Abstract
Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) is an mRNA-binding protein that has an oncofetal pattern of expression. It is also expressed in intestinal tissue, suggesting that it has a possible role in intestinal homeostasis. To investigate this possibility, here we generated Villin CreERT2:Igf2bp1flox/flox mice, which enabled induction of an IGF2BP1 knockout specifically in intestinal epithelial cells (IECs) of adult mice. Using gut barrier and epithelial permeability assays and several biochemical approaches, we found that IGF2BP1 ablation in the adult intestinal epithelium causes mild active colitis and mild-to-moderate active enteritis. Moreover, the IGF2BP1 deletion aggravated dextran sodium sulfate–induced colitis. We also found that IGF2BP1 removal compromises barrier function of the intestinal epithelium, resulting from altered protein expression at tight junctions. Mechanistically, IGF2BP1 interacted with the mRNA of the tight-junction protein occludin (Ocln), stabilizing Ocln mRNA and inducing expression of occludin in IECs. Furthermore, ectopic occludin expression in IGF2BP1-knockdown cells restored barrier function. We conclude that IGF2BP1-dependent regulation of occludin expression is an important mechanism in intestinal barrier function maintenance and in the prevention of colitis.
Keywords: insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1), occludin, intestinal epithelium, permeability, mRNA decay, RNA-binding proteins, inflammatory bowel disease (IBD), colitis, epithelial barrier, intestinal mucosa, RNA-binding protein, intestinal mucosa
Insulin-like growth factor 2 mRNA–binding protein 1 (IGF2BP1, also known as CRD-BP/ZBP1/IMP1/VICKZ1) is a protein with an oncofetal pattern of expression exhibiting higher levels in fetal and neonatal tissues and low levels in adult tissues and is re-expressed in a variety of cancers (1, 2). IGF2BP1 binds a diverse set of mRNAs, affecting their stability and subcellular localization (3, 4). Well-established mRNA targets of IGF2BP1 include c-MYC, βTrCP1, GLI1, MITF, MDR1, H19, CD44, and PTGS2 (5–16). IGF2BP1 is also known to regulate cellular polarity and cell migration by escorting its target mRNAs to their proper subcellular site of protein synthesis (8, 10, 17). Photoactivatable ribonucleoside–enhanced cross-linking and immunoprecipitation (PAR-CLIP) and enhanced cross-linking and immunoprecipitation (eCLIP) experiments identified additional IGF2BP1 targets, suggesting its role in a multitude of cellular processes (18, 19). Identification of IGF2BP1 as a reader of N6-methyladenosine modification further added a new dimension to its regulatory function (20, 21).
Intestinal epithelial cells (IECs) establish and maintain a barrier to separate the submucosa from the intestinal lumen (22). This epithelial integrity is controlled by the junction complex comprised of the tight junction (TJ) and the adherent junction (23). The principle determinant of intestinal permeability and trans-epithelial transport is the TJ complex. Disrupted TJ and increased intestinal permeability may lead to intestinal inflammation (23, 24). Occludin is one of the key TJ proteins that contributes to the maintenance of an intact intestinal epithelium. Interestingly, Ocln mRNA is unstable and has been shown to undergo post-transcriptional regulation (23, 25, 26).
Inflammatory bowel disease (IBD) is a chronic inflammatory condition of the intestine and can manifest severe chronic active mucosal injury with restricted therapeutic options (27, 28). There are two main types of IBD, Crohn's disease and ulcerative colitis (29). One of the major factors in the development of IBD is the loss of intestinal epithelial barrier function, which elicits an inflammatory response that contributes to further barrier disruption (29). IGF2BP1 hypomorphic mice manifested various developmental defects and embryonic lethality (2). The intestine of these mice displayed abnormal crypt and villous architecture indicating a role for IGF2BP1 in intestinal development. Moreover, the role of IGF2BP1 was also implicated in healing of a mechanical wound across an epithelial monolayer of cells (11, 30). Interestingly, the deletion of IGF2BP1 in intestinal epithelial cells was recently shown to ameliorate experimental colitis in mice (31). Together, these findings hint to the function of IGF2BP1 in maintaining intestinal homeostasis.
For the current study, we generated mice that allow inducible knockout of IGF2BP1 specifically in the adult intestinal epithelium to study pathological processes independent of potential developmental defects. Here, we demonstrate that the ablation of Igf2bp1 in adult IECs leads to acute colitis in mice, and IGF2BP1 deficiency in IECs decreases occludin levels, resulting in a defective barrier function. These findings uncover an important function of IGF2BP1 in IECs to protect against colitis.
Results and discussion
Deletion of IGF2BP1 in IECs leads to acute colitis in mice
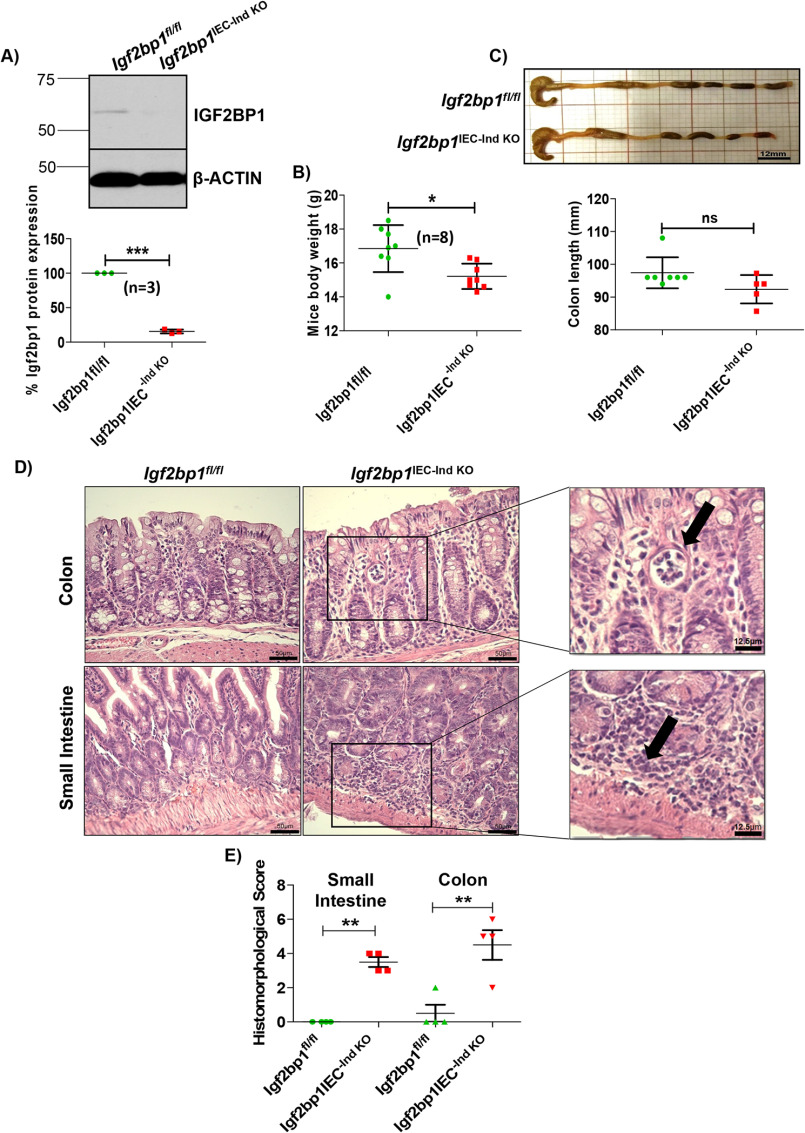
To investigate the role of IGF2BP1 in intestinal homeostasis, we generated Villin-CreERT2-Igf2bp1fl/fl transgenic mice. In these mice, transient knockout of Igf2bp1 (hereafter, Igf2bp1IEC-Ind KO) is induced via tamoxifen. Immunoblotting with anti-IGF2BP1 antibody was performed to confirm the ablation of Igf2bp1 from intestinal epithelium cells of Igf2bp1IEC-Ind KO mice (Fig. 1A). Intriguingly, adult Igf2bp1IEC-Ind KO mice displayed loss in body weight compared with Igf2bp1fl/fl littermates (Fig. 1B). Next, we examined the gross appearance of intestine and length of colon to inspect colon shortening, which is considered as the indicator of inflammation. In our analysis, we did not observe any significant change in gross appearance of intestine and in colon length of Igf2bp1IEC-Ind KO mice (Fig. 1C). Histological analysis of small intestines revealed that all Igf2bp1IEC-Ind KO mice exhibited increased lamina propria cellularity with mild to moderate active enteritis and patchy to diffuse neutrophilic and lymphocytic infiltration, when compared with control mice (Fig. 1D and Fig. S1B). Likewise, the colons of Igf2bp1IEC-Ind KO mice displayed mild active colitis with increased lamina propria cellularity and mild to moderate patchy to diffuse neutrophilic and lymphocytic infiltration. At places, there were loss of crypts with abscess formation (Fig. 1D and Fig. S1A). Based on the hematoxylin/eosin (H&E)-stained sections, analyzed by the pathologist in blinded manner, the samples were scored on two major histomorphological criteria: (i) severity of inflammatory cell infiltrates and (ii) epithelial changes (cryptitis, crypt abscesses, and erosion) (Fig. 1E). These results illustrate the importance of epithelial IGF2BP1 for normal intestinal physiology.
