Abstract
Prions are lipidated proteins that interact with endogenous lipids and metal ions. They also assemble into multimers and propagate into the infectious scrapie form known as PrPSc. The high-resolution structure of the infectious PrPSc state remains unknown, and its analysis largely relies on detergent-based preparations devoid of endogenous ligands. Here we designed polymers that allow isolation of endogenous membrane:protein assemblies in native nanodiscs without exposure to conventional detergents that destabilize protein structures and induce fibrillization. A set of styrene–maleic acid (SMA) polymers including a methylamine derivative facilitated gentle release of the infectious complexes for resolution of multimers, and a thiol-containing version promoted crystallization. Polymer extraction from brain homogenates from Syrian hamsters infected with Hyper prions and WT mice infected with Rocky Mountain Laboratories prions yielded infectious prion nanoparticles including oligomers and microfilaments bound to lipid vesicles. Lipid analysis revealed the brain phospholipids that associate with prion protofilaments, as well as those that are specifically enriched in prion assemblies captured by the methylamine-modified copolymer. A comparison of the infectivity of PrPSc attached to SMA lipid particles in mice and hamsters indicated that these amphipathic polymers offer a valuable tool for high-yield production of intact, detergent-free prions that retain in vivo activity. This native prion isolation method provides an avenue for producing relevant prion:lipid targets and potentially other proteins that form multimeric assemblies and fibrils on membranes.
Keywords: membrane protein, nanotechnology, prion, lipid, lipid binding protein, neurodegenerative disease, lipid-protein interaction, protein misfolding, native nanodisc, SMALP, styrene maleic acid copolymer, styrene–maleic acid lipid particle (SMALP)
Introduction
Neurodegenerative diseases, including Alzheimer's disease, Parkinson's disease, and transmissible spongiform encephalopathies, involve pliable membrane-associated proteins that adopt multiple conformational states. The critical role of the membrane is a confounding variable in mechanistic studies of these systems. Bound lipids are typically lost during membrane protein separation with detergents. This can lead to deleterious artifacts because these lipids usually stabilize otherwise labile structures and modulate sensitive functions (1). Preserving the relevant target state is important for developing accurate diagnostic assays and therapeutic agents, which are growing priorities given the rising incidence and impacts of these neurodegenerative conditions (2, 3). There is a particular need to find alternatives for presenting and studying prions because of the complexity of their states including monomers, multimers, protofibrils, and fibrils that are modulated by various modifications, ligands, detergents, and surfaces. Native nanodiscs are increasingly used to prepare assemblies of endogenous membrane:protein assemblies (“memteins”) with lipid ligands and post-translational modifications in their biologically intact states (1) but have limitations including excessive negative charge and polydispersity. Moreover, they have not yet been used to tackle prions. Here, SMA copolymer derivatives optimized for stabilizing and isolating memteins were developed to address the challenges presented by transient lipid-bound infectious prion multimers.
Prions are normally associated with membranes through GPI anchors, although their endogenous lipid complement is unclear. Prion analysis has relied on maintaining high concentrations of conventional detergents such as sarkosyl (4, 5), despite induced alterations of structural properties (6), compromised interactions with physiological partners including heparin (7), and fibrilization of prions including proteinase K (PK)– and phosphotungstic acid (PTA)–treated Rocky Mountain Laboratories (RML) strain-infected mouse brain homogenates (8). These studies prevail because of the lack of milder substitutes that could more gently release intact prion:lipid assemblies for analysis (9). Addressing this limitation could lead to resolving the structural identities of the infectious prion forms and the mechanisms underlying their toxicity, lipid perturbations (10–13), GPI anchoring, and PK-sensitive oligomerization during prion propagation (14). Toward this end, we have developed and tested a series of SMA polymers for the detergent-free isolation of infectious prions in native nanodiscs. These are used for in vitro structural and in vivo infectivity assays of protease-resistant prion (PrPres) assemblies from the brains of Hyper strain–infected hamsters and RML strain–infected FVB mice.
A custom polymer was designed to overcome anticipated challenges with prions. Amphipathic polymers that contain statistical distributions of styrene and maleic acid monomers in nonalternating ratios are known to insert into virtually any cellular membrane for spontaneous formation of native nanodiscs (16–18). However, the heterogeneity of their sequences precludes atomic resolution of the polymers or bound lipid headgroups in 3D structures (19, 20). Alternating polymers are more homogenous because of their regular 1:1 repeating pattern of styrene and maleic acid subunits but are relatively ineffectual at membrane solubilization (21–23). To synthesize alternating SMA(1:1) derivatives, which are more homodispersed and better suited to solubilizing metal-dependent proteins, including prions, we used less charged maleimide groups rather than maleic acid groups. Clusters of lipid-inserting styrenes can also mediate undesirable nonspecific interactions with fibrils, and hence the styrene ratio was halved with a compensatory methyl added onto the maleimide. Crystallization and structural analysis would benefit from more regularized belts of polymer around nanodiscs that encapsulate protein–phospholipid complexes. Hence we incorporated thiol groups to offer hydrogen bonding and cross-linking potential. This SMA series is optimized for prions but has broad implications for biochemical, structural, and lipidomic analyses of diverse membrane proteins, many of which remain intractable (24). Here, we investigated the strengths and weaknesses of different formulations of SMA copolymers in vivo and in vitro applications as alternatives for detergents for resolving infectious prions, with potential applicability to virtually any membrane-associated target.
Results
Comparison of SMA copolymers
A series of four SMA copolymers was synthesized to contrast the effects of polymer charge, hydrophobicity, and reactivity on native prion solubilization and activity. The SMA(2:1) and SMA(3:1) copolymers contain nonalternating sequences with 2:1 and 3:1 ratios of styrene to maleic acid monomers, respectively, and offer distinct membrane interaction and solubilization profiles. They were activated by alkaline hydrolysis of the respective styrene–maleic anhydride forms (Fig. 1A) for subsequent analysis of prion solubilization and activity. A derivative with free thiol groups was synthesized from SMA polymer by grafting cysteamine to SMA(2:1) polymer (Fig. 1B), with the thiol groups in the resulting SMA-SH polymer (16) offering handles for cross-linking. A methylamine derivative termed “SMA(1:1)ma” was synthesized that offers an equimolar ratio of comonomers (Fig. 1C) and less charge and hence increased in predicted polycation compatibility and alternating hydrophobic subunits for greater homogeneity and potential for structural resolution.
