Abstract
Synapse formation is a dynamic process essential for the development and maturation of the neuronal circuitry in the brain. At the synaptic cleft, trans-synaptic protein–protein interactions are major biological determinants of proper synapse efficacy. The balance of excitatory and inhibitory synaptic transmission (E-I balance) stabilizes synaptic activity, and dysregulation of the E-I balance has been implicated in neurodevelopmental disorders, including autism spectrum disorders. However, the molecular mechanisms underlying the E-I balance remain to be elucidated. Here, using single-cell transcriptomics, immunohistochemistry, and electrophysiology approaches to murine CA1 pyramidal neurons obtained from organotypic hippocampal slice cultures, we investigate neuroligin (Nlgn) genes that encode a family of postsynaptic adhesion molecules known to shape excitatory and inhibitory synaptic function. We demonstrate that the NLGN3 protein differentially regulates inhibitory synaptic transmission in a splice isoform–dependent manner at hippocampal CA1 synapses. We also found that distinct subcellular localizations of the NLGN3 isoforms contribute to the functional differences observed among these isoforms. Finally, results from single-cell RNA-Seq analyses revealed that Nlgn1 and Nlgn3 are the major murine Nlgn genes and that the expression levels of the Nlgn splice isoforms are highly diverse in CA1 pyramidal neurons. Our results delineate isoform-specific effects of Nlgn genes on the E-I balance in the murine hippocampus.
Keywords: neurobiology, neuron, neurotransmitter receptor, synapse, GABA receptor, glutamate receptor, hippocampus, CA1 pyramidal neuron, excitatory and inhibitory balance, neuroligin 3 (NLGN3), splice variants, trans-synaptic cell adhesion, synaptic transmission
Neuroligin proteins (NLGNs) were the first identified binding partners of α-latrotoxin receptors, neurexin proteins (NRXs), and localize at postsynaptic sites to regulate synapse maturation and function (1). Four Nlgn genes (Nlgn1, Nlgn2, Nlgn3, and Nlgn4) encode trans-synaptic adhesion proteins (NLGN1, NLGN2, NLGN3, and NLGN4) that contain extracellular cholinesterase-like domains and transmembrane and PDZ-binding motif–containing intracellular domains. Whereas the intracellular domain is important for NLGN binding with postsynaptic scaffold molecules, the extracellular domain confers the formation of excitatory and inhibitory synapses with NRX, its sole presynaptic binding partner. Therefore, precise combinations of NRX-NLGN interactions allow NLGN to diversify synapse identity.
NLGN1 and NLGN2 are postsynaptic adhesion molecules localized to excitatory and inhibitory synapses, respectively. Overexpression, knockdown, and knockout approaches have revealed that NLGN1 is important for excitatory synaptic structure and transmission and synaptic plasticity, but not for inhibitory synaptic function (2–6). NLGN2 protein has specific functional roles in inhibitory synaptic transmission in the hippocampus (6–8). In contrast, it has been reported that NLGN3 protein localizes at both excitatory and inhibitory synaptic sites and regulates both synaptic functions (7–12). This unique ability alludes to a NLGN3 protein-specific molecular code that promotes its translocation to both excitatory and inhibitory sites.
Splice insertion in Nlgn genes differentially regulates E-I balance and alters their binding affinity with presynaptic NRX proteins. In the extracellular cholinesterase-like domain, Nlgn genes have one or two splice insertion sites, Nlgn1 (A and B sites), Nlgn2 (A), and Nlgn3 (A1 and A2), leading to 2-4 theoretical splice isoforms. The splice insertion site B in NLGN1 determines its binding preference to NRXs (13, 14) and excitatory synaptic function (15). Similarly, NLGN2 contains a splice insertion at site A, which regulates inhibitory synaptic function (8). However, to the best of our knowledge, the splice isoform–specific function of NLGN3 and the transcript levels of Nlgn splice isoforms at the single-cell level have not been addressed.
In the present study, we assess the function of NLGN3 splice isoforms on excitatory and inhibitory synaptic transmission in CA1 pyramidal neurons in mouse organotypic slice cultures. Our results suggest that NLGN3 up- or down-regulates inhibitory synaptic transmission in a splice isoform–dependent manner. Furthermore, our single-cell RNA-Seq analysis reveals that Nlgn1 and Nlgn3 are the major Nlgn genes, and the expressions of Nlgn splice variants are highly distinct in hippocampal CA1 pyramidal neurons.