Figure 1.
Induced deletion of IGF2BP1 in IECs confers acute mild colitis in mice. A, immunoblot in the top panel showing depletion of Igf2bp1 from intestinal epithelium cells of Villin-CreERT2-Igf2bp1fl/fl mice upon tamoxifen treatment. Signals are quantified in the bottom panel from three independent Igf2bp1 immunoblot analyses using ImageJ software. B, body weight of age-matched female (10–12-week) littermates after the induction of IGF2BP1 knockout from IECs. The data are means ± S.D. (error bars) (n = 8 for each genotype). C, colon length comparison after knocking out Igf2bp1. Top, representative image of colon from mentioned genotypes. Scale bar, 12 mm. Bottom, quantification of colon length. The data are means ± S.D. (n = 7 for Igf2bp1fl/fl mice and n = 5 for Villin-CreERT2-Igf2bp1fl/fl mice). D, H&E-stained sections of colon and small intestine tissue from Villin-CreERT2-Igf2bp1fl/fl and control mice showing mild acute colitis and mild to medium acute enteritis. Scale bars, 50 μm (micrograph) and 12.5 μm (inset). E, histomorphological scores given to H&E-stained small intestine and colon tissue sections of Igf2bp1fl/fl and Villin-CreERT2-Igf2bp1fl/fl mice by a sample-blinded pathologist for colitis and enteritis. Scores are provided to each sample based on general criteria of histomorphological characters reported previously (33, 59). *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
IGF2BP1 deficiency sensitizes mice to dextran sodium sulfate-induced colitis
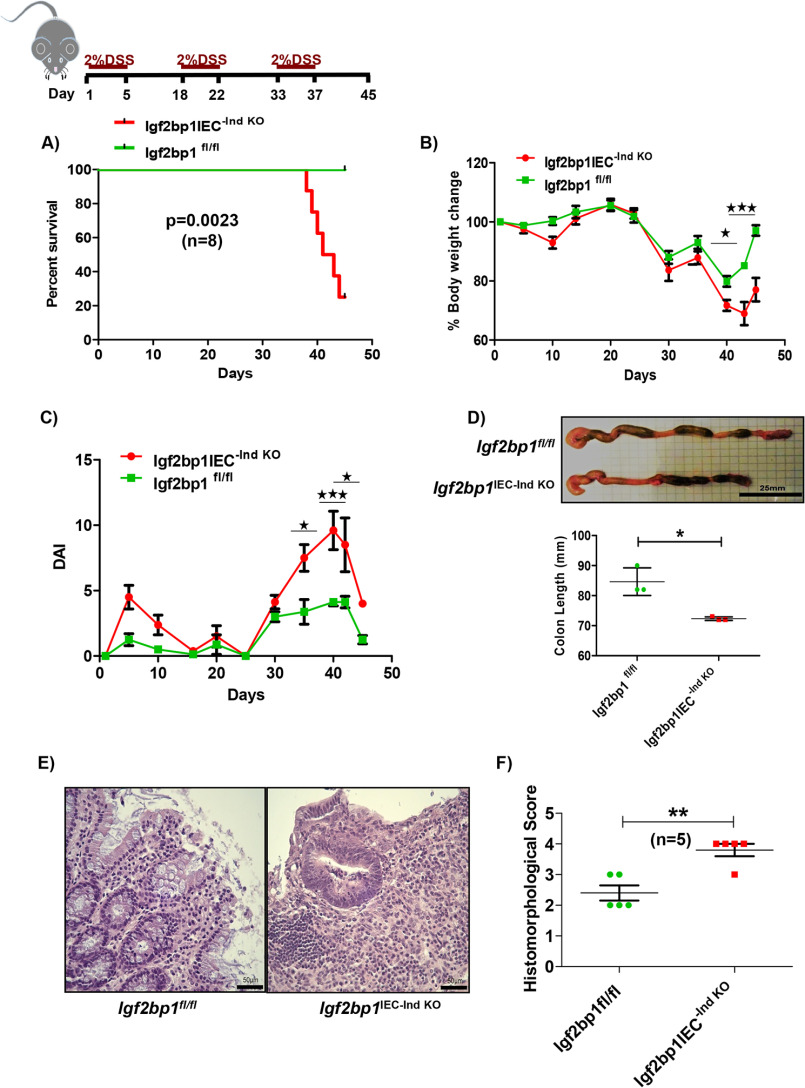
Next, we examined whether induced intestinal epithelial IGF2BP1 knockout affects sensitivity of mice to DSS, which is a toxic chemical that when delivered in the drinking water causes colitis by disrupting the intestinal epithelium. Here we performed chronic DSS treatment as it is the most relevant model for experimental colitis in mice. In the first cycle of this experiment, mice were treated with 2% DSS for 5 days, followed by intermittent periods of normal drinking water. This cycle was repeated two more times, and mice were then euthanized and examined. This experiment conferred 75% mortality in Igf2bp1IEC-Ind KO mice (Fig. 2A). We also observed significant loss in weight and increases in disease activity index (DAI) scores (32) in Igf2bp1IEC-Ind KO mice when compared with control littermates (Fig. 2, B and C). As severe chronic inflammation is known to decrease colon lengths, we evaluated colons at the conclusion of the experiment and found significantly shorter colons in Igf2bp1IEC-Ind KO mice than in the control littermates (Fig. 2D). We have also analyzed the H&E-stained sections of these colons and observed that the colon sections from DSS-treated Igf2bp1IEC-Ind KO mice showed increased disruption of colonic mucosa along with crypt abscesses, loss of colonic surface epithelium and crypts, and diffuse infiltration of inflammatory cells when compared with control mice (Fig. 2E and Fig. S1C). These tissue sections were analyzed and histomorphologically scored based on established criteria (Fig. 2F) (33, 59). These results indicate that IGF2BP1IEC-Ind KO mice are more sensitive to DSS challenge and develop more disease-related phenotypes in the intestine.
Figure 2.
