Abstract
The transcriptional coactivator YAP1 (yes-associated protein 1) regulates cell proliferation, cell–cell interactions, organ size, and tumorigenesis. Post-transcriptional modifications and nuclear translocation of YAP1 are crucial for its nuclear activity. The objective of this study was to elucidate the mechanism by which the steroid hormone androgen regulates YAP1 nuclear entry and functions in several human prostate cancer cell lines. We demonstrate that androgen exposure suppresses the inactivating post-translational modification phospho–Ser-127 in YAP1, coinciding with increased YAP1 nuclear accumulation and activity. Pharmacological and genetic experiments revealed that intact androgen receptor signaling is necessary for androgen's inactivating effect on phospho–Ser-127 levels and increased YAP1 nuclear entry. We also found that androgen exposure antagonizes Ser/Thr kinase 4 (STK4/MST1) signaling, stimulates the activity of protein phosphatase 2A, and thereby attenuates the phospho–Ser-127 modification and promotes YAP1 nuclear localization. Results from quantitative RT-PCR and CRISPR/Cas9–aided gene knockout experiments indicated that androgen differentially regulates YAP1-dependent gene expression. Furthermore, an unbiased computational analysis of the prostate cancer data from The Cancer Genome Atlas revealed that YAP1 and androgen receptor transcript levels correlate with each other in prostate cancer tissues. These findings indicate that androgen regulates YAP1 nuclear localization and its transcriptional activity through the androgen receptor–STK4/MST1–protein phosphatase 2A axis, which may have important implications for human diseases such as prostate cancer.
Keywords: signal transduction, androgens, androgen receptor, YAP1, Hippo/MST1/STK4, protein phosphatase PP2A, protein–protein interaction, post-translational modification, phosphorylation, nuclear translocation, gene transcription, androgen, cell signaling, Hippo pathway, yes-associated protein (YAP), protein phosphorylation, protein serine/threonine phosphatase (PSP), gene transcription, androgen/AR signaling, YAP1 nuclear localization
YAP1 (yes-associated protein 1) and its paralog WWTR1 (WW domain–containing protein) are transcriptional coactivators (1, 2). YAP1 is a well-characterized nuclear effector of the Hippo pathway in mammals (2–4). The STK4/MST1, STK3/MST2, and LATS1/2 protein kinases are core components of the Hippo pathway (5). YAP1 was initially identified from the protein complexes of the Src family kinases (6). YAP1 regulates diverse cellular activities, including cell proliferation, cell survival, cell differentiation, stem cell maintenance, cell–cell interaction, organ size, and tumorigenesis (2). Nuclear localization of YAP1 is critical for its transcriptional-dependent biological functions, even though a majority of YAP1 proteins are present in the cytoplasm (7, 8). YAP1 exerts its transcriptional-dependent biological activity by interacting with transcription factors. The family of the TEAD transcription factors is a critical mediator of the YAP1-dependent gene transcription. Mounting evidence indicates that interaction between the YAP1 and TEAD proteins is mutual because YAP1 functions as a coactivator for TEAD-dependent gene expression (3, 8, 9).
Although the exact mechanisms are mostly unknown, YAP1 shuttles between the cytoplasm and nucleus. Protein–protein interactions (8, 10) and nuclear import or export signaling are possible mechanisms that control the translocations of YAP1 between the cytoplasm and nucleus (11–14). Post-translational modification through phosphorylation is one of the best-characterized mechanisms that modulate the nuclear–cytoplasmic translocation of YAP1 (15). The MST1/2 and LATS1/2 kinase cascade phosphorylate Ser-127 and inactivate YAP1 (3, 7, 16). In addition to the covalent modification, a mechanical force could also increase YAP1 nuclear import (14, 17). For example, the PDLIM5/7 family of PDZ and LIM domain–containing proteins increased YAP1 nuclear import through integrin-mediated mechanotransduction (18). Regardless, TEAD or 14-3-3 protein binding mediates YAP1 nuclear localization (11, 19, 20). Our published study suggests that androgen hormone signaling regulates the nuclear–cytoplasmic translocation of YAP1 (7), but the mechanism is unknown.
In this study, we have demonstrated that androgen exposure can suppress the inhibitory phospho–Ser-127 on YAP1, which causes increases in YAP1 protein levels and nuclear abundance. We showed that androgen receptor (AR) signaling was critical for the regulation of YAP1 by androgen. We also showed that androgen exposure antagonized the Hippo/STK4 signaling and promoted PP2A activity. Our data further demonstrate that androgen signaling regulates the YAP1-dependent gene expression. Thus, our study uncovers a new mechanism of YAP1 regulation that involves androgen hormone signaling.
Results
Expression of YAP1 protein and transcript varies in prostate cancer cell models
To identify suitable cell lines that allow studying the effects of androgen on YAP1, we analyzed the levels of YAP1, WWTR1, and AR in the established prostate cancer cell models. We found that the levels of YAP1, WWTR1, and AR protein varied among the cell lines tested (Fig. 1, A and B). Surprisingly, LNCaP and its androgen-insensitive LN-81 and C4-2 sublines showed no detectable WWTR1 protein. Also, the 22Rv1 cell line was YAP1-deficient compared with LNCaP and C4-2. The ARCaP and PC3 cell lines expressed YAP1, but they were AR-negative. Similarly, the expression of YAP1 transcripts also varied in the select cell lines (Fig. 1C). Thus, LNCaP is the ideal cell model to study the mechanism of YAP1 regulation by androgen because it harbors an intact AR and YAP1 signaling pathway (7).
Figure 1.
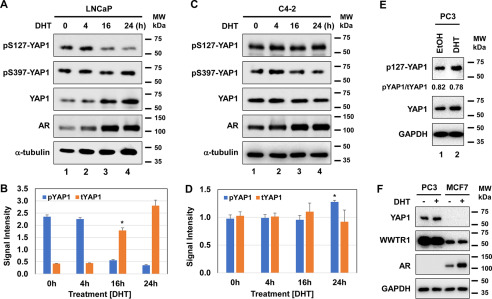
Expression of YAP1, WWTR1, and AR in prostate cancer cell models. A, total proteins were isolated from cells at 80% confluence in steady-state growth conditions and analyzed by Western blotting for YAP1, WWTR1, or AR protein abundance. β-Actin was used as a loading control in immunoblots. The membranes were blotted with the protein-specific antibody. B, quantification of YAP1 protein was normalized to the β-actin. ImageJ software was used to quantify YAP1 and β-actin protein blots. C, expression of YAP1 transcripts in the select cell lines. Total RNA was isolated from cells at 80% confluence in steady-state conditions 24 h after cell seeding and analyzed by quantitative RT-PCR for the abundance of YAP1 transcripts, *,**, p < 0.01. YAP1 mRNA expression was normalized to 18S rRNA transcript. The data ± S.D. are from three independent experiments. MW, molecular mass.
