Abstract
The Ebola virus (EBOV) VP40 matrix protein (eVP40) orchestrates assembly and budding of virions in part by hijacking select WW-domain–bearing host proteins via its PPxY late (L)-domain motif. Angiomotin (Amot) is a multifunctional PPxY-containing adaptor protein that regulates angiogenesis, actin dynamics, and cell migration/motility. Amot also regulates the Hippo signaling pathway via interactions with the WW-domain–containing Hippo effector protein Yes-associated protein (YAP). In this report, we demonstrate that endogenous Amot is crucial for positively regulating egress of eVP40 virus-like particles (VLPs) and for egress and spread of authentic EBOV. Mechanistically, we show that ectopic YAP expression inhibits eVP40 VLP egress and that Amot co-expression rescues budding of eVP40 VLPs in a dose-dependent and PPxY-dependent manner. Moreover, results obtained with confocal and total internal reflection fluorescence microscopy suggested that Amot's role in actin organization and dynamics also contributes to promoting eVP40-mediated egress. In summary, these findings reveal a functional and competitive interplay between virus and host proteins involving the multifunctional PPxY-containing adaptor Amot, which regulates both the Hippo pathway and actin dynamics. We propose that our results have wide-ranging implications for understanding the biology and pathology of EBOV infections.
Keywords: Ebola virus, angiomotin (Amot), filovirus, budding, virus-like particle (VLP), PPxY motif, L-domain, Hippo pathway, WW-domain protein, Yes-associated protein (YAP), virology, virus, actin, angiomotin, budding, filovirus, virus-like particle (VLP)
EBOV is a zoonotic and emerging pathogen that continues to cause severe outbreaks of hemorrhagic fever in animals and humans. A more comprehensive understanding of EBOV-host interactions that regulate and/or influence transmission and pathogenesis of this deadly virus is critical for the development of future strategies and countermeasures for therapeutic intervention. EBOV VP40 (eVP40) is the major structural protein that independently directs assembly and egress of virus-like particles (eVP40 VLPs) and drives the budding process of infectious virus. To accomplish this, VP40 possesses two late (L) budding domains (PTAP and PPxY) that hijack select host proteins to facilitate late stages of virus-cell separation (1–6). Specifically, the PPxY L-domain recruits several host HECT-family E3 ubiquitin ligases via one or more of their cognate WW-domains, and these interactions, along with the host ESCRT machinery, facilitate both VLP and virus egress (2, 3, 7, 8).
We recently identified host YAP1, the downstream transcriptional effector of the Hippo pathway, as a novel WW-domain–bearing protein that interacts with the PPxY motifs of filovirus VP40 (9). The cellular localization and function of YAP1 is itself regulated via strong, modular interactions with the PPxY motifs present at the N terminus of full-length angiomotin (Amot-p130) (10, 11). Interestingly, Amot-p130 has been reported to influence assembly and budding of non-PPxY-containing viruses such, as HIV-1 (12) and paramyxoviruses (13, 14), and most recently the PPxY-containing Marburg virus (MARV) (9). Because two of the three PPxY-like motifs present within Amot-p130 have core sequences that precisely match the PPEY motif within eVP40, we postulated that a potential competitive interplay between binding of these host and viral PPEY-containing proteins to WW-domain–containing YAP could influence eVP40-mediated egress of VLPs and EBOV.
Here, we demonstrate that egress of eVP40 VLPs and authentic EBOV was reduced significantly in a stable Amot knockdown cell line (shAmot cells) compared with egress of VLPs and EBOV from control cells (shCtrl cells). As we recently demonstrated for MARV and mVP40 VLPs, ectopic expression of YAP1 inhibited egress of eVP40 VLPs, whereas co-expression of Amot-p130 rescued YAP1 inhibition of eVP40-mediated egress in a dose- and PPxY-dependent manner. Indeed, the N-terminally truncated isoform of Amot (Amot-p80), which lacks all PPxY motifs and the F-actin–binding domain, failed to rescue YAP1-mediated inhibition of eVP40 VLP egress. Moreover, images of eVP40 VLP formation and egress obtained using TIRF microscopy of transfected shCtrl or shAmot cells show that in the absence of Amot, the actin cytoskeleton appears less organized and is misaligned with eVP40 VLPs budding at the plasma membrane, whereas actin filaments observed in shCtrl cells appear to align with, and form a foundation for, budding eVP40 VLPs. In sum, our findings identify EBOV as a PPxY-containing virus whose egress and spread in vitro is regulated by host Amot-p130 via a mechanism involving competitive PPxY-mediated interactions with YAP1 and actin cytoskeleton dynamics at the plasma membrane.