Deletion of IGF2BP1 sensitizes mice to DSS colitis. A, survival curve for chronic DSS treatment showing significant loss of mice in the Villin-CreERT2-Igf2bp1fl/fl group. Age-matched (8–10-week) male and female Villin-CreERT2-Igf2bp1fl/fl (n = 8) and Igf2bp1fl/fl (n = 8) mice were treated with 2% DSS in drinking water for 5 days followed by two additional cycles of 2% DSS with a 10–14-day interval between cycles. Mice were sacrificed on the 45th day of the experiment. B, percentage weight profile of mice during the experiment for chronic DSS treatment. C, DAI score plotted against time for chronic DSS treatment. D, representative pictures of colon and colon length comparison shown in the top. Scale bar, 25 mm. The lengths of age-matched female (14-16-week) Villin-CreERT2-Igf2bp1fl/fl (n = 3) and Igf2bp1fl/fl (n = 3) mouse colons are quantified in the bottom panel. The data are mean ± S.D. (error bars). E, H&E-stained sections of colon from chronic DSS-treated Villin-CreERT2-Igf2bp1fl/f mice showing severe loss of colonic surface epithelium and crypts when compared with control mice. Scale bar, 50 μm. F, histomorphological scores were plotted for H&E-stained colon sections from the chronic DSS experiment. Colon sections were scored by a pathologist who was blinded for sample identity. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
IGF2BP1 deficiency increases intestinal permeability by affecting the occludin expression in IECs
The intact epithelium imparts a protective barrier against entry of foreign antigens from the intestinal lumen into the lamina propria. The aforementioned data suggest that IGF2BP1 may be important for maintaining barrier function of intestinal epithelium. To assess the permeability of intestine, we extracted serum from blood for quantifying specific IgG levels by ELISA. We found elevated levels of IgG to LPS (serological bacterial marker) in the serum of Igf2bp1IEC-Ind KO mice, which was indicative of increased intestinal permeability (Fig. 3A). To confirm these results, we used the FITC-dextran permeability assay, which is a classical method for evaluating the status of intestinal barrier function. Four hours after oral administration of FITC-dextran, we isolated serum and examined the level of FITC-dextran in the samples. Significantly higher levels of FITC-dextran were found in Igf2bp1IEC-Ind KO mice compared with Igf2bp1fl/fl littermates (Fig. 3B). These results indicate that IGF2BP1 is involved in the maintenance of intestinal barrier function, and its absence disrupted the barrier.
Figure 3.
Deletion of IGF2BP1 from IECs increases the intestinal permeability in mice. A, LPS-specific IgG levels in the serum of Villin-CreERT2-Igf2bp1fl/fl (n = 5) and Igf2bp1fl/fl (n = 5) mice. Rag1 knockout mice (n = 5) were taken as a negative experimental control. The data are means ± S.D. (error bars). B, analysis of intestinal epithelial permeability of mice by measuring FITC-dextran levels in the serum of mice Villin-CreERT2-Igf2bp1fl/fl (n = 7) and Igf2bp1fl/fl (n = 7). The data are means ± S.D. C, immunoblot of tight-junction proteins in mouse intestinal epithelium (representative picture of three independent experiments). Shown is whole-cell extract from IECs of Igf2bp1fl/fl (n = 3) and Villin-CreERT2-Igf2bp1fl/fl (n = 3) mice analyzed for occludin, CLAUDIN-1, CLAUDIN-2, ZO-1, and ZO-3 protein expression. β-Actin was evaluated as loading control. D, colon sections with occludin staining (green) showing the level of occludin expression in Igf2bp1fl/fl (n = 5) and Villin-CreERT2-Igf2bp1fl/fl (n = 3) mice. Scale bar, 10 μm. Images are quantified for occludin by ImageJ software, and data are means ± S.D. **, p < 0.01; ***, p < 0.001. N.D., not detected; RFU, relative fluorescence units.
A disrupted TJ is a major cause of intestine barrier dysfunction (34), and this motivated us to investigate the levels of major TJ proteins in IECs of Igf2bp1IEC-Ind KO mice. We found decrease specifically in the levels of occludin TJ protein in the IECs from the intestines of Igf2bp1IEC-Ind KO mice, whereas other TJ proteins, CLAUDIN-1, CLAUDIN-2, ZO-1, and ZO-3, remained unaffected when compared with control littermates (Fig. 3C). Immunostaining analysis revealed significantly lower levels of occludin in colon IECs of Igf2bp1IEC-Ind KO mice when compared with control mice (Fig. 3D). These results suggest that IGF2BP1 affects the expression of occludin TJ protein and maintains intestine barrier function.
IGF2BP1 regulates occludin expression by directly binding to and stabilizing its mRNA
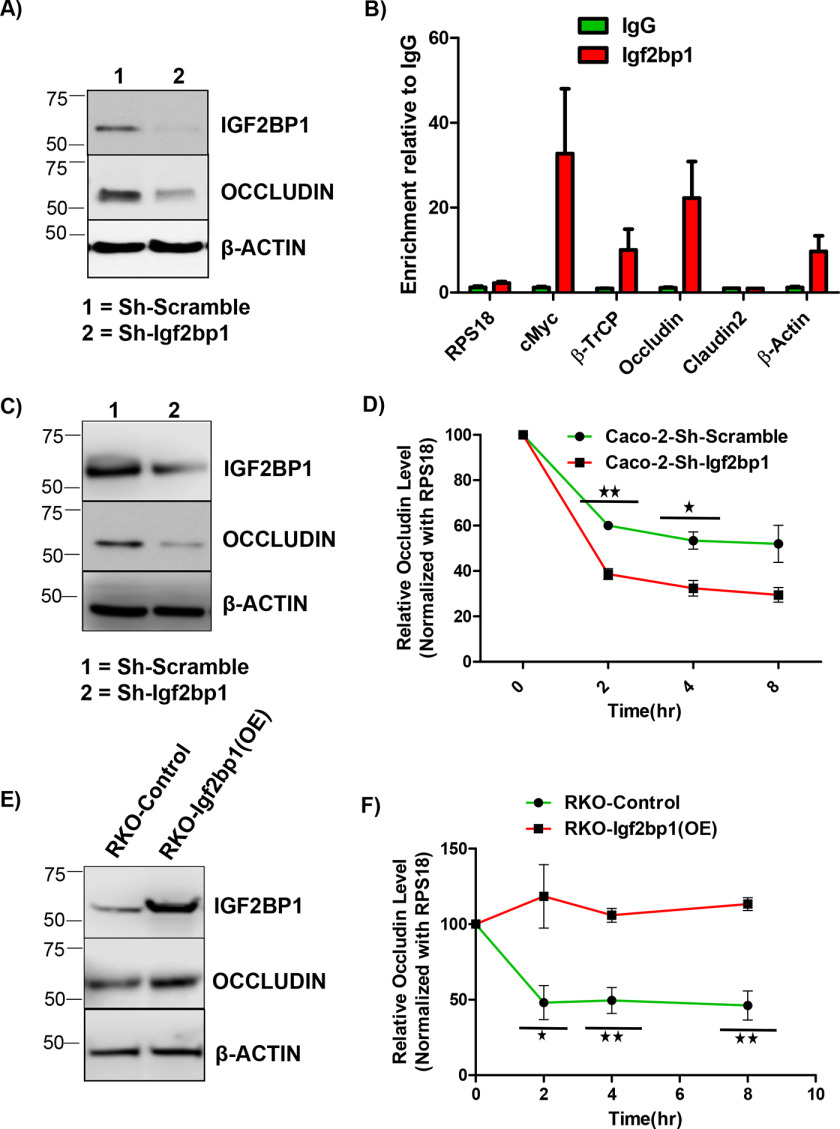
To investigate the mechanism of regulation of occludin TJ expression by IGF2BP1, we chose the human CCD841-CoTr normal colon epithelial cell line with moderate expression of Igf2bp1. Knockdown of IGF2BP1 using shRNAs in this cell line resulted in significant inhibition of occludin expression (Fig. 4A). IGF2BP1 is an RNA-binding protein (RBP) that often stabilizes its mRNA targets (6, 14), and to evaluate whether IGF2BP1 binds to the mRNA of occludin directly, we performed the CLIP assay. In our CLIP analysis, we found enrichment of OCLN mRNA to a similar extent as bona fide IGF2BP1 targets, such as MYC and β-TRCP1 (Fig. 4B). To elucidate whether IGF2BP1 regulates the turnover of OCLN mRNAs, we performed an actinomycin D chase experiment in a pair of colorectal cell lines. The loss-of-function experiments (by knocking down IGF2BP1) were performed in Caco-2 cells characterized by high expression of IGF2BP1. The knockdown of IGF2BP1 and its effect on occludin expression were confirmed by immunoblotting (Fig. 4C). The actinomycin D chase experiments in Caco-2 cells showed that the knockdown of IGF2BP1 resulted in accelerated rate of OCLN mRNA degradation (Fig. 4D) but not of Claudin2 mRNA (Fig. S3A). The gain-of-function experiments (by overexpressing IGF2BP1) were performed in RKO cells that have negligible expression of endogenous IGF2BP1. Overexpression of IGF2BP1 protein in this cell line resulted in increased levels of occludin protein compared with control cells (Fig. 4E). Additional mRNA stability assays using these cells detected an increased t1/2 of OCLN mRNA upon overexpression of IGF2BP1 (Fig. 4F), whereas there was no significant change in t1/2 of Claudin2 mRNA (Fig. S3B). These results show that IGF2BP1 protein regulates occludin expression by directly interacting with OCLN mRNA and stabilizing it, establishing OCLN mRNA as a novel direct target of IGF2BP1.