Androgen attenuates the inhibitory phospho–Ser-127 modification on YAP1
To determine whether androgen signaling post-transcriptionally regulates phospho–Ser-127, a potent inhibitor of YAP1 nuclear translocation (3, 7, 16), we examined the levels of phospho–Ser-127 and the total amount of the YAP1 protein in LNCaP and its hormone-independent C4-2 subline. Both cell lines were exposed to dihydrotestosterone (DHT) at varying times (0, 4, 16, or 24 h). We also examined the levels of phospho–Ser-397 on YAP1 as a control. Androgen significantly inhibited phospho–Ser-127 in LNCaP cells but without affecting phospho–Ser-397 (Fig. 2, A and B), suggesting that androgen specifically regulates phospho–Ser-127 on YAP1. Notably, the inhibition of phospho–Ser-127 by androgen correlated with increases in the total amount of YAP1 protein (13, 21, 22), which overlapped with AR activity (Fig. 2A). Surprisingly, androgen did not significantly alter the levels of the phospho–Ser-127 and the total amount of YAP1 protein in C4-2 cells compared with that of LNCaP (23) under the same experimental conditions (Fig. 2, C and D).
Figure 2.
Steroid hormone androgen regulates phospho–Ser-127 and YAP1 protein levels in cultures. A–D, Western blotting and quantification of phospho–Ser-127 and total YAP1 proteins in androgen-dependent LNCaP (*, p < 0.01) and androgen-independent C4-2 (*, p > 0.05) cells, respectively. The cells were exposed to DHT (10 nm) at varying times. The data in B and C were normalized to α-tubulin. The phospho–Ser-127 blot was included as a control. E and F, Western blotting analysis of YAP1 and WWTR1 proteins in the PC3 prostate cancer and MCF7 breast cancer cell lines that were also treated with vehicle (ethanol) or DHT overnight (16–18 h). DHT treatment was conducted in 5% CSS-fed growth conditions. ImageJ software was used to quantify the intensity of phospho–Ser-127, total YAP, and α-tubulin protein bands. YAP1 and AR blots were included as a positive control in F. The α-tubulin or GAPDH protein blot was incorporated as a loading control. The membranes were blotted with the protein-specific antibody. The data ± S.D. are representative of two independent experiments. MW, molecular mass.
To further confirm the specificity of the effects of androgen on YAP1, we analyzed the levels of phospho–Ser-127 and total YAP1 in the PC3 cell line, which is AR-negative (24), and the MCF7 cell line, which is AR-positive and responsive to androgen (25, 26). Treatment of these cells with DHT did not alter the phospho–Ser-127 in PC3 cells (Fig. 2E). Surprisingly, MCF7 cells did not show detectable YAP1 protein, although expressing its paralog WWTR1 protein (Fig. 2F). Androgen did not alter the levels of the WWTR1 protein in both PC3 and MCF7 cells. Taken together, the effects of androgen on phospho–Ser-127 are specific.
Androgen attenuates phospho–Ser-127 through AR and Hippo/SKT4 signaling
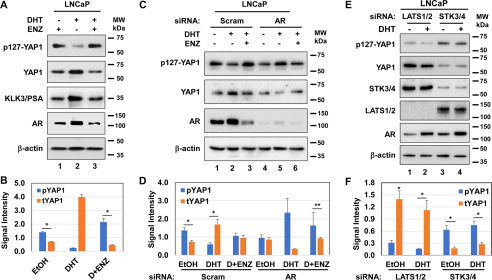
To demonstrate whether the AR activity is critical for the inhibition of phospho–Ser-127 by androgen, we performed a series of experiments. First, we evaluated the levels of phospho–Ser-127 and total YAP1 protein in LNCaP cells after treatment with a vehicle, DHT, and DHT plus enzalutamide (ENZ), a direct pharmacological inhibitor of the AR (Fig. 3, A and B). Compared with DHT, ENZ exposure reversed the inhibitory effects of androgen on phospho–Ser-127, accompanied by a reduction in the total YAP1 protein, which likely occurred because of the induction of phospho–Ser-127 (Fig. 3A, lane 2 versus lane 3). Second, we also examined the levels of phospho–Ser-127 and total YAP1 protein in LNCaP with or without AR knockdown by siRNA (Fig. 3, C and D). The results showed that androgen was unable to reduce phospho–Ser-127 in LNCaP cells with AR silencing compared with the scrambled siRNA control (lane 4 versus lane 5), which overlapped with a diminished total amount of YAP1 protein. The treatment of cells with ENZ had the opposite effect (Fig. 3C, lane 6 versus lane 5). Nevertheless, AR knockdown reduced the total YAP1 protein relative to the control siRNA (Fig. 3, C and D).
Figure 3.
AR and STK4/3 signaling is necessary for the inhibition of phospho–Ser-127 by androgen. A and B, Western blotting and quantification of phospho–Ser-127 and total YAP1 proteins, respectively. *,**, p < 0.01. KLK3/prostate specific antigen (PSA) blot was included as a positive control to assess the activity of DHT. C and D, Western blotting and quantification of phospho–Ser-127 and total YAP1 protein in LNCaP cells after transient transfection with the pool of scrambled (Scram) or the AR gene-specific siRNA for 36 h, followed by treatment with or without DHT and ENZ overnight in 5% CSS-fed conditions. E and F, Western blotting and quantification of phospho–Ser-127 and total YAP1 in LNCaP cells transfected with Scram siRNA or the LATS1/2 and STK4/3 (MST1/2) gene-specific siRNA for 36 h, followed by treatment with or without DHT overnight in 5% CSS-fed growth conditions. β-Actin was used as a loading control in immunoblots. The membranes were blotted with the protein-specific antibody. ImageJ software was used to quantify the intensity of the phospho–Ser-127 and the total YAP1 signal. The data ± S.D. are representative of three independent experiments. MW, molecular mass.
