Abstract
The selective pressure imposed by extrinsic death signals and stressors adds to the challenge of isolating and interpreting the roles of proteins in stress-activated signaling networks. By expressing a kinase with activating mutations and a caged lysine blocking the active site, we can rapidly switch on catalytic activity with light and monitor the ensuing dynamics. Applying this approach to MAP kinase 6 (MKK6), which activates the p38 subfamily of MAPKs, we found that decaging active MKK6 in fibroblasts is sufficient to trigger apoptosis in a p38-dependent manner. Both in fibroblasts and in a murine melanoma cell line expressing mutant B-Raf, MKK6 activation rapidly and potently inhibited the pro-proliferative extracellular signal–regulated kinase (ERK) pathway; to our surprise, this negative cross-regulation was equally robust when all p38 isoforms were inhibited. These results position MKK6 as a new pleiotropic signal transducer that promotes both pro-apoptotic and anti-proliferative signaling, and they highlight the utility of caged, light-activated kinases for dissecting stress-activated signaling networks.
Keywords: optogenetics, signal transduction, cell signaling, mitogen-activated protein kinases, apoptosis, cell death, cell stress, mitogen-activated protein kinase kinase 6 (MAP2K6), mitogen-activated protein kinase (MAPK)
Cells respond to their dynamically changing and chemically diverse surroundings through highly regulated, intracellular signal transduction networks. Some cellular responses, such as growth and cell cycle progression, may be viewed as short-term, whereas responses such as terminal differentiation and programmed cell death (apoptosis) may be viewed as decisive and cell fate-determining, with limited or no plasticity. Core modules in signal transduction networks include the mitogen-activated protein kinase (MAPK) cascades, notably the extracellular signal–regulated kinase (ERK), c-Jun N-terminal kinase (JNK), and p38 pathways in mammals (1, 2). In each of these cascades, three kinases are sequentially activated: a serine/threonine kinase classified as a MAPK kinase kinase (MAPKKK or M3K) phosphorylates and activates a dual-specificity MAPK kinase (MAPKK or M2K), which in turn phosphorylates and activates the MAPK. Whereas the ERK cascade controls cell proliferation in response to growth factor stimulation (3, 4), JNK and p38 are considered stress-activated protein kinases (SAPKs) that commonly promote apoptosis and other responses to environmental insults and inflammatory cytokines (5–7). Consistent with these opposing roles that influence normal tissue development, homeostasis, and immune responses, aberrations in MAPK signal transduction are found in the vast majority of human cancers and are broadly implicated in tumorigenesis (8, 9).
Parsing the roles of specific nodes in a signal transduction network, many of which are protein kinases, is plagued by multiple challenges that are often unacknowledged, because traditional methods frequently fail to address them. For one, cytokines and environmental stressors generally activate multiple pathways simultaneously, making it difficult to isolate the effect of a particular signaling protein. Adding to the difficulty of this issue are the natural interactions between canonical pathways (i.e. cross-talk). For example, the MAPK cascades affect one another by influencing the expression of dual-specificity phosphatases that dephosphorylate and thus deactivate the various MAPKs with differing specificities (10, 11). Another, more specific example is found in the regulation of ERK signaling by p38. This interaction was first suggested by the evidence that ERK1/2 co-immunoprecipitates with p38 (12); activation of p38 by constitutively active MKK3 resulted in potent inhibition of ERK and its upstream MAPKK, MEK. In another study, arsenite treatment induced p38 activation and prevented phorbol ester–triggered activation of MEK and ERK, and this inhibitory effect was attributed to enhanced activity of protein phosphatase 2A (PP2A) (13). Activation of p38 induces dephosphorylation of MEK and apoptosis in various cell types (14, 15).
In such studies, pharmacological inhibitors are commonly used to help deduce a protein's function(s), but such an approach only probes loss of function and often lacks complete kinase specificity (16). To target gain-of-function, cells are typically transfected with a constitutively active variant of the signaling protein, but this approach allows ample time for the cell to alter its gene expression profile. This issue is especially problematic for a pathway that mediates stress responses, as its activation can introduce selective pressure to adapt; obviously, this issue is further magnified for pathways that directly and rapidly promote cell death, such as the p38 signaling cascade (17, 18).
Various strategies have been implemented to address the limitations outlined above, most commonly by inducing the association or dissociation of a protein-protein interaction via the addition of a small-molecule dimerizer or through light exposure (19–22). Typically, this strategy involves the translocation of a signaling protein to the plasma membrane to enhance its activity or the release of a protein from a sequestering interaction (23–26). Although induction or release of membrane association is often sufficient to promote signal transduction, it is not a definite “on” switch for catalytic activity. In this study, we develop and apply a photodecaging strategy to rapidly introduce active MKK6, a MAPKK specific to the p38 pathway (27, 28), in cells. This optogenetic approach, site-directed unnatural amino acid mutagenesis for the introduction of light-removable protecting groups—so-called “caging groups”—has enabled the development of genetically encoded caged kinases (29–34), phosphatases (35), and other light-activatable proteins (34, 36). With the ability to rapidly generate active MKK6 in cells, we have studied the dynamics of the p38 pathway in relation to p38-mediated apoptosis and the cross-regulation of the ERK pathway. In normal fibroblasts, we found that MKK6 activation is sufficient, and that p38 activation is necessary, for triggering the various phases of apoptosis (caspase activation, cytochrome c release, and cell death) within hours of light-triggered decaging. Consistent with reports on apoptotic responses to inflammatory cytokines and environmental stressors (37–39), the timing of the critical cytochrome c release varied widely in the cell population. MKK6 activation also rapidly induces potent inhibition of the ERK pathway; surprisingly, we discovered that this negative cross-talk is essentially p38-independent. This finding was replicated in a murine melanoma line with mutant B-Raf, and optical triggering of MKK6 in these oncogene-addicted cells abrogated ERK-dependent cell division. These results position MKK6 as a pleiotropic signal transducer that both promotes cell death and inhibits pro-proliferative signaling, and they demonstrate the utility of caged kinases as unique tools for dissecting signaling networks—stress-activated pathways in particular.
Results
Genetically encoded, caged MKK6 isolates kinase-dependent signaling through optical control
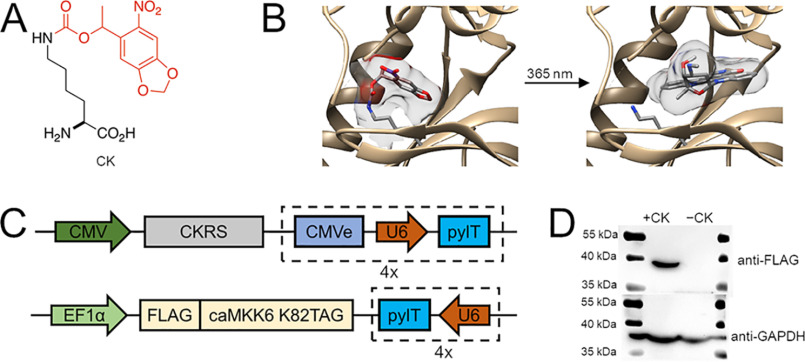
To parse the functions of a stress-activated signaling pathway in cells, we developed a genetically encoded, optically controlled MKK6. The incorporation of the caged lysine (CK) (Fig. 1A) through site-directed, unnatural amino acid mutagenesis has enabled the development of light-activatable proteins. This approach utilizes an orthogonal aminoacyl-tRNA synthetase/tRNA pair to selectively incorporate an unnatural amino acid (UAA) into proteins, in response to a recoded UAG amber codon, introduced at a desired site into a gene of interest. Supplementing the protein biosynthetic machinery of cells and animals with an engineered Methanosarcina barkeri pyrrolysyl-tRNA synthetase (pylRS) and its cognate tRNACUA (pylT) enables the incorporation of a wide range of UAAs, including photocaged amino acids (36). Photocaged amino acids, such as the CK (31), contain a light-removable protecting group that imposes steric demand on an enzyme active site and alters the electronic and nucleophilic characteristics of amino acid side chains. This renders the protein of interest inactive until a brief light exposure (e.g. 365-nm irradiation) removes the caging group and generates the protein's WT catalytic site. This is a generalizable approach for the study of protein kinases (32, 40, 41), because of the conserved lysine residue that coordinates ATP and catalyzes phosphotransfer in almost all protein kinases (42). Site-specific incorporation of CK at the conserved Lys82 of constitutively active MKK6 (43) renders the kinase enzymatically inactive, until light exposure removes the caging group (Fig. 1B). Expression of caged MKK6 in mammalian cells was achieved by introducing pylRS, eight expression cassettes encoding pylT, and constitutively active MKK6-K82TAG (referred to hereafter as caged MKK6) from two plasmids (Fig. 1C). In HEK293T cells co-transfected with the plasmids and supplemented with or without 2 mm CK, caged protein expression was detected only in the presence of the UAA (Fig. 1D).
Figure 1.
Construction of genetically encoded caged MKK6. A, structure of the photocaged lysine (CK), with the light-removable caging group shown in red. B, caging of Lys82 in MKK6 blocks the ATP-binding site until the caging group is removed through UV exposure, restoring the native, constitutively active kinase (Protein Data Bank entry 3FME). C, two-plasmid system to express caged MKK6 in mammalian cells. The TAG mutation is introduced at the Lys82 position of MKK6. Two other mutations (S207E and T211E) transform the kinase into a constitutively active form. pylRS for CK (CKRS) is driven by a CMV promoter, and FLAG-caMKK6 K82TAG is driven by an EF1α promoter. U6, an RNA polymerase III promoter, controls the expression of pylT on both plasmids in the form of four (4×) repetitive cassettes. CMVe (CMV 5′ enhancer fragment) is coupled with each U6 promoter on one of the plasmids for higher pylT yield. D, Western blotting shows CK-dependent expression of caged MKK6 in HEK 293T cells.