Figure 4.
IGF2BP1 deletion alters the expression of tight-junction proteins. A, immunoblot analysis of occludin proteins. CCD-841-CoTr cells transduced with scramble or Igf2bp1-specific shRNA were analyzed for occludin protein expression. Igf2bp1 immunoblotting demonstrates the extent of knockdown. The picture is representative of three independent experiments. β-Actin was evaluated as a loading control. B, CLIP in CCD-841-CoTr cells shows -fold enrichment of the indicated RNA. RNAs were co-immunoprecipitated with anti-IGF2BP1 antibodies with an isotype IgG serving as a control. Two independent repeats were performed, and the data are representative of the mean of two repeats with error bars showing S.D. RPS18 was evaluated as a negative control, whereas MYC and β-TRCP served as positive controls. C, immunoblotting of Caco-2 cells transduced with scramble or Igf2bp1-specific shRNA with anti-occludin antibody. The anti-IGF2BP1 immunoblot shows the efficiency of IGF2BP1 knockdown. The picture shown is representative of three independent experiments. β-Actin was evaluated as loading control. D, mRNA degradation assay in Caco-2 cells transduced with doxycycline-inducible scramble or Igf2bp1-specific shRNA. Cells were grown for 4 days with doxycycline treatment to induce knockdown of IGF2BP1. Actinomycin D was added at time 0, and cell samples were collected at 0-, 2-, 4-, and 8-h time points from the same plate. Occludin mRNA levels were evaluated via quantitative RT-PCR. The data are means ± S.D. (error bars) of three independent experimental repeats. E, immunoblot analysis of occludin in RKO cells transduced with control or IGF2BP1 overexpression (Igf2bp1-OE) constructs. β-Actin was shown as loading control. F, the mRNA degradation assay in RKO cells transduced with control or IGF2BP1 overexpression (Igf2bp1-OE) constructs to show the stabilization of OCLN mRNA upon IGF2BP1 overexpression. Actinomycin D was treated at 0 h, and cells were collected at 0, 2, 4, and 8 h from the same plate. Total RNA was isolated followed by quantitative RT-PCR to analyze the occludin mRNA levels. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Re-expression of occludin in IGF2BP1 deficient cells restores the tight-junction barrier function
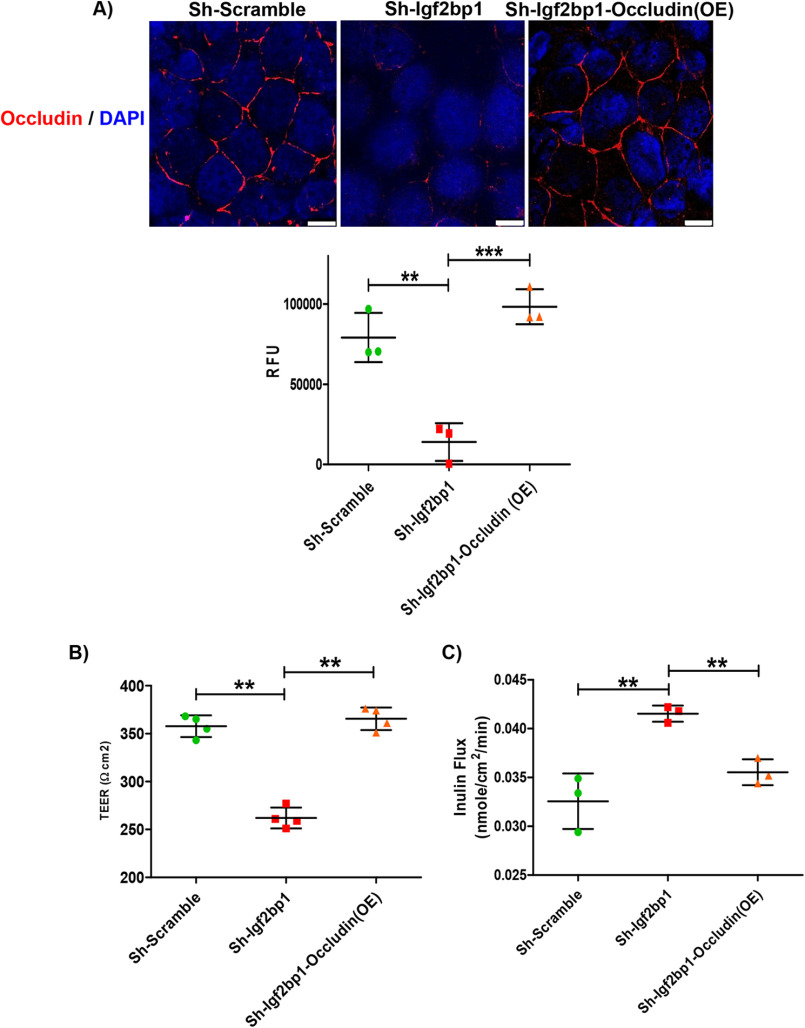
Occludin plays an important role in the formation of the tight-junction seal by regulating macromolecule flux across the barrier (23, 26). To determine whether disregulation of occludin contributes to altered barrier function induced by down-regulation of IGF2BP1, we analyzed the permeability of Caco-2 cells grown as a monolayer. As predicted, IGF2BP1 knockdown resulted in decreased occludin expression in Caco-2 cells (Figs. 4C and 5A). To analyze the barrier, we grew Caco-2, with or without knockdown of IGF2BP1, on transwell plates. We found that IGF2BP1 knockdown caused a significant decrease in trans-epithelial electrical resistance (TER), indicating increased permeability in the Caco-2 monolayer (Fig. 5B). The TER levels were completely restored after re-expressing occludin in IGF2BP1 knockdown cells (Fig. 5B). We had also found increase flux in IGF2BP1 knockdown cells while this flux was restored after re-expression of occludin in IGF2BP1 knockdown cells (Fig. 5C).
Figure 5.
Restoration of barrier function upon re-expression of occludin. A, monolayer of Caco-2 cells, transduced as indicated, were stained with occludin (red) antibody. Five images, each from a different area of the same plate, were captured and quantified using ImageJ software. The data shown below the images represent three biological replicates. Scale bar, 10 μm. B, determination of paracellular permeability by TER. The TER was measured after growing Caco-2 cells transduced as indicated for 15 days in chambered plates. The data are mean ± S.D. (n = 3) with p = 0.0030 and p = 0.0024, respectively. C, the apical flux of the paracellular macromolecular probe, inulin (molecular radius, 15 Å) was measured and plotted for filter-grown Caco-2 cells transduced as indicated. **, p < 0.01; ***, p < 0.001; RFU, relative fluorescence units.
Occludin knockout mice displayed retarded growth and chronically inflamed gastrointestinal tracts (35). Studies on Ocln knockout mice had revealed that occludin is an important factor in regulating the leak pathway in the mouse intestine (36–39). Our results demonstrate that deletion of Igf2bp1 from intestinal epithelium leads to the disruption of occludin, which in turn increases the permeability of intestine. Re-expression of occludin restores the barrier function in the IGF2BP1-depleted monolayer of Caco-2 cells. These results suggest that IGF2BP1 maintains the intestinal barrier function at least partially by binding to and stabilizing occludin mRNAs.
