Abstract
Mitochondrial DNA gene expression is coordinately regulated both pre- and post-transcriptionally, and its perturbation can lead to human pathologies. Mitochondrial rRNAs (mt-rRNAs) undergo a series of nucleotide modifications after release from polycistronic mitochondrial RNA precursors, which is essential for mitochondrial ribosomal biogenesis. Cytosine N4-methylation (m4C) at position 839 (m4C839) of the 12S small subunit mt-rRNA was identified decades ago; however, its biogenesis and function have not been elucidated in detail. Here, using several approaches, including immunofluorescence, RNA immunoprecipitation and methylation assays, and bisulfite mapping, we demonstrate that human methyltransferase-like 15 (METTL15), encoded by a nuclear gene, is responsible for 12S mt-rRNA methylation at m4C839 both in vivo and in vitro. We tracked the evolutionary history of RNA m4C methyltransferases and identified a difference in substrate preference between METTL15 and its bacterial ortholog rsmH. Additionally, unlike the very modest impact of a loss of m4C methylation in bacterial small subunit rRNA on the ribosome, we found that METTL15 depletion results in impaired translation of mitochondrial protein-coding mRNAs and decreases mitochondrial respiration capacity. Our findings reveal that human METTL15 is required for mitochondrial function, delineate the evolution of methyltransferase substrate specificities and modification patterns in rRNA, and highlight a differential impact of m4C methylation on prokaryotic ribosomes and eukaryotic mitochondrial ribosomes.
Keywords: m4C methylation, 12S mt-rRNA, mitochondrial translation, mitoribosome biogenesis, methyltransferase-like 15 (METTL15), rsmH, m4C839, post-transcriptional modification, RNA methylation, RNA methyltransferase, RNA modification, mitochondria, ribosomal ribonucleic acid (rRNA) (ribosomal RNA), m4C methylation, mitochondrial translation
Mitochondrial gene expression requires a series of interconnected processes encompassing mitochondrial DNA (mtDNA) replication and repair, mitochondrial RNA transcription, maturation, and mitoribosome assembly (1, 2). The mt-RNAs, especially rRNAs and tRNAs, are subjected to extensive enzyme-mediated modifications, which play key roles in RNA stability, RNA structure, and mitochondrial ribosome assembly (3, 4). Some of these modifications are deposited co-transcriptionally or immediately after transcription, whereas others occur when the rRNA is assembled into the preribosomal particle (3–5).
Prokaryotic and eukaryotic cytoplasmic rRNAs contain more than 30 and 200 modified sites, respectively, but only ∼10 modifications are found in the mitochondrial rRNAs (4, 5). These modifications are located at the functionally important regions of the mitoribosome, such as the decoding center of the small subunit (SSU), suggesting that these modifications might be preserved because of their essential roles (5, 6). The best-characterized example is the TFB1M-mediated dimethylation on the two highly conserved sites, A936 and A937, at the 3′-end of the 12S mt-rRNA, which is necessary for the assembly of the SSU (7, 8). The NOP2/Sun RNA Methyltransferase 4 (NSUN4) forms a complex with MTERF4 to catalyze m5C methylation at position 841 in 12S mt-rRNA and to coordinate the mitoribosome assembly (9, 10). However, enzymes for m4C and m5U (uracil) methylation in mammalian mitochondrial rRNAs remain to be identified (6, 11).
Mitochondrial diseases may be caused by mutations in mtDNA (12), but growing evidence suggests that defects in the nuclear genes involved in mitochondrial RNA modifications can also lead to human mitochondrial diseases. For instance, loss of TFB1M results in mitochondrial dysfunction that leads to impaired insulin secretion and diabetes (13). A missense mutation in pseudouridylate synthase 1 (PUS1), which converts uridine to pseudouridine at several mitochondrial tRNA positions, has been reported to be associated with myopathy, lactic acidosis, and sideroblastic anemia (14). Moreover, a defect in the mitochondrial rRNA methyltransferase MRM2 that causes loss of 2′-O-methyl modification at position U1369 in the human mitochondrial 16S rRNA leads to mitochondrial encephalomyopathy, lactic acidosis, and stroke-like clinical syndrome in patients (15). In addition, a 11p14.1 microdeletion was identified to be highly associated with attention-deficit/hyperreactive disorder, autism, developmental delay, and obesity (16). Intriguingly, the microdeletion region always encompasses the METTL-family protein METTL15. More recently, a transancestral meta-analysis of genome-wide association studies uncovered a completely novel SNP (rs10835310) in METTL15 associated with childhood obesity (17), further implicating the involvement of METTL15 in this human syndrome, although the underlying molecular mechanisms for these associations are still unclear.
In this current study, we demonstrate that human METTL15 protein, encoded by a nuclear gene, is localized in mitochondria and is responsible for methylation of the 12S mt-RNA at C839 in vivo and in vitro. Furthermore, we demonstrate that METTL15-dependent modification of 12S mt-rRNA is necessary for the proper function of mitoribosome. Our study reveals that methylation of 12S mt-rRNA m4C839 by METTL15 is an important epitranscriptomic modification, critical for efficient mitochondrial protein synthesis and respiratory function.