Figure 1.
Chemical structures and synthesis of the SMA(2:1) and SMA(3:1) (A), SMA-SH (B), and SMA(1:1)ma (C) copolymers, which have average m:n ratios of styrene to maleic acid groups of 2:1, 3:1, 2:1, and 1:1, respectively.
Before proceeding to the solubilization of prions from the mammalian brain, we compared the abilities of the various polymers to solubilize multilayer vesicles composed of dimyristoyl phosphatidylcholine lipid. The SMA(1:1)ma form was able to solubilize membranes at a concentration of 1%, which is similar to conventional nonalternating SMA polymers, despite having a lower amount membrane-binding styrene groups. This was expected because of the alkylamine derivatization reducing the net negative charge and increasing the overall polymer hydrophobicity, which is needed for efficient membrane insertion. The membranes were solubilized by SMA(1:1)ma at up to 10 mm calcium chloride, whereas SMA(2:1) precipitates above 5 mm; SMA(1:1) is active from pH 5 to 10 (Fig. S1), and other SMA polymers have narrower pH ranges and are optimal at pH 8. Thus SMA(1:1)ma has a broader solution compatibility, reduced charge, sufficient hydrophobicity, and lower sequence heterogeneity while retaining comparable membrane solubilization activity. Hence it was included in our polymer panel to solubilize native-state prion particles.
Isolation and partial purification of infectious PrP (PrPSc) using SMALP
Like established prion purification methods, the SMA-based approach also utilizes PTA to bind PrPSc assemblies and separate them from other brain components (8). Optimization of the SMA protocol to achieve high recovery of multimeric PrPSc reduced the duration of the incubation with PTA to 1 h. Unlike detergents that need to be maintained throughout the protocol, a minimal concentration of SMA polymer (1% w/v) was added only during the initial incubation with brain homogenate (Fig. S2) to reduce the heterogeneity of nanodiscs for fractionation and transmission electron microscopy imaging. Because of the stability of the nanodiscs, no further polymer needed to be added downstream of the initial solubilization.
Resistance to proteolytic digestion is a hallmark of many prion strains found in tissues of organisms exhibiting transmissible spongiform encephalopathies. Both proteinase K and Pronase E were used, because the latter retains the GPI-anchoring needed for stable membrane interactions of prions (25). Either protease could be used to prepare PrPSc from brain homogenate with similar results. However, the yield of PrPSc filaments in electron micrographs from Pronase E–treated samples was noticeably higher. Consequently, the Pronase E–resistant prion (hence PrPSc) was prioritized for preparation of membrane:prion assemblies in native SMA nanodiscs.
Native prion proteins are variably glycosylated and are present as three characteristic bands on immunoblots, corresponding to diglycosylated, monoglycosylated, and unglycosylated forms. All SMA-isolated PrPSc samples, whether isolated from Hyper strain–infected hamsters or RML strain–infected mice, share a similar profile of glycoforms with distinct molecular weights that match those of sarkosyl-purified PrPSc samples (Fig. 2, B and C). However, the purity of PrPSc extracts, as well as their physical appearances, differed between the various SMA-purified samples (Fig. S2). Prions solubilized by SMA(2:1) treatment were the least turbid, whereas SMA(1:1)ma-purified prions appeared as a brown waxy pellet, which suggested the inclusion of more lipid- or heme-containing proteins such as ferritin or cytoskeletal proteins like keratin (Fig. S2) (26). Despite the use of benzonase nuclease (18) and high-speed centrifugation of all samples before the gel electrophoresis, high-molecular-weight aggregates appeared in both SMA(1:1)ma and SMA(2:1) preparations. Immunoblotting experiments confirmed that the size of aggregates does not correlate with PrPSc (data not shown). Because of the presence of apparent keratin fibers in EM images, we attributed the high-molecular-weight aggregates to cytoskeletal protein fibers.
Figure 2.
A, Prion purification of detergent-free isolation of protease-resistant prions from infected brain tissues. B, immunoblots of SMA-purified PrPSc from Hyper-infected Syrian hamsters (left panel) and RML-infected WT mice (right panel) using anti-PrP primary antibody, showing similar molecular masses (Mw) and glycosylation patterns for the SMA-purified PrPSc preparation compared with samples purified with sarkosyl. C, comparison between glycosylation patterns of sarkosyl-purified and SMA(2:1)-isolated PrPSc derived from Hyper infected Syrian hamster (left panel) and RML-infected WT mice (right panel).
Given the milder membrane solubilization provided by SMA polymers, we speculated that more prion multimers might be retained. The distribution of low-density SMA-purified PrPSc complexes from hamster and mouse strains was examined via equilibrium sedimentation in stepwise sucrose density ultracentrifugations (Fig. 3). The majority of PrPSc protein extracted by SMA from Hyper-infected hamsters was buoyant and found in the top fraction and hence represented lipid-associated states, whereas the levels of dense aggregates that sedimented to the bottom of the gradient were relatively insignificant. The SMA(1:1)ma treatment of PrPSc derived from brains of the Hyper (HY) strain showed a distribution of oligomeric states that migrated faster in the sedimentation three-step sucrose gradients than complexes obtained with the other SMAs. Together, the migration behavior in the sucrose gradients indicates that the various SMA polymers preferentially solubilize the lipid–prion complexes.
Figure 3.
Multimeric states of prion isolates. Protease-treated SMA-PrPSc particles isolated from Hyper-infected Syrian hamsters and RML-infected FVB mice were separated according to their densities using two-step (40 and 80%) and three-step (40, 55 and 80%) sucrose gradient ultracentrifugations, with fractions collected from the top to the bottom of tubes for immunoblotting. The majority of prion particles remained in the top layer of the gradient (40%), implying less aggregated, lipid-bound states of PrPSc in SMA nanodiscs. Mw, molecular mass.