Results
NLGN3 splice isoform-dependent regulation of inhibitory synaptic transmission
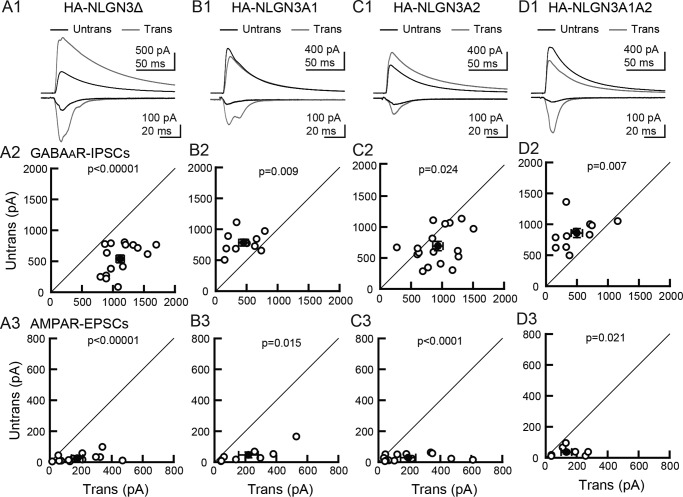
The Nlgn3 gene contains two splice insertion sites (A1 and A2) that can yield four NLGN3 splice isoforms (Nlgn3Δ, A1, A2, and A1A2). NLGN3Δ lacks all splice insertions, whereas NLGN3A1, 3A2, and 3A1A2 receive insertion of A1, A2, or both A1A2 cassettes, respectively. To examine the potential roles of NLGN3 splice isoforms on excitatory and inhibitory synapse function, we biolistically transfected the Nlgn3 splice isoform genes in CA1 pyramidal cells of organotypic hippocampal slice cultures (Fig. 1). Transfection of NLGN3 splice variants did not alter gross cell morphology or levels of overexpressed NLGN3 splice variants identified by HA immunoreactivity (Fig. S1, A and B). Simultaneous electrophysiological recordings were made from transfected and neighboring untransfected neurons. CA1 pyramidal neurons overexpressing NLGN3Δ or 3A2 showed increased evoked inhibitory postsynaptic currents (IPSCs) compared with neighboring untransfected control neurons and a marked increase in excitatory postsynaptic currents (EPSCs), as reported previously (Fig. 1, A and C) (7, 12). In contrast, overexpression of NLGN3A1 or 3A1A2 resulted in reduced amplitude of IPSCs and increased amplitude of EPSCs compared with neighboring untransfected cells (Fig. 1, B and D). Paired stimulation of input fibers with a short interval (50 ms) induced paired-pulse facilitation (PPF) and depression (PPD) of EPSCs and IPSCs, respectively. NLGN3Δ or 3A2 transfection displayed both reduced AMPAR-PPF and GABAAR-PPD compared with untransfected neurons, consistent with previous work (5, 7) (Fig. S1, C–E). As paired-pulse ratio (PPR) inversely correlates with presynaptic release probability, these results suggest that overexpression of NLGN3Δ and 3A2 can modulate presynaptic release probability. NLGN3 increased or decreased inhibitory synaptic transmission in a splice isoform–dependent manner, whereas all NLGN3 splice isoforms enhanced excitatory synaptic transmission. Next, we tested whether NLGN3 overexpression changes membrane excitability. The input-output relationship of CA1 pyramidal neurons transfected with any NLGN3 splice isoforms exhibited no significant difference compared with untransfected control neurons (Fig. S2). The findings above suggest that NLGN3 splice isoforms regulate synaptic but not membrane functions.
Figure 1.
NLGN3 splice isoforms differentially encode synapse specificity. Effect of NLGN3 splice isoform overexpression on GABAAR-IPSCs and AMPAR-EPSCs in hippocampal CA1 pyramidal cells. A1–D1, top row, superimposed sample IPSCs and EPSCs from pairs of transfected (gray) and untransfected (black) cells. Stimulus artifacts were truncated. A, NLGN3Δ; B, 3A1; C, 3A2; D, 3A1A2. IPSC (middle row, A2–D2) and EPSC (bottom row, A3–D3) amplitudes were plotted for each pair of transfected (Trans) and neighboring untransfected (Untrans) cells (open symbols). Filled symbols, the mean ± S.E. Numbers of cell pairs were as follows: NLGN3Δ (IPSCs/EPSCs: 16/14); 3A1 (10/8); 3A2 (16/14); 3A1A2 (11/8). The number of tested slice cultures is the same as the number of cell pairs. Results in all figures are reported as mean ± S.E.M., Mann–Whitney U test.
