Abstract
Background
TP53 mutations are common in breast cancer. There is currently no large‐scale cohort study to investigate the TP53 landscape in breast cancer patients from China. The predictive value of TP53 mutations for the efficacy of human epidermal growth factor receptor 2 (HER2)‐targeted therapy in breast cancer remains controversial. In the present study, we aimed to analyze the clinical spectrum and prognostic value of TP53 mutations in circulating tumor DNA (ctDNA) from breast cancer patients in China.
Methods
We retrospectively analyzed the clinical data and TP53 mutation features in ctDNA samples from 804 patients with metastatic breast cancer. TP53 mutations were detected by target region capture‐based next‐generation sequencing. The relationship between TP53 mutation status and disease‐free survival (DFS) was analyzed in 444 patients with metastatic breast cancer. Moreover, the relationship between TP53 mutation status and progression‐free survival (PFS) was analyzed in 55 HER2‐positive patients treated with first‐line trastuzumab‐based therapy. Kaplan‐Meier analysis was performed to estimate the survival curves of the different subgroups, and the log‐rank test was used to compare the curves. A Cox regression model was used to estimate multivariable‐adjusted hazard ratios and their 95% confidence intervals (CIs) associated with the DFS and PFS.
Results
Among the 804 investigated patients, 431 (53.6%) patients harbored TP53 mutations. TP53 mutations were differentially distributed among different molecular subtypes of breast cancer (P < 0.05). Patients with TP53 mutations had a shorter DFS than those with wild‐type TP53 (hazard ratio = 1.32, 95% CI = 1.09‐1.61, P = 0.005). TP53 mutations in exons 5‐8 were associated with worse outcome (hazard ratio = 1.50, 95% CI = 1.11‐2.03, P = 0.009). However, TP53 mutation status was not significantly associated with PFS in HER2‐positive patients who received first‐line trastuzumab‐based therapy (P = 0.966). Interestingly, in the taxane combination group, patients with TP53 mutations exhibited longer PFS than those without TP53 mutations (hazard ratio = 0.08, 95% CI = 0.02‐0.30, P < 0.001). However, in the non‐taxane combination group, patients with TP53 mutations displayed shorter PFS than those with wild‐type TP53 (hazard ratio = 4.84, 95% CI = 1.60‐14.66, P = 0.005).
Conclusions
TP53 mutations in exons 5‐8 may be an independent prognostic marker for short DFS in patients with metastatic breast cancer. TP53 mutations had opposite effects on trastuzumab‐treated patients treated with and without taxanes.
Keywords: breast cancer, TP53 mutation, circulating tumor DNA, next‐generation sequencing, Chinese, prognosis, trastuzumab, taxanes
Abbreviations
- CI
confidence interval
- ctDNA
circulating tumor DNA
- DFS
disease‐free survival
- gDNA
genomic DNA
- HER2
human epidermal growth factor receptor 2
- HR
hormone receptor
- IDC
infiltrating ductal carcinoma
- IHC
immunohistochemistry
- ILC
infiltrating lobular carcinoma
- PFS
progression‐free survival
- SNV
single‐nucleotide variant
- TNBC
triple‐negative breast cancer
1. BACKGROUND
TP53 is a tumor suppressor gene that plays a key role in many cellular pathways and regulates essential cell activities, such as proliferation, differentiation, cell death, DNA repair, and the formation of blood vessels [1, 2]. Mutations in TP53 are common in cancers [3, 4] and present in approximately 25%‐30% of all breast cancer cases [5, 6]. Several studies have investigated the TP53 mutational spectrum in various subtypes of breast cancer [5, 6, 7, 8]. However, the TP53 genetic landscape in Chinese breast cancer patients remains unclear.
Numerous studies have indicated that TP53 is a biomarker for predicting poor prognosis in breast cancer patients [5, 9, 10, 11]. TP53 mutations located in the regions encoded TP53 DNA‐binding domains have been reported to be associated with poor prognosis in patients with breast cancer [10, 12]. However, their predictive value for the efficacy of human epidermal growth factor receptor 2 (HER2)‐targeted therapy in breast cancer remains controversial. Several studies most of which relied upon immunohistochemistry (IHC) to assess p53 alterations have indicated that TP53 mutations were not associated with the response to anticancer therapy [8, 13‐16]. Nevertheless, IHC cannot be used to detect all gene mutations, as many mutations do not lead to p53 protein accumulation. One early study indicated that p53 status, as determined by IHC, was not associated with the response to trastuzumab‐based treatment [15]. However, two clinical studies have indicated that patients with TP53 mutations exhibited good responses to trastuzumab [14, 17]. Since most of the patients in these studies received chemotherapy in addition to trastuzumab, these findings must be confirmed by further studies.
