A major hurdle to the development of an effective HIV-1 vaccine is the elicitation of serum responses capable of neutralizing circulating strains of HIV, which are extraordinarily diverse in sequence and often highly neutralization resistant. Recently, we showed how sera with 20 to 30% neutralization breadth could, nevertheless, be elicited in standard vaccine test animals by priming with the most prevalent N-terminal 8 residues of the HIV-1 fusion peptide (FP8), followed by boosting with a stabilized BG505-envelope (Env) trimer. Here, we show that subsequent boosting with a 3mut-apex-stabilized consensus C-Env trimer, modified to have the second most prevalent FP8 sequence, elicits higher neutralization breadth than that induced by continued boosting with the stabilized BG505-Env trimer. With increased neutralizing breadth elicited by boosting with a heterologous trimer containing the second most prevalent FP8 sequence, the fusion peptide-directed immune-focusing approach moves a step closer toward realizing an effective HIV-1 vaccine regimen.
KEYWORDS: HIV-1, fusion peptide, neutralization, vaccine
ABSTRACT
HIV-1 envelope (Env) trimers, stabilized in a prefusion-closed conformation, can elicit humoral responses capable of neutralizing HIV-1 strains closely matched in sequence to the immunizing strain. One strategy to increase elicited neutralization breadth involves vaccine priming of immune responses against a target site of vulnerability, followed by vaccine boosting of these responses with prefusion-closed Env trimers. This strategy has succeeded at the fusion peptide (FP) site of vulnerability in eliciting cross-clade neutralizing responses in standard vaccine-test animals. However, the breadth and potency of the elicited responses have been less than optimal. Here, we identify three mutations (3mut), Met302, Leu320, and Pro329, that stabilize the apex of the Env trimer in a prefusion-closed conformation and show antigenically, structurally, and immunogenically that combining 3mut with other approaches (e.g., repair and stabilize and glycine-helix breaking) yields well-behaved clade C-Env trimers capable of boosting the breadth of FP-directed responses. Crystal structures of these trimers confirmed prefusion-closed apexes stabilized by hydrophobic patches contributed by Met302 and Leu320, with Pro329 assuming canonically restricted dihedral angles. We substituted the N-terminal eight residues of FP (FP8, residues 512 to 519) of these trimers with the second most prevalent FP8 sequence (FP8v2, AVGLGAVF) and observed a 3mut-stabilized consensus clade C-Env trimer with FP8v2 to boost the breadth elicited in guinea pigs of FP-directed responses induced by immunogens containing the most prevalent FP8 sequence (FP8v1, AVGIGAVF). Overall, 3mut can stabilize the Env trimer apex, and the resultant apex-stabilized Env trimers can be used to expand the neutralization breadth elicited against the FP site of vulnerability.
IMPORTANCE A major hurdle to the development of an effective HIV-1 vaccine is the elicitation of serum responses capable of neutralizing circulating strains of HIV, which are extraordinarily diverse in sequence and often highly neutralization resistant. Recently, we showed how sera with 20 to 30% neutralization breadth could, nevertheless, be elicited in standard vaccine test animals by priming with the most prevalent N-terminal 8 residues of the HIV-1 fusion peptide (FP8), followed by boosting with a stabilized BG505-envelope (Env) trimer. Here, we show that subsequent boosting with a 3mut-apex-stabilized consensus C-Env trimer, modified to have the second most prevalent FP8 sequence, elicits higher neutralization breadth than that induced by continued boosting with the stabilized BG505-Env trimer. With increased neutralizing breadth elicited by boosting with a heterologous trimer containing the second most prevalent FP8 sequence, the fusion peptide-directed immune-focusing approach moves a step closer toward realizing an effective HIV-1 vaccine regimen.
INTRODUCTION
The HIV-1 Env trimer is a type 1 fusion machine capable of accessing multiple conformational states, with some related to structural rearrangements required for the fusion of viral and target cell membranes during virus cell entry (1, 2) and some related to immune evasion (3, 4). Several prefusion states have been defined, and these have been classified into “open” and “closed” conformational states (5). Closed conformations of Env are recognized by most broadly neutralizing antibodies but not by nonneutralizing antibodies or weakly neutralizing antibodies capable of neutralizing only laboratory-adapted strains of HIV-1; in contrast, open conformations of Env are recognized by most antibodies, except for broadly neutralizing antibodies targeting the trimer apex.
Extensive efforts have been made to produce soluble Env trimers, stabilized in prefusion-closed conformations, as these have been shown to be capable of eliciting neutralizing responses against HIV-1 strains matched in sequence to the vaccine strain (6–9). Sanders et al. developed the soluble, cleaved Env trimer named BG505 SOSIP.664 by truncating strain BG505 at residue 664, adding a disulfide between residues 501 and 605, substituting proline for isoleucine at residue 559, and utilizing a 6-Arg sequence to enhance subunit cleavage (10), and they have described additional mutations that further stabilize the SOSIP trimer (7, 9, 11, 12). Other groups have also developed mutations or methods to stabilize further the BG505 SOSIP trimer or to improve its yield, including single-chain versions (13–15), glycine mutations to disrupt the formation of postfusion helices (16), computational redesigns of a largely disordered portion of the heptad region 1 (15), chemical crosslinks to fix conformation (17), and multiple mutations identified by mammalian cell surface display (18). Stabilized diverse strains of Env have also been developed by mutating critical residues identified from BG505 SOSIP structures (19) by replacing the gp41 subunit and the N and C termini of gp120 with those from BG505 (20), or by replacing rare residues with consensus amino acid types (21). We have previously developed BG505 DS-SOSIP.664 (Trimer 4571) with an engineered disulfide bond between residues 201 and 433 (DS). The DS-SOSIP-stabilized Env trimer binds only a single CD4 molecule and does not transition to the three CD4-bound conformation accessed by both wild-type trimers and by the BG505 SOSIP.664 (22). In addition to DS, we have also identified disulfides that lock the movement between gp120 inner and outer domains of the SOSIP trimer, resulting in impaired CD4 binding (23). We and others have also previously developed mutations that reduced the weak or nonneutralizing V3 antibody antigenicity of Env trimers by stabilizing the interface between V1/V2 and V3 (6, 8).
The strain-specific neutralizing responses elicited by prefusion-closed Env trimers are similar in many respects to the strain-specific responses elicited against other diverse pathogens, such as influenza A virus or coronaviruses (reviewed in references 24 and 25), and expansion of elicited neutralizing breadth against these pathogens has been a major outstanding challenge of the vaccine field. Recently, we have found that an epitope-focused vaccine strategy targeting the exposed N terminus of the fusion peptide of gp41 could elicit humoral responses capable of neutralizing 20 to 30% of a diverse panel of 208 HIV-1 strains (26), with antibodies isolated from vaccinated rhesus macaques capable of neutralizing up to almost 60% of this diverse panel (26, 27). Boosting with the BG505 DS-SOSIP.664 trimer significantly enhanced neutralizing titers (26), and subsequent boost with a diverse HIV-1-Env trimer increased the consistency of inducing broad cross-clade neutralizing responses (28), highlighting the utility of sequential boosting with a heterologous trimer.
In this study, we developed a set of three mutations (3mut) to stabilize the apex of BG505 DS-SOSIP.664 and to improve its antigenic specificity, thermostability, and immunogenic specificity. We combined 3mut with previously published glycine-helix breaking and repair-and-stabilize approaches (16, 21) to produce stabilized clade C-Env trimers from a consensus clade C Env (21) and from a week 34 Env from donor CAP256 (29) and further determined the crystal structure of the stabilized variant from CAP256 by utilizing an antibody-facilitated lattice (30). We swapped the FP8 sequence of the apex-stabilized Env trimers with the second most prevalent FP8 sequence (FP8v2) and tested the ability of the resultant FP8v2-containing trimer to boost the breadth and potency of FP-directed responses in guinea pigs that had previously been immunized with FP8 carrier and BG505 Env trimer immunogens, which had the most prevalent FP8 sequence (FP8v1). Overall, our results provide a developmental framework outlining how a specific type of Env stabilization (apex fixation) can be honed to produce an Env trimer immunogen able to boost the breadth of neutralizing responses directed at the FP site of vulnerability.
RESULTS
Design and crystal structure of 3mut that stabilizes BG505 DS-SOSIP.664 apex.
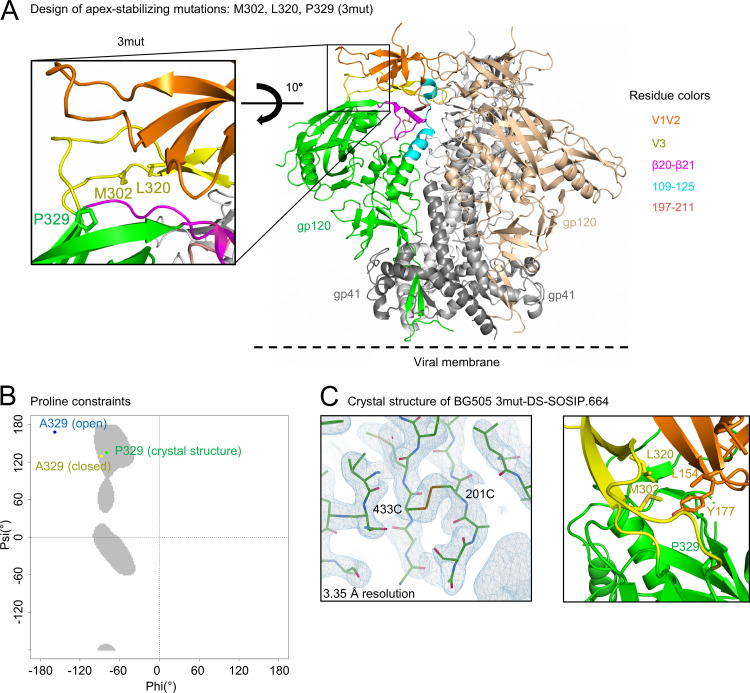
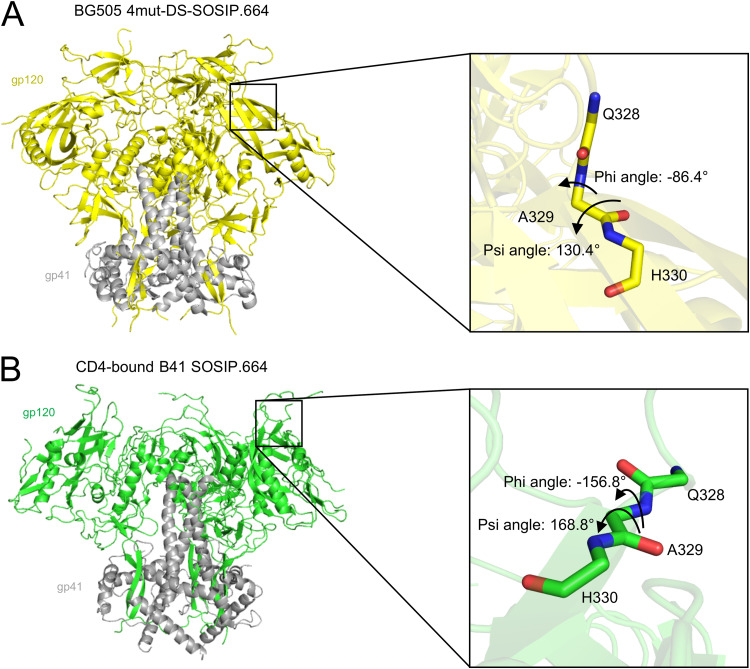
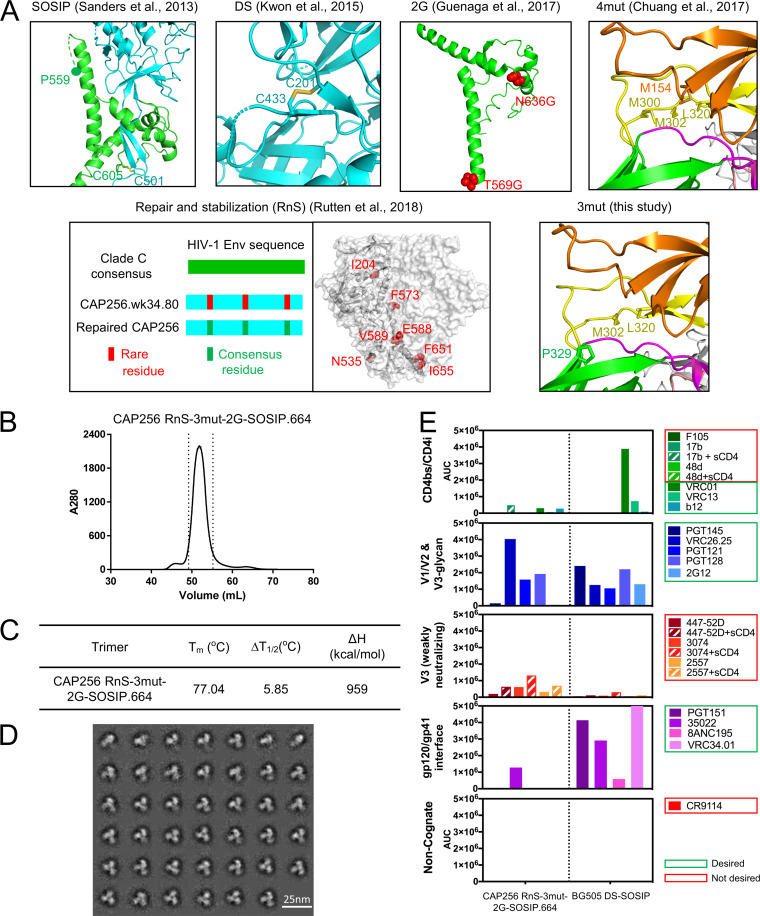
The apex of the Env trimer is highly accessible geometrically and thus an important vaccine target, although it is shielded by dense glycosylation and highly variable loops. We designed three mutations to BG505 DS-SOSIP.664 involving apex residues N302M, T320L, and A329P (BG505 3mut-DS-SOSIP.664) to stabilize the structure in a prefusion-closed conformation (Fig. 1A). Notably, the A329P mutation was expected to stabilize the prefusion-closed conformation because the backbone dihedral angles of residue 329 in the prefusion-closed conformation (6), but not in the CD4-bound open conformation (31), are compatible with a trans-proline (Fig. 1B and 2).
