Abstract
Estrogen receptor α (ERα) modulates gene expression by interacting with chromatin regions that are frequently distal from the promoters of estrogen-regulated genes. Active chromatin-enriched “super-enhancer” (SE) regions, mainly observed in in vitro culture systems, often control production of key cell type–determining transcription factors. Here, we defined super-enhancers that bind to ERα in vivo within hormone-responsive uterine tissue in mice. We found that SEs are already formed prior to estrogen exposure at the onset of puberty. The genes at SEs encoded critical developmental factors, including retinoic acid receptor α (RARA) and homeobox D (HOXD). Using high-throughput chromosome conformation capture (Hi-C) along with DNA sequence analysis, we demonstrate that most SEs are located at a chromatin loop end and that most uterine genes in loop ends associated with these SEs are regulated by estrogen. Although the SEs were formed before puberty, SE-associated genes acquired optimal ERα-dependent expression after reproductive maturity, indicating that pubertal processes that occur after SE assembly and ERα binding are needed for gene responses. Genes associated with these SEs affected key estrogen-mediated uterine functions, including transforming growth factor β (TGFβ) and LIF interleukin-6 family cytokine (LIF) signaling pathways. To the best of our knowledge, this is the first identification of SE interactions that underlie hormonal regulation of genes in uterine tissue and optimal development of estrogen responses in this tissue.
Keywords: estrogen receptor, gene expression, chromatin modification, transcription, reproduction, chromatin conformation capture, chromatin looping, epigenetic regulation, female reproduction, super-enhancer
Introduction
Estrogen hormones are intricately involved in key molecular events that underlie development of female reproductive tract tissues, optimizing responses to ovarian hormones essential for successful reproduction later in life (1). Many studies of estrogen response mechanisms have utilized in vitro cell culture models, which have advanced understanding but are limited in their application to more biological contexts. In vivo models of hormone response include the rodent female reproductive tract, which is highly hormone-responsive, facilitating study of mechanisms of tissue development and hormone response. For example, female reproductive tissues of estrogen receptor α (ERα)-null and estrogen-deficient Cyp19 (aromatase) KO mouse models, both of which resemble their WT littermates at birth, lack pubertal uterine development, and thus exhibit a hypoplastic uterine phenotype (1–4). These experimental characteristics duplicate those found clinically in patients with insensitivity syndromes due to mutations in their ESR1 or CYP19 genes (5–8). Estrogen's activity involves interaction with its nuclear receptor, ERα, which localizes to enhancer regions of chromatin, driven by high affinity for estrogen response element (ERE) DNA motifs. Enhancer regions that ERα interacts with are often distal from promoters of estrogen-regulated genes (1, 9), and current models incorporate a mechanism of chromatin looping to facilitate contacts between enhancer and promoter regions (10, 11). Earlier studies have noted that some enhancers have particularly high enrichment of indicators of active chromatin; these are classified as “super-enhancers” (12, 13). Frequently, these super-enhancers control production of key transcription factors that characterize a particular cell type (12–14). In mice, prior to production of ovarian hormones, which begins with the onset of estrus cyclicity at puberty (postnatal day 28), the prepubertal uterus is hypoplastic, has ERα in all cells, and exhibits rapid transcriptional and growth responses of all uterine cell types to exogenous estradiol (E2) (15–17). The response, however, differs from what is observed in an ovariectomized adult mouse uterus, where the uterus response is restricted to only the epithelial cells (15, 17). Through the process of pubertal maturation, the reproductive tract acquires optimal reproductive functionality (3, 15, 16, 18, 19). The molecular details of this important process have not been explored; therefore, we sought to define the estrogen-dependent enhancer landscape of the developing mouse uterus by analyzing uterine chromatin isolated from prepubertal (21-day-old) or from adult (10-week-old) mice that were ovariectomized (ovexed) to remove the source of endogenous E2 and then treated for 1 h with vehicle (V) or with E2 to gain insights into molecular components and responses impacted during development of a hormonally regulated tissue.
Results
Defining uterine super-enhancers that bind ERα
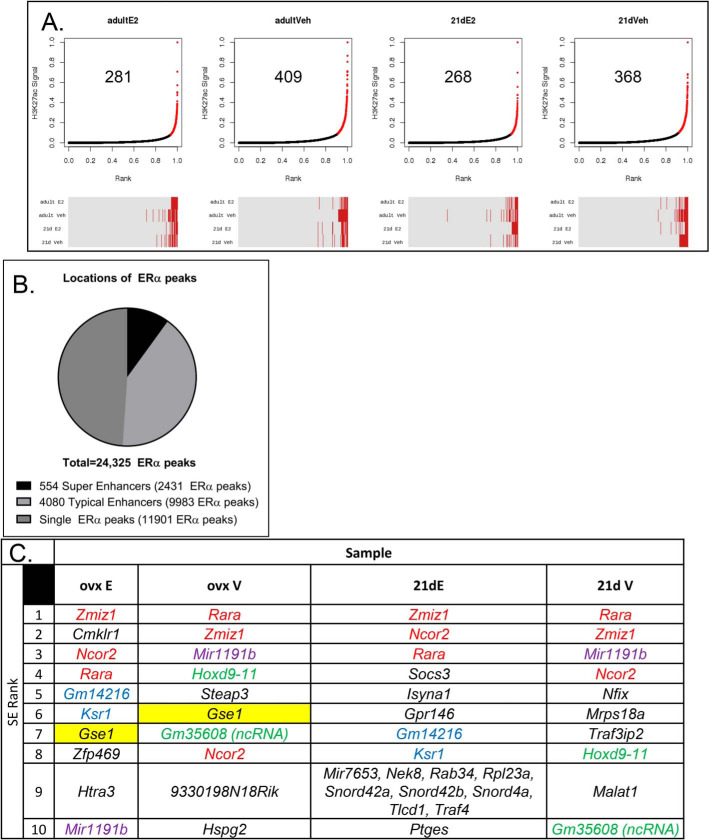
We defined super-enhancers (SEs) that bind ERα-in prepubertal uterine samples from mice treated for 1 h with vehicle (V) or with E2, as well as in uterine samples from ovexed adult mice treated for 1 h with V or E2 as described under “Experimental procedures.” Briefly, ERα-binding enhancers were identified using ERα ChIP-Seq data by stitching together regions with more than one ERα peak within 12,500 bp in at least one of the samples. The 4634 enhancer stretches containing multiple ERα-binding sites were then classified as “typical” or “super” enhancers (TEs or SEs, respectively) by plotting ranked H3K27Ac signal of each; enhancers beyond the slope = 1 elbow of the curve (Fig. 1A) were considered SEs. Therefore, locations of ERα ChIP-Seq peaks from all four sample types were either classified as single peaks (Fig. 1B) or were within enhancer regions with multiple peaks (Fig. 1B). The multipeak enhancer regions were then classified as super-enhancers or typical enhancers (Fig. 1B). Several hundred SEs were identified in each sample type (Fig. 1A). Comparison of the enhancers called in each sample indicates that many SEs overlap across the sample types (Fig. 1A and Fig. 2), and most that are SEs, but not in all samples, are ranked in the top 25% of enhancers (Fig. 1A, red lines in the heatmap below each curve). The SEs in the prepubertal V-treated mice are present before the cells have been exposed to ovarian E2 beginning at puberty, whereas the SEs in the ovexed adult samples represent super-enhancers after the uterus has undergone pubertal maturation. For each super-enhancer curve, genes located at the top 10 SEs are listed. We noted that three of the genes at the SEs (Zmiz1, Ncor2, and Rara) were among the 10 highest ranked in all samples (Fig. 1C). Comparison of the SEs using heatmaps of ERα and H3K27Ac ChIP-Seq signals centered on each of the 2431 ERα-binding sites that is within a SE in at least one of the four samples (Fig. 2) indicates that the H3K27Ac signal intensities of the adult and the 21-day-old are similar in V- and E2-treated samples, respectively, showing that the SEs seen in prepubertal samples, prior to pubertal uterine development, are largely unchanged by pubertal development. E2 leads to increased ERα binding at each site, as well as increased H3K27Ac signal flanking SE ERα-binding locations. Interestingly, although E2 also increases ERα binding at the 21,894 non-SE ERα-binding sites (TE and single ERα-binding sites), H3K27Ac signal is more static (Fig. 2), with only the highest ranked ERα binding sites exhibiting E2-dependent H3K27Ac signal increase. Some researchers use H3K4Me1 ChIP-Seq signal intensity, also associated with active enhancers, to classify SEs. Using H3K4Me1 signal to classify uterine enhancers revealed that 65–87% of SEs by H3K4Me1 were also classified as SEs using H3K27Ac (Table S1). Additionally, >98% of SEs by H3K4Me1 signal were within the top quartile of the H3K27Ac signal. Locations of SEs called using H3K27Ac in the adult ovexed 1-h E2 uterus relative to genes and chromosomes are illustrated in Fig. S1.
Figure 1.
