2, are located upstream of gag. (F) Functional model of m5C modification in the MLV life cycle. m5C is added to MLV RNAs in the nucleus and interacts with the m5C reader protein ALYREF, which is known to function in mRNA export, resulting in higher cellular Gag levels and increased viral release.
Concluding remarks.
Together, our results support a functional model of m5C modifications in the MLV life cycle (Fig. 7F). m5C is installed in viral RNA transcripts in the nucleus, and they are recognized by the m5C reader ALYREF, which facilitates their export to the cytoplasm. In tRNA, m5C is known to exert its function through stabilization of the RNA structure that reduces nuclease cleavage in the cell (22). In contrast, m5C function in mRNA and, in our model, retroviral RNA parallels the function of its reader protein.
In summary, we mapped the m5C topology of MLV genomic RNA at single-nucleotide resolution and suggest a nuclear-cytoplasmic export function of viral m5C through its reader protein ALYREF. Our study adds m5C RNA modification to the expanding universe of topology and function of viral RNA epitranscriptomes.
MATERIALS AND METHODS
Cells, plasmids, and virus infection.
Mouse embryonic fibroblasts (NIH 3T3 cells; ATCC), a line of chronically infected 3T3 cells (a gift from Sandra Wolin, National Cancer Institute [23]), and HEK293T cells (ATCC) were cultured in DMEM (Thermo Fisher Scientific) containing 100 U/ml penicillin-streptomycin, 10% fetal bovine serum, and 2 mM l-glutamine. To produce viral particles for infection of mouse fibroblasts, HEK293T cells were transfected with pNCA (Addgene plasmid number 17363), which harbors the wild-type MLV genome sequence, using polyethylenimine (PEI; PolySciences) (23). Virus titer was determined via the Retro-X quantitative RT-PCR (qRT-PCR) titration kit (TaKaRa Bio), and 3T3 cells were infected at a 20:1 multiplicity of infection in media containing 8 μg/ml hexadimethrine bromide (Polybrene; American Bioanalytical). Plasmids encoding mouse ALYREF (catalog no. MR220236) and NSUN2 (catalog no. MR208356) were purchased from OriGene, as well as a control vector (catalog no. PS100001). The ALYREF-K171A plasmid was manufactured by GenScript. The NSUN2 mutant vector (pNSUN2-C35A) was designed based on alignment of the mouse NSUN2 protein sequence with a known catalytically dead human NSUN2 sequence (24) and produced by GenScript.
Bisulfite sequencing.
To prepare virus for bisulfite sequencing, viral particles were first concentrated by sedimenting filtered medium from infected cells through 2-ml cushions of 20% sucrose, as previously described (23). Viral RNAs were isolated using TRIzol (Invitrogen) and DNase treated. For infected cellular samples, total RNA was DNase treated and ribosomal RNAs depleted using the RiboMinus transcriptome isolation kit (Thermo). The Zymogen EZ RNA methylation kit protocol was followed to convert unmethylated cytosines to uracil. Recovered RNA was used to generate sequencing libraries with the TruSeq stranded mRNA kit (Illumina). cDNA libraries were amplified by 12 cycles of PCR, loaded onto a HiSeq 2000 Illumina sequencer, and sequenced for 100 bases at The University of Chicago Genomics Facility. After adapter removal (FASTX tool kit), reads were aligned to a bisulfite-converted version of the pNCA proviral sequence (i.e., all cytosines were changed to thymines). Reads were mapped using Bowtie (25), allowing up to three mismatches. As a positive control for m5C detection, reads were aligned to the 28S mouse rRNA sequence (accession number X00525.1). Candidate m5C sites were identified using the bam-readcount tool and visually inspected on the Integrative Genomics Viewer (IGV) browser.
siRNA transfections, overexpression, and rescue.