In the present study, we used target region capture‐based next‐generation sequencing to analyze the TP53 mutation spectrum of circulating tumor DNA (ctDNA) from 804 Chinese breast cancer patients and analyzed the relationships between TP53 mutations, prognosis, and the efficacy of first‐line trastuzumab‐based therapy.
2. MATERIALS AND METHODS
2.1. Patients
TP53 mutation status was determined in the plasma of patients with metastatic breast cancer treated at National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (Beijing, China) between March 2015 and October 2018. The inclusion criteria were as follows: (1) patients who were at least 18 years old; (2) patients who had a histologic/cytologic diagnosis of invasive breast cancer; (3) patients who had received radical surgery for primay tumor; (4) patients who were female; (5) patients who had complete clinicopathological data; and (6) patients who had sufficient blood samples for ctDNA analysis. The exclusion criteria were as follows: (1) patients who were male; and (2) patients who had received chemotherapy within the past month before the peripheral blood samples collection. This study was approved by the Ethics Committee of National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (ref.: 16‐038/1117). All patients provided written informed consent.
To analyze the association between TP53 mutations and trastuzumab responses, we retrospectively calculated the progression‐free survival (PFS) of human epidermal growth factor receptor 2‐positive (HER2+) patients who received trastuzumab (6 mg/kg every 3 weeks intravenously after an initial loading dose of 8 mg/kg until disease progress) combined with chemotherapy as first‐line therapy after being diagnosed with recurrent or metastatic breast cancer. Chemotherapy was chosen by the treating physician according to prior treatments and the patient's clinical characteristics. Patients were excluded if they had received previous targeted therapy, endocrine therapy, or any previous systemic chemotherapy for advanced disease. We did correlative analyses of PFS in patient subgroups based on combined chemotherapy drugs. Patietns who received trastuzumab combined with paclitaxel and docetaxel treatment were defined as taxane combination group; patients who received trastuzumab combined with capecitabine, vinorelbine, gemcitabine and cisplatin were defined as non‐taxane combination group.
We compared the TP53 mutation frequency in our cohort with the MSK‐IMPACT cohort [18]. The gene mutation status and clinical information of 1,746 patients with breast cancer were downloaded from the MSK‐IMPACT dataset using the cBioPortal database (http://www.cbioportal.org).
2.2. Blood sample collection
The testing for patients was ordered by the treating physician to identify clinically relevant genomic alterations that could potentially help to determine prognosis or to make treatment decisions. Peripheral blood samples for ctDNA analysis were collected in Streck tubes (Streck, Omaha, NE, USA) and centrifuged (1,600 × g at 4°C for 10 min, followed by transfer to new microcentrifuge tubes, and centrifugation at 16,000 × g at 4°C for 10 min to remove remaining cell debris) within 72 hours to separate the plasma from the peripheral blood.
2.3. DNA extraction and quantification
A QIAamp Circulating Nucleic Acid Kits (Qiagen, Hilden, Germany) was used to isolate circulating DNA from plasma samples. Genomic DNA (gDNA) was isolated from peripheral lymphocytes using the QIAamp DNA Blood Mini Kit (Qiagen) as a control. All DNA extractions were performed according to the manufacturer's protocols.
The DNA concentration was assessed using a Qubit fluorometer (Invitrogen, Carlsbad, CA, USA) and the Qubit dsDNA High‐sensitivity (HS) Assay Kit (Invitrogen). The size distribution of the circulating DNA was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and the DNA HS kit (Agilent Technologies).
2.4. Target capture and next‐generation sequencing
Library preparation, hybrid capture, and circulating DNA and gDNA sequencing were performed as previously described [19, 20]. The targeted region for sequencing covered ∼1.1 Mb, including 1,021 genes [21], and encompassed all introns and exons of TP53. DNA sequencing was performed using the HiSeq 3000 Sequencing System (Illumina, San Diego, CA, USA) with 2 × 101 bp paired‐end reads.