MKK6 is a dual-specificity kinase that activates p38 by phosphorylation (Fig. 2A) (27, 44). To confirm the optically controlled signaling function of caged MKK6, NIH 3T3 fibroblasts were co-transfected with plasmids expressing caged MKK6 and a plasmid encoding a p38 kinase activity reporter construct, which is exported from the nucleus upon phosphorylation (45), and the transfected cells were supplemented with or without CK (2 mm). After washing the cells and replacing the medium with imaging buffer, reporter localization was monitored by live-cell epifluorescence microscopy. In cells expressing caged MKK6 and incubated with CK, a brief (<2-s) exposure to UV light (DAPI excitation filter) rapidly induced p38 kinase activity, as shown by the live-cell reporter (45) (Fig. 2, B and C), confirming that MKK6 photoactivation and activation of p38 (the canonical MKK6 substrate) was substantial. No such UV-induced translocation responses were observed in the negative controls: cells incubated without CK (Fig. 2C) or in cells incubated with CK but expressing mCherry in place of constitutively active MKK6 (Fig. S1, A–C). These controls indicate that p38 is not activated by a brief UV exposure alone or by decaging of lysines that might have been incorporated in an inert protein or the endogenous proteome. The translocation response in caged MKK6-expressing cells was promptly and completely reversed following the addition of the p38α/β-specific inhibitor SB 239063 (46) (10 μm; Fig. 2D). The extent and kinetics of the p38 reporter response elicited by MKK6 decaging were comparable with those elicited by the chemical stressor, anisomycin (Fig. 2E). We also confirmed that brief UV exposure does not induce significant translocation of the analogous JNK activity reporter, either with or without expression of caged MKK6 (Fig. 2F and Fig. S1D). For Fig. 2 (C–F), variability among the time courses of the individual cells is shown in Fig. S2. Taken together, these results establish an experimental system in which canonical MKK6 → p38 signaling is readily and specifically induced by a brief and benign exposure to low-energy UV light.
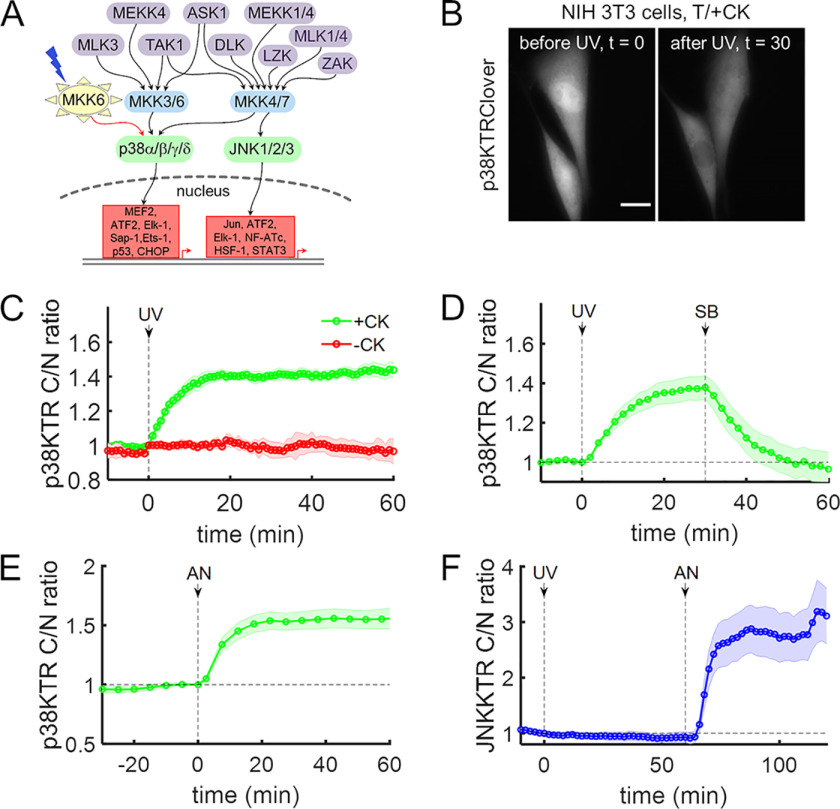
Figure 2.
Light-activated MKK6 specifically activates p38 in cells. A, simplified scheme showing the activation and downstream transcription factors of two mammalian stress-activated MAPKs, JNK and p38, by their respective MAPKKs and MAPKKKs. B–D, NIH 3T3 cells were transiently transfected with plasmids to express caged MKK6 and p38KTRClover, serum-starved, and briefly exposed to UV light (DAPI excitation filter, 365 nm, <2 s) as indicated. B, time-course montage of representative cells, before and after MKK6 light activation. Scale bar, 10 μm. C, quantification of p38 activity in cells grown in medium supplemented with or without CK (2 mm) (n = 10 each). D, cells (n = 12) were briefly exposed to UV light and then treated with the p38α/β-specific inhibitor SB 239063 (10 μm). E, analysis of NIH 3T3 cells (n = 27) transiently expressing p38KTRClover and treated with anisomycin (50 ng/ml). F, NIH 3T3 cells (n = 11) co-expressing caged MKK6 and the JNK activity reporter JNKKTRClover were briefly exposed to UV light and then treated with anisomycin (50 ng/ml). The solid lines in C–F show the mean kinase translocation reporter translocation, and the shaded regions show the 95% confidence intervals. All data are representative of at least three independent experiments.
Decaging of MKK6 induces p38-dependent apoptosis
Stress-activated signaling pathways orchestrate pronounced cellular responses, such as autophagy and apoptosis, which are difficult to study in traditional genetics experiments. JNK and p38 signaling have been strongly implicated in apoptosis of normal and cancer cells through a variety of mechanisms (5–7, 47–50). We asked whether an acutely active MKK6 upstream of p38 is sufficient to switch on the apoptotic program. Within hours after brief UV exposure of serum-starved fibroblasts co-expressing caged MKK6 and the p38 activity reporter, morphological changes associated with apoptosis, namely shrinkage, blebbing, and nuclear condensation or fragmentation (51, 52), were observed in a significant fraction of cells (Fig. 3A and Movie S1). Although the timing of cell death varied greatly, as seen in other studies (39, 53), the tendency of each cell to undergo apoptosis correlated well with the intensity and duration of p38 activity (Fig. 3B). To quantify apoptotic responses more precisely, we monitored the release of fluorescent protein-tagged cytochrome c from mitochondria, a hallmark of apoptosis (54, 55). This event manifests as an abrupt transition from punctate to diffuse fluorescence (Fig. 3 (C and D) and Movie S2). Optical activation of MKK6 induced cytochrome c release in a substantial fraction of the cells within ∼5 h; in contrast, almost none of the cells responded within that time period when CK was not supplied (Fig. 3E). Whereas detection of cytochrome c release stereotypically lagged MKK6 decaging by hours, we observed accumulation of a fluorescent cleavage product reporting caspase-3/-7 protease activity (56) much earlier (Fig. S3).
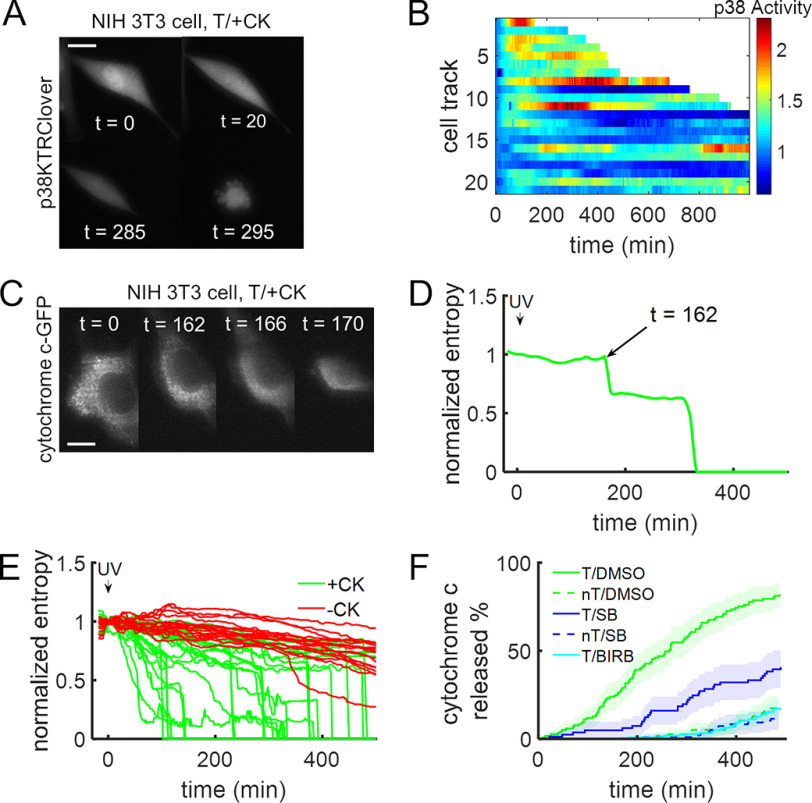
Figure 3.
Decaging active MKK6 triggers cellular apoptosis. A and B, NIH 3T3 cells expressing the p38 activity reporter along with caged MKK6 were briefly irradiated with UV light (365 nm, ∼2s), and p38 activity was monitored for an extended period, leading to cell death. A, time-course montage of a representative cell. Scale bar, 10 μm; times are in minutes. B, heatmap of p38 activity over time in individual cells following light activation of MKK6. Truncated cell tracks indicate when cells showed catastrophic shrinkage. C, time-course montage of a representative NIH 3T3 cell stably expressing cytochrome c-GFP. Cytochrome c is released from mitochondria following decaging of constitutively active MKK6. Scale bar, 10 μm; times are in minutes. D, determination of the timing of cytochrome c release for the cell in C, quantified as the first abrupt drop in entropy, a measure of image texture. E, NIH 3T3 cells stably expressing cytochrome c-GFP were transfected with plasmids to express caged MKK6 and grown in the presence or absence of 2 mm CK. MKK6 was activated by brief UV irradiation (365 nm, <2 s). F, percentage of cells having released cytochrome c as a function of time following brief UV exposure. Cells were transiently transfected (T, solid lines) or not transfected (nT, dashed lines) with plasmids to express caged MKK6 and then preincubated with DMSO control (green; n = 117 and n = 115), SB 239063 (10 μm; blue; n = 74 and n = 92), or BIRB 796 (10 μm; cyan; n = 105) before UV exposure. The shaded regions represent 95% confidence intervals. All analyses include data replicated in three independent experiments.