Defining super-enhancers in mouse uterine tissue. A, ranked H3K27Ac signal at 4634 regions with >1 ERα-binding site within 12,500-bp regions from uterine samples of 21-day-old or adult ovexed mice treated for 1 h with V or E2 (1 h). The plots show the distribution of the rescaled scores per sample. The red points beyond the elbow of the curve, where slope = 1, are classified as super-enhancers. The enhancer status (typical (gray) or super (red)) for each peak in each of the four samples in the same order as the corresponding plot above it is shown below each curve. B, proportion of ERα ChIP-Seq peaks that are single peaks or that are in regions with multiple ERα ChIP-Seq peaks within 12.5 kb that were classified as super-enhancers in at least one sample or typical enhancers (not a super-enhancer in any sample) in the above analysis. C, genes that are colocalized with the 10 top-ranked SE regions of each sample are listed. The top 10 SEs shared by all four samples are in red text, SEs that are in the top 10 in all samples except 21-day E2 are in purple text, SEs in the top 10 only in ovexed samples are highlighted in yellow, SEs that are in the top 10 only in V sample are in green text, SEs that are in the top 10 only in E2 samples are in blue text.
Figure 2.
Estrogen increases ERα and H3K27Ac enrichment at super-enhancers. Shown is a heatmap of ERα ChIP-Seq and H3K27Ac ChIP-Seq signal centered on 2431 ERα peaks that are at super-enhancers (top) or 21,894 ERα peaks that are not at super-enhancers but are at typical enhancers or are single ERα-binding sites (bottom) in uterine chromatin samples from adult ovexed or 21-day-old female mice treated with saline V or with E2 for 1 h. For each set, enhancers are in the order of ERα signal in the adult E2 sample. Each panel shows ChIP-Seq signal at ±1 kb relative to the ERα peak midpoint, with signal depth-normalized to 20 million uniquely mapped nonduplicate reads/sample.
Chromatin interactions in uterine tissue
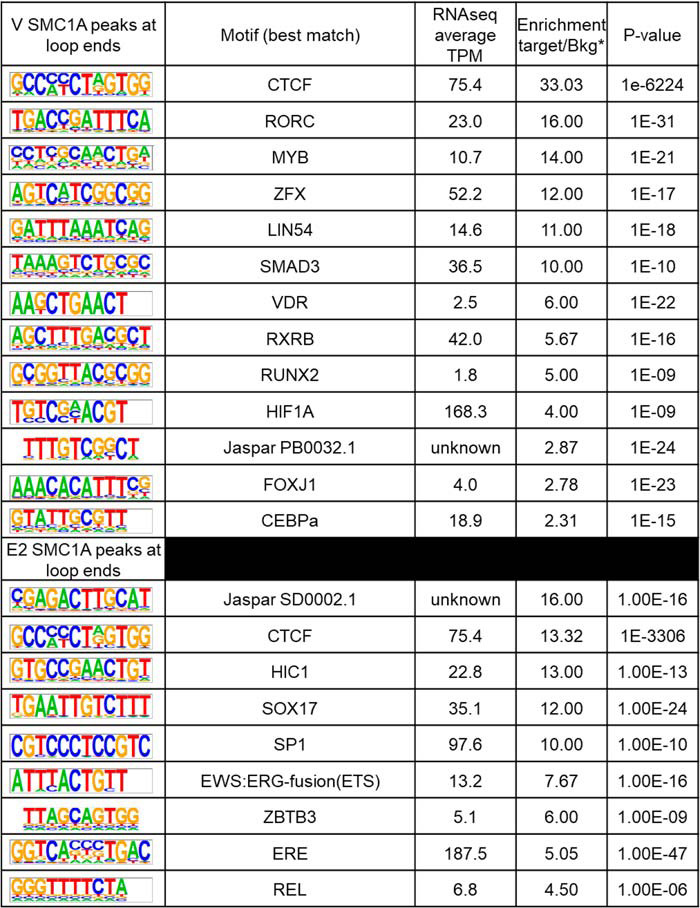
Distal enhancers can exert transcriptional control by physically contacting gene promoters through “looping” mechanisms (10). It is not known whether contacts are dynamically formed in order to exert regulatory signals between distal enhancers and promoters, whether loops are preformed and the activity of the promoters is mediated via dynamic transcription factor interactions, or whether a combination of mechanisms occurs (20). Assigning ERα-interacting regions to regulated genes is important for understanding how ERα binding to distal enhancers regulates gene promoters; therefore, we analyzed ovariectomized adult mouse uterus 1 h after treatment with V, E2, or progesterone (P4) using Hi-C. We used the Juicer tool to call loops from the Hi-C data of each treatment as well as differential loops in pairwise comparisons (Fig. 3A). We observed little variability between replicates or between different treatments, suggesting that loops are not altered globally by the hormone treatments we administered (Fig. 3A). Therefore, we combined the loops from all WT samples into an “atlas” of uterine loops. Then we used cohesin subunit SMC1a ChIP-Seq to assess interactions and potential impacts of hormone treatment. Because cohesin slides along chromatin until it contacts a pair of converging CTCF sites (10, 21), we compared SMC1a peaks at loop anchors selected to have cohesin at both ends as a surrogate for the presence of a loop in the sample. We examined SMC1A signal intensity, both at selected loop anchors and at other locations, and compared V– and E2–treated samples (Fig. 3B). When we compared V versus E2 SMC1a signal intensity, we did not observe a notable impact of E2 treatment (Fig. 3B) at selected loop anchors or at other locations, suggesting once again that loops are preformed and not impacted by our hormone treatments. To assess whether specific transcription factors might be associated with V versus E2 loops, we analyzed the DNA motifs associated with SMC1a peaks that were located at both ends of loops. The CTCF motif is highly enriched in SMC1a peaks at loop anchors of both V and E2 samples (Table 1). The ERα motif estrogen response element and ERα “tethering” factor motif SP1 are both selectively enriched at SMC1a peaks of the E2 sample. These observations suggest E2-induced mechanisms in which ERα and associated transcription factors such as SP1 interact with preformed chromatin loops. Consistent with this mechanism, we note that Hi-C analysis of uterine tissue from mice lacking ERα shows that loops are present and do not differ from WT samples any more than the variation observed between replicates (Fig. 3A).
Figure 3.
E2/ERα does not alter uterine loops. A, Juicer output showing the number of loops called from each sample in parentheses and differential loops called between the indicated pairs. Minimal differences are seen between replicates (V-1, V-2, V-3; P4-1, P4–2) as well as between V and hormone treatment (E2 or P4) or between mice lacking ERα (ERαKO) and WT litter mates (V-2). B, an “atlas” of uterine loops was built from the six combined WT sample Hi-C data sets. This collapsed set of loops was then filtered to retain only those with SMC1a ChIP-Seq peaks at both loop ends using either V- or E2-treated SMC1a ChIP-Seq data. The heatmap shows SMC1a signal centered on V (left) or E2 (right) SMC1a peaks that are (top) or are not (bottom) at loop ends of the selected loops. Each panel shows ChIP-Seq signal at ±1 kb relative to the SMC1a peak midpoint, with signal depth-normalized to 20 million uniquely mapped nonduplicate reads/sample
Table 1.
HOMER analysis showing de novo motifs in SMC1a peaks that overlap with anchors of loops with SMC1a at both ends (Fig. 3B) in V-treated (top) or E2-treated (bottom) samples
Shown is the average TPM (average of V, E2 2-h, and E2 6-h transcripts per million) of transcription factors that bind to motifs. *, ratio of the percentage of target with motif to the percentage of background with motif.