Smartpool siRNAs for ALYREF (catalog no. L-046521-01-0005), NSUN2 (catalog no. L-059976-01-0005), DNMT2 (catalog no. L-060459-01-0005), and nontargeting control siRNA 3 (D-001210-03-05) were purchased from Dharmacon. An individual siRNA-targeting NSUN2, referred to as NSUN2 10 (catalog no. J-060459-10), was also obtained (Dharmacon). Transfections were carried out as previously described (23) using Lipofectamine RNAiMAX (Invitrogen) and 20 nM siRNAs. At 24 h after transfection, cells were infected with MLV; medium was harvested between 48 and 72 h, and cells were harvested at 72 h after transfection. Viral release was determined by assessing MLV capsid levels in viral lysates. To confirm knockdown of DNMT2 mRNA, the following primers were used for qRT-PCR: DNMT2_fwd, 5′-CAGTAGTTGAAGAAAGTCAGCCA-3′; DNMT2_rev, 5′-AGCCTGTAACACGGAGCCTG-3′.
For overexpression experiments, 3T3 cells seeded in 6-well dishes were transfected with 3 μg of plasmid DNA (pALYREF-FLAG, pNSUN2-FLAG, the C35A mutant pNSUN2-FLAG, or vector control) following the guidelines for Lipofectamine 2000 (Thermo Fisher). Cells were infected with MLV on the next day and harvested ∼48 h later.
For ALYREF rescue experiments, 3T3 cells were reverse transfected with an siRNA targeting the 3′ untranslated region (UTR) of ALYREF (referred to as ALYREF 12; Dharmacon catalog no. J-046521-12) using the Lipofectamine RNAiMAX reagent (Thermo Fisher). After 24 h, cells were cotransfected with 3 μg of pNCA and 4 μg of a plasmid encoding wild-type ALYREF, the ALYREF-K171A mutant, or the control vector using Lipofectamine 2000. Cells were harvested ∼48 h later, and protein levels were determined by Western blotting. MLV capsid (CA) levels were quantified and normalized to the GAPDH loading control.
Immunoblotting.
For immunoblotting experiments, cell pellets were resuspended in CelLytic M (Sigma-Aldrich). Cleared lysates were electrophoresed through 4 to 12% bis-Tris protein gels (NuPage; Thermo) and transferred to polyvinylidene difluoride (PVDF) membranes (EMD Millipore). Antibodies were anti-MLV (ATCC), anti-ALYREF (Abcam), anti-NSUN2 (Sigma), anti-GAPDH (Neta Scientific), anti-lamin B1 (Abcam), and anti-tubulin (Abcam).
Cell fractionation and Sanger validation.
Cell fractionation of MLV-infected fibroblasts was performed using the PARIS isolation system (Thermo Fisher). To evaluate fractionation success, a portion of the samples was saved for protein analysis and probed for the nuclear marker lamin B1 and the cytoplasmic marker tubulin. RNA samples from the nuclear and cytoplasmic fractions were subjected to DNase treatment, bisulfite conversion (Zymogen), and cDNA synthesis (SuperScript III) with random hexamers. Next, ZymoTaq DNA polymerase (Zymogen) was used to amplify a target region of the MLV genome containing three high-confidence m5C sites. PCR products of the expected size were excised from an 8% polyacrylamide gel and subjected to Sanger sequencing. The same RT-PCR approach was followed to confirm candidate m5C sites in MLV-infected fibroblasts and virus.
Primer sequences for RT-PCR of bisulfite-treated samples are as follows: MLV_cluster1_bs_fwd-T7, 5′-taatacgactcactatagGTATTAGTTAGTTAATTAGTTTTGTATTTGG-3′; MLV_cluster1_bs_rev, 5′-CAAATACAAAAACACAATCAAACAAAAACAACAC-3′.
LC-MS analysis.
To purify viral genomic RNA for LC-MS, virions were isolated as described above on a sucrose cushion and resuspended in TRIzol, and RNA was extracted. Viral gRNA was then size selected on a low-melting-point agarose gel and isolated using the Zymoclean gel RNA recovery kit (Zymo Research). To purify cellular mRNA, total RNA samples were DNase treated and poly(A) selected (Dynabeads mRNA purification kit; Invitrogen), and rRNAs were removed (RiboMinus kit; Thermo Fischer). Purified viral and cellular RNAs were then digested with nuclease P1 (1 U; Sigma) in buffer containing 20 mM NH4OAc (pH 5.3) for 2 h at 42°C, followed by the addition of NH4HCO3 (100 mM) and alkaline phosphatase (1 U; Sigma) and incubated at 37°C for 2 h. After the samples had been filtered (0.22-μm pore size, 4-mm diameter; Millipore), 10 μl was injected into a C18 reverse-phase column with mass spectrometry detection using an Agilent 6410 QQQ (triple-quadrupole) LC-MS/MS spectrophotometer in positive electrospray ionization mode. Nucleosides were quantified by using retention time and nucleoside-to-base ion mass transitions.