Subcellular localization of NLGN3 splice isoforms in the dendritic segment of CA1 pyramidal neurons
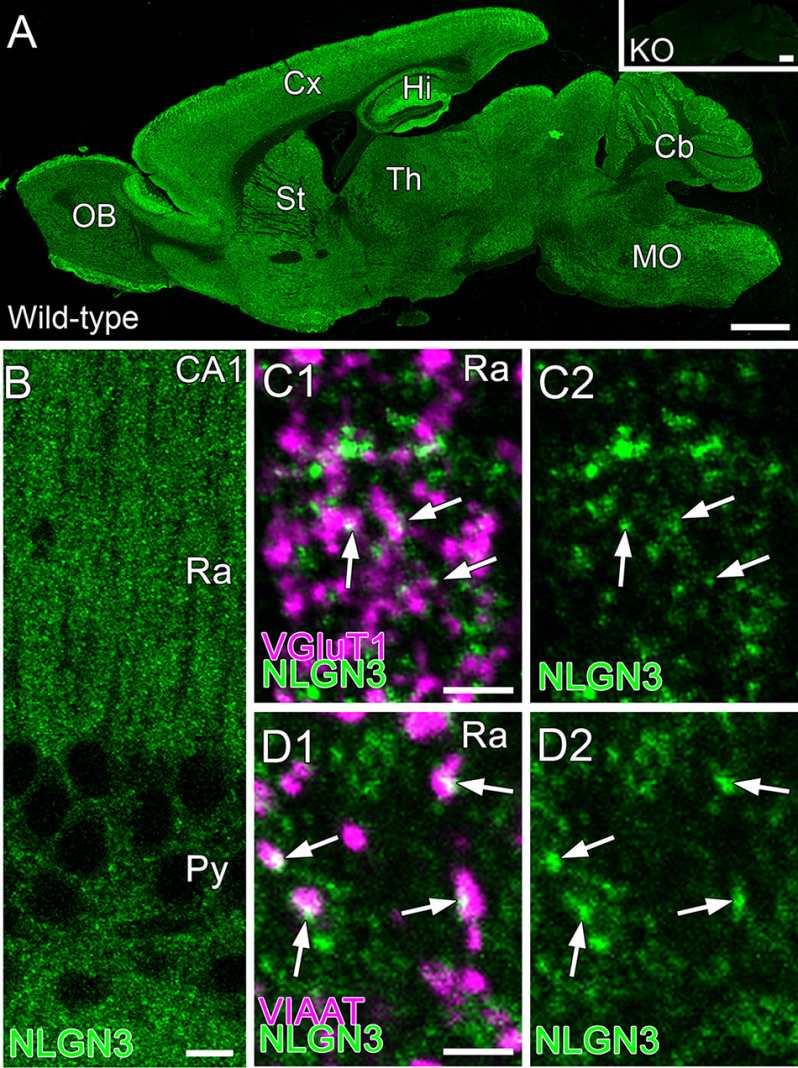
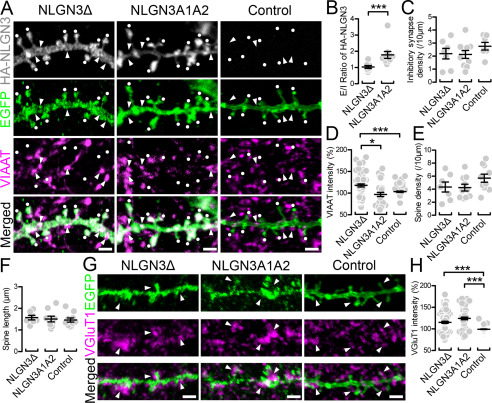
Expression of NLGN3 at excitatory and inhibitory synapses has been observed in primary neurons, but in vivo NLGN3 expression has been studied only in the cerebellum and striatum (11, 16, 17). To ensure the expression of NLGN3 in the hippocampus, we performed immunohistochemistry against NLGN3 with the markers for excitatory and inhibitory synapses, vesicular glutamate transporter type 1 (VGluT1) and vesicular inhibitory amino acid transporter (VIAAT), respectively (Fig. 2). Our NLGN3 antibody, validated by Nlgn3 knockout tissue, detected punctate signals in the hippocampus (Fig. 2, A and B). The signals overlapped with VGluT1 and VIAAT puncta, indicating that NLGN3 proteins are targeted to both excitatory and inhibitory synapses, respectively (Fig. 2, C and D). To understand the mechanistic roles of NLGN3 splice isoforms in inhibitory synaptic transmission, we next performed immunocytochemistry to elucidate the subcellular localization of NLGN3Δ and 3A1A2, which displayed strong enhancement and suppression of IPSC, respectively. Excitatory synaptic sites were characterized by spine or VGluT1. Inhibitory synaptic sites were identified by the dendritic shaft proximal to VIAAT puncta. HA immunoreactivity illustrated that NLGN3A1A2 is highly concentrated in spines. In contrast, NLGN3Δ showed more diffuse expression in both spines and dendrites (Fig. 3A). The ratio of NLGNA1A2 signals between excitatory and inhibitory synapses was significantly higher than that of 3Δ (Fig. 3B). Next, we addressed whether these NLGN3 splice isoforms differentially promote excitatory and inhibitory synapses by comparing NLGN3/GFP and GFP (control) transfected neurons. Importantly, inhibitory synapse density was comparable among NLGN3Δ, 3A1A2, and control, whereas VIAAT signal intensity in 3Δ-expressing neurons was significantly higher than that of 3A1A2 and control (Fig. 3, C and D). The spine density or length was comparable among NLGN3Δ, 3A1A2, and control neurons (Fig. 3, E and F). The signal intensities of VGluT1 were markedly elevated in neurons overexpressing NLGN3Δ or 3A1A2 compared with control neurons (Fig. 3, G and H). These results suggest that differences in the subcellular localization of NLGN3Δ and 3A1A2 contribute to their distinct inhibitory synaptic functions. Last, we tested whether NLGN3Δ or 3A1A2 overexpression changes endogenous expression of NLGN2 protein, an inhibitory synapse-specific NLGN protein (3, 18). Immunohistochemistry against GFP, VIAAT, and NLGN2 revealed that transfection of NLGN3 splice isoform has no effect on the level of endogenous NLGN2 at inhibitory synapses (Fig. S3). This result suggests that the physiological and anatomical phenotypes we observed in Figs. 1 and 3 are mediated by overexpressed NLGN3 and not due to an indirect detrimental side effect of transgene overexpression.