Results and discussion
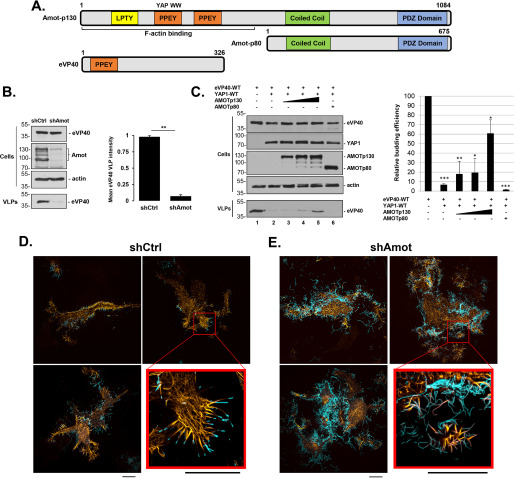
Full-length Amot-p130 possesses three PPxY-like motifs at its N terminus, two of which have the core sequence PPEY, which precisely matches that conserved at the N terminus of eVP40 (Fig. 1A). To determine whether endogenous expression of Amot affected budding of eVP40 VLPs, shCtrl or shAmot cells were transfected with eVP40, and both cell extracts and VLPs were harvested and analyzed by Western blotting (Fig. 1B). We observed robust inhibition of eVP40 VLP egress from shAmot cells compared with that from shCtrl cells (Fig. 1B), suggesting that expression of endogenous Amot is important for efficient egress of eVP40 VLPs.
Figure 1.
eVP40 VLP budding from shCtrl or shAmot cells. A, schematic diagrams of full-length Amot-p130, N-terminally truncated isoform Amot-p80, and eVP40 with key domains highlighted. Both the PPEY motif that binds to the WW-domain of YAP and the F-actin–binding domain of Amot-p130 are indicated. Numbers represent amino acids. B, representative Western blotting for eVP40 VLP budding assay in shCtrl and shAmot cells (0.25 µg of eVP40 plasmid/transfection). The indicated proteins were quantified in cells and VLPs using National Institutes of Health ImageJ software. The bar graph represents data from at least three independent experiments. **, p < 0.01. C, representative Western blotting for YAP1 inhibition and Amot-p130 rescue of eVP40 VLP budding. HEK293T cells were transfected with the indicated combinations of plasmids (0.25 µg of eVP40; 0.25 µg of YAP1-WT; 0.25, 0.5, or 1.0 µg of Amot-p130; and 1.0 µg of Amot-p80). The indicated proteins were quantified in cells and VLPs using National Institutes of Health ImageJ software. The bar graph represents data from at least three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001. shCtrl (D) or shAmot cells (E) were transfected with 0.5 µg each of pCMV-LifeAct-RFP and YFP-eVP40 plasmids, and cells were imaged by SIM-TIRF microscopy at 24 h post-transfection. Three representative images of F-actin (yellow) and YFP-eVP40 (cyan) are shown along with an enlarged view of the area indicated by a red box. All scale bars, 10 μm. Error bars, S.D.