Low‐quality reads and terminal adaptor sequences were filtered out. Burrows‐Wheeler Aligner (BWA, version 0.7.12‐r1039) was carried out to align the clean reads to the reference human genome (hg19), and Genome Analysis Toolkit (GATK, version 3.4‐46‐gbc02625) was used for realignment and recalibration. Single‐nucleotide variants (SNVs) were characterized by MuTect (version 1.1.4) and NChot [22]. Small insertions and deletions (indels) were identified by GATK.
2.5. Follow‐up
All patients were regularly followed‐up at the out‐patient clinic every 3 months within 2 years and every 6 months 2‐5 years after the surgery. The efficacy, safety, and survival data of advanced breast cancer patients were followed‐up every 2‐3 months. Last follow‐up time was October 30, 2018.
2.6. Statistical analysis
The chi‐square test and Fisher's exact test were used to compare categorical variables. Disease‐free survival (DFS) was defined as the duration from radical resection of breast cancer to the time of recurrence or death of any cause. PFS was calculated from the date of treatment initiation to the date of disease progression or death of any cause. Patients without an endpoint (progression or death) were censored at the date of the last follow‐up. Kaplan‐Meier survival plots were generated based on gene mutation status, and curves were compared using log‐rank tests. A Cox proportional hazards model was used to analyze the association between DFS or PFS and gene mutations and clinical characteristics. All statistical tests used were two‐sided, and results with P values less than 0.05 were considered significant. All statistical analyses were performed using SPSS version 23.0 (IBM Corporation, Armonk, NY, USA) or GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA, USA).
3. RESULTS
3.1. Patient characteristics
A total of 804 female patients with metastatic breast cancer treated between March 2015 and October 2018 were retrospectively investigated. Patient characteristics are summarized in Table 1. The median age at which the patients were initially diagnosed with breast cancer was 42 years (range: 22 to 78 years) in all of the patients. The median age was 43 years (range: 22 to 78 years) in the TP53 mutant group versus 42 years (range: 26 to 78 years) in the TP53 wild‐type group (P = 0.568). Fifty‐five (6.8%) patients received trastuzumab plus chemotherapy as the first‐line treatment after being diagnosed with metastatic breast cancer.
TABLE 1.
Clinical characteristics of 804 patients with metastatic breast cancer
| Characteristic | Whole cohort [cases (%)] | TP53 mutant [cases (%)] | TP53 wild‐type [cases (%)] | P value |
|---|---|---|---|---|
| Total | 804 | 431 | 373 | |
| Menopausal status | ||||
| Premenopausal | 469 (58.3) | 247 (57.3) | 222 (59.5) | 0.526 |
| Postmenopausal | 335 (41.7) | 184 (42.7) | 151 (40.5) | |
| Tumor stage at diagnosis | ||||
| Stage 1‐2 | 495 (61.6) | 255 (59.2) | 240 (64.3) | 0.132 |
| Stage 3 | 309 (38.4) | 176 (40.8) | 133 (35.7) | |
| Histology at diagnosis | ||||
| ILC | 42 (5.2) | 23 (5.3) | 19 (5.1) | 0.877 |
| IDC | 762 (94.8) | 408 (94.7) | 354 (94.9) | |
| Molecular subtype | ||||
| HR+/HER2− | 397 (49.4) | 170 (39.4) | 227 (60.9) | <0.001 |
| HR+/HER2+ | 138 (17.2) | 69 (16.0) | 69 (18.5) | |
| HR−/HER2+ | 121 (15.0) | 84 (19.5) | 37 (9.9) | |
| HR−/HER2− | 148 (18.4) | 108 (25.1) | 40 (10.7) | |
| HR status † | ||||
| Positive | 535 (66.5) | 239 (55.5) | 296 (79.4) | <0.001 |
| Negative | 269 (33.5) | 192 (44.5) | 77 (20.6) | |
| HER2 status | ||||
| Positive | 259 (32.2) | 153 (35.5) | 106 (28.4) | 0.032 |
| Negative | 545 (67.8) | 278 (64.5) | 267 (71.6) | |
| Line of chemotherapy | ||||
| 0 | 16 (2.0) | 5 (1.2) | 11 (2.9) | 0.051 |
| 1 | 194 (24.1) | 96 (22.3) | 98 (26.3) | |
| ≥2 | 594 (73.9) | 330 (76.6) | 264 (70.8) | |
| Line of endocrine therapy for HR+ patients | ||||
| 0 | 221 (41.3) | 111 (46.4) | 110 (37.2) | 0.057 |
| 1 | 146 (27.3) | 64 (26.8) | 82 (27.7) | |
| ≥2 | 168 (31.4) | 64 (26.8) | 104 (35.1) | |
| Line of anti‐HER2 therapy for HER2+ patients | ||||
| 0 | 116 (44.8) | 76 (49.7) | 40 (37.7) | 0.057 |
| ≥1 | 143 (55.2) | 77 (50.3) | 66 (62.3) | |
Positive HR status was defined as positive for estrogen receptor and/or progesterone receptor.