FIG 1.
Design and structure of BG505 3mut-DS-SOSIP.664 reveal a stabilized apex with enhanced hydrophobic interactions and proline-specific conformational constraints. (A) 3mut mutations (N302M, T320L, and A329P) are highlighted on the BG505 SOSIP.664 structure. (B) Ramachandran plot of allowable proline phi-psi angles (gray), with phi-psi angles of A329 (closed) from a prefusion-closed trimer (PDB 5UTY) labeled in yellow, phi-psi angles of A329 (open) from a CD4-bound open trimer (PDB 5VN3) labeled in blue, and phi-psi angles of P329 (crystal structure) described in panel C (PDB 6W03) labeled in green. (C) Structure of BG505 3mut-DS-SOSIP.664 at 3.35-Å resolution. Left, well-defined electron density was observed in the vicinity of DS mutation (201C-433C disulfide) in the BG505 3mut-DS-SOSIP.664 structure. Right, close-up view of the hydrophobic core between L320 and M302, two of the mutations from 3mut, with L154 and Y177 from the BG505 3mut-DS-SOSIP.664 structure. The electron density map is contoured at 1.5 sigma.
FIG 2.
Design of A329P based upon phi-psi angles of prefusion-closed and CD4-bound structures. (A) Phi-psi angle of residue 329 in the prefusion-closed conformation of BG505 4mut-DS-SOSIP.664 (PDB 5UTY). (B) Phi-psi angle of residue 329 in the CD4-bound conformation of B41 SOSIP.664 (PDB 5VN3).
We assessed a panel of over 100 differently stabilized variants of BG505 in a 96-well antigenic screening platform (see Materials and Methods) and found BG505 3mut-DS-SOSIP.664 to display the best antigenicity based on an antigenic score calculated from enzyme-linked immunosorbent assay (ELISA) antibody binding responses as PGT145*VRC26.25/447-52D (see Data Set S1 in the supplemental material). In this score, the numerator was defined by the binding of antibodies that specifically recognize the prefusion-closed conformation at the trimer apex (PGT145 [32, 33] and CAP256-VRC26.25 [VRC26.25] [34, 35]), and the denominator was defined by the binding of antibody 447-52D, which only binds V3 region of Env in open configurations. Compared with DS-SOSIP.664, the incorporation of A329P showed only slightly reduced antigenicity toward nonneutralizing or weakly neutralizing V3 antibodies, while constructs containing the N302M-T320L mutations generally had only weak recognition by nonneutralizing or weakly neutralizing V3 antibodies.
We determined the crystal structure of BG505 3mut-DS-SOSIP.664 with two engineered crystallization chaperones, 35O22_scFv and 3H109L_MM (30), and obtained a crystal structure at a nominal resolution of 3.35 Å (Table 1). The crystal structure was well defined, as exemplified by the electron density around the vicinity of the DS mutation (Fig. 1C, left). As expected, the P329 phi-psi angle was in the allowed region for trans-proline and close to that of A329 in the prefusion-closed conformation (Fig. 1B). The V3 mutations, M302 and L320, formed hydrophobic interaction with residues L154 and Y177 on V1/V2 loop, stabilizing the Env trimer in the prefusion-closed conformation (Fig. 1C, right).
TABLE 1.
Data collection and refinement statistics for BG505 3mut-DS-SOSIP.664 and CAP256 RnS-3mut-2G-SOSIP.664a
| Statistics | BG505 3mut-DS-SOSIP.664 (PDB 6W03) | CAP256 RnS-3mut-2G-SOSIP.664 (PDB 6VZI) |
|---|---|---|
| Data collection | ||
| Space group | P63 | P63 |
| Cell dimensions | ||
| a, b, c (Å) | 131.4, 131.4, 315.3 | 133.9, 133.9, 315.1 |
| α = β = γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution range (Å) | 50.00–3.35 (3.47–3.35)b,c ; 50.00–2.40 (2.70–2.59, 2.59–2.49, 2.49–2.40)d | 50.00–3.50 (3.56–3.50)b ,c ; 50.00–2.70 (3.04–2.91, 2.91–2.80, 2.80–2.70)d |
| Rsym or Rmerge (%) | 14.7 (44.2); 14.9 (77.5, 120.3, 38.9) | 9.8 (46.8); 12.8 (152.2, 156.9, 206.7) |
| I/σI | 16.5 (3.3); 11.6 (0.7, 0.4, 0.2) | 16.1 (2.1); 11.7 (0.4, 0.3, 0.5) |
| Completeness (%) | 88.8 (56.7); 42.3 (11.4, 7.5, 2.3) | 88.2 (50.0); 63.3 (28.8, 21.2, 14.6) |
| Redundancy | 3.9 (2.9); 3.3 (1.2, 1.0, 1.1) | 7.5 (5.8); 7.1 (4.8, 3.9, 3.2) |
| Refinement | ||
| Resolution range (Å) | 39.8–2.4 | 43.8–2.7 |
| No. of reflections | 36,659 | 31,204 |
| Rwork/Rfree (%) | 23.5/27.7 | 22.8/27.8 |
| No. of atoms | 10,113 | 9,930 |
| Protein | 9,540 | 9,468 |
| Ligand/ion | 573 | 462 |
| Water | 0 | 0 |
| B-factors (Å2) | 46.70 | 47.30 |
| Protein | 45.01 | 46.04 |
| Ligand/ion | 74.86 | 73.09 |
| Water | NA | NA |
| RMSD | ||
| Bond length (Å) | 0.004 | 0.003 |
| Bond angle (°) | 0.69 | 0.72 |
NA, not available; RMSD, root mean square deviation.
Data processing statistics based on the overall resolution cutoff determined as completeness greater than 50% and I/σI greater than 2, as mentioned in Materials and Methods.
Statistics for the highest-resolution shell are shown in parentheses.
The diffraction limits were determined by the UCLA anisotropy server. Anisotropic data statistics are shown in italics. Statistics for the three highest-resolution shells are shown in parentheses.
BG505 3mut-DS-SOSIP.664 showed improved antigenicity, thermostability, and immunogenicity.
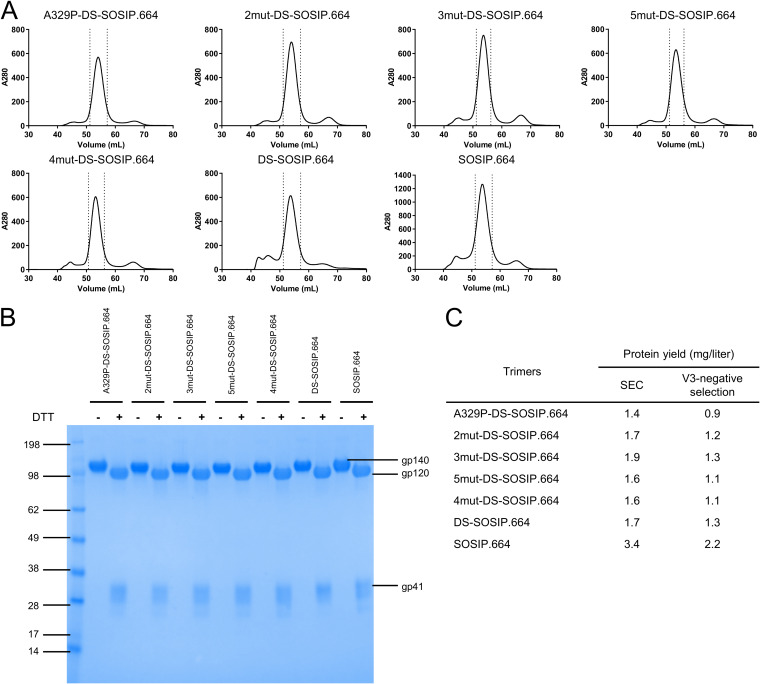
We compared various properties of BG505 3mut-DS-SOSIP.664 to BG505 SOSIP.664, BG505 DS-SOSIP.664, and other variants with mutations derived from 3mut or 4mut (6), such as BG505 A329P-DS-SOSIP.664, BG505 DS-SOSIP.664 N302M/T320L (BG505 2mut-DS-SOSIP.664), BG505 4mut-DS-SOSIP.664 (6), and BG505 A329P-4mut-DS-SOSIP.664 (BG505 5mut-DS-SOSIP.664), to evaluate the effects from different mutations. BG505 3mut-DS-SOSIP.664 had a size exclusion chromatography (SEC) profile similar to those of other BG505 SOSIP variants (Fig. 3A), with all variants showing similar yields (0.9 to 1.3 mg/liter) (Fig. 3B and C). To evaluate differences in antigenicity, we compared the antigenicity of BG505 3mut-DS-SOSIP.664 with that of the other BG505 SOSIP variants, using Meso Scale Discovery (MSD) and a panel of antibodies; BG505 3mut-DS-SOSIP.664 exhibited improved antigenicity over DS-SOSIP.664, with little to no measurable binding to V3 antibodies, even in the presence of soluble CD4 (Fig. 4A).
FIG 3.
Properties of stabilized BG505 DS-SOSIP.664 Env trimers. (A) Size exclusion chromatography (SEC) profiles of stabilized DS-SOSIP.664 variants before V3-negative selection. (B) SDS-PAGE of stabilized DS-SOSIP.664 variants after V3-negative selection, with and without the presence of dithiothreitol (DTT). The unit of measure of the SDS-PAGE marker is kDa. (C) Expression yield of stabilized DS-SOSIP.664 variants after SEC alone or after SEC with V3-negative selection.
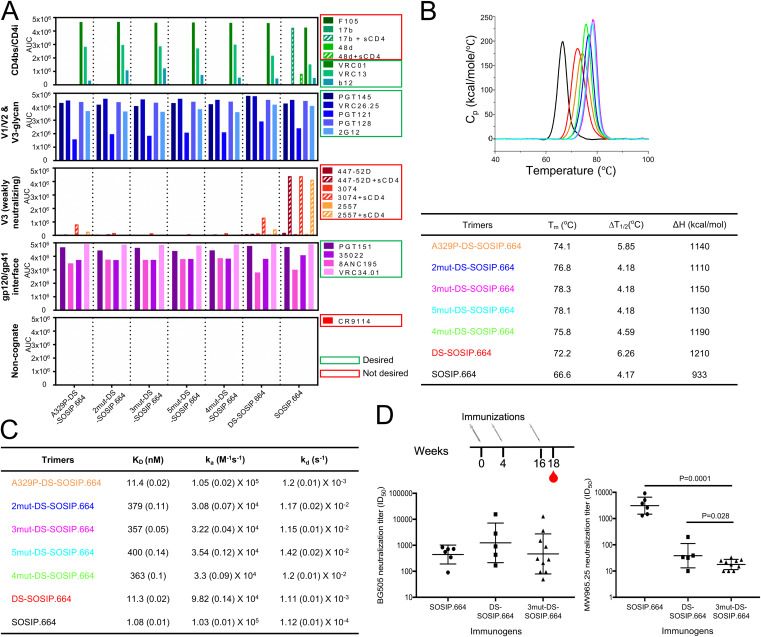
FIG 4.
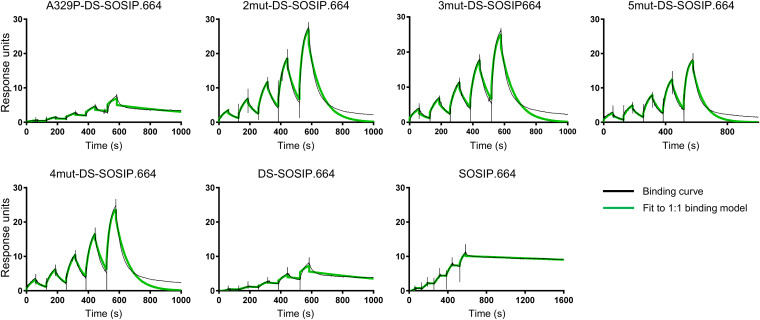
BG505 3mut-DS-SOSIP.664 shows enhanced antigenic specificity, thermostability, and immunogenic specificity. (A) Antigenicity, as determined by MSD, of stabilized BG505 DS-SOSIP.664 trimers after V3-negative selection assessed on a panel of antibodies, including nonneutralizing/weakly neutralizing CD4-induced antibodies (17b and 48d with and without soluble CD4, and F105), CD4-binding site antibodies (VRC01, VRC13, and b12), broadly neutralizing V1/V2 antibodies (PGT145, CAP256-VRC26.25), broadly neutralizing glycan-V3 antibodies (PGT121, PGT128, and 2G12), nonneutralizing/weakly neutralizing V3-directed antibodies (447-52D, 3074, and 2557, with and without soluble CD4), broadly neutralizing gp41-gp120 interface antibodies (35O22 and 8ANC195), and fusion peptide antibody (VRC34.01 and PGT151). (B) Thermostability of stabilized BG505 Env trimers assessed by DSC. Top, raw data from DSC shown in solid lines, with A329P-DS-SOSIP.664 shown in orange, 2mut-DS-SOSIP.664 shown in blue, 3mut-DS-SOSIP.664 shown in magenta, 5mut-DS-SOSIP.664 shown in cyan, 4mut-DS-SOSIP.664 shown in green, DS-SOSIP.664 shown in red, and BG505 SOSIP.664 shown in black. Bottom, Tm, ΔT1/2 (width at half-peak height), and enthalpy of unfolding from DSC (ΔH). (C) Binding parameters of stabilized HIV-1 Env trimers to the soluble CD4 D1D2 domain. Values in parentheses report standard errors from fitting data to a 1:1 Langmuir binding model. (D) Autologous neutralization titer (BG505, left) and MW965.26 neutralization titer (right) of guinea pig sera at week 18 (after three trimer injections). Statistical differences between BG505 3mut-DS-SOSIP.664 and BG505 SOSIP.664 or BG505 DS-SOSIP.664 were calculated using a one-tailed Mann-Whitney test. AUC, area under the curve; Cp, heat capacity; KD, equilibrium dissociation constant; ka, absorption rate constant; kd, dissociation constant.