We also wanted to know whether androgen antagonizes STK3/4 signaling in a way that suppresses phospho–Ser-127, given that the STK3/4 kinase mediates phospho–Ser-127 on YAP1 (16). To test this possibility, we assessed the levels of phospho–Ser-127 and total YAP1 protein in LNCaP cells with LATS1/2 and STK3/4 knockdown. Compared with the vehicle, androgen exposure slightly decreased phospho–Ser-127 in LATS1/2 knockdown, but without altering the levels of phospho–Ser-127 in STK3/4 knockdown cells (Fig. 3, E and F).
Androgen antagonizes the Hippo/STK4 signaling through the Ser/Thr phosphatases
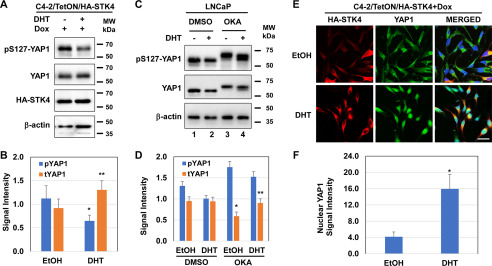
Previously, we reported that controlled, ectopic expression of MST1/STK4 in the engineered C4-2/HA-MST1/STK4 cell line prevented YAP1 nuclear localization (7). We utilized this cell line to demonstrate whether androgen antagonizes Hippo/STK4 signaling in a way that inhibits phospho–Ser-127. In addressing this idea, we analyzed the levels of phospho–Ser-127 and total YAP1 protein in C4-2/HA-STK4 cells that were exposed to doxycycline (Dox) along with the vehicle, or DHT. Dox was used to induce ectopic MST1/STK4 expression. Compared with vehicle control, androgen exposure significantly reduced phospho–Ser-127, which resulted in increases in total YAP1 protein (Fig. 4, A and B).
Figure 4.
Androgen negatively regulates the Hippo/STK4 signaling. A and B, Western blotting and quantification of phospho–Ser-127 and total YAP1 proteins in C4-2/HA-STK4/MST1 cells that express tetracycline- or Dox-inducible HA-STK4 protein (45), respectively. C and D, Western blotting and quantification of phospho–Ser-127 and total YAP1 protein in LNCaP cells that were exposed to DMSO (vehicle) or OKA followed by treatment with or without DHT overnight in 5% CSS-fed growth condition. β-Actin was used as a loading control in immunoblots. E, immunofluorescence analysis of ectopic HA-STK4 (red), and native YAP1 (green) proteins and nuclei (DAPI, blue) in Dox-treated C4-2/HA-STK4 cells. Scale bar, 20 μm. Micrographs are the representation of multiple images. F, quantification of nuclear YAP1 protein by ImageJ from multiple images and nuclear YAP1 signals were normalized to the total number of cells subjected to image quantification. The ectopic expression of HA-STK4/MST1 protein was assessed using an HA tag antibody. The cells were treated with EtOH or DHT (10 nm) and Dox (1 μg/ml) in serum-depleted conditions overnight before analysis. *,**, p < 0.05. MW, molecular mass.
The Ser/Thr phosphatases PP1/PP2A are known to inactivate the STK4/3 signaling (27–29). Okadaic acid (OKA), a potent inhibitor of the PP1/PP2A phosphatases, activates STK4/MST1 (30). Thus, we proposed that androgen positively regulates PP2A, which, in turn, attenuates Hippo/STK4 signaling. To test this possibility, we assessed the levels of phospho–Ser-127 and total YAP1 protein in LNCaP cells after treatment with DMSO (mock) control and OKA. Compared with DMSO treatment, OKA reduced the mobility of phospho–Ser-127 and YAP1 protein in the reduced SDS-PAGE (Fig. 4, C and D). We suggest that a reduction in the mobility of YAP1 protein accounts for its persistent multiple phosphorylation sites that increased the molecular weight of the YAP1 protein. This is likely due to the inactivation of the Ser/Thr phosphatases by OKA (30). However, compared with the vehicle, androgen slightly accelerated the YAP1 mobility, most likely by activating the Ser/The phosphatases, which in turn decreased the phosphomodifications on YAP1 (29, 31). Furthermore, immunofluorescence imaging showed that androgen exposure enhanced YAP1 nuclear abundance in Dox-treated C4-2/HA-STK4 cells compared with vehicle treatment (Fig. 4, E and F). Hence, androgen may activate the Ser/Thr phosphatases to attenuate the Hippo/STK4 activity.
Androgen promotes YAP1 nuclear localization through AR and PP2A
To examine whether AR activity is necessary for the androgen-induced nuclear accumulation of YAP1, we conducted a coimmunofluorescence analysis of native AR and YAP1 proteins in LNCaP cells after treatment with vehicle, DHT, or DHT plus ENZ (Fig. 5A). Compared with the mock treatment, androgen exposure significantly increased the colocalization of YAP1 and AR protein in the cell nuclei. ENZ, however, reversed the effects of androgen on YAP1 and AR nuclear abundance (Fig. 5B). Consistently, coimmunoprecipitation and Western blotting experiments demonstrated that androgen enhanced the interaction of YAP1 with AR and the catalytic C subunit of the PP22A, particularly in the nuclear fraction, under the same experimental conditions (Fig. 5C). These findings indicate that AR, in concert with PP2A, mediates the reduction of phospho–Ser-127 and induction of YAP1 nuclear localization by androgen.
Figure 5.
Androgen promotes YAP1 nuclear localization. A and B, coimmunofluorescence imaging of YAP1 and AR proteins in LNCaP cells. The cells were treated with or without DHT (10 nm) and ENZ (20 μm) overnight in 5% CSS-fed growth conditions. Images of AR (Alexa 647, red), YAP1 (Alexa 488, green), and nuclei (DAPI, blue) were acquired using confocal microscopy. Scale bar, 20 μm. The micrographs are the representation of multiple images. B, ImageJ software was used to quantify the intensity of the nuclear AR and YAP1 protein from multiple images. The data ± S.D. normalized to the total cell number from multiple images of three independent experiments. *,**, p < 0.0001. C, coimmunoprecipitation and Western blotting analysis of the cytoplasmic and nuclear YAP1, PP2A C, and AR proteins in LNCaP cells after treatment with or without DHT overnight in 5% CSS-fed growth condition. Topoisomerase I (TOPI) was used as a nuclear marker. The blots are representative of three independent experiments. MW, molecular mass.