Abbreviations: HR, hormone receptor; HER2, human epidermal growth factor receptor 2; ILC, infiltrating lobular carcinoma; IDC, infiltrating ductal carcinoma.
3.2. Characterization of TP53 mutations
We performed target region capture‐based next‐generation sequencing to evaluate the status of TP53 mutations in the whole cohort. We found that 431 (53.6%) patients harbored at least one TP53 mutation. Among them, 65 (15.1%) patients carried two different TP53 mutations. The TP53 mutations p.R213*, p.R248Q, p.R273H, p.Y220C, and p.R175H were the top five mutations according to frequency and were present in 17 (3.9%), 15 (3.5%), 14 (3.2%), 12 (2.8%), and 11 (2.6%) patients, respectively. The majority of the mutations were missense mutations (55.7%), followed by frameshift (17.3%) and nonsense (15.4%) mutations. The distribution of mutations was nonuniform across the gene, with 75.1% of the mutations clustering in exons 5‐8, mostly spanning the sequence encoding the DNA‐binding domain. Exons 4 and 10 also harbored a substantial number of mutations, accounting for 10.4% and 4.2% of all mutations, respectively. Only 2.5% of the mutations were in exon 9, and the remaining 7.8% were in exon 3 and the intervening sequences (Supplementary Table S1).
3.3. Spectra of TP53 mutations in different molecular subtypes
TP53 mutations were differentially distributed among different molecular subtypes of breast cancer (P < 0.01; Figure 1A). TP53 mutation frequency was lower in the patients with luminal‐type breast cancer (hormone receptor‐positive [HR+]HER2+/−; 44.7% [239/535];) than in the patients with HR−HER2+ breast cancer (69.4% [84/121]; P < 0.001) and triple‐negative breast cancer (TNBC) (HR−HER2−; 73.0% [108/148]; P < 0.001).
FIGURE 1.
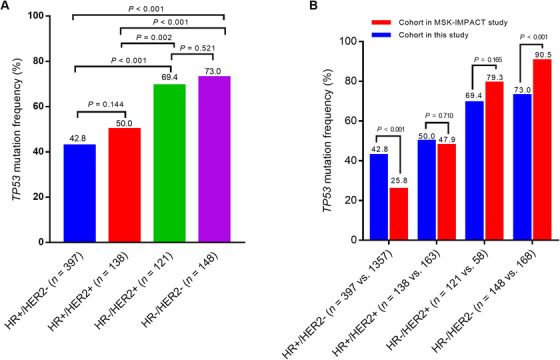
Frequencies of TP53 mutations in different molecular subtypes of breast cancer. A. Proportion of patients with TP53 mutations according to molecular subtype. B. Comparison of TP53 mutation frequencies between our data and the MSK‐IMPACT data. Abbreviations: HER2, human epidermal growth factor receptor 2; HR, hormone receptor
Patients with HR−HER2+ tumors showed a higher proportion of span mutations as compared with non‐HR−HER2+ patients (17.1% [18/105] versus 8.0% [33/414]; P = 0.005), whereas those with HR+HER2+ cancer showed a higher proportion of missense mutations compared with patients with non‐HR+HER2+ cancer (67.1% [51/76] versus 53.7% [238/443]; P = 0.030). We did not find any differences in mutational hotspots among the molecular subtypes (P > 0.05; Table 2).
TABLE 2.