Results
METTL15 is a mitochondrial protein associated with 12S mt-rRNA
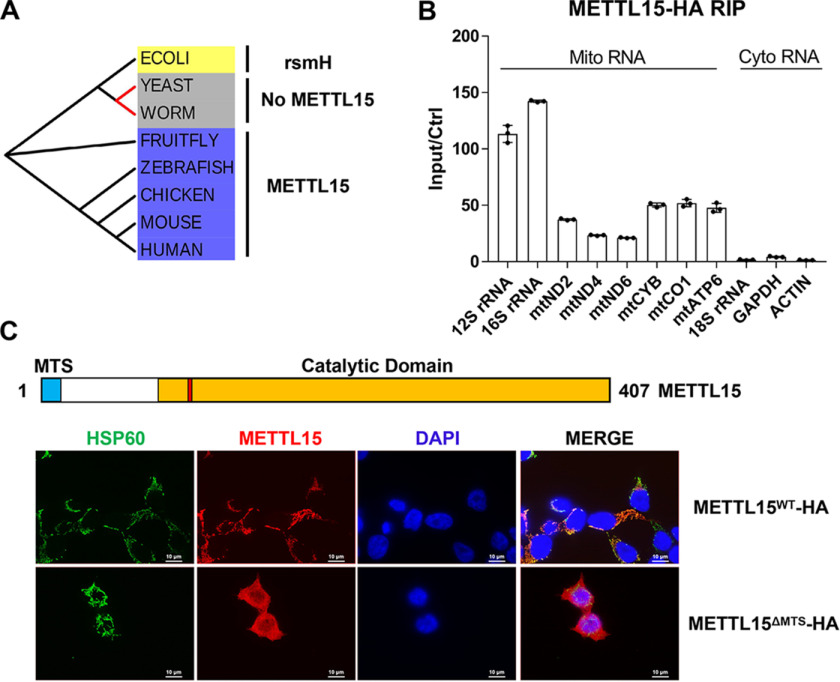
METTL15 is a member of the methyltransferase like (METTL) family, characterized by the presence of a binding domain for SAM, which is a methyl-group donor for methylation reactions (18, 19). Through phylogenetic analysis, we found that METTL15 is highly conserved during evolution and is an ortholog of the bacterial methyltransferase, rsmH (Fig. 1A and Data File S1), which is responsible for the N4-methylation of m4Cm1402 in the 16S rRNA in almost all species of bacteria (Fig. S1A) (20). Given its similarity with rsmH (Fig. 1A and Data File S1), we asked whether METTL15 is also a m4Cm methyltransferase for rRNA. We first purified the SSU rRNA fragments containing C1402 or its equivalent nucleotide from four representative species and measured the levels of m4Cm by HPLC-MS/MS. We did not detect any meaningful levels of m4Cm in the cytoplasmic SSU rRNAs from fruit fly, zebrafish, or human; however, a varied but significant amount of Cm was readily detectable (Fig. S1B). This finding is consistent with previous reports that the SSU rRNAs of those eukaryotic cells were abundantly modified by 2′-O-methylated cytosine (Fig. S1C) (21).
Figure 1.
METTL15 localizes in the mitochondria. A, phylogenetic analysis demonstrating that METTL15 is likely an evolutionarily conserved m4C methyltransferase from fruit fly to human but absent in worm and yeast. B, RIP-qPCR analysis of the interactions between METTL15-HA and the indicated RNA species. C, fluorescence confocal analysis of the subcellular location of WT, mitochondria-targeting signal deletion METTL15 (red), and HSP60 (green) was used as a marker for mitochondria.
In eukaryotic cells, mitochondria has its own ribosomes translating mitochondrial mRNAs, which prompted us to investigate whether METTL15 is a mitoribosome-specific methyltransferase. Indeed, a considerable amount of mitochondrial genome-encoded RNAs, especially 12S and 16S mt-rRNA, but not cytoplasmic RNAs such as 18S rRNAs, are found to be associated with the HA-tagged METTL15 in an RNA immunoprecipitation (RIP) experiment (Fig. 1B). Consistently, immunofluoresence experiments showed that METTL15 is exclusively localized in the mitochondria, which depends on its putative mitochondria-targeting signals (Fig. 1C) (22), suggesting that METTL15 is a bona fide mitochondria protein that interacts with mitoribosome rRNAs.
METTL15 is responsible for methylation of 12S mt-rRNA m4C839 in vivo
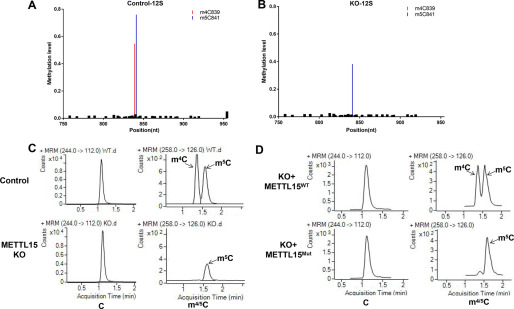
To unambiguously identify the in vivo methylation sites modified by METTL15, we profiled the mitochondrial RNA methylome in WT and METTL15 knockout (KO) cells using RNA bisulfite sequencing (BS-seq), which detects both m5C and m4C cytosine modifications in RNAs (23). The RNA BS-seq revealed that in the absence of METTL15, the methylation level of 12S mt-C839 is drastically decreased from 58% to a near background level (0.9%), which suggests that methylation of 12S mt-C839 may be mediated by METTL15 (Fig. 2, A and B). To validate the BS-seq results, we designed sequence-specific primers to amplify a 145-nucleotide region surrounding C839 from bisulfite-treated RNA samples and employed targeted sequencing (detailed procedures can be found under “Experimental procedures”) to examine the methylation levels of C839. In agreement with the BS-seq result, methylation of C839 was found to almost completely disappear in the METTL15 KO cells (Fig. S2C). Importantly, the methylation level of m4C839 can be fully rescued by WT METTL15 but not a catalytically compromised mutant METTL15 (GA mutant: 108GSGG112 to 108ASAA112) (Fig. S2C) (24), which strongly supports the hypothesis that METTL15 is responsible for m4C839 on 12S mt-rRNA in vivo and is consistent with a very recent study (25). Interestingly, the neighboring methylation site, m5C841, which is catalyzed by NSUN4 (9), is also reduced (but not eliminated) upon METTL15 deletion. In addition, the m5C841 reduction could be fully restored by reintroducing WT METTL15 and partially restored by enzymatically inactive METTL15, suggesting that METTL15 might influence the installation of m5C841 by NSUN4 in both enzymatic activity–dependent and –independent manners (Fig. S2C).