Transmission EM of SMALP-isolated PrPSc
We investigated the morphology of the prion fibrils that were obtained by SMA extraction from the brain. The SMA isolates of PrPSc contain mainly two protofilaments that have diameters of ∼20 nm in negative stain EM images (Fig. 4), whereas entangled long rods were seen in sarkosyl-treated samples. Moreover, lipid vesicles were found in PrPSc preparations obtained with SMA and are particularly enriched in SMA(1:1)ma isolates, which show protofilament–vesicle complexes. The lipid vesicles can be removed by treatment of SMA-PrPSc preparations with polyethylene glycol (PEG)6000 (4% w/v, 4 °C, overnight), which precipitates the PrPSc for EM analysis as relatively homogenous protein fibrils. Large discs with diameters of 45 ± 5 nm containing 2D crystals of PrPSc can be obtained from PK- and SMA-SH–treated Hyper strain brains (Fig. 5A) and are strongly reminiscent of those seen in earlier studies (27–29). The available thiols of SMA-SH and the higher prion yield from prion-infected hamster samples may both contribute to the formation of these PrPSc 2D crystals, which were rarely seen in other SMA brain isolates. The top fractions of the sucrose gradients of SMA(2:1)- and SMA(1:1)ma HY PrPSc contain fibrils within the low-density lipid raft microdomains, which is consistent with native PrPSc fibril association with the plasma membrane (Fig. 5, B and C).
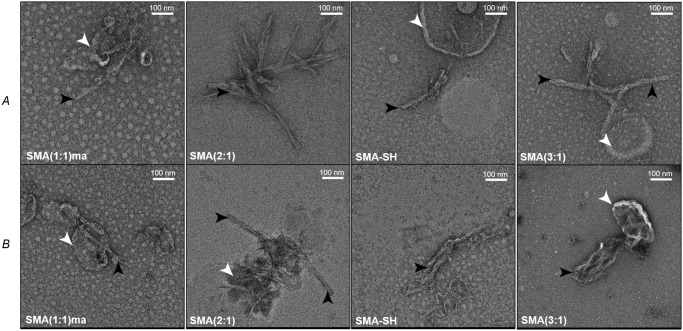
Figure 4.
Negative stain electron micrographs of PrPSc fibrils from Hyper-infected Syrian hamsters (A) and RML-infected WT mice (B) using the indicated SMA copolymers. Black arrowheads point to isolated PrPSc fibrils, and white arrowheads highlight the copurified lipid vesicles.
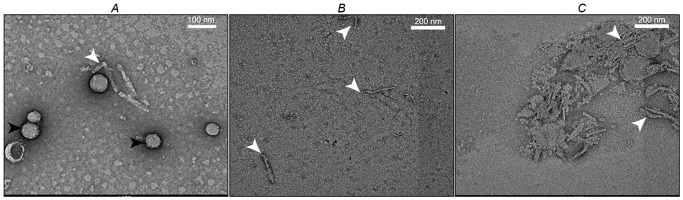
Figure 5.
Prion isolate structures. A, negative stained electron micrographs of PrPSc fibrils (white arrowhead) and small 2D crystals from Hyper-infected Syrian hamster following SMA-SH and PK treatment (black arrowheads). B and C, protofilaments and vesicle from the top-most fractions of sucrose gradients of SMA(2:1)-treated (B) and SMA(1:1)ma-treated (C) Hyper-infected hamster brain.
Lipid profile of the infectious PrPSc in SMALPs versus sarkosyl
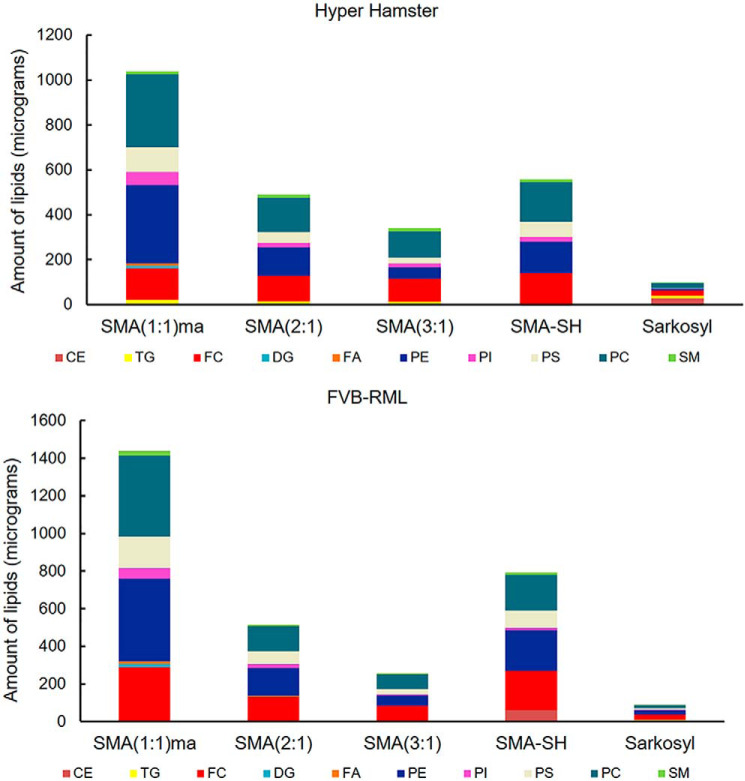
Copurified lipids of PrPSc are also key for the biological transformation of cellular PrP to the scrapie isoform (11, 13, 30, 31). Unlike some detergents that dissociate protein-bound lipids, SMA polymers have been shown to be useful tools for studies of lipids bound to membrane proteins (32) and amyloids (14, 33). In this regard, however, the use of detergents and the infectivity associated with high titer PrPSc pellets have limited the investigation of the lipid profile of infectious prions from endogenous sources (34). The lipids associated with PrPSc assemblies extracted from brains of infected and healthy hamsters and mice were identified and quantified, allowing the relative levels of 11 types of lipid to be compared (Fig. 6). Using an internal standard (batyl alcohol) added to the sample during lipid extraction, the amount of each lipid was determined by HPLC (Table S1). The findings demonstrate that SMA(1:1)ma discs provide the highest capacity for isolation of various types of lipids, a finding in accordance with the waxy appearance of PrPSc pellet purified using this polymer. The next highest prion-associated lipid capacity levels are offered by SMA(2:1) and SMA-SH polymers, respectively. Interestingly, HY hamster and RML strains have a relatively distinctive lipid profile; for instance, triglyceride is solely present in HY hamster samples. The lipid profiles also reveal the presence of critical signaling lipids including cholesterol, phosphatidylinositol, and sphingomyelin (35) in close proximity to PrPSc molecules. This is consistent with reports of other misfolding diseases such as Alzheimer's disease and type 2 diabetes (36, 37).
Figure 6.