Androgen promotes YAP1-dependent gene expression
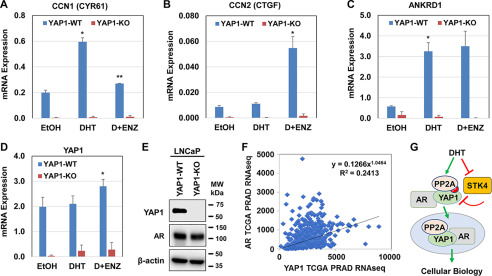
The CCN1-encoded CYR61, the CCN2-encoded CTGF, and the ANKRD1 genes are well-characterized YAP1 targets (32, 33). To test whether androgen regulates the transacting functions of YAP1, we assessed the levels of CYR61, CTGF, and ANKRD1 transcripts, including YAP1 in the YAP1-WT (WT) and YAP1–knockout (KO) LNCaP cell lines after treatment with the vehicle, DHT, or DHT plus ENZ (Fig. 6, A–E). Quantitative PCR (qPCR) demonstrated that androgen signaling differentially modulated the expression of CYR61, CTGF, and ANKRD1 transcripts in YAP1-WT cells. Regulation of these genes by androgen was YAP1-dependent because the silencing of YAP1 by CRISPR/Cas9 gene KO technology completely abolished the expression of CYR61, CTGF, and ANKRD1, including YAP1.
Figure 6.
Androgen signaling regulates the transcriptional activity of YAP1. A–C, quantitative RT-PCR analysis of YAP1 and the YAP1 target gene expression in YAP1-WT and YAP1-KO LNCaP cell models after treatment with EtOH, DHT, and DHT plus ENZ overnight in 5% CSS-fed growth condition. D and E, quantitative RT-PCR and Western blotting analysis YAP1 transcripts and protein expression in YAP1-WT and YAP1-KO LNCaP cell models, respectively. The data ± S.D. are from three independent experiments. *,**, p < 0.01. F, coexpression and correlation analysis of AR and YAP1 mRNA levels in TCGA Pan-Cancer prostate adenocarcinoma data set (PRAD) from 493 patients (46). Pearson correlation = 0.45, p = 1.57e−14). The data were accessed using the cBioPortal website (RRID:SCR_014555). G, a model illustrates the regulation of YAP1 nuclear localization by androgen. In this model, androgen antagonizes the Hippo/STK4/MST1 in a way that attenuates phospho-YAP and induces the YAP1 nuclear localization. Androgen also enhanced YAP1 nuclear localization by promoting protein–protein interaction between YAP1, PP2A, and AR, leading to cellular biology. MW, molecular mass.
AR and YAP1 activity correlates in prostate tumor tissues
To verify the physiological significance of our observations in cultures, we conducted a computational analysis of prostate cancer data sets from The Cancer Genome Atlas (TCGA), which is accessible via the cBioPortal website (RRID:SCR_014555). The results revealed that the expression of AR and YAP1 transcripts were significantly positively correlated (Pearson correlation = 0.45, p = 1.57e−14) in 493 prostate cancer patients (Fig. 6F). Overall, our data indicate that the regulation of YAP1 activity by androgen is physiologically and clinically relevant.
Discussion
Here, we have demonstrated that androgen attenuates the phospho–Ser-127 modification to promote YAP1 nuclear localization and activity. We have also shown that the AR-Hippo/STK4-PP2A axis mediates the effects of androgen on YAP1. Our findings suggest a new mechanism of YAP1 regulation that involves androgen hormone signaling.
Our data have revealed that androgen signaling suppressed phospho–Ser-127 on YAP1 in an AR-dependent manner because the disruption of AR activity by genetic and pharmacological methods attenuated the inhibition of phospho–Ser-127 modification by androgen. Also, inhibition of phospho–Ser-127 resulted in increases in the total YAP protein and nuclear abundance. Our current and published (7) studies demonstrated that androgen promoted protein–protein interaction between YAP1 and AR in the androgen-sensitive LNCaP cell line. Unlike LNCaP cells, the YAP1 and AR interaction occurred independently of androgen exposure in the androgen-independent C4-2 cell model that mainly expresses nuclear AR and YAP1 proteins (7). Based on these observations, we suggest that AR modulates YAP1 activity by at least three mechanisms. First, the AR may function as a chaperone for YAP1 through the protein binding, which increases the stability of YAP1. Second, the AR binding to the YAP1 may prevent the phosphorylation of Ser-127 by the kinases. Finally, the AR may serve as a cargo protein to import YAP1 into the nucleus via protein–protein interaction. Therefore, we suggest that protein–protein interaction provides critical mechanistic insights into the regulation of YAP1 by androgen in the cell.
In addition, we have shown that androgen may antagonize Hippo/STK4 signaling to attenuate phospho–Ser-127 and enhance YAP nuclear abundance, possibly by activating the Ser/Thr phosphatase PP1/PP2A. Our findings are consistent with the literature that PP2A was shown to inactivate the Hippo/STK4 activity (29, 31). In a published study, we demonstrated that androgen enhanced the protein complex formation between AR and the full-length MST1 (MST1-FL) (34). We also found that the MST1-FL protein localized to the cell nuclei, where the MST1-FL was devoid of phospho–Thr-183 in the activation loop, a critical site of the MST1 activity (34). It is possible that the AR binding restricts the ability of the kinases to phosphorylate Ser-127 on YAP1. In addition, androgen might antagonize STT4/MST1 by enhancing phospho–Thr-120 modification (34). Phospho–Thr-120 was shown to reduce phospho–Thr-183, which in turn results in the inactivation of STK4/MST1 activity (35).