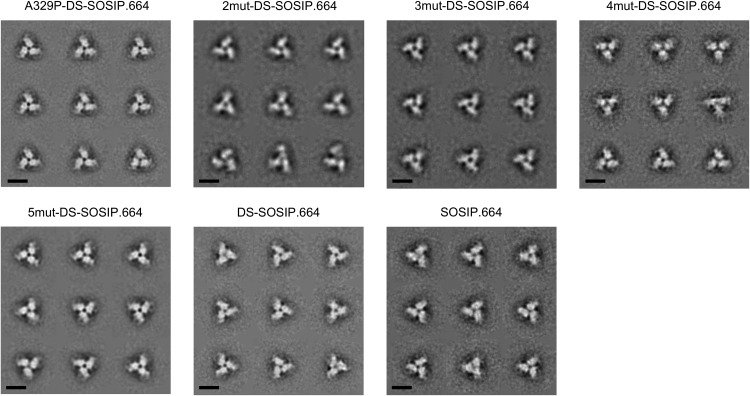
To evaluate the thermostability of BG505 3mut-DS-SOSIP.664, we performed differential scanning calorimetry (DSC) on BG505 3mut-DS-SOSIP.664 and other DS-SOSIP.664 variants (Fig. 4B). We observed that A329P alone increased the melting temperature (Tm) by about 2°C, while BG505 3mut-DS-SOSIP.664 gave the highest Tm (78.3°C). We also evaluated the binding of BG505 3mut-DS-SOSIP.664 and other DS-SOSIP.664 variants to the first two domains of CD4 (D1D2), and we observed BG505 A329P-DS-SOSIP.664 to have CD4-binding affinity very similar to that of DS-SOSIP.664, while the CD4 affinities for BG505 DS-SOSIP.664 variants containing the N302M-T320L mutations were much lower (Fig. 4C and 5). In addition, negative-stain electron microscopy (EM) on the purified DS-SOSIP.664 variants showed that BG505 3mut-DS-SOSIP.664 and other DS-SOSIP.664 variants formed uniform trimers in a prefusion-closed conformation without monomers or aggregates (Fig. 6).
FIG 5.
Binding curves for soluble CD4 to stabilized BG505 Env trimers as measured by SPR with single-cycle kinetics. For BG505 SOSIP.664, DS-SOSIP.664, and A329P-DS-SOSIP.664, the concentrations of CD4 injected were 180, 90, 45, 22.5, and 11.25 nM. For other DS-SOSIP.664 mutants, the concentrations of CD4 injected were 1,000, 500, 250, 125, and 62.5 nM.
FIG 6.
Negative-stain electron microscopy class averages of stabilized BG505 Env trimers. Reference-free classification and averaging produced symmetrical propeller-like classes typical of the prefusion-closed conformation of the HIV-1 envelope trimer, indicative of high homogeneity and correct folding/assembly of the proteins. Scale bars = 10 nm.
To evaluate the immunogenicity of BG505 3mut-DS-SOSIP.664, we immunized guinea pigs at weeks 0, 4, and 16 with BG505 SOSIP.664, BG505 DS-SOSIP.664, or BG505 3mut-DS-SOSIP.664 and evaluated serum neutralization at week 18 (Fig. 4D). While there was no statistically significant difference in autologous neutralization levels among the three groups (Fig. 4D, left), the neutralization of a V3-sensitive tier 1 strain, MW965.26, by sera from BG505 3mut-DS-SOSIP.664-immunized animals was significantly lower than that by sera from animals immunized with BG505 SOSIP.664 or BG505 DS-SOSIP.664 (Fig. 4D, right).
Stabilization of a clade C Env trimer from donor CAP256.
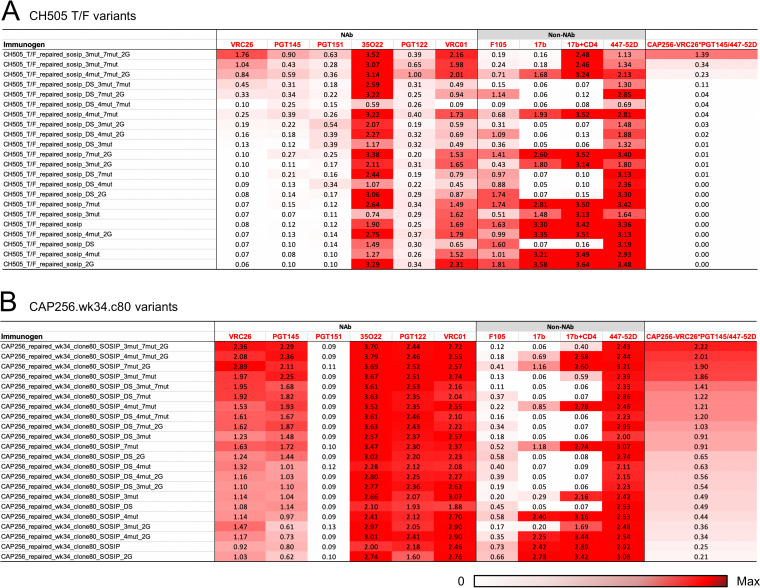
We had previously shown that boosting FP-directed broadly neutralizing responses with a heterologous clade C trimer, derived from transmitted founder virus of donor CH505, could increase the consistency of inducing broad cross-clade neutralizing responses (28), highlighting the utility of sequential boosting with a heterologous trimer. The CH505 trimer that we used for boosting, however, was a chimera, with the sequences of gp41 as well as the N- and C termini of gp120 being swapped with that of BG505. As such swapping might have decreased the benefit of heterologous boosting, we decided to test the ability of 3mut, in combination with other forms of stabilization, to allow correctly folded soluble Env trimers in which the gp41 had also been switched from BG505. We chose to stabilize two clade C variants, the previously tested transmitted founder virus from donor CH505, as well as the week 34 (clone 80) version of the superinfecting strains of CAP256 (CAP256.wk34.c80), a strain which likely triggered the development of the broadly neutralizing CAP256-VRC26 antibody lineage (29). We designed soluble CH505 and CAP256.wk34.c80 variants incorporating combinations of the 3mut mutations developed in this study with other stabilizing methods, including SOSIP mutations (10), DS mutations (36), glycine helix-breaking mutations (16), repair-and-stabilize (RnS) mutations (21), and 4mut mutations (6) (Fig. 7A). We used a 96-well antigenic screening platform to assess these constructs based on an antigenic score calculated from ELISA antibody binding outputs as (VRC26*PGT145)/447-52D (Fig. 8). Notably, only one of the CH505 variants showed good antigenicity, whereas good antigenicity was observed for a number of CAP256 variants; in both cases, the highest antigenic score was observed for the RnS-3mut-2G-SOSIP.664 variant, which combined 3mut with RnS and two glycine substitutions (2G).
FIG 7.
Development of a stabilized clade C trimer utilizing a week 34 (wk34) virus from donor CAP256. (A) Stabilization strategies employed to screen for stabilized CAP256.wk34.80 (Chuang et al. [6], Sanders et al. [10], Guenaga et al. [16], Rutten et al. [21], and Kwon et al. [36]). The stabilizing mutations used in RnS were 204I, 535N, 573F, 588E, 589V, 651F, and 655I. (B) SEC profile of CAP256 RnS-3mut-2G-SOSIP.664. (C) Thermostability of CAP256_RnS-3mut-2G SOSIP trimers assessed by DSC. (D) Negative-stain electron microscopy class averages of CAP256 RnS-3mut-2G-SOSIP.664. (E) Antigenicity of CAP256 RnS-3mut-2G-SOSIP.664 as determined by MSD. BG505 DS-SOSIP.664 was used as a control.
FIG 8.
Antigenic screening of soluble clade C trimer variants using a 96-well microplate ELISA. The variants are sorted by (CAP256 VRC26*PGT145)/447-25D, a metric aimed to identify prefusion-closed Env variants that had high affinity to prefusion-closed specific V1/V2 antibodies and low binding to weak neutralizing V3 antibodies. (A) CH505 variants. (B) CAP256.wk34.c80 variants. 7mut, 204I, 535N, 573F, 588E, 589V, 651F, and 655I; 2G, 569G and 636G.
When expressed at the 1-liter scale, CH505 RnS-3mut-2G-SOSIP.664 had low yield, whereas CAP256 RnS-3mut-2G-SOSIP.664 had high yield and showed a single-peak SEC profile after 2G12 affinity chromatography (Fig. 7B). The CAP256 RnS-3mut-2G-SOSIP.664 trimer showed good thermostability, with a Tm of 77°C (Fig. 7C), and formed uniform trimers in the closed prefusion conformation without monomers or aggregates (Fig. 7D). The CAP256 RnS-3mut-2G-SOSIP.664 also showed high binding affinity toward CAP256-VRC26.25 when evaluated with MSD, suggesting the trimer to be stabilized in the prefusion-closed form (Fig. 7E), despite the low binding to PGT145 (somewhat surprising in light of the high ELISA output from the 96-well antigenic assessment). The reduced binding of CAP256 RnS-3mut-2G-SOSIP.664 trimer to a number of broadly neutralizing antibodies, such as 2G12, VRC01, PGT151, and 8ANC195, might be related to the intrinsic resistance of the CAP256 viral strain toward these antibodies (Fig. 9); the reduced binding of 2G12 may be especially problematic in the MSD-formatted assays, as labeled 2G12 is used for detection (see Materials and Methods). Thus, while CAP256 RnS-3mut-2G-SOSIP.664 showed decent trimer expression and melting temperature, its antigenicity indicated some V3 exposure, especially in the presence of soluble CD4, suggesting this clade C trimer to be less fixed than BG505 DS-SOSIP.664 in the prefusion-closed state.
FIG 9.
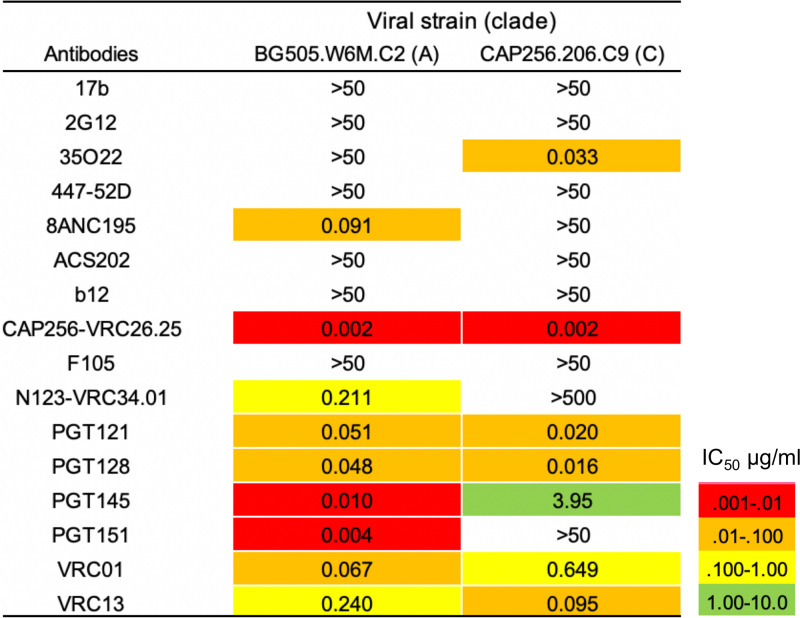
Neutralization 50% inhibitory concentrations (IC50s) of BG505.W6M.C2 and CAP256.206.C9 strains for antibodies used in MSD analyses.
Structure of the stabilized clade C Env trimer from donor CAP256.
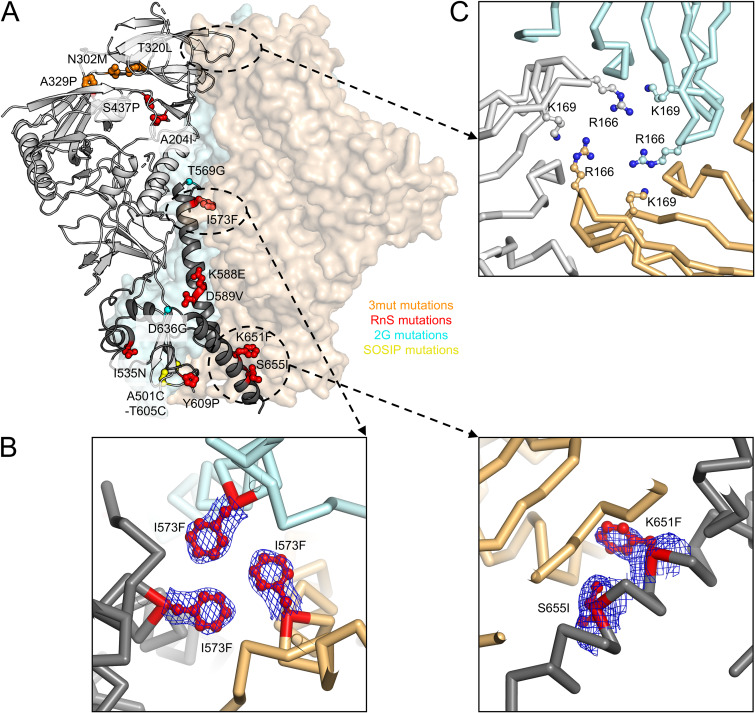
We again used the engineered crystallization chaperones 35O22_scFv and 3H109L_MM (30) to obtain a crystal structure of the CAP256 RnS-3mut-2G-SOSIP.664 trimer at a nominal resolution of 3.5 Å (Fig. 10A and Table 1). Electron density was well defined for most regions of the CAP256 RnS-3mut-2G-SOSIP.664 trimer. Several mutations introduced at the interface between the three protomers were clearly defined by electron density (Fig. 10B), consistent with them forming stable interactions.