ALYREF RNA immunoprecipitation.
3T3 cells were grown in 10-cm dishes and infected with MLV. After reaching ∼85% confluence, cells were washed twice with ice-cold 1× phosphate-buffered saline (PBS), harvested with a cell scraper, and lysed by sonication in NET-2 buffer (40 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.1% NP-40, 1 mM MgCl2) containing 1 mM phenylmethylsulfonyl fluoride (PMSF), 1× protease inhibitor cocktail (Thermo Fisher), and 400 U/ml RNase inhibitor (Thermo Fisher). After centrifugation of the lysate for 30 min at 15,000 rpm, the supernatant was precleared with 20 μl of Dynabeads protein A (Invitrogen) and incubated for 1 h at 4°C with rotation. Antibody-bead complexes were prepared by first washing 50 μl of Dynabeads protein A twice with NET-2 lysis buffer and then resuspending in 200 μl. Next, 4 μg of antibody (anti-ALYREF [Abcam] or anti-IgG [Cell Signaling Technology]) was added to the resuspended beads and incubated for 2 h at 4°C. After the conjugated beads had been washed three times with NET-2 buffer, the precleared lysate was added and incubated overnight at 4°C with rotation. The supernatant was discarded; the beads were then washed five times with NET-2 buffer and resuspended in TRIzol. After extraction of the RNA, the samples were DNase treated and cDNA synthesized using the iScript kit (Bio-Rad). MLV regions of interest were amplified using AccuPrime Pfx SuperMix (Thermo Fisher).
Primer sequences used for RT-PCR were as follows: MLV_cluster1_fwd, 5′-CTAACTAGCTCTGTATCTGGCGGAC-3′; MLV_cluster1_rev, 5′-ACCAGAACCACATATCCCTCCTC-3′.
Data availability.
The RNA-BisSeq data generated by this study have been deposited in the NCBI GEO database under the accession number GSE139606.
ACKNOWLEDGMENTS
This work was supported by the NIH (RM1 HG008935 to T.P.; F32GM126745 and 5T32HL007381 to M.E.), the University of Chicago Yen Fellowship (M.E.), and the China Scholarship Council (grant number 201906320410 to R.X.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank Q. Dai and J. Quin for technical advice on using LC-MS.
M.E. and T.P. designed and initiated the study. M.E. performed and analyzed sequencing data. M.E., R.X., J.M., W.Z., P.P., and Z.C. performed experiments and analyzed data. M.E. and T.P. wrote the paper.
REFERENCES
- 1.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. 2011. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res 39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roundtree IA, Evans ME, Pan T, He C. 2017. Dynamic RNA modifications in gene expression regulation. Cell 169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanco S, Frye M. 2014. Role of RNA methyltransferases in tissue renewal and pathology. Curr Opin Cell Biol 31:1–7. doi: 10.1016/j.ceb.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hussain S, Aleksic J, Blanco S, Dietmann S, Frye M. 2013. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol 14:215. doi: 10.1186/gb4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krug RM, Morgan MA, Shatkin AJ. 1976. Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methylguanosine in cap structures. J Virol 20:45–53. doi: 10.1128/JVI.20.1.45-53.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beemon K, Keith J. 1977. Localization of N6-methyladenosine in the Rous sarcoma virus genome. J Mol Biol 113:165–179. doi: 10.1016/0022-2836(77)90047-x. [DOI] [PubMed] [Google Scholar]