Because the second PPxY motif of Amot-p130 interacts strongly with the WW-domain of YAP1 (15), and we recently identified YAP1 as a novel WW-domain interactor with filovirus VP40 PPxY motifs (9), we sought to determine whether competitive PPxY/WW-domain interplay among eVP40/Amot-p130/YAP1 would affect VLP egress. Toward this end, HEK293T cells were transfected with the indicated plasmid combinations (Fig. 1C), and both cell extracts and VLPs were harvested and analyzed by Western blotting. As observed previously for mVP40 (9), ectopic expression of YAP1 significantly inhibited egress of eVP40 VLPs (Fig. 1C, compare lanes 1 and 2). Interestingly, co-expression of Amot-p130 relieved the inhibitory effect of YAP1 and rescued budding of eVP40 VLPs in a dose-dependent manner (Fig. 1C, lanes 3-5). Importantly, co-expression of Amot-p80 did not relieve the inhibitory effect of YAP1 and did not rescue egress of eVP40 VLPs (Fig. 1C, lane 6). These results indicate that YAP1 negatively regulates egress of eVP40 VLPs, as it does for mVP40 VLPs, and suggest that the PPxY/WW-domain interaction between Amot-p130 and YAP1 may outcompete the PPxY/WW-domain interaction between eVP40 and YAP1, thus relieving YAP1's inhibitory effect on VLP budding. The potential interplay among eVP40/Amot/YAP1 in EBOV-infected cells could have wide-ranging pathological implications because Amot-YAP1 interactions regulate a wide array of cellular processes, including actin polymerization, filopodia formation, tight junction formation, and cell adhesion/migration (15–21).
Because Amot-p130 is an F-actin–binding protein and affects many cellular processes, such as cell morphology, migration, and plasma membrane dynamics, that are probably biologically relevant for the egress and spread of filoviruses, we sought to determine whether the observed differences in eVP40 VLP budding from shAmot and shCtrl cells may be related to Amot's role in actin cytoskeletal dynamics at the plasma membrane. Toward this end, we co-transfected either shCtrl or shAmot cells with plasmids expressing fluorescently tagged F-actin (pCMV-LifeAct-RFP) and eVP40 (YFP-eVP40) and used TIRF microscopy to visualize these proteins at the cell surface (Fig. 1, D and E). In the majority of shCtrl cells visualized, we observed pronounced localization of both F-actin and eVP40 in cell surface projections, and F-actin appeared to be most abundant at the base or foundation of the short, well-defined eVP40 VLP projections (Fig. 1D). Indeed, the actin cytoskeleton appeared to serve as a platform from which eVP40 VLPs could form and pinch off efficiently. In contrast, F-actin filaments were not as well-aligned with eVP40 projections in shAmot cells, and the eVP40 projections themselves appeared longer, tangled, more abundant, and less-defined than those observed in the shCtrl cells (Fig. 1E). These results highlight the importance of endogenous Amot as an actin-binding and organizing scaffold that contributes mechanistically in regulating efficient formation and egress of eVP40 VLPs.
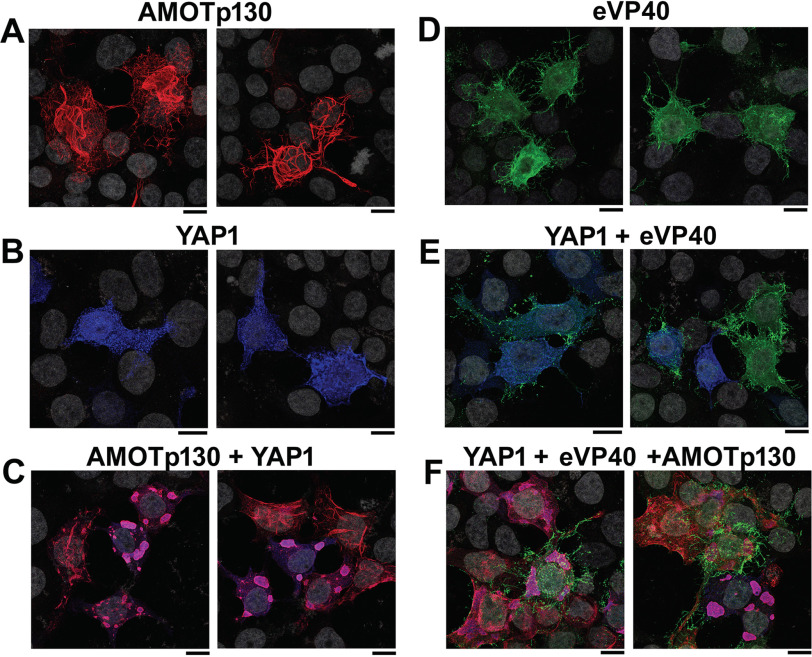
In a complementary approach, we sought to use confocal microscopy to investigate the localization patterns of eVP40, YAP1, and/or Amot-p130 alone or in combinations in transfected HEK293T cells (Fig. 2). Amot-p130 displayed a distinct tubular pattern throughout the cytoplasm (Fig. 2A), whereas YAP1 displayed a more punctate cytoplasmic pattern (Fig. 2B), and eVP40 displayed the typical filamentous projections at the plasma membrane (Fig. 2D). We observed colocalization of Amot-p130 and YAP1 in large globular regions within the cytoplasm when co-expressed (Fig. 2C), indicative of their ability to interact in a PPxY/WW-domain manner. We also observed that eVP40 colocalized partially with YAP1 in puncta in the cytoplasm with a concomitant reduction in the levels of eVP40 at the plasma membrane and in the number and sharpness of VLP projections (Fig. 2E). These observations are consistent with the ability of YAP1 to physically interact with eVP40 and functionally reduce egress of eVP40 VLPs. Last, we found that the patterns of eVP40 membrane localization and filamentous VLP projections in cells co-expressing YAP1 + eVP40 + Amot-p130 (Fig. 2F) were similar to those observed in cells expressing eVP40 alone (Fig. 2D). This observation is consistent with the ability of exogenously expressed Amot-p130 to rescue the inhibitory effect of YAP1 on eVP40 VLP egress.
