Abstract
N-Methyl-d-aspartate type glutamate receptors (NMDARs) are key mediators of synaptic activity-regulated gene transcription in neurons, both during development and in the adult brain. Developmental differences in the glutamate receptor ionotropic NMDA 2 (GluN2) subunit composition of NMDARs determines whether they activate the transcription factor cAMP-responsive element-binding protein 1 (CREB). However, whether the developmentally regulated GluN3A subunit also modulates NMDAR-induced transcription is unknown. Here, using an array of techniques, including quantitative real-time PCR, immunostaining, reporter gene assays, RNA-Seq, and two-photon glutamate uncaging with calcium imaging, we show that knocking down GluN3A in rat hippocampal neurons promotes the inducible transcription of a subset of NMDAR-sensitive genes. We found that this enhancement is mediated by the accumulation of phosphorylated p38 mitogen-activated protein kinase in the nucleus, which drives the activation of the transcription factor myocyte enhancer factor 2C (MEF2C) and promotes the transcription of a subset of synaptic activity-induced genes, including brain-derived neurotrophic factor (Bdnf) and activity-regulated cytoskeleton-associated protein (Arc). Our evidence that GluN3A regulates MEF2C-dependent transcription reveals a novel mechanism by which NMDAR subunit composition confers specificity to the program of synaptic activity-regulated gene transcription in developing neurons.
Keywords: N‐methyl‐d‐aspartate receptor (NMDA receptor, NMDAR), p38 MAPK, excitation-transcription coupling, neurodevelopment, brain-derived neurotrophic factor (BDNF), gene regulation, synaptic activity, mitogen-activated protein kinase (MAPK) signaling, glutamate receptor ionotropic NMDA (GluN), ionotropic glutamate receptor, MEF2 transcription factors
N-Methyl-d-aspartate type glutamate receptors (NMDARs) are essential for coupling sensory experience with brain development. In addition to functioning as ligand-gated ionotropic receptors, NMDARs exert long-lasting effects on neuronal biology by activating intracellular signaling cascades that subsequently impact synapse formation, maturation, and function. These inductive effects of NMDARs are mediated at least in part by the regulation of programs of gene transcription. The targets of NMDAR-regulated transcriptional signaling cascades in neurons include genes like Bdnf and Arc, which directly alter synapse development and function, providing a compelling mechanism for activity-induced synaptic development and plasticity (1).
NMDARs are heterotetrameric receptors, all of which contain the obligatory GluN1 pore-forming subunit in various stoichiometries with the glutamate-binding GluN2 (A–D) subunits and the modulatory GluN3 (A–B) subunits (2). In addition to determining the biophysical properties of NMDA receptors (e.g. conductance, magnesium sensitivity, and calcium permeability), subunit composition influences the specificity of the downstream signaling cascades initiated following receptor activation largely via protein–protein interactions with the highly variable subunit C-terminal tails (3, 4). Unlike the ubiquitous expression of the GluN1 subunit, the GluN2 and GluN3 subunits show substantial regional and developmental regulation of their expression in the brain, suggesting a mechanism for NMDAR functional diversity. The ratio of GluN2A to GluN2B subunits is dynamic during postnatal development (5) and has been shown to modulate the activation of transcriptional programs by NMDARs as neurons age (6, 7). Specifically, GluN2A, which predominates at synaptic receptors and in older neurons, promotes cell survival via the phosphorylation/activation of the transcription factor CREB, whereas GluN2B, which is more widely expressed in younger neurons and preferentially found at extrasynaptic receptors, activates signaling cascades that lead to CREB dephosphorylation and cell death (8, 9).
Neurons in the developing postnatal brain also transiently express high levels of the GluN3A subunit, which is incorporated into functional GluN1-GluN2-GluN3 glutamate-activated NMDARs (10–13). Genetic deletion of GluN3A is associated with dysregulation of the plasticity of glutamatergic synapses in the developing brain, implicating the expression of this subunit in NMDAR-dependent aspects of brain development (14, 15). Functionally, excitatory synapses show evidence of premature maturation in GluN3A knockout mice (15, 16), whereas prolonging the expression of GluN3A into adulthood causes excitatory synapses to persist in a juvenile state (17). Although these data indicate that GluN3A is a negative regulator of synapse maturation, the mechanism of its action remains to be fully understood.
We identified the transcription factor CaRF as an upstream regulator of GluN3A expression (18). CaRF knockout mice show enhanced NMDAR-induced transcription of Bdnf both in culture and in vivo in a manner that correlates with reduced GluN3A expression (18), raising the possibility that GluN3A functions to transiently inhibit the NMDAR-dependent induction of Bdnf and other neuronal activity-regulated genes in developing neurons. Here, to determine the function of GluN3A in NMDAR-dependent transcription, we knocked down the expression of GluN3A in primary rat hippocampal neurons and determined the consequences for NMDAR-induced programs of gene expression. We find that GluN3A limits the NMDAR activation of a select group of genes, including Bdnf and Arc. GluN3A impacts downstream gene transcription by inhibiting the nuclear translocation of the p38 mitogen-activated protein kinase (MAPK), which is required for the activation of genes that depend on the transcription factor MEF2C. The net effect of this pathway is to confer specificity upon the set of synaptic signaling pathways that can induce activity-dependent transcription in developing neurons. These data bring new understanding to the mechanisms by which NMDARs regulate activity-dependent brain development.
Results
Knockdown of GluN3A potentiates NMDAR-dependent induction of Bdnf
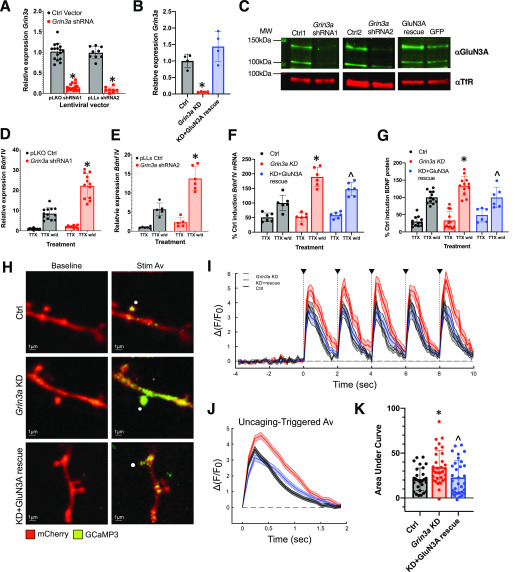
To test the requirement for GluN3A in NMDAR-inducible transcription of Bdnf, we characterized two independent shRNAs in lentivirus that robustly knock down the expression of Grin3a mRNA and GluN3A protein in cultured embryonic rat hippocampal neurons and generated a GluN3A rescue construct that is resistant to shRNA1 (Fig. 1, A–C). To activate NMDARs, we silenced neurons with tetrodotoxin (TTX) for 48 h and then acutely withdrew TTX (TTX w/d), which induces rebound firing, glutamate release, and NMDAR-dependent changes in gene transcription (Fig. S1) (19). As we reported previously in mouse cortical neurons (18), TTX w/d from rat hippocampal neurons significantly induces the NMDAR-dependent expression of exon IV-containing forms of Bdnf, which are produced by the activation of Bdnf promoter IV (Fig. S1) (20). Knockdown (KD) of GluN3A with either of the two shRNAs had no significant effect on Bdnf IV levels in TTX silenced neurons but resulted in a significant potentiation of Bdnf IV mRNA transcription upon TTX w/d for each shRNA compared with its paired control infected neurons (Fig. 1, D and E). The potentiation of Bdnf promoter IV activation in GluN3A KD neurons was selective for transcription induced by NMDARs, because there were no significant differences in the fold change of Bdnf IV mRNA induced in GluN3A KD versus control neurons following KCl-mediated membrane depolarization (Fig. S2A), which activates transcription selectively via the opening of l-type voltage-gated calcium channels (LVGCCs) and does not require the activation of NMDARs (18).
Figure 1.