TP53 mutation characteristics in different molecular subtypes of breast cancer
| TP53 mutation characteristic | Total | HR+HER2− | HR+HER2+ | HR+HER2− | HR−HER2− | P value |
|---|---|---|---|---|---|---|
| Number of mutations | 519 | 206 | 76 | 105 | 132 | |
| Variant classification [number of mutations (%)] | ||||||
| Missense | 289 (55.7) | 120 (58.3) | 51 (67.1) | 47 (44.8) | 71 (53.8) | 0.020 |
| Frame shift | 90 (17.3) | 27 (13.1) | 11 (14.5) | 21 (20.0) | 31 (23.5) | 0.072 |
| Nonsense | 80 (15.4) | 37 (18.0) | 10 (13.2) | 17 (16.2) | 16 (12.1) | 0.481 |
| Span | 51 (9.8) | 16 (7.8) | 4 (5.3) | 18 (17.1) | 13 (9.8) | 0.028 |
| In‐frame Del/Ins | 9 (1.7) | 6 (2.9) | 0 (0) | 2 (1.9) | 1 (0.8) | 0.371 |
| Mutational hotspots [number of mutations (%)] | ||||||
| R213* | 17 (3.3) | 9 (4.4) | 1 (1.3) | 5 (4.8) | 2 (1.5) | 0.291 |
| R248Q | 15 (2.9) | 7 (3.4) | 1 (1.3) | 1 (1.0) | 6 (4.5) | 0.196 |
| R273H | 14 (2.7) | 4 (1.9) | 3 (3.9) | 4 (3.8) | 3 (2.3) | 0.283 |
| Y220C | 12 (2.3) | 3 (1.5) | 2 (2.6) | 1 (1.0) | 6 (4.5) | 0.050 |
| R175H | 11 (2.1) | 4 (1.9) | 3 (3.9) | 2 (1.9) | 2 (1.5) | 0.648 |
| Others | 450 (86.7) | 179 (86.9) | 66 (86.8) | 92 (87.6) | 113 (85.6) | <0.001 |
Abbreviations: HR, hormone receptor; HER2, human epidermal growth factor receptor 2. Del, deletion; Ins, insertion.
We further compared the data in this study (from a Chinese cohort) with those from the MSK‐IMPAC cohort. A total of 1746 breast cancer patients from the MSK‐IMPACT cohort were included. Among them, 626 (35.9%) patients harbored TP53 mutations. The frequency of TP53 mutations in the Chinese cohort was significantly higher than that in the MSK‐IMPACT cohort (P < 0.001). We further investigated the TP53 mutation frequencies among different molecular subtypes and compared them between the Chinese and the MSK‐IMPACT cohorts. We observed significant differences between these two cohorts: in the HR+HER2− subgroup, the TP53 mutation frequency was higher in the Chinese cohort (P < 0.001); in contrast, in the TNBC subgroup, the frequency of TP53 mutation was higher in the MSK‐IMPACT cohort (P < 0.001; Figure 1B).
3.4. Association of TP53 mutation status and the DFS of breast cancer patients
To analyze the association between TP53 mutation and DFS, we retrospectively calculated the DFS of the eligible patients. A total of 444 patients recorded DFS, including 243 (54.7%) patients with TP53 mutations. We found that patients with TP53 mutations had a shorter DFS than those with wild‐type TP53. The median DFS for patients with TP53 mutations was 28.5 months (95% confidence interval [CI] = 23.6‐33.3 months) versus 40.6 months (95% CI = 33.5‐47.7 months) for patients without TP53 mutations (hazard ratio = 1.32; 95% CI = 1.09‐1.61; P = 0.005; Figure 2).
FIGURE 2.
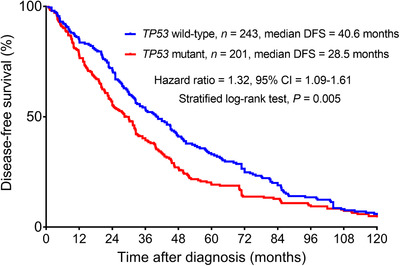
Kaplan‐Meier analyses of DFS in 444 patients with metastatic breast cancer. Abbreviations: DFS, disease‐free survival; CI, confidence interval
The results of the univariate Cox regression analysis with clinical characteristics and TP53 mutation status are shown in Table 3. HER2 status, HR status, tumor stage at diagnosis, histology at diagnosis, TP53 mutation status, and exons 5‐8 mutation status were related to DFS of patients with metastatic breast cancer (all P < 0.05). A multivariate Cox regression analysis was conducted to investigate the association between these characteristics and DFS. TP53 mutation status was not significantly associated with DFS (hazard ratio = 1.18, 95% CI = 0.97‐1.45, P = 0.101). However, the DFS of patients with TP53 mutations in exons 5‐8 was significantly shorter than the DFS of patients with wild‐type TP53 (hazard ratio = 1.50, 95% CI = 1.11‐2.03, P = 0.009, Table 3).