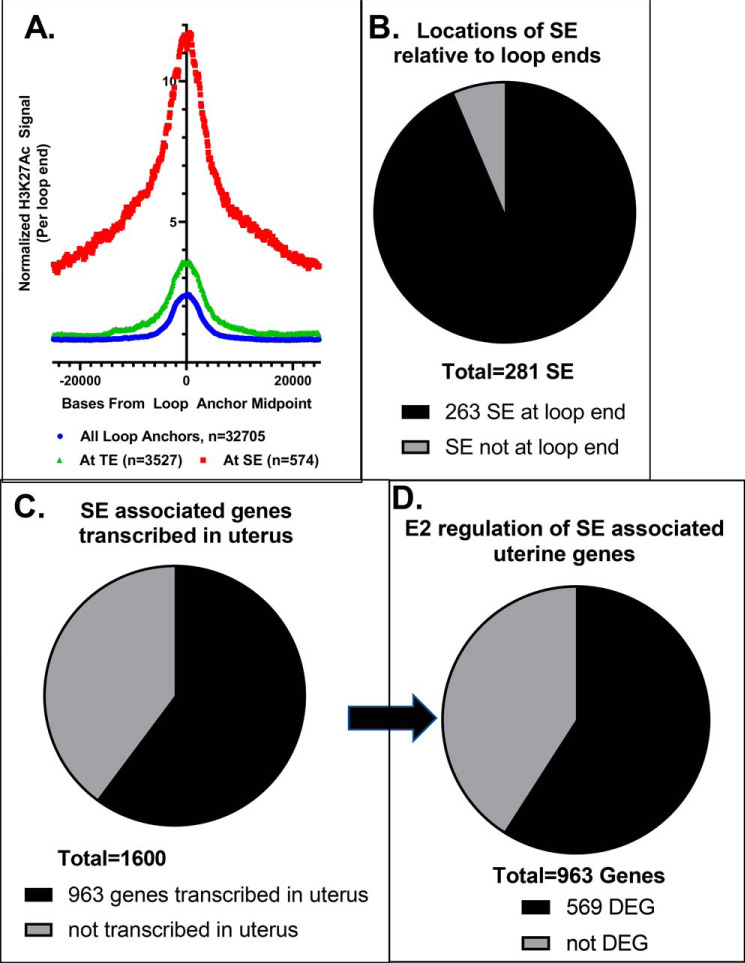
Super-enhancers are predominantly located in loop ends and are associated with uterine genes
Next, we evaluated the relationships between SEs and chromatin loops and whether SE and uterine genes are brought into proximity in 3D space. For this part of our analysis, we focused on the adult ovexed E2-treated samples. First, we examined average H3K27Ac signal per loop at all loop ends or “anchors” (Fig. 4A), which indicated that this histone modification was centered on the ends of loops. By further comparing loop anchors overlapping TEs versus SEs, we observed significantly more H3K27Ac signal at TE-associated loop anchors than at all loop anchors (p < 0.0001; Fig. 4A). Signal was further enriched at SE-associated loop anchors relative to all or to TE-associated loop anchors (p < 0.0001), reflecting the more robust enhancer activity and confirming these regions as SEs. Of the 281 SEs identified in adult E2-treated tissue, 94% (263 SEs) were at the end of a loop, suggesting that they were likely to contact genes in distal regions (Fig. 4B). To get an indication of the gene regulation that might occur by looping between genes and SEs, we selected genes that either were at one of the SEs in a loop end or were at the other end of a loop that formed from a SE. Altogether, we found this entailed 1600 genes (Fig. 4C). Our previous studies have indicated that ERα interaction with chromatin occurs within 1 h and that resulting gene regulation occurs subsequently (9, 22). Therefore, we used RNA-Seq of uterine RNA from ovariectomized mice treated with V or treated with E2 for 2 or 6 h to determine whether these 1600 ERα-binding SE-associated genes were transcribed and E2-regulated in the uterus. Indeed, 963 of these 1600 SE-associated genes are expressed (transcripts per million (TPM) ≥ 1 in at least one treatment condition) in uterine cells (Fig. 4C). Overall, 9975 uterine genes are regulated by E2 (TPM ≥ 1, adjusted p value <0.01) in at least one treatment condition (Table S2). 569 of the 963 uterine SE-associated genes are regulated by estrogen (Fig. 4D and Table S3). This suggests that ERα-binding SEs may be involved in regulation of 5–6% of E2-responsive uterine genes. The remaining E2-regulated uterine genes are likely to be mediated by ERα interactions at TE or at single ERα-binding sites, as we have previously demonstrated that the ERα is required for uterine E2 genomic response (4, 22). To assess how the 569 SE-associated estrogen-regulated uterine genes impact biological functions, we used INGENUITY PATHWAY ANALYSIS. Genes involved with the promotion of gene transcription, cell survival, and genitourinary tract development and suppression of mortality and apoptosis were apparent (Table 2). Notably, upstream regulators with known roles in uterine function, such as estrogen and cytokines, including LIF, a critical mediator of embryo implantation (23), as well as IGF1 and TGFβ (24–32), are identified as activating upstream regulators (Table 3). Therefore, these ERα-binding SE-associated genes are enriched for important uterine processes.
Figure 4.
Association of SEs with uterine gene expression. A, plot of normalized H3K27Ac signal in 1-h E2 adult ovexed sample per called loop end from the same sample. Curves are from all called loop ends, loop ends at TEs, and loop ends at SEs. B, proportion of adult E2 1-h SEs that are in E2 1-h called loop ends. C, proportion of 1600 SE-associated genes (at adult E2 1-h SEs that overlap a loop end or at the other end of the SE loop) that are detected in uterine V, E2 2-h, or E2 6-h samples (TPM ≥ 1). D, proportion of 963 SE-associated uterine genes differentially expressed (DEG; p < 0.01) 2 or 6 h after E2 treatment versus V treatment.
Table 2.
Biological functions and diseases associated with 569 E2-regulated uterine genes identified in Fig. 4D
| Disease or function annotation | p | Activation z-score, 2 h | Activation z-score, 6 h |
|---|---|---|---|
| Morbidity or mortality | 7.47E−08 | −5.82 | −3.22 |
| Activation of DNA endogenous promoter | 9.63E−11 | 4.30 | 2.47 |
| Cell movement | 3.50E−10 | 3.99 | 3.86 |
| Migration of cells | 3.89E−08 | 3.92 | 3.41 |
| Microtubule dynamics | 8.93E−07 | 3.85 | 2.41 |
| Development of body trunk | 5.71E−10 | 3.48 | 2.30 |
| Outgrowth of cells | 1.07E−07 | 3.28 | |
| Cell survival | 4.54E−08 | 3.07 | |
| Organization of cytoskeleton | 7.52E−09 | 3.06 | |
| Organization of cytoplasm | 9.02E−09 | 3.06 | |
| Size of body | 2.39E−07 | 3.06 | |
| Cell movement of endothelial cells | 8.57E−07 | 2.92 | 2.62 |
| Formation of vessel | 4.97E−06 | 2.73 | 2.25 |
| Phosphorylation of protein | 1.19E−06 | 2.71 | 2.25 |
| Apoptosis | 1.70E−10 | −2.53 | −2.28 |
| Development of genitourinary system | 1.30E−09 | 2.51 | 2.18 |
Table 3.
Upstream regulators enriched in 569 E2-regulated uterine genes identified in Fig. 4D
| Upstream regulator | Activation z-score, 2 h | Activation z-score, 6 h | p value of overlap |
|---|---|---|---|
| TGFB1a | 5.32 | 5.27 | 1.77E−11 |
| β-Estradiol | 4.63 | 4.54 | 1.16E−12 |
| LIF | 3.12 | 2.29 | 3.01E−05 |
| Cytokineb | 3.06 | 3.06 | 1.3E−06 |
| PGR | 2.92 | 2.92 | 5.14E−07 |
| Ige | 2.62 | 2.07 | 2.86E−05 |
| GDF2 | 2.61 | 2.61 | 2.44E−06 |
| ERK1/2c | 2.50 | 2.62 | 2.84E−05 |
| IGF1 | 2.35 | 4.41 | 3.49E−06 |
| α-Catenin | −2.51 | −2.11 | 1.22E−10 |
a Includes TGFB1, TGFB3, and SMAD3.
b Includes CSF1, CSF2, EDN1, IFNG, IL1, IL17A, IL1A, IL1B, IL2, Il3, IL4, IL6, OSM, TNF, and TNFSF11.
c Includes EGF, ERBB2, MAPK3, MAPK8, MAPK9, and p38 MAPK.
Super-enhancer–associated genes acquire estrogen responsiveness during postpubertal development
To assess whether postpubertal development impacted E2 regulation of the SE-associated genes identified above, we analyzed gene regulation in 21-day-old and ovexed adult samples from WT mice and mice with deletion of the ERα gene (ERαKO). We treated the mice for 2 h with V or E2 and isolated uterine RNA for microarray. We observed that 245 SE-associated genes were differentially expressed after 2-h treatment with E2. Hierarchical clustering reveals that the responses are ERα-dependent (Fig. S2 and Table S4). What is surprising is that, despite the presence of the SEs prior to pubertal development (Figs. 1 and 2), estrogen regulation is much more robust in adult ovexed samples (Fig. S2 and Table S4), indicating that these genes acquire the transcriptional response as a result of pubertal development.
Super-enhancers are associated with key uterine factors
Our analysis has indicated an association between ERα-mediated responses and looping between SEs and estrogen-regulated genes. Because genes at the SE often encode factors critical for specific cell types (14), we evaluated examples of four genes at the SE that are potentially important for uterine development and E2 transcriptional response and that are among the top-ranked SEs (Fig. 1A). Retinoic acid receptor a (Rara) is a ligand-dependent transcription factor in the nuclear receptor superfamily (33). Homeobox (Hox) transcription factors are key mediators of anterior-to-posterior developmental patterning of tissues (34). Ncor2 (SMRT; nuclear receptor corepressor 2) interacts with and activates histone deacetylases, thereby reducing transcriptional activity (35). Zinc finger MIZ domain-containing protein 1 (Zmiz1) is a novel transcription factor. We first used RT-PCR of RNA samples from prepubertal (21-day-old) or adult ovariectomized uteri to examine the impact of pubertal development on expression of these four genes. All genes showed increases in E2 response in adult tissues versus prepubertal tissues (Fig. 5A).
Figure 5.
Uterine factors at SEs. A, RT-PCR of uterine RNA isolated from 21-day-old or ovexed adult mice treated for 2 h with V or with E2. a, Rara; b, Hoxd11; c, Ncor2; d, Zmiz1; *, p < 0.05 versus V; +, p < 0.01 versus 21-day-old. B, chromatin loops between genes at SE and distal transcripts. SE are indicated by black blocks. Chromatin loops from uterine Hi-C samples are indicated by thin black lines. Shown are UCSC Genome Browser screenshots of the following. a, Rara (Chr11;mm10; chr11:98,854,783–99,160,103) near the SE (yellow highlight) at the Rara transcript. b, Igf1 (Chr10; mm10 chr10:87,635,437–88,361,810) near the SE (yellow highlight) distal from Igf1. The ERα-binding site that was deleted in IGF1enh4KO is outlined in red. C, disrupting the ERα-binding site in SE distal from Igf1 prevents E2 induction of Igf1 but does not alter expression of other genes that contact the SE or that are within the loop. Shown is RT-PCR of RNA from ovariectomized IGF1enh4KO or WT littermates that were treated with V (0) or with E2 for 2, 6, or 24 h. a, Dram1; b, Igf1; c, Nup37; d, Parpbp. *, p < 0.05 versus V; +, p < 0.05 versus WT.