To assess the role of p38 in the apoptotic response to MKK6 decaging, we pretreated cells with SB 239063, the pan-p38 inhibitor BIRB 796 (57), or a DMSO vehicle control prior to UV exposure, and cytochrome c localization was monitored. For each cell, the time of the punctate-to-diffuse transition was recorded, and cumulative distributions of cytochrome c release were constructed. Control cells were not transfected with caged MKK6 but were incubated with CK as usual. The results show that p38 inhibition delayed the onset of cytochrome c release, almost to the same extent as in cells lacking caged MKK6 (Fig. 3F). Collectively, these results demonstrate that light activation of MKK6 through active site decaging is sufficient to trigger the hallmarks of apoptosis—caspase activation, release of cytochrome c, and cell morphology changes—and that the response is largely, if not completely, p38-dependent.
Photoactivation of MKK6 reveals p38-independent cross-regulation of ERK
The optogenetic system developed here also affords the opportunity to elucidate how individual kinases influence cross-talk between signaling pathways. MKK6 signaling is thought to flow exclusively through activation of p38, which has been implicated in negative cross-regulation of the Raf/MEK/ERK kinase cascade (12, 13, 15). To assess this pathway dynamically, we used an ERK kinase translocation reporter (45). A robust translocation response was observed in serum-starved NIH 3T3 cells stimulated with platelet-derived growth factor-BB (PDGF; 1 nm), and the response was reversed by the addition of the MEK inhibitor U0126 (20 μm) (Fig. 4 (A and B) and Movie S3). To assess pathway cross-talk, we co-expressed caged MKK6 and the ERK reporter. Cells were incubated with a moderate concentration of serum (3% fetal bovine serum (FBS)) to foster basal ERK activity; upon optical decaging of active MKK6, the ERK reporter was rapidly imported to the nucleus, indicating an acute reduction of ERK kinase activity and net dephosphorylation of the reporter construct. After the subsequent addition of PDGF, ERK activity was only partially and transiently restored (Fig. 4C and Movie S4). Using compartment-specific FRET biosensors (58), we confirmed that MKK6 decaging reduced ERK activity in both the nucleus and the cytosol (Fig. S4). When cells were not supplied with CK, or in cells incubated with CK but expressing mCherry in place of MKK6-K82TAG, UV exposure had no discernible effect on ERK activity, whereas the subsequent addition of a MEK inhibitor dramatically reduced ERK reporter translocation, as expected (Fig. S5). In a different stimulation protocol, the cells were serum-starved and then stimulated with PDGF to elicit ERK activation. In this context, decaging of active MKK6 also reduced ERK reporter translocation (Fig. 4D).
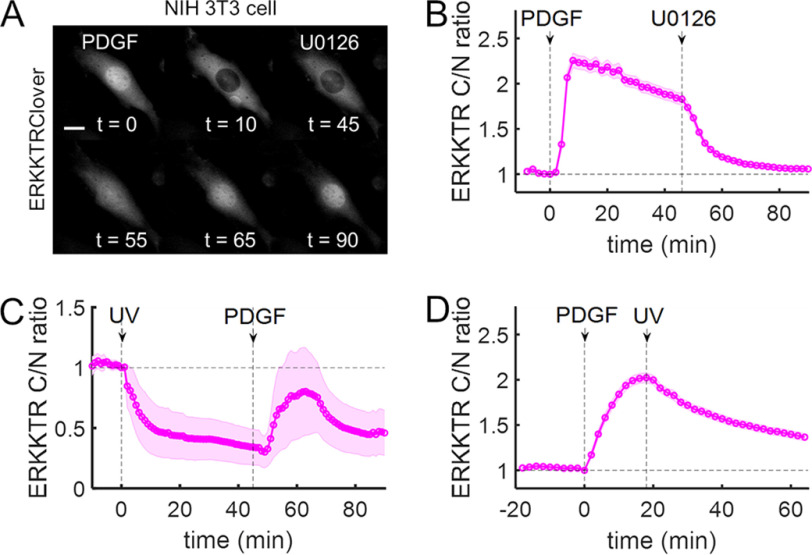
Figure 4.
Light-activated MKK6 negatively regulates ERK kinase activity. A, time-course montage of a representative NIH 3T3 cell expressing ERK kinase activity reporter ERKKTRClover that was first (t = 0 min) stimulated with PDGF (1 nm) and then (t = 45 min) treated with the MEK inhibitor, U0126 (20 μm). Scale bar, 10 μm; times are in minutes. B, quantification of the cell responses represented in A (n = 32). C, quantification of NIH 3T3 cells (n = 9) expressing caged MKK6 and ERKKTRClover that were first (t = 0 min) irradiated with UV (365 nm, <2 s) and then (t = 45 min) stimulated with PDGF (1 nm). D, NIH 3T3 cells (n = 83) co-expressing caged MKK6 and ERKKTRClover were serum-starved and then stimulated with PDGF, followed by brief exposure to UV light. The solid lines in B–D show the mean kinase translocation reporter translocation, and the shaded regions show the 95% confidence intervals. All data are representative of at least three independent experiments.
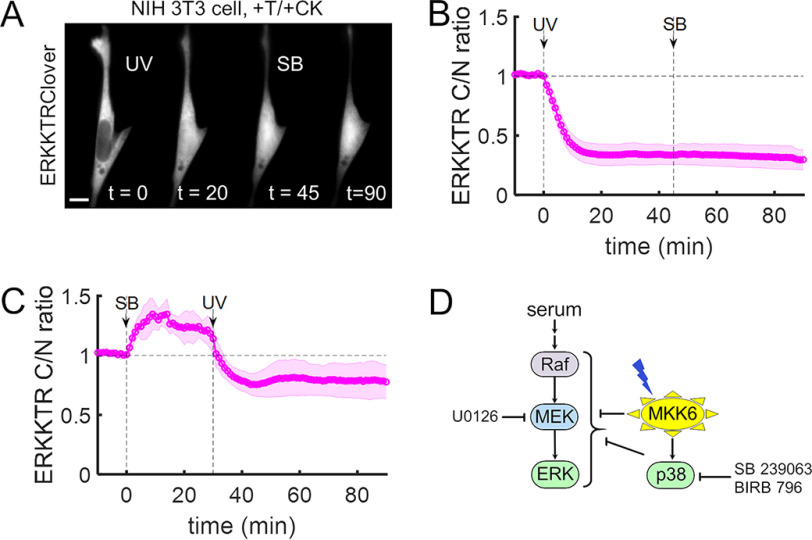
To further test this newly discovered role of MKK6 in the inhibition of ERK signaling through pathway cross-talk, we performed the MKK6-decaging experiment and subsequently added the p38 inhibitor SB 239063. Surprisingly, no significant recovery of ERK activity was observed; inhibition of p38 did not even partially reverse the negative regulation of ERK induced by light activation of MKK6 (Fig. 5 (A and B) and Movie S5). Faced with this evidence, we considered the possibility that the negative regulation of ERK could be p38-dependent, but inherently irreversible. To rule this out, we performed the experiment with the treatments in reverse order. With p38 inhibited by SB 239063, we observed a significant increase in ERK activity, consistent with the known cross-regulation (59); however, even with p38 activity blocked, subsequent decaging of MKK6 abrogated ERK activity as before (Fig. 5C). We repeated these experiments using the pan-p38 inhibitor BIRB 796, at a concentration (10 μm) reported to block all p38 isoforms (57) and obtained similar results (Fig. S6). These findings establish that active MKK6 rapidly and potently abrogates the ERK pathway in a p38-independent manner, in parallel with the previously established p38-dependent mechanisms (Fig. 5D).
Figure 5.
MKK6-mediated regulation of ERK is independent of p38 kinase activity. A, NIH 3T3 cells expressing caged MKK6 and ERKKTRClover were pre-incubated in medium containing 3% FBS. The time-course montage shows a representative cell that was first (t = 0 min) irradiated with UV (365 nm, <2 s) and then (t = 45 min) treated with p38α/β-specific inhibitor SB 239063 (10 μm). Scale bar, 10 μm; times are in minutes. B, quantification of the experiments represented in A (n = 9). C, cells prepared as in A and B were first treated with SB 239063 and then irradiated with UV (n = 8). The solid lines in B and C show the mean kinase translocation reporter translocation, and the shaded regions show the 95% confidence intervals. All data are representative of at least three independent experiments. D, simplified schematic illustrating known and putative regulatory links between MKK6 and the ERK pathway.