Moreover, our coimmunoprecipitation and Western blotting experiments demonstrated that in addition to the AR, androgen enhanced the protein–complex formation between the catalytic C subunit of PP2A and YAP1 in cell nuclei. This observation suggests that androgen maintains nuclear YAP1 abundance by promoting the protein–protein interaction between YAP1 and PP2A, possibly via an AR-dependent manner because androgen also augmented YAP–AR interaction under the same experimental conditions. Nevertheless, the precise mechanism of how androgen regulates the nuclear YAP–PP2A interaction and whether it is biologically functional warrants further investigation, which is not the subject of the current study. Our finding is consistent with the literature that YAP1 interacts with PP2A in cell nuclei (36, 37).
Furthermore, our proteomic analysis of the YAP1 proteome indicated that 14-3-3 was part of the YAP1 proteome (not shown). Our finding suggests the possibility that androgen also regulates phospho–Ser-127 through the 14-3-3 protein binding. This notion is consistent with the literature that the 14-3-3 protein was shown to bind the phosphorylated serine residues, including phospho–Ser-127 on YAP1 (38). Also, the 14-3-3 binding caused the ubiquitination and proteasomal degradation of YAP1 (39). The LATS and CK1δ/ε protein kinase signaling cascade phosphorylates and leads to the ubiquitination and proteasomal degradation of YAP1 through SCF (β-TRCP) E3 ubiquitin ligase (22). LATS could reduce the stability and transcriptional activity of YAP1 via the Amot130–AIP4 complex in serum-starved conditions (40).
Nevertheless, we were unable to demonstrate androgen-regulated YAP1 ubiquitination, although we made several attempts (not shown). Evidence suggests that other kinases and also the phosphorylation sites on YAP1 could also regulate YAP1 nuclear localization (3, 21, 22, 41, 42). However, it remains unknown whether androgen influences the activity of other kinases, such as Src family kinases (6), to modulate YAP nuclear import (22, 43). Our work also revealed that androgen hormone signaling regulates the YAP1-dependent gene expression. Therefore, further research is necessary to precisely determine the mechanism of how androgen signaling modulates the stability, nuclear localization, and transcriptional activity of YAP1 in future studies.
Based on our observations, we have provided a model (Fig. 6G), in which androgen modulates the Hippo/STK4-AR-PP2A axis to attenuate phospho–Ser-127 and to promote YAP1 nuclear abundance and activity. Our findings are biologically crucial because androgen regulates YAP1-dependent gene expression by possibly enhancing the interaction of YAP1 with AR and the PP2A protein phosphatase. Our results are also are physiologically and clinically significant because the YAP1 and AR transcripts correlate in the subset of prostate cancer tissues. Overall, our study uncovers a new mechanism of YAP1 regulation that is mediated by the AR–Hippo/MST1–PP2A axis. Our findings have important implications for human diseases, given that the Hippo–YAP pathway regulates a range of cellular events, including cell differentiation, stem cell biology, and immune responses.
Experimental procedures
Cell culture
LNCaP, LNCaP-81, C4-2, C4-2B, 22Rv1, PC3, and ARCaP cells were grown in RPMI 1640 cell culture medium at 37 °C in 5% CO2 incubator. 5% fetal bovine serum and 1% penicillin and streptomycin antibiotics were added to the cell culture medium. LNCaP, C4-2, C4-2B, 22Rv1, and PC3 cell lines were purchased from the American Type Culture Collection.
YAP1-knockout cell line
LNCaP cells (3 × 105 cells/well in 6-well plate) were seeded overnight before transfection. The cells were cotransfected with 1–3 μg of human YAP1 HDR plasmid (Santa Cruz Biotechnology, Inc., sc-400040-HDR) and human YAP1 CRISPR/Cas9 KO plasmid (Santa Cruz Biotechnology, Inc., sc-400040) or control CRISPR/Cas9 plasmid (Santa Cruz Biotechnology, Inc., sc-418922). Plasmids were transfected using Lipofectamine 3000 reagents according to the manufacturer's instructions (Thermo Fisher Scientific, L3000001). After 72 h of transfection, the cells were exposed to an increasing dose of puromycin (2–10 μg/ml) to select puromycin-resistant clones. Individual clones were transferred to new tissue culture plates and grown in a medium supplemented with puromycin (2 μg/ml). Western blotting and quantitative PCR analyses were performed to verify the loss of YAP1 expression in selected clones. All protocols and procedures were conducted according to the manufacturer's instructions (Santa Cruz Biotechnology, Inc.).
RNA isolation and quantitative PCR
Total RNA was isolated at 80% confluency using TRIzol RNA isolation reagent according to the manufacturer's instructions (Thermo Fisher Scientific). The GoTaq one-step RT-qPCR system (Promega, A6020) was used to carry out quantitative qPCR according to the manufacturer's instructions. The gene expression was determined using a 2-ΔCt method (44) normalized to the 18S rRNA control in qPCR results. The gene-specific primers used in qPCR are listed in Table 1.
Table 1.
Primer set used in qPCRs
| Gene ID | Primer set (5′ → 3′) |
|
|---|---|---|
| Forward | Reverse | |
| CCN1 | GGCAAGAAATGCAGCAAGAC | CAGTACTTGGGCCGGTATTT |
| CCN2 | CATTCTCCAGCCATCAAGAGAC | CCACAAGCTGTCCAGTCTAATC |
| ANKRD1 | CAAGACTCCTTCAGCCAACA | GGAGCAGAGATGTGGGAATG |
| YAP1 | TGCGTAGCCAGTTACCAACACTG | TCGAGAGTGATAGGTGCCACTG |
| 18S rRNA | GCTTAATTTGACTCAACACGGGA | AGCTATCAATCTGTCAATCCTGTC |
Protein analysis
Total proteins were extracted from cells using ice-cold lysis buffer (20 mm HEPES, pH 7.4, 150 mm NaCl, 0.5% Nonidet P-40, 1 mm EDTA, and 1× protease inhibitors and phosphatase inhibitors; Calbiochem). Cytoplasmic and nuclear extracts were prepared according to the published protocol (45). The cells were washed with an ice-cold PBS three times before protein isolation. Coimmunoprecipitation was performed with anti-YAP1 at 4 °C as described (7). The YAP1 immune complexes were captured with protein A–Sepharose conjugate (GE Healthcare) and washed with a lysis buffer three times. Bound proteins were eluted and analyzed by 8% SDS–PAGE and Western blotting. The membranes were blocked with PBS containing 0.1% Tween 20 and 5% (w/v) skim milk, followed by incubation with the protein-specific antibody to YAP1 (Cell Signaling Technology (8418 and 12395S, 1:1000 or Santa Cruz Biotechnology, Inc. (sc-376830, 1:100), phospho–Ser-127-YAP1 (Cell Signaling Technology, 4911, 1:1000), phospho–Ser-397–YAP1 (Cell Signaling Technology, 13619, 1:1000), STK4/MST1 (Abnova, H00006789-M01, 1:2000), AR (EMD Millipore, 06-680, 1:1000), HA tag (Cell Signaling Technology, 3724, 1:1000), KLK3/prostate specific antigen (PSA) (Cell Signaling Technology, 332475P, 1:1000), β-actin (Sigma–Aldrich, A2228, 1:3000), GAPDH (Cell Signaling Technology, 5174, 1:2000), α-tubulin (Cell Signaling Technology, 2144, 1:2000), PP2A C subunit (Cell Signaling Technology, 2028S, 1:1000), or TOPI (Cell Signaling Technology, 79971, 1:1000). Protein signals were detected using a Luminata Forte Western HRP substrate (Millipore Sigma, WBLUFO500) and Bio-Rad ChemiDoc MP Imaging System.