Hi-C analysis indicates distal chromatin loops with SEs at Rara (Fig. 5Ba), the Hoxd cluster (Fig. S3), Ncor2 (Fig. S4A), and Zmiz1 (Fig. S4B) genes. We wondered whether the expression of genes that either directly coincide with the SEs or are within loops would be impacted by E2. Therefore, we looked at expression and estrogen regulation of genes within loops formed at these four SEs. We used our RNA-Seq data to evaluate the levels and E2 regulation of these genes (Table 4). We assessed the relative level of expression of each gene by computing the average TPM of nine uterine RNA samples (3 replicates each of uterine RNA from animals treated with V or with E2 for 2 or 6 h). We examined the E2 regulation of each gene based on fold change of E2 2-h RNA versus V RNA or E2 6-h RNA versus V RNA. The Rara SE forms loops that include several genes (Table 4 and Fig. 5Ba), but only Rara and Igfbp4 were regulated by E2 (TPM ≥ 1, FDR p value <0.01; Table 4). The Hoxd cluster SE coincides with the coding genes for Hoxd8, -9, -10, and -11 and contacts two additional SEs (Fig. S3A). Some of the genes within loops that include the SE are regulated by E2 (Table 4). There is a loop between the Hoxd SE and a third SE nearly 30 megabases away, in the Wilms tumor 1 (Wt1) gene (Fig. S3B). Thus, the Hoxd cluster in the uterus appears to be at the core of a very large 3D chromatin structure of SEs. Two SEs are localized at the Ncor2 coding sequence (Fig. S4A). Ncor2 is one of the most highly expressed genes in the loops that include the SE within it (Fig. S4A and Table 4). The Zmiz1 SE loops with two distal SEs (Fig. S4B). Zmiz1 and several other genes within the loop are expressed (Fig. S4B and Table 4).
Table 4.
RNA-Seq values for genes within loops of each SE
SE genes are highlighted in yellow. Boldface type shows FDR <0.01 versus V.
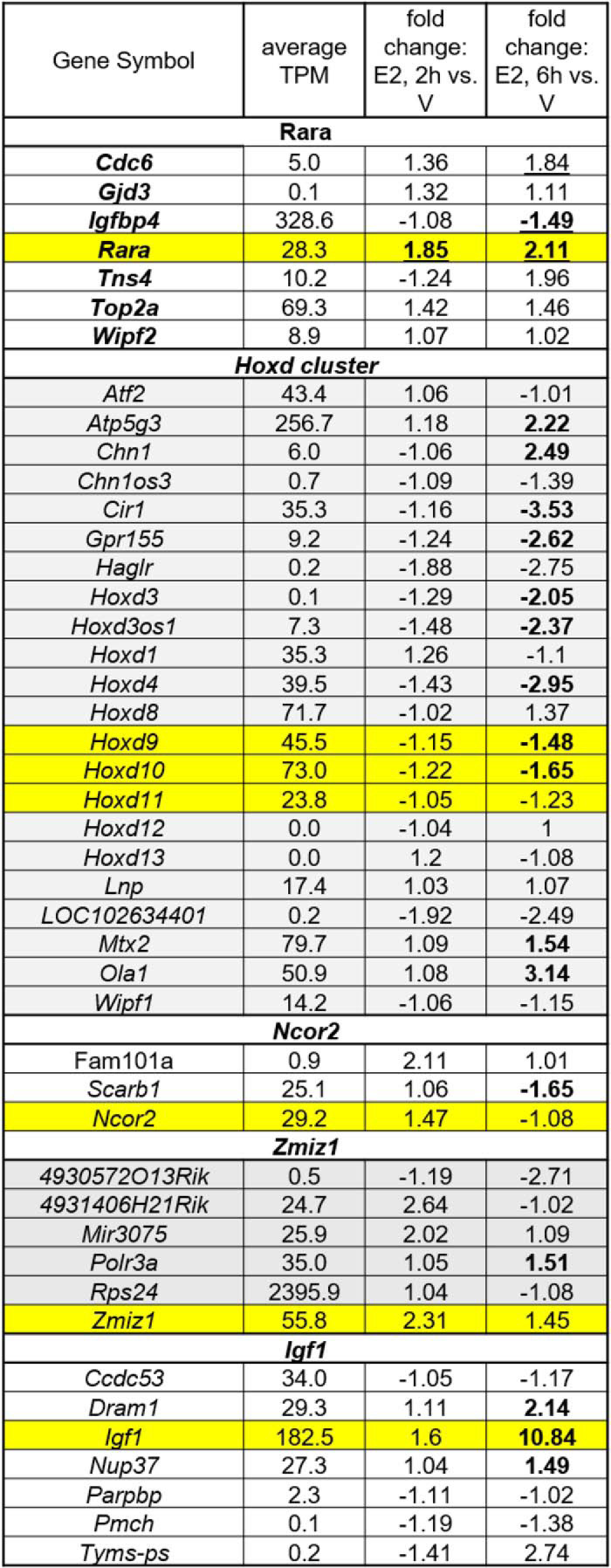
To experimentally determine whether a SE ERα-binding site can impact expression of genes within Hi-C defined chromatin loops, we examined expression levels of genes within loops from a previously described SE distal from the Igf1 gene (36). The Igf1 SE forms a loop that includes Tyms-ps, Dram1, Pmch, Parpbp, Nup37, and Ccdc53 within it (Fig. 5Bb). Pmch and Tym-ps are not expressed (average TPM < 1); Dram1, Igf1, and Nup37 are regulated by E2 (Table 4). In our previous study, we disrupted the Igf1 distal SE by deleting one of its ERα-binding sites (36); using this IGF1enh4KO model, we examined whether the distal SE is important for E2 regulation of genes in the loops that emanate from it. RT-PCR analysis of uterine RNA samples from IGF1enh4KO and WT littermates shows that although E2 regulation of Igf1 itself was affected by disrupting the SE (Fig. 5C), no change in expression of Dram1, Nup37, or Parpbp occurred, indicating that the deleted site selectively impacts only Igf1 expression.
Mouse uterus super-enhancers differ from those in breast cancer cells
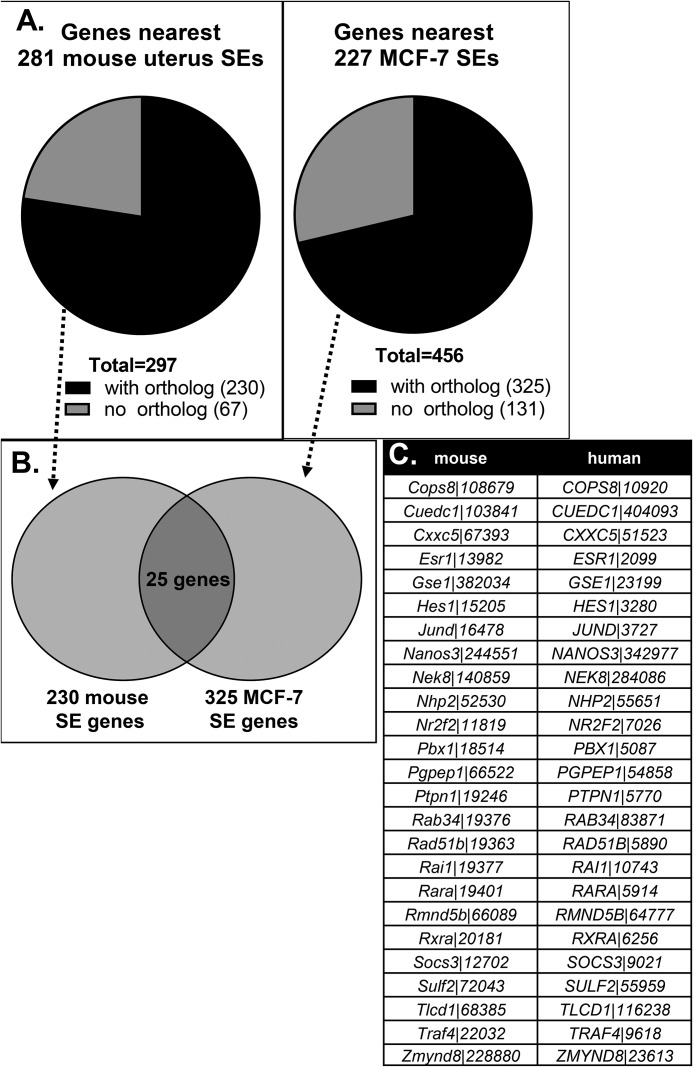
To evaluate how the mouse uterine SEs might be related to SEs in other E2-responsive cells, we compared our findings with a similar analysis that defined SEs that bind ERα in the MCF-7 breast cancer cell line grown in hormone-deprived medium (37). We compared the genes that are closest to 227 MCF-7 SEs with a list of genes closest to 281 uterine tissue SEs (Fig. 6). After removing genes that do not have both mouse and human orthologs (Fig. 6A), about 10% of the SE genes were common between the mouse uterus and MFC-7 cell systems (Fig. 6B). The small number of SEs located at genes shared in MCF7 cells and uterine tissue is likely due to differences in the biological responses E2 elicits in a normal E2-responsive tissue (mouse uterus) versus an immortalized breast cancer–derived cell line.
Figure 6.
Genes closest to SEs in the uterus differ from those in MCF-7 breast cancer cells. A, genes closest to 281 mouse uterus or 227 MCF-7 cell (human) SEs were evaluated for whether they had an ortholog in the other species. Only genes with both mouse and human orthologs were used in the subsequent comparison. B, 25 of the genes closest to mouse uterus or MCF-7 cell SEs were shared and are listed in C. C, table listing the gene symbols and Entrez gene IDs of the 25 shared genes from the analysis in B.