Figure 2.
METT15 is the methyltransferase responsible for the m4C839 on mitochondrial 12S rRNA in vivo. A and B, relative methylation levels of 12S rRNA determined after sequencing of cDNA obtained from bisulfite-treated RNA from control (A) and METTL15 KO (B) cells. C, LC–MS/MS chromatograms of C, m4C, and m5C in the corresponding 12S rRNA fragments purified from total RNA. Samples from control, METTL15 KO cells were analyzed. D, m4C methylation is readily restored by re-expression of WT METTL15 but not the catalytic mutant.
Given that both m4C(m) and m5C are able to block the C-to-T transition by bisulfite treatment and therefore cannot be distinguished in the BS-seq analysis (23), we turned to an optimized LC–MS/MS method to efficiently separate different forms of methyl cytosines to define the exact type of methylation in C839 (Fig. S2B). As shown in Fig. 2C, m4C was detected in WT cell lines but reduced to a background level in the METTL15 KO cells, indicating that METTL15 may be a m4C methyltransferase for C839. Consistent with the BS-seq data, HPLC-MS/MS analysis also found a modest reduction of m5C at C841 caused by METTL15 depletion, which again points to a potential cross-talk between C839 and C841 methylation. m4C methylation of C839 is mediated by the intrinsic enzymatic activity of METTL15 because reintroduction of WT, but not the catalytically inactive METLL15, back into the METTL15 KO cells restored the methylation level of m4C839 (Fig. 2D and Fig. S2, A–C). Collectively, these findings demonstrate that METTL15 is likely the enzyme responsible for methylation of 12S mt-m4C839 in vivo (Fig. S2D).
METTL15 methylates 12S mt-rRNA m4C839 in vitro
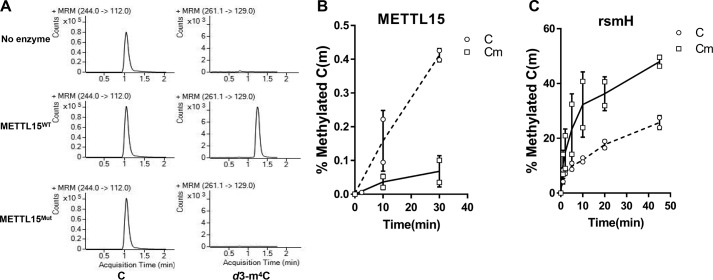
We next asked whether recombinant METTL15 mediates 12S mt-rRNA methylation at C839. C-terminally FLAG-tagged METTL15 was expressed in 293T cells and purified using an anti-FLAG M2 affinity column (Fig. S3A). Recombinant METTL15 was incubated with 12S mt-rRNA oligonucleotides (nucleotides 832–846) in the presence of d3-SAM (S-(5′-adenosyl)-l-methionine-d3) as a methyl group donor, and the resulting rRNAs oligonucleotides were isolated for LC–MS measurement. As shown in Fig. 3A, m4 methylation of C839 was successfully detected, whereas the catalytically compromised METTL15 failed to mediate C839 methylation.
Figure 3.
METT15 is the methyltransferase responsible for the m4C839 on mitochondrial 12S rRNA in vitro. A, in vitro methylation assay indicates that recombinant WT, but not catalytic mutant METTL15, is able to deposit a methyl group onto the N4 position of C839 in 12S mt-rRNA. B, human METTL15 prefers RNA oligonucleotides with unmodified C839. C, bacterial rsmH methyltransferase shows stronger activity toward 2′-O-methylated substrates. All data are represented as means ± S.D. from two biological replicates.
As described before, a main difference between the m4C methylation site of bacterial ribosome and human mitoribosome is that cytosine in bacterial ribosome is mainly m4Cm, whereas in the human mitoribosome at the equivalent cytosine residue, it is m4C without the 2′-O-methylation. This prompted us to determine whether human METTL15 displays any preference for unmodified cytosine versus 2′-O-methylated cytosine. Consistently, unmodified cytosine in the 12S mt-rRNA appears to be a better substrate for human METT15 in vitro (C versus Cm in Fig. 3B). In contrast, rsmH shows a higher apparent activity toward Cm compared with C of 12S mt-rRNAs (solid line in Fig. 3C; Fig. S3C). Interestingly, we found the aromatic amino acid (Trp173) in rsmH, which has been changed to valine (Val242) in the eukaryotic orthologs (METTL15) and might potentially mediate the interaction of 2′-O-methyl group of Cm with rsmH based on the published rsmH structure (Fig. S3B) (26). These results confirmed that human METTL15 is a bona fide m4C methyltransferase and has a higher activity toward unmodified cytosine than 2′-O-methylated cytosine in vitro.