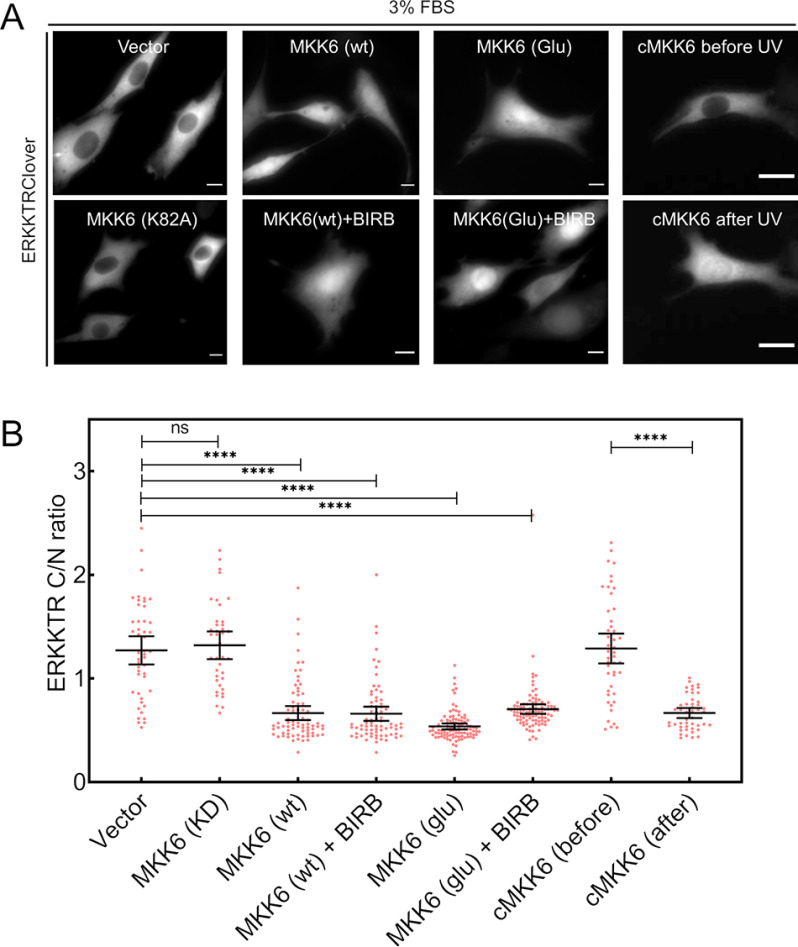
To corroborate the results obtained with caged MKK6 and its dependence on MKK6 activity, we co-expressed the ERK reporter with various MKK6 variants (43) and assessed ERK activity in the presence of 3% FBS (Fig. 6). Whereas expression of a kinase-dead MKK6 (K82A) had no apparent effect on ERK activity, expression of WT MKK6 or constitutively active MKK6 (S207E/T211E) diminished ERK reporter translocation whether or not p38 was inhibited (BIRB 796; Fig. 6A). Quantification of these data show that these comparisons are significant (Fig. 6B). Immunoblotting was performed to confirm that transfection of kinase-competent MKK6 yielded phosphorylation of p38 and of the p38 substrates, ATF-2/7, and that the latter response was blocked by BIRB 796 (Fig. S7). For the aforementioned conditions where ERK was inhibited, PDGF stimulation elicited little or no increase in ERK signaling in the majority of the cells (Fig. S8).
Figure 6.
Expression of kinase-competent MKK6 reduces ERK kinase activity. A, representative images of NIH 3T3 cells expressing ERKKTRClover along with either empty vector, kinase-dead MKK6 (K82A), WT MKK6 (with or without BIRB 796, 10 μm), constitutively active MKK6 (Glu, S207E/Y211E; with or without BIRB 796, 10 μm), or caged MKK6 (before and after UV irradiation) in medium containing 3% FBS. Scale bars, 10 μm. B, quantification of ERKKTRClover cytosol/nucleus (C/N) ratio for the conditions shown in A. Data are pooled from three independent experiments. ****, p < 10−4; ns, p > 0.05 (unpaired t test).
Whereas introduction of active MKK6 clearly regulates ERK kinase activity, as observed through the application of multiple biosensors, we did not find evidence implicating perturbation of the Raf/MEK/ERK phosphorylation cascade. MKK6 decaging did not perturb the nuclear localization of a GFP-tagged ERK2 construct, and transfection of kinase-active MKK6 did not elicit a reduction in MEK or ERK phosphorylation measured by immunoblotting (Fig. S9).
Light-activated MKK6 inhibits ERK signaling and abrogates mitosis in oncogene-addicted cells
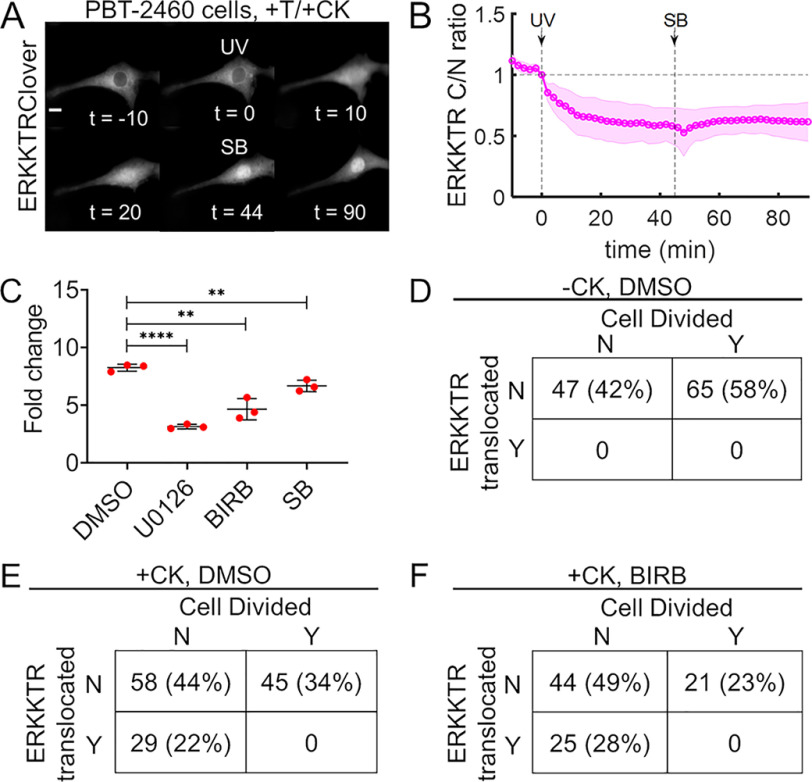
To confirm that MKK6-mediated regulation of ERK is not cell line–specific, we repeated the experiments of MKK6 decaging followed by p38 inhibition (as in Fig. 5 (A and B)) in serum-starved PBT-2460 cells, a genetically engineered murine melanoma line with constitutively elevated ERK signaling due to expression of active B-Raf (V600E) (60, 61). As seen in untransformed fibroblasts, light-activated MKK6 inhibited ERK signaling in PBT-2460 cells, even with p38 inhibited (Fig. 7 (A and B) and Movie S6; see Fig. S10 for the corresponding controls). Unlike untransformed cells, PBT-2460 cells proliferate in low serum (3% FBS), but they are addicted to V600E B-Raf signaling; their proliferation is abrogated by inhibition of MEK (Fig. 7C and Fig. S11). In the same set of experiments, PBT-2460 proliferation was modestly reduced by inhibition of p38, consistent with reports that p38 is required for efficient G2-M transition (62). In live-cell imaging experiments, PBT-2460 cells with stable expression of the ERK kinase translocation reporter and transfected with caged MKK6 were irradiated and monitored for ∼16 h. When the cells had not been incubated with CK beforehand, none exhibited a detectable decrease in ERK activity upon UV treatment, and 58% of the observed cells divided (Fig. 7D). In cells that had been incubated with CK, 22% of the cells exhibited a rapid drop in ERK activity following UV treatment (consistent with the transfection efficiency for PBT-2460). ERK activity remained depressed in those cells, and none of them divided, during the 16-h post-light exposure. Among the cells in this cohort that did not show a rapid drop in ERK activity, a substantial fraction divided (Fig. 7E and Movie S7). The same relationship between ERK signaling and mitosis was observed when the experiment was performed in the presence of the p38 inhibitor BIRB 796, except with somewhat reduced overall proliferation consistent with the results shown in Fig. 7C (Fig. 7F). These observations confirm that p38-independent regulation of ERK signaling by rapid introduction of active MKK6 prevents mitosis of cells addicted to the ERK pathway. As a whole, this study demonstrates how optical control of kinase function through unnatural amino acid mutagenesis with photocaged lysine can be used to study the dynamic regulation of normal and dysfunctional signaling networks.
Figure 7.
MKK6-mediated regulation of ERK blocks proliferation of oncogene-addicted cells. A, time-course montage of a representative, serum-starved PBT-2460 mouse melanoma cell expressing caged MKK6 and ERKKTRClover that was first (t = 0 min) irradiated with UV (365 nm, <2 s) and then (t = 45 min) treated with p38α/β-specific inhibitor SB 239063 (10 μm). Scale bar, 10 μm; times are in minutes. B, quantification of the experiments represented in A (n = 8; representative of three independent experiments). The solid line shows the mean kinase translocation reporter translocation, and the shaded regions show the 95% confidence intervals. C, proliferation assay of murine melanoma PBT-2460 cells in low-serum (3% FBS and essential and nonessential amino acids) medium containing DMSO vehicle (0.1%, v/v), MEK1/2 inhibitor U0126 (10 μm), or either p38 inhibitor BIRB 796 (10 μm) or SB 239063 (10 μm). Proliferation is quantified as the -fold change in cell density after 48 h, and the error bars show mean ± S.D. (n = 3 independent experiments); ****, p < 10−4; **, p < 0.01 (unpaired t test). D, PBT-2460 cells stably expressing ERKKTRClover were transiently transfected with caged MKK6, but CK was withheld. After 24 h, cells were switched to imaging buffer supplemented with nonessential amino acids and DMSO vehicle control (0.1% v/v), irradiated with UV on our microscope, and monitored for ∼16 h. Cells were classified according to whether they did/did not show translocation of the ERK reporter following UV treatment and whether they did/did not divide during the experiment, and the numbers and percentages of cells are tabulated (pooled from three independent experiments). E, same as D except that 2 mm CK was supplied following transfection. F, same as E except the imaging buffer contained the pan-p38 inhibitor, BIRB 796 (10 μm).