Figure 2.
Confocal microscopy of HEK293T cells expressing YAP1, AMOT-p130, and/or eVP40. Two independent images are shown for the following conditions: AMOT-p130 alone (A), YAP1 alone (B), AMOT-p130 + YAP1 (C), eVP40 alone (D), YAP1 + eVP40 (E), and YAP1 + eVP40 + AMOT-p130 (F). Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (gray). Scale bars, 10 μm.
Next, we sought to determine whether the dramatic visual and functional differences observed for eVP40 VLP formation and egress in shCtrl versus shAmot cells would be evident during live EBOV infection. Toward this end, shCtrl or shAmot cells were infected with EBOV at MOIs of either 0.01 or 0.05, and the cells were fixed at either 24, 48, or 72 h post-infection to assess virus spreading. We observed that spread of EBOV throughout the shAmot cells was inhibited significantly compared with the degree of spread observed in the shCtrl cells (Fig. 3, A and C). We also quantified release of infectious EBOV from both shCtrl and shAmot cells, and we observed significant inhibition of virion release from shAmot cells compared with that measured from shCtrl cells (Fig. 3, B and D). These live virus experiments clearly indicate that expression of endogenous Amot plays a key role in regulating efficient egress and spread of authentic EBOV in cell culture. Moreover, these results support the mechanisms proposed for Amot-mediated regulation of EBOV budding, as they correlate well with data obtained using eVP40 VLPs. Overall, our results are in excellent agreement with those reported recently for MARV (9), and because the PPxY motif is physically and functionally conserved in a wide array of matrix proteins encoded by viruses belonging to the Filoviridae, Arenaviridae, Retroviridae, and Rhabdoviridae families, our results may have wide-ranging implications for the biology and pathogenesis of a plethora of viral pathogens.
Figure 3.
Spread and release of EBOV from shCtrl or shAmot cells. shCtrl or shAmot cells were infected with EBOV-EGFP at an MOI of either 0.01 (A and B), or 0.05 (C and D), and both virus spread (A and C) and release (B and D) were quantified at 24, 48, and 72 h post-infection. Green, EBOV; blue, Hoechst 33342–stained nuclei. Bar graphs represent data from three independent experiments. Scale bars, 200 μm. ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. Error bars, S.D.
Moving forward, it will be of interest to investigate additional Amot PPxY-mediated interactions that may be influenced or disrupted by the presence of VP40 during virus infection. For example, Amot contributes to the formation and maintenance of cellular tight junctions and to the regulation of cellular adhesion/migration/invasion. Interestingly, another member of the Amot family, Amotl2, localizes to and regulates formation of podosomes, mechanosensitive adhesive structures used by macrophages and dendritic cells to migrate through tissues (22, 23). These cells are primary targets of EBOV and believed to harbor and disseminate infection in the host (24). Thus, it is tempting to speculate that the interplay among VP40, Amot, and perhaps other Hippo pathway components (e.g. YAP, TAZ, and LATS kinases) may influence how EBOV is spread to various tissues during infection and how EBOV may cross or breach cellular barriers/junctions to enter and establish persistence in immunologically privileged sites, such as the central nervous system and eye. In sum, our findings serve as a foundation for a new area of investigation into the biological and pathological intersection of the Hippo pathway, the angiomotin family of proteins, and PPxY-mediated virus egress and transmission.