Immunofluorescence and microscopy
Immunofluorescence imaging was conducted according to a published protocol (7). Briefly, LNCaP cells seeded in a chamber slide were androgen-starved overnight, followed by treatment with EtOH, 10 nm DHT, or DHT plus ENZ in 5% CSS-fed condition for 16–18 h. The cells were fixed with 4% paraformaldehyde for 20 min, permeabilized with 0.3% Triton X-100 for 10 min, and blocked with blocking buffer (2% BSA and 0.2% Triton X-100 prepared in PBS) for 1 h at room temperature. The cells were incubated with the primary YAP1 mouse monoclonal (G-6) antibody (Santa Cruz Biotechnology, Inc., sc-376830, 1:50 dilution), AR rabbit polyclonal antibody (Millipore Sigma, 06-680, 1:400), or HA tag rabbit monoclonal (C29F4) antibody (Cell Signaling Technology, 3724, 1:100) at 4 °C overnight. The cells were incubated with the secondary antibody conjugated to Alexa Fluor 488 (Invitrogen, anti-mouse A32723, or anti-rabbit A32731) or conjugated to Alexa Fluor 647 (Invitrogen, anti-mouse A32728 or anti-rabbit A32733) for 1 h at room temperature. Antibody dilutions were prepared in a blocking buffer. The cells were washed with PBS three times after each procedure. The slides were mounted using Prolong Gold-Antifade reagent containing DAPI (Cell Signaling Technology, 89615). YAP1, AR, or HA-STK4 signals were captured using confocal microscopy (Zeiss SP5-X) at 40× magnification.
RNAi and plasmids
Scramble (control), or AR, STK4/3, or LATS1/2 siRNA were purchased from Dharmacon/Fisher Scientific (Pittsburgh, PA, USA). Construction of tetracycline or doxycycline-inducible and HA-tagged STK4/MST1 expression plasmid was described previously (45). Briefly, PCR-amplified HA tag STK4/MST1-WT cDNA was inserted into the BamHI and MluI enzyme sites in the pRetro-X-Pur vector (Clontech Laboratories, Inc.), and the resulting plasmid was designated as pRXTP–HA–STK4/MST1. The gene-specific siRNA were transfected using DharmaFect-2 or Lipofectamine RNAi MAX reagent (Thermo Fisher Scientific, 13778150) in Opti-MEM I reduced serum medium (Thermo Fisher Scientific, 31985070) according to the manufacturer's instructions.
Data mining
Prostate cancer gene expression data were mined from the Pan-Cancer data set from TCGA for patients with prostate adenocarcinoma (46). Batch-normalized Illumina RNAseq V2 data from 493 patients were accessed via the cBioPortal website (RRID:SCR_014555) (47, 48). Coexpression and correlation analysis of AR and YAP1 mRNA levels was performed using cBioPortal coexpression tools (Pearson correlation = 0.45, p = 1.57e−14).
Data availability
All data described in the article are contained within the article.
Acknowledgments
We thank Drs. Kenneth Moberg and Zhengxin Wang for critically reviewing the manuscript and Pooneh Amin and Esma Alp for technical support.
Author contributions—B. C. conceptualization; B. C. and S. A. K. resources; B. C. and C. S. M. data curation; B. C., M. M. A.-M., and C. S. M. formal analysis; B. C. supervision; B. C. funding acquisition; B. C. and M. M. A.-M. validation; B. C. and M. M. A.-M. investigation; B. C., M. M. A.-M., and C. S. M. visualization; B. C., M. M. A.-M., and S. A. K. methodology; B. C. writing-original draft; B. C. and S. A. K. project administration; B. C., M. M. A.-M., S. A. K., and C. S. M. writing-review and editing.
Funding and additional information—This work was supported in part by National Science Foundation–Division of Molecular and Cellular Biosciences Grant 1832022 (to B. C.) and by NIMHD, National Institutes of Health Grant 2U54MD007590-32 (to the Center for Cancer Research and Therapeutic Program, Clark Atlanta University). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare no competing conflicts of interest with the content of this article.
- qPCR
- quantitative PCR
- TCGA
- The Cancer Genome Atlas
- PP2A
- protein phosphatase 2A
- AR
- androgen receptor
- DHT
- dihydrotestosterone
- Dox
- doxycycline
- OKA
- okadaic acid
- KO
- knockout
- HA
- hemagglutinin
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ENZ
- enzalutamide
- DAPI
- 4′,6′-diamino-2-phenylindole
- CSS
- charcoal stripped-serum.