Depletion of METTL15 inhibits the function of mitoribosomes
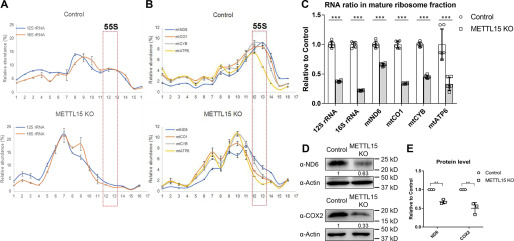
Because METTL15 is localized in mitochondria, we first investigated the effect of METTL15 deletion on mtDNA copy number and transcription of mitochondrial genome-encoded genes. We found METTL15 deletion only causes minor changes of mtDNA copy number and transcription (Fig. S4, A and B). Given that the methylation site lies in the critical region of the mitoribosome, we next asked whether loss of METTL15 affects the function of mitoribosome. We performed mitochondrial ribosome profiling in a 10–30% sucrose gradient to determine whether there was any difference in the assembly of mitoribosome. The distribution of SSU and large subunit in the sucrose gradient was detected by the presence of 12S and 16S mt-rRNA, respectively. According to the protein complex density, the first peak of 12S rRNA (fraction 8) represents SSU, whereas the first peak of 16S rRNA (fraction 9) represents large the subunit, and the co-fractionated peaks (fractions 12 and 13) represent the mature ribosome. The co-fractionation ratio of 12S and 16S mt-rRNAs in METTL15 KO cells was significantly reduced (compared with factions 12 and 13 in WT cells), thus identifying a major defect in mitoribosome assembly. In addition, the ratio of mRNA encoded by mitochondrial genome was also significantly reduced in the 55S mature monosomes (fractions 12 and 13), indicating compromised translation efficiency, which was consistent with the observed mitoribosome assembly defects (Fig. 4, A–C). Western blotting results of two representative mitochondrial protein-coding genes, COX2 and ND6, showed that the levels of the protein products were also reduced significantly (Fig. S4C and Fig. 4, D and E). Importantly, the translational defects of multiple mitochondria-encoded genes could be rescued by WT METTL15 but not the catalytic mutant (Fig. S4D), suggesting that the impact of METTL15 on mitoribosome is likely to be dependent on N4-methylation of C839. These data thus demonstrate that the methylation mediated by METTL15 is important for the proper function of mitoribosomes and mt-mRNA translation.
Figure 4.
METTL15 is required for proper mitochondrial ribosome assembly and the translation of genes encoded in the mitochondrial genome. A, the distribution of the mitochondrial ribosome small and large subunits in the indicated sucrose gradient fractions, examined by 12S and 16S rRNA RT-qPCR. B, the distribution of the mRNAs of the mitochondrial coding genes in the indicated sucrose gradient fractions examined by RT-qPCR. C, the bar graph shows the statistical analysis of the enrichment of the indicated RNA species in the mature ribosome fraction (fractions 12 and 13 in A and B), normalized to the control. D and E, Western blotting analyses of ND6 and COX2 protein levels in the control and METTL15 KO cells. The bar graph represents the quantification results of three replicate experiments.
Mitochondrial functions are affected by METTL15 depletion
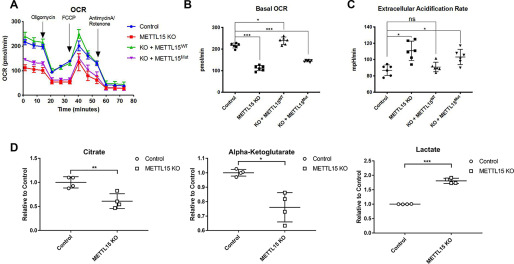
The most prominent role of mitochondria is to produce ATP through respiration and to regulate cellular metabolism. Most of the ATP synthesized during glucose metabolism is produced in the mitochondria through oxidative phosphorylation (OxPhos) powered by the electron transport chain complex, which consists essentially of ∼70 nuclear-encoded proteins and 13 mtDNA-encoded proteins translated by mitoribosome in mitochondria. To determine the impact of METTL15 loss on respiratory activity, we measured the respiratory activity of METTL15 KO cells using a Seahorse XF96 analyzer. The oxygen consumption rate (OCR) of the METTL15 KO cells was substantially lower than that of WT cells, and this effect depends on the enzymatic activity (Fig. 5, A and B), indicating that METTL15-mediated m4C839 on 12S mt-rRNA is likely to be required for proper oxidative phosphorylation function. After 2 days in culture, the medium of METTL15 KO cells turned yellower, indicating a lower pH and more lactate secretion, although the cell numbers are comparable between WT and KO (Fig. S5A). Consistently, the extracellular acidification rate, which approximates glycolytic activity, was significantly up-regulated in the METTL15 KO cells, likely to compensate for dysfunction of the mitochondria (Fig. 5C). Furthermore, metabolites profiling shows a decline of citrate and α-ketoglutarate, the intermediates of the TCA cycle, which is closely coupled with OxPhos to generate ATP. It is also known that an essential function of respiration in proliferating cells is to support aspartate biosynthesis (27, 28). The decline of the aspartate level in METTL15 KO cell is consistent with the compromised respiration function. At the same time, the up-regulated level of lactate suggests that cells use more anaerobic glycolysis to compensate for impaired mitochondrial function (Fig. 5D and S5, A–C). These results indicate that METTL15 is important for maintaining mitochondrial function and cellular metabolic homeostasis.
Figure 5.
METTL15 deletion facilitates the transformation from aerobic glycolysis to anaerobic glycolysis. A, OCR of the control, METTL15 KO cells, or 1the METTL15 KO cells with the WT or catalytic mutation of METTL15-FLAG-HA rescuing constructs measured by Seahorse XF96 machine. B, quantification of the basal OCR for the indicated cells. C, quantification of extracellular acidification rate for the indicated cells at the basal condition. D, cellular metabolite concentration determined by LC–MS/MS in the control and METTL15 KO cells. All data are represented as means ± S.D. from four biological replicates. ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001, t test.