Discussion
The importance of E2 and ERα in uterine maturation is emphasized by the observation of hypoplastic uterine tissue in adult-aged ERα-null mice (4), which circulate high levels of E2 but have no receptor protein, and in mice lacking Cyp19, which are therefore unable to synthesize E2 (2, 38) but have normal levels of ERα. We defined the estrogen-dependent enhancer landscape of the developing mouse uterus and observed that overall, most of the 21-day-old and adult SEs had similar characteristics; in general, both ERα and H3K27Ac signals are increased by E2 at the SE (Fig. 2). Although E2 increases ERα signal of TE and single ERα-binding sites as well, H3K27Ac is less impacted by E2 (Fig. 2), with only the highest-ranked ERα-binding sites showing an increase. Overall, this suggests that the enhancer landscape necessary for uterine maturation is already in place by postnatal day 21. It was clear, however, from our gene regulation analyses, that pubertal development leads to more robust regulation of genes associated with these SEs (Fig. S2).
Comprehensive analysis of ERα interaction with chromatin using ChIP-Seq has revealed that ERα-binding regions are often distal from coding genes, which has led to a need to understand the 3D structure of chromatin within ERα-expressing cells. We noted that all ERα-binding loop ends had significantly more H3K27Ac signal, centered on the loop anchor, than loop anchors in general (Fig. 4A), especially those at super-enhancers. This highlights the importance of ERα in the mechanism of activation via these 3D structures. We did note that H3K27Ac signal is greater at loop ends with TE (Fig. 4A), indicating a significant role of these ERα-binding regions in E2 response as well. How responses mediated by single ERα-binding sites or by TE versus SE impact uterine development and biological processes will be an important focus of future study. Our previous work characterized one such gene mechanism by deleting an ERα-binding site in a super-enhancer distal from the Igf1 gene, resulting in loss of E2 induction of the coding transcript (36). Deleting one of the five distal ERα-binding sites did not impact E2-dependent ERα binding to the four remaining distal sites or to a site near the promoter; however, E2-dependent recruitment of the histone acetyltransferase p300 and the cohesin subunit SMC1a was decreased at neighboring sites, and E2-dependent induction of enhancer RNA and coding mRNA transcription was lost (36). Thus, E2-dependent recruitment of factors necessary to optimize enhancer-promoter–mediated Igf1 transcription is governed by a critical site in the distal enhancer. The Igf1 distal SE also forms chromatin loops that include other genes, some of which are also regulated by E2; however, deletion of the ERα-binding site did not change the E2 response of these other genes, indicating that the primary target of the SE in this case is the Igf1 gene. Whether this relationship between SEs and regulated distal genes is generally seen, or whether there are cases in which expression of multiple genes is impacted by a single SE remains to be studied.
In this study, we identified SEs either in or looping to other well-known highly E2-responsive genes, including Greb1, Lif, Fos, Cebpb, Cyr61, and Inhbb (Table S3). The Greb1-interacting super-enhancer identified here was the focus of a previous study indicating the potential role of distal ERα-binding regions in Greb1 regulation (39). The same ERα-binding region was utilized to model mechanisms of distal interaction in a recent study (40), highlighting a role for the steroid receptor coactivator, SRC-3, in optimizing contacts between enhancer and promoter regions via interaction with an intronic SRC-3–binding sequence (40). Prior to E2 treatment, the enhancer and promoter are held in proximity to each other through their contacts with SRC-3 bound to the intron. E2 treatment leads to SRC-3–dependent formation of the enhancer-promoter transcriptional complex (40) and, consequently, increased Greb1 transcription. The formation of structures, such as the SRC-3/intron/enhancer/promoter complex, may be a mechanism utilized by enhancer-promoter interactions.
We note that most of the SEs identified in adult ovexed E2-treated samples were at a loop end (263 of 281; Fig. 4B) and were likely to be at or form a loop to genes expressed in the uterus (Fig. 4C). Many of these SE-associated genes are involved in transcription and in pathways important for uterine function, such as p53 (41) and Tgfβ (26) signaling; therefore, these SEs are associated with important mediators of uterine maturation and function. We did not observe a SE at the Esr1 gene, although ERα is detected in all uterine cells and SE-associated genes are often characteristic of a particular cell type. Perhaps we should have expected that ERα is not driven by a SE, as we know that the ERα-null mouse develops all uterine cell types normally (4, 42). We did see that Esr1 was found to be one of the genes closest to an ERα-binding SE in both the MCF-7 cell and our uterus data sets (Fig. 6C). When we examined the impact of E2 on expression of the 963 SE-associated uterine genes, 569 were regulated by E2, a small portion of the 9975 uterine genes that are regulated by E2 (Fig. 4D and Tables S2 and S3). These SE-associated differentially expressed genes (DEG) are involved with activation of TGFβ and estrogen responses and with cytokine pathways, including LIF, a cytokine that is critical for embryo implantation (23) (Table 3).
Our finding of a SE overlapping the uterine Rara gene confirms the association of SE with key genes that determine cell fate (12, 13), as critical roles for retinoic acid (RA) signaling in uterine development have been described. RARα is activated by retinol derivatives (Vitamin A), and disruption of retinol signaling, either by disrupting the Rara gene or with a retinol-deficient diet, impacts female reproductive tract development (43, 44). RA is required for Mullerian duct development and for maintenance of epithelia of adult female reproductive tracts (45). Mechanistically, retinoic acid signaling influences uterine versus vaginal epithelial differentiation (46); therefore, precise regulation of RA signaling mediators is important for normal development and function. This is partly achieved via regulation of enzymes involved in retinoic acid synthesis and degradation (47), but clearly RAR expression is needed to mediate the signal, suggesting that SE in the Rara gene plays an important role in female reproductive tract development and function. Additionally, inappropriate perinatal estrogen exposures are known to disrupt the normal patterning of female reproductive tract mesenchyme, but this effect can be prevented by co-treatment with RA (45), illustrating an interplay between estrogen and RA signaling during female reproductive tract development. Evaluation of the Rara SE showed that it is within chromatin loops that include two members of the DNA replication complex, Cdc6 and Top2a (Fig. 5Ba and Table 4), which may indicate a role for the Rara SE in expression of genes involved in growth responses to E2.
There are four Hox clusters in the mouse genome (Hoxa, Hoxb, Hoxc, and Hoxd), and Hox9–13 are believed to have arisen from a single ancestral gene (48). The Hoxa cluster, in particular, has been shown to play a key role in reproductive tract development and function (34). We were interested, then, to note that one of the top ranked prepubertal V SEs was located in the Hoxd cluster. Hoxa factors are expressed in a progressive pattern in the developing female reproductive tract, with Hoxa9 in the oviduct, Hoxa10 in the uterus, and Hoxa11 in the cervix (34). Hoxd9, -10, and -11 are expressed similarly to their Hoxa counterparts in the female reproductive tract (49) (Fig. S5A). A role for Hoxd in uterine function has been described using complementation studies with Hoxa- and Hoxd-deleted mice (49). In addition, a recent study evaluated the impact of deletion of one allele each of Hoxa9,10,11, Hoxc9,10,11, and Hoxd9,10,11 on uterine development. This study indicated that the combined deletion of one allele each of these nine Hox genes greatly decreased uterine gland development (50), further showing a role for Hoxd genes in uterine function. Although the Hoxa cluster has been very thoroughly described and characterized for its role in uterine function, we did not find any SEs at this gene cluster (Fig. S5B). We do observe chromatin loops between the Hoxa cluster and a region more than 1 megabase from the Hoxa cluster that includes SEs (Fig. S5B).
Ncor2, also called SMRT (silencing mediator of retinoic acid and thyroid hormone receptor), was originally described as a corepressor for RAR and TR, mainly through its interactions with these receptors and its associated histone deacetylase activity (35). Further study has revealed its dual role as a gene-selective co-activator and co-repressor for ERα (51, 52). A role for NCOR2 in the uterus has not been investigated, but considering its potential to regulate responses of TR, RAR, and ERα, it is noteworthy that we see two SEs in the Ncor2 gene (Fig. S4A). Recently, it was shown that removing Ncor2 disrupts formation of a retinoid gradient in mouse embryos and leads to perturbation of HOXC expression patterns, greatly impacting proper development (53). The disruptions observed incorporate developmental pathways involving three SE-associated uterine genes (Ncor2, Rara, and Hoxd), suggesting that these SEs might underlie processes important for pubertal development of uterine cells.
Zmiz1 is a member of the PIAS (protein inhibitor of activated STAT) family and was originally identified due to its role in prostate cancer cells as a co-activator that facilitates androgen receptor sumoylation (54). ZMIZ1 regulates activities of certain transcription factors, several of which are important for uterine development and function, including p53 (41), Notch (55), SMAD (24), STAT (56), and androgen receptor (57). Its essential role is highlighted by the finding that its deletion in mice causes embryonic lethality (12). Embryonic fibroblasts isolated from Zmiz1 null mouse embryos fail to proliferate normally (58, 59), emphasizing its critical cellular role. A missense mutation in ZMIZ1 has been identified from endometroid cancer samples (Catalogue of Somatic Mutations in Cancer; RRID:SCR_002260 (60)), and additionally, decreased ZMIZ1 expression (61) has been reported in adenomyosis biopsies, indicating an essential role for ZMIZ1 in human uterine health as well. We could readily detect Zmiz1 in the mouse uterus (Fig. 5A and Table 4). Its expression was recently noted in rat uterine stromal cells, with down-regulation observed in response to circadian synchronization using the glucocorticoid agonist dexamethasone (62). SE-dependent expression of Zmiz1 could ensure optimal uterine responses mediated by pathways it regulates; however, the embryonic lethality of the global deletion prevents evaluation of uterine function with current models. We plan to examine its role in uterine tissue via conditional deletion of its expression.