Figure 2.
Subcellular localization of NLGN3 in the hippocampal CA1 area. A, immunohistochemical staining for NLGN3 in whole brains of 8-week-old WT and Nlgn3-knockout mice (Nlgn3-KO, inset). B, immunohistochemical staining for NLGN3 in the hippocampal CA1 area. C and D, double immunohistochemical staining for NLGN3 (green) and VGluT1 (C, magenta) or VIAAT (D, magenta) in the stratum radiatum of the hippocampal CA1 area. Cb, cerebellum; Cx, cortex; Hi, hippocampus; MO, medulla oblongata; OB, olfactory bulb; Py, stratum pyramidale; Ra, stratum radiatum; St, striatum; Th, thalamus. Scale bars, 1 mm (A), 10 μm (B), 2 μm (C and D).
Figure 3.
Synaptic targeting of NLGN3 splice isoforms in CA1 pyramidal neurons. A, maximum projection images of dendritic segments labeled for HA-NLGN3 (gray), EGFP (green), and VIAAT (magenta) in CA1 pyramidal cells overexpressing HA-NLGN3Δ (left), A1A2 (middle), and EGFP control (right). White dots and arrows indicate EGFP+ spines and VIAAT+ inhibitory synapses, respectively. Scale bars, 2 μm. B–F, summary scatter plot showing the E/I ratio of HA signals (B), inhibitory synapse density (C), VIAAT intensity (D), spine density (E), and spine length (F) for individual dendritic segments of CA1 pyramidal cells overexpressing HA-NLGN3Δ (left, n = 7 dendrites), A1A2 (middle, n = 10), and control (right, n = 10). VIAAT intensity is normalized to the averaged VIAAT intensity in neighboring inhibitory terminals on the same image. G, maximum projection images of dendritic segments labeled for EGFP (green) and VGluT1 (magenta) in CA1 pyramidal cells overexpressing HA-NLGN3Δ (left), A1A2 (middle), and control (right). Arrowheads indicate VGluT1-labeled terminals contacting transfected dendrites. H, summary scatter plot showing VGluT1 intensity for individual dendritic segments of CA1 pyramidal cells overexpressing HA-NLGN3Δ (left, n = 7 dendrites), A1A2 (middle, n = 6), and control (left, n = 7). VGluT1 intensity is normalized to the averaged VGluT1 intensity in neighboring excitatory terminals on the same image. Scale bars, 2 μm. *, p < 0.05; ***, p < 0.001 (one-way analysis of variance followed by Sidak's multiple-comparison test or U test). Error bars, S.E.M.
Endogenous expression of nlgn genes and splice isoforms in hippocampal CA1 pyramidal neurons
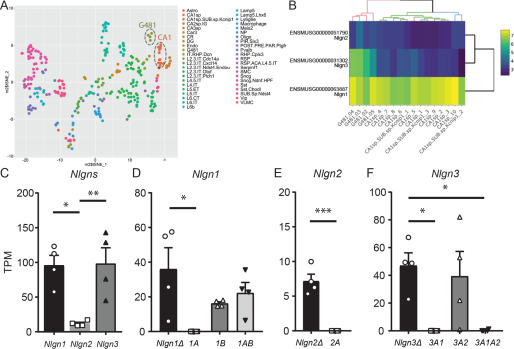
Finally, to understand the expression of endogenous Nlgn genes in CA1 pyramidal neurons, we harvested cytosol from four neurons and performed single-cell deep RNA-Seq. The t-SNE plot indicates that the four cell transcripts (G418) were clustered together and close to that of adult hippocampal CA1-3 pyramidal neurons derived from the Allen Brain Atlas (Fig. 4A). The expression of Nlgn genes was clustered and well-correlated with the single-cell RNA-Seq data in the RNA-Seq data sets provided by the Allen Institute for Brain Science (Fig. 4B). The quantification of Nlgn genes (Fig. 4C) indicates that the expression of Nlgn1 and Nlgn3 are comparable but that of Nlgn2 is significantly lower than the other two genes. We also compared the expression of Nlgn splice isoforms in each Nlgn gene. Six Nlgn splice isoforms, Nlgn1A, 1B, 2Δ, 3Δ, and 3A1, that were not annotated were manually modified (Fig. S4A), and their expression was compared. Nlgn1Δ, 1B, and 1AB were the most highly expressed Nlgn1 splice isoforms (Fig. 4D). Nlgn2Δ was the only isoform counted in the Nlgn2 gene (Fig. 4E). Nlgn1A and 2A transcripts were not detected in any of the four CA1 pyramidal neurons. Importantly, Nlgn3Δ and 3A2, which exhibited increased inhibitory synaptic transmission, were the dominant Nlgn3 splice isoforms in CA1 pyramidal neurons (Fig. 4F). The expressions of Nlgn3A1 and 3A1A2 were significantly lower than that of Nlgn3Δ (TPM: Nlgn3A1: 0.004 ± 0.004, 3A1A2: 0.3 ± 0.3). The expression of these single-cell Nlgn splice isoforms was also confirmed by semi-quantitative PCR (Fig. S4B).