Discussion
Here we describe the identification of METTL15 as the methyltransferase that generates m4C839 in human 12S mt-rRNA. METTL15 orthologs exist in most eukaryotes, which implies the importance of this methyltransferase for proper mitoribosome functions. Consistent with this hypothesis, mitochondrial translation is inhibited, and oxidative phosphorylation is remarkably compromised in the METTL15 KO cells, identifying an important function of METTL15 in regulating mitochondria functions, likely by methylating 12S mt-rRNA.
Cross-talk between m4C839 and m5C841 on 12S mt-rRNA
Ribosomal maturation involves multiple steps of subunit assembly and incorporation of chemical modifications into the rRNA (3). The assembly of protein components and rRNAs has been well characterized through high-resolution cryo-EM, whereas the process of modification deposition is still largely unclear (29). For the bacterial 16S rRNA, quantitative analysis of rRNA modifications finds that the modification events seem to occur in a 5′-to-3′ sequential order: from the 5′ body domain, to the 3′ head domain, to the 3′ minor domain (30). In this current study, we found that m4C839 methylation appears to precede m5C841 and is important for the nearby m5C841 methylation, suggesting cross-talk between modifications of the two nearby residues. Furthermore, the enzymatically inactive METTL15 can partially restore the m5C841 methylation decrease in METTL15-null cells, raising the question of whether the cross-talk is mediated by physical interactions between METTL15 and the m5C methyltransferase, NSUN4. Undoubtedly, the investigation of how modifications of mt-rRNA are coordinately deposited in an orderly manner will significantly increase our understanding of mitoribosome maturation (31).
Co-evolution of rRNA methyltransferases and rRNA functions
Unlike the universally conserved rRNA modifications (such as m6,6A) (5), the N4-methylation of SSU rRNA is only maintained in prokaryotes and mitochondria of eukaryotic cells. In bacteria, rsmI and rsmH (which is the bacterial METTL15 ortholog) install 2′-O-methylation and N4-methylation, respectively, on the equivalent cytosine to generate m4Cm1402 in the bacterial 16S rRNAs (20). There are two notable differences between methylation of human mt-rRNAs versus bacterial rRNA. First, at the molecular level, our in vitro enzymatic assays showed that although human METTL15 mediates N4-methylation of cytosine, bacterial homolog rsmH prefers 2′-O-methyl cytosine as a substrate, and m4C methylation at C1402 in bacteria may occur subsequent to the 2′-O-methylation. Interestingly, the aromatic pocket in the rsmH enzymatic domain appears to have degenerated in its eukaryotic ortholog METTL15 proteins during evolution, and this might be coupled with the loss of rsmI in eukaryotic organisms. Second, the m4C methylation is essential for the mitoribosome assembly and maturation, whereas in contrast, m4Cm loss in bacteria only generates a rather modest phenotype (20). These findings suggest that m4C appears to have gained importance in regulating mitochondrial functions during evolution.
METTL15 and human diseases
Previous studies demonstrate that mitochondrial dysfunctions in mature adipocytes cause defects in fatty acid oxidation, secretion of adipokines, and dysregulation of glucose homeostasis (32). Importantly, a reduction in the oxidative capacity of brown adipocytes results in impaired thermogenesis and has been linked to diet-induced obesity (33). In this context, it is important to note that microdeletion and SNPs in METTL15 gene are highly associated with obesity (16, 17). Therefore, we speculate that if altered METTL15 activity impacts obesity onset, it is likely to be due to the ability of METTL15 to regulate mitochondrial functions by methylating 12S mt-rRNA.
In summary, we identify METTL15 as a m4C methyltransferase that specifically mediates m4C methylation of 12S mt-rRNA at residue C839. Interestingly, our HPLC-MS/MS and enzymological analyses reveal a methylation pattern shift during evolution, which is likely a consequence of degeneration of an ancestral aromatic pocket present only in the bacterial METTL15 orthologs, which recognizes 2′-O-methyl cytosine. This pocket is absent in eukaryotic METTL15, which is probably why human METTL15 prefers C839 but not Cm839 as a substrate. Importantly, METTL15 depletion affects mitoribosome assembly, inhibits translation of mitochondria encoded proteins, and compromises the oxidative phosphorylation, which underlies the importance of METTL15 in maintaining mitochondrial functions. Future experiments will determine whether METTL15 plays a role in human diseases such as obesity through its ability to mediate methylation of mitochondrial rRNA.
Experimental procedures
Constructs and antibodies
For the expression of METTL15 protein, METTL15 ORF cDNA was cloned into pHAGE or pET28a expression vector (Invitrogen). METTL15 and rsmH mutants were generated using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. Anti-FLAG (M2) beads and antibody (F1804) were purchased from Sigma. Total OxPhos human WB antibody mixture (ab110411) and anti-ND6 (ab81212) antibodies were purchased from Abcam. Anti-COX2 (55070-1-AP) antibody was purchased from Proteintech.
Immunofluorescence
The cells were seeded in 24-well plates at a density of 20,000 cells/well 1 day before immunofluorescence examination. Through standard fixation, permeabilization, and antibody incubation procedures, confocal imaging was taken by Yokogawa spinning disk confocal on an inverted Nikon Ti fluorescence microscope and then processed by ImageJ.
Generation of METTL15 knockout cell lines
The CRISPR-Cas9 targeting system was utilized as previously described (34). The guide RNA sequence is 5′-TTGAGATCTGTGTAACTCCT-3′, targeting exon 3 of the NCBI METTL15 reference sequence NM_001113528.