Prion-associated lipids. Different lipid species in each SMA-isolated PrPSc pellet from Hyper-infected hamster (top panel) and RML-infected WT mice (bottom panel) were quantified, and their cumulative amounts are shown in stack plots including levels of cholesteryl esters (CE), triacylglycerol (TG), free cholesterol (FC), diacylglycerol (DG), free fatty acid (FA), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylserine (PS), phosphatidylcholine (PC), and sphingomyelin (SM).
Bioassay of SMALP-PrPSc particles
The symptoms of mice and hamsters inoculated with PrPSc extracted from brain with different SMA polymers or sarkosyl were compared with those inoculated with brain homogenate from infected animals (Table 1). Prion disease presents as ataxia, scruffy coats, loss of gait, weight loss, and head bobbing at the time of euthanasia (38). The average incubation times from SMALP-PrPSc particle inoculation until terminal disease was consistently ∼85–90 days in Hyper hamsters, with SMA(1:1)ma, SMA(2:1), and SMA-SH-purified PrPSc showing similar periods (Table 1) as sarkosyl purified PrPSc. In contrast, inoculation with SMA(3:1)-purified PrPSc yielded the longest incubation times. As expected, inoculation with 1% prion 263K brain homogenate from infected animals was most efficient. Given the 153 days for average incubation time of RML prions in FVB mice, mice incubated with SMA-SH and SMA(3:1) purified PrPSc showed the shortest and longest incubation times, i.e. 163 and 187 days, respectively (Table 1). Despite the variability of incubation period for different SMA-purified samples, animals in SMA(3:1), SMA(2:1), SMA(1:1)-methylamine, and SMA-SH groups show the same clinical symptoms, suggesting that the isolation method did not alter prion strain characteristics. Brain homogenates of animals infected with isolated SMALP-PrPSc particles were used for a second passage into healthy hamsters (Table 2). The incubation periods display, to a great extent, the same trend in both incubation period and symptoms at terminal stage as first passage, with SMA(3:1) having the longest incubation periods. Protease digestion of post-mortem brain tissues reveals that all animals, regardless of the incubation period and clinical symptoms, display the characteristic profile of PK-resistant PrP (Fig. S3), again supporting our conclusion that each purification protocol did not alter the strain characteristics.
Table 1.
A summary of the infectivity of SMA-treated PrPSc samples in samples from Hyper-infected hamsters and RML-infected mice
In both Hyper-infected hamsters and RML-infected mice, 10% brain homogenates and Sarkosyl-purified PrPSc were used as controls.
| Sample | Incubation period ± S.E. | N/N0 |
|---|---|---|
| days | ||
| Samples from Hyper-infected hamsters | ||
| 1% 263K brain homogenate (control) | 71 ± 0.5 | 4/4 |
| Treated with 1% sarkosyl-PrPSc (control) | 76 ± 5 | 12/12 |
| Treated with 1% SMA(1:1)ma-PrPSc | 84 ± 8 | 12/12 |
| Treated with 1% SMA(2:1)-PrPSc | 87 ± 3 | 12/12 |
| Treated with 1% SMA-SH-PrPSc | 88 ± 4a | 12/12a |
| Treated with 1% SMA(3:1)-PrPSc | 94 ± 3a | 12/12a |
| Samples from RML-infected mice | ||
| Treated with 1% sarkosyl-PrPSc (control) | 158 ± 19 | 19/19 |
| Treated with 1% SMA(1:1)ma-PrPSc | 174 ± 27 | 9/9 |
| Treated with 1% SMA(2:1)-PrPSc | 172 ± 21 | 15/15 |
| Treated with 1% SMA-SH-PrPSc | 163 ± 15 | 14/14 |
| Treated with 1% SMA(3:1)-PrPSc | 187 ± 33.5a | 17/17a |
a Significant difference (p value < 0.001) between the incubation periods of SMA-treated samples and sarkosyl-purified PrPSc.
Table 2.
The second passage of brain homogenates of animals infected with SMA-isolated Hyper PrPSc into healthy Syrian hamsters
| Reinoculation sample (hamster) | Incubation period ± S.E. | N/N0 |
|---|---|---|
| days | ||
| 1% 263K brain homogenate (control) | 71 ± 0.5 | 4/4 |
| Treated with 1% SMA(1:1)ma-PrPSc | 87 ± 3a | 10/10a |
| Treated with 1% SMA(2:1)-PrPSc | 74 ± 3 | 6/6 |
| Treated with 1% SMA-SH-PrPSc | 97 ± 2a | 6/6a |
| Treated with 1% SMA(3:1)-PrPSc | 115 ± 0.5a | 6/6a |
a Significant difference (p value < 0.001) between the incubation periods of SMA-treated samples and the control. The largest differences were in SMA-SH-PrPSc and SMA(3:1)-PrPSc, as seen in the first passage (Table 1).
Discussion
We have shown that infectious membrane-associated prions can be directly isolated intact from rodent brains using custom SMA copolymers, providing a viable route to prepare one of the most challenging complexes with no exposure to detergent and minimal use of PTA or added polymer. The density-based fractionation of the resulting particles yields the lipid-bound state of PrPSc protofilaments with the least aggregation (8) and multimers that copurify with SMALP nanodiscs.
The choice of protease is a key element in the isolation of nonfibrillar, membrane-bound, and infectious PrPSc. To this end, the copolymer-based strategy in combination with high-performance proteases such as thermolysin (39) could yield a gentle and minimal perturbing preparative approach for further structural analysis. Our data show that the SMALP system is compatible with proteases such as PK and phosphatidylethanolamine for isolation of truncated and intact PrPSc assemblies, respectively. Structural investigation of GPI anchor–dependent propagation of PrPSc and hence its lipid raft related signaling would require the intact native assemblies of PrPSc lipids, and to this date the amphipathic polymers are the only available tools for this purpose (40). This discovery that SMA(1:1)ma is particularly effective at intact membrane:prion isolation provides an avenue for production of nanodiscs with reduced nonspecific interactions, polydispersity, and calcium sensitivity and thus overcomes multiple limitations of other SMA polymers. The notable increase in 2D crystals that resulted from the PrPSc solubilization with SMA-SH demonstrated its utility as a tool for the structural characterization of PrPSc in the form of nonfibrillar assemblies (27–29).