Experimental procedures
Cell lines, plasmids, and antibodies
HEK293T, HEK293T-based shCtrl, and shAmot cells (kindly provided by J. Kissil, Scripps Research Institute, Jupiter, FL, USA) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, penicillin (100 units/ml)/streptomycin (100 μg/ml) at 37°C in a humidified 5% CO2 incubator. eVP40– and YFP-eVP40–expressing plasmids were described previously (25, 26). pcDNA4-AMOT-p130-Myc– and pcDNA4-AMOT-p80-Myc–expressing plasmids were kindly provided by D. McCollum (University of Massachusetts Medical School). pCMV FLAG-YAP1 was obtained from Addgene (catalog no. 66853). Anti-AMOT antibody was obtained from EMD Millipore (catalog no. ABS1024), and anti-β-actin antibody was from Gene Tex (catalog no. GT5512). Anti-rabbit IgG-HRP (NA934V) and anti-mouse IgG-HRP(NA931V) were obtained from GE Healthcare.
VP40 VLP budding assay
The eVP40 VLP budding assay has been described previously (6, 26).
Confocal and TIRF microscopy
For confocal imaging, HEK293T cells on glass slides were transfected with 0.5 μg of the appropriate plasmids for 20 h. Cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 and then mounted with Prolong gold anti-fade mountant with 4′,6-diamidino-2-phenylindole. Images were obtained on a Leica SP5 inverted confocal microscope with a ×100 (numerical aperture 1.46) objective lens. The confocal images were subsequently deconvolved with Huygens Essential deconvolution software. For TIRF imaging, cells were seeded onto coverglass-bottom 35-mm dishes (MatTek) and co-transfected with 0.5 μg each of the appropriate plasmids in Opti-MEM. Dishes were placed onto the stage of a GE DeltaVision OMX SR microscope and maintained at 37 °C and 5% CO2 in a stage-top incubator. Approximately 100 cells/group were randomly chosen and imaged in SIM-TIRF mode. SIM superresolution images were reconstructed from the acquired TIRF images.
EBOV stocks
All experiments with replication competent EBOV were performed in the biosafety level 4 (BSL-4) laboratory at the Texas Biomedical Research Institute (San Antonio, TX). The recombinant EBOV variant Mayinga expressing enhanced GFP (EBOV-EGFP) (NCBI accession number KF_990213) was kindly provided by Heinz Feldmann (NIH, Hamilton, MT). The virus stock was generated and characterized as previously described (27).
EBOV spread and release
shCtrl and shAmot cells were grown in 96-well plates and incubated with EBOV at an MOI of 0.01 or 0.05. Three separate plates were challenged in triplicate for each MOI. After 1 h, inoculum was removed, cells were washed with medium, and new medium was added to wells. Cells were fixed 24, 48, or 72 h after virus challenge, stained with Hoechst 33342, and photographed using a Nikon Ti-Eclipse microscope running high content analysis software (Nikon, Tokyo, Japan). The numbers of nuclei and EGFP-positive (infected) cells were counted using Cell Profiler software (Broad Institute, Boston, MA). Virus release from shCtrl or shAmot cells was determined by overlaying supernatants of infected cells onto monolayers of Vero cells (American Type Culture Collection; ATCC, Manassas, VA) for 24 h and analysis as above.
Statistical analysis
Absolute intensities for immunoblot lanes were normalized to the intensity of the first lane and averaged across at least three replicates (collected from three or more separate transfections). Mean normalized intensity ± S.D. is shown. Because of observed inequality of variance between the groups (determined by F-test p value <0.05), Welch's t test was used to determine statistical significance. For live virus data, mean percentage ± S.D. is shown from three independent experiments. Virus spreading was compared using Student's t test. All p values are as follows: ns, p > 0.05; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
Data availability
All data are contained within the article.