RNA immunoprecipitation
A total of 5 million METTL15-HA stable cells were cross-linked with 1% formaldehyde and then harvested and lysed in 3 volumes of lysis buffer (50 mm Tris-Cl, pH 7.5, 300 mm NaCl, 0.5 mm DTT, 0.25% Nonidet P-40 with protease inhibitor) on ice for 15 min. After centrifugation at 14,000 rpm for 15 min at 4 °C, the supernatant was used for IP. For each RIP reaction, 5 million cells and 10 μl of HA beads were used. After washing, the precipitated RNA samples were extracted by TRIzol directly and reverse transcribed, followed by qPCR detection.
In vitro RNA methylation assay
Recombinant rsmH WT and mutant proteins were expressed in the Rosetta (DE3) bacterial cells, which were incubated at 37 °C until A600 reached ∼0.6–1 and then cooled down to 16 °C. isopropyl β-d-thiogalactopyranoside was added to 0.2 mm final concentration, and the cells were further incubated at 16 °C for 16 h. Cell pellets were lysed in a buffer containing 300 mm NaCl, 25 mm Tris, pH 7.5, 10% glycerol, 0.5% Nonidet P-40. Total lysate was incubated with HisPur nickel–nitrilotriacetic acid resin at 4 °C for 5 h. His-rsmH protein was eluted with elution buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 200 mm imidazole) in 0.5-ml aliquots until color change was no longer observed (Bradford assay).
Sucrose gradient sedimentation
Sucrose gradient sedimentation was performed as previously described with minor modifications (35). For each sample, ∼1 mg of mitochondria protein lysate was used, and the lysates were loaded on a 10-ml 10 to 30% uncontinuous sucrose gradient (50 mm Tris-Cl, 100 mm KCl, 10 mm MgCl2) and centrifuged at 32,000 rpm for 130 min in a Beckman SW60-Ti rotor. A total of 16 fractions were collected from the top and used for RT-qPCR analyses.
Seahorse assay
An XF96 extracellular flux analyzer (Seahorse Bioscience) was used to determine the OCR between WT and METTL15 KO cells, which were seeded at 15,000 cells/well. The concentrations of oligomycin, carbonyl cyanide p-trifluoromethoxyphenylhydrazone, rotenone, and antimycin A are 1, 1.5, 0.5, and 0.5 μm, respectively.
Isolation of a defined rRNA fragment
To isolate the corresponding fragments of 12S rRNA containing known modified residues, we refer to the method as previously described (36) with minor modifications. Briefly, we used 200 pmol of biotin-tagged synthetic DNA probe and 100 μg of total RNA for one experiment, and the sequences of the probes are listed in Table S1. After annealing and digestion with mung bean nuclease (NEB) and RNase A, the duplex of the probe and corresponding RNA fragment is purified with streptavidin C1.
Processing of RNA samples for MS
The RNA sample is digested with 0.5 unit of nuclease P1 (Sigma) in 80 μl of NP1 buffer at 42 °C for 2 h. To dephosphorylate the single nucleotides, 1 unit of CIP (NEB) is added and incubated for 1 h at 37 °C. The samples are filtered with Millex-GV 0.22-μm filters before being loaded onto the column of a mass spectrometer machine. 5 μl of solution was loaded into LC–MS/MS (Agilent6410 QQQ triple-quadrupole mass spectrometer). Nucleosides were quantified by using retention time and nucleoside to base ion mass transitions of 244 to 112 (C), 258 to 126 (m4C and m5C), 258 to 112 (Cm), and 272 to 126 (m4Cm).
Bisulfite mapping of mC residues in mitochondrial RNA
For purified mitochondria, mitochondrial RNA for bisulfite treatment was isolated with TRIzol and treated with TURBO DNase (Ambion) to remove mitochondrial DNA. Bisulfite treatment was performed with the EZ RNA methylation kit (Zymo Research). Half of the treated RNA was used for bisulfite RNA-seq library preparation with NEBNext Ultra II directional RNA library prep kit (NEB), according to the manufacturer's instructions. For the targeted bisulfite analysis, the bisulfite-treated RNA was directly converted to cDNA using PrimeScript RT reagent kit (Takara Bio, Inc.), followed by PCR amplification using primers specific for the region surrounding C839 of 12S mt-rRNA. The PCR products were separated from unincorporated primers using low-melting agarose and submitted for Amplicon-EZ sequencing (Genewiz).
BS RNA-seq was carried out on Illumina NextSeq platform with single-end 75-bp read length. Raw reads were stripped of adaptor sequences and removed of low quality bases using Cutadapt. The processed reads were aligned to human mitochondrial genome with meRanT align (meRanTK, version 1.2.0), and the methylation ratio was calculated by meRanCall (meRanTK, version 1.2.0) (37).
Data availability
All data described in the article are contained within the article. Additional data are available upon request, Yang Shi, Division of Newborn Medicine and Epigenetics Program, Department of Medicine, Boston Children's Hospital, Boston, Massachusetts 02115, USA. Yang.Shi@childrens.harvard.edu.
Supplementary Material
Acknowledgments
We thank Alison Ringel and Kiran Kurmi for assistance with setting up the Seahorse experiments; Phillip A. Dumesic, Marwan Shinawi, and Alberto Fernández Jaén for insightful advice; and Noa Liberman-Isakov in Eric Greer's lab for providing the N4-methyl-CTP standard. We also thank Santa Cruz Biotechnology, Inc., and Abclonal, Inc., for providing antibodies.
This article contains supporting information.