FIG 10.
Structure of CAP256 RnS-3mut-2G-SOSIP.664 provides details on how multiple mutations stabilize a clade C soluble trimer. Previous studies have shown that CAP256 Env sequence cannot be expressed as soluble trimer by DS-SOSIP stabilization. (A) Crystal structure of the stabilized CAP256 trimer (CAP256 RnS-3mut-2G-SOSIP.664, with stabilizing mutations highlighted in stick-and-ball representation and colored according by corresponding mutation categories) in one protomer shown as ribbons (gp120 in light gray and gp41 in dark gray). The other two protomers are shown as surface representations in orange and cyan, respectively. (B) RnS mutations, highlighted in red, likely stabilize the trimerization of gp41. Electron density maps (contoured at 1.5 sigma) are shown for the mutated side chains. (C) R166 and K169 from each protomer form a positively charged patch at the trimer apex that can interact with negatively charged CDRH3 of VRC26.25 and other V1/V2 antibodies.
We also analyzed the trimer apex to gain insight into the especially tight recognition by VRC25.26. Residues R166 and K169 formed a positive patch, made especially cationic by their close proximity to the trimer 3-fold (Fig. 10C). The sulfated anionic tips of antibodies PGT145 and VRC26.25 have recently been found to penetrate the hole formed at the trimer apex (33, 37, 38), providing an explanation for the very tight binding of VRC25.26, though not of the weaker binding of PGT145.
Development of stabilized Env trimers with the second most prevalent fusion peptide.
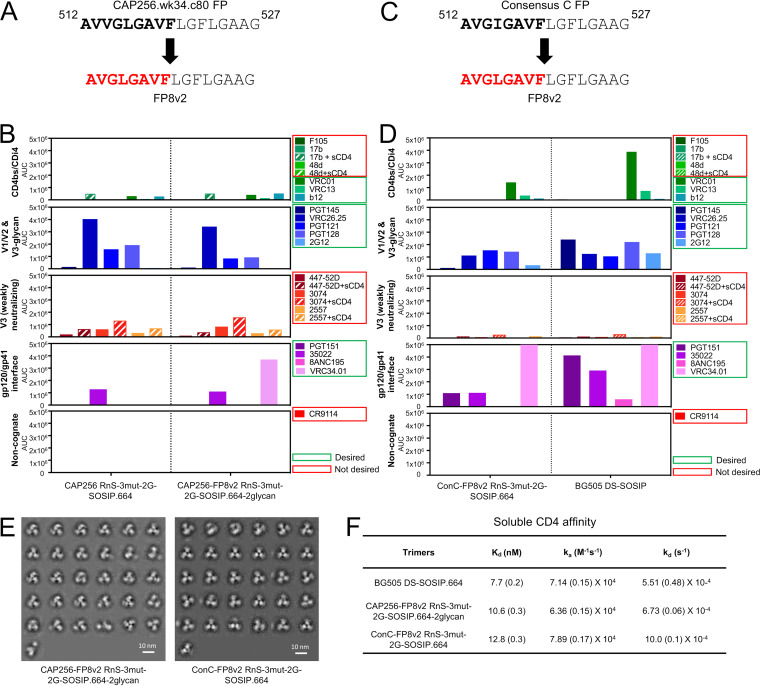
The CAP256 RnS-3mut-2G-SOSIP.664 trimer showed very poor recognition of the fusion peptide-directed antibody VRC34.01 (Fig. 7E). Analysis of the CAP256 FP sequence revealed an unusual insertion (Fig. 11A); however, the crystal structure of CAP256 RnS-3mut-2G-SOSIP.664 indicated residues 512 to 517 to be disordered, while CAP256 FP in a recently published cryo-EM structure was associated with some helical density (39). As a key factor governing the elicited neutralization breadth appeared to be variation in the FP sequence itself, to expand the neutralization breadth elicited by fusion peptide and BG505 trimer with the most prevalent first eight residues of the fusion peptide (FP8v1, AVGIGAVF), we substituted the native fusion peptide sequence of CAP256 RnS-3mut-2G-SOSIP.664 with the second most prevalent FP8 (FP8v2, AVGLGAVF) (Fig. 11A). Two glycan sequon mutations (R502N and R504T, and L660N and A662T) that introduced glycans to mask the exposed membrane-facing base were also incorporated to this construct (CAP256-FP8v2 RnS-3mut-2G-SOSIP.664-2glycan). The expression of the CAP256-FP8v2 RnS-3mut-2G-SOSIP.664-2glycan was lower than that of CAP256 RnS-3mut-2G-SOSIP.664; nonetheless, when evaluated by MSD, the purified CAP256-FP8v2 RnS-3mut-2G-SOSIP.664-2glycan showed antigenicity similar to that of the native FP version, with the notable exception that it was now capable of binding VRC34.01, unlike its native FP counterpart (Fig. 11B).
FIG 11.
Identified stabilizations yield well-behaved CAP256 and consensus-clade C trimers with FP8v2 (AVGLGAVF). (A) Replacement of native CAP256.wk34.c80 FP8 with FP8v2. (B) Antigenicity, as determined by MSD, of CAP256-FP8v2 RnS-3mut-2G-SOSIP.664. (C) Replacement of native consensus C FP8 with FP8v2. (D) Antigenicity, as determined by MSD, of ConC-FP8v2 RnS-3mut-2G-SOSIP.664. (E) Negative-stain electron microscopy class averages of CAP256-FP8v2 RnS-3mut-2G-SOSIP.664 (left) and ConC-FP8v2 RnS-3mut-2G-SOSIP.664 (right). (F) Binding parameters of stabilized clade C HIV-1 Env trimers to soluble CD4. Values in parentheses report standard errors from fitting data to a 1:1 Langmuir binding model.
We also developed another stabilized FP8v2 HIV-1 Env trimer from consensus clade C sequence by the incorporation of SOSIP.664, RnS, 3mut, and 2G (ConC-FP8v2 RnS-3mut-2G-SOSIP.664 [Trimer 6931]) (Fig. 11C and D). Both CAP256-FP8v2 RnS-3mut-2G-SOSIP.664-2glycan and ConC-FP8v2 RnS-3mut-2G-SOSIP.664 assumed prefusion-closed conformation based on negative-stain EM (Fig. 11E). Despite not having the “DS” mutations, CAP256-FP8v2 RnS-3mut-2G-SOSIP.664-2glycan and ConC-FP8v2 RnS-3mut-2G-SOSIP.664 had an affinity toward soluble CD4 similar to that of BG505 DS-SOSIP.664 (Fig. 11F) and lower than that of BG505 SOSIP.664 (6, 36), consistent with the prefusion-closed conformation.
Boosting immunization with ConC-FP8v2 RnS-3mut-2G-SOSIP.664 expands elicited neutralization breadth in guinea pigs.
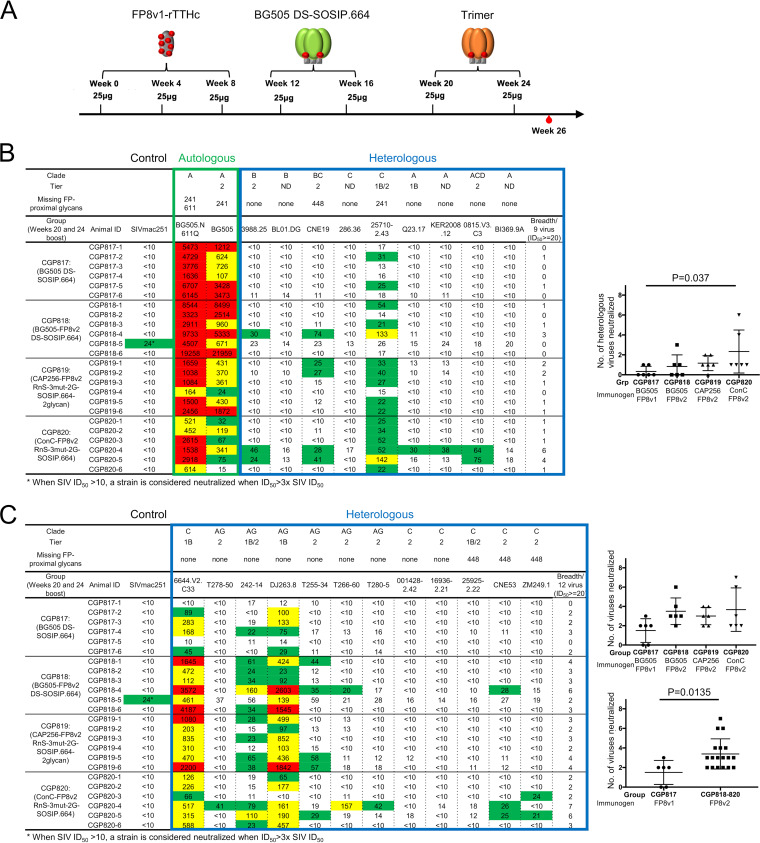
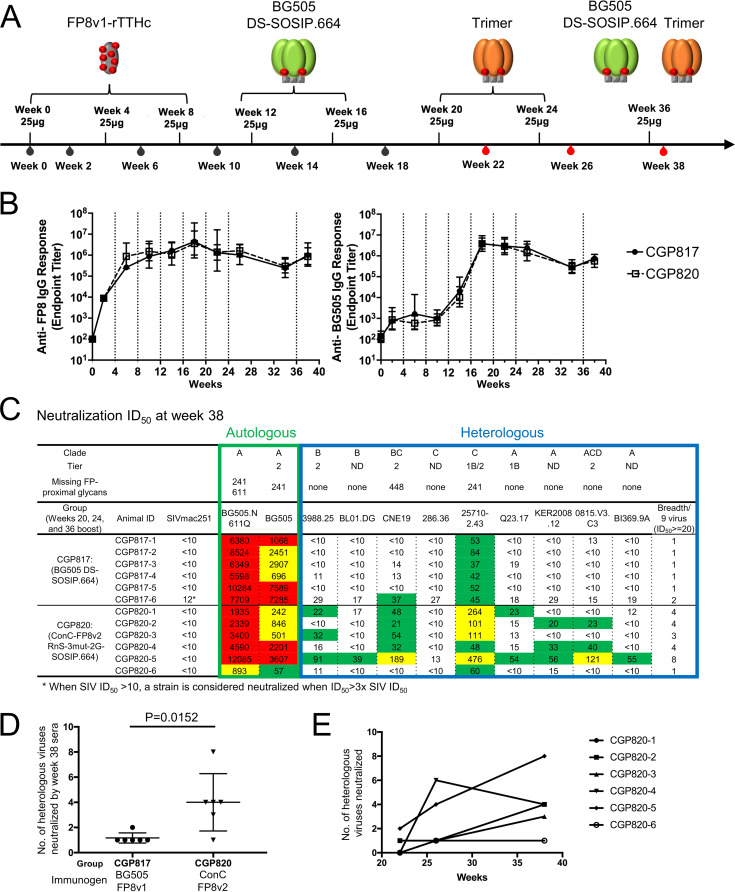
To evaluate the utility of CAP256-FP8v2 RnS-3mut-2G-SOSIP.664-2glycan and ConC-FP8v2 RnS-3mut-2G-SOSIP.664 as boosting reagents for FP-targeting immunizations, we used them to immunize guinea pigs that have been preimmunized with three injections of FP8v1 conjugated to a recombinant tetanus toxoid heavy-chain (rTTHC) fragment (FP8v1-rTTHC) at weeks 0, 4, and 8 and two boosts with BG505 DS-SOSIP.664 at weeks 12 and 16. These guinea pigs were further boosted at weeks 20 and 24 with CAP256-FP8v2 RnS-3mut-2G-SOSIP.664-2glycan, ConC-FP8v2 RnS-3mut-2G-SOSIP.664, BG505 DS-SOSIP.664, or BG505 DS-SOSIP-FP8v2 (Fig. 12A). When evaluated against a 9-strain panel of non-BG505 strains containing the FP8v1 sequence, week 26 serum from the group that was boosted with ConC-FP8v2 RnS-3mut-2G-SOSIP.664 (CGP820) showed the highest neutralization breadth, significantly higher than that of the serum from the group boosted with BG505 DS-SOSIP.664 (CGP817, P = 0.037) (Fig. 12B). When evaluated on a 12-strain panel of FP8v2 strains, sera from all three groups that were boosted with FP8v2 trimers (CGP818 to CGP820) showed substantially higher neutralization breadth than did sera from group CGP817, for which an FP8v1 trimer was used as the final boosts (Fig. 12C). Meta-analysis by combining groups CGP818 to CGP820 showed that boosting with FP8v2 trimer elicited significantly higher FP8v2 neutralization breadth than did boosting with FP8v1 trimer (P = 0.0135) (Fig. 12C, bottom right). To examine if the elicited neutralization was targeting the HIV-1 Env fusion peptide, we performed competition neutralization assays. In the presence of a peptide containing the first nine residues of fusion peptide (AVGIGAVFL, FP9), guinea pig serum neutralization against strains 25710-2.43 and CNE19 was substantially reduced (Table 2), indicating that the majority of the immunization-elicited neutralization response was targeting the fusion peptide.