Discussion
Protein kinases form the backbone of signal transduction networks, and several genetic and proteomic approaches have been developed and refined to study their regulation and function (63–65). Considering that signaling networks are rife with cross-talk and feedback interactions, isolating the function of a particular signaling protein is challenging. The general, genetically encoded photodecaging approach applied here to MKK6 allows for introduction of a particular kinase activity in an otherwise quiescent background in a dynamic fashion. It combines the complete specificity of genetic tools with the temporal precision of pharmacological inhibitors. In contrast to the latter, abrupt gain-of-function is achieved, enabling studies that are otherwise difficult to perform. Given that MKK6 phosphorylates and thus activates the p38 SAPKs, which are functionally associated with apoptosis in many cell types, the abrupt introduction of activity is absolutely necessary, because the long-term activation of stress-activated signaling inherently forces selection and/or adaptation within the cell population. Indeed, in cells transfected with either WT or constitutively active MKK6 (∼60% transfection efficiency), we found that the cells initially survived in high-serum growth medium, but after 48 h, the cell population showed substantial cell death relative to empty-vector control (Fig. S12).
In the context of apoptosis, simple transfection of active MKK6 or p38 only allows one to evaluate overall cell death; it is not possible to study the dynamics of pre- and post-mitochondrial apoptotic processes, nor is it possible to cleanly isolate the effect of a single kinase by incubating cells with pro-apoptotic factors or stressors. Here, we found that decaging active MKK6 in a quiescent background was sufficient to induce a complete and kinetically staged apoptotic response, which required the activation of endogenous p38. The early activation of p38 following MKK6 decaging was accompanied by cleavage of a caspase-3/7 biosensor. This early emergence of executioner caspase activity is consistent with the reported ability of p38 to mediate activation of the initiator caspase-8, either in concert with or independent of death receptor signaling (66, 67).
Loss of mitochondrial integrity, marked by release of cytochrome c into the cytoplasm, is a pivotal occurrence during apoptosis that was typically observed within several hours of MKK6 activation through light-induced decaging. The timing of this event varied broadly in the cell population, consistent with natural apoptotic responses to pro-death signals and environmental insults. The aforementioned induction of caspases-3 and -7 likely contributes to the late-stage response, considering that recombinant caspase-3 is sufficient to induce cytochrome c release from isolated mitochondria, accompanied by cleavage of the anti-apoptotic regulator, Bcl-2 (55). In parallel, p38-mediated inhibition of Bcl-2 and the related Bcl-xL (68) and/or potentiation of the pro-apoptotic regulators Bax and Bim (69, 70) might also contribute to late-stage loss of mitochondrial membrane potential and release of cytochrome c.
Our experiments also showed that after a reasonably consistent p38 activation response following MKK6 decaging (Fig. 2C), cells typically showed a further, sudden increase in p38 activity after a variable lag (Fig. 3B). This delayed increase in p38 activity reporter translocation was not due to collapse of the nuclear envelope, and it was often transient. The phenomenon has hallmarks of an excitable system and could be driven by positive feedback, which might or might not be intertwined with the apoptotic cascade. For example, activation of caspase-3 has been implicated in p38 activation through cleavage of the upstream kinase MEKK1 to produce a constitutively active, truncated form (71, 72). Another pivotal player in the p38 and apoptotic signaling networks is the tumor suppressor p53 (73, 74), which responds to p38-catalyzed phosphorylation (75, 76) and can enhance p38 activity through increased reactive oxygen species production (77). Future studies will reveal the dynamics of these intertwined networks; the general approach demonstrated here offers a systematic way to do so.
Decaging MKK6 also rapidly and potently abrogated ERK activity, theretofore maintained by a moderate concentration of serum. Although negative cross-talk among MAPK cascades has been described for some time (12, 78, 79), and despite strong indications that p38 enhances the activity of PP2A as a regulator of the Raf/MEK/ERK pathway (13, 80), we unexpectedly found that acute activation of MKK6 rapidly mediates the inhibition of ERK1/2 in a p38 kinase activity–independent fashion (as demonstrated through conditional blocking of p38 activity). In the context of B-Raf-transformed cells, which are reliant on ERK signaling for proliferation, this regulation blocked cell division. Because our understanding of MKK6 function is currently limited to its activation of p38 kinase activities by phosphorylation, these results force a broader view of MKK6 as a signaling branch point affecting pro-death and anti-proliferative pathways by mechanisms that are apparently different from those previously thought. We assert that a deeper investigation of the mechanisms by which MKK6 regulates the ERK pathway will be necessary, as might be a general re-examination of MAPKK functions.
Regarding the mechanism of ERK1/2 regulation, we did not detect significant MKK6-mediated inhibition of the upstream pathway at the level of ERK localization or MEK/ERK phosphorylation. Although stasis of ERK localization or MEK/ERK phosphorylation can belie changes in the pathway input (81), we are inclined to consider other mechanisms. An intriguing observation in that regard is that an MKK6-based docking-site peptide inhibits in vitro ERK2 activity with potency comparable with that of MEK1/2-based peptides (82). If active MKK6 were able to sequester ERK1/2, presumably MKK6 would be in competition with MEK1/2; however, protein scaffolds may bias MEK-ERK interactions toward inactive ERK (83), indirectly biasing the hypothetical MKK6-ERK interactions toward active ERK.
Our results using caged MKK6 illustrate how acute and precise activation of a single kinase can affect signaling and cell response outcomes. MKK6 activation blocked pro-proliferative signaling while promoting apoptosis, suggesting that MKK6 orchestrates a definitive switch in cell priorities and fate. Such cell fate switches are likely to vary across contexts of cell type and environmental conditions and are critically important for understanding outcomes of targeted cancer therapies, for example. Here, in the context of cancer, we have scratched the surface of the application of genetically encoded, caged kinases, which could be used to probe dynamics of signaling networks with aberrant or reprogrammed kinomes (84).
Materials and methods
Cell culture and other commercial reagents
HEK293T cells and NIH 3T3 mouse fibroblast cells were acquired from American Type Culture Collection. PBT-2460 tumor cells (60, 61) were a gift from James E. Bear (University of North Carolina, Chapel Hill, NC, USA). Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS, 2 mm l-glutamine, and the antibiotics penicillin and streptomycin in a 37 °C, 5% CO2 incubator. All cell culture reagents were purchased from Thermo Fisher Scientific/Invitrogen. Transient transfections were performed using Lipofectamine and PLUS reagents (Thermo Fisher Scientific).
Human recombinant PDGF-BB was purchased from Peprotech (catalog no. 100-14B). Antibodies against p38 MAPK (catalog no. 9212), p38 MAPK Thr180/Tyr182 (catalog no. 4511), ATF-2 pThr71/ATF-7 pThr53 (catalog no. 27934), ERK1/2 (catalog no. 9107), ERK pThr202/pTyr204 (catalog no. 9101), MEK1/2 (catalog no. 9126), MEK pSer217/pSer221 (catalog no. 9121), and β-actin (catalog no. 4970) were purchased from Cell Signaling Technologies. Pharmacological inhibitors U0126 (catalog no. 662005), SB 239063 (catalog no. S0569-5MG), and BIRB 796 (catalog no. 506172) were obtained from MilliporeSigma. All other reagents, except where otherwise noted, were from MilliporeSigma.
CK
The photocaged lysine amino acid was synthesized as reported previously (31).
Plasmids
The plasmid encoding CKRS and 4 copies of the CMV enhancer-U6-pylT expression cassette was previously constructed (85). The original coding sequence for FLAG-caMKK6 was obtained from Addgene (no. 13518). The K82TAG mutation was subsequently introduced by site-directed mutagenesis (forward primer, 5′-ggcagtgTAGcggatccgagccacagtaaatagccaggaacagaaacgg-3′; reverse primer, 5′-ggatccgCTAcactgccatgatctgcccgctgggcac-3′). The original pE363 plasmid encoding sfGFP and four copies of the U6-pylT expression cassette was kindly provided by the Chin laboratory (31). After sfGFP was removed by HindIII/KpnI double digestion, the FLAG-caMKK6 K82TAG coding sequence was inserted between the two restriction sites (forward primer, 5′-GCCGAAGCTTATGGACTATAAGGACGATGA-3′; reverse primer, 5′-GCGCGGTACCTTAGTCTCCAAGAATCAGTT-3′).
The following live-cell fluorescence reporters were obtained from Addgene: kinase translocation reporters for ERK (pLentiCMV Puro DEST ERKKTRClover; Addgene no. 59150), p38 (pLentiPGK Puro DEST p38KTRClover; Addgene no. 59152) and JNK (pLentiPGK Blast DEST JNKKTRmRuby2; Addgene no. 59154) (45); GFP-ERK2 (Addgene no. 118296) (86), pBabe(LTR)-Cytochrome C-GFP (Addgene no. 41183) (54); and the caspase-3/7 cleavage reporter CA-GFP (Addgene no. 32748) (56). The MKK6 mutants (43) were also acquired from Addgene: pCDNA3-FLAG MKK6 (Addgene no. 13517), pCDNA3-FLAG MKK6 (K82A) (Addgene no. 13519), pCDNA3-FLAG MKK6(glu) (Addgene no. 13518), and pCDNA3-FLAG-HA (Addgene no. 10792).
Transient transfection, live-cell imaging, and image analysis
NIH 3T3 cells plated at 50–60% confluence were transiently transfected with Lipofectamine 3000 and P3000 reagent in normal growth medium with no antibiotics, following the manufacturer's protocol. Where applicable, CK (2 mm) was added to the growth medium immediately after the transfection. 24 h after transient transfection, cells were replated on fibronectin (10 µg/ml)-coated glass-bottom dishes (MatTek) and were allowed to spread in the same growth medium for 3-4 h. For serum-starved conditions, the cells were incubated for an additional 4 h in imaging buffer (20 mm HEPES, 125 mm NaCl, 5 mm KCl, 1.5 mm MgCl2, 1.5 mm CaCl2, 10 mm glucose, and 2 mg/ml fatty acid-free BSA, pH 7.4) supplemented with 0.1% FBS and penicillin-streptomycin-glutamine. The buffer was maintained at 37 °C during experiments, and mineral oil was layered on top to prevent evaporation.