Figure 4.
Endogenous Nlgn expression in hippocampal CA1 pyramidal neurons. A, single-cell t-SNE plot of four single-cells (G481.O2, .03, .04, and.05) and Allen Brain Atlas single cells. B, heat map of Nlgn gene expression in hippocampal CA1 primary neurons. A hierarchical clustering demonstrated a segregation of G481 cells from adult CA1 pyramidal neurons for Nlgn3, potentially due to relatively higher expression of Nlgn3 in G481 cells. Scale bar, log2 + 1–transformed TPM values. C, summary bar graph of Nlgn gene (Nlgn1, 2, and 3) expression. D–F, summary graphs of splice isoforms of Nlgn1 (D), 2 (E), and 3 (F). *, p < 0.05, one-way analysis of variance followed by Sidak's multiple-comparison test or U test, n = 4. Error bars, S.E.M.
Discussion
Trans-synaptic protein–protein interactions are fundamental biological events for synapse formation, maturation, and function. NLGNs are critical postsynaptic adhesion molecules that regulate excitatory and inhibitory synaptic transmission. Here we demonstrated that NLGN3 regulates inhibitory synaptic transmission and excitatory and inhibitory synapse localization in a splice isoform–dependent manner. Our single-cell transcriptome analysis revealed that Nlgn3Δ and 3A2 are the highest-expressed Nlgn3 splice isoforms in hippocampal CA1 pyramidal neurons.
The distinct subcellular localization of NLGN3Δ and 3A1A2 suggests intriguing mechanisms regarding how splice isoforms influence synapse specificity. Given that the intracellular and transmembrane domains are identical between NLGN3 splice isoforms, each isoform exerts their synapse coding effect through their unique extracellular domains. Similarly, the extracellular domain of NLGN2 mediates changes in inhibitory synaptic function (6). Based on our current knowledge, NRXs are the only trans-synaptic family proteins that directly bind to NLGNs. Trans-synaptic interactions between NLGN3Δ, but not 3A1A2, and NRXs modulate inhibitory synaptic transmission in pyramidal neurons. We previously reported that postsynaptic NLGN2 can couple with presynaptic αNRX1 but not with βNRX1 to form functional inhibitory synapses (6), suggesting that the specific binding of NLGN and NRX isoforms regulates functional synapse formation. It is possible that inhibitory interneurons do not express NRX isoforms that can bind to NLGN3A1A2. Therefore, NLGN3A1A2 may exclusively localize to excitatory synapses. Further studies should be performed to identify specific NRX-NLGN3 isoform interactions that affect inhibitory synaptic function. Another possible mechanism is cis-cis protein interactions between NLGN3 and postsynaptic proteins. It has been reported that protein complexes formed between the extracellular domain of NLGN1 and NMDAR are important for synaptic function (19). NLGN3 splice variants may have different binding interactions with postsynaptic proteins that provide distinct functions on inhibitory synapses. It has been suggested that the relative levels of NLGNs and their postsynaptic scaffold complex at excitatory and inhibitory synapses determine E-I balance (20). Additional studies are required, but it is interesting to address the hypothesis that postsynaptic NLGN3A1 and A1A2 are strong and specific regulators at excitatory synaptic sites and sequester the necessary protein interactions (i.e. NLGN2-mediated scaffold complex) from inhibitory synapses to reduce inhibitory synaptic transmission.