The epithelial barrier function is required to maintain intestinal homeostasis, and any alterations contribute to diseases such as IBD (23). Actively inflamed tissue was found to be leaky in various studies (22). However, occludin was the only tight-junction protein found to be down-regulated even in nonactively inflamed tissue in ulcerative colitis (40). This supports the importance of occludin in barrier function and disease. OCLN was shown to be regulated post-transcriptionally by RBPs and miRNAs (41, 42). The miRNA-429 and RBP CUGBP1 regulate OCLN negatively by binding to its 3′-UTR, whereas another RBP, HuR, regulates OCLN expression positively by binding to its 3′-UTR (25, 43). Our results provide further evidence for the importance of post-transcriptional regulation of OCLN mRNA at the level of mRNA stability. Interestingly, the reported miR-429–binding site (at positions 139-145 in the 3′-UTR of the OCLN mRNA) is in close proximity to one of the IGF2BP1 consensus-binding sequences, CA(A/U)(C/U)A (positions 133-137) (19). It is therefore plausible that IGF2BP1 protects OCLN mRNA from microRNA-dependent degradation. This mechanism of action of IGF2BP1 has been reported previously for other IGF2BP1 targets (12, 13, 20, 44)
It is plausible that other targets of IGF2BP1 contribute to the observed intestinal phenotype. For example, knockout of one of the previously identified IGF2BP1 target genes, β-TrCP1 (14), together with heterozygous deletion of its paralog, Fbxw11, in mouse gut epithelium causes lethal mucosal inflammation (45); however, we did not observe significant changes in mRNA expression of β-TrCP1 and Fbxw11 genes in IECs Igf2bp1IEC-Ind KO mice compared with control littermates (Fig. S2, B and C). We also examined the expression level of Myc, another IGF2BP1 target gene, shown to maintain intestinal crypt numbers in juvenile mice (46), but we did not detect significant changes in the mRNA expression of Myc between control and Igf2bp1IEC-Ind KO mice (Fig. S2A).
Wnt signaling and its importance in maintaining stem cell populations in the intestinal crypts has been reviewed extensively by Gehart and Clevers (47). IGF2BP1 expression is transcriptionally regulated by Wnt/β-catenin signaling (14), and the mRNAs of several Wnt target genes (e.g. MYC and MITF) were shown to be stabilized by IGF2BP1 in various cancer cell lines (5, 13). These findings hint at a possible role that IGF2BP1 plays in supporting some functions of Wnt signaling and suggests that it might be a potentially important component of Wnt-regulated intestinal homeostasis.
Deletion of Igf2bp1 from mouse intestinal epithelium has been shown to mitigate experimental colitis (31). Therefore, one might think that deletion of Igf2bp1 would probably protect intestinal epithelial barrier function. However, our results show that Igf2bp1 loss exacerbates experimental colitis in mice, implying a protective role for this protein. The contrast in results could potentially be explained by the different knockout models used. Utilization of inducible knockouts in our studies is probably more relevant for modeling intestinal inflammation in adults, whereas constitutive knockouts used by Chatterji et al. (31) are more useful to model development of intestinal epithelia and intestinal abnormalities during childhood.
Our data imply that in adult animals, the IGF2BP1 regulates occludin and hence protects the barrier function of intestinal epithelia. The experimental data summarized in this article provide novel evidence indicating the importance of IGF2BP1 in post-transcriptional regulation of occludin, via regulating OCLN mRNA stability.
Experimental procedures
Cell culture and reagents
Normal human colon epithelium CCD-841-CoTr cells, human colon cancer RKO cells, and Caco-2 cells were cultured and maintained as monolayers in Dulbecco's modified Eagle's medium (VWR International), supplemented with 10% (v/v) fetal bovine serum (Gibco, Life Technologies, Inc.) and 100 units/ml penicillin and streptomycin (Corning). The cells were incubated at 37 °C, 5% CO2. All cell lines were tested for mycoplasma contamination with the MycoAlert PLUS mycoplasma detection kit (Lonza).
Generation of IGF2BP1 knockdown, IGF2BP1 overexpression, and occludin expression cell lines
IGF2BP1 was knocked down using doxycycline-inducible shRNA constructs (purchased from GE Dharmacon). The IGF2BP1 doxycycline-inducible overexpression lentiviral plasmid was constructed by cloning PCR-amplified human IGF2BP1 ORF into the pInducer-20 vector (48). Lentivirus particles were produced from the above plasmids in Lenti-X 293T cells and were used to transduce CCD-CoTr, Caco-2, and RKO cells to make stable cell lines. Constitutively occludin-expressing Caco-2 cell lines in an IGF2BP1 knockdown background were constructed by transducing Caco-2-Sh-Igf2bp1 cells with pDUAL OCLN (GFP) lentiviral plasmid, which was gifted from Joe Grove (Addgene plasmid 86982; RRID:Addgene_86982).
Animals
Animals were maintained and procedures were performed in accordance with conditions approved by the Institutional Animal Care and Use Committee, Pennsylvania State University College of Medicine. Villin-CreERT2-Igf2bp1fl/fl mice were generated by crossing Villin-CreERT2 (49) mice with Igf2bp1f/fl mice. Igf2bp1fl/fl mice were generated previously and maintained on C57Bl/6 background (50). The Igf2bp1fl/fl mice were used as experimental controls. To knock out Igf2bp1, Villin-CreERT2-Igf2bp1f/fl mice were treated with a tamoxifen chow diet (Envigo-TD-130857) on weekdays, whereas a regular diet was given on weekends. Igf2bp1f/fl mice were also treated with tamoxifen chow in all experiment to serve as experimental control. During all animal experiments, Igf2bp1fl/fl and Villin-CreERT2-Igf2bp1fl/fl mice were co-housed for the reproducibility of results and to avoid potential contribution of differences in microbiota in observed phenotypes (51).
Tissue staining
Colonic tissues were formalin-fixed, paraffin-embedded, and H&E-stained. Immunofluorescent and/or immunohistochemical staining was performed on frozen tissue sections as described (52). Imaging was performed at the Penn State Medical Center Cell Imaging Core. In brief, after blocking, the colon sections were incubated overnight with primary antibodies against occludin (Thermo Fisher OC-3F10). Fluorescence-labeled secondary antibodies (Jackson Laboratories) were used in conjunction with nuclear dye 4′,6-diamidino-2-phenylindole to visualize the distribution of occludin in the colon tissues. Images were quantified using ImageJ software.
Gut barrier assay
Relative levels of serum LPS-specific immunoreactivity were examined by ELISA, as described previously (53). Briefly, microtiter plates were coated with purified Escherichia coli LPS (2 μg/well; from E. coli 0128: B12, Sigma, catalog no. 2887). Serum samples were serially diluted 1:200, 1:600, and 1:1200 and applied to wells coated with LPS. After incubation and washing, the wells were incubated with anti-mouse IgG (1:1000) coupled to horseradish peroxidase. Immunoreactivity to LPS was captured using the colorimetric peroxidase substrate tetramethylbenzidine, and absorbance was read at 450 nm with the use of an ELISA plate reader. Data are reported as optical density corrected by subtracting background (determined by readings in blank samples). Serum samples from Rag1-deficient mice were used as negative controls.
DSS colitis
Sex- and age-matched littermates (8–10 weeks) received DSS (2%) in drinking water for 5 days. Mouse body weights were recorded daily. The disease activity index was determined as described (32). Mice that died or were euthanized prior to the study end point because of impaired condition were included in weight loss and DAI calculations where the data points were available.
In vivo epithelial permeability assay
The in vivo intestinal epithelial permeability was measured as described previously (54). Briefly, age- and sex-matched littermates were orally administered (0.6 mg/g of body weight) with FITC-dextran solution (4 kDa, 80 mg/ml). After 4 h, the mice were sacrificed, blood was obtained by cardiac puncture, and plasma was separated for determination of FITC by a fluorimeter at 488 nm.