Epifluorescence microscopy was performed with a modified microscope (Axioskop 2 FS) equipped with a 50-watt mercury lamp, ×40/0.8 NA and ×60/1.0 NA Achroplan water-dipping objectives (Carl Zeiss), a Hamamatsu ORCA ER charge-coupled device camera, and MetaMorph image acquisition software (Universal Imaging). The microscope is outfitted with an automated multistage and CRISP autofocus control system (Applied Scientific Instrumentation). Band-pass excitation filters were 350/50 nm for UV decaging, 436/20 nm for Cerulean (FRET reporters), and 488/10 nm for Clover and EGFP constructs. Band-pass emission filters were 480/40 and 535/30 nm for Cerulean and Venus, respectively (FRET reporters) and 525/50 nm for Clover and EGFP constructs. UV irradiation was from the mercury lamp passed through a DAPI filter. The duration of exposure (1-2 s) was controlled using a shutter (Uniblitz). All filters were obtained from Chroma Technology.
The mean intensity of an acellular region of each image was considered as background and was subtracted from the intensity of each pixel before further analysis. For nucleocytoplasmic translocation analyses, nuclear and cytosolic masks were generated for each cell using MATLAB-based Microscopy Image Browser software (87). Mean nuclear and cytosolic intensities were calculated for each frame, accounting for modest photobleaching during the experimental time period. To quantify cytochrome c release, a custom MATLAB code was written to track each individual cell across consecutive image frames, and the entropy (a measure of randomness commonly used to quantify image texture) of each cellular region of each frame was calculated using the entropy function. For FRET biosensors, the cellular regions were segmented from the background, and the ratio of acceptor (Venus) to donor (Cerulean) fluorescence intensities was calculated for each frame. For all live-cell fluorescence measurements, the value for each cell was normalized by the value prior to stimulation. For cell proliferation measurements, equal densities of cells were plated in each dish, and one dish was used to measure the initial cell density; 48 h after treatment, cell density was measured for each condition. Nuclei were stained with Hoechst 33258 (1 µg/ml) before imaging at ×10 magnification. Images of 10-15 fields/dish were acquired to estimate cell density. The -fold change in cell density was calculated using average cell density of all imaging fields of each condition relative to that of the initial condition. For the cell division assay, individual cells were tracked using Lineage Mapper (88), which automatically detects cell division events. The data were reviewed by eye to confirm the events.
Immunoblotting
SDS-PAGE, wet electrophoretic transfer to polyvinylidene difluoride membranes, and immunoblotting were performed by standard methods. After primary and horseradish peroxidase–conjugated secondary antibody incubations and intermittent washes, SuperSignal West Femto Maximum Sensitivity substrate (Pierce/Fisher Scientific, catalog no. PI34095) was applied, and chemiluminescence was imaged using a digital imaging system (Syngene G:BOX, Synoptics Group).
Data Availability
Data not contained in the paper are available by request from the corresponding author (Jason M. Haugh, jason_haugh@ncsu.edu).
Supplementary Material
This article contains supporting information.
Author contributions—S. M. T. R., A. D., and J. M. H. conceptualization; S. M. T. R. and W. Z. data curation; S. M. T. R. and W. Z. formal analysis; S. M. T. R. and J. M. H. supervision; S. M. T. R., A. D., and J. M. H. funding acquisition; S. M. T. R., W. Z., and J. M. H. writing-original draft; S. M. T. R., W. Z., A. D., and J. M. H. writing-review and editing; W. Z, A. D., and J. M. H. resources; W. Z., A. D., and J. M. H. methodology.
Funding and additional information—This work was supported by National Science Foundation Grants MCB-1330746 (to A. D. and J. M. H.) and CBET-1603930 (to A. D.) and a UNC Lineberger Developmental Grant from the University Cancer Research Fund.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- MAPK
- mitogen-activated protein kinase
- MAPKK
- MAPK kinase
- MAPKKK
- MAPKK kinase
- ERK
- extracellular signal–regulated kinase
- JNK
- c-Jun N-terminal kinase
- SAPK
- stress-activated protein kinase
- PP2A
- protein phosphatase 2A
- CK
- caged lysine
- UAA
- unnatural amino acid
- pylRS
- pyrrolysyl-tRNA synthetase
- pylT
- tRNACUA
- DAPI
- 4′,6-diamidino-2-phenylindole
- PDGF
- platelet-derived growth factor
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase
- FBS
- fetal bovine serum
- NA
- numerical aperture
- CMV
- cytomegalovirus.
References
- 1. Chang L., and Karin M. (2001) Mammalian MAP kinase signalling cascades. Nature 410, 37–40 10.1038/35065000 [DOI] [PubMed] [Google Scholar]
- 2. Johnson G. L., and Lapadat R. (2002) Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298, 1911–1912 10.1126/science.1072682 [DOI] [PubMed] [Google Scholar]
- 3. Pearson G., Robinson F., Gibson T. B., Xu B. E., Karandikar M., Berman K., and Cobb M. H. (2001) Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 22, 153–183 10.1210/edrv.22.2.0428 [DOI] [PubMed] [Google Scholar]
- 4. Wei Z., and Liu H. T. (2002) MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 12, 9–18 10.1038/sj.cr.7290105 [DOI] [PubMed] [Google Scholar]
- 5. Mansouri A., Ridgway L. D., Korapati A. L., Zhang Q., Tian L., Wang Y., Siddik Z. H., Mills G. B., and Claret F. X. (2003) Sustained activation of JNK/p38 MAPK pathways in response to cisplatin leads to Fas ligand induction and cell death in ovarian carcinoma cells. J. Biol. Chem. 278, 19245–19256 10.1074/jbc.M208134200 [DOI] [PubMed] [Google Scholar]
- 6. Sui X., Kong N., Ye L., Han W., Zhou J., Zhang Q., He C., and Pan H. (2014) P38 and JNK MAPK pathways control the balance of apoptosis and autophagy in response to chemotherapeutic agents. Cancer Lett. 344, 174–179 10.1016/j.canlet.2013.11.019 [DOI] [PubMed] [Google Scholar]
- 7. Wada T., and Penninger J. M. (2004) Mitogen-activated protein kinases in apoptosis regulation. Oncogene 23, 2838–2849 10.1038/sj.onc.1207556 [DOI] [PubMed] [Google Scholar]
- 8. Dhillon A. S., Hagan S., Rath O., and Kolch W. (2007) MAP kinase signalling pathways in cancer. Oncogene 26, 3279–3290 10.1038/sj.onc.1210421 [DOI] [PubMed] [Google Scholar]
- 9. Wagner E. F., and Nebreda Á. R. (2009) Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer 9, 537–549 10.1038/nrc2694 [DOI] [PubMed] [Google Scholar]
- 10. Caunt C. J., and Keyse S. M. (2013) Dual-specificity MAP kinase phosphatases (MKPs): Shaping the outcome of MAP kinase signalling. FEBS J. 280, 489–504 10.1111/j.1742-4658.2012.08716.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Finch A. R., Caunt C. J., Perrett R. M., Tsaneva-Atanasova K., and McArdle C. A. (2012) Dual specificity phosphatases 10 and 16 are positive regulators of EGF-stimulated ERK activity: indirect regulation of ERK signals by JNK/p38 selective MAPK phosphatases. Cell. Signal. 24, 1002–1011 10.1016/j.cellsig.2011.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang H., Shi X., Hampong M., Blanis L., and Pelech S. (2001) Stress-induced inhibition of ERK1 and ERK2 by direct interaction with p38 MAP kinase. J. Biol. Chem. 276, 6905–6908 10.1074/jbc.C000917200 [DOI] [PubMed] [Google Scholar]
- 13. Westermarck J., Li S.-P., Kallunki T., Han J., and Kähäri V.-M. (2001) p38 mitogen-activated protein kinase-dependent activation of protein phosphatases 1 and 2A inhibits MEK1 and MEK2 activity and collagenase 1 (MMP-1) gene expression. Mol. Cell Biol. 21, 2373–2383 10.1128/MCB.21.7.2373-2383.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grethe S., and Pörn-Ares M. I. (2006) p38 MAPK regulates phosphorylation of Bad via PP2A-dependent suppression of the MEK1/2-ERK1/2 survival pathway in TNF-α induced endothelial apoptosis. Cell. Signal. 18, 531–540 10.1016/j.cellsig.2005.05.023 [DOI] [PubMed] [Google Scholar]
- 15. Li S. P., Junttila M. R., Han J., Kähäri V. M., and Westermarck J. (2020) P38 mitogen-activated protein kinase pathway suppresses cell survival by inducing dephosphorylation of mitogen-activated protein/extracellular signal-regulated kinase kinase1,2. Cancer Res. 80, 364 10.1158/0008-5472.CAN-19-3320 [DOI] [PubMed] [Google Scholar]
- 16. Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., Mclauchlan H., Klevernic I., Arthur J. S. C., Alessi D. R., and Cohen P. (2007) The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297–315 10.1042/BJ20070797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Díaz-Martínez L. A., Karamysheva Z. N., Warrington R., Li B., Wei S., Xie X., Roth M. G., and Yu H. (2014) Genome‐wide siRNA screen reveals coupling between mitotic apoptosis and adaptation. EMBO J. 33, 1960–1976 10.15252/embj.201487826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weaver B. A. A., and Cleveland D. W. (2005) Decoding the links between mitosis, cancer, and chemotherapy: the mitotic checkpoint, adaptation, and cell death. Cancer Cell 8, 7–12 10.1016/j.ccr.2005.06.011 [DOI] [PubMed] [Google Scholar]