References
- 1. Chen Y. A., Lu C. Y., Cheng T. Y., Pan S. H., Chen H. F., and Chang N. S. (2019) WW domain–containing proteins YAP and TAZ in the Hippo pathway as key regulators in stemness maintenance, tissue homeostasis, and tumorigenesis. Front. Oncol. 9, 60 10.3389/fonc.2019.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ma S., Meng Z., Chen R., and Guan K. L. (2019) The Hippo pathway: biology and pathophysiology. Annu. Rev. Biochem. 88, 577–604 10.1146/annurev-biochem-013118-111829 [DOI] [PubMed] [Google Scholar]
- 3. Zhao B., Li L., Lei Q., and Guan K. L. (2010) The Hippo–YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 24, 862–874 10.1101/gad.1909210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu A. M., Wong K. F., Jiang X., Qiao Y., and Luk J. M. (2012) Regulators of mammalian Hippo pathway in cancer. Biochim. Biophys. Acta 1826, 357–364 10.1016/j.bbcan.2012.05.006 [DOI] [PubMed] [Google Scholar]
- 5. Yu F. X., and Guan K. L. (2013) The Hippo pathway: regulators and regulations. Genes Dev. 27, 355–371 10.1101/gad.210773.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sudol M. (1994) Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene 9, 2145–2152 [PubMed] [Google Scholar]
- 7. Kuser-Abali G., Alptekin A., Lewis M., Garraway I. P., and Cinar B. (2015) YAP1 and AR interactions contribute to the switch from androgen-dependent to castration-resistant growth in prostate cancer. Nat. Commun. 6, 8126 10.1038/ncomms9126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vassilev A., Kaneko K. J., Shu H., Zhao Y., and DePamphilis M. L. (2001) TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 15, 1229–1241 10.1101/gad.888601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Z., Zhao B., Wang P., Chen F., Dong Z., Yang H., Guan K. L., and Xu Y. (2010) Structural insights into the YAP and TEAD complex. Genes Dev. 24, 235–240 10.1101/gad.1865810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Badouel C., Gardano L., Amin N., Garg A., Rosenfeld R., Le Bihan T., and McNeill H. (2009) The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev. Cell 16, 411–420 10.1016/j.devcel.2009.01.010 [DOI] [PubMed] [Google Scholar]
- 11. Ege N., Dowbaj A. M., Jiang M., Howell M., Hooper S., Foster C., Jenkins R. P., and Sahai E. (2018) Quantitative analysis reveals that actin and Src-family kinases regulate nuclear YAP1 and its export. Cell Syst. 6, 692–708.e13 10.1016/j.cels.2018.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kofler M., Speight P., Little D., Di Ciano-Oliveira C., Szászi K., and Kapus A. (2018) Mediated nuclear import and export of TAZ and the underlying molecular requirements. Nat. Commun. 9, 4966 10.1038/s41467-018-07450-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Manning S. A., Dent L. G., Kondo S., Zhao Z. W., Plachta N., and Harvey K. F. (2018) Dynamic fluctuations in subcellular localization of the Hippo pathway effector Yorkie in vivo. Curr. Biol. 28, 1651–1660.e4 10.1016/j.cub.2018.04.018 [DOI] [PubMed] [Google Scholar]
- 14. Elosegui-Artola A., Andreu I., Beedle A. E. M., Lezamiz A., Uroz M., Kosmalska A. J., Oria R., Kechagia J. Z., Rico-Lastres P., Le Roux A. L., Shanahan C. M., Trepat X., Navajas D., Garcia-Manyes S., and Roca-Cusachs P. (2017) Force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell 171, 1397–1410.e14 10.1016/j.cell.2017.10.008 [DOI] [PubMed] [Google Scholar]
- 15. Oka T., and Sudol M. (2009) Nuclear localization and pro-apoptotic signaling of YAP2 require intact PDZ-binding motif. Genes Cells 14, 607–615 10.1111/j.1365-2443.2009.01292.x [DOI] [PubMed] [Google Scholar]
- 16. Zhou D., Conrad C., Xia F., Park J. S., Payer B., Yin Y., Lauwers G. Y., Thasler W., Lee J. T., Avruch J., and Bardeesy N. (2009) Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell 16, 425–438 10.1016/j.ccr.2009.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elosegui-Artola A., Oria R., Chen Y., Kosmalska A., Pérez-Gonzalez C., Castro N., Zhu C., Trepat X., and Roca-Cusachs P. (2016) Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat. Cell Biol. 18, 540–548 10.1038/ncb3336 [DOI] [PubMed] [Google Scholar]
- 18. Elbediwy A., Vanyai H., Diaz-de-la-Loza M. D., Frith D., Snijders A. P., and Thompson B. J. (2018) Enigma proteins regulate YAP mechanotransduction. J. Cell Sci. 131, jcs221788 10.1242/jcs.221788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chan S. W., Lim C. J., Loo L. S., Chong Y. F., Huang C., and Hong W. (2009) TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J. Biol. Chem. 284, 14347–14358 10.1074/jbc.M901568200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lin K. C., Moroishi T., Meng Z., Jeong H. S., Plouffe S. W., Sekido Y., Han J., Park H. W., and Guan K. L. (2017) Regulation of Hippo pathway transcription factor TEAD by p38 MAPK-induced cytoplasmic translocation. Nat. Cell Biol. 19, 996–1002 10.1038/ncb3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moon S., Kim W., Kim S., Kim Y., Song Y., Bilousov O., Kim J., Lee T., Cha B., Kim M., Kim H., Katanaev V. L., and Jho E. H. (2017) Phosphorylation by NLK inhibits YAP-14-3-3-interactions and induces its nuclear localization. EMBO Rep. 18, 61–71 10.15252/embr.201642683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao B., Li L., Tumaneng K., Wang C. Y., and Guan K. L. (2010) A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(β-TRCP). Genes Dev. 24, 72–85 10.1101/gad.1843810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thalmann G. N., Anezinis P. E., Chang S. M., Zhau H. E., Kim E. E., Hopwood V. L., Pathak S., von Eschenbach A. C., and Chung L. W. (1994) Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer Res. 54, 2577–2581 [PubMed] [Google Scholar]
- 24. Kaighn M. E., Narayan K. S., Ohnuki Y., Lechner J. F., and Jones L. W. (1979) Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Invest. Urol. 17, 16–23 [PubMed] [Google Scholar]