In vitro permeability assay and determination of caco-2 paracellular permeability
Caco-2 cells were cultured on transwells with polyester membrane insert (Corning, 3421) allowing proper cellular polarization with formation of an apical (top compartment) and basolateral face (bottom compartment). The insert was pretreated with Dulbecco's modified Eagle's medium overnight before cell plating. Caco-2 cells were seeded at a density of 0.5 × 105 cells/insert. The medium was replaced with fresh medium every 2 days. The TER of the filter-grown Caco-2 cells was measured by an epithelial Voltohmmeter (World Precision Instruments, Sarasota, FL, USA). Beside the measurements of TER, Caco-2 paracellular permeability was determined using the inulin (−14 °C, Mr = 5000) as paracellular marker for the determination of apical flux rates. Known concentrations (1.5 μm) of this paracellular marker were added to the apical solution, and radioactivity was measured in basal solution using a scintillation counter, as described previously (55).
CLIP
CLIP was carried out with minor modifications in accordance with the previously published protocols (56, 57). Briefly, CCD 841 CoTr cells were cultured for 3 days and then fixed with 3% formaldehyde in PBS for 10 min followed by lysis through sonication (10 pulses for 10 s). Overnight immunoprecipitation was performed at 4 °C using anti-IGF2BP1 antibodies (MBL, catalog no. RN007P) linked to Dynabeads (Life Technologies, Inc.). After collection by magnetic separation and washing five times with lysis buffer, the RNA-protein complexes were reverse-cross-linked, and RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Isolated RNA was treated with RNase-free DNase I (Thermo Scientific) to remove traces of genomic DNA. First-strand cDNA was generated using a cDNA synthesis kit from Bio-Rad according to the manufacturer's instructions, followed by real-time PCR using an iTaq Universal SYBR Green kit (Bio-Rad). The conditions for PCR amplification was as follows: initial denaturation for 3 minutes at 95°C, PCR amplification for 40 cycles with denaturation for the 20 seconds at 95°C, annealing for 20 seconds at 58°C and elongation at 72°C for 20 seconds. Analysis was done to obtain fold change in expression of particular gene by using CFX software from BioRad.
SDS-PAGE and immunoblotting analysis
Immunoblotting was done according to protocols described previously (58). Briefly, CCD-841-CoTr and Caco-2 cells were harvested, and whole-cell extract (WCE) was made by adding 1 ml of cell lysis buffer per 5 × 107 cells. The WCE was quantified using a Bio-Rad protein estimation kit. The WCE was analyzed by SDS-PAGE (10%) followed by immunoblotting. Blots were probed with the following antibodies: IGF2BP1, ZO-1, ZO-3, and β-actin from Cell Signaling, Claudin1 and occludin from Thermo Fisher, and Claudin2 from Abcam. Either anti-mouse or anti-rabbit horseradish peroxidase–conjugated secondary IgGs were used to detect their respective primary antibody. Quantitative luminescence of immunoblots was performed using ImageJ software.
Real-time PCR
Quantitative real-time PCR was conducted to quantify the mRNA levels. Total RNA was extracted using an RNeasy minikit from Qiagen, and cDNA was generated using an iScript cDNA synthesis kit (Bio-Rad) as per the manufacturer's instructions. Real-time PCR was performed using a CFX-96 RT-PCR machine with Bio-Rad SYBR Green mix. The conditions for PCR amplification were as follows: initial denaturation for 3 min at 95 °C, PCR amplification for 40 cycles with denaturation for the 20 s at 95 °C, annealing for 20 s at 58 °C, and elongation at 72 °C for 20 s. Analysis was done to obtain -fold change in expression of a particular gene by using CFX software from Bio-Rad. Three independent experiments were performed, and each sample was run in triplicate.
Data availability
All data relevant to these studies are contained within the article.
Supplementary Material
Acknowledgments
We thank Dr. Matam Vijay Kumar for invaluable input and suggestions to improve the manuscript. We also thank Dr. Joe Grove for the generous gift of reagent.
This article contains supporting information.
Author contributions—Vikash Singh and V. S. S. conceptualization; Vikash Singh, C. P. G., Vishal Singh, A. S. G., D. M. K., M. A. E., G. S. Y., P. N., and V. S. S. data curation; Vikash Singh, Vishal Singh, A. S. G., D. M. K., M. A. E., P. N., and V. S. S. formal analysis; Vikash Singh, C. P. G., A. S. G., G. S. Y., P. N., and V. S. S. validation; Vikash Singh, Vishal Singh, A. S. G., D. M. K., M. A. E., G. S. Y., and P. N. investigation; Vikash Singh, C. P. G., Vishal Singh, A. S. G., D. M. K., M. A. E., G. S. Y., P. N., and V. S. S. methodology; Vikash Singh writing-original draft; Vikash Singh, Vishal Singh, G. S. Y., P. N., and V. S. S. writing-review and editing; C. P. G., A. S. G., and D. M. K. visualization; G. S. Y. and V. S. S. resources; G. S. Y. and V. S. S. supervision; P. N. software; V. S. S. funding acquisition; V. S. S. project administration.
Funding and additional information—This study was supported in part by NCI, National Institutes of Health, Grants CA191550 and CA243167 (to V. S. S.). This work was also supported in part by NIDDK, National Institutes of Health, Grant DK114024 (to P. N.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- IGF2BP1
- insulin-like growth factor 2 mRNA-binding protein 1
- CLIP
- cross-linking and RNA immunoprecipitation
- IEC
- intestinal epithelial cell
- TJ
- tight junction
- IBD
- inflammatory bowel disease
- DSS
- dextran sodium sulfate
- DAI
- disease activity index
- RBP
- RNA-binding protein
- TER
- trans-epithelial electrical resistance
- miRNA
- microRNA
- shRNA
- short hairpin RNA
- WCE
- whole-cell extract
- H&E
- hematoxylin and eosin
- Ocln
- occludin.