TABLE 3.
Univariate and multivariate Cox regression analyses for DFS in patients with breast cancer
| Variable | Univariate Cox regression analysis | Multivariate Cox regression analysis | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | |
| HER2 status (positive vs. negative) | 1.42 (1.15‐1.77) | 0.001 | 1.28 (1.02‐1.60) | 0.031 |
| HR status † (positive vs. negative) | 0.69 (0.56‐0.85) | 0.001 | 0.76 (0.62‐0.95) | 0.015 |
| Tumor stage at diagnosis (stages 1‐2 vs. stage 3) | 0.71 (0.59‐0.86) | 0.001 | 0.47 (0.24‐0.91) | 0.005 |
| Histology at diagnosis (ILC vs. IDC) | 0.46 (0.24‐0.88) | 0.020 | 0.76 (0.62‐0.92) | 0.026 |
| TP53 mutation status (mutation vs. wild‐type) | 1.32 (1.09‐1.61) | 0.005 | 1.18 (0.97‐1.45) | 0.101 |
| TP53 exons 5‐8 mutation status (mutation vs. wild‐type) | 1.61 (1.20‐2.17) | 0.002 | 1.50 (1.11‐2.03) | 0.009 |
Positive HR status was defined as positive for estrogen receptor and/or progesterone receptor.
Abbreviations: DFS, disease‐free survival; HER2, human epidermal growth factor receptor 2; HR, hormone receptor; ILC, infiltrating lobular carcinoma; IDC, infiltrating ductal carcinoma.
3.5. Association of TP53 mutation status and the response to first‐line trastuzumab‐based therapy in breast cancer patients
A total of 55 metastatic breast cancer patients who received trastuzumab‐based treatment as first‐line therapy were selected to evaluate the relationship between TP53 mutation status and the response to trastuzumab. As shown in Supplementary Table S2, there were no significant differences in clinical characteristics between TP53 mutant and wild‐type groups (all P > 0.05). Additionally, TP53 mutation status was not significantly associated with PFS. The median PFS for patients with TP53 mutation was 9.1 months versus 5.4 months for those without TP53 mutation (hazard ratio = 1.01; 95% CI = 0.59‐1.75; P = 0.966; Figure 3A).
FIGURE 3.
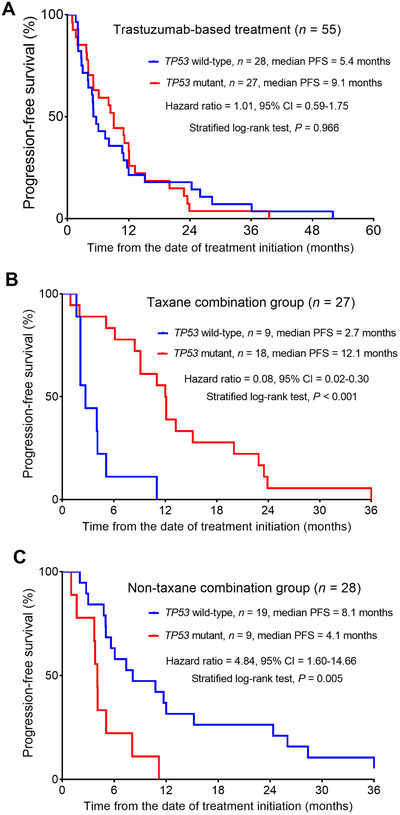
Impact of TP53 mutation on the response to trastuzumab‐based first‐line therapy. A. PFS of patients underwent trastuzumab‐based first‐line treatment based on the presence of TP53 mutation. B. PFS of patients in the taxane combination group based on the presence of TP53 mutation. C. PFS of patients in the non‐taxane combination group based on the presence of TP53 mutation. Abbreviations: PFS, progression‐free survival; CI, confidence interval
To further explore the relationship between TP53 mutation status and taxanes (including paclitaxel and docetaxel) treatment response in the breast cancer patients who received trastuzumab‐based treatment as first‐line therapy, we divided these patients into the taxane and non‐taxane combination groups (Supplementary Table S3). Interestingly, we found that in the taxane combination group, patients with TP53 mutations had longer median PFS than those without TP53 mutations (12.1 months vs. 2.7 months; hazard ratio = 0.08, 95% CI = 0.02‐0.30, P < 0.001; Figure 3B). However, in the non‐taxane combination group, the opposite pattern was observed. Patients with TP53 mutations had shorter median PFS than those with wild‐type TP53 (4.1 months vs. 8.1 months, (hazard ratio = 4.84, 95% CI = 1.60‐14.66, P = 0.005; Figure 3C).