GluN3A knockdown selectively enhances NMDAR-dependent Bdnf transcription. A, Levels of Grin3a mRNA in rat hippocampal neurons infected with lentiviruses encoding shRNAs targeting Grin3a (shRNA1 and shRNA2) or their paired control vectors (pLKO or pLLx3.8). Grin3a mRNA levels are scaled to levels in cells infected with the paired control viruses. Two-way ANOVA for vector, F(1, 45) = 1.033, p = 0.32; and shRNA, F(1, 45) = 425.6, p < 0.0001. For pLKO shRNA1 versus Ctrl1, n = 16/virus, p < 0.0001. For pLLx3.8 shRNA2 versus Ctrl2, n = 9 Ctrl2, 8 shRNA2, p < 0.0001. B, Expression of Grin3a mRNA in neurons infected with the Grin3a KD and shRNA-resistant GluN3A rescue viruses. C, Membrane fraction from cultured hippocampal neurons infected with the indicated lentiviruses, analyzed by Western blotting using an antibody that detects the GluN3A protein (green). Transferrin receptor is shown as a loading control (red). The predicted molecular weight (MW) of GluN3A is 130 kDa. D and E, Levels of Bdnf IV mRNA in hippocampal neurons infected with the indicated lentiviruses in the presence of TTX or 6 h following TTX w/d. Values are scaled to Ctrl TTX for comparison. D, pLKO. Two-way ANOVA for treatment, F(1, 41) = 49.53, p < 0.0001; for virus, F(1, 41) = 172.5, p < 0.0001; and for treatment–virus interaction, F(1, 41) = 36.12, p < 0.0001. Ctrl TTX, n = 11; Ctrl TTX w/d, n = 13; shRNA TTX, n = 9; shRNA TTX w/d, n = 12. Ctrl TTX versus TTX w/d, p < 0.0001; shRNA TTX versus TTX w/d, p < 0.0001. Ctrl TTX w/d versus shRNA TTX w/d, p < 0.0001. Ctrl TTX versus shRNA TTX is not significant, p = 0.90. E, pLLx3.8. Two-way ANOVA for treatment, F(1, 18) = 32.97, p < 0.0001; for virus, F(1, 18) = 97.67, p < 0.0001; and for treatment–virus interaction, F(1, 18) = 16.98, p = 0.0006. Ctrl TTX, n = 6; Ctrl TTX w/d, n = 6; shRNA TTX, n = 6; shRNA TTX w/d, n = 5. Ctrl TTX versus TTX w/d, p = 0.0036; shRNA TTX versus TTX w/d, p < 0.0001. Ctrl TTX w/d versus shRNA TTX w/d, p < 0.0001. Ctrl TTX versus shRNA TTX is not significant, p = 0.67. F, Levels of TTX w/d-induced Bdnf IV mRNA in hippocampal neurons infected with the indicated lentiviruses relative to the control condition (Ctrl), whose induced average is scaled to 100%. Two-way ANOVA for treatment, F(1, 30) = 160.5, p < 0.0001; for virus, F(2, 30) = 13.33, p = 0.0001; and for treatment–virus interaction, F(2, 30) = 12.25, p = 0.0001. n = 6/condition. For TTX w/d, Ctrl versus KD, p < 0.0001; KD versus rescue, p = 0.0064; Ctrl versus rescue, p = 0.0019. G, Levels of BDNF protein in hippocampal neurons infected with the indicated lentiviruses in the presence of TTX or 6 h following TTX w/d. Control condition (Ctrl) TTX w/d induced average is scaled to 100%. Two-way ANOVA for treatment, F(1, 54) = 144.8, p < 0.0001; for virus, F(2, 54) = 4.103, p = 0.022; and for treatment–virus interaction, F(2, 54) = 5.547, p = 0.0064. n = 12/condition for Ctrl and KD, 6/condition for rescue. For TTX w/d Ctrl versus Grin3a KD, p = 0.0016; KD versus rescue, p = 0.012. H, Representative 2-photon images of hippocampal CA1 neurons expressing GCaMP3 (green) and mCherry (red). (Left) Baseline images corresponding to the average projection for the first 31 frames of imaging, prior to uncaging. (Right) Average projection images of the first two frames after each of the five uncaging pulses (10 total imaging frames). White circles indicate the uncaging point. (I) Average ΔF/F0 traces showing response to glutamate uncaging (black triangles) for control (n = 29 spines; 3 cells), Grin3a KD (n = 32 spines, 3 cells), and KD plus GluN3A rescue (n = 32 spines, 3 cells) conditions. Data correspond to mean ± S.E. J, Uncaging-triggered average of the responses shown in panel I. Traces correspond to the average baseline-subtracted response to each glutamate uncaging pulse, where the baseline is the frame immediately before each uncaging stimulus. Error corresponds to S.E. of the five uncaging pulses for each condition. K, Average integrated GCaMP3 signal for each condition. The average area under the curve was calculated for each spine and then averaged for a given condition. Asterisks indicate statistical significance as determined by ANOVA and a post hoc test using Fisher's least significant difference. Ctrl versus KD, p = 0.003; ctrl versus rescue, p = 0.6; KD versus rescue, p = 0.009. *, p < 0.05 for Grin3a KD versus Ctrl; ∧, p < 0.05 GluN3A rescue versus Grin3a KD.
To determine whether the enhanced activation of Bdnf IV transcription in GluN3A KD neurons is a direct result of the absence of GluN3A, as opposed to an off-target effect of the shRNAs, we reexpressed the shRNA-resistant form of GluN3A in neurons that had endogenous GluN3A knocked down (Fig. 1, B and C). Whereas GluN3A KD led to potentiation of Bdnf IV induction by TTX w/d relative to the control, reexpression of GluN3A in KD neurons was sufficient to significantly reduce the fold change of Bdnf IV induction toward control levels (Fig. 1F). To determine whether TTX w/d-induced changes in Bdnf mRNA induction are sufficient to result in changes in BDNF protein, we performed BDNF ELISAs. TTX w/d induced the expression of BDNF protein in all samples (Fig. 1G). Similar to the effects of GluN3A KD on Bdnf exon IV mRNA induction, GluN3A KD neurons showed a significantly greater induction of BDNF protein after TTX w/d than with the control, whereas the coexpression of the GluN3A rescue restored BDNF protein induction to control levels.
In the context of GluN1/GluN2/GluN3 NMDARs, GluN3 has been reported to lower calcium permeability (15). To determine whether our manipulations of GluN3A expression in hippocampal neurons were affecting glutamate-induced calcium influx at synapses, we performed two-photon glutamate uncaging and dendritic spine calcium imaging in organotypic rat hippocampal slices. Local glutamate uncaging led to a transient increase in spine calcium, as detected by the calcium indicator GCaMP3 (Fig. 1H). GluN3A KD neurons showed a significantly greater increase in GCaMP3 fluorescence upon uncaging than control pLKO vector-transfected neurons, whereas coexpression of the GluN3A rescue plasmid with the Grin3a shRNA restored the calcium signal to control levels (Fig. 1, I–K). This evidence that GluN3A levels regulate glutamate-dependent changes in spine calcium is consistent with the possibility that GluN3A modulates calcium-regulated transcriptional signaling pathways.
Knockdown of GluN3A modulates the NMDAR-induced transcriptional program
We tested other activity-regulated genes as candidate GluN3A targets and found that the potentiation of NMDAR-dependent transcription in GluN3A KD neurons was limited to a subset of NMDAR-regulated genes. Specifically, TTX w/d-induced Fos expression did not differ between GluN3A KD and control neurons; however, the TTX w/d-induced expression of Arc was also potentiated in the GluN3A KD neurons compared with the control, indicating that the potentiation was not limited to Bdnf IV (Fig. S2B). Thus, to determine the extent of GluN3A-regulated gene expression, we performed RNA-Seq on GluN3A KD and control infected neurons either in the presence of TTX silencing or 6 h following TTX w/d. Expression of 10,731 genes was detected at a fragments per kilobase of exon per million fragments mapped (FPKM) of >1 across triplicate replicate samples for each condition (Table S2).
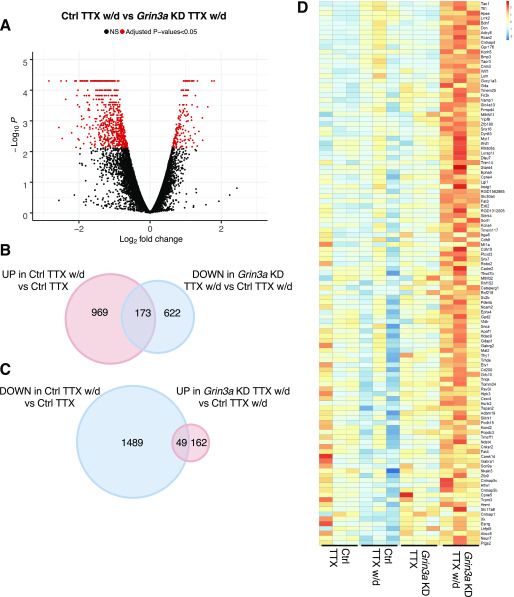
232 genes showed differential expression (false discovery rate [FDR]-adjusted p < 0.05) in the presence of TTX comparing GluN3A KD with the control. The expression of 196 genes, including Grin3a, was significantly reduced in GluN3A KD compared with the control, and expression of 36 genes was significantly elevated (Table S2, Fig. S3A). In contrast, 815 genes were differentially expressed 6 h after TTX w/d compared with the GluN3A KD and control samples. 642 of these showed significantly reduced expression in the GluN3A KD samples relative to the control, whereas 173, including Bdnf, were expressed more highly (Fig. 2, A and B). Of the genes that were differentially expressed between the Grin3a KD and the control neurons after TTX w/d, only 30 genes were also differentially expressed in the presence of TTX (Fig. S3, B–C, Table S2). These data suggest that the vast majority of differences between GluN3A KD and control samples arise as a result of TTX w/d-dependent changes in gene expression. To focus on these stimulus-dependent changes in gene expression, we removed the 30 stimulus-independent, GluN3A-regulated genes from subsequent analyses.
Figure 2.
GluN3A contributes to the program of NMDAR-inducible transcription. A, Volcano plot showing genes significantly different at an FDR-adjusted p < 0.05 (red dots) comparing Ctrl TTX w/d with Grin3a KD TTX w/d. B, Venn diagram showing genes significantly induced by TTX w/d in Ctrl neurons (red) compared with genes reduced in Grin3a KD versus Ctrl after TTX w/d (blue). C, Venn diagram showing genes significantly repressed by TTX w/d in Ctrl neurons (blue) compared with genes with higher expression in Grin3a KD compared with Ctrl after TTX w/d (red). D, Heat map showing the relative expression of the 113 genes is higher after TTX w/d in Grin3a KD compared with Ctrl that are either unchanged or induced by TTX w/d in Ctrl. n = 3 biological replicates per sample. Heat map shows the z-score for FPKM.
To determine whether there are common biological processes among genes dysregulated in the absence of GluN3A, we subjected the differentially regulated gene lists to gene ontology (GO) analysis. The program of gene expression induced by TTX w/d in control neurons is enriched for a number of categories, suggesting inflammatory and oxidative stress responses (Table S3). Genes that showed significantly lower expression in GluN3A KD neurons than the control after TTX w/d were enriched for categories that include inflammatory responses and signaling cascades (Table S3). Two of these GO categories (GO:0007568∼aging and GO:0071222∼cellular response to lipopolysaccharide) are shared with the ontologies of genes that are induced by TTX w/d in control neurons. Furthermore, a substantial fraction (∼18%) of the overall set of TTX w/d-inducible genes show impaired poststimulus expression in the absence of GluN3A (Fig. 2B). These data suggest that GluN3A positively contributes to the regulation of a subset of the transcriptional response induced upon TTX w/d.
We next turned to the set of 162 genes that, like Bdnf, showed elevated expression in GluN3A KD compared with control neurons after TTX w/d. These genes were enriched for two GO categories, one of which, GO:0007268∼chemical synaptic transmission, contains a set of genes that mediate synaptic communication between neurons (Grm5, Hcrtr2, Gabrg2, Gabra1, Grm6, P2rx2, Gabra5, Snca, Grin2a, Tac1, and Slitrk5). Mirroring the inverse relationship we discussed above between GluN3A KD and TTX w/d, about 30% (49/162) of the genes that show elevated expression in GluN3A KD neurons are among the large set of genes significantly repressed by TTX w/d in the control (Fig. 2C, Table S2). Overall, the set of genes that show reduced expression in control neurons after TTX w/d include many categories with synaptic functions, suggesting homeostatic repression of synaptic function by TTX w/d-induced neuronal firing (Table S3). These data suggest that GluN3A also positively contributes to the regulation of a subset of the transcriptional responses repressed upon TTX w/d.