The single-cell sequencing results demonstrate a comprehensive unbiased gene expression profile of Nlgn splice isoforms in hippocampal CA1 pyramidal neurons obtained from neonatal mice. Our transcriptome findings are highly correlated with the expression pattern of the three Nlgns genes provided by the Allen Brain Atlas single-cell database from adult neurons, indicating that the expression ratio of Nlgns are stable throughout development. Interestingly, NLGN2 has been well-characterized at inhibitory synapses, yet its transcript levels were significantly lower than Nlgn1 and Nlgn3 (Fig. 4, B and C). NLGN2 may have unique post-translational modifications and turnover mechanisms compared with NLGN1 and NLGN3. The expression of Nlgn3A1 and 3A1A2 were much lower than of Nlgn3Δ and 3A2, suggesting that these two splice isoforms are not the major functional Nlgn3 molecules under basal conditions. Further work is necessary to address whether modifications to neuronal activity such as synaptic plasticity can alter the expression of Nlgn splice isoforms.
A shift in E-I balance has been considered a pathophysiological hallmark of neurodevelopmental disorders and repeatedly reported in corresponding mouse models (21). Mutations and deletions of Nlgn3 loci are associated with autism spectrum disorders (22), and mutant mice that mimic the human autism Nlgn3 mutation exhibit E-I imbalance and abnormal synaptic plasticity (1). A closer examination of specific NLGN3 splice isoform functions will elucidate their role in producing critical molecular outcomes that may influence neuropsychiatric disease pathogenesis.
Experimental procedures
Animal and organotypic slice culture preparation
All animal protocols were approved and reviewed by the Institutional Animal Care and Use Committee at the University of Massachusetts Medical School and the Hokkaido University. The Nlgn3 KO mouse line was a gift from Dr. Tanaka (23). Organotypic hippocampal slice cultures were prepared from postnatal 6–7-day-old C57BL6 mice of both sexes as described previously (24).
Single-cell sequencing and analysis
The cytosol of four CA1 neurons were harvested using the whole-cell patch-clamp technique described previously (6). Library preparation, mRNA sequencing, and data analysis were done based on our established protocol. The single-cell RNA-Seq procedure is described in detail in the supporting Experimental procedures.
Immunohistochemistry and immunocytochemistry
Mice were transcardially perfused with 4% paraformaldehyde, 0.1 m phosphate buffer. Brains were dehydrated and embedded with paraffin. Paraffin sections (2 μm in thickness) were generated using a sliding microtome (Leica). Prior to immunoreactions, paraffin sections were boiled with Immunosaver (Nisshin EM) for 30 min. Organotypic slice cultures transfected with NLGN3 splice isoforms were fixed with 4% paraformaldehyde and 4% sucrose in 0.01 m PBS. Organotypic slices were permeabilized with 0.1-0.5% Triton X-100/PBS (PBST), followed by blocking with 10% goat serum. A mixture of primary antibodies against enhanced GFP (EGFP (chicken); Millipore), human influenza hemagglutinin (HA; rabbit, Cell Signals), NLGN2 (rabbit (16)), NLGN3 (guinea pig (16)), VGluT1 (rabbit and guinea pig) (25), and VIAAT (guinea pig and goat) (26) and a mixture of species-specific secondary antibodies conjugated with Alexa 488, 555, and 647 (Thermo Fisher Scientific) were used for immunostaining. Images were taken with a confocal microscope (FV1200, Olympus) equipped with a ×60 silicone oil immersion objective (UPLSAPO 60XS) and analyzed with ImageJ software.
Electrophysiology
Organotypic hippocampal slice cultures were prepared from postnatal 6-7-day-old mice of either sex. Neurons at days in vitro 4-6 were transfected using a biolistic gene gun (Bio-Rad) and were assayed 3 days after transfection (days in vitro 7-9) as described previously (5, 6, 27, 28). Whole-cell voltage clamp recordings were made simultaneously from a pair of CA1 pyramidal neurons, one transfected (visualized by co-transfecting GFP) and one untransfected neighbor. GABAA receptor–mediated IPSCs were evoked with a stimulating electrode placed in the stratum radiatum and measured at Vhold ± 0 mV. AMPAR-mediated EPSCs were evoked at Vhold −70 mV in the presence of picrotoxin (0.1 mm, Sigma). Further electrophysiological procedures are described in the supporting Experimental procedures.
Statistical analyses
Results are reported as mean ± S.E. Mann–Whitney U test and Student's t test were used for two-group comparison. Statistical significance was set at p < 0.05 for Student's t test and U test.
Data availability
The accession number for the RNA-Seq and processed data reported in this paper is GEO: GSE143295. Other experimental procedures are described in the supporting Experimental procedures.
Supplementary Material
Acknowledgments
The authors thank Naoe Watanabe for skillful technical assistance. We thank Dr. Paul D. Gardner for comments on the manuscript.
This article contains supporting information.