- 19. Kennedy M. J., Hughes R. M., Peteya L. A., Schwartz J. W., Ehlers M. D., and Tucker C. L. (2010) Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975 10.1038/nmeth.1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Levskaya A., Weiner O. D., Lim W. A., and Voigt C. A. (2009) Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001 10.1038/nature08446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Umeda N., Ueno T., Pohlmeyer C., Nagano T., and Inoue T. (2011) A photocleavable rapamycin conjugate for spatiotemporal control of small GTPase activity. J. Am. Chem. Soc. 133, 12–14 10.1021/ja108258d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yazawa M., Sadaghiani A. M., Hsueh B., and Dolmetsch R. E. (2009) Induction of protein-protein interactions in live cells using light. Nat. Biotechnol. 27, 941–945 10.1038/nbt.1569 [DOI] [PubMed] [Google Scholar]
- 23. Pathak G. P., Vrana J. D., and Tucker C. L. (2013) Optogenetic control of cell function using engineered photoreceptors. Biol. Cell 105, 59–72 10.1111/boc.201200056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Toettcher J. E., Weiner O. D., and Lim W. A. (2013) Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–1434 10.1016/j.cell.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee S., Park H., Kyung T., Kim N. Y., Kim S., Kim J., and Heo W. D. (2014) Reversible protein inactivation by optogenetic trapping in cells. Nat. Methods 11, 633–636 10.1038/nmeth.2940 [DOI] [PubMed] [Google Scholar]
- 26. Guntas G., Hallett R. A., Zimmerman S. P., Williams T., Yumerefendi H., Bear J. E., and Kuhlman B. (2015) Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. U.S.A. 112, 112–117 10.1073/pnas.1417910112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Enslen H., Raingeaud J., and Davis R. J. (1998) Selective activation of p38 mitogen-activated protein (MAP) kinase isoforms by the MAP kinase kinases MKK3 and MKK6. J. Biol. Chem. 273, 1741–1748 10.1074/jbc.273.3.1741 [DOI] [PubMed] [Google Scholar]
- 28. Han J., Lee J. D., Jiang Y., Li Z., Feng L., and Ulevitch R. J. (1996) Characterization of the structure and function of a novel MAP kinase kinase (MKK6). J. Biol. Chem. 271, 2886–2891 10.1074/jbc.271.6.2886 [DOI] [PubMed] [Google Scholar]
- 29. Edwards W. F., Young D. D., and Deiters A. (2009) Light-activated Cre recombinase as a tool for the spatial and temporal control of gene function in mammalian cells. ACS Chem. Biol. 4, 441–445 10.1021/cb900041s [DOI] [PubMed] [Google Scholar]
- 30. Chou C., Young D. D., and Deiters A. (2010) Photocaged T7 RNA polymerase for the light activation of transcription and gene function in pro- and eukaryotic cells. ChemBioChem 11, 972–977 10.1002/cbic.201000041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gautier A., Nguyen D. P., Lusic H., An W., Deiters A., and Chin J. W. (2010) Genetically encoded photocontrol of protein localization in mammalian cells. J. Am. Chem. Soc. 132, 4086–4088 10.1021/ja910688s [DOI] [PubMed] [Google Scholar]
- 32. Gautier A., Deiters A., and Chin J. W. (2011) Light-activated kinases enable temporal dissection of signaling networks in living cells. J. Am. Chem. Soc. 133, 2124–2127 10.1021/ja1109979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Q., and Deiters A. (2014) Optochemical control of deoxyoligonucleotide function via a nucleobase-caging approach. Acc. Chem. Res. 47, 45–55 10.1021/ar400036a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ankenbruck N., Courtney T., Naro Y., and Deiters A. (2018) Optochemical control of biological processes in cells and animals. Angew. Chem. Int. Ed. Engl. 57, 2768–2798 10.1002/anie.201700171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Courtney T. M., and Deiters A. (2019) Optical control of protein phosphatase function. Nat. Commun. 10, 1–10 10.1038/s41467-018-07882-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Courtney T., and Deiters A. (2018) Recent advances in the optical control of protein function through genetic code expansion. Curr. Opin. Chem. Biol. 46, 99–107 10.1016/j.cbpa.2018.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goldstein J. C., Muñoz-Pinedo C., Ricci J. E., Adams S. R., Kelekar A., Schuler M., Tsien R. Y., and Green D. R. (2005) Cytochrome c is released in a single step during apoptosis. Cell Death Differ. 12, 453–462 10.1038/sj.cdd.4401596 [DOI] [PubMed] [Google Scholar]
- 38. Muñoz-Pinedo C., Guío-Carrión A., Goldstein J. C., Fitzgerald P., Newmeyer D. D., and Green D. R. (2006) Different mitochondrial intermembrane space proteins are released during apoptosis in a manner that is coordinately initiated but can vary in duration. Proc. Natl. Acad. Sci. U.S.A. 103, 11573–11578 10.1073/pnas.0603007103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Spencer S. L., Gaudet S., Albeck J. G., Burke J. M., and Sorger P. K. (2009) Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature 459, 428–432 10.1038/nature08012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liaunardy-Jopeace A., Murton B. L., Mahesh M., Chin J. W., and James J. R. (2017) Encoding optical control in LCK kinase to quantitatively investigate its activity in live cells. Nat. Struct. Mol. Biol. 24, 1155–1163 10.1038/nsmb.3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu J., Hemphill J., Samanta S., Tsang M., and Deiters A. (2017) Genetic code expansion in zebrafish embryos and its application to optical control of cell signaling. J. Am. Chem. Soc. 139, 9100–9103 10.1021/jacs.7b02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saraste M., Sibbald P. R., and Wittinghofer A. (1990) The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15, 430–434 10.1016/0968-0004(90)90281-F [DOI] [PubMed] [Google Scholar]
- 43. Raingeaud J., Whitmarsh A. J., Barrett T., Dérijard B., and Davis R. J. (1996) MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol. Cell Biol. 16, 1247–1255 10.1128/MCB.16.3.1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Remy G., Risco A. M., Iñesta-Vaquera F. A., González-Terán B., Sabio G., Davis R. J., and Cuenda A. (2010) Differential activation of p38MAPK isoforms by MKK6 and MKK3. Cell. Signal. 22, 660–667 10.1016/j.cellsig.2009.11.020 [DOI] [PubMed] [Google Scholar]
- 45. Regot S., Hughey J. J., Bajar B. T., Carrasco S., and Covert M. W. (2014) High-sensitivity measurements of multiple kinase activities in live single cells. Cell 157, 1724–1734 10.1016/j.cell.2014.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Underwood D. C., Osborn R. R., Kotzer C. J., Adams J. L., Lee J. C., Webb E. F., Carpenter D. C., Bochnowicz S., Thomas H. C., Hay D. W. P., and Griswold D. E. (2000) SB 239063, a potent p38 MAP kinase inhibitor, reduces inflammatory cytokine production, airways eosinophil infiltration, and persistence. J. Pharmacol. Exp. Ther. 293, 281–288 [PubMed] [Google Scholar]
- 47. Xia Z., Dickens M., Raingeaud J., Davis R. J., and Greenberg M. E. (1995) Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science 270, 1326–1331 10.1126/science.270.5240.1326 [DOI] [PubMed] [Google Scholar]
- 48. Hoppe J., Kilic M., Hoppe V., Sachinidis A., and Kagerhuber U. (2002) Formation of caspase-3 complexes and fragmentation of caspase-12 during anisomycin-induced apoptosis in AKR-2B cells without aggregation of Apaf-1. Eur. J. Cell Biol. 81, 567–576 10.1078/0171-9335-00276 [DOI] [PubMed] [Google Scholar]
- 49. Joo S. S., and Yoo Y. M. (2009) Melatonin induces apoptotic death in LNCaP cells via p38 and JNK pathways: therapeutic implications for prostate cancer. J. Pineal Res. 47, 8–14 10.1111/j.1600-079X.2009.00682.x [DOI] [PubMed] [Google Scholar]
- 50. Tobiume K., Matsuzawa A., Takahashi T., Nishitoh H., Morita K. I., Takeda K., Minowa O., Miyazono K., Noda T., and Ichijo H. (2001) ASK1 is required for sustained activations of JNK/p38 MAP kinases and apoptosis. EMBO Rep. 2, 222–228 10.1093/embo-reports/kve046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Coleman M. L., Sahai E. A., Yeo M., Bosch M., Dewar A., and Olson M. F. (2001) Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat. Cell Biol. 3, 339–345 10.1038/35070009 [DOI] [PubMed] [Google Scholar]
- 52. Häcker G. (2000) The morphology of apoptosis. Cell Tissue Res. 301, 5–17 10.1007/s004410000193 [DOI] [PubMed] [Google Scholar]
- 53. Albeck J. G., Burke J. M., Aldridge B. B., Zhang M., Lauffenburger D. A., and Sorger P. K. (2008) Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol. Cell. 30, 11–25 10.1016/j.molcel.2008.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goldstein J. C., Waterhouse N. J., Juin P., Evan G. I., and Green D. R. (2000) The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat. Cell Biol. 2, 156–162 10.1038/35004029 [DOI] [PubMed] [Google Scholar]