GluN3A inhibits NMDAR-dependent activation of MEF2C
Given that TTX w/d-induced gene expression is mediated by the activation of NMDARs, it is not surprising that loss of the NMDAR subunit GluN3A would blunt the cellular response to this stimulus. However, 70% (113/162) of genes that show elevated expression in Grin3a KD neurons after TTX w/d are either not regulated by TTX w/d in control neurons or are inducible in control neurons but more strongly induced by TTX w/d in the GluN3A KD neurons (Fig. 2D). These data suggest that GluN3A actively opposes a mechanism of gene expression that can be induced by NMDAR activation only in the absence of this subunit. In addition to Bdnf, the set of genes that are “superinduced” in GluN3A KD neurons includes the metalloproteinase Tll1, the endoglucuronidase enzyme Hpse, the neuropeptide Tac1, the extracellular matrix protein Dcn, the calcium-stimulated adenylate cyclase Adcy8, the leucine-rich repeat kinase Lrrk2, the regulator of calcineurin Rcan2, and the synaptic adhesion protein Cntnap4. Although we cannot rule out that some of the differentially expressed genes in GluN3A KD neurons arise from off-target effects of the shRNA, the fact that we found other genes coregulated with Bdnf encouraged us to pursue possible transcriptional mechanisms of this effect.
Prior studies have shown that the incorporation of GluN2B subunits in NMDARs represses NMDAR-dependent activation of the prosurvival transcription factor CREB (9). To determine whether GluN3A has a similar repressive effect on specific NMDAR-regulated transcription factors, we first used Gene Set Enrichment Analysis (GSEA) to identify common transcription factor binding sites found among the promoters of the 113 genes that show elevated induction after TTX w/d in GluN3A KD neurons compared with the control. Among known TF motifs, these data identified enriched binding sites for the MEF2 and NFAT transcription factors (Table 1). Both of these transcription factors are known to mediate synaptic activity-regulated gene transcription in neurons, although NFAT is thought to be selectively dependent on somatic L-type calcium channels for its activation, even following direct synaptic glutamate receptor stimulation (21, 22).
Table 1.
GSEA top transcription factor motifs identified in the set of 113 genes from Fig. 2D
| Gene set name | No. of genes in gene set (K) | No. of genes in overlap (k) | k/K | p value | FDR q value |
|---|---|---|---|---|---|
| AACTTT_UNKNOWN | 1890 | 33 | 0.0175 | 1.15E-20 | 7.05E-18 |
| CAGGTG_E12_Q6 | 2485 | 31 | 0.0125 | 2.57E-15 | 7.90E-13 |
| YTATTTTNR_MEF2_O2 | 697 | 17 | 0.O244 | 3.96E-13 | 8.12E-11 |
| TGGAAA_NFAT_Q4_01 | 1896 | 24 | 0.0127 | 4.93E-12 | 7.57E-10 |
| TAATTA_CH× 10_01 | 810 | 16 | 0.0198 | 4.71E-11 | 5.79E-09 |
| TTGTTT_FOXO4_01 | 2061 | 23 | 0.0112 | 1.78E-10 | 1.83E-08 |
| GGGAGGRR_MAZ_Q6 | 2274 | 21 | 0.0092 | 3.40E-08 | 2.65E-06 |
| TTANTCA_UNKNOWN | 952 | 14 | 0.0147 | 3.45E-08 | 2.65E-06 |
| AMEF2_Q6 | 259 | 8 | 0.0309 | 1.48E-07 | 9.39E-06 |
| CTGCAGY_UNKNOWN | 765 | 12 | 0.0157 | 1.78E-07 | 9.39E-06 |
Both Bdnf promoter IV and Arc are targets of regulation by MEF2, which works in collaboration with the activity-dependent transcription factor CREB on these genes (23, 24). To monitor the activity of these transcription factor families, we transfected GluN3A KD or control infected neurons with luciferase reporter plasmids in which the expression of luciferase is under the control of binding sites for either the CREB (CREB response elements; CRE) or MEF2 (MEF2 response elements; MRE) families of transcription factors.
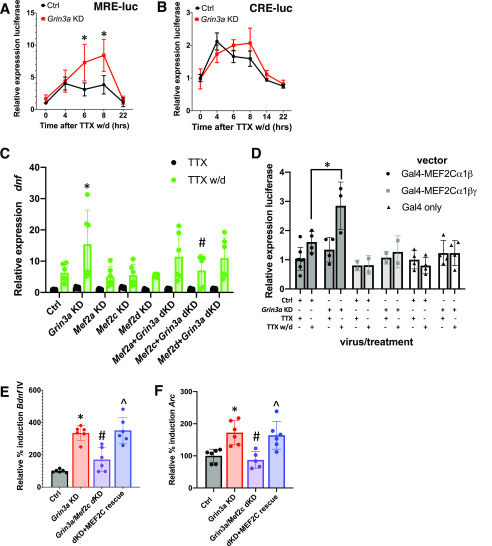
In control infected neurons, TTX w/d induced a significant increase in luciferase expression from both the CRE and MRE reporters (Fig. 3, A and B), demonstrating NMDAR-dependent activation of CREB and MEF2 family transcription factors. However, in GluN3A KD neurons, we detected significantly more luciferase expression from the MRE reporter after 6 h and 8 h of TTX w/d than with the expression in control infected neurons. In contrast, CRE reporter activity was not different in GluN3A KD neurons from that with controls at any time point (Fig. 3, A and B). These data suggest that the potentiation of NMDAR-dependent transcription in GluN3A KD neurons arises from enhanced activation of MEF2 transcription factors.
Figure 3.
GluN3A knockdown enhances MEF2C-dependent transcription. A, Luciferase expression in hippocampal neurons transfected with MRE-Luc plasmid and infected with either Grin3a shRNA1 or control (Ctrl) lentiviruses. Neurons were stimulated with TTX w/d for the indicated amounts of time prior to lysis. Data are expressed as fold induction over TTX (0 h). Two-way ANOVA for time, F(4,62) = 34.65, p < 0.0001; for virus, F(1, 62) = 32.16, p < 0.0001; and for virus–time interaction, F(4, 62) = 7.602, p < 0.001. n = 11 at 0 h, 6 at 4 h, 7 at 6 h, 7 at 8 h, and 6 at 22 h. Grin3a shRNA versus Ctrl at 6 h and 8 h, p < 0.0001. B, Luciferase expression in hippocampal neurons transfected with CRE-Luc plasmid and infected with either Grin3a shRNA1 or Ctrl lentiviruses. Neurons were stimulated as described for panel A. Two-way ANOVA for time, F(5, 24) = 23.00, p < 0.0001; virus, F(1, 24) = 1.407, p = 0.2471. n = 4 at 0 h, 4 at 4 h, 2 at 6 h, 4 at 8 h, 2 at 14 h, and 2 at 22 h. C, Levels of Bdnf IV mRNA in hippocampal neurons infected both with lentiviruses containing Grin3a shRNA1 or its paired control and lentiviruses expressing shRNAs targeting individual MEF2 family members. Two-way ANOVA for treatment, F(1, 77) = 68.53, p < 0.001; for virus, F(7, 77) = 2.520, p = 0.022; and for treatment–virus interaction, F(7, 77) = 2.254, p = 0.039. n = 6/condition, except Ctrl TTX, n = 4, and Mef2c+Grin3a double KD TTX w/d, n = 5. For each viral condition, TTX w/d was significantly different from TTX, and for TTX, there were no significant differences between viral conditions. Significant TTX w/d post hoc comparisons: Ctrl versus Grin3a KD, p = 0.0093; Grin3a KD versus Mef2c+Grin3a double KD, p = 0.041. D, Luciferase expression in hippocampal neurons cotransfected with the pUAS-Luc plasmid, expression vectors for Gal4 fusions of the indicated MEF2C splice variants, and Grin3a shRNA1 or its paired Ctrl1 vector. Data are shown as relative luciferase expression compared with control conditions (Ctrl1 and TTX). Two-way ANOVA for vector, F(2, 31) = 12.67, p < 0.0001; for virus/treatment, F(3, 31) = 6.803, p = 0.0012; and for vector–virus/treatment interaction, F(6, 31) = 3.318, p = 0.012. For Gal4-MEF2Cα1β, Grin3a KD TTX versus TTX w/d, p = 0.0003; Grin3a KD TTX w/d versus Ctrl TTX w/d, p = 0.0027, n = 4 for Ctrl TTX w/d and Grin3a KD TTX and 3 for Grin3a KD TTX w/d. E and F, Levels of TTX w/d-induced Bdnf IV mRNA (E) or Arc mRNA (F) in hippocampal neurons infected with the indicated lentiviruses. Arc mRNA was measured following 30 min TTX w/d. Induced mRNA levels are shown as percentages of induction relative to control conditions (Ctrl). Bdnf IV ANOVA for virus, F(3, 20) = 25.85, p < 0.0001, n = 6/sample; Ctrl versus KD, p < 0.0001; KD versus dKD, p = 0.0007; dKD versus rescue, p = 0.0002. Arc ANOVA for virus, F(3, 19) = 9.641, p = 0.0004. n = 6/sample, except double KD, n = 5. Ctrl versus KD, p = 0.0066; KD versus dKD, p = 0.0024; double KD versus rescue, p = 0.006. *, p < 0.05 for Grin3a KD versus Ctrl; #, p < 0.05 for dKD versus Grin3a KD; ∧, p < 0.05 for rescue versus dKD.
To determine whether MEF2 family transcription factors are required for the potentiation of NMDAR-induced gene transcription in GluN3A KD neurons, we used lentiviral shRNAs to knockdown the expression of MEF2A, MEF2C, and MEF2D, the three major MEF2 family members expressed in cultured hippocampal neurons (23). Previously, we showed that knockdown of MEF2C but not MEF2A or MEF2D impairs the LVGCC-dependent induction of Bdnf exon IV (23). Here, we found that whereas knocking down any of the three MEF2 family members (Fig. S4, A–C) did not affect TTX w/d-induced Bdnf IV expression in control neurons, knockdown of MEF2C, but not MEF2A or MEF2D, eliminated the potentiation of Bdnf IV expression upon TTX w/d in GluN3A KD neurons (Fig. 3C).