- 7.Kennedy EM, Bogerd HP, Kornepati AV, Kang D, Ghoshal D, Marshall JB, Poling BC, Tsai K, Gokhale NS, Horner SM, Cullen BR. 2016. Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 19:675–685. doi: 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L. 2016. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife 5:e15528. doi: 10.7554/eLife.15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE, Rana TM. 2016. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol 1:16011. doi: 10.1038/nmicrobiol.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, Hopcraft SE, Quicke KM, Vazquez C, Willer J, Ilkayeva OR, Law BA, Holley CL, Garcia-Blanco MA, Evans MJ, Suthar MS, Bradrick SS, Mason CE, Horner SM. 2016. N6-methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20:654–665. doi: 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lichinchi G, Zhao BS, Wu Y, Lu Z, Qin Y, He C, Rana TM. 2016. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe 20:666–673. doi: 10.1016/j.chom.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courtney DG, Chalem A, Bogerd HP, Law BA, Kennedy EM, Holley CL, Cullen BR. 2019. Extensive epitranscriptomic methylation of A and C residues on murine leukemia virus transcripts enhances viral gene expression. mBio 10:e01209-19. doi: 10.1128/mBio.01209-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A, Swanstrom R, Holley CL, Cullen BR. 2019. Epitranscriptomic addition of m(5)C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe 26:217–227 E6. doi: 10.1016/j.chom.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, Li A, Wang X, Bhattarai DP, Xiao W, Sun HY, Zhu Q, Ma HL, Adhikari S, Sun M, Hao YJ, Zhang B, Huang CM, Huang N, Jiang GB, Zhao YL, Wang HL, Sun YP, Yang YG. 2017. 5-Methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res 27:606–625. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckwahl MJ, Telesnitsky A, Wolin SL. 2016. Host RNA packaging by retroviruses: a newly synthesized story. mBio 7:e02025-15. doi: 10.1128/mBio.02025-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckwahl MJ, Arnion H, Kharytonchyk S, Zang T, Bieniasz PD, Telesnitsky A, Wolin SL. 2016. Analysis of the human immunodeficiency virus-1 RNA packageome. RNA 22:1228–1238. doi: 10.1261/rna.057299.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, Kellner S, Hölter SM, Garrett L, Wurst W, Becker L, Klopstock T, Fuchs H, Gailus-Durner V, Hrabĕ de Angelis M, Káradóttir RT, Helm M, Ule J, Gleeson JG, Odom DT, Frye M. 2014. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J 33:2020–2039. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bohnsack KE, Hobartner C, Bohnsack MT. 2019. Eukaryotic 5-methylcytosine (m(5)C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes (Basel) 10:102. doi: 10.3390/genes10020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dev RR, Ganji R, Singh SP, Mahalingam S, Banerjee S, Khosla S. 2017. Cytosine methylation by DNMT2 facilitates stability and survival of HIV-1 RNA in the host cell during infection. Biochem J 474:2009–2026. doi: 10.1042/BCJ20170258. [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X, Chen RX, Wei WS, Liu Y, Gao CC, Chen YS, Zhang M, Ma XD, Liu ZW, Luo JH, Lyu C, Wang HL, Ma J, Zhao YL, Zhou FJ, Huang Y, Xie D, Yang YG. 2019. 5-Methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol 21:978–990. doi: 10.1038/s41556-019-0361-y. [DOI] [PubMed] [Google Scholar]
- 21.Yang Y, Wang L, Han X, Yang WL, Zhang M, Ma HL, Sun BF, Li A, Xia J, Chen J, Heng J, Wu B, Chen YS, Xu JW, Yang X, Yao H, Sun J, Lyu C, Wang HL, Huang Y, Sun YP, Zhao YL, Meng A, Ma J, Liu F, Yang YG. 2019. RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol Cell 75:1188–1202 e11. doi: 10.1016/j.molcel.2019.06.033. [DOI] [PubMed] [Google Scholar]
- 22.Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Cortes-Garrido R, Gkatza N, Dietmann S, Frye M. 2016. Stem cell function and stress response are controlled by protein synthesis. Nature 534:335–340. doi: 10.1038/nature18282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckwahl MJ, Sim S, Smith D, Telesnitsky A, Wolin SL. 2015. A retrovirus packages nascent host noncoding RNAs from a novel surveillance pathway. Genes Dev 29:646–657. doi: 10.1101/gad.258731.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, Frye M. 2013. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep 4:255–261. doi: 10.1016/j.celrep.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The RNA-BisSeq data generated by this study have been deposited in the NCBI GEO database under the accession number GSE139606.