Author contributions—M. U., Y. I. K., and K. F. data curation; M. U. and K. F. funding acquisition; M. U., M. L., T. W., A. C., T. L., S. P., A. L. M. D., Y. I. K., and K. F. investigation; M. U., T. W., Y. I. K., and K. F. visualization; M. U., T. W., A. C., M. W., Y. I. K., and K. F. writing-original draft; M. U., T. W., A. C., M. W., Y. I. K., and K. F. writing-review and editing; M. U., M. L., T. W., Y. I. K., and K. F. formal analysis; M. U., M. W., Y. I. K., and K. F. validation; M. U., M. W., Y. I. K., and K. F. resources; M. U., Y. I. K., and K. F. software; M. U., M. W., Y. I. K., and K. F. supervision; M. U., Y. I. K., and K. F. methodology; Y. I. K. and K. F. project administration.
Funding and additional information—This work was supported by National Institutes of Health Grants R01NS085215 (to K. F.) and T32 GM107000 (to A. C.) and Grant-in-Aid for Scientific Research 15K06732 (to M. U.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- NLGN
- neuroligin
- NRX
- neurexin
- E-I
- excitatory-inhibitory
- IPSC
- inhibitory postsynaptic current
- EPSC
- excitatory postsynaptic current
- PPF
- paired-pulse facilitation
- PPD
- paired-pulse depression
- EGFP
- enhanced GFP
- VIAAT
- vesicular inhibitory amino acid transporter
- VGluT1
- vesicular glutamate transporter type 1
- AMPAR
- AMPA receptor.
References
- 1. Sudhof T. C. (2017) Synaptic neurexin complexes: a molecular code for the logic of neural circuits. Cell 171, 745–769 10.1016/j.cell.2017.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jiang M., Polepalli J., Chen L. Y., Zhang B., Südhof T. C., and Malenka R. C. (2017) Conditional ablation of neuroligin-1 in CA1 pyramidal neurons blocks LTP by a cell-autonomous NMDA receptor-independent mechanism. Mol. Psychiatry 22, 375–383 10.1038/mp.2016.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chubykin A. A., Atasoy D., Etherton M. R., Brose N., Kavalali E. T., Gibson J. R., and Südhof T. C. (2007) Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron 54, 919–931 10.1016/j.neuron.2007.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shipman S. L., and Nicoll R. A. (2012) A subtype-specific function for the extracellular domain of neuroligin 1 in hippocampal LTP. Neuron 76, 309–316 10.1016/j.neuron.2012.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Futai K., Kim M. J., Hashikawa T., Scheiffele P., Sheng M., and Hayashi Y. (2007) Retrograde modulation of presynaptic release probability through signaling mediated by PSD-95-neuroligin. Nat. Neurosci. 10, 186–195 10.1038/nn1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Futai K., Doty C. D., Baek B., Ryu J., and Sheng M. (2013) Specific trans-synaptic interaction with inhibitory interneuronal neurexin underlies differential ability of neuroligins to induce functional inhibitory synapses. J. Neurosci. 33, 3612–3623 10.1523/JNEUROSCI.1811-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shipman S. L., Schnell E., Hirai T., Chen B. S., Roche K. W., and Nicoll R. A. (2011) Functional dependence of neuroligin on a new non-PDZ intracellular domain. Nat. Neurosci. 14, 718–726 10.1038/nn.2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nguyen Q. A., Horn M. E., and Nicoll R. A. (2016) Distinct roles for extracellular and intracellular domains in neuroligin function at inhibitory synapses. eLife 5, 10.7554/eLife.19236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Etherton M., Foldy C., Sharma M., Tabuchi K., Liu X., Shamloo M., Malenka R. C., and Sudhof T. C. (2011) Autism-linked neuroligin-3 R451C mutation differentially alters hippocampal and cortical synaptic function. Proc. Natl. Acad. Sci. U.S.A. 108, 13764–13769 10.1073/pnas.1111093108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Foldy C., Malenka R. C., and Sudhof T. C. (2013) Autism-associated neuroligin-3 mutations commonly disrupt tonic endocannabinoid signaling. Neuron 78, 498–509 10.1016/j.neuron.2013.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Budreck E. C., and Scheiffele P. (2007) Neuroligin-3 is a neuronal adhesion protein at GABAergic and glutamatergic synapses. Eur. J. Neurosci. 26, 1738–1748 10.1111/j.1460-9568.2007.05842.x [DOI] [PubMed] [Google Scholar]
- 12. Shipman S. L., and Nicoll R. A. (2012) Dimerization of postsynaptic neuroligin drives synaptic assembly via transsynaptic clustering of neurexin. Proc. Natl. Acad. Sci. U.S.A. 109, 19432–19437 10.1073/pnas.1217633109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ichtchenko K., Hata Y., Nguyen T., Ullrich B., Missler M., Moomaw C., and Südhof T. C. (1995) Neuroligin 1: a splice site-specific ligand for β-neurexins. Cell 81, 435–443 10.1016/0092-8674(95)90396-8 [DOI] [PubMed] [Google Scholar]