Acknowledgments
We thank J. Kissil, D. McCollum, and H. Feldmann for kindly providing reagents, L. King for critical reading of the manuscript, and the Penn Vet Imaging Core for SIM-TIRF microscopy.
Author contributions— Z. H, R. N. H., and O. S. conceptualization; Z. H., G. R., S. D., C. T. B., and O. S. data curation; Z. H., G. R., S. D., C. T. B., B. D. F., R. N. H., and O. S. formal analysis; Z. H., G. R., and O. S. investigation; Z. H., G. R., S. D., C. T. B., and O. S. methodology; Z. H., G. R., S. D., C. T. B., B. D. F., R. N. H., and O. S. writing-review and editing; G. R., B. D. F., and R. N. H. resources; B. D. F. and R. N. H. supervision; R. N. H. funding acquisition; R. N. H. writing-original draft; R. N. H. and O. S. project administration.
Funding and additional information—Funding was provided in part by a University Research Foundation Award (Penn) and National Institutes of Health Grants AI138052 and AI139392 (to R. N. H.). Biosafety level-4 live virus studies were supported by the Texas Biomedical Research Institute (to O. S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- EBOV
- Ebola virus
- eVP40
- EBOV VP40 matrix protein
- VLP
- virus-like particle
- MARV
- Marburg virus
- TIRF
- total internal reflection fluorescence
- MOI
- multiplicity of infection
- EGFP
- enhanced GFP
- HRP
- horseradish peroxidase
- Amot
- angiomotin.
References
- 1. Noda T., Ebihara H., Muramoto Y., Fujii K., Takada A., Sagara H., Kim J. H., Kida H., Feldmann H., and Kawaoka Y. (2006) Assembly and budding of Ebolavirus. PLoS Pathog. 2, e99 10.1371/journal.ppat.0020099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hartlieb B., and Weissenhorn W. (2006) Filovirus assembly and budding. Virology 344, 64–70 10.1016/j.virol.2005.09.018 [DOI] [PubMed] [Google Scholar]
- 3. Jasenosky L. D., and Kawaoka Y. (2004) Filovirus budding. Virus Res. 106, 181–188 10.1016/j.virusres.2004.08.014 [DOI] [PubMed] [Google Scholar]
- 4. Yasuda J., Nakao M., Kawaoka Y., and Shida H. (2003) Nedd4 regulates egress of Ebola virus-like particles from host cells. J. Virol. 77, 9987–9992 10.1128/JVI.77.18.9987-9992.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Timmins J., Schoehn G., Ricard-Blum S., Scianimanico S., Vernet T., Ruigrok R. W., and Weissenhorn W. (2003) Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 326, 493–502 10.1016/S0022-2836(02)01406-7 [DOI] [PubMed] [Google Scholar]
- 6. Harty R. N., Brown M. E., Wang G., Huibregtse J., and Hayes F. P. (2000) A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. U.S.A. 97, 13871–13876 10.1073/pnas.250277297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bieniasz P. D. (2006) Late budding domains and host proteins in enveloped virus release. Virology 344, 55–63 10.1016/j.virol.2005.09.044 [DOI] [PubMed] [Google Scholar]
- 8. Chen B. J., and Lamb R. A. (2008) Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372, 221–232 10.1016/j.virol.2007.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Han Z., Dash S., Sagum C. A., Ruthel G., Jaladanki C. K., Berry C. T., Schwoerer M. P., Harty N. M., Freedman B. D., Bedford M. T., Fan H., Sidhu S. S., Sudol M., Shtanko O., and Harty R. N. (2020) Modular mimicry and engagement of the Hippo pathway by Marburg virus VP40: implications for filovirus biology and budding. PLoS Pathog. 16, e1008231 10.1371/journal.ppat.1008231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lv M., Shen Y., Yang J., Li S., Wang B., Chen Z., Li P., Liu P., and Yang J. (2017) Angiomotin family members: oncogenes or tumor suppressors? Int. J. Biol. Sci. 13, 772–781 10.7150/ijbs.19603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moleirinho S., Guerrant W., and Kissil J. L. (2014) The angiomotins–from discovery to function. FEBS Lett. 588, 2693–2703 10.1016/j.febslet.