Demonstration of the solubilization of the biologically active multimer prion protein with SMA(1:1)ma with a 2-fold improvement in tolerance to cation and membrane-binding capacity indicates wider utility for diverse memteins that are often cation- and lipid-dependent. The advantages of SMA(1:1)ma indicate significant potential for lipidomic and metabolomic analyses of membrane assemblies including high-titer PrPSc complexes from infected organisms. Such polymers provide an advantage over conventional detergents that dissociate physiologically important lipid molecules or ligands, except those that are tightly bound and buried between core fibers, such as the unresolved hydrophobic ligands in structures of Tau fibers (41). Determination of ligand identities and quantification of bound lipids allows analysis of the native membrane environment and enables mechanistic studies of the lipid-dependent transformation of prions from the normal cellular state to the scrapie form. This analysis will rely on fractionation and time-resolved studies of the SMA-treated brain homogenate. This separation of transient and irreversible states could be facilitated by differences in their hydrodynamic sizes and conformations, which we speculate could be lipid-dependent. Stabilization and resolution of the intermediate state most critical for infectivity is becoming increasingly feasible with the SMA derivatives presented here. In addition, other solutions may be developed, with cellulose ethers and methylcellulose having been shown to prolong the incubation periods of PrPSc in prion disease animal models (42), providing a broader context for the use of polymers to stabilize and study critical prion states.
In addition to their utility for in vitro structural and lipid-binding studies of membrane proteins, SMA polymers have in vivo applications as drug delivery vehicles (43, 44). Here we used a polymer concentration of ∼1% (w/v) to prepare prion nanoparticles for structural and infectivity studies, although even lower concentrations (0.5% w/v) are also effective in liberating the PrP–lipid complexes. Because the prion-containing SMALP was added to brain homogenate in only one initial step, the purified PrPSc–lipid assembly was injected into animals with little if any free SMA polymer present. The incubation periods of SMA-purified PrPSc strains can be explained in light of previous studies on the anti-prion activity of dendrimers (45), which suggest that polymer surface charge does not correlate with the mechanism of action of these polymers, including prolonging of the incubation time of PrPSc. Moreover, several studies confirmed that low-toxicity polymers can disaggregate PrPSc polymers, thus giving rise to longer incubation times in murine models (46–48). The application of SMA(1:1)ma would reduce the net charge and hydrophobic clusters of copolymers and nanoparticles, thus allowing specific target interactions to be favored over nonspecific electrostatic and hydrophobic interactions. Hence the SMA derivative polymers presented here could offer insights and utility for structural biology, drug discovery, and delivery applications for a wide array of memtein targets.
Experimental procedures
Polymer synthesis
SMA(2:1) and SMA(3:1) (Polyscope) were each modified from the anhydride version to their corresponding acid forms by hydrolysis in 1 m NaOH while refluxing at 70 °C for 3 h (14) and dried under vacuum. A cysteamine-grafted derivative of SMA(2:1) (SMA-SH) was synthesized using established methods (49) and stored with 5 mm DTT. SMA(1:1)ma was synthesized from SMA(1:1) as described for Fig. 1. Stock solutions (8%, w/v, pH 7.5) of each polymer were prepared in Dulbecco's PBS (DPBS) with 5% glycerol (v/v), FT-IR spectra were collected (data not shown), and samples were stored frozen until use.
Prion isolation from brains
Brain homogenates (20% w/v) were generated from clinically ill Hyper strain–infected Syrian hamster brains and RML-infected FVB mouse brains and prepared in DPBS + 5% glycerol, mixed with 8% (w/v) SMA stock solution to a final concentration of 1% (w/v) and Pronase E (Sigma). Each mixture was then incubated at 37 °C for 30 min, after which the protease was inactivated by EDTA addition (50). Sodium phosphotungstic acid (PTA, 200 μl of 10% w/v, pH 7.2) was added, and the mixtures were incubated for 1 h at 37 °C. The solutions were centrifuged at 16,500 × g, and the pellets were resuspended in DPBS and 5% (v/v) glycerol and stored at −20 °C for later assays.
Equilibrium sucrose ultracentrifugation
Sucrose gradients in DPBS were prepared in 3.5-ml Beckman ultracentrifuge tubes to resolve prion multimers. Three independent sets of SMA-purified samples were overlaid on top of each gradient and spun at 130,000 × g in a SWTi55 swinging-bucket rotor (Beckman Coulter) for at least 17 h at 4 °C. Fractions (200 μl) were collected from the top to the bottom of tubes, and 40-μl aliquots were mixed with an equal volume of sample buffer (Bio-Rad) and heated for 10 min at 100 °C as per Western blotting and SDS-PAGE gels.
Negative-stain transmission electron microscopy
Carbon-coated copper grids (400 mesh) were charged using an EMS Pelco Easy Glow 100× glow discharge unit (Ted Pella Inc.) for 30 s. Microliter amounts of each SMA-purified prion sample were loaded on the grids and adsorbed for 30 s. The grids were washed three times (3 × 50 μl) with ammonium acetate (100 and 10 mm, pH 6.8) and stained with filtered 2% uranyl acetate. Excess dye was removed using a filter paper, and the grid was air-dried for at least 5 min before transmission electron microscopy imaging. Micrographs were collected using a Tecnai G20 transmission electron microscope equipped with an Eagle 4 k × 4 k CCD camera (FEI Company) using an acceleration voltage of 200 kV.
Infectivity assays
FVB mice were obtained by in-house breeding and 23-day-old golden Syrian hamsters were purchased from Envigo. The SMA-isolated PrPSc samples and controls were serially diluted 100 times in DPBS and normalized by volume to brain equivalent. Likewise, brain homogenates of infected animals were subsequently used for the second passage. Diluted samples were then used to intracerebrally inoculate three independent groups, each with four animals, of healthy FVB mice (30 μl/animal) and/or Syrian hamsters (50 μl/animal). Their behavior was monitored on a daily basis, and after clinical diagnosis of prion disease, the brains of euthanized animals were isolated and stored at −80 °C for analysis. The infectivity data were analyzed by SigmaPlot 14.0. All experiments in mice and Syrian hamsters were performed in accordance with guidelines set by the Canadian Council on Animal Care and approved by the animal care use committee for Health Sciences 2 at the University of Alberta (protocol AUP00000884).