To test whether knockdown of GluN3A potentiates the transcriptional activity of MEF2C, we cotransfected neurons with plasmids encoding a Gal4-MEF2C fusion protein and a UAS-luciferase reporter along with either Grin3a shRNA1 or the paired control vector. Neuronal MEF2C is comprised of two major splice variants. Both isoforms contain exons encoding the α1 and β domains, but the alternatively spliced exon encoding the γ domain is present in only about 50% of Mef2c mRNA transcripts in hippocampus and cortex (23, 25). MEF2C splice variants lacking the γ-domain are the most highly activated of the MEF2C isoforms following LVGCC activation (23). Here, we found that only Gal4-MEF2C fusion proteins that lack the γ domain are activated upon TTX w/d (Fig. 3D). Furthermore, only the γ-lacking isoform of MEF2C (MEF2Cα1β) showed potentiation upon TTX w/d in GluN3A KD neurons compared with control neurons (Fig. 3D). Importantly, because these studies were done with cotransfection of the knockdown and the Gal4 reporter plasmids, which leads to the expression of both constructs in only about ∼1% of the neurons in our cultures, these experiments demonstrate that the effects of GluN3A knockdown on the activation of MEF2C are cell autonomous.
To test whether the expression of the MEF2Cα1β isoform is sufficient to drive NMDAR-dependent Bdnf IV transcription, we reexpressed human MEF2Cα1β, which is resistant to the rat Mef2c shRNA, in GluN3A and MEF2C double KD neurons (Fig. S4D). Whereas the knockdown of MEF2C eliminated the potentiation of both Bdnf IV and Arc transcription in GluN3A KD neurons, replacement of human MEF2Cα1β in GluN3A and MEF2C double KD neurons was sufficient to restore induction of expression to the levels seen in GluN3A KD neurons (Fig. 3, E and F).
Enhanced NMDAR-dependent nuclear activation of p38 MAPK in GluN3A knockdown neurons
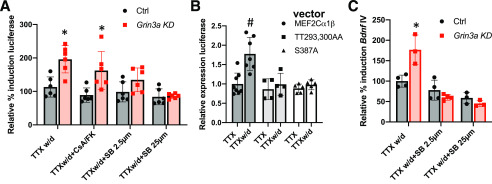
To identify the signaling mechanisms underlying the potentiation of NMDAR-induced transcription in GluN3A KD neurons, we first asked whether pharmacological blockade of known MEF2 regulatory pathways (26) would inhibit the potentiation of Gal4-MEF2Cα1β activity upon TTX w/d. Although calcineurin-dependent dephosphorylation of MEF2s is required for the membrane depolarization-dependent activation of MEF2-dependent transcription in cultured hippocampal neurons (27), we found that inhibition of calcineurin with cyclosporin A and FK506 had no effect on Gal4-MEF2Cα1β activation following TTX w/d in either control or GluN3A KD neurons (Fig. 4A). In contrast, the pretreatment of neurons with the p38 MAPK inhibitor SB203580 inhibited the induction of Gal4-MEF2Cα1β activity in GluN3A KD neurons (Fig. 4A). Interestingly, p38 MAPK blockade had no significant effect on TTX w/d-induced Gal4-MEF2Cα1β activity in control infected neurons, indicating a selective requirement for the p38 pathway in MEF2C activation in GluN3A KD neurons.
Figure 4.
p38 MAP kinase activity is required for Bdnf IV potentiation. A, Luciferase expression in hippocampal neurons cotransfected with the UAS-Luc plasmid, a Gal4-MEF2Cα1β expression vector, and either the Grin3a shRNA1 plasmid or its paired control. Hippocampal neurons were stimulated for 6 h with TTX w/d in the absence of presence of cyclosporin A plus FK506 (CsA/FK) or SB203580 at the doses indicated. Induced luciferase levels are shown as percentages of induction relative to the control at 100%. Two-way ANOVA for virus, F(1, 40) = 25.07, p < 0.0001; for treatment, F(3, 40) = 8.724, p = 0.0001; and for virus–treatment interaction, F(3, 40) = 3.620, p = 0.021. n = 6/condition. Grin3a KD versus Ctrl for TTX w/d, p = 0.0005; for TTX w/d+CsA/FK, p = 0.0021. B, Luciferase expression in hippocampal neurons cotransfected with the pUAS-Luc plasmid, expression plasmids encoding the indicated Gal4-MEF2Cα1β vectors, and the Grin3a shRNA1 plasmid. Data are shown as fold change in luciferase levels compared with the WT MEF2Cα1β in TTX. Two-way ANOVA for vector, F(2, 27) = 10.53, p = 0.0004; for treatment, F(1, 27) = 10.41, p = 0.0033; and treatment–vector interaction, F(2, 27) = 5.590, p = 0.0093. n = 8/group for WT plasmid, 4 for TT293,300AA, and 5 for S387A. For Gal4-MEF2Cα1β TTX versus TTX w/d, p = 0.0002. C, Levels of Bdnf IV mRNA in hippocampal neurons infected with the indicated lentiviruses and then stimulated with TTX w/d. During TTX w/d, neurons were left untreated (Ctrl) or SB203580 was added. mRNA levels are reported as percentages of induction relative to the control. Two-way ANOVA for virus, F(1, 15) = 3.09, p = 0.01; for treatment, F(2, 15) = 36.48, p < 0.0001; and for treatment–virus interaction, F(2, 15) = 12.88, p = 0.0006. n = 3, except for Ctrl TTX w/d, n = 4. Ctrl versus Grin3a KD for TTX w/d, p = 0.0014. *, p < 0.05 Grin3a KD versus Ctrl; #, p < 0.05 TTX w/d versus TTX.
p38 MAPK activates MEF2C by inducing its phosphorylation at three sites, Thr293, Thr300, and Ser387, with the numbering referring to amino acid positions in human MEF2Cα1β (28). To test the functional importance of these phosphorylation sites in neurons, we mutated each of these residues to Ala in the context of the Gal4-MEF2Cα1β fusion protein and assessed the effects on TTX w/d-induced luciferase expression in neurons cotransfected with the GluN3A KD shRNA plasmid. Mutating MEF2C at either Thr293/Thr300 or Ser387 rendered the Gal4 fusion protein unresponsive to TTX w/d in GluN3A knockdown neurons (Fig. 4B). These data suggest that p38 MAPK-dependent phosphorylation of MEF2C is required for the activation of MEF2C in GluN3A knockdown neurons.
If p38 MAPK-dependent phosphorylation of MEF2C is selectively mediating the potentiation of NMDAR-dependent transcription in GluN3A KD neurons, then pharmacological inhibition of the p38 MAPK pathway in GluN3A KD neurons should restore transcriptional activation following TTX w/d to the levels observed in control neurons. Indeed, pretreatment of neurons with the p38 MAPK inhibitor SB203580 had no effect on the magnitude of TTX w/d-induced Bdnf IV expression in control neurons, but it abolished the potentiation of Bdnf IV induction in GluN3A KD neurons (Fig. 4C). Taken together, these data suggest that GluN3A inhibits NMDAR-dependent transcription by opposing the p38 MAPK-dependent phosphorylation and activation of MEF2C.
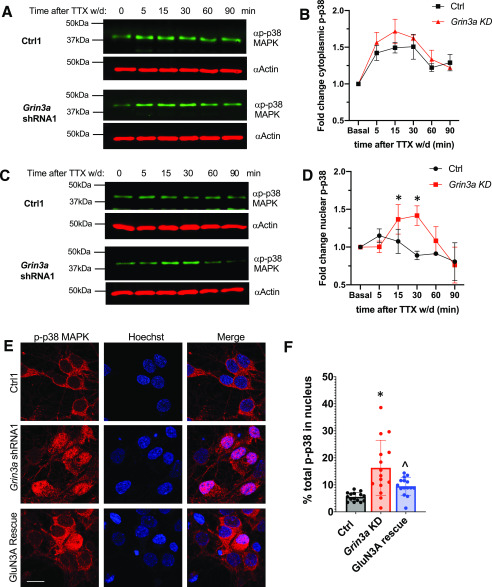
To determine whether the differential p38 MAPK-dependent activation of MEF2C we observed in GluN3A KD neurons reflects differential NMDAR-dependent activation of the p38 MAPK pathway, we subjected GluN3A KD or control infected neurons to TTX w/d and measured the activation of p38 MAPK by Western blotting with an antibody selective for the MKK3/6 phosphorylation sites on p38 at Thr180/Tyr18. When we assayed cytoplasmic fractions from our neuronal cultures, we found that p38 MAPK was phosphorylated within 5 min after TTX w/d in both control and GluN3A KD neurons, and we saw that the magnitude and time course of activation of p38 MAPK did not differ between these cultures (Fig. 5, A and B). Thus, GluN3A does not inhibit the activation of cytoplasmic p38 MAPK.
Figure 5.
Nuclear p38 MAPK phosphorylation is selectively enhanced upon NMDAR activation in GluN3A knockdown neurons. A, Western blotting of phosphorylated p38/MAPK (p-p38) in the cytoplasm fraction of hippocampal neurons infected with the indicated shRNA virus or paired control virus and then stimulated with TTX w/d for the indicated amounts of time prior to lysis. Actin is shown as a loading control. B, Quantification of two independent experiments as described in panel A. Band density in each lane was quantified using ImageJ. Expression was normalized to actin for each sample. p-p38 levels are reported as fold induction relative to the unstimulated (0 min) condition. Two-way ANOVA for time, F(5, 24) = 30.85, p < 0.0001; for virus, F(1, 24) = 6.8, p = 0.015; and for time–virus interaction, F(5, 24) = 1.6, p = 0.20. C, Western blotting of phosphorylated p38/MAPK (p-p38) in the nuclear fraction of hippocampal neurons after TTX w/d. D, Quantification of three independent experiments as described for panel C. Two-way ANOVA for time, F(5, 22) = 6.2, p = 0.001; for virus, F(1, 22) = 6.7, p = 0.017; and for time–virus interaction, F(5, 22) = 3.8, p = 0.012. For Grin3a KD, values differed from basal levels at 15 min (p = 0.031) and 30 min (p = 0.012). E, Representative images of hippocampal neurons (DIV7) immunostained with antibodies against p-p38 (red) and Hoechst nuclear dye (blue). Neurons were infected with the indicated lentiviruses and then stimulated with TTX w/d for 30 min. F, Quantification of the fraction of total p-p38 in the nucleus from (E). ANOVA for virus, F(2, 42) = 11.31, p = 0.0001. n = 15/virus. Ctrl versus KD, p < 0.001, KD versus rescue, p = 0.014. *, p < 0.05 for Grin3a KD versus Ctrl; ∧, p < 0.05 for rescue versus Grin3a KD.