FIG 12.
The FP8v2 version of consensus-clade C trimer elicits increased neutralization breadth when used to boost FP-directed vaccine responses in guinea pigs. (A) Immunization scheme. (B) Week 26 serum neutralization of autologous BG505 viruses (boxed in green) and nine heterologous FP8v1 viruses (boxed in blue). Statistical comparison was performed between CGP817 and CGP818, CGP819, or CGP820 using a Kruskal-Wallis test with P values calculated with a post hoc Dunn’s multiple-comparison test. A P value of less than 0.05 is displayed. (C) Week 26 serum neutralization of 12 FP8v2 viruses. Statistical comparison was performed between CGP817 and CGP818-820 using two-tailed Mann-Whitney test. A strain is considered neutralized by the sera when the ID50 is equal to or greater than the higher number between 20 and 3× the simian immunodeficiency virus in macaques (SIVmac) ID50 of the given sample. ID, identifier.
TABLE 2.
Competition assay suggests heterologous HIV-1 strains to be neutralized by fusion peptide-directed plasma responses
| Group | CGP ID | Virus | Plasma dilution or antibody concn | Antibody concn (μg/ml) | Medium %Neut | FP9 |
Scrambled FP9 |
||
|---|---|---|---|---|---|---|---|---|---|
| %Neut | Reduction in Neut (%)a | %Neut | Reduction in Neut (%) | ||||||
| Guinea pig sera | 817-2 | 25710-2.43 | 20 | 77.3 | 48.8 | 37 | 78.3 | −1 | |
| 817-5 | 25710-2.43 | 20 | 53.6 | 35.0 | 35 | 55.4 | −3 | ||
| 820-1 | 25710-2.43 | 20 | 61.7 | 56.7 | 8 | 59.6 | 3 | ||
| 820-2 | 25710-2.43 | 20 | 68.4 | 29.0 | 58 | 66.0 | 3 | ||
| 820-3 | 25710-2.43 | 20 | 65.0 | 60.7 | 6 | 67.7 | −4 | ||
| 820-4 | 25710-2.43 | 20 | 71.2 | 42.2 | 41 | 72.1 | −1 | ||
| CNE19 | 20 | 55.3 | 31.9 | 42 | 55.4 | 0 | |||
| 820-5 | 25710-2.43 | 20 | 88.6 | 7.0 | 92 | 82.0 | 7 | ||
| CNE19 | 20 | 57.6 | 4.8 | 92 | 53.3 | 7 | |||
| 820-6 | 25710-2.43 | 20 | 50.2 | 22.6 | 55 | 49.1 | 2 | ||
| FP antibody | VRC34 | 25710-2.43 | 50 | 45.2 | 8.4 | 81 | 42.2 | 7 | |
| CNE19 | 50 | 73.0 | 53.1 | 27 | 70.4 | 4 | |||
| CD4bs antibody | VRC01 | 25710-2.43 | 2 | 75.0 | 75.5 | −1 | 74.8 | 0 | |
| CNE19 | 0.9 | 75.6 | 74.3 | 2 | 75.1 | 1 | |||
Values in reduction in neutralization of greater than the average plus three standard deviations of the scramble FP9 values are shown in bold.
To examine the longitudinal development of neutralization breadth for the guinea pig group (CGP820) that was boosted with ConC-FP8v2 RnS-3mut-2G-SOSIP.664, we evaluated the neutralization levels of the nine FP8v1 viruses at week 22 and week 38. The latter time point was 2 weeks after the animals were boosted with a cocktail of BG505 DS-SOSIP.664 and ConC-FP8v2 RnS-3mut-2G-SOSIP.664 (Fig. 13A). Longitudinal development of responses to both fusion peptide and BG505 trimer were similar across groups, regardless of the Env trimer used for additional boosting (Fig. 13B). FP ELISA titers increased through the first two FP8v1-rTTHC immunizations and plateaued after week 6 (Fig. 13B, left). Anti-BG505 trimer ELISA titers increased after the first BG505 DS-SOSIP.664 immunization and peaked after the second BG505 DS-SOSIP.664 immunization (Fig. 13B, right). Five out of the six sera from week 38 neutralized at least three out of the nine viruses, higher than what was observed from week 26, where only two of the six sera neutralized at least three out of the nine viruses (Fig. 12B and 13C). Similar to the week 26 data, at week 38, group CGP820 showed higher serum neutralization breadth than did CGP817, the guinea pig group boosted with only BG505 DS-SOSIP.664 (Fig. 13C and D). Finally, all six animals in group CGP820 showed higher neutralization breadth at week 38 than at week 22, and four of the six animals showed higher neutralization breadth at week 38 than week 26 (Fig. 13E).
FIG 13.
Longitudinal analyses of FP8v2-consensus-clade C trimer elicited responses reveals increased neutralization of heterologous HIV-1 strains. (A) Immunization scheme. Black blood symbol indicates time points for which serum ELISA was conducted. Red blood symbol indicates time points for which both serum ELISA was conducted and neutralization was assessed. (B) Longitudinal FP-specific serum immunogenicity (left), as measured by ELISA against FP8 peptide, and trimer-specific serum immunogenicity (right), as measured by ELISA against HIV-1 BG505 DS-SOSIP.664, are shown for groups CGP817 and CGP820. Immunization time points are indicated by vertical dotted lines. (C) Week 38 serum neutralization of autologous BG505 viruses (boxed in green) and nine heterologous FP8v1 viruses (boxed in blue). (D) Statistical comparison of week 38 breadth between groups CGP817 and CGP820 using two-tailed Mann-Whitney test. (E) Longitudinal development of neutralization breadth of each animal in group CGP820.
DISCUSSION
In this study, we identify a set of three mutations, N302M, T320L, and A329P (3mut), that increased thermostability while reducing CD4 binding, reducing antigenic responses to nonneutralizing or weakly neutralizing V3 antibodies, and reducing the elicitation of tier 1-only neutralizing titers. These mutations stabilize the apex of the Env trimer in a prefusion-closed conformation. Two of the mutations, N302M and T320L, which are part of the published 4mut mutations (6), create an energetically favorable hydrophobic patch between V1/V2 and V3. The A329P mutation restricts the backbone in the prefusion-closed conformation, as the dihedral angles of residue 329 in the CD4-bound conformation is not compatible with a proline. The use of proline to restrict viral antigen conformations has been reported frequently in the recent literature (36, 40–43).
The structures of Env trimers with 3mut determined here provide atomic-level information on these mutations in the context of clade A BG505 and clade C CAP256.wk34.c80. As for BG505 3mut-DS-SOSIP.664, residues M302 and L320 formed a hydrophobic patch with I154 (L in BG505 3mut-DS-SOSIP.664) and Y177 in CAP256 RnS-3mut-2G-SOSIP.664 structure, and the two structures have very similar phi-psi angles at residue P329 (−78.5° and 136.4° for BG505 3mut-DS-SOSIP.664 and −76.2° and 151.9° for the CAP256 RnS-3mut-2G-SOSIP.664 structure, respectively). Overall, our results indicate that the 3mut-apex-stabilizing mutations developed here should be generally applicable to other Env trimers as a means to enhance their prefusion-closed antigenic and physical properties. We also calculated the root mean square deviation (RMSD) between a unliganded BG505 SOSIP.664 structure (PDB 4ZMJ) and a BG505 SOSIP.664 structure in complex with the crystallization chaperone used in this study (PDB 6MTN), and the overall RMSD between these two structures was only 2.0 Å, suggesting the crystallization chaperone to have a minimal effect on the Env conformation.
In this study, we showed that boosting with FP8v2 trimers following prior immunizations with FP8v1 immunogens could expand the elicited breadth against FP8v2 isolates, suggesting the utility of these trimers in the FP immunization regimens to cover additional antigenic variations. It was also interesting to see that boosting with a heterologous trimer harboring FP8v2 sequence following BG505 DS-SOSIP.664 boosts elicited higher FP8v1 neutralization than that with continuous boosting with BG505 DS-SOSIP.664, suggesting that guiding the immune system to recognize the sequence diversity of the non-FP region of the Env trimer is a critical step for the development of the heterologous neutralization breadth.
While boosting with ConC-FP8v2 RnS-3mut-2G-SOSIP.664 showed statistically significant improvement of neutralization breadth over boosting with BG505 DS-SOSIP.664 only at week 26 (Fig. 12B), the neutralization titer and breadth were modest. The consistency, breadth, and potency of neutralization from CGP820 sera were further enhanced after being boosted with a cocktail of ConC-FP8v2 RnS-3mut-2G-SOSIP.664 and BG505 DS-SOSIP.664 (Fig. 13). By using an ID50 (reciprocal plasma dilutions causing 50% reduction in virus entry) threshold of 90, which was implicated in an NHP study as the titer required to achieve a 50% chance of protection against infection by a challenge strain (44), serum from one animal neutralized four out of nine heterologous viruses tested, and sera from three other animals neutralized one virus. A neutralization titer of greater than 500, which was implicated as an requirement to achieve a 90% chance of protection against an autologous strain (44), was observed only against tier 1B FP8v2 viruses (Fig. 12C). Further optimization of the immunogens, immunization interval, adjuvants, and immunization scheme is necessary to achieve the elicitation of neutralization titers sufficient for protection from HIV-1.
The RnS-3mut-2G-SOSIP combination was able to stabilize CAP256 and ConC in the prefusion-closed conformation. However, with the CAP256 variant, the antigenicity in the presence of CD4 was not as good as that of BG505 DS-SOSIP.664, suggesting that the addition of DS might be helpful in stabilizing the prefusion-closed conformation in the presence of CD4. It would be interesting to test the RnS-3mut-2G combination with DS-SOSIP.664 on a large panel of strains to see if such a stabilization strategy is improved over chimeric variants of DS-SOSIP.664 (21) or if it is even universal.
MATERIALS AND METHODS
High-throughput antigenic analysis of HIV-1 Env variants in a 96-well microplate by ELISA.
D7324 antibody-coated 96-well ELISA plates were prepared by incubating 2 μg/ml affinity-purified sheep anti-HIV-1 gp120 antibody (catalog no. D7324; Aalto Bio Reagents, Ireland) in 100 μl phosphate-buffered saline (PBS) in a 96-well flat-bottom Nunc-Immuno plate (Thermo, IL) overnight at 4°C, followed by the removal of the coating solution and incubation of the wells with 200 μl of 2% (wt/vol) dry milk in PBS overnight at 4°C. The wells were then washed 5 times with PBS plus 0.05% Tween 20. Each of the HIV-1 Env trimer constructs contained a C-terminal D7324 epitope tag (APTKAKRRVVQREKR). Thirty microliters of supernatant expressed in each well of the 96-well microplate was incubated with 70 μl of PBS in each well of a D7324 antibody-coated 96-well ELISA plate for 1 h at 60°C, after which the wells were washed 5 times with PBS plus 0.05% Tween 20. One hundred milliliters of anti-specific epitope primary antibody at a concentration of 10 μg/ml in PBS with 0.2% (wt/vol) dry milk and 0.2% Tween 20 was added to each well and incubated for 1 h at room temperature (RT), after which the wells were washed 5 times with PBS plus 0.05% Tween 20. One hundred milliliters of horseradish peroxidase (HRP)-conjugated goat anti-human IgG antibody (Jackson ImmunoResearch Laboratories, Inc., PA) at 1:10,000 in PBS with 1.0% (wt/vol) dry milk and 0.2% Tween 20 was added to each well and incubated for 30 min at RT, after which the wells were washed 5 times with PBS plus 0.05% Tween 20. The wells were developed with 3,3′,5,5′-tetramethylbenzidine (TMB) at RT for 10 min, after which the reaction was stopped with 180 mM HCl. The readout was measured at a wavelength of 450 nm. All samples were read in duplicate. For BG505 variants, the antigenicity was assessed for the following antibodies: VRC01, VRC13, CAP256-VRC26, PGT145, F105, 3074, 447-52D, and CD4-Ig. For CAP256 variants, the antigenicity for the following antibodies were assessed: CAP256-VRC26.25 (34), PGT145 (45), PGT151 (46), 35O22 (47), PGT121 (45), VRC01 (48), F105 (49), 17b (50), 17b+CD4, and 447-52D (51).
Env expression and purification.
The various BG505, CAP256, and ConC SOSIP trimer mutants were produced in transiently transfected 293 FreeStyle cells, as described previously (52). Briefly, 600 μg of Env trimer construct was cotransfected with 150 μg of furin plasmid DNA into 1 liter of cells and cultured for 6 days. The transfected supernatants were harvested, filtered, and loaded over a VRC01 affinity column (BG505 and ConC) or a 2G12 affinity column (CAP256). The bound proteins were eluted with 3 M MgCl2 and 30 mM Tris at a pH of 7.0, followed by concentrating with Amicon Ultracel-50K (Millipore) and size exclusion chromatography using a Superdex 200 16/600 column (GE Healthcare). The peak corresponding to trimeric HIV-1 Env was collected and subjected to negative selection with 447-52D (PDB ID 4M1D) and V3 cocktail columns to remove aberrant trimer species. The V3 cocktail column contains 6 V3-directed antibodies, 1006-15D (53), 2219 (54), 2557 (53), 2558 (55), 3074 (53), and 50.1 (56). BG505 DS-SOSIP.664 (FP8v1) and ConC trimers used in immunizations were produced in stable CHO cell lines and purified using nonaffinity chromatography. The 293F- and CHO-produced trimers exhibit nearly identical antigenicity, as assessed by MSD.