Our approach of identifying super-enhancers in developing uterine tissue is validated by observations that some SEs are at genes encoding factors known to be critical for uterine development and function, such as RARα. The location of most super-enhancers at the ends of chromatin loops and the uterine expression and developmental acquisition of E2 regulation of genes either at SEs or connected by looping to a SE indicate an important role for these enhancers. Indeed, regulatory pathways known to be involved in uterine development and function are impacted by these SE-associated E2-responsive genes. These important regulatory regions likely serve to optimize appropriate, efficient, and timely responses to circulating E2 during each estrous cycle and in preparation for establishing pregnancy.
Experimental procedures
Animals
All mice were used in accordance with an NIEHS-approved animal study protocol and using the 2015 edition of the Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals. For E2 response experiments, intact 21-day-old or ovexed adult (10+-week-old) female C57bl6/J mice were purchased from Charles River Laboratories (Raleigh, NC). ERαKO mice (4) (also called Ex3αERKO) and their WT and heterozygous (ERα +/−) littermates were produced in our contract colony at Taconic Farms (Albany, NY). IGF1enh4 KO mice and their WT littermates were produced in our colony at NIEHS and ovariectomized at 10 weeks of age. Genotypes were determined from an ear biopsy by Transnetyx (Cordova, TN). 21-Day-old females were used the week they were received; ovariectomized females were housed for 10–14 days before the experiments to allow endogenous ovarian hormones to diminish. There was no blinding, and mice were randomly assigned to treatment groups. Mice were given a single intraperitoneal injection of 250 ng of E2 (Research Plus Inc., Barnegat, NJ) dissolved in 0.1 ml of normal saline, which is a 10 μg/kg dose. Some were injected subcutaneously with 1 mg of P4 (Sigma) dissolved in 100 μl of sesame oil (Sigma). Control V animals were injected with 0.1 ml of normal saline. Uterine tissue was collected 1, 2, 6, or 24 h after the injections and was snap-frozen in liquid nitrogen. RNA was isolated and cDNA was synthesized for real-time RT-PCR as described previously (22, 36). Primer sequences are listed in Table S5. For gene expression studies, at least three animals per group were used based on an at least 2-fold change, with a coefficient of variation of 0.2 and 90% power, tested by two-way analysis of variance.
Identification of super-enhancers
Super-enhancers were identified based on the method described by Bojcsuk et al. (63). First, 1-h V and 1-h E2 ERα peak calls with high stringency were called by HOMER (parameters: -fdr 0.00001 -F 12 -style factor) for the ovexed adult and 21-day-old samples and then combined via BEDtools mergeBed (version 2.24.0), entailing more than 24,000 ERα-binding locations altogether. BEDtools mergeBed (version 2.24.0) was then rerun, this time to merge all peak calls within 12.5 kb. This set was subsequently filtered to retain only those regions with more than one contributing called peak, resulting in 4634 ERα-binding enhancer regions, each with multiple ERα-binding locations. Each enhancer region was scored by counting the number of overlapping uniquely mapped nonduplicate reads with BEDtools multiBamCov (version 2.24.0); reads in this calculation were H3K27Ac or H3K4Me1 ChIP-Seq data after extension of mapped read length to 200 nucleotides. After normalizing to 20 million reads, the input-subtracted signal was determined per region. The signal curve was plotted as H3K27Ac or H3K4Me1 signal versus region rank and rescaled to span 0–1 on both axes. Definition of each enhancer region containing multiple ERα-binding sites as a “typical enhancer” or “super-enhancer” was determined according to the elbow of the signal curve where slope = 1.
SMC1a ChIP-Seq analysis
Processing of the SMC1a ChIP-Seq data was performed as described previously (36). Peak calls were made by HOMER version 4.10.3 via findPeaks with parameter “-style factor”. The de novo motif analysis was done with HOMER version 4.10.3 findMotifsGenome.pl with parameter “-size given”; for consistency, query regions (SMC1a peaks at loop ends) were resized to 300 bp centered on the midpoint of the called peaks.
RNA-Seq analysis
Processing of the RNA-Seq data was performed as described previously (28). Read counts per gene were determined by Subread featureCounts version 1.5.0-p1 (64) with parameters “-s2 -Sfr -p” for gene models based on RefSeq transcripts as downloaded from the UCSC Table Browser on February 9, 2015. Differential gene analysis was performed with DESeq2 version 1.15.1 (65). Genes were considered expressed in the uterus if TPM ≥ 1 in at least one condition. Genes were considered differentially expressed by applying a FDR cutoff of ≤0.01. SE-associated genes that were expressed or differentially expressed (FDR < 0.01) in the uterus were identified using the Partek List Manager Tool to find RefSeq gene symbols common to both sets.
Microarray
Two sets of uterine RNA samples were isolated. The first set was obtained from ovariectomized adult ERαKO females and their WT littermates treated for 2 h with V or E2 as above. The second set was obtained from 21-day-old ERαKO females and their WT littermates (produced by timed matings of heterozygous (ERα +/−) males and females) treated for 2 h with V or E2. RNA was assessed by microarray as described previously (66) and analyzed in Partek using analysis of variance to find DEG and then filtering for signal intensity >100 in at least one sample and then filtering for super-enhancer–associated genes. Data from ovariectomized adults (68) (GSE100131) and data from 21-day-old mice (GSE148006) have been deposited in GEO.
Hi-C and data analysis
The Hi-C analysis was based on samples described previously (36) (V-1, E2, and P4-1 in Fig. 3A) together with additional samples. Samples V-2 and ERαKO V were processed by Arima Genomics (San Diego, CA), and libraries were sequenced by the NIEHS Molecular Genomics Core, as described for the previous study (36). Samples V-3 and P4-2 were processed and sequenced by Active Motif, Inc. (Carlsbad, CA) using the Arima Genomics Hi-C kit. The Juicer (version 1.5.6) platform was used for processing the Hi-C samples as described previously (36, 67). The quality control metrics from the Juicer output are shown in Table S6. Chromatin loops were identified with the Juicer hiccup utility, an algorithm for finding chromatin loops, by searching for clusters of contact matrix entries with enriched contact frequency relative to local background at default parameters (67). Simple overlap assessment via BEDtools intersectBed was used to determine localization of super-enhancers and SMC1a peaks at loop ends. The “atlas” of mouse uterine chromatin loops was generated by combining Juicer loop calls from the six individual WT samples, whereby all loops with both ends overlapping were collapsed.
Data availability
Hi-C and microarray data from samples are deposited in GEO under GSE147843 and GSE148006.
Supplementary Material
Acknowledgments
We are grateful to Drs. Alicia Chi and Joseph Rodriguez for critical reading of the manuscript. We thank the NIEHS Molecular Genomics Core for sequencing and the NIEHS Microarray Group for microarray. We greatly appreciate the animal care and surgical expertise provided by individuals in the NIEHS Comparative Medicine Branch. We thank Active Motif Inc. for providing complementary processing and sequencing of Hi-C samples V-3 and P4-2.
This article contains supporting information.
Author contributions—S. C. H. and F. J. D. conceptualization; S. C. H. and S. A. G. formal analysis; S. C. H. and S.-P. W. validation; S. C. H., S.-P. W., and F. J. D. investigation; S. C. H. and S. A. G. visualization; S. C. H. and S. A. G. methodology; S. C. H. writing-original draft; S. C. H., S. A. G., S.-P. W., and K. S. K. writing-review and editing; S. A. G. data curation; F. J. D. and K. S. K. resources; K. S. K. funding acquisition; K. S. K. project administration.
Funding and additional information—This work was supported in part by the Intramural Research Program of NIEHS, National Institutes of Health, Grants 1ZIAES070065 (to K. S. K.) and 1ZIAES103311 (to F. J. D.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- ERα
- estrogen receptor α
- KO
- knockout
- ovexed
- ovariectomized
- V
- vehicle
- SE
- super-enhancer
- TE
- typical enhancer
- H3K4Me1
- histone H3 Lys-4 monomethylation
- H3K27Ac
- histone H3 Lys-27 acetylation
- E2
- estradiol
- P4
- progesterone
- LIF
- leukemia-inhibitory factor interleukin-6 family cytokine
- FDR
- false discovery rate
- DEG
- differentially expressed genes
- RA
- retinoic acid
- RAR
- retinoic acid receptor
- SMRT
- silencing mediator of retinoic acid and thyroid hormone receptor.