Immunoblotting
The prion samples were mixed with an equal volume of 2× sample buffer containing 700 μm β-mercaptoethanol and heated at 100 °C for 10 min. Before loading on precast 10 or 12% stain-free SDS-PAGE gels (Bio-Rad), the samples were centrifuged for 2 min at 14,000 × g. Electrophoresis was performed at 100 V for ∼ 2 h at room temperature. The proteins were electrotransferred to polyvinylidene difluoride membranes (Millipore) at 100 V for 1 h at room temperature in 25 mm Tris, pH 8.3, 192 mm glycine, 20% methanol (v/v). The membranes were blocked in Tris-buffered saline with 0.1% (v/v) Tween and 5% (w/v) BSA for 1 h at room temperature, followed by incubation overnight at 4 °C with an in-house mouse anti-prion antibody. The membrane was washed three times for 10 min in Tris-buffered saline with 0.1% Tween and further incubated with a secondary alkaline phosphatase–conjugated goat anti-mouse antibody (Bio-Rad) for 1 h. After three 10-min washes, the membrane was incubated with 1 ml of alkaline phosphatase substrate (Bio-Rad), and the immunoblots were imaged using ImageQuant (GE Life Science).
Proteinase K resistance assay and silver staining
The brain tissue of each infected animal was removed post mortem, and a brain homogenate (10% w/v) was prepared in DPBS with 5% glycerol. To assess prion protease sensitivity, 160 μl of 10% brain homogenate was incubated with varying concentrations of PK (0, 5, 50, and 200 μg/ml) at 37 °C for 60 min with constant agitation. PK was inactivated by adding 0.5 mm phenylmethylsulfonyl fluoride, and the samples were mixed with 2 × sample buffer, heated to 100 °C, run on SDS-PAGE, and immunoblotted using an anti-prion antibody. Silver-staining methods (29) included fixing and washing gels in ethanol to remove excess SDS. After treatment with Farmer's reducer (0.05% sodium carbonate, 0.15% potassium hexacyanoferrate (III) and, 0.3% sodium thiosulfate), the gels were incubated with AgNO3 for 20 min and developed in formaldehyde sodium carbonate solution.
Lipid analysis
The total lipid was isolated from high-titer PTA complexes of PrPSc solubilized in SMA polymer or sarkosyl according to established methods (51) using methanol:chloroform (1:2 v/v) in a BSL-2 lab biosafety cabinet following decontamination by incubation of PTA pellets with 5 m guanidinium thiocyanate for 1 h at room temperature (52). Total lipids were analyzed by HPLC based on Ref. 15, lipid classes were identified according to their retention time and comparison with commercial standards, and the amounts were quantified using calibration curves for each lipid class.
Data availability
All data are contained within the article and the supporting information.
Supplementary Material
Acknowledgments
We thank Claudia Acevedo-Morantes for assistance with EM and Xinli Tang for providing a primary anti-prion antibody.
This article contains supporting information.
Author contributions—M. E., L. M. C., V. L. S., H. W., and M. O. conceptualization; M. E., B. P. T., X. W., and A. M. data curation; M. E., B. P. T., and X. W. formal analysis; M. E. and H. W. validation; M. E., B. P. T., X. W., and M. O. investigation; M. E., A. M., and M. O. visualization; M. E., B. P. T., X. W., A. M., and H. W. methodology; M. E. and M. O. writing-original draft; M. E., L. M. C., V. L. S., H. W., and M. O. writing-review and editing; A. M., L. M. C., V. L. S., H. W., and M. O. resources; H. W. and M. O. supervision; H. W. and M. O. funding acquisition; H. W. and M. O. project administration.
Funding and additional information—This work was supported by Alberta Prion Research Institute Exploration Grant 201600018 (to M. O.), NSERC Discovery Grant RGPIN-2018-04994 (to M. O.), Campus Alberta Innovates Program Grant RCP-12-002C (to M. O.), and Alberta Prion Research Institute Team Program Award 201600029 (to H. W.).
Conflict of interest—M. O. and M. E. have filed a patent on related SMA copolymers and methods.
- SMA
- styrene–maleic acid
- RML
- Rocky Mountain Laboratories
- SMALP
- SMA lipid particle
- GPI
- glycosylphosphatidylinositol
- PK
- proteinase K
- PTA
- phosphotungstic acid
- HY
- Hyper
- DPBS
- Dulbecco's PBS.
References
- 1. Overduin M., and Esmaili M. (2019) Memtein: the fundamental unit of membrane-protein structure and function. Chem. Phys. Lipids 218, 73–84 10.1016/j.chemphyslip.2018.11.008 [DOI] [PubMed] [Google Scholar]
- 2. Chiti F., and Dobson C. M. (2006) Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 10.1146/annurev.biochem.75.101304.123901 [DOI] [PubMed] [Google Scholar]
- 3. Giles K., Woerman A. L., Berry D. B., and Prusiner S. B. (2017) Bioassays and inactivation of prions. Cold Spring Harb. Perspect. Biol. 9, a023499 10.1101/cshperspect.a023499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McKinley M. P., Taraboulos A., Kenaga L., Serban D., Stieber A., DeArmond S. J., Prusiner S. B., and Gonatas N. (1991) Ultrastructural localization of scrapie prion proteins in cytoplasmic vesicles of infected cultured cells. Lab. Invest. 65, 622–630 [PubMed] [Google Scholar]
- 5. Riesner D. (2003) Biochemistry and structure of PrP(C) and PrP(Sc). Br. Med. Bull. 66, 21–33 10.1093/bmb/66.1.21 [DOI] [PubMed] [Google Scholar]
- 6. Breyer J., Wemheuer W. M., Wrede A., Graham C., Benestad S. L., Brenig B., Richt J. A., and Schulz-Schaeffer W. J. (2012) Detergents modify proteinase K resistance of PrP Sc in different transmissible spongiform encephalopathies (TSEs). Vet. Microbiol. 157, 23–31 10.1016/j.vetmic.2011.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shaked Y., Engelstein R., and Gabizon R. (2002) The binding of prion proteins to serum components is affected by detergent extraction conditions. J. Neurochem. 82, 1–5 10.1046/j.1471-4159.2002.00995.x [DOI] [PubMed] [Google Scholar]