Antigenic analysis of DS-SOSIP.664 variants by MSD-ECLIA.
Standard 96-well bare Multi-Array plates (catalog no. L15XA-3; MSD) were coated with all or a subset of a panel of HIV-neutralizing (VRC01 [48], b12 [57], VRC13 [58], PGT121 [45], PGT128 [45], 2G12 [59], PGT145 [45], CAP256-VRC26.25 [34], 35O22 [47], 8ANC195 [60], PGT151 [46], and VRC34.01 [61]) and nonneutralizing or weakly neutralizing monoclonal (F105 [49], 17b [50] [± soluble CD4 {sCD4}], 48D [62] [±sCD4], 447-52D [51] [±sCD4], 3074 [53, 63] [±sCD4], 2557 [53] [±sCD4]) and noncognate (anti-influenza antibody CR9114 [64]) antibodies in duplicate (30 μl/well) at a concentration of 4 μg/ml diluted in 1× PBS by incubating overnight at 4°C. The following day, the plates were washed (wash buffer, 0.05% Tween 20 plus 1× PBS) and blocked with 150 μl of blocking buffer (5% [wt/vol] blocker A [catalog no. R93BA-4; MSD]) by incubating for 1 h on a vibrational shaker (Heidolph Titramax 100, catalog no. 544-11200-00) at 650 rpm. All incubations were performed at room temperature except for the coating step. During the incubation, trimers were titrated in serial 2× dilutions starting at a concentration of 5 μg/ml of the trimer in the assay diluent (1% [wt/vol] MSD blocker A plus 0.05% Tween 20). For sCD4 induction, the trimer was combined with sCD4 at a constant molar concentration of 1 μM before being added to the MSD plate. After the incubation with blocking buffer was complete, the plates were washed, and the diluted trimer was transferred (25 μl/well) to the MSD plates and incubated for 2 h on the vibrational shaker at 650 rpm. After the 2-h incubation with trimer, the plates were washed again and 2G12 antibody labeled with Sulfo-Tag (catalog no. R91AO-1; MSD) at a conjugation ratio of 1:15 (2G12:Sulfo-Tag), which was diluted in assay diluent at 2 μg μg/ml, added to the plates (25 μl/well), and incubated for 1 h on the vibrational shaker at 650 rpm. The plates were washed and read using read buffer (Read Buffer T, catalog no. R92TC-1; MSD) on the MSD Sector Imager 2400 or equivalent instrument.
Negative-stain electron microscopy.
Samples were diluted to ∼0.01 to 0.02 mg/ml with a buffer containing 10 mM HEPES (pH 7) and 150 mM NaCl and adsorbed to a freshly glow-discharged carbon-coated grid for 15 s. The grid was washed with the above-described buffer, and adsorbed proteins were stained with 0.75% uranyl formate. Micrographs were collected using SerialEM (65) on a FEI Tecnai T20 microscope operated at 200 kV and equipped with a 2K-by-2K Eagle charge-coupled-device (CCD) camera. The pixel size was 0.22 nm. Particles were picked automatically using in-house written software (Y. Tsybovsky, unpublished data) and extracted using a box size of 128 by 128 pixels. Reference-free two-dimensional (2D) classification was performed with EMAN2 (66) and Relion 1.4 (67).
Surface plasmon resonance analysis.
Single-cycle kinetics analysis was performed to access the binding affinities and kinetics of soluble CD4 (sCD4) to HIV-1 DS-SOSIP.664 various trimers on Biacore T-200 (GE Healthcare) at 25°C with HBS-EP+ buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 3 mM EDTA, and 0.05% surfactant P-20). First, 2G12 antibody was immobilized on flow cells of a CM5 chip at ∼2,000 response units. Next, 200 nM trimer was captured onto the sample flow cell at a flow rate of 5 μl/min for 120 s. Finally, sCD4 at five concentrations (180 nM, 90 nM, 45 nM, 22.5 nM, and 11.25 nM for BG505 SOSIP.664 and BG505 DS-SOSIP.664, and 500 nM, 250 nM, 125 nM, 62.5 nM, and 31.25 nM for optimized DS-SOSIP.664 trimers) was injected incrementally in a single cycle, starting from the lowest concentration at a flow rate of 50 μl/min for 60 s, which was followed by a dissociation phase of 30 min. Blank sensorgrams were obtained by injection of the same volume of HBS-EP+ buffer in place of sCD4. Sensorgrams of the concentration series were corrected with corresponding blank curves and fitted globally with Biacore T200 evaluation software with a 1:1 Langmuir model of binding.
Differential scanning calorimetry.
The heat capacity of the trimers was measured by a high-precision differential scanning VP-DSC microcalorimeter (GE Healthcare/MicroCal). All HIV-1 Env trimers were diluted to 0.3 mg/ml with PBS. Thermal denaturation scans were performed from 30°C to 110°C at a rate of 1°C/min.
HIV-1 Env trimer crystallization, X-ray data collection, structure solution, and model building.
HIV-1 trimers were expressed in cells lacking N-acetylglucosaminyltransferase I (GnTi− cells; catalog no. CRL-3022; ATCC) to ensure a high-mannose composition of glycan. Trimers were purified by a 2G12 affinity column, followed by size exclusion chromatography. Purified trimers were mixed with a variant of 3H+109L Fab and a variant of 35O22 scFv in a 1:3.2:3.2 molar ratio (gp140 protomer/Fab/scFv) and incubated overnight at room temperature. The 3H+109L variant contained two methionine substitutions in the light chain, and the 35O22 variant three contained threonine and two serine mutations in the heavy chain. These alterations were designed to enhance the crystallization lattice (30). Glycans on the Env and antibody fragments were preserved without treatment with glycosidase. Complexes were further purified by SEC and concentrated to 10 to 15 mg/ml for crystallization. Crystals of the BG505 DS-SOSIP.664 3mut in complex with 3H+109L Fab and 35O22 scFv were grown in 75 mM imidazole (pH 6.5), 150 mM LiSO4, 3.8% polyethylene glycol (PEG) MPD, and 6.2% PEG 3350, with a protein-to-reservoir ratio of 1:1 μl. Crystals of the CAP256 RnS-3mut-2G-SOSIP.664 in complex with 3H+109L Fab and 35O22 scFv were grown in 60 mM sodium acetate (pH 4.6), 120 mM ammonium sulfate, and 6.3% PEG 4000, with a protein-to-reservoir ratio of 0.5 μl/0.5 μl.
Crystals were harvested and incubated under crystallization conditions supplemented with 15 to 20% 2R-3R butanediol as a cryo-protectant for 10 s before freezing in liquid nitrogen, for both BG505 3mut-DS-SOSIP.664 and CAP256 RnS-3mut-2G-SOSIP.664 crystals. Diffraction data were collected at SER-CAT beamline ID22 of the Advanced Photo Source at the Argonne National Laboratory. HKL-2000 (68) was used to process the diffraction data; the overall resolution was determined as the highest resolution for which the completeness was greater than 50% and the I/σI was greater than 2.0. Data were then truncated using the University of California, Los Angeles (UCLA) anisotropy server (https://services.mbi.ucla.edu/anisoscale/). A structure of BG505 SOSIP.664 in complex with PGT122 and 35O22 (PDB 4TVP) was modified by using Sculptor (69) to change the BG505 sequence in the model to the corresponding CAP256 RnS-3mut-2G-SOSIP.664 sequence and to change PGT122 Fab to 3H+109L Fab; the constant domain in 35O22 Fab in the 4TVP.pdb was removed to match to 35O22 scFv used for crystallization. Molecular replacement was carried out by using Phaser (70), utilizing the modified 4TVP.pdb as the search model. Refinement and structure evaluation were carried out in Coot (71) and Phenix (72).
Animal protocols and immunization.
All animal experiments were reviewed and approved by the Animal Care and Use Committee of the Vaccine Research Center at the NIAID (NIH). The animal work was covered under protocol VRC no. 16-552 amendment 8. Animals were housed and cared for in accordance with local, state, federal, and institute policies in an American Association for Accreditation of Laboratory Animal Care-accredited facility at the Vaccine Research Center.
Female Hartley guinea pigs with body weights of more than 300 g were obtained from Charles River Laboratories (Wilmington, MA) and used for immunization studies. For each immunization, 0.4 ml of immunogen mix, containing 25 μg of specified, filter-sterilized immunogen and 80 μl of Adjuplex (Advanced BioAdjuvants LLC, Omaha, NE) in PBS was injected via a needle syringe to the caudle thigh of the two hind legs. Blood was collected for serological analyses.
Neutralization.
The neutralization protocol was modified from reference 73. Sera were 5-fold serially diluted in complete Dulbecco’s modified Eagle’s medium (cDMEM), and 10 μl of diluted serum was incubated with 40 μl of diluted HIV-1 Env-pseudotyped virus, in duplicate, in a 96-well CulturPlate (PerkinElmer). The mixes were incubated for 30 min at 37°C. Twenty microliters of TZM-bl cells (10,000 cells/well) with or without 70 μg/ml DEAE-dextran was then added and incubated overnight at 37°C. Each experiment plate also had a column of cells only (no antibody [Ab] or virus) and a column of virus only (no Ab) as controls for background TZM-bl luciferase activity and maximal viral entry, respectively. Serial dilutions were performed with a change of tips at each dilution step to prevent carryover. The following day, all wells received 100 μl of fresh cDMEM and were incubated overnight at 37°C. The following day, 50 μl of the steadylite plus reporter gene assay system (PerkinElmer) was added to all wells, and plates were shaken at 600 rpm for 15 min. Luminometry was then performed on a SpectraMax L (Molecular Devices) luminometer. Percent neutralization was determined by calculating the difference in average relative light units (RLU) between virus-only wells (cells plus virus column) and test wells (cells plus serum/Ab sample plus virus), dividing this result by the average RLU of virus-only wells (cells plus virus column), and multiplying by 100. Background is subtracted from all test wells using the average RLU from the uninfected control wells (cells-only column) before calculating the percent neutralization. Neutralizing serum antibody titers are expressed as the antibody concentration required to achieve 50% neutralization and calculated using a dose-response curve fit with a 5-parameter nonlinear function.
Neutralization with peptide competition was performed as described previously (26, 61). Briefly, guinea pig plasma was tested at a single-point dilution that resulted in 50 to 80% neutralization of the virus. Ten microliters of plasma was mixed with 5 μl of control medium, PEGylated FP9 (AVGIGAVFL), or PEGylated scrambled FP9 (VLAGFAIGV), and the mixture was incubated at 37°C for 30 min; 35 μl of each virus was then added, and incubation at 37°C was continued for 30 min. The final peptide concentration was 12.5 pM. TZM-bl cells were added, incubated, fed, and lysed, and luciferase activity was assessed as described above. The assay was performed in duplicate wells and repeated three times. The reduction in neutralization was calculated as (1 − %Neut[peptide]/%Neut[medium]), where %Neut is the average of the results from three independent assays with duplicates in each assay. For example, a value of 100% indicates complete inhibition of neutralization, whereas a value of 0% indicates no effect by peptide.
Anti-BG505 DS-SOSIP.664 ELISA.
Lectin capture ELISA was carried out as previously described (13). Ninety-six-well plates (Costar half-area high binding; Corning, Kennebunk, ME) were coated overnight at 4°C with 50 μl/well of 2 μg/ml lectin (from Galanthus nivalis; Sigma-Aldrich) in 1× PBS. Between each subsequent step, plates were washed five times with PBS plus 0.05% Tween (PBS-T). After being coated, plates were blocked with 5% skim milk in PBS, followed by trimer capture with BG505 DS-SOSIP.664 in 10% fetal bovine serum (FBS)-PBS. Serially diluted (5-fold; starting dilution, 1:100) guinea pig serum in 0.2% Tween-PBS buffer was added, and goat anti-guinea pig IgG secondary antibody conjugated to horseradish peroxidase (KPL, Gaithersburg, MD) diluted 1:5,000 in 0.2% Tween-PBS buffer was added. Plates were developed with 50 μl/well tetramethylbenzidine (TMB) substrate (SureBlue; KPL) for 10 min before the addition of 1 N sulfuric acid (Fisher Chemical, Fair Lawn, NJ) to stop the reaction. Plates were read at 450 nm (SpectraMax using SoftMax Pro software, version 5; Molecular Devices, Sunnyvale, CA), and the optical density (OD) was analyzed following subtraction of the nonspecific horseradish peroxidase background activity. The endpoint titer was defined as the reciprocal of the greatest dilution with an OD above 0.1 (2 times average raw plate background) or, when the OD of the dilution was greater than 0.2, the reciprocal of the midpoint between this dilution and the subsequent dilution.
Anti-FP8-peptide ELISA.