References
- 1. Hewitt S. C., and Korach K. S. (2018) Estrogen receptors: new directions in the new millennium. Endocr. Rev. 39, 664–675 10.1210/er.2018-00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fisher C. R., Graves K. H., Parlow A. F., and Simpson E. R. (1998) Characterization of mice deficient in aromatase (ArKO) because of targeted disruption of the cyp19 gene. Proc. Natl. Acad. Sci. U.S.A. 95, 6965–6970 10.1073/pnas.95.12.6965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nanjappa M. K., Medrano T. I., March A. G., and Cooke P. S. (2015) Neonatal uterine and vaginal cell proliferation and adenogenesis are independent of estrogen receptor 1 (ESR1) in the mouse. Biol. Reprod. 92, 78 10.1095/biolreprod.114.125724 [DOI] [PubMed] [Google Scholar]
- 4. Hewitt S. C., Kissling G. E., Fieselman K. E., Jayes F. L., Gerrish K. E., and Korach K. S. (2010) Biological and biochemical consequences of global deletion of exon 3 from the ER α gene. FASEB J. 24, 4660–4667 10.1096/fj.10-163428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quaynor S. D., Stradtman E. W. Jr, Kim H. G., Shen Y., Chorich L. P., Schreihofer D. A., and Layman L. C. (2013) Delayed puberty and estrogen resistance in a woman with estrogen receptor α variant. N. Engl. J. Med. 369, 164–171 10.1056/NEJMoa1303611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grumbach M. M., and Auchus R. J. (1999) Estrogen: consequences and implications of human mutations in synthesis and action. J. Clin. Endocrinol. Metab. 84, 4677–4694 10.1210/jcem.84.12.6290 [DOI] [PubMed] [Google Scholar]
- 7. Bernard V., Kherra S., Francou B., Fagart J., Viengchareun S., Guéchot J., Ladjouze A., Guiochon-Mantel A., Korach K. S., Binart N., Lombès M., and Christin-Maitre S. (2017) Familial multiplicity of estrogen insensitivity associated with a loss-of-function ESR1 mutation. J. Clin. Endocrinol. Metab. 102, 93–99 10.1210/jc.2016-2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin L., Ercan O., Raza J., Burren C. P., Creighton S. M., Auchus R. J., Dattani M. T., and Achermann J. C. (2007) Variable phenotypes associated with aromatase (CYP19) insufficiency in humans. J. Clin. Endocrinol. Metab. 92, 982–990 10.1210/jc.2006-1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hewitt S. C., Li L., Grimm S. A., Chen Y., Liu L., Li Y., Bushel P. R., Fargo D., and Korach K. S. (2012) Research resource: whole-genome estrogen receptor α binding in mouse uterine tissue revealed by ChIP-seq. Mol. Endocrinol. 26, 887–898 10.1210/me.2011-1311 GEO, GSE56501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rowley M. J., and Corces V. G. (2018) Organizational principles of 3D genome architecture. Nat. Rev. 19, 789–800 10.1038/s41576-018-0060-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang J., Meng X., Chen H., Yuan C., Li X., Zhou Y., and Chen M. (2016) Exploring the mechanisms of genome-wide long-range interactions: interpreting chromosome organization. Brief. Funct. Genomics 15, 385–395 10.1093/bfgp/elv062 [DOI] [PubMed] [Google Scholar]
- 12. Pott S., and Lieb J. D. (2015) What are super-enhancers? Nat. Genet. 47, 8–12 10.1038/ng.3167 [DOI] [PubMed] [Google Scholar]
- 13. Sengupta S., and George R. E. (2017) Super-enhancer-driven transcriptional dependencies in cancer. Trends Cancer 3, 269–281 10.1016/j.trecan.2017.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hnisz D., Abraham B. J., Lee T. I., Lau A., Saint-André V., Sigova A. A., Hoke H. A., and Young R. A. (2013) Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 10.1016/j.cell.2013.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Binder A. K., Winuthayanon W., Hewitt S. C., Couse J. F., and Korach K. S. (2015) Steroid receptors in the uterus and ovary. in Knobil and Neill's Physiology of Reproduction (Plant T. M., and Zeleznik A. J., eds) pp. 1099–1193, Elsevier, Amsterdam [Google Scholar]
- 16. Katzenellenbogen B. S., and Greger N. G. (1974) Ontogeny of uterine responsiveness to estrogen during early development in the rat. Mol. Cell. Endocrinol. 2, 31–42 10.1016/0303-7207(74)90010-0 [DOI] [PubMed] [Google Scholar]
- 17. Quarmby V. E., and Korach K. S. (1984) The influence of 17β-estradiol on patterns of cell division in the uterus. Endocrinology 114, 694–702 10.1210/endo-114-3-694 [DOI] [PubMed] [Google Scholar]
- 18. Kelleher A. M., DeMayo F. J., and Spencer T. E. (2019) Uterine glands: developmental biology and functional roles in pregnancy. Endocr. Rev. 40, 1424–1445 10.1210/er.2018-00281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stewart C. A., Fisher S. J., Wang Y., Stewart M. D., Hewitt S. C., Rodriguez K. F., Korach K. S., and Behringer R. R. (2011) Uterine gland formation in mice is a continuous process, requiring the ovary after puberty, but not after parturition. Biol. Reprod. 85, 954–964 10.1095/biolreprod.111.091470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Le Dily F., and Beato M. (2018) Signaling by steroid hormones in the 3D nuclear space. Int. J. Mol. Sci. 19, E306 10.3390/ijms19020306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gassler J., Brandão H. B., Imakaev M., Flyamer I. M., Ladstätter S., Bickmore W. A., Peters J. M., Mirny L. A., and Tachibana K. (2017) A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J. 36, 3600–3618 10.15252/embj.201798083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hewitt S. C., Deroo B. J., Hansen K., Collins J., Grissom S., Afshari C. A., and Korach K. S. (2003) Estrogen receptor-dependent genomic responses in the uterus mirror the biphasic physiological response to estrogen. Mol. Endocrinol. 17, 2070–2083 10.1210/me.2003-0146 [DOI] [PubMed] [Google Scholar]
- 23. Chen J. R., Cheng J. G., Shatzer T., Sewell L., Hernandez L., and Stewart C. L. (2000) Leukemia inhibitory factor can substitute for nidatory estrogen and is essential to inducing a receptive uterus for implantation but is not essential for subsequent embryogenesis. Endocrinology 141, 4365–4372 10.1210/endo.141.12.7855 [DOI] [PubMed] [Google Scholar]
- 24. Gao Y., Duran S., Lydon J. P., DeMayo F. J., Burghardt R. C., Bayless K. J., Bartholin L., and Li Q. (2015) Constitutive activation of transforming growth factor β receptor 1 in the mouse uterus impairs uterine morphology and function. Biol. Reprod. 92, 34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Q., Kannan A., Wang W., Demayo F. J., Taylor R. N., Bagchi M. K., and Bagchi I. C. (2007) Bone morphogenetic protein 2 functions via a conserved signaling pathway involving Wnt4 to regulate uterine decidualization in the mouse and the human. J. Biol. Chem. 282, 31725–31732 10.1074/jbc.M704723200 [DOI] [PubMed] [Google Scholar]
- 26. Li Q. (2019) Tumor-suppressive signaling in the uterus. Proc. Natl. Acad. Sci. U.S.A. 116, 3367–3369 10.1073/pnas.1821606116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Monsivais D., Peng J., Kang Y., and Matzuk M. M. (2019) Activin-like kinase 5 (ALK5) inactivation in the mouse uterus results in metastatic endometrial carcinoma. Proc. Natl. Acad. Sci. U.S.A. 116, 3883–3892 10.1073/pnas.1806838116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kriseman M., Monsivais D., Agno J., Masand R. P., Creighton C. J., and Matzuk M. M. (2019) Uterine double-conditional inactivation of Smad2 and Smad3 in mice causes endometrial dysregulation, infertility, and uterine cancer. Proc. Natl. Acad. Sci. U.S.A. 116, 3873–3882 10.1073/pnas.1806862116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Monsivais D., Clementi C., Peng J., Titus M. M., Barrish J. P., Creighton C. J., Lydon J. P., DeMayo F. J., and Matzuk M. M. (2016) Uterine ALK3 is essential during the window of implantation. Proc. Natl. Acad. Sci. U.S.A. 113, E387–E395 10.1073/pnas.1523758113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peng J., Monsivais D., You R., Zhong H., Pangas S. A., and Matzuk M. M. (2015) Uterine activin receptor-like kinase 5 is crucial for blastocyst implantation and placental development. Proc. Natl. Acad. Sci. U.S.A. 112, E5098–E5107 10.1073/pnas.1514498112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagashima T., Li Q., Clementi C., Lydon J. P., DeMayo F. J., and Matzuk M. M. (2013) BMPR2 is required for postimplantation uterine function and pregnancy maintenance. J. Clin. Invest. 123, 2539–2550 10.1172/JCI65710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clementi C., Tripurani S. K., Large M. J., Edson M. A., Creighton C. J., Hawkins S. M., Kovanci E., Kaartinen V., Lydon J. P., Pangas S. A., DeMayo F. J., and Matzuk M. M. (2013) Activin-like kinase 2 functions in peri-implantation uterine signaling in mice and humans. PLoS Genet. 9, e1003863 10.1371/journal.pgen.1003863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Das B. C., Thapa P., Karki R., Das S., Mahapatra S., Liu T. C., Torregroza I., Wallace D. P., Kambhampati S., Van Veldhuizen P., Verma A., Ray S. K., and Evans T. (2014) Retinoic acid signaling pathways in development and diseases. Bioorg. Med. Chem. 22, 673–683 10.1016/j.bmc.2013.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Du H., and Taylor H. S. (2015) The role of Hox genes in female reproductive tract development, adult function, and fertility. Cold Spring Harbor Perspect. Med. 6, a023002 10.1101/cshperspect.a023002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cunliffe V. T. (2008) Eloquent silence: developmental functions of Class I histone deacetylases. Curr. Opin. Genet. Dev. 18, 404–410 10.1016/j.gde.2008.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hewitt S. C., Lierz S. L., Garcia M., Hamilton K. J., Gruzdev A., Grimm S. A., Lydon J. P., Demayo F. J., and Korach K. S. (2019) A distal super enhancer mediates estrogen-dependent mouse uterine-specific gene transcription of Igf1 (insulin-like growth factor 1). J. Biol. Chem. 294, 9746–9759 10.1074/jbc.RA119.008759 GEO, GSE125972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miano V., Ferrero G., Rosti V., Manitta E., Elhasnaoui J., Basile G., and De Bortoli M. (2018) Luminal lncRNAs regulation by ERα-controlled enhancers in a ligand-independent manner in breast cancer cells. Int. J. Mol. Sci. 19, E593 10.3390/ijms19020593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Santen R. J., and Simpson E. (2019) History of estrogen: its purification, structure, synthesis, biologic actions, and clinical implications. Endocrinology 160, 605–625 10.1210/en.2018-00529 [DOI] [PubMed] [Google Scholar]