However, many signaling proteins, including those in the p38 MAPK pathway, must undergo rapid nuclear translocation to induce gene transcription following extracellular stimulation (29, 30). To determine whether the nuclear activation of p38 MAPK is affected by the presence of GluN3A in NMDARs, we harvested nuclei from GluN3A KD and control neurons and ran them for Western blot analysis. In contrast to the activation of p38 MAPK in the cytoplasm, we saw TTX w/d-induced phosphorylation of p38 MAPK only in the nuclei of GluN3A KD neurons, with no significant increase in phosphorylation in the control-infected neurons (Fig. 5, C and D). To confirm the differential localization of phospho-p38 MAPK after TTX w/d in GluN3A KD and control neurons, we used immunostaining with the phospho-p38 MAPK antibody to localize the activated kinase in dissociated hippocampal neurons (Fig. 5E). Confocal images through control-infected cells show a robust TTX w/d-induced signal in the cytoplasm that is absent from the nucleus. In contrast, in GluN3A KD neurons the phospho-p38 MAPK signal fills the cell, including the nucleus. The quantification of the relative distributions of phospho-p38 MAPK in these two compartments demonstrates that there is a significant increase in the nuclear:cytoplasmic ratio of phospho-p38 MAPK immunostaining signal in neurons lacking GluN3A (Fig. 5F). This difference in the localization of activated p38 MAPK in the GluN3A knockdown neurons is due to the lack of GluN3A, because we were able to restore the control distribution by reexpressing shRNA-resistant GluN3A in the GluN3A KD neurons (Fig. 5, E and F). Taken together, these data suggest that GluN3A inhibits the subset of NMDAR-dependent transcription that is driven by MEF2C by blocking the activation of nuclear p38 MAPK. Our model is shown in Fig. S5.
Discussion
Our data describe a novel mechanism that regulates the specificity of NMDAR-dependent gene expression in the developing brain. We show that the NMDAR subunit GluN3A inhibits the ability of NMDARs to induce activity-dependent gene transcription. Because GluN3A is directly incorporated into NMDARs, it provides a means to selectively inhibit the ability of NMDARs to induce activity-regulated genes while leaving LVGCC-induced transcription intact. This is important, because many activity-regulated neuronal genes have pleiotropic functions that are differentially coupled to the activation of distinct upstream calcium signals. For example, although Bdnf is induced by both LVGCCs and NMDARs, LVGGC-induced BDNF expression is strongly linked to neuronal survival pathways (31), where NMDAR-induced BDNF is implicated in GABAergic synapse maturation (32). It is likely that the consequences of BDNF expression are defined by the context in which it is induced. Thus, by inhibiting the coupling of activity-regulated genes to NMDAR activation, GluN3A can limit NMDAR-dependent effects on synapse development without impairing the activation of these genes by stimuli, such as spontaneous action potentials, that promote neuronal survival and neurite outgrowth (33).
GluN3A is highly expressed only during a brief period of early postnatal forebrain development (3, 11), and the downregulation of GluN3A prior to the onset of the critical period for sensory-driven cortical plasticity is required for the maturation of excitatory synapse development (17). However, the mechanisms that allow GluN3A to exert this inhibitory effect on cortical plasticity have been unknown. Our evidence that GluN3A limits NMDAR-dependent Bdnf and Arc transcription suggests that the GluN3A-dependent inhibition of NMDAR-dependent gene transcription inhibits premature synaptic maturation by restricting the activation of plasticity-inducing gene products. In the visual cortex, Arc is required for the experience-dependent establishment of normal ocular dominance (34), and BDNF promotes the maturation of GABAergic inhibition in the developing cortex, which drives closure of the critical period (32). The developmental downregulation of GluN3A would permit the robust induction of this transcriptional program, promoting the onset of the later stages of critical period plasticity.
Although NMDARs pass current and contribute to synaptic potentials, it is primarily the ability of these channels to function as synaptic activity-regulated signaling receptors that underlies their unique roles in brain development and synaptic plasticity. All functional glutamate-sensing NMDARs are comprised of both GluN1 and GluN2 subunits, with some receptors also containing GluN3s, and the biophysical and biochemical properties of specific subtypes of these receptors vary based on their subunit composition. Notably, GluN3A can form excitatory glycine receptors with GluN1 in the absence of the glutamate-binding GluN2 subunits (2). The contribution of these receptors to stimulus-regulated transcription remains unknown, and it is possible that the loss of GluN1/GluN3A receptors contributes to the regulation of genes that show impaired TTX w/d induction in the GluN3A KD neurons (Fig. 2B). However, with respect to the potentiation of NMDAR-inducible gene expression in GluN3A KD neurons, we suggest that it is more likely that GluN3A is modulating calcium influx (Fig. 1K) or protein–protein interactions to influence the activation of transcriptional signaling cascades.
GluN2A- and GluN2B-containing NMDARs have been particularly highly studied, given the evidence linking these two subtypes of receptors to distinct biological functions (7). The distinct signaling consequences of activating these two classes of NMDARs have been shown to depend on the ability of their intracellular C-terminal tails to differentially recruit signaling complexes to the NMDARs (9). Similar to the GluN2s, the long C-tail of GluN3A provides unique NMDAR docking sites for intracellular signaling proteins that can influence the functional impact of NMDAR activation. For example, GluN3A-dependent association with the endocytic protein Pacsin1 (Syndapin1) enhances the membrane trafficking of NMDARs (35), and the association of GluN3A with the phosphatase PP2A promotes the dephosphorylation of the GluN1 subunit at Ser897 (36). In addition to effects on NMDAR itself, protein associations with the GluN3A C-terminal tail regulate local synaptic signaling in dendritic spines (37). Our data further indicate that the incorporation of GluN3A can modulate the ability of NMDARs to signal to the nucleus. Interestingly, whereas GluN2A and GluN2B differentially regulate NMDAR-dependent activation of the transcription factor CREB, we find that GluN3A selectively inhibits the activation of MEF2 transcription factors. Thus, in addition to controlling the likelihood of transcriptional activation, the differential use of NMDAR subunits can confer specificity on the set of downstream transcription factors that are activated by synaptic stimulation.
Although the activation of any single activity-regulated transcription factor is sufficient to induce transcription of a reporter gene (e.g. Fig. 3, A and B), the regulation of most neuronal immediate-early genes (IEGs) is under the control of multiple activity-regulated transcription factors (38). The presence of multiple transcription factor binding sites in the promoters and enhancers of neuronal activity-dependent genes may allow for specificity in the coupling of subsets of activity-responsive genes to distinct sources of upstream activation (39). Consistent with this model, we find that GluN3A selectively inhibits the component of NMDAR-induced transcription that depends on MEF2C while leaving LVGCC- and CREB-dependent transcription intact. The selective regulation of these pathways results in the activation of MEF2C/CREB coregulated genes, like Bdnf and Arc, being driven to two different levels, either high levels of transcription through the coordinate activation of MEF2C and CREB or a lower level of activation, presumably mediated by CREB in the absence of MEF2C activation. Importantly, substantial data support the hypothesis that the levels to which these stimulus-regulated genes are induced is important for their function in the brain. The spatial and temporal induction of BDNF is particularly tightly regulated in the brain, and even mild increases or decreases in BDNF levels are associated with developmental and functional neuronal abnormalities (20, 40–42). More recently, we have shown that the modulation of the bursting properties of Fos transcription is sufficient to change membrane properties of hippocampal neurons, demonstrating a direct link between the magnitude of activity-inducible gene expression and neuronal function (43).
One of the most interesting questions raised by this study is how the incorporation of GluN3A into NMDARs at the cell surface leads to the regulation of gene transcription in the nucleus. Our data indicate that a key step in this process is the ability of GluN3A to inhibit the activation of nuclear p38 MAPK. p38 MAPK is rapidly activated by glutamate-induced calcium influx through NMDARs in neurons (44), and in this context p38 has attracted interest for its involvement in the recycling of AMPA-type glutamate receptors that underlies activity-dependent changes in synaptic strength (45). NMDARs activate p38 MAPK via the regulation of the small GTPases Ras, Rac1, and/or Rap1, and the differential association of GTPase exchange factors and GTPase activating proteins with NMDARs that have distinct GluN2 subunit composition has been suggested to contribute to the ability of NMDARs to induce long-term potentiation (LTP) versus long-term depression (LTD) (46). Like the GluN2 subunits, GluN3A regulates the NMDAR-dependent activation of small GTPases by binding directly to the small GTPase Rheb and indirectly inhibiting the activation of Rac1 (37). However, our data indicate that GluN3A has no effect on the time course or magnitude of p38 MAPK activation; instead, the primary consequence of GluN3A is to restrict the subcellular distribution of activated p38 MAPK to the cytoplasm, whereas NMDARs lacking GluN3A can also drive the appearance of activated p38 MAPK in neuronal nuclei.
Nuclear translocation of p38 requires the phosphorylation-dependent association of p38 with β-like importins in the cytoplasm, followed by active transport through the nuclear pore that is mediated by the small GTPase Ran (29). The nuclear export of p38 MAPK is also regulated, either as a consequence of dephosphorylation in the nucleus (47) or via the physical association of p38 MAPK with a downstream substrate bearing a nuclear export signal (48). One mechanism that controls the localization of phospho-p38 MAPK is the mechanism of its upstream activation. In addition to phosphorylation by the canonical upstream MAPK kinases MKK3/6, p38 MAPK can be activated by autophosphorylation in complex with the TAB-1 scaffold protein (49). In cardiomyocytes, when TAB-1 binds to and induces the autophosphorylation of p38, it prevents the nuclear translocation of the activated kinase and antagonizes the activation of gene transcription driven by MKK-dependent p38 activation (50). Future investigation of the mechanisms of NMDAR-induced p38 MAPK activation in neurons may expand our understanding of the regulatory processes that control this important kinase signaling cascade and regulate its impact on activity-dependent brain development.
Materials and methods
Plasmids
Two independent shRNAs were used to knock down rat Grin3a, and each was paired with a vector-matched control. Grin3a shRNA1 (TRCN0000100220; 5′-GCTCCATGACAAGTGGTACAA-3′) was purchased from Thermo Scientific, and the empty pLKO.1 vector was used as Ctrl1. Grin3a shRNA2 (5′-GTATCCGGCAGATATTTGAAA-3′) was cloned into the pLLx3.8 vector, and a scrambled version of the shRNA2 sequence (5′-GCCTGCAGTATGACTCAGTAA-3′) in pLLx3.8 was used as Ctrl2. The shRNA-resistant GluN3A expression plasmid was constructed by using PCR to introduce silent mutations indicated by the lower case bolded letters (GCTaCATGACAAGTtGTACAA) into the region of rat Grin3a targeted by Grin3a shRNA1 and then placing the Grin3a coding sequence under the control of the ubiquitin promoter in the lentiviral expression vector pFUIGW (51). The Gal4 DNA binding domain fusions with human MEF2Cα1β and MEF2Cα1βγ were described previously (25). The TT293/300AA and S387A mutations of Gal4-MEF2Cα1β were generated by site-directed mutagenesis PCR. The viral expression plasmid for the rescue of MEF2 expression in neurons infected with the shRNA targeting rat Mef2c was generated by cloning the full coding sequence of human MEF2Cα1β, which is not targeted by the rat Mef2c shRNA used, into the vector pFUIGW. We purchased the CRE luciferase reporter (Agilent, 219075) and TK-renilla luciferase plasmid (Promega, E2231). The following plasmids were reported previously: MRE luciferase reporter plasmid (27), the UAS luciferase reporter plasmid (52), and shRNAs targeting rat Mef2a, Mef2c, and Mef2d in the vector pLKO.1 (TRCN0000095959, TRCN0000012068, and TRCN0000085268, respectively) (Thermo Scientific) (23).