Anti-FP8-peptide ELISA was carried out as previously described (28). Ninety-six-well streptavidin-coated plates (Pierce high binding capacity; Thermo Scientific) were coated overnight at 4°C with FP8-PEG12-biotin (AVGIGAVF-PEG12-Lys biotin; GenScript, Piscataway, NJ) in 1× PBS. Plates were washed with PBS-T and blocked with B3T buffer (150 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA, 3.3% fetal bovine serum, 2% bovine albumin, 0.07% Tween 20, 0.02% thimerosal). Serially diluted (5-fold; starting dilution, 1:100) guinea pig serum in B3T buffer was added. Afterward, goat anti-guinea pig IgG antibody conjugated to horseradish peroxidase (KPL, Gaithersburg, MD) diluted 1:5,000 in B3T buffer was added. Plates were washed with PBS-T and developed with TMB substrate (SureBlue; KPL) for 10 min before the addition of 1 N sulfuric acid (Fisher Chemical, Fair Lawn, NJ) to stop the reaction. Plates were read at 450 nm (SpectraMax using SoftMax Pro software, version 5; Molecular Devices, Sunnyvale, CA), and the OD was analyzed following subtraction of the nonspecific horseradish peroxidase background activity. The endpoint titer was defined as the reciprocal of the greatest dilution with an OD value above 0.1 (2 times average raw plate background) or, when OD of the dilution was greater than 0.2, the reciprocal of the midpoint between this dilution and the subsequent dilution.
Data availability.
The atomic coordinates and structure factors of the BG505 3mut-DS-SOSIP.664 and CAP256 RnS-3mut-2G-SOSIP.664 X-ray crystal structures have been deposited in the Protein Data Bank (PDB) under accession numbers 6W03 and 6VZI, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Stuckey for assistance with figures and the Structural Biology Section, Structural Bioinformatics Core, Humoral Immunology Section, and Biosafety Level 3 (BSL-3) Core at the NIH Vaccine Research Center (VRC) for helpful discussions and comments on the manuscript. We thank the Vaccine Production Program of the VRC for providing FP8v1-rTTHC, BG505 DS-SOSIP.664, and ConC-FP8v2 RnS-3mut-2G-SOSIP.664 immunogens. The Vaccine Production Program includes N. Amharref, F. J. Arnold, N. Barefoot, C. Barry, B. Boonyaratanakornkit, E. Carey, R. Caringal, K. Carlton, N. Chalamalsetty, A. L. Chamberlain, A. Charlton, R. Chaudhuri, M. Chen, P. Chen, Y. Chen, N. Cibelli, J. W. Cooper, H. Dahodwala, M. Fleischman, J. C. Frederick, H. Fuller, J. Gall, M. Ghosh, I. Godfroy, D. Gollapudi, D. Gowetski, K. Gulla, J. Horwitz, A. Hussain, V. Ivleva, T. Khin, L. Kueltzo, G. Lagos, Q. P. Lei, Y. Li, S. Manceva, V. Mangalampalli, G. Moxey, S. O’Connell, A. Patel, E. Rosales-Zavala, E. Scheideman, N. A. Schneck, Z. Schneiderman, A. Shaddeau, W. Shadrick, S. Shetty, B. Tippett, J. Varriale, A. Vinitsky, H. Wang, X. E. Wang, C. Webber, S. Witter, G. J. Yang, L. Yang, Y. Yang, and Y. Zhang.
Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, by the Office of AIDS Research, National Institutes of Health, and by the International AIDS Vaccine Initiative (IAVI) Neutralizing Antibody Consortium. This project has been funded in part with federal funds from the Frederick National Laboratory for Cancer Research, NIH, under contract HHSN261200800001E (Y. Tsybovsky). The use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38.
G.-Y. C. and J.C.B. developed the BG505 3mut-DS-SOSIP.664 trimer. G.-Y.C., Y.-T.L., and R.R. developed the stabilized clade C trimers. C.C., K.X., A.R.C., and E.K.S. performed the guinea pig immunization studies. H.G. and L.O. purified the Env trimers. S.N. and M.C. performed the MSD analyses. Y.T. performed negative-stain EM. R.V. expressed the Env trimers. B.Z. purified the HIV-1 antibodies. Y.Y. performed the 96-well ELISA assay. Y.-T.L. determined the structure of BG505 3mut-DS-SOSIP.664 and CAP256 RnS-3mut-2G-SOSIP.664. H.G. and A.S.O. determined Env affinity toward CD4. S.D.S., R.K., W.W., and N.A.D.-R. assessed serum neutralization. A.C. coordinated with the Vaccine Production Program Laboratory, Vaccine Research Center, NIAID, for immunogen production. G.-Y.C., Y.-T.L., S.W., and P.D.K. wrote the manuscript, with all authors providing comments and revisions.
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Harrison SC. 2015. Viral membrane fusion. Virology 479–480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rey FA, Lok SM. 2018. Common features of enveloped viruses and implications for immunogen design for next-generation vaccines. Cell 172:1319–1334. doi: 10.1016/j.cell.2018.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu M, Ma X, Castillo-Menendez LR, Gorman J, Alsahafi N, Ermel U, Terry DS, Chambers M, Peng D, Zhang B, Zhou T, Reichard N, Wang K, Grover JR, Carman BP, Gardner MR, Nikic-Spiegel I, Sugawara A, Arthos J, Lemke EA, Smith AB III, Farzan M, Abrams C, Munro JB, McDermott AB, Finzi A, Kwong PD, Blanchard SC, Sodroski JG, Mothes W. 2019. Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 568:415–419. doi: 10.1038/s41586-019-1101-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB III, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chuang G-Y, Geng H, Pancera M, Xu K, Cheng C, Acharya P, Chambers M, Druz A, Tsybovsky Y, Wanninger TG, Yang Y, Doria-Rose NA, Georgiev IS, Gorman J, Joyce MG, O'Dell S, Zhou T, McDermott AB, Mascola JR, Kwong PD. 2017. Structure-based design of a soluble prefusion-closed HIV-1 Env trimer with reduced CD4 affinity and improved immunogenicity. J Virol 91:e02268-16. doi: 10.1128/JVI.02268-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Taeye SW, Ozorowski G, Torrents de la Pena A, Guttman M, Julien JP, van den Kerkhof TL, Burger JA, Pritchard LK, Pugach P, Yasmeen A, Crampton J, Hu J, Bontjer I, Torres JL, Arendt H, DeStefano J, Koff WC, Schuitemaker H, Eggink D, Berkhout B, Dean H, LaBranche C, Crotty S, Crispin M, Montefiori DC, Klasse PJ, Lee KK, Moore JP, Wilson IA, Ward AB, Sanders RW. 2015. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell 163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kulp DW, Steichen JM, Pauthner M, Hu X, Schiffner T, Liguori A, Cottrell CA, Havenar-Daughton C, Ozorowski G, Georgeson E, Kalyuzhniy O, Willis JR, Kubitz M, Adachi Y, Reiss SM, Shin M, de Val N, Ward AB, Crotty S, Burton DR, Schief WR. 2017. Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat Commun 8:1655. doi: 10.1038/s41467-017-01549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torrents de la Peña A, Julien JP, de Taeye SW, Garces F, Guttman M, Ozorowski G, Pritchard LK, Behrens AJ, Go EP, Burger JA, Schermer EE, Sliepen K, Ketas TJ, Pugach P, Yasmeen A, Cottrell CA, Torres JL, Vavourakis CD, van Gils MJ, LaBranche C, Montefiori DC, Desaire H, Crispin M, Klasse PJ, Lee KK, Moore JP, Ward AB, Wilson IA, Sanders RW. 2017. Improving the immunogenicity of native-like HIV-1 envelope trimers by hyperstabilization. Cell Rep 20:1805–1817. doi: 10.1016/j.celrep.2017.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sliepen K, Han BW, Bontjer I, Mooij P, Garces F, Behrens AJ, Rantalainen K, Kumar S, Sarkar A, Brouwer PJM, Hua Y, Tolazzi M, Schermer E, Torres JL, Ozorowski G, van der Woude P, de la Pena AT, van Breemen MJ, Camacho-Sanchez JM, Burger JA, Medina-Ramirez M, Gonzalez N, Alcami J, LaBranche C, Scarlatti G, van Gils MJ, Crispin M, Montefiori DC, Ward AB, Koopman G, Moore JP, Shattock RJ, Bogers WM, Wilson IA, Sanders RW. 2019. Structure and immunogenicity of a stabilized HIV-1 envelope trimer based on a group-M consensus sequence. Nat Commun 10:2355. doi: 10.1038/s41467-019-10262-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Taeye SW, Go EP, Sliepen K, de la Pena AT, Badal K, Medina-Ramirez M, Lee WH, Desaire H, Wilson IA, Moore JP, Ward AB, Sanders RW. 2019. Stabilization of the V2 loop improves the presentation of V2 loop-associated broadly neutralizing antibody epitopes on HIV-1 envelope trimers. J Biol Chem 294:5616–5631. doi: 10.1074/jbc.RA118.005396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Georgiev IS, Joyce MG, Yang Y, Sastry M, Zhang B, Baxa U, Chen RE, Druz A, Lees CR, Narpala S, Schon A, Van Galen J, Chuang GY, Gorman J, Harned A, Pancera M, Stewart-Jones GB, Cheng C, Freire E, McDermott AB, Mascola JR, Kwong PD. 2015. Single-chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV-1 Env. J Virol 89:5318–5329. doi: 10.1128/JVI.03451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, Dubrovskaya V, Ward AB, Wyatt RT. 2015. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep 11:539–550. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong L, He L, de Val N, Vora N, Morris CD, Azadnia P, Sok D, Zhou B, Burton DR, Ward AB, Wilson IA, Zhu J. 2016. Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Nat Commun 7:12040. doi: 10.1038/ncomms12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guenaga J, Garces F, de Val N, Stanfield RL, Dubrovskaya V, Higgins B, Carrette B, Ward AB, Wilson IA, Wyatt RT. 2017. Glycine substitution at helix-to-coil transitions facilitates the structural determination of a stabilized subtype C HIV envelope glycoprotein. Immunity 46:792–803.e3. doi: 10.1016/j.immuni.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiffner T, Pallesen J, Russell RA, Dodd J, de Val N, LaBranche CC, Montefiori D, Tomaras GD, Shen X, Harris SL, Moghaddam AE, Kalyuzhniy O, Sanders RW, McCoy LE, Moore JP, Ward AB, Sattentau QJ. 2018. Structural and immunologic correlates of chemically stabilized HIV-1 envelope glycoproteins. PLoS Pathog 14:e1006986. doi: 10.1371/journal.ppat.1006986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steichen JM, Kulp DW, Tokatlian T, Escolano A, Dosenovic P, Stanfield RL, McCoy LE, Ozorowski G, Hu X, Kalyuzhniy O, Briney B, Schiffner T, Garces F, Freund NT, Gitlin AD, Menis S, Georgeson E, Kubitz M, Adachi Y, Jones M, Mutafyan AA, Yun DS, Mayer CT, Ward AB, Burton DR, Wilson IA, Irvine DJ, Nussenzweig MC, Schief WR. 2016. HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity 45:483–496. doi: 10.1016/j.immuni.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guenaga J, Dubrovskaya V, de Val N, Sharma SK, Carrette B, Ward AB, Wyatt RT. 2015. Structure-guided redesign increases the propensity of HIV Env to generate highly stable soluble trimers. J Virol 90:2806–2817. doi: 10.1128/JVI.02652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joyce MG, Georgiev IS, Yang Y, Druz A, Geng H, Chuang GY, Kwon YD, Pancera M, Rawi R, Sastry M, Stewart-Jones GBE, Zheng A, Zhou T, Choe M, Van Galen JG, Chen RE, Lees CR, Narpala S, Chambers M, Tsybovsky Y, Baxa U, McDermott AB, Mascola JR, Kwong PD. 2017. Soluble prefusion closed DS-SOSIP.664-Env trimers of diverse HIV-1 strains. Cell Rep 21:2992–3002. doi: 10.1016/j.celrep.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 21.Rutten L, Lai YT, Blokland S, Truan D, Bisschop IJM, Strokappe NM, Koornneef A, van Manen D, Chuang GY, Farney SK, Schuitemaker H, Kwong PD, Langedijk J. 2018. A universal approach to optimize the folding and stability of prefusion-closed HIV-1 envelope trimers. Cell Rep 23:584–595. doi: 10.1016/j.celrep.2018.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Q, Acharya P, Dolan MA, Zhang P, Guzzo C, Lu J, Kwon A, Gururani D, Miao H, Bylund T, Chuang GY, Druz A, Zhou T, Rice WJ, Wigge C, Carragher B, Potter CS, Kwong PD, Lusso P. 2017. Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer. Nat Struct Mol Biol 24:370–378. doi: 10.1038/nsmb.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang P, Gorman J, Geng H, Liu Q, Lin Y, Tsybovsky Y, Go EP, Dey B, Andine T, Kwon A, Patel M, Gururani D, Uddin F, Guzzo C, Cimbro R, Miao H, McKee K, Chuang GY, Martin L, Sironi F, Malnati MS, Desaire H, Berger EA, Mascola JR, Dolan MA, Kwong PD, Lusso P. 2018. Interdomain stabilization impairs CD4 binding and improves immunogenicity of the HIV-1 envelope trimer. Cell Host Microbe 23:832–844.e6. doi: 10.1016/j.chom.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cavanagh D. 2007. Coronavirus avian infectious bronchitis virus. Vet Res 38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 25.Kreijtz JH, Fouchier RA, Rimmelzwaan GF. 2011. Immune responses to influenza virus infection. Virus Res 162:19–30. doi: 10.1016/j.virusres.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 26.Xu K, Acharya P, Kong R, Cheng C, Chuang G-Y, Liu K, Louder MK, O'Dell S, Rawi R, Sastry M, Shen C-H, Zhang B, Zhou T, Asokan M, Bailer RT, Chambers M, Chen X, Choi CW, Dandey VP, Doria-Rose NA, Druz A, Eng ET, Farney SK, Foulds KE, Geng H, Georgiev IS, Gorman J, Hill KR, Jafari AJ, Kwon YD, Lai Y-T, Lemmin T, McKee K, Ohr TY, Ou L, Peng D, Rowshan AP, Sheng Z, Todd J-P, Tsybovsky Y, Viox EG, Wang Y, Wei H, Yang Y, Zhou AF, Chen R, Yang L, Scorpio DG, McDermott AB, Shapiro L, et al. 2018. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat Med 24:857–867. doi: 10.1038/s41591-018-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong R, Duan H, Sheng Z, Xu K, Acharya P, Chen X, Cheng C, Dingens AS, Gorman J, Sastry M, Shen C-H, Zhang B, Zhou T, Chuang G-Y, Chao CW, Gu Y, Jafari AJ, Louder MK, O'Dell S, Rowshan AP, Viox EG, Wang Y, Choi CW, Corcoran MM, Corrigan AR, Dandey VP, Eng ET, Geng H, Foulds KE, Guo Y, Kwon YD, Lin B, Liu K, Mason RD, Nason MC, Ohr TY, Ou L, Rawi R, Sarfo EK, Schön A, Todd JP, Wang S, Wei H, Wu W, NISC Comparative Sequencing Program, Mullikin JC, Bailer RT, Doria-Rose NA, Karlsson Hedestam GB, et al. 2019. Antibody lineages with vaccine-induced antigen-binding hotspots develop broad HIV neutralization. Cell 178:567–584.e19. doi: 10.1016/j.cell.2019.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng C, Xu K, Kong R, Chuang G-Y, Corrigan AR, Geng H, Hill KR, Jafari AJ, O'Dell S, Ou L, Rawi R, Rowshan AP, Sarfo EK, Sastry M, Saunders KO, Schmidt SD, Wang S, Wu W, Zhang B, Doria-Rose NA, Haynes BF, Scorpio DG, Shapiro L, Mascola JR, Kwong PD. 2019. Consistent elicitation of cross-clade HIV-neutralizing responses achieved in guinea pigs after fusion peptide priming by repetitive envelope trimer boosting. PLoS One 14:e0215163. doi: 10.1371/journal.pone.0215163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O’Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, NISC Comparative Sequencing Program, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR, Becker J, Benjamin B, Blakesley R, Bouffard G. 2014. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai Y-T, Wang T, O'Dell S, Louder MK, Schön A, Cheung CSF, Chuang G-Y, Druz A, Lin B, McKee K, Peng D, Yang Y, Zhang B, Herschhorn A, Sodroski J, Bailer RT, Doria-Rose NA, Mascola JR, Langley DR, Kwong PD. 2019. Lattice engineering enables definition of molecular features allowing for potent small-molecule inhibition of HIV-1 entry. Nat Commun 10:47. doi: 10.1038/s41467-018-07851-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozorowski G, Pallesen J, de Val N, Lyumkis D, Cottrell CA, Torres JL, Copps J, Stanfield RL, Cupo A, Pugach P, Moore JP, Wilson IA, Ward AB. 2017. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature 547:360–363. doi: 10.1038/nature23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng C, Pancera M, Bossert A, Schmidt SD, Chen RE, Chen X, Druz A, Narpala S, Doria-Rose NA, McDermott AB, Kwong PD, Mascola JR. 2015. Immunogenicity of a prefusion HIV-1 envelope trimer in complex with a quaternary-structure-specific antibody. J Virol 90:2740–2755. doi: 10.1128/JVI.02380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JH, Andrabi R, Su CY, Yasmeen A, Julien JP, Kong L, Wu NC, McBride R, Sok D, Pauthner M, Cottrell CA, Nieusma T, Blattner C, Paulson JC, Klasse PJ, Wilson IA, Burton DR, Ward AB. 2017. A broadly neutralizing antibody targets the dynamic HIV envelope trimer apex via a long, rigidified, and anionic beta-hairpin structure. Immunity 46:690–702. doi: 10.1016/j.immuni.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doria-Rose NA, Bhiman JN, Roark RS, Schramm CA, Gorman J, Chuang G-Y, Pancera M, Cale EM, Ernandes MJ, Louder MK, Asokan M, Bailer RT, Druz A, Fraschilla IR, Garrett NJ, Jarosinski M, Lynch RM, McKee K, O’Dell S, Pegu A, Schmidt SD, Staupe RP, Sutton MS, Wang K, Wibmer CK, Haynes BF, Abdool-Karim S, Shapiro L, Kwong PD, Moore PL, Morris L, Mascola JR. 2016. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J Virol 90:76–91. doi: 10.1128/JVI.01791-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLellan JS, Pancera M, Carrico C, Gorman J, Julien J-P, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-Ul-Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang G-Y, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang Z-Y, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang L-X, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, Soto C, Terry DS, Yang Y, Zhou T, Ahlsen G, Bailer RT, Chambers M, Chuang G-Y, Doria-Rose NA, Druz A, Hallen MA, Harned A, Kirys T, Louder MK, O'Dell S, Ofek G, Osawa K, Prabhakaran M, Sastry M, Stewart-Jones GBE, Stuckey J, Thomas PV, Tittley T, Williams C, Zhang B, Zhao H, Zhou Z, Donald BR, Lee LK, Zolla-Pazner S, Baxa U, Schön A, Freire E, Shapiro L, Lee KK, Arthos J, Munro JB, Blanchard SC, Mothes W, Binley JM, McDermott AB, Mascola JR, Kwong PD. 2015. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol 22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Acharya P, Dogo-Isonagie C, LaLonde JM, Lam SN, Leslie GJ, Louder MK, Frye LL, Debnath AK, Greenwood JR, Luongo TS, Martin L, Watts KS, Hoxie JA, Mascola JR, Bewley CA, Kwong PD. 2011. Structure-based identification and neutralization mechanism of tyrosine sulfate mimetics that inhibit HIV-1 entry. ACS Chem Biol 6:1069–1077. doi: 10.1021/cb200068b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorman J, Soto C, Yang MM, Davenport TM, Guttman M, Bailer RT, Chambers M, Chuang GY, DeKosky BJ, Doria-Rose NA, Druz A, Ernandes MJ, Georgiev IS, Jarosinski MC, Joyce MG, Lemmin TM, Leung S, Louder MK, McDaniel JR, Narpala S, Pancera M, Stuckey J, Wu X, Yang Y, Zhang B, Zhou T, NISC Comparative Sequencing Program, Mullikin JC, Baxa U, Georgiou G, McDermott AB, Bonsignori M, Haynes BF, Moore PL, Morris L, Lee KK, Shapiro L, Mascola JR, Kwong PD. 2016. Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design. Nat Struct Mol Biol 23:81–90. doi: 10.1038/nsmb.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorman J, Chuang GY, Lai YT, Shen CH, Boyington JC, Druz A, Geng H, Louder M, McKee K, Rawi R, Verardi R, Yang Y, Zhang B, Doria-Rose NA, Lin B, Moore PL, Morris L, Shapiro L, Mascola JR, Kwong PD. 2020. Structure of super-potent antibody CAP256-VRC26.25 in complex with HIV-1 envelope reveals a combined mode of trimer-apex recognition. Cell Rep 31:107488. doi: 10.1016/j.celrep.2020.03.052. [DOI] [PubMed] [Google Scholar]
- 40.Krarup A, Truan D, Furmanova-Hollenstein P, Bogaert L, Bouchier P, Bisschop IJM, Widjojoatmodjo MN, Zahn R, Schuitemaker H, McLellan JS, Langedijk J. 2015. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat Commun 6:8143. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pallesen J, Wang N, Corbett KS, Wrapp D, Kirchdoerfer RN, Turner HL, Cottrell CA, Becker MM, Wang L, Shi W, Kong WP, Andres EL, Kettenbach AN, Denison MR, Chappell JD, Graham BS, Ward AB, McLellan JS. 2017. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci U S A 114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Battles MB, Mas V, Olmedillas E, Cano O, Vazquez M, Rodriguez L, Melero JA, McLellan JS. 2017. Structure and immunogenicity of pre-fusion-stabilized human metapneumovirus F glycoprotein. Nat Commun 8:1528. doi: 10.1038/s41467-017-01708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 76:8875–8889. doi: 10.1128/jvi.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pauthner MG, Nkolola JP, Havenar-Daughton C, Murrell B, Reiss SM, Bastidas R, Prevost J, Nedellec R, von Bredow B, Abbink P, Cottrell CA, Kulp DW, Tokatlian T, Nogal B, Bianchi M, Li H, Lee JH, Butera ST, Evans DT, Hangartner L, Finzi A, Wilson IA, Wyatt RT, Irvine DJ, Schief WR, Ward AB, Sanders RW, Crotty S, Shaw GM, Barouch DH, Burton DR. 2019. Vaccine-induced protection from homologous tier 2 SHIV challenge in nonhuman primates depends on serum-neutralizing antibody titers. Immunity 50:241–252.e6. doi: 10.1016/j.immuni.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol G Principal Investigators, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, McBride R, von Bredow B, Shivatare SS, Wu CY, Chan-Hui PY, Liu Y, Feizi T, Zwick MB, Koff WC, Seaman MS, Swiderek K, Moore JP, Evans D, Paulson JC, Wong CH, Ward AB, Wilson IA, Sanders RW, Poignard P, Burton DR. 2014. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang GY, Druz A, Doria-Rose NA, Laub L, Sliepen K, van Gils MJ, de la Pena AT, Derking R, Klasse PJ, Migueles SA, Bailer RT, Alam M, Pugach P, Haynes BF, Wyatt RT, Sanders RW, Binley JM, Ward AB, Mascola JR, Kwong PD, Connors M. 2014. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu X, Yang Z-Y, Li Y, Hogerkorp C-M, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen L, Kwon YD, Zhou T, Wu X, O'Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang Z-Y, Zhang M-Y, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Killikelly A, Zhang HT, Spurrier B, Williams C, Gorny MK, Zolla-Pazner S, Kong XP. 2013. Thermodynamic signatures of the antigen binding site of mAb 447-52D targeting the third variable region of HIV-1 gp120. Biochemistry 52:6249–6257. doi: 10.1021/bi400645e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang X, Burke V, Totrov M, Williams C, Cardozo T, Gorny MK, Zolla-Pazner S, Kong XP. 2010. Conserved structural elements in the V3 crown of HIV-1 gp120. Nat Struct Mol Biol 17:955–961. doi: 10.1038/nsmb.1861. [DOI] [PubMed] [Google Scholar]
- 54.Stanfield RL, Gorny MK, Zolla-Pazner S, Wilson IA. 2006. Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity. J Virol 80:6093–6105. doi: 10.1128/JVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gorny MK, Sampson J, Li H, Jiang X, Totrov M, Wang X-H, Williams C, O'Neal T, Volsky B, Li L, Cardozo T, Nyambi P, Zolla-Pazner S, Kong X-P. 2011. Human anti-V3 HIV-1 monoclonal antibodies encoded by the VH5-51/VL lambda genes define a conserved antigenic structure. PLoS One 6:e27780. doi: 10.1371/journal.pone.0027780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rini JM, Stanfield RL, Stura EA, Salinas PA, Profy AT, Wilson IA. 1993. Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen. Proc Natl Acad Sci U S A 90:6325–6329. doi: 10.1073/pnas.90.13.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, Ernandes MJ, Kong R, Longo NS, Louder MK, McKee K, O'Dell S, Schmidt SD, Tran L, Yang Z, Druz A, Luongo TS, Moquin S, Srivatsan S, Yang Y, Zhang B, Zheng A, Pancera M, Kirys T, Georgiev IS, Gindin T, Peng H-P, Yang A-S, NISC Comparative Sequencing Program4, Mullikin JC, Gray MD, Stamatatos L, Burton DR, Koff WC, Cohen MS, Haynes BF, Casazza JP, Connors M, Corti D, Lanzavecchia A, Sattentau QJ, Weiss RA, West AP, Bjorkman PJ, Scheid JF, Nussenzweig MC, et al. 2015. Structural repertoire of HIV-1-neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell 161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 60.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 61.Kong R, Xu K, Zhou T, Acharya P, Lemmin T, Liu K, Ozorowski G, Soto C, Taft JD, Bailer RT, Cale EM, Chen L, Choi CW, Chuang G-Y, Doria-Rose NA, Druz A, Georgiev IS, Gorman J, Huang J, Joyce MG, Louder MK, Ma X, McKee K, O'Dell S, Pancera M, Yang Y, Blanchard SC, Mothes W, Burton DR, Koff WC, Connors M, Ward AB, Kwong PD, Mascola JR. 2016. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352:828–833. doi: 10.1126/science.aae0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol 67:3978–3988. doi: 10.1128/JVI.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gorny MK, Williams C, Volsky B, Revesz K, Wang XH, Burda S, Kimura T, Konings FA, Nadas A, Anyangwe CA, Nyambi P, Krachmarov C, Pinter A, Zolla-Pazner S. 2006. Cross-clade neutralizing activity of human anti-V3 monoclonal antibodies derived from the cells of individuals infected with non-B clades of human immunodeficiency virus type 1. J Virol 80:6865–6872. doi: 10.1128/JVI.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin O, Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RH. 2012. Highly conserved protective epitopes on influenza B viruses. Science 337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 66.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 67.Scheres SH. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 69.Bunkóczi G, Read RJ. 2011. Improvement of molecular-replacement models with Sculptor. Acta Crystallogr D Biol Crystallogr 67:303–312. doi: 10.1107/S0907444910051218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 72.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin CL, Bilska M, Greene KM, Gao H, Todd CA, Ozaki DA, Seaman MS, Mascola JR, Montefiori DC. 2014. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods 409:131–146. doi: 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors of the BG505 3mut-DS-SOSIP.664 and CAP256 RnS-3mut-2G-SOSIP.664 X-ray crystal structures have been deposited in the Protein Data Bank (PDB) under accession numbers 6W03 and 6VZI, respectively.