- 14. Boucard A. A., Chubykin A. A., Comoletti D., Taylor P., and Südhof T. C. (2005) A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to α- and β-neurexins. Neuron 48, 229–236 10.1016/j.neuron.2005.08.026 [DOI] [PubMed] [Google Scholar]
- 15. Chih B., Gollan L., and Scheiffele P. (2006) Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron 51, 171–178 10.1016/j.neuron.2006.06.005 [DOI] [PubMed] [Google Scholar]
- 16. Uchigashima M., Ohtsuka T., Kobayashi K., and Watanabe M. (2016) Dopamine synapse is a neuroligin-2-mediated contact between dopaminergic presynaptic and GABAergic postsynaptic structures. Proc. Natl. Acad. Sci. U.S.A. 113, 4206–4211 10.1073/pnas.1514074113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baudouin S. J., Gaudias J., Gerharz S., Hatstatt L., Zhou K., Punnakkal P., Tanaka K. F., Spooren W., Hen R., De Zeeuw C. I., Vogt K., and Scheiffele P. (2012) Shared synaptic pathophysiology in syndromic and nonsyndromic rodent models of autism. Science 338, 128–132 10.1126/science.1224159 [DOI] [PubMed] [Google Scholar]
- 18. Varoqueaux F., Jamain S., and Brose N. (2004) Neuroligin 2 is exclusively localized to inhibitory synapses. Eur J. Cell Biol. 83, 449–456 10.1078/0171-9335-00410 [DOI] [PubMed] [Google Scholar]
- 19. Budreck E. C., Kwon O. B., Jung J. H., Baudouin S., Thommen A., Kim H. S., Fukazawa Y., Harada H., Tabuchi K., Shigemoto R., Scheiffele P., and Kim J. H. (2013) Neuroligin-1 controls synaptic abundance of NMDA-type glutamate receptors through extracellular coupling. Proc. Natl. Acad. Sci. U.S.A. 110, 725–730 10.1073/pnas.1214718110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Levinson J. N., and El-Husseini A. (2005) Building excitatory and inhibitory synapses: balancing neuroligin partnerships. Neuron 48, 171–174 10.1016/j.neuron.2005.09.017 [DOI] [PubMed] [Google Scholar]
- 21. Nelson S. B., and Valakh V. (2015) Excitatory/inhibitory balance and circuit homeostasis in autism spectrum disorders. Neuron 87, 684–698 10.1016/j.neuron.2015.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jamain S., Quach H., Betancur C., Rastam M., Colineaux C., Gillberg I. C., Soderstrom H., Giros B., Leboyer M., Gillberg C., Bourgeron T., and Paris Autism Research International Sibpair Study (2003) Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 34, 27–29 10.1038/ng1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tanaka K. F., Ahmari S. E., Leonardo E. D., Richardson-Jones J. W., Budreck E. C., Scheiffele P., Sugio S., Inamura N., Ikenaka K., and Hen R. (2010) Flexible accelerated STOP tetracycline operator-knockin (FAST): a versatile and efficient new gene modulating system. Biol. Psychiatry 67, 770–773 10.1016/j.biopsych.2009.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stoppini L., Buchs P. A., and Muller D. (1991) A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods 37, 173–182 10.1016/0165-0270(91)90128-M [DOI] [PubMed] [Google Scholar]
- 25. Kawamura Y., Fukaya M., Maejima T., Yoshida T., Miura E., Watanabe M., Ohno-Shosaku T., and Kano M. (2006) The CB1 cannabinoid receptor is the major cannabinoid receptor at excitatory presynaptic sites in the hippocampus and cerebellum. J. Neurosci. 26, 2991–3001 10.1523/JNEUROSCI.4872-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miyazaki T., Fukaya M., Shimizu H., and Watanabe M. (2003) Subtype switching of vesicular glutamate transporters at parallel fibre-Purkinje cell synapses in developing mouse cerebellum. Eur. J. Neurosci. 17, 2563–2572 10.1046/j.1460-9568.2003.02698.x [DOI] [PubMed] [Google Scholar]
- 27. Hasegawa Y., Mao W., Saha S., Gunner G., Kolpakova J., Martin G. E., and Futai K. (2017) Luciferase shRNA presents off-target effects on voltage-gated ion channels in mouse hippocampal pyramidal neurons. eNeuro 4, ENEURO.0186-17.2017 10.1523/ENEURO.0186-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mao W., Salzberg A. C., Uchigashima M., Hasegawa Y., Hock H., Watanabe M., Akbarian S., Kawasawa Y. I., and Futai K. (2018) Activity-induced regulation of synaptic strength through the chromatin reader L3mbtl1. Cell Rep. 23, 3209–3222 10.1016/j.celrep.2018.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van der Maaten L., and Hinton G. (2008) Visualizing data using t-SNE. J. Machine Learning Res. 9, 2579–2605 [Google Scholar]
- 30. Bray N. L., Pimentel H., Melsted P., and Pachter L. (2016) Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-Seq and processed data reported in this paper is GEO: GSE143295. Other experimental procedures are described in the supporting Experimental procedures.