Author contributions—H. C. conceptualization; H. C., Z. S., and J. G. data curation; H. C. formal analysis; H. C., Z. S., K.-J. C., Q. C., and C.-H. Y. investigation; H. C. and Y. S. writing-original draft; H. C., Z. S., and Y. S. writing-review and editing; M. C. H. and Y. S. supervision; Y. S. funding acquisition.
Funding and additional information—This work was supported by Boston Children's Hospital funds. Y. S. is an American Cancer Society Research Professor.
Conflict of interest—Y. S. is a co-founder/equity holder of Constellation Pharmaceuticals, Inc. and Athelas Therapeutics, Inc., an equity holder of Imago Biosciences and a consultant for Active Motif.
- mtDNA
- mitochondrial DNA
- mt-rRNA
- mitochondrial rRNA
- m4C
- N4-methylation
- m4C839
- m4C at position 839
- SSU
- small subunit
- HA
- hemagglutinin
- RIP
- RNA immunoprecipitation
- KO
- knockout
- BS-seq
- bisulfite sequencing
- OxPhos
- oxidative phosphorylation
- OCR
- oxygen consumption rate
- qPCR
- quantitative PCR.
References
- 1. Costanzo M. C., and Fox T. D. (1990) Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annu. Rev. Genet. 24, 91–113 10.1146/annurev.ge.24.120190.000515 [DOI] [PubMed] [Google Scholar]
- 2. Taanman J. W. (1999) The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta 1410, 103–123 10.1016/S0005-2728(98)00161-3 [DOI] [PubMed] [Google Scholar]
- 3. Pearce S. F., Rebelo-Guiomar P., D'Souza A. R., Powell C. A., Van Haute L., and Minczuk M. (2017) Regulation of mammalian mitochondrial gene expression: recent advances. Trends Biochem. Sci. 42, 625–639 10.1016/j.tibs.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bohnsack M. T., and Sloan K. E. (2018) The mitochondrial epitranscriptome: the roles of RNA modifications in mitochondrial translation and human disease. Cell. Mol. Life Sci. 75, 241–260 10.1007/s00018-017-2598-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sergiev P. V., Aleksashin N. A., Chugunova A. A., Polikanov Y. S., and Dontsova O. A. (2018) Structural and evolutionary insights into ribosomal RNA methylation. Nat. Chem. Biol. 14, 226–235 10.1038/nchembio.2569 [DOI] [PubMed] [Google Scholar]
- 6. Dubin D. T. (1974) Methylated nucleotide content of mitochondrial ribosomal RNA from hamster cells. J. Mol. Biol. 84, 257–273 10.1016/0022-2836(74)90584-1 [DOI] [PubMed] [Google Scholar]
- 7. Metodiev M. D., Lesko N., Park C. B., Camara Y., Shi Y., Wibom R., Hultenby K., Gustafsson C. M., and Larsson N. G. (2009) Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 9, 386–397 10.1016/j.cmet.2009.03.001 [DOI] [PubMed] [Google Scholar]
- 8. Seidel-Rogol B. L., McCulloch V., and Shadel G. S. (2003) Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat. Genet. 33, 23–24 10.1038/ng1064 [DOI] [PubMed] [Google Scholar]
- 9. Metodiev M. D., Spahr H., Loguercio Polosa P., Meharg C., Becker C., Altmueller J., Habermann B., Larsson N. G., and Ruzzenente B. (2014) NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 10, e1004110 10.1371/journal.pgen.1004110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Spahr H., Habermann B., Gustafsson C. M., Larsson N. G., and Hallberg B. M. (2012) Structure of the human MTERF4–NSUN4 protein complex that regulates mitochondrial ribosome biogenesis. Proc. Natl. Acad. Sci. U.S.A. 109, 15253–15258 10.1073/pnas.1210688109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dubin D. T., Taylor R. H., and Davenport L. W. (1978) Methylation status of 13S ribosomal RNA from hamster mitochondria: the presence of a novel riboside, N4-methylcytidine. Nucleic Acids Res. 5, 4385–4397 10.1093/nar/5.11.4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Taylor R. W., and Turnbull D. M. (2005) Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 6, 389–402 10.1038/nrg1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sharoyko V. V., Abels M., Sun J., Nicholas L. M., Mollet I. G., Stamenkovic J. A., Gohring I., Malmgren S., Storm P., Fadista J., Spegel P., Metodiev M. D., Larsson N. G., Eliasson L., Wierup N., et al. (2014) Loss of TFB1M results in mitochondrial dysfunction that leads to impaired insulin secretion and diabetes. Hum. Mol. Genet. 23, 5733–5749 10.1093/hmg/ddu288 [DOI] [PubMed] [Google Scholar]
- 14. Fernandez-Vizarra E., Berardinelli A., Valente L., Tiranti V., and Zeviani M. (2006) Nonsense mutation in pseudouridylate synthase 1 (PUS1) in two brothers affected by myopathy, lactic acidosis and sideroblastic anaemia (MLASA). J. Med. Genet. 44, 173–180 10.1136/jmg.2006.045252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garone C., D'Souza A. R., Dallabona C., Lodi T., Rebelo-Guiomar P., Rorbach J., Donati M. A., Procopio E., Montomoli M., Guerrini R., Zeviani M., Calvo S. E., Mootha V. K., DiMauro S., Ferrero I., et al. (2017) Defective mitochondrial rRNA methyltransferase MRM2 causes MELAS-like clinical syndrome. Hum. Mol. Genet. 26, 4257–4266 10.1093/hmg/ddx314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shinawi M., Sahoo T., Maranda B., Skinner S. A., Skinner C., Chinault C., Zascavage R., Peters S. U., Patel A., Stevenson R. E., and Beaudet A. L. (2011) 11p14.1 microdeletions associated with ADHD, autism, developmental delay, and obesity. Am. J. Med. Genet. A 155, 1272–1280 10.1002/ajmg.a.33878 [DOI] [PubMed] [Google Scholar]