- 25. Macedo L. F., Guo Z., Tilghman S. L., Sabnis G. J., Qiu Y., and Brodie A. (2006) Role of androgens on MCF-7 breast cancer cell growth and on the inhibitory effect of letrozole. Cancer Res. 66, 7775–7782 10.1158/0008-5472.CAN-05-3984 [DOI] [PubMed] [Google Scholar]
- 26. Cochrane D. R., Bernales S., Jacobsen B. M., Cittelly D. M., Howe E. N., D'Amato N. C., Spoelstra N. S., Edgerton S. M., Jean A., Guerrero J., Gómez F., Medicherla S., Alfaro I. E., McCullagh E., Jedlicka P., et al. (2014) Role of the androgen receptor in breast cancer and preclinical analysis of enzalutamide. Breast Cancer Res. 16, R7 10.1186/bcr3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Couzens A. L., Knight J. D., Kean M. J., Teo G., Weiss A., Dunham W. H., Lin Z. Y., Bagshaw R. D., Sicheri F., Pawson T., Wrana J. L., Choi H., and Gingras A. C. (2013) Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase–phosphatase interactions. Sci. Signal. 6, rs15 10.1126/scisignal.2004712 [DOI] [PubMed] [Google Scholar]
- 28. Deng Y., Pang A., and Wang J. H. (2003) Regulation of mammalian STE20-like kinase 2 (MST2) by protein phosphorylation/dephosphorylation and proteolysis. J. Biol. Chem. 278, 11760–11767 10.1074/jbc.M211085200 [DOI] [PubMed] [Google Scholar]
- 29. O'Neill E., Rushworth L., Baccarini M., and Kolch W. (2004) Role of the kinase MST2 in suppression of apoptosis by the proto-oncogene product Raf-1. Science 306, 2267–2270 10.1126/science.1103233 [DOI] [PubMed] [Google Scholar]
- 30. Hata Y., Timalsina S., and Maimaiti S. (2013) Okadaic Acid: a tool to study the Hippo pathway. Mar. Drugs 11, 896–902 10.3390/md11030896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ory S., Zhou M., Conrads T. P., Veenstra T. D., and Morrison D. K. (2003) Protein phosphatase 2A positively regulates Ras signaling by dephosphorylating KSR1 and Raf-1 on critical 14-3-3 binding sites. Curr. Biol. 13, 1356–1364 10.1016/S0960-9822(03)00535-9 [DOI] [PubMed] [Google Scholar]
- 32. Wang K. C., Yeh Y. T., Nguyen P., Limqueco E., Lopez J., Thorossian S., Guan K. L., Li Y. J., and Chien S. (2016) Flow-dependent YAP/TAZ activities regulate endothelial phenotypes and atherosclerosis. Proc. Natl. Acad. Sci. U.S.A. 113, 11525–11530 10.1073/pnas.1613121113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zanconato F., Forcato M., Battilana G., Azzolin L., Quaranta E., Bodega B., Rosato A., Bicciato S., Cordenonsi M., and Piccolo S. (2015) Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 17, 1218–1227 10.1038/ncb3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Collak F. K., Yagiz K., Luthringer D. J., Erkaya B., and Cinar B. (2012) Threonine-120 phosphorylation regulated by phosphoinositide-3-kinase/Akt and mammalian target of rapamycin pathway signaling limits the antitumor activity of mammalian sterile 20-like kinase 1. J. Biol. Chem. 287, 23698–23709 10.1074/jbc.M112.358713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deleted in proof.
- 36. Wang P., Bai Y., Song B., Wang Y., Liu D., Lai Y., Bi X., and Yuan Z. (2011) PP1A-mediated dephosphorylation positively regulates YAP2 activity. PLoS One 6, e24288 10.1371/journal.pone.0024288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hein A. L., Brandquist N. D., Ouellette C. Y., Seshacharyulu P., Enke C. A., Ouellette M. M., Batra S. K., and Yan Y. (2019) PR55α regulatory subunit of PP2A inhibits the MOB1/LATS cascade and activates YAP in pancreatic cancer cells. Oncogenesis 8, 63 10.1038/s41389-019-0172-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Muslin A. J., Tanner J. W., Allen P. M., and Shaw A. S. (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84, 889–897 10.1016/S0092-8674(00)81067-3 [DOI] [PubMed] [Google Scholar]
- 39. Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., Zheng P., Ye K., Chinnaiyan A., Halder G., Lai Z. C., et al. (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Adler J. J., Johnson D. E., Heller B. L., Bringman L. R., Ranahan W. P., Conwell M. D., Sun Y., Hudmon A., and Wells C. D. (2013) Serum deprivation inhibits the transcriptional co-activator YAP and cell growth via phosphorylation of the 130-kDa isoform of Angiomotin by the LATS1/2 protein kinases. Proc. Natl. Acad. Sci. U.S.A. 110, 17368–17373 10.1073/pnas.1308236110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. He M., Zhou Z., Shah A. A., Hong Y., Chen Q., and Wan Y. (2016) New insights into posttranslational modifications of Hippo pathway in carcinogenesis and therapeutics. Cell Div 11, 4 10.1186/s13008-016-0013-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hong A. W., Meng Z., Yuan H. X., Plouffe S. W., Moon S., Kim W., Jho E. H., and Guan K. L. (2017) Osmotic stress-induced phosphorylation by NLK at Ser128 activates YAP. EMBO Rep. 18, 72–86 10.15252/embr.201642681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Meng Z., Moroishi T., Mottier-Pavie V., Plouffe S. W., Hansen C. G., Hong A. W., Park H. W., Mo J. S., Lu W., Lu S., Flores F., Yu F. X., Halder G., and Guan K. L. (2015) MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat. Commun. 6, 8357 10.1038/ncomms9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 45. Cinar B., Collak F. K., Lopez D., Akgul S., Mukhopadhyay N. K., Kilicarslan M., Gioeli D. G., and Freeman M. R. (2011) MST1 is a multifunctional caspase-independent inhibitor of androgenic signaling. Cancer Res. 71, 4303–4313 10.1158/0008-5472.CAN-10-4532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sanchez-Vega F., Mina M., Armenia J., Chatila W. K., Luna A., La K. C., Dimitriadoy S., Liu D. L., Kantheti H. S., Saghafinia S., Chakravarty D., Daian F., Gao Q., Bailey M. H., Liang W. W., et al. (2018) Oncogenic signaling pathways in The Cancer Genome Atlas. Cell 173, 321–337.e10 10.1016/j.cell.2018.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., and Schultz N. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., and Schultz N. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data described in the article are contained within the article.