4. DISCUSSION
In the present study, we performed a TP53 mutation analysis within a large cohort of 804 Chinese patients with metastatic breast cancer and further compared our data to those from the MSK‐IMPACT cohort. Our results indicated that TP53 mutation was a prognostic marker for poor outcomes in breast cancer. Additionally, it may affect the response of breast cancer patients to first‐line therapy based on trastuzumab.
In the present study, the frequency of TP53 mutation was 53.6%, which was higher than that in the MSK‐IMPACT cohort (35.9%) [18]. Additionally, our data indicated that the TP53 mutation frequency was higher in TNBC and HER2+/HR− breast cancer than in luminal‐type breast cancer. We also found that the frequency of TP53 mutation in HER2−/HR+ breast cancer was higher in the MSK‐IMPACT cohort than in our cohort, while the opposite was true for the TNBC subgroup. Further study is needed to investigate these differences between cohorts. A striking feature of the patient cohort in the present study was that the median age at diagnosis was very young (42 years). This corresponds to increased proportions of HER2+ (32.2%) and TNBC (18.4%) patients in the study population. In addition, all patients in the study population had metastatic disease at the time of consent and blood drawn for the study. Moreover, most patients had received at least 2 lines of chemotherapy. It is our understanding that metastatic breast cancer are also associated with a higher frequency of TP53 mutations than cancers at earlier stages. Both of these features of the patient population may have contributed to the higher percentage of TP53 mutations found in this cohort when compared with that in the MSK‐IMPACT study. Another possibility is that this discrepancy reflects differences between the ethnicities of the patients.
Previous studies have shown that the TP53 sequence encoding the DNA‐binding domain was the most frequently mutated region in the TP53 gene in breast cancer [6, 8]. Our study also found that most mutations occurred in the DNA‐binding domain and that p.R213*, p.R248Q, p.R273H, p.Y220C, and p.R175H were common mutations of TP53. The majority (55.7%) of the mutations were missense mutations. Previous studies have shown that TP53 mutation was a prognostic marker for poor breast cancer outcomes. Several studies have investigated the impact of different types of TP53 somatic mutations on breast cancer, with conflicting results [5, 10, 12, 23, 24]. Some studies [10, 12] have shown that only missense mutations in the DNA‐binding domain affected the outcome of breast cancer patients, but this was not confirmed by other studies [10, 12, 24, 25]. However, most previous studies have failed to investigate whether the prognostic impact of TP53 mutation is independent of other clinicopathologic factors [10, 12, 24]. In the present study, univariate Cox regression analysis indicated that TP53 mutation was related to short DFS. However, in the multivariate analysis, TP53 mutation status was not significantly associated with DFS. Further analysis showed that TP53 mutation in exons 5‐8 was significantly associated with shorter DFS than wild‐type TP53. TP53 mutations located in exons 5‐8 may predict shorter DFS of breast cancer patients than wild‐type TP53. Most of the TP53 mutations found in breast cancer were missense substitutions located in the DNA binding domain (exons 5‐8). The difference in prognosis of TP53 DNA‐binding domain mutations and mutations in other domains is still under debate. This difference is hypothesed to be related to the degree to which the mutants function as dominant negative mutants [24]. However, this hypothesis needs to be confirmed by further studies.