References
- 1. Degrauwe N., Suva M. L., Janiszewska M., Riggi N., and Stamenkovic I. (2016) IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev. 30, 2459–2474 10.1101/gad.287540.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hansen T. V., Hammer N. A., Nielsen J., Madsen M., Dalbaeck C., Wewer U. M., Christiansen J., and Nielsen F. C. (2004) Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol. Cell Biol. 24, 4448–4464 10.1128/MCB.24.10.4448-4464.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Farina K. L., Hüttelmaier S., Musunuru K., Darnell R., and Singer R. H. (2003) Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation, and cytoskeletal attachment. J. Cell Biol. 160, 77–87 10.1083/jcb.200206003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hüttelmaier S., Zenklusen D., Lederer M., Dictenberg J., Lorenz M., Meng X., Bassell G. J., Condeelis J., and Singer R. H. (2005) Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438, 512–515 10.1038/nature04115 [DOI] [PubMed] [Google Scholar]
- 5. Leeds P., Kren B. T., Boylan J. M., Betz N. A., Steer C. J., Gruppuso P. A., and Ross J. (1997) Developmental regulation of CRD-BP, an RNA-binding protein that stabilizes c-myc mRNA in vitro. Oncogene 14, 1279–1286 10.1038/sj.onc.1201093 [DOI] [PubMed] [Google Scholar]
- 6. Doyle G. A., Betz N. A., Leeds P. F., Fleisig A. J., Prokipcak R. D., and Ross J. (1998) The c-myc coding region determinant-binding protein: a member of a family of KH domain RNA-binding proteins. Nucleic Acids Res 26, 5036–5044 10.1093/nar/26.22.5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ross A. F., Oleynikov Y., Kislauskis E. H., Taneja K. L., and Singer R. H. (1997) Characterization of a β-actin mRNA zipcode-binding protein. Mol. Cell Biol. 17, 2158–2165 10.1128/MCB.17.4.2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vikesaa J., Hansen T. V., Jønson L., Borup R., Wewer U. M., Christiansen J., and Nielsen F. C. (2006) RNA-binding IMPs promote cell adhesion and invadopodia formation. EMBO J. 25, 1456–1468 10.1038/sj.emboj.7601039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Noubissi F. K., Goswami S., Sanek N. A., Kawakami K., Minamoto T., Moser A., Grinblat Y., and Spiegelman V. S. (2009) Wnt signaling stimulates transcriptional outcome of the Hedgehog pathway by stabilizing GLI1 mRNA. Cancer Res. 69, 8572–8578 10.1158/0008-5472.CAN-09-1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yisraeli J. K. (2005) VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol. Cell 97, 87–96 10.1042/BC20040151 [DOI] [PubMed] [Google Scholar]
- 11. Manieri N. A., Drylewicz M. R., Miyoshi H., and Stappenbeck T. S. (2012) Igf2bp1 is required for full induction of Ptgs2 mRNA in colonic mesenchymal stem cells in mice. Gastroenterology 143, 110–121.e110 10.1053/j.gastro.2012.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elcheva I., Goswami S., Noubissi F. K., and Spiegelman V. S. (2009) CRD-BP protects the coding region of βTrCP1 mRNA from miR-183-mediated degradation. Mol Cell 35, 240–246 10.1016/j.molcel.2009.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goswami S., Tarapore R. S., Poenitzsch Strong A. M., TeSlaa J. J., Grinblat Y., Setaluri V., and Spiegelman V. S. (2015) MicroRNA-340-mediated degradation of microphthalmia-associated transcription factor (MITF) mRNA is inhibited by coding region determinant-binding protein (CRD-BP). J. Biol. Chem. 290, 384–395 10.1074/jbc.M114.590158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Noubissi F. K., Elcheva I., Bhatia N., Shakoori A., Ougolkov A., Liu J., Minamoto T., Ross J., Fuchs S. Y., and Spiegelman V. S. (2006) CRD-BP mediates stabilization of βTrCP1 and c-myc mRNA in response to β-catenin signalling. Nature 441, 898–901 10.1038/nature04839 [DOI] [PubMed] [Google Scholar]
- 15. Sparanese D., and Lee C. H. (2007) CRD-BP shields c-myc and MDR-1 RNA from endonucleolytic attack by a mammalian endoribonuclease. Nucleic Acids Res. 35, 1209–1221 10.1093/nar/gkl1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Runge S., Nielsen F. C., Nielsen J., Lykke-Andersen J., Wewer U. M., and Christiansen J. (2000) H19 RNA binds four molecules of insulin-like growth factor II mRNA-binding protein. J. Biol. Chem. 275, 29562–29569 10.1074/jbc.M001156200 [DOI] [PubMed] [Google Scholar]
- 17. Gu W., Katz Z., Wu B., Park H. Y., Li D., Lin S., Wells A. L., and Singer R. H. (2012) Regulation of local expression of cell adhesion and motility-related mRNAs in breast cancer cells by IMP1/ZBP1. J. Cell Sci. 125, 81–91 10.1242/jcs.086132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr., Jungkamp A. C., Munschauer M., Ulrich A., Wardle G. S., Dewell S., Zavolan M., and Tuschl T. (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Conway A. E., Van Nostrand E. L., Pratt G. A., Aigner S., Wilbert M. L., Sundararaman B., Freese P., Lambert N. J., Sathe S., Liang T. Y., Essex A., Landais S., Burge C. B., Jones D. L., and Yeo G. W. (2016) Enhanced CLIP uncovers IMP protein-RNA targets in human pluripotent stem cells important for cell adhesion and survival. Cell Rep. 15, 666–679 10.1016/j.celrep.2016.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Müller S., Glaß M., Singh A. K., Haase J., Bley N., Fuchs T., Lederer M., Dahl A., Huang H., Chen J., Posern G., and Hüttelmaier S. (2019) IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 47, 375–390 10.1093/nar/gky1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., Zhao B. S., Mesquita A., Liu C., Yuan C. L., Hu Y. C., Hüttelmaier S., Skibbe J. R., Su R., Deng X., et al. (2018) Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peterson L. W., and Artis D. (2014) Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nature reviews. Immunology 14, 141–153 10.1038/nri3608 [DOI] [PubMed] [Google Scholar]
- 23. Turner J. R. (2009) Intestinal mucosal barrier function in health and disease. Nature reviews. Immunology 9, 799–809 10.1038/nri2653 [DOI] [PubMed] [Google Scholar]
- 24. Marchiando A. M., Graham W. V., and Turner J. R. (2010) Epithelial barriers in homeostasis and disease. Annual review of pathology 5, 119–144 10.1146/annurev.pathol.4.110807.092135 [DOI] [PubMed] [Google Scholar]
- 25. Yu T. X., Rao J. N., Zou T., Liu L., Xiao L., Ouyang M., Cao S., Gorospe M., and Wang J. Y. (2013) Competitive binding of CUGBP1 and HuR to occludin mRNA controls its translation and modulates epithelial barrier function. Mol. Biol. Cell 24, 85–99 10.1091/mbc.e12-07-0531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cummins P. M. (2012) Occludin: one protein, many forms. Mol. Cell. Biol. 32, 242–250 10.1128/MCB.06029-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim D. H., and Cheon J. H. (2017) Pathogenesis of inflammatory bowel disease and recent advances in biologic therapies. Immune Netw. 17, 25–40 10.4110/in.2017.17.1.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rutgeerts P., Vermeire S., and Van Assche G. (2009) Biological therapies for inflammatory bowel diseases. Gastroenterology 136, 1182–1197 10.1053/j.gastro.2009.02.001 [DOI] [PubMed] [Google Scholar]
- 29. Abraham C., and Cho J. H. (2009) Inflammatory bowel disease. N. Engl. J. Med. 361, 2066–2078 10.1056/NEJMra0804647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hamilton K. E., Chatterji P., Lundsmith E. T., Andres S. F., Giroux V., Hicks P. D., Noubissi F. K., Spiegelman V. S., and Rustgi A. K. (2015) Loss of stromal IMP1 promotes a tumorigenic microenvironment in the colon. Mol. Cancer Res. 13, 1478–1486 10.1158/1541-7786.MCR-15-0224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chatterji P., Williams P. A., Whelan K. A., Samper F. C., Andres S. F., Simon L. A., Parham L. R., Mizuno R., Lundsmith E. T., Lee D. S., Liang S., Wijeratne H. S., Marti S., Chau L., Giroux V., et al. (2019) Posttranscriptional regulation of colonic epithelial repair by RNA binding protein IMP1/IGF2BP1. EMBO Rep. 20 10.15252/embr.201847074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rath E., Berger E., Messlik A., Nunes T., Liu B., Kim S. C., Hoogenraad N., Sans M., Sartor R. B., and Haller D. (2012) Induction of dsRNA-activated protein kinase links mitochondrial unfolded protein response to the pathogenesis of intestinal inflammation. Gut 61, 1269–1278 10.1136/gutjnl-2011-300767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wirtz S., Neufert C., Weigmann B., and Neurath M. F. (2007) Chemically induced mouse models of intestinal inflammation. Nat. Protoc. 2, 541–546 10.1038/nprot.2007.41 [DOI] [PubMed] [Google Scholar]