Antibodies
Primary antibodies used in this study for Western blotting were mouse anti-Actin, 1:5000 (Millipore, catalog number MAB1501, RRID:AB_2223041); rabbit anti-GluN3A, 1:1000 (Millipore, catalog number 07-356, RRID:AB_2112620); mouse anti-Transferrin Receptor, 1:2000 (Thermo Fisher Scientific, catalog number 13-6800, RRID:AB_2533029); rabbit anti-phospho-Thr180/Tyr182 p38 MAPK, 1:1000 (Cell Signaling Technology, catalog number 4511, RRID:AB_2139682); and rabbit-anti-MEF2C 1:1000 (Abcam, catalog number ab64644, RRID:AB_2142861). The primary antibody used in this study for immunocytochemistry was mouse anti-phospho-Tyr182 p38 MAPK, 1:50 (Santa Cruz Biotechnology, catalog number sc-7973, RRID:AB_670359).
Dissociated neuron cultures
Neuron-enriched cultures were generated from the hippocampus of male and female embryonic day 18.5 CD IGS rat embryos (Charles River Laboratories) and cultured as previously described (40, 52). The activation of NMDAR-dependent transcription by withdrawal from tetrodotoxin (TTX w/d) (18, 19) was done by treating neurons starting on the fifth day in vitro (DIV5) for 48 h with 1 μM TTX (Tocris) prior either to harvesting cells (for the control condition) or washing out the TTX with neurobasal medium with B27 supplements (Invitrogen) for 6 h, unless otherwise indicated in the text, on DIV7. Isotonic membrane depolarization with 55 mm extracellular KCl was done as previously described for 6 h on DIV7 (23). Pharmacological blockers were added 2 min prior to TTX w/d or KCl addition and maintained throughout the period of stimulation. TTX (1 μm), FK506 (1 μm), cyclosporin A (1 μm), SB203580 (2.5 μm or 25 μm), and CNQX (10 μm) were purchased from Tocris. Nimodipine (5 μm) was purchased from Sigma, and APV (100 μm) was purchased from Fisher Scientific. All experiments were conducted in accordance with an animal protocol approved by the Duke University Institutional Animal Care and Use Committee.
Lentiviral infection
For viral infection of neurons, shRNA or viral expression constructs were packaged as lentiviruses in HEK 293T cells by following standard procedures. Titers of concentrated viruses were determined on cultured neurons, and rat hippocampal neurons were infected for 6 h on DIV1 or DIV2 at a multiplicity of infection of 1 in β-mercaptoethanol medium (Sigma) with 0.4 µg/ml added Polybrene (Sigma).
Multi-electrode array (MEA) recordings
Rat hippocampal neurons were plated in a 48-well MEA plate (Lumos 48, Axion Biosystems) coated with poly-d-lysine and laminin at a density of 60,000 cells/well. Extracellular recordings were performed at 37 °C with 5% CO2 using a Maestro MEA system and AxIS software (Axion Biosystems). Data were acquired at 12.5 kHz and filtered with a Butterworth bandpass filter at 200 Hz–3kHz. A spike detector was used to detect action potentials with a 6× standard deviation. TTX w/d was conducted as described above. Recordings were conducted immediately prior to TTX washout, 30 min after TTX washes, and immediately after the addition of 10 μm CNQX (Tocris, 1045) and 100 μm APV (Fisher Scientific, 01-051-0). Data were displayed and analyzed using Axion Biosystem's Neural Metrics Tool.
Quantitative PCR
RNA was harvested on DIV7 following 90 min or 6 h of KCl-mediated membrane depolarization or TTX w/d as described above. RNA was harvested using the Absolutely RNA Miniprep kit (Agilent), and cDNA was synthesized by Superscript II (Invitrogen). Quantitative SYBR green PCR was performed on an ABI 7300 real-time PCR machine (Applied Biosystems) using intron-spanning primers (IDT) listed in Table S1. Data were normalized to the expression of the housekeeping gene Gapdh to control for sample size and processing. In some cases, as described above, we reported mRNA levels as relative expression or relative fold induction in neurons infected with any single independent shRNA compared with its paired control shRNA (Ctrl1 or Ctrl2).
Western blotting
Cells were homogenized in homogenization buffer (320 mm sucrose, 10 mm HEPES, pH 7.4, 2 mm EDTA, 1 mm DTT, and protease inhibitors). The nuclear pellet was pelleted by centrifuging at 1500 × g for 15 min. Cytosolic and membrane fractions were separated from the supernatant by centrifuging at 200,000 × g for 20 min. Membrane, cytoplasmic, and nuclear extracts were run for SDS-PAGE and transferred to nitrocellulose for Western blotting by following standard procedures. Bands were visualized with fluorescent secondary antibodies (Biotium) using the Odyssey imaging system (LI-COR Bioscience) and quantified using ImageJ. Actin was used as a loading control.
Bdnf ELISA
Two-site BDNF ELISA was performed as previously described, using acid extraction of BDNF from cultured neurons (40, 42). Total protein concentration in the lysate was measured by a BCA protein assay kit (Pierce), and BDNF protein concentrations were measured by the BDNF Emax ImmunoAssay System (Promega) or the mature BDNF rapid ELISA kit (Biosensis). Neurons were infected with lentiviral constructs and treated with TTX, as described for RNA studies, and then harvested for ELISA 6 h after TTX w/d.
Two-photon glutamate uncaging and calcium imaging
Hippocampal slices were prepared from postnatal day 5–7 rats and cut into 350-μm sections using a McIlwain tissue chopper. Slices were plated on tissue culture inserts (Millicell) fed by tissue medium (for 2.5 liters: 20.95 g MEM, 17.9 g HEPES, 1.1 g NaHCO3, 5.8 g d-glucose, 120 μl 25% ascorbic acid, 12.5 ml l-glutamine, 2.5 ml insulin, 500 ml horse serum, 5 ml 1 M MgSO4, 2.5 ml 1 M CaCl2) and incubated at 35 °C in 3% CO2. After 1–2 weeks in culture, CA1 pyramidal neurons were transfected with the control, Grin3a shRNA KD, or KD plus GluN3A rescue plasmids, as well as the cell fill mCherry and the calcium indicator GCaMP3, with ballistic gene transfer using gold beads (8–12 mg) coated with plasmids containing 30 μg of total cDNA. Cells were imaged 2–5 days after transfection. Two-photon imaging was performed using a Ti-sapphire laser (MaiTai, Spectraphysics) tuned to a wavelength of 920 nm, allowing the simultaneous excitation of GCaMP3 and mCh. Imaging experiments were performed at 8-Hz collection speed (frames/second). All samples were imaged using <2-mW laser power measured at the objective. Fluorescence emission was collected using an immersion objective (60×, numerical aperture of 0.9, Olympus), divided with a dichroic mirror (565 nm), and detected with two separate photoelectron multiplier tubes (PMTs) placed downstream of two wavelength filters (Chroma, HQ510-2p to select for green and HQ620/90-2p to select for red). The green channel was fitted with a PMT having a low-transfer-time spread (H7422-40p; Hamamatsu), while the red channel was fitted with a wide-aperture PMT (R3896; Hamamatsu). The green and red channel signals were acquired using a data acquisition board (PCI-6110) controlled with ScanImage software. A second Ti-sapphire laser tuned at a wavelength of 720 nm was used to uncage 4-methoxy-7-nitroindolinyl-caged-l-glutamate (MNI-caged glutamate) in extracellular solution with a train of 4–6-ms, 4–5-mW pulses (5 times at 0.5 Hz) near a spine of interest. Experiments were performed in Mg2+ free artificial cerebral spinal fluid (ACSF; 127 mm NaCl, 2.5 mm KCl, 4 mm CaCl2, 25 mm NaHCO3, 1.25 mm NaH2PO4, and 25 mm glucose) containing 1 μm TTX and 4 mm MNI-caged l-glutamate aerated with 95% O2 and 5% CO2 at 30 °C, as described previously. Anywhere from 1–10 spines were stimulated per cell.
Calcium trace analysis
GCaMP3 intensity was calculated as the background-subtracted integrated fluorescence intensity over a region of interest around the dendritic spine head (fluorescence, F). Changes in the intensity of GCaMP3 were calculated as ΔF/F0, where F0 is the average fluorescence intensity before stimulation. To account for potential drift in the focal plane during imaging, the GCaMP3 signal was normalized to the red channel signal, smoothed by a factor of 16 frames (2 s). Uncaging-triggered averages were calculated by first averaging the GCaMP3 traces from all spines from a particular condition together. Stimulation periods then were extracted according to uncaging onset timing, resulting in 2-s blocks corresponding to the time point immediately before uncaging through the frame immediately before the next stimulus for each of the five stimuli delivered. The fluorescence intensity value at the beginning of each of the five 2-s blocks was subtracted from that block to account for the incomplete decay of GCaMP3. Blocks were then averaged together to generate an average response to the uncaging stimulus, here referred to as the uncaging-triggered average. As such, error in this measurement is indicative of the variability between the successive uncaging-evoked GCaMP3 responses. All analyses were performed using custom MATLAB software.