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mercenne G., Alam S. L., Arii J., Lalonde M. S., and Sundquist W. I. (2015) Angiomotin functions in HIV-1 assembly and budding. eLife 4, 10.7554/eLife.03778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pei Z., Bai Y., and Schmitt A. P. (2010) PIV5 M protein interaction with host protein angiomotin-like 1. Virology 397, 155–166 10.1016/j.virol.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ray G., Schmitt P. T., and Schmitt A. P. (2019) Angiomotin-like 1 links paramyxovirus M proteins to NEDD4 family ubiquitin ligases. Viruses 11, 128 10.3390/v11020128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moleirinho S., Hoxha S., Mandati V., Curtale G., Troutman S., Ehmer U., and Kissil J. L. (2017) Regulation of localization and function of the transcriptional co-activator YAP by angiomotin. eLife 6, e23966 10.7554/eLife.23966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chan S. W., Lim C. J., Chong Y. F., Pobbati A. V., Huang C., and Hong W. (2011) Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J. Biol. Chem. 286, 7018–7026 10.1074/jbc.C110.212621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao B., Li L., Lu Q., Wang L. H., Liu C. Y., Lei Q., and Guan K. L. (2011) Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 25, 51–63 10.1101/gad.2000111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Spadaro D., Tapia R., Pulimeno P., and Citi S. (2012) The control of gene expression and cell proliferation by the epithelial apical junctional complex. Essays Biochem. 53, 83–93 10.1042/bse0530083 [DOI] [PubMed] [Google Scholar]
- 19. Chan S. W., Lim C. J., Guo F., Tan I., Leung T., and Hong W. (2013) Actin-binding and cell proliferation activities of angiomotin family members are regulated by Hippo pathway-mediated phosphorylation. J. Biol. Chem. 288, 37296–37307 10.1074/jbc.M113.527598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hong W. (2013) Angiomotin'g YAP into the nucleus for cell proliferation and cancer development. Sci. Signal. 6, p e27 10.1126/scisignal.2004573 [DOI] [PubMed] [Google Scholar]
- 21. Mana-Capelli S., Paramasivam M., Dutta S., and McCollum D. (2014) Angiomotins link F-actin architecture to Hippo pathway signaling. Mol. Biol. Cell 25, 1676–1685 10.1091/mbc.e13-11-0701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Proszynski T. J., and Sanes J. R. (2013) Amotl2 interacts with LL5β, localizes to podosomes and regulates postsynaptic differentiation in muscle. J. Cell Sci. 126, 2225–2235 10.1242/jcs.121327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van den Dries K., Linder S., Maridonneau-Parini I., and Poincloux R. (2019) Probing the mechanical landscape—new insights into podosome architecture and mechanics. J. Cell Sci. 132, jcs236828 10.1242/jcs.236828] [DOI] [PubMed] [Google Scholar]
- 24. Zeng X., Blancett C. D., Koistinen K. A., Schellhase C. W., Bearss J. J., Radoshitzky S. R., Honnold S. P., Chance T. B., Warren T. K., Froude J. W., Cashman K. A., Dye J. M., Bavari S., Palacios G., Kuhn J. H., and Sun M. G. (2017) Identification and pathological characterization of persistent asymptomatic Ebola virus infection in rhesus monkeys. Nat. Microbiol. 2, 17113 10.1038/nmicrobiol.2017.113 [DOI] [PubMed] [Google Scholar]
- 25. Liu Y., Lee M. S., Olson M. A., and Harty R. N. (2011) Bimolecular complementation to visualize filovirus VP40-host complexes in live mammalian cells: toward the identification of budding inhibitors. Adv. Virol. 2011, 341816 10.1155/2011/341816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Licata J. M., Simpson-Holley M., Wright N. T., Han Z., Paragas J., and Harty R. N. (2003) Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 77, 1812–1819 10.1128/JVI.77.3.1812-1819.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shtanko O., Sakurai Y., Reyes A. N., Noel R., Cintrat J. C., Gillet D., Barbier J., and Davey R. A. (2018) Retro-2 and its dihydroquinazolinone derivatives inhibit filovirus infection. Antiviral Res. 149, 154–163 10.1016/j.antiviral.2017.11.016 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the article.