- 8. Levine D. J., Stöhr J., Falese L. E., Ollesch J., Wille H., Prusiner S. B., and Long J. R. (2015) Mechanism of scrapie prion precipitation with phosphotungstate anions. ACS Chem. Biol. 10, 1269–1277 10.1021/cb5006239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Terry C., Wenborn A., Gros N., Sells J., Joiner S., Hosszu L. L., Tattum M. H., Panico S., Clare D. K., Collinge J., Saibil H. R., and Wadsworth J. D. (2016) Ex vivo mammalian prions are formed of paired double helical prion protein fibrils. Open Biol. 6, 160035 10.1098/rsob.160035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bode D. C., Freeley M., Nield J., Palma M., and Viles J. H. (2019) Amyloid-β oligomers have a profound detergent-like effect on lipid membrane bilayers, imaged by atomic force and electron microscopy. J. Biol. Chem. 294, 7566–7572 10.1074/jbc.AC118.007195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gursky O. (2015) Role of lipids in protein misfolding. Adv. Exp. Med. Biol. 855, v–vii [PubMed] [Google Scholar]
- 12. Korshavn K. J., Satriano C., Lin Y., Zhang R., Dulchavsky M., Bhunia A., Ivanova M. I., Lee Y.-H., La Rosa C., Lim M. H., and Ramamoorthy A. (2017) Reduced lipid bilayer thickness regulates the aggregation and cytotoxicity of amyloid-β. J. Biol. Chem. 292, 4638–4650 10.1074/jbc.M116.764092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang F., and Ma J. (2013) Role of lipid in forming an infectious prion? Acta Biochim. Biophys. Sin (Shanghai) 45, 485–493 10.1093/abbs/gmt038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ambadi Thody S., Mathew M. K., and Udgaonkar J. B. (2018) Mechanism of aggregation and membrane interactions of mammalian prion protein. Biochim. Biophys. Acta Biomembr. 1860, 1927–1935 10.1016/j.bbamem.2018.02.031 [DOI] [PubMed] [Google Scholar]
- 15. Graeve M., and Janssen D. (2009) Improved separation and quantification of neutral and polar lipid classes by HPLC-ELSD using a monolithic silica phase: application to exceptional marine lipids. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877, 1815–1819 10.1016/j.jchromb.2009.05.004 [DOI] [PubMed] [Google Scholar]
- 16. Dörr J. M., Scheidelaar S., Koorengevel M. C., Dominguez J. J., Schäfer M., van Walree C. A., and Killian J. A. (2016) The styrene–maleic acid copolymer: a versatile tool in membrane research. Eur. Biophys. J. 45, 3–21 10.1007/s00249-015-1093-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Knowles T. J., Scott-Tucker A., Overduin M., and Henderson I. R. (2009) Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat. Rev. Microbiol. 7, 206–214 10.1038/nrmicro2069 [DOI] [PubMed] [Google Scholar]
- 18. Lee S. C., Knowles T. J., Postis V. L., Jamshad M., Parslow R. A., Lin Y.-P., Goldman A., Sridhar P., Overduin M., Muench S. P., and Dafforn T. R. (2016) A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc. 11, 1149–1162 10.1038/nprot.2016.070 [DOI] [PubMed] [Google Scholar]
- 19. Qiu W., Fu Z., Xu G. G., Grassucci R. A., Zhang Y., Frank J., Hendrickson W. A., and Guo Y. (2018) Structure and activity of lipid bilayer within a membrane-protein transporter. Proc. Natl. Acad. Sci. U.S.A. 115, 12985–12990 10.1073/pnas.1812526115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun C., Benlekbir S., Venkatakrishnan P., Wang Y., Hong S., Hosler J., Tajkhorshid E., Rubinstein J. L., and Gennis R. B. (2018) Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Nature 557, 123–126 10.1038/s41586-018-0061-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grethen A., Oluwole A. O., Danielczak B., Vargas C., and Keller S. (2017) Thermodynamics of nanodisc formation mediated by styrene/maleic acid (2:1) copolymer. Sci. Rep. 7, 11517 10.1038/s41598-017-11616-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Korotych O., Mondal J., Gattás-Asfura K. M., Hendricks J., and Bruce B. D. (2018) Evaluation of commercially available styrene–co-maleic acid polymers for the extraction of membrane proteins from spinach chloroplast thylakoids. Eur. Polym. J. 114, 485–500 10.1016/j.eurpolymj.2018.10.035 [DOI] [Google Scholar]
- 23. Morrison K. A., Akram A., Mathews A., Khan Z. A., Patel J. H., Zhou C., Hardy D. J., Moore-Kelly C., Patel R., Odiba V., Knowles T. J., Javed M.-U., Chmel N. P., Dafforn T. R., and Rothnie A. J. (2016) Membrane protein extraction and purification using styrene–maleic acid (SMA) copolymer: effect of variations in polymer structure. Biochem. J. 473, 4349–4360 10.1042/BCJ20160723 [DOI] [PubMed] [Google Scholar]
- 24. Overduin M., and Esmaili M. (2019) Native nanodiscs and the convergence of lipidomics, metabolomics, interactomics and proteomics. Appl. Sci. 9, 1230 10.3390/app9061230 [DOI] [Google Scholar]
- 25. D'Castro L., Wenborn A., Gros N., Joiner S., Cronier S., Collinge J., and Wadsworth J. D. (2010) Isolation of proteinase K–sensitive prions using pronase E and phosphotungstic acid. PLoS One 5, e15679 10.1371/journal.pone.0015679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Priola S. A., and McNally K. L. (2009) The role of the prion protein membrane anchor in prion infection. Prion 3, 134–138 10.4161/pri.3.3.9771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wille H., and Prusiner S. B. (1999) Ultrastructural studies on scrapie prion protein crystals obtained from reverse micellar solutions. Biophys. J. 76, 1048–1062 10.1016/S0006-3495(99)77270-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wille H., Michelitsch M. D., Guenebaut V., Supattapone S., Serban A., Cohen F. E., Agard D. A., and Prusiner S. B. (2002) Structural studies of the scrapie prion protein by electron crystallography. Proc. Natl. Acad. Sci. U.S.A. 99, 3563–3568 10.1073/pnas.052703499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wille H., Shanmugam M., Murugesu M., Ollesch J., Stubbs G., Long J. R., Safar J. G., and Prusiner S. B. (2009) Surface charge of polyoxometalates modulates polymerization of the scrapie prion protein. Proc. Natl. Acad. Sci. U.S.A. 106, 3740–3745 10.1073/pnas.0812770106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fantini J., Garmy N., Mahfoud R., and Yahi N. (2002) Lipid rafts: structure, function and role in HIV, Alzheimer's and prion diseases. Expert Rev. Mol. Med. 4, 1–22 10.1017/S1462399402005392 [DOI] [PubMed] [Google Scholar]