RNA-Seq
Rat hippocampal neurons were infected with the Ctrl1 or Grin3a shRNA1 lentivirus on DIV1, treated with 1 μm TTX on DIV5, and then, on DIV7, cells were either harvested for RNA (Ctrl TTX and KD TTX conditions) or TTX was withdrawn for 6 h prior to RNA harvest (Ctrl TTX w/d and KD TTX w/d). Three independent triplicate replicate samples were obtained for each condition. RNA was submitted to the Duke University sequencing core facility for poly(A) selection, library generation, and single-end 50-bp sequencing on an Illumina HiSeq2500. Following quality score-based trimming and adaptor filtering, reads were aligned to the UCSC rn5 reference transcriptome using Tophat2.0. Cuffdiff was used for pairwise tests of differential expression between normalized gene read counts of FPKM with a default significance threshold of an FDR of <0.05 used for all analyses. Heat maps and volcano plots were generated using the R biostatistics packages. We used the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (53) to find enriched biological process gene ontologies and the Molecular Signatures Database v6.2 (MSigDB) of Gene Set Enrichment Analysis (54) to search a database of transcription factor binding sites (55) with our gene sets. RNA-seq data were deposited at GEO with the accession number GSE133917.
Neuronal transfection for luciferase assays
Neuron cultures were transfected with calcium phosphate on DIV3–5 (52). Cotransfection of pTK-renilla luciferase (Promega) was used to control for transfection efficiency and sample handling. For the MRE-luc and CRE-luc reporters, luciferase reporter plasmids were transfected into cultured embryonic rat hippocampal neurons that had previously been infected with either Grin3a shRNA1 or Ctrl1 lentivirus. Lysates were harvested on DIV9–10 after 0, 4, 6, 8, 14, or 22 h of TTX w/d. For the UAS-luc reporter, luciferase reporter and Gal4 fusions were cotransfected with either Grin3a shRNA1 or Ctrl1 plasmid. Lysates were harvested on DIV7 plus 6 h TTX w/d as described above. Luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega).
Immunofluorescence
Embryonic rat hippocampal neurons were cultured on PDL/laminin-coated glass coverslips (Bellco) and fixed in 4% paraformaldehyde at room temperature for 10 min. Neurons were blocked in 10% normal goat serum and permeabilized in 0.3% Triton X-100 prior to antibody incubation. Coverslips were incubated in primary antibodies overnight at 4 °C. Secondary antibodies (1:500) were incubated at room temperature for 1 h. Hoechst dye (0.1 µg/ml, Sigma) was used to label nuclei. Images were captured on a Leica SP8 confocal microscope with a 1-μm-thick optical section. Nuclear and cytoplasmic total pixel intensities were quantified by the ImageJ macro Intensity Ratio Nuclei Cytoplasm Tool (RRID:SCR_018573). Briefly, this plugin first identifies cells based on their diameter and threshold signal above background. Once set, these parameters are held constant for all images in the set. ∼300 cells in 15 images per treatment group were quantified.
Statistical analyses
Unless otherwise indicated, all data presented are the averages of at least three biological replicates from each of at least two independent experiments. Also, unless otherwise indicated, data were analyzed in GraphPad Prism by one- or two-way analysis of variance (ANOVA) as appropriate to the data structure, followed by post hoc comparisons with Tukey's or Sidak's multiple-comparison tests. For the calcium imaging data, multiple comparisons between the control, shRNA, and rescue conditions were performed using ANOVA followed by Fisher's least significant difference test. p < 0.05 was considered significant. Bar and line graphs show mean values, and all error bars show standard deviation.
Data availability
Raw data used to generate the figures in this paper are stored on a secure Duke University School of Medicine server and are available upon request to the corresponding author. RNA-seq data were deposited at GEO under accession number GSE133917.
Supplementary Material
This article contains supporting information.
Author contributions—L.-F. C., M. R. L., and A. E. W. conceptualization; L.-F. C., M. R. L., and F. L. data curation; L.-F. C., M. R. L., F. L., M. V. G., N. G. H., A. B. W., A. N., R. Y., and A. E. W. formal analysis; L.-F. C., M. R. L., F. L., M. V. G., N. G. H., A. B. W., and A. N. investigation; L.-F. C., M. R. L., and F. L. methodology; L.-F. C., M. R. L., and A. E. W. writing-original draft; L.-F. C., M. R. L., F. L., and A. E. W. writing-review and editing; R. Y. and A. E. W. supervision; A. E. W. validation; A. E. W. project administration.
Funding and additional information—This work was supported by the Ruth K. and Shepherd Broad Foundation and NIH grants R01NS098804 (to L. -F. C., F. L., M. V. G., A. B. W., A. N., and A. E. W.), DP1NS096787 (to N. G. H. and R. Y.) and R01MH080047 (to N. G. H. and R. Y.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- NMDARs
- N-methyl-d-aspartate type glutamate receptors
- LVGCCs
- L-type voltage gated calcium channels
- FPKM
- fragments per kilobase of exon per million fragments mapped
- p38 MAPK
- p38 mitogen-activated protein kinase
- FDR
- false discovery rate
- w/d
- withdrawal
- KD
- knockdown
- DIV5
- fifth day in vitro
- ANOVA
- analysis of variance
- APV
- amino-5-phosphonovaleric acid
- TTX
- tetrodotoxin
- Nim
- nimodipine
- KCl
- potassium chloride
- MRE
- Mef2 response element
- CRE
- Creb response element
- MEF2
- myocyte enhancer factor 2
- shRNA
- short hairpin RNA
- GluN
- glutamate receptor ionotropic NMDA.
References
- 1. Yap E. L., and Greenberg M. E. (2018) Activity-regulated transcription: bridging the gap between neural activity and behavior. Neuron 100, 330–348 10.1016/j.neuron.2018.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Paoletti P., Bellone C., and Zhou Q. (2013) NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci. 14, 383–400 10.1038/nrn3504 [DOI] [PubMed] [Google Scholar]
- 3. Sasaki Y. F., Rothe T., Premkumar L. S., Das S., Cui J., Talantova M. V., Wong H. K., Gong X., Chan S. F., Zhang D., Nakanishi N., Sucher N. J., and Lipton S. A. (2002) Characterization and comparison of the NR3A subunit of the NMDA receptor in recombinant systems and primary cortical neurons. J. Neurophysiol. 87, 2052–2063 10.1152/jn.00531.2001 [DOI] [PubMed] [Google Scholar]
- 4. Lau C. G., and Zukin R. S. (2007) NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat. Rev. Neurosci. 8, 413–426 10.1038/nrn2153 [DOI] [PubMed] [Google Scholar]
- 5. Sheng M., Cummings J., Roldan L. A., Jan Y. N., and Jan L. Y. (1994) Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature 368, 144–147 10.1038/368144a0 [DOI] [PubMed] [Google Scholar]
- 6. Sala C., Rudolph-Correia S., and Sheng M. (2000) Developmentally regulated NMDA receptor-dependent dephosphorylation of cAMP response element-binding protein (CREB) in hippocampal neurons. J. Neurosci. 20, 3529–3536 10.1523/JNEUROSCI.20-10-03529.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wyllie D. J., Livesey M. R., and Hardingham G. E. (2013) Influence of GluN2 subunit identity on NMDA receptor function. Neuropharmacology 74, 4–17 10.1016/j.neuropharm.2013.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hardingham G. E., Fukunaga Y., and Bading H. (2002) Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat. Neurosci. 5, 405–414 10.1038/nn835 [DOI] [PubMed] [Google Scholar]
- 9. Martel M. A., Ryan T. J., Bell K. F., Fowler J. H., McMahon A., Al-Mubarak B., Komiyama N. H., Horsburgh K., Kind P. C., Grant S. G., Wyllie D. J., and Hardingham G. E. (2012) The subtype of GluN2 C-terminal domain determines the response to excitotoxic insults. Neuron 74, 543–556 10.1016/j.neuron.2012.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sucher N. J., Akbarian S., Chi C. L., Leclerc C. L., Awobuluyi M., Deitcher D. L., Wu M. K., Yuan J. P., Jones E. G., and Lipton S. A. (1995) Developmental and regional expression pattern of a novel NMDA receptor-like subunit (NMDAR-L) in the rodent brain. J. Neurosci. 15, 6509–6520 10.1523/JNEUROSCI.15-10-06509.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Al-Hallaq R. A., Jarabek B. R., Fu Z., Vicini S., Wolfe B. B., and Yasuda R. P. (2002) Association of NR3A with the N-methyl-D-aspartate receptor NR1 and NR2 subunits. Mol. Pharmacol. 62, 1119–1127 10.1124/mol.62.5.1119 [DOI] [PubMed] [Google Scholar]
- 12. Pilli J., and Kumar S. S. (2012) Triheteromeric N-methyl-D-aspartate receptors differentiate synaptic inputs onto pyramidal neurons in somatosensory cortex: involvement of the GluN3A subunit. Neuroscience 222, 75–88 10.1016/j.neuroscience.2012.07.020 [DOI] [PubMed] [Google Scholar]
- 13. Ciabarra A. M., Sullivan J. M., Gahn L. G., Pecht G., Heinemann S., and Sevarino K. A. (1995) Cloning and characterization of chi-1: a developmentally regulated member of a novel class of the ionotropic glutamate receptor family. J. Neurosci. 15, 6498–6508 10.1523/JNEUROSCI.15-10-06498.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Larsen R. S., Corlew R. J., Henson M. A., Roberts A. C., Mishina M., Watanabe M., Lipton S. A., Nakanishi N., Perez-Otano I., Weinberg R. J., and Philpot B. D. (2011) NR3A-containing NMDARs promote neurotransmitter release and spike timing-dependent plasticity. Nat. Neurosci. 14, 338–344 10.1038/nn.2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Das S., Sasaki Y. F., Rothe T., Premkumar L. S., Takasu M., Crandall J. E., Dikkes P., Conner D. A., Rayudu P. V., Cheung W., Chen H. S., Lipton S. A., and Nakanishi N. (1998) Increased NMDA current and spine density in mice lacking the NMDA receptor subunit NR3A. Nature 393, 377–381 10.1038/30748 [DOI] [PubMed] [Google Scholar]