- 17. Bradfield J. P., Vogelezang S., Felix J. F., Chesi A., Helgeland Ø., Horikoshi M., Karhunen V., Lowry E., Cousminer D. L., Ahluwalia T. S., Thiering E., Boh E. T., Zafarmand M. H., Vilor-Tejedor N., Wang C. A., et al. (2019) A trans-ancestral meta-analysis of genome-wide association studies reveals loci associated with childhood obesity. Hum. Mol. Genet. 28, 3327–3338 10.1093/hmg/ddz161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schubert H. L., Blumenthal R. M., and Cheng X. (2003) Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 28, 329–335 10.1016/S0968-0004(03)00090-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martin J. L., and McMillan F. M. (2002) SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 12, 783–793 10.1016/S0959-440X(02)00391-3 [DOI] [PubMed] [Google Scholar]
- 20. Kimura S., and Suzuki T. (2010) Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 38, 1341–1352 10.1093/nar/gkp1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Decatur W. A., and Fournier M. J. (2002) rRNA modifications and ribosome function. Trends Biochem. Sci. 27, 344–351 10.1016/S0968-0004(02)02109-6 [DOI] [PubMed] [Google Scholar]
- 22. Fukasawa Y., Tsuji J., Fu S. C., Tomii K., Horton P., and Imai K. (2015) MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell. Proteomics 14, 1113–1126 10.1074/mcp.M114.043083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schaefer M., Pollex T., Hanna K., and Lyko F. (2008) RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 37, e12 10.1093/nar/gkn954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kozbial P. Z., and Mushegian A. R. (2005) Natural history of S-adenosylmethionine–binding proteins. BMC Struct. Biol. 5, 19 10.1186/1472-6807-5-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haute L. V., Hendrick A. G., D'Souza A. R., Powell C. A., Rebelo-Guiomar P., Harbour M. E., Ding S., Fearnley I. M., Andrews B., and Minczuk M. (2019) METTL15 introduces N4-methylcytidine into human mitochondrial 12S rRNA and is required for mitoribosome biogenesis. Nucleic Acids Res. 47, 10267–10281 10.1093/nar/gkz735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wei Y., Zhang H., Gao Z. Q., Wang W. J., Shtykova E. V., Xu J. H., Liu Q. S., and Dong Y. H. (2012) Crystal and solution structures of methyltransferase RsmH provide basis for methylation of C1402 in 16S rRNA. J. Struct. Biol. 179, 29–40 10.1016/j.jsb.2012.04.011 [DOI] [PubMed] [Google Scholar]
- 27. Sullivan L. B., Gui D. Y., Hosios A. M., Bush L. N., Freinkman E., and Vander Heiden M. G. (2015) Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162, 552–563 10.1016/j.cell.2015.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang T., Birsoy K., Hughes N. W., Krupczak K. M., Post Y., Wei J. J., Lander E. S., and Sabatini D. M. (2015) Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 10.1126/science.aac7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Klinge S., and Woolford J. L. Jr. (2018) Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 20, 116–131 10.1038/s41580-018-0078-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Popova A. M., and Williamson J. R. (2014) Quantitative analysis of rRNA modifications using stable isotope labeling and mass spectrometry. J. Am. Chem. Soc. 136, 2058–2069 10.1021/ja412084b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shi Z., Xu S., Xing S., Yao K., Zhang L., Xue L., Zhou P., Wang M., Yan G., Yang P., Liu J., Hu Z., and Lan F. (2019) Mettl17, a regulator of mitochondrial ribosomal RNA modifications, is required for the translation of mitochondrial coding genes. FASEB J. 33, 13040–13050 10.1096/fj.201901331R [DOI] [PubMed] [Google Scholar]
- 32. Bournat J. C., and Brown C. W. (2010) Mitochondrial dysfunction in obesity. Curr. Opin. Endocrinol. Diabetes Obes. 17, 446–452 10.1097/MED.0b013e32833c3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schulz T. J., and Tseng Y. H. (2013) Brown adipose tissue: development, metabolism and beyond. Biochem. J. 453, 167–178 10.1042/BJ20130457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., Mikkelsen T. S., Heckl D., Ebert B. L., Root D. E., Doench J. G., and Zhang F. (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kehrein K., Schilling R., Möller-Hergt B. V., Wurm C. A., Jakobs S., Lamkemeyer T., Langer T., and Ott M. (2015) Organization of mitochondrial gene expression in two distinct ribosome-containing assemblies. Cell Reports 10, 843–853 10.1016/j.celrep.2015.01.012 [DOI] [PubMed] [Google Scholar]
- 36. Ma H., Wang X., Cai J., Dai Q., Natchiar S. K., Lv R., Chen K., Lu Z., Chen H., Shi Y. G., Lan F., Fan J., Klaholz B. P., Pan T., Shi Y., et al. (2019) N6-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 15, 88–94 10.1038/s41589-018-0184-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rieder D., Amort T., Kugler E., Lusser A., and Trajanoski Z. (2016) meRanTK: methylated RNA analysis ToolKit. Bioinformatics 32, 782–785 10.1093/bioinformatics/btv647 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described in the article are contained within the article. Additional data are available upon request, Yang Shi, Division of Newborn Medicine and Epigenetics Program, Department of Medicine, Boston Children's Hospital, Boston, Massachusetts 02115, USA. Yang.Shi@childrens.harvard.edu.