- 39. Sun J., Nawaz Z., and Slingerland J. M. (2007) Long-range activation of GREB1 by estrogen receptor via three distal consensus estrogen-responsive elements in breast cancer cells. Mol. Endocrinol. 21, 2651–2662 10.1210/me.2007-0082 [DOI] [PubMed] [Google Scholar]
- 40. Panigrahi A. K., Foulds C. E., Lanz R. B., Hamilton R. A., Yi P., Lonard D. M., Tsai M. J., Tsai S. Y., and O'Malley B. W. (2018) SRC-3 Coactivator governs dynamic estrogen-induced chromatin looping interactions during transcription. Mol. Cell 70, 679–694.e7 10.1016/j.molcel.2018.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu W., Feng Z., Teresky A. K., and Levine A. J. (2007) p53 regulates maternal reproduction through LIF. Nature 450, 721–724 10.1038/nature05993 [DOI] [PubMed] [Google Scholar]
- 42. Lubahn D. B., Moyer J. S., Golding T. S., Couse J. F., Korach K. S., and Smithies O. (1993) Alteration of reproductive function but not prenatal sexual development after insertional disruption of the mouse estrogen receptor gene. Proc. Natl. Acad. Sci. U.S.A. 90, 11162–11166 10.1073/pnas.90.23.11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mark M., Ghyselinck N. B., and Chambon P. (2009) Function of retinoic acid receptors during embryonic development. Nucl. Recept. Signal. 7, e002 10.1621/nrs.07002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wilson J. G., and Warkany J. (1948) Malformations in the genito-urinary tract induced by maternal vitamin A deficiency in the rat. Am. J. Anat. 83, 357–407 10.1002/aja.1000830303 [DOI] [PubMed] [Google Scholar]
- 45. Nakajima T., Sato T., Iguchi T., and Takasugi N. (2019) Retinoic acid signaling determines the fate of the uterus from the mouse Mullerian duct. Reprod. Toxicol. 86, 56–61 10.1016/j.reprotox.2019.03.006 [DOI] [PubMed] [Google Scholar]
- 46. Nakajima T., Iguchi T., and Sato T. (2016) Retinoic acid signaling determines the fate of uterine stroma in the mouse Mullerian duct. Proc. Natl. Acad. Sci. U.S.A. 113, 14354–14359 10.1073/pnas.1608808113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nakajima T., Yamanaka R., and Tomooka Y. (2019) Elongation of Mullerian ducts and connection to urogenital sinus determine the borderline of uterine and vaginal development. Biochem. Biophys. Rep. 17, 44–50 10.1016/j.bbrep.2018.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McGinnis W., and Krumlauf R. (1992) Homeobox genes and axial patterning. Cell 68, 283–302 10.1016/0092-8674(92)90471-N [DOI] [PubMed] [Google Scholar]
- 49. Raines A. M., Adam M., Magella B., Meyer S. E., Grimes H. L., Dey S. K., and Potter S. S. (2013) Recombineering-based dissection of flanking and paralogous Hox gene functions in mouse reproductive tracts. Development 140, 2942–2952 10.1242/dev.092569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mucenski M. L., Mahoney R., Adam M., Potter A. S., and Potter S. S. (2019) Single cell RNA-seq study of wild type and Hox9,10,11 mutant developing uterus. Sci. Rep. 9, 4557 10.1038/s41598-019-40923-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blackmore J. K., Karmakar S., Gu G., Chaubal V., Wang L., Li W., and Smith C. L. (2014) The SMRT coregulator enhances growth of estrogen receptor-α-positive breast cancer cells by promotion of cell cycle progression and inhibition of apoptosis. Endocrinology 155, 3251–3261 10.1210/en.2014-1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Peterson T. J., Karmakar S., Pace M. C., Gao T., and Smith C. L. (2007) The silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) corepressor is required for full estrogen receptor α transcriptional activity. Mol. Cell. Biol. 27, 5933–5948 10.1128/MCB.00237-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hong S. H., Fang S., Lu B. C., Nofsinger R., Kawakami Y., Castro G. L., Yin Y., Lin C., Yu R. T., Downes M., Izpisúa Belmonte J. C., Shilatifard A., and Evans R. M. (2018) Corepressor SMRT is required to maintain Hox transcriptional memory during somitogenesis. Proc. Natl. Acad. Sci. U.S.A. 115, 10381–10386 10.1073/pnas.1809480115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sharma M., Li X., Wang Y., Zarnegar M., Huang C. Y., Palvimo J. J., Lim B., and Sun Z. (2003) hZimp10 is an androgen receptor co-activator and forms a complex with SUMO-1 at replication foci. EMBO J. 22, 6101–6114 10.1093/emboj/cdg585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Afshar Y., Jeong J. W., Roqueiro D., DeMayo F., Lydon J., Radtke F., Radnor R., Miele L., and Fazleabas A. (2012) Notch1 mediates uterine stromal differentiation and is critical for complete decidualization in the mouse. FASEB J. 26, 282–294 10.1096/fj.11-184663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pawar S., Starosvetsky E., Orvis G. D., Behringer R. R., Bagchi I. C., and Bagchi M. K. (2013) STAT3 regulates uterine epithelial remodeling and epithelial-stromal crosstalk during implantation. Mol. Endocrinol. 27, 1996–2012 10.1210/me.2013-1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Walters K. A., McTavish K. J., Seneviratne M. G., Jimenez M., McMahon A. C., Allan C. M., Salamonsen L. A., and Handelsman D. J. (2009) Subfertile female androgen receptor knockout mice exhibit defects in neuroendocrine signaling, intraovarian function, and uterine development but not uterine function. Endocrinology 150, 3274–3282 10.1210/en.2008-1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Beliakoff J., and Sun Z. (2006) Zimp7 and Zimp10, two novel PIAS-like proteins, function as androgen receptor coregulators. Nucl. Recept. Signal. 4, e017 10.1621/nrs.04017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Beliakoff J., Lee J., Ueno H., Aiyer A., Weissman I. L., Barsh G. S., Cardiff R. D., and Sun Z. (2008) The PIAS-like protein Zimp10 is essential for embryonic viability and proper vascular development. Mol. Cell. Biol. 28, 282–292 10.1128/MCB.00771-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tate J. G., Bamford S., Jubb H. C., Sondka Z., Beare D. M., Bindal N., Boutselakis H., Cole C. G., Creatore C., Dawson E., Fish P., Harsha B., Hathaway C., Jupe S. C., Kok C. Y., et al. (2019) COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 47, D941–D947 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Herndon C. N., Aghajanova L., Balayan S., Erikson D., Barragan F., Goldfien G., Vo K. C., Hawkins S., and Giudice L. C. (2016) Global transcriptome abnormalities of the eutopic endometrium from women with adenomyosis. Reprod. Sci. 23, 1289–1303 10.1177/1933719116650758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tasaki H., Zhao L., Isayama K., Chen H., Nobuhiko Y., Yasufumi S., Hashimoto S., and Hattori M. A. (2013) Profiling of circadian genes expressed in the uterus endometrial stromal cells of pregnant rats as revealed by DNA microarray coupled with RNA interference. Front. Endocrinol. 4, 82 10.3389/fendo.2013.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bojcsuk D., Nagy G., and Balint B. L. (2017) Inducible super-enhancers are organized based on canonical signal-specific transcription factor binding elements. Nucleic Acids Res. 45, 3693–3706 10.1093/nar/gkw1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liao Y., Smyth G. K., and Shi W. (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 65. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hewitt S. C., Li R., Adams N., Winuthayanon W., Hamilton K. J., Donoghue L. J., Lierz S. L., Garcia M., Lydon J. P., DeMayo F. J., Adelman K., and Korach K. S. (2019) Negative elongation factor is essential for endometrial function. FASEB J. 33, 3010–3023 10.1096/fj.201801752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Durand N. C., Shamim M. S., Machol I., Rao S. S., Huntley M. H., Lander E. S., and Aiden E. L. (2016) Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 10.1016/j.cels.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hewitt S. C., Winuthayanon W., Lierz S. L., Hamilton K. J., Donoghue L. J., Ramsey J. T., Grimm S. A., Arao Y., and Korach K. S. (2017) Role of ERα in mediating female uterine transcriptional responses to IGF1. Endocrinology 158, 2427–2435 10.1210/en.2017-00349 GEO, GSE100131 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Hi-C and microarray data from samples are deposited in GEO under GSE147843 and GSE148006.