- 16. Henson M. A., Larsen R. S., Lawson S. N., Perez-Otano I., Nakanishi N., Lipton S. A., and Philpot B. D. (2012) Genetic deletion of NR3A accelerates glutamatergic synapse maturation. PLoS ONE 7, e42327 10.1371/journal.pone.0042327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roberts A. C., Diez-Garcia J., Rodriguiz R. M., Lopez I. P., Lujan R., Martinez-Turrillas R., Pico E., Henson M. A., Bernardo D. R., Jarrett T. M., Clendeninn D. J., Lopez-Mascaraque L., Feng G., Lo D. C., Wesseling J. F., et al. (2009) Downregulation of NR3A-containing NMDARs is required for synapse maturation and memory consolidation. Neuron 63, 342–356 10.1016/j.neuron.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lyons M. R., Chen L. F., Deng J. V., Finn C., Pfenning A. R., Sabhlok A., Wilson K. M., and West A. E. (2016) The transcription factor calcium-response factor limits NMDA receptor-dependent transcription in the developing brain. J. Neurochem. 137, 164–176 10.1111/jnc.13556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saha R. N., Wissink E. M., Bailey E. R., Zhao M., Fargo D. C., Hwang J. Y., Daigle K. R., Fenn J. D., Adelman K., and Dudek S. M. (2011) Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat. Neurosci. 14, 848–856 10.1038/nn.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. West A. E., Pruunsild P., and Timmusk T. (2014) Neurotrophins: transcription and translation. Handb. Exp. Pharmacol. 220, 67–100. 10.1007/978-3-642-45106-5_4 [DOI] [PubMed] [Google Scholar]
- 21. Flavell S. W., Kim T. K., Gray J. M., Harmin D. A., Hemberg M., Hong E. J., Markenscoff-Papadimitriou E., Bear D. M., and Greenberg M. E. (2008) Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60, 1022–1038 10.1016/j.neuron.2008.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wild A. R., Sinnen B. L., Dittmer P. J., Kennedy M. J., Sather W. A., and Dell'Acqua M. L. (2019) Synapse-to-nucleus communication through NFAT is mediated by L-type Ca(2+) channel Ca(2+) spike propagation to the soma. Cell Rep. 26, 3537–3550 10.1016/j.celrep.2019.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lyons M. R., Schwarz C. M., and West A. E. (2012) Members of the myocyte enhancer factor 2 transcription factor family differentially regulate BDNF transcription in response to neuronal depolarization. J. Neurosci. 32, 12780–12785 10.1523/JNEUROSCI.0534-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kawashima T., Okuno H., Nonaka M., Adachi-Morishima A., Kyo N., Okamura M., Takemoto-Kimura S., Worley P. F., and Bito H. (2009) Synaptic activity-responsive element in the Arc/Arg3.1 promoter essential for synapse-to-nucleus signaling in activated neurons. Proc. Natl. Acad. Sci. U S A 106, 316–321 10.1073/pnas.0806518106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhu B., and Gulick T. (2004) Phosphorylation and alternative pre-mRNA splicing converge to regulate myocyte enhancer factor 2C activity. Mol. Cell. Biol. 24, 8264–8275 10.1128/MCB.24.18.8264-8275.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McKinsey T. A., Zhang C. L., and Olson E. N. (2002) MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27, 40–47 10.1016/S0968-0004(01)02031-X [DOI] [PubMed] [Google Scholar]
- 27. Flavell S. W., Cowan C. W., Kim T. K., Greer P. L., Lin Y., Paradis S., Griffith E. C., Hu L. S., Chen C., and Greenberg M. E. (2006) Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311, 1008–1012 10.1126/science.1122511 [DOI] [PubMed] [Google Scholar]
- 28. Han J., Jiang Y., Li Z., Kravchenko V. V., and Ulevitch R. J. (1997) Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature 386, 296–299 10.1038/386296a0 [DOI] [PubMed] [Google Scholar]
- 29. Zehorai E., and Seger R. (2014) Beta-like importins mediate the nuclear translocation of mitogen-activated protein kinases. Mol. Cell. Biol. 34, 259–270 10.1128/MCB.00799-13 This article is marked in PubMed as a Retraction!] [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30. Plotnikov A., Zehorai E., Procaccia S., and Seger R. (2011) The MAPK cascades: signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim. Biophys. Acta 1813, 1619–1633 10.1016/j.bbamcr.2010.12.012 [DOI] [PubMed] [Google Scholar]
- 31. Ghosh A., Carnahan J., and Greenberg M. E. (1994) Requirement for BDNF in activity-dependent survival of cortical neurons. Science 263, 1618–1623 10.1126/science.7907431 [DOI] [PubMed] [Google Scholar]
- 32. Huang Z. J., Kirkwood A., Pizzorusso T., Porciatti V., Morales B., Bear M. F., Maffei L., and Tonegawa S. (1999) BDNF regulates the maturation of inhibition and the critical period of plasticity in mouse visual cortex. Cell 98, 739–755 10.1016/S0092-8674(00)81509-3 [DOI] [PubMed] [Google Scholar]
- 33. West A. E., and Greenberg M. E. (2011) Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb. Perspect. Biol. 3, a005744 10.1101/cshperspect.a005744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCurry C. L., Shepherd J. D., Tropea D., Wang K. H., Bear M. F., and Sur M. (2010) Loss of Arc renders the visual cortex impervious to the effects of sensory experience or deprivation. Nat. Neurosci. 13, 450–457 10.1038/nn.2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Perez-Otano I., Lujan R., Tavalin S. J., Plomann M., Modregger J., Liu X. B., Jones E. G., Heinemann S. F., Lo D. C., and Ehlers M. D. (2006) Endocytosis and synaptic removal of NR3A-containing NMDA receptors by PACSIN1/syndapin1. Nat. Neurosci. 9, 611–621 10.1038/nn1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chan S. F., and Sucher N. J. (2001) An NMDA receptor signaling complex with protein phosphatase 2A. J. Neurosci. 21, 7985–7992 10.1523/JNEUROSCI.21-20-07985.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fiuza M., Gonzalez-Gonzalez I., and Perez-Otano I. (2013) GluN3A expression restricts spine maturation via inhibition of GIT1/Rac1 signaling. Proc. Natl. Acad. Sci. U S A 110, 20807–20812 10.1073/pnas.1312211110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lyons M. R., and West A. E. (2011) Mechanisms of specificity in neuronal activity-regulated gene transcription. Prog. Neurobiol. 94, 259–295 10.1016/j.pneurobio.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Joo J. Y., Schaukowitch K., Farbiak L., Kilaru G., and Kim T. K. (2016) Stimulus-specific combinatorial functionality of neuronal c-fos enhancers. Nat. Neurosci. 19, 75–83 10.1038/nn.4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McDowell K. A., Hutchinson A. N., Wong-Goodrich S. J., Presby M. M., Su D., Rodriguiz R. M., Law K. C., Williams C. L., Wetsel W. C., and West A. E. (2010) Reduced cortical BDNF expression and aberrant memory in Carf knock-out mice. J. Neurosci. 30, 7453–7465 10.1523/JNEUROSCI.3997-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen Z. Y., Jing D., Bath K. G., Ieraci A., Khan T., Siao C. J., Herrera D. G., Toth M., Yang C., McEwen B. S., Hempstead B. L., and Lee F. S. (2006) Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science 314, 140–143 10.1126/science.1129663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hong E. J., McCord A. E., and Greenberg M. E. (2008) A biological function for the neuronal activity-dependent component of Bdnf transcription in the development of cortical inhibition. Neuron 60, 610–624 10.1016/j.neuron.2008.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen L. F., Lin Y. T., Gallegos D. A., Hazlett M. F., Gomez-Schiavon M., Yang M. G., Kalmeta B., Zhou A. S., Holtzman L., Gersbach C. A., Grandl J., Buchler N. E., and West A. E. (2019) Enhancer histone acetylation modulates transcriptional bursting dynamics of neuronal activity-inducible genes. Cell Rep. 26, 1174–1188 10.1016/j.celrep.2019.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kawasaki H., Morooka T., Shimohama S., Kimura J., Hirano T., Gotoh Y., and Nishida E. (1997) Activation and involvement of p38 mitogen-activated protein kinase in glutamate-induced apoptosis in rat cerebellar granule cells. J. Biol. Chem. 272, 18518–18521 10.1074/jbc.272.30.18518 [DOI] [PubMed] [Google Scholar]
- 45. Thomas G. M., and Huganir R. L. (2004) MAPK cascade signalling and synaptic plasticity. Nat. Rev. Neurosci. 5, 173–183 10.1038/nrn1346 [DOI] [PubMed] [Google Scholar]
- 46. Zhu Y., Pak D., Qin Y., McCormack S. G., Kim M. J., Baumgart J. P., Velamoor V., Auberson Y. P., Osten P., van Aelst L., Sheng M., and Zhu J. J. (2005) Rap2-JNK removes synaptic AMPA receptors during depotentiation. Neuron 46, 905–916 10.1016/j.neuron.2005.04.037 [DOI] [PubMed] [Google Scholar]
- 47. Gong X., Ming X., Deng P., and Jiang Y. (2010) Mechanisms regulating the nuclear translocation of p38 MAP kinase. J. Cell. Biochem. 110, 1420–1429 10.1002/jcb.22675 [DOI] [PubMed] [Google Scholar]
- 48. Ben-Levy R., Hooper S., Wilson R., Paterson H. F., and Marshall C. J. (1998) Nuclear export of the stress-activated protein kinase p38 mediated by its substrate MAPKAP kinase-2. Curr. Biol. 8, 1049–1057 10.1016/S0960-9822(98)70442-7 [DOI] [PubMed] [Google Scholar]
- 49. Ge B., Gram H., Di Padova F., Huang B., New L., Ulevitch R. J., Luo Y., and Han J. (2002) MAPKK-independent activation of p38alpha mediated by TAB1-dependent autophosphorylation of p38alpha. Science 295, 1291–1294 10.1126/science.1067289 [DOI] [PubMed] [Google Scholar]
- 50. Lu G., Kang Y. J., Han J., Herschman H. R., Stefani E., and Wang Y. (2006) TAB-1 modulates intracellular localization of p38 MAP kinase and downstream signaling. J. Biol. Chem. 281, 6087–6095 10.1074/jbc.M507610200 [DOI] [PubMed] [Google Scholar]
- 51. Lois C., Hong E. J., Pease S., Brown E. J., and Baltimore D. (2002) Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295, 868–872 10.1126/science.1067081 [DOI] [PubMed] [Google Scholar]
- 52. Tao X., West A. E., Chen W. G., Corfas G., and Greenberg M. E. (2002) A calcium-responsive transcription factor, CaRF, that regulates neuronal activity-dependent expression of BDNF. Neuron 33, 383–395 10.1016/S0896-6273(01)00561-X [DOI] [PubMed] [Google Scholar]
- 53. Huang D. W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols 4, 44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 54. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., and Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A 102, 15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xie X., Lu J., Kulbokas E. J., Golub T. R., Mootha V., Lindblad-Toh K., Lander E. S., and Kellis M. (2005) Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature 434, 338–345 10.1038/nature03441 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data used to generate the figures in this paper are stored on a secure Duke University School of Medicine server and are available upon request to the corresponding author. RNA-seq data were deposited at GEO under accession number GSE133917.