- 31. Taylor D. R., and Hooper N. M. (2006) The prion protein and lipid rafts. Mol. Membr. Biol. 23, 89–99 10.1080/09687860500449994 [DOI] [PubMed] [Google Scholar]
- 32. Camargo M., Intasqui Lopes P., Del Giudice P. T., Carvalho V. M., Cardozo K. H., Andreoni C., Fraietta R., and Bertolla R. P. (2013) Unbiased label-free quantitative proteomic profiling and enriched proteomic pathways in seminal plasma of adult men before and after varicocelectomy. Hum. Reprod. 28, 33–46 10.1093/humrep/des357 [DOI] [PubMed] [Google Scholar]
- 33. Sciacca M. F. M., Tempra C., Scollo F., and Milardi D. L. (2018) Amyloid growth and membrane damage: current themes and emerging perspectives from theory and experiments on Aβ and hIAPP. Biochim. Biophys. Acta Biomembr. 1860, 1625–1638 10.1016/j.bbamem.2018.02.022 [DOI] [PubMed] [Google Scholar]
- 34. Klein T. R., Kirsch D., Kaufmann R., and Riesner D. (1998) Prion rods contain small amounts of two host sphingolipids as revealed by thin-layer chromatography and mass spectrometry. Biol. Chem. 379, 655–666 [DOI] [PubMed] [Google Scholar]
- 35. Simons K., and Toomre D. (2000) Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1, 31–39 10.1038/35036052 [DOI] [PubMed] [Google Scholar]
- 36. Guo B. B., Bellingham S. A., and Hill A. F. (2015) The neutral sphingomyelinase pathway regulates packaging of the prion protein into exosomes. J. Biol. Chem. 290, 3455–3467 10.1074/jbc.M114.605253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hannun Y. A., and Obeid L. M. (2008) Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9, 139–150 10.1038/nrm2329 [DOI] [PubMed] [Google Scholar]
- 38. Gonzalez-Romero D., Barria M. A., Leon P., Morales R., and Soto C. (2008) Detection of infectious prions in urine. FEBS Lett. 582, 3161–3166 10.1016/j.febslet.2008.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cronier S., Gros N., Tattum M. H., Jackson G. S., Clarke A. R., Collinge J., and Wadsworth J. D. (2008) Detection and characterization of proteinase K-sensitive disease-related prion protein with thermolysin. Biochem. J 416, 297–305 10.1042/BJ20081235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Angelisová P., Ballek O., Sýkora J., Benada O., Čajka, Pokorná T. J., Pinkas D., and Hoøejší V. (2019) The use of styrene–maleic acid copolymer (SMA) for studies on T cell membrane rafts. Biochim. Biophys. Acta Biomembr. 1861, 130–141 10.1016/j.bbamem.2018.08.006 [DOI] [PubMed] [Google Scholar]
- 41. Falcon B., Zivanov J., Zhang W., Murzin A. G., Garringer H. J., Vidal R., Crowther R. A., Newell K. L., Ghetti B., Goedert M., and Scheres S. H. W. (2019) Novel Tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature 568, 420–423 10.1038/s41586-019-1026-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Teruya K., Oguma A., Nishizawa K., Kawata M., Sakasegawa Y., Kamitakahara H., and Doh-Ura K. (2016) A single subcutaneous injection of cellulose ethers administered long before infection confers sustained protection against prion diseases in rodents. PLoS Pathog. 12, e1006045 10.1371/journal.ppat.1006045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tonge S. R., and Tighe B. J. (2001) Responsive hydrophobically associating polymers: a review of structure and properties. Adv. Drug Deliv. Rev 53, 109–122 10.1016/S0169-409X(01)00223-X [DOI] [PubMed] [Google Scholar]
- 44. Tsukigawa K., Liao L., Nakamura H., Fang J., Greish K., Otagiri M., and Maeda H. (2015) Synthesis and therapeutic effect of styrene–maleic acid copolymer-conjugated pirarubicin. Cancer Sci. 106, 270–278 10.1111/cas.12592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McCarthy J. M., Rasines Moreno B., Filippini D., Komber H., Maly M., Cernescu M., Brutschy B., Appelhans D., and Rogers M. S. (2013) Influence of surface groups on poly(propylene imine) dendrimers antiprion activity. Biomacromolecules 14, 27–37 10.1021/bm301165u [DOI] [PubMed] [Google Scholar]
- 46. Lim Y. B., Mays C. E., Kim Y., Titlow W. B., and Ryou C. (2010) The inhibition of prions through blocking prion conversion by permanently charged branched polyamines of low cytotoxicity. Biomaterials 31, 2025–2033 10.1016/j.biomaterials.2009.11.085 [DOI] [PubMed] [Google Scholar]
- 47. Fischer M., Appelhans D., Schwarz S., Klajnert B., Bryszewska M., Voit B., and Rogers M. (2010) Influence of surface functionality of poly(propylene imine) dendrimers on protease resistance and propagation of the scrapie prion protein. Biomacromolecules 11, 1314–1325 10.1021/bm100101s [DOI] [PubMed] [Google Scholar]
- 48. Supattapone S., Nguyen H. O., Cohen F. E., Prusiner S. B., and Scott M. R. (1999) Elimination of prions by branched polyamines and implications for therapeutics. Proc. Natl. Acad. Sci. U.S.A. 96, 14529–14534 10.1073/pnas.96.25.14529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lindhoud S., Carvalho V., Pronk J. W., and Aubin-Tam M. E. (2016) SMA-SH: Modified styrene–maleic acid copolymer for functionalization of lipid nanodiscs. Biomacromolecules 17, 1516–1522 10.1021/acs.biomac.6b00140 [DOI] [PubMed] [Google Scholar]
- 50. Wenborn A., Terry C., Gros N., Joiner S., D'Castro L., Panico S., Sells J., Cronier S., Linehan J. M., Brandner S., Saibil H. R., Collinge J., and Wadsworth J. D. (2015) A novel and rapid method for obtaining high titre intact prion strains from mammalian brain. Sci. Rep. 5, 10062 10.1038/srep10062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dörr J. M., Koorengevel M. C., Schäfer M., Prokofyev A. V., Scheidelaar S., van der Cruijsen E. A., Dafforn T. R., Baldus M., and Killian J. A. (2014) Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs. Proc. Natl. Acad. Sci. U.S.A. 111, 18607–18612 10.1073/pnas.1416205112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Botsios S., Tittman S., and Manuelidis L. (2015) Rapid chemical decontamination of infectious CJD and scrapie particles parallels treatments known to disrupt microbes and biofilms. Virulence 6, 787–801 10.1080/21505594.2015.1098804 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and the supporting information.