- 34. Catalioto R. M., Maggi C. A., and Giuliani S. (2011) Intestinal epithelial barrier dysfunction in disease and possible therapeutical interventions. Curr. Med. Chem. 18, 398–426 10.2174/092986711794839179 [DOI] [PubMed] [Google Scholar]
- 35. Saitou M., Furuse M., Sasaki H., Schulzke J. D., Fromm M., Takano H., Noda T., and Tsukita S. (2000) Complex phenotype of mice lacking occludin, a component of tight junction strands. Molecular biology of the cell 11, 4131–4142 10.1091/mbc.11.12.4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Al-Sadi R., Khatib K., Guo S., Ye D., Youssef M., and Ma T. (2011) Occludin regulates macromolecule flux across the intestinal epithelial tight junction barrier. Am. J. Physiol. Gastrointest. Liver Physiol. 300, G1054–G1064 10.1152/ajpgi.00055.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Van Itallie C. M., Fanning A. S., Holmes J., and Anderson J. M. (2010) Occludin is required for cytokine-induced regulation of tight junction barriers. J. Cell Sci. 123, 2844–2852 10.1242/jcs.065581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marchiando A. M., Shen L., Graham W. V., Weber C. R., Schwarz B. T., Austin J. R. 2nd, Raleigh D. R., Guan Y., Watson A. J., Montrose M. H., and Turner J. R. (2010) Caveolin-1-dependent occludin endocytosis is required for TNF-induced tight junction regulation in vivo. J. Cell Biol. 189, 111–126 10.1083/jcb.200902153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Buschmann M. M., Shen L., Rajapakse H., Raleigh D. R., Wang Y., Wang Y., Lingaraju A., Zha J., Abbott E., McAuley E. M., Breskin L. A., Wu L., Anderson K., Turner J. R., and Weber C. R. (2013) Occludin OCEL-domain interactions are required for maintenance and regulation of the tight junction barrier to macromolecular flux. Mol. Biol. Cell 24, 3056–3068 10.1091/mbc.e12-09-0688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Feldman G. J., Mullin J. M., and Ryan M. P. (2005) Occludin: structure, function and regulation. Adv. Drug Deliv. Rev. 57, 883–917 10.1016/j.addr.2005.01.009 [DOI] [PubMed] [Google Scholar]
- 41. Martinez C., Rodino-Janeiro B. K., Lobo B., Stanifer M. L., Klaus B., Granzow M., Gonzalez-Castro A. M., Salvo-Romero E., Alonso-Cotoner C., Pigrau M., Roeth R., Rappold G., Huber W., Gonzalez-Silos R., Lorenzo J., et al. (2017) miR-16 and miR-125b are involved in barrier function dysregulation through the modulation of claudin-2 and cingulin expression in the jejunum in IBS with diarrhoea. Gut 66, 1537–1538 10.1136/gutjnl-2016-311477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xiao L., and Wang J. Y. (2014) RNA-binding proteins and microRNAs in gastrointestinal epithelial homeostasis and diseases. Curr. Opin. Pharmacol. 19, 46–53 10.1016/j.coph.2014.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yu T., Lu X. J., Li J. Y., Shan T. D., Huang C. Z., Ouyang H., Yang H. S., Xu J. H., Zhong W., Xia Z. S., and Chen Q. K. (2016) Overexpression of miR-429 impairs intestinal barrier function in diabetic mice by down-regulating occludin expression. Cell Tissue Res. 366, 341–352 10.1007/s00441-016-2435-5 [DOI] [PubMed] [Google Scholar]
- 44. Müller S., Bley N., Glaß M., Busch B., Rousseau V., Misiak D., Fuchs T., Lederer M., and Hüttelmaier S. (2018) IGF2BP1 enhances an aggressive tumor cell phenotype by impairing miRNA-directed downregulation of oncogenic factors. Nucleic Acids Res. 46, 6285–6303 10.1093/nar/gky229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kanarek N., Grivennikov S. I., Leshets M., Lasry A., Alkalay I., Horwitz E., Shaul Y. D., Stachler M., Voronov E., Apte R. N., Pagano M., Pikarsky E., Karin M., Ghosh S., and Ben-Neriah Y. (2014) Critical role for IL-1β in DNA damage-induced mucositis. Proc. Natl. Acad. Sci. U.S.A. 111, E702–E711 10.1073/pnas.1322691111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bettess M. D., Dubois N., Murphy M. J., Dubey C., Roger C., Robine S., and Trumpp A. (2005) c-Myc is required for the formation of intestinal crypts but dispensable for homeostasis of the adult intestinal epithelium. Mol. Cell. Biol. 25, 7868–7878 10.1128/MCB.25.17.7868-7878.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gehart H., and Clevers H. (2019) Tales from the crypt: new insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 16, 19–34 10.1038/s41575-018-0081-y [DOI] [PubMed] [Google Scholar]
- 48. Meerbrey K. L., Hu G., Kessler J. D., Roarty K., Li M. Z., Fang J. E., Herschkowitz J. I., Burrows A. E., Ciccia A., Sun T., Schmitt E. M., Bernardi R. J., Fu X., Bland C. S., Cooper T. A. et al. (2011) The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 108, 3665–3670 10.1073/pnas.1019736108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. el Marjou F., Janssen K. P., Chang B. H., Li M., Hindie V., Chan L., Louvard D., Chambon P., Metzger D., and Robine S. (2004) Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 10.1002/gene.20042 [DOI] [PubMed] [Google Scholar]
- 50. Hamilton K. E., Noubissi F. K., Katti P. S., Hahn C. M., Davey S. R., Lundsmith E. T., Klein-Szanto A. J., Rhim A. D., Spiegelman V. S., and Rustgi A. K. (2013) IMP1 promotes tumor growth, dissemination and a tumor-initiating cell phenotype in colorectal cancer cell xenografts. Carcinogenesis 34, 2647–2654 10.1093/carcin/bgt217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Franklin C. L., and Ericsson A. C. (2017) Microbiota and reproducibility of rodent models. Lab Anim. 46, 114–122 10.1038/laban.1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen Y., Zhang H. S., Fong G. H., Xi Q. L., Wu G. H., Bai C. G., Ling Z. Q., Fan L., Xu Y. M., Qin Y. Q., Yuan T. L., Sun H., and Fang J. (2015) PHD3 stabilizes the tight junction protein occludin and protects intestinal epithelial barrier function. J. Biol. Chem. 290, 20580–20589 10.1074/jbc.M115.653584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ziegler T. R., Luo M., Estivariz C. F., Moore D. A. 3rd, Sitaraman S. V., Hao L., Bazargan N., Klapproth J. M., Tian J., Galloway J. R., Leader L. M., Jones D. P., and Gewirtz A. T. (2008) Detectable serum flagellin and lipopolysaccharide and upregulated anti-flagellin and lipopolysaccharide immunoglobulins in human short bowel syndrome. Am. J. Physiol. Regul. Integr. Comp. Physiol. 294, R402–R410 10.1152/ajpregu.00650.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tambuwala M. M., Cummins E. P., Lenihan C. R., Kiss J., Stauch M., Scholz C. C., Fraisl P., Lasitschka F., Mollenhauer M., Saunders S. P., Maxwell P. H., Carmeliet P., Fallon P. G., Schneider M., and Taylor C. T. (2010) Loss of prolyl hydroxylase-1 protects against colitis through reduced epithelial cell apoptosis and increased barrier function. Gastroenterology 139, 2093–2101 10.1053/j.gastro.2010.06.068 [DOI] [PubMed] [Google Scholar]
- 55. Nighot P. K., Hu C. A., and Ma T. Y. (2015) Autophagy enhances intestinal epithelial tight junction barrier function by targeting claudin-2 protein degradation. J. Biol. Chem. 290, 7234–7246 10.1074/jbc.M114.597492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vogt M., and Taylor V. (2013) Cross-linked RNA immunoprecipitation. Bio-protoc. 3, e398 10.21769/BioProtoc.398 [DOI] [Google Scholar]
- 57. Knuckles P., Vogt M. A., Lugert S., Milo M., Chong M. M., Hautbergue G. M., Wilson S. A., Littman D. R., and Taylor V. (2012) Drosha regulates neurogenesis by controlling neurogenin 2 expression independent of microRNAs. Nat. Neurosci. 15, 962–969 10.1038/nn.3139 [DOI] [PubMed] [Google Scholar]
- 58. Harlow E. (1988) Immunoblotting. in Antibodies: A Laboratory Manual (Harlow H., and Lane D., eds) pp. 471–510, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 59. Erben U., Loddenkemper C., Doerfel K., Spieckermann S., Haller D., Heimesaat M. M., Zeitz M., Siegmund B., and Kühl A. A. (2014) A guide to histomorphological evaluation of intestinal inflammation in mouse models. Int. J. Clin. Exp. Pathol. 7, 4557–4576 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data relevant to these studies are contained within the article.