- 55. Chen Q., Gong B., and Almasan A. (2000) Distinct stages of cytochrome c release from mitochondria: evidence for a feedback amplification loop linking caspase activation to mitochondrial dysfunction in genotoxic stress induced apoptosis. Cell Death Differ. 7, 227–233 10.1038/sj.cdd.4400629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nicholls S. B., Chu J., Abbruzzese G., Tremblay K. D., and Hardy J. A. (2011) Mechanism of a genetically encoded dark-to-bright reporter for caspase activity. J. Biol. Chem. 286, 24977–24986 10.1074/jbc.M111.221648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kuma Y., Sabio G., Bain J., Shpiro N., Márquez R., and Cuenda A. (2005) BIRB796 inhibits all p38 MAPK isoforms in vitro and in vivo. J. Biol. Chem. 280, 19472–19479 10.1074/jbc.M414221200 [DOI] [PubMed] [Google Scholar]
- 58. Harvey C. D., Ehrhardt A. G., Cellurale C., Zhong H., Yasuda R., Davis R. J., and Svoboda K. (2008) A genetically encoded fluorescent sensor of ERK activity. Proc. Natl. Acad. Sci. U.S.A. 105, 19264–19269 10.1073/pnas.0804598105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shimo T., Matsumura S., Ibaragi S., Isowa S., Kishimoto K., Mese H., Nishiyama A., and Sasaki A. (2007) Specific inhibitor of MEK-mediated cross-talk between ERK and p38 MAPK during differentiation of human osteosarcoma cells. J. Cell Commun. Signal. 1, 103–111 10.1007/s12079-007-0010-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Asokan S. B., Johnson H. E., Rahman A., King S. J., Rotty J. D., Lebedeva I. P., Haugh J. M., and Bear J. E. (2014) Mesenchymal chemotaxis requires selective inactivation of Myosin II at the leading edge via a noncanonical PLCγ/PKCα pathway. Dev. Cell. 31, 747–760 10.1016/j.devcel.2014.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hanna S. C., Krishnan B., Bailey S. T., Moschos S. J., Kuan P. F., Shimamura T., Osborne L. D., Siegel M. B., Duncan L. M., O'Brien E. T., Superfine R., Miller C. R., Simon M. C., Wong K. K., and Kim W. Y. (2013) HIF1α and HIF2α independently activate SRC to promote melanoma metastases. J. Clin. Invest. 123, 2078–2093 10.1172/JCI66715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bulavin D. V., Higashimoto Y., Popoff I. J., Gaarde W. A., Basrur V., Potapova O., Appella E., and Fornace A. J. (2001) Initiation of a G2/M checkpoint after ultraviolet radiation requires p38 kinase. Nature 411, 102–107 10.1038/35075107 [DOI] [PubMed] [Google Scholar]
- 63. Gstaiger M., and Aebersold R. (2009) Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat. Rev. Genet. 10, 617–627 10.1038/nrg2633 [DOI] [PubMed] [Google Scholar]
- 64. Wolf-Yadlin A., Hautaniemi S., Lauffenburger D. A., and White F. M. (2007) Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U.S.A. 104, 5860–5865 10.1073/pnas.0608638104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shi Q., Qin L., Wei W., Geng F., Fan R., Shin Y. S., Guo D., Hood L., Mischel P. S., and Heath J. R. (2012) Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proc. Natl. Acad. Sci. U.S.A. 109, 419–424 10.1073/pnas.1110865109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yoon C. H., Kim M. J., Park M. T., Byun J. Y., Choi Y. H., Yoo H. S., Lee Y. M., Hyun J. W., and Lee S. J. (2009) Activation of p38 mitogen-activated protein kinase is required for death receptor-independent caspase-8 activation and cell death in response to sphingosine. Mol. Cancer Res. 7, 361–370 10.1158/1541-7786.MCR-08-0069 [DOI] [PubMed] [Google Scholar]
- 67. Tourian L., Zhao H., and Srikant C. B. (2004) p38α but not p38β, inhibits the phosphorylation and presence of c-FLIPs in DISC to potentiate Fas-mediated caspase-8 activation and type I apoptotic signaling. J. Cell Sci. 117, 6459–6471 10.1242/jcs.01573 [DOI] [PubMed] [Google Scholar]
- 68. Farley N., Pedraza-Alva G., Serrano-Gomez D., Nagaleekar V., Aronshtam A., Krahl T., Thornton T., and Rincón M. (2006) p38 mitogen-activated protein kinase mediates the Fas-induced mitochondrial death pathway in CD8+ T cells. Mol. Cell Biol. 26, 2118–2129 10.1128/MCB.26.6.2118-2129.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu J., Wu N., Ma L. N., Zhong J. T., Liu G., Zheng L. H., and Lin X. K. (2014) p38 MAPK signaling mediates mitochondrial apoptosis in cancer cells induced by oleanolic acid. Asian Pacific J. Cancer Prev. 15, 4519–4525 10.7314/APJCP.2014.15.11.4519 [DOI] [PubMed] [Google Scholar]
- 70. Gomez-Lazaro M., Galindo M. F., Melero-Fernandez De Mera R. M., Fernandez-Gómez F. J., Concannon C. G., Segura M. F., Comella J. X., Prehn J. H. M., and Jordan J. (2007) Reactive oxygen species and p38 mitogen-activated protein kinase activate bax to induce mitochondrial cytochrome c release and apoptosis in response to malonate. Mol. Pharmacol. 71, 736–743 10.1124/mol.106.030718 [DOI] [PubMed] [Google Scholar]
- 71. Nakagami H., Morishita R., Yamamoto K., Yoshimura S. I., Taniyama Y., Aoki M., Matsubara H., Kim S., Kaneda Y., and Ogihara T. (2001) Phosphorylation of p38 mitogen-activated protein kinase downstream of bax-caspase-3 pathway leads to cell death induced by high d-glucose in human endothelial cells. Diabetes 50, 1472–1481 10.2337/diabetes.50.6.1472 [DOI] [PubMed] [Google Scholar]
- 72. Lee S. R., and Lo E. H. (2003) Interactions between p38 mitogen-activated protein kinase and caspase-3 in cerebral endothelial cell death after hypoxia-reoxygenation. Stroke 34, 2704–2709 10.1161/01.STR.0000096540.40826.BA [DOI] [PubMed] [Google Scholar]
- 73. Ryan K. M., Phillips A. C., and Vousden K. H. (2001) Regulation and function of the p53 tumor suppressor protein. Curr. Opin. Cell Biol. 13, 332–337 10.1016/S0955-0674(00)00216-7 [DOI] [PubMed] [Google Scholar]
- 74. Schmitt C. A., Fridman J. S., Yang M., Baranov E., Hoffman R. M., and Lowe S. W. (2002) Dissecting p53 tumor suppressor functions in vivo. Cancer Cell 1, 289–298 10.1016/S1535-6108(02)00047-8 [DOI] [PubMed] [Google Scholar]
- 75. Sanchez-Prieto R., Rojas J. M., Taya Y., and Gutkind J. S. (2000) A role for the p38 mitogen-activated protein kinase pathway in the transcriptional activation of p53 on genotoxic stress by chemotherapeutic agents. Cancer Res. 60, 2464–2472 [PubMed] [Google Scholar]
- 76. She Q. B., Bode A. M., Ma W. Y., Chen N. Y., and Dong Z. (2001) Resveratrol-induced activation of p53 and apoptosis is mediated by extracellular-signal-regulated protein kinases and p38 kinase. Cancer Res. 61, 1604–1610 [PubMed] [Google Scholar]
- 77. Bragado P., Armesilla A., Silva A., and Porras A. (2007) Apoptosis by cisplatin requires p53 mediated p38α MAPK activation through ROS generation. Apoptosis 12, 1733–1742 10.1007/s10495-007-0082-8 [DOI] [PubMed] [Google Scholar]
- 78. Shen Y. H., Godlewski J., Zhu J., Sathyanarayana P., Leaner V., Birrer M. J., Rana A., and Tzivion G. (2003) Cross-talk between JNK/SAPK and ERK/MAPK pathways: Sustained activation of JNK blocks ERK activation by mitogenic factors. J. Biol. Chem. 278, 26715–26721 10.1074/jbc.M303264200 [DOI] [PubMed] [Google Scholar]
- 79. Lee J., Hong F., Kwon S., Kim S. S., Kim D. O., Kang H. S., Lee S. J., Ha J., and Kim S. S. (2002) Activation of p38 MAPK induces cell cycle arrest via inhibition of Raf/ERK pathway during muscle differentiation. Biochem. Biophys. Res. Commun. 298, 765–771 10.1016/S0006-291X(02)02562-7 [DOI] [PubMed] [Google Scholar]
- 80. Liu Q., and Hofmann P. A. (2004) Protein phosphatase 2A-mediated cross-talk between p38 MAPK and ERK in apoptosis of cardiac myocytes. Am. J. Physiol. Circ. Physiol. 286, H2204–H2212 10.1152/ajpheart.01050.2003 [DOI] [PubMed] [Google Scholar]
- 81. Ahmed S., Grant K. G., Edwards L. E., Rahman A., Cirit M., Goshe M. B., and Haugh J. M. (2014) Data‐driven modeling reconciles kinetics of ERK phosphorylation, localization, and activity states. Mol. Syst. Biol. 10, 718 10.1002/msb.134708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bardwell A. J., Frankson E., and Bardwell L. (2009) Selectivity of docking sites in MAPK kinases. J. Biol. Chem. 284, 13165–13173 10.1074/jbc.M900080200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kolch W. (2005) Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 6, 827–837 10.1038/nrm1743 [DOI] [PubMed] [Google Scholar]
- 84. Graves L. M., Duncan J. S., Whittle M. C., and Johnson G. L. (2013) The dynamic nature of the kinome. Biochem. J. 450, 1–8 10.1042/BJ20121456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Engelke H., Chou C., Uprety R., Jess P., and Deiters A. (2014) Control of protein function through optochemical translocation. ACS Synth. Biol. 3, 731–736 10.1021/sb400192a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Horgan A. M., and Stork P. J. S. (2003) Examining the mechanism of Erk nuclear translocation using green fluorescent protein. Exp. Cell Res. 285, 208–220 10.1016/S0014-4827(03)00037-5 [DOI] [PubMed] [Google Scholar]
- 87. Belevich I., Joensuu M., Kumar D., Vihinen H., and Jokitalo E. (2016) Microscopy image browser: a platform for segmentation and analysis of multidimensional datasets. PLoS Biol. 14, e1002340 10.1371/journal.pbio.1002340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chalfoun J., Majurski M., Dima A., Halter M., Bhadriraju K., and Brady M. (2016) Lineage mapper: a versatile cell and particle tracker. Sci. Rep. 6, 36984 10.1038/srep36984 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data not contained in the paper are available by request from the corresponding author (Jason M. Haugh, jason_haugh@ncsu.edu).