Data on the association between the presence of TP53 mutations and clinical treatment response are conflicting. Although a number of prior studies of breast cancer have suggested that the presence of TP53 mutations was associated with a lack of response to a variety of chemotherapy agents, including anthracyclines and the combination of cyclophosphamide, methotrexate, and fluorouracil [8, 26, 27], some other studies have found that TP53 mutation was associated with a better response to docetaxel and trastuzumab [13, 14, 28, 29, 30]. The association between TP53 mutation and trastuzumab efficacy is also controversial. One early study indicated that p53 status, as determined by IHC, is not associated with the response to trastuzumab‐based treatment [15]. Nevertheless, two clinical studies have indicated that patients with TP53 mutations may exhibit a better response to trastuzumab [14, 17]. However, trastuzumab is used in combination with chemotherapy in most cases during clinical practice. Therefore, we investigated the efficacy of trastuzumab in metastatic breast cancer. To reduce bias from other factors, we chose patients who received trastuzumab as first‐line therapy. We did not find any association between TP53 mutation and the efficacy of trastuzumab‐based first‐line therapy. We further analyzed the impact of TP53 mutation in different chemotherapy subgroups. Interestingly, we found that in the taxane combination group (including paclitaxel and docetaxel), the presence of TP53 mutation was a sensitive biomarker of efficacy. In contrast, in the non‐taxane combination group, patients with TP53 mutations exhibited a poor response. Different types of chemotherapeutics have different mechanisms of action, which may explain why patients with TP53 mutations showed a better response to taxanes. A preclinical study indicated that in vitro, TP53 mutant tumor cell lines are more sensitive to mitotic poisons, such as taxanes, than are wild‐type TP53 cell lines, presumably because mutant cells lack the p53‐dependent capacity to induce G1 and G2 phase arrest and cannot defend themselves from taxane‐induced mitotic blockade and apoptosis [29]. A phase III clinical trial assessed the impact of TP53 status on taxane‐ and non‐taxane‐based chemotherapy regimens [31]. However, this study showed no effect of TP53 mutation on the response to taxane‐ and non‐taxane combination treatments. This may be because this study enrolled patients with all subtypes of breast cancer, and the impact of TP53 mutation may be distinct for different subtypes of breast cancer. This study also enrolled locally advanced breast cancer patients who received neoadjuvant chemotherapy, and the dosage and intensity of chemotherapy are of crucial importance in inducing a response and a survival benefit in TP53‐mutated tumors [8]. In our study, we investigated the impact of TP53 mutation specifically in HER2+ patients who received first‐line trastuzumab‐based therapy. Another difference between the previous study and our study is that we used next‐generation sequencing to detect TP53 mutations in ctDNA, which may limit the impacts of temporal and spatial heterogeneity among tumors.
Some limitations of this study must be acknowledged. First, DFS was retrospectively calculated, which may influence the survival analysis, as TP53 mutation status may change after distant metastasis. Second, the retrospective design for the analysis of trastuzumab‐based first‐line therapy did not provide sufficient power to arrive at statistically sound conclusions. The predictive value of TP53 mutation among different chemotherapy agents needs to be verified in prospective studies.
5. CONCLUSIONS
In conclusion, our data indicate that TP53 somatic mutations are common in Chinese patients with breast cancer and are more frequently observed in HER2+/HR− patients and those with TNBC than in those with other subtypes of breast cancer. TP53 mutation in exons 5‐8 may be an independent prognostic marker for short DFS. TP53 mutation has opposite effects in patients treated with trastuzumab‐based chemotherapy in combination with and without taxanes. Large‐scale prospective studies are needed to verify our findings.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study protocol was reviewed and approved by the Institutional Review Board of National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (16‐038/1117). Each participant signed the informed consent before participating to this study.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data used to support the findings of this study are available from the corresponding authors upon request.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDING
This work was supported by the grants from the Chinese Academy of Medical Sciences (CAMS) Initiative for Innovative Medicine (CAMS‐12 M‐1‐010, 2017‐I2M‐3‐004) and the National Natural Science Foundation of China (81874122).
AUTHOR CONTRIBUTIONS
Z.Y. designed and performed experiments, analyzed data, drafted and provided critical revision of the manuscript. F.M. contributed substantially to the conception of the study, acquired the funding, designed and performed experiments, drafted and provided critical revision of the manuscript. G.R. analyzed data and co‐wrote the manuscript. Y.G. and C.L. designed and performed experiments and drafted the manuscript. B.X. contributed substantially to the conception of the study, supervised the research, acquired the funding, and drafted and provided critical revision of the manuscript. All authors read and approved the final manuscript
Supporting information
Supporting Information
ACKNOWLEDGMENTS
We thank the patients, their families, and the study personnel across all sites for participating in this study.
Yi Z, Ma F, Rong G, Guan Y, Li C, Xu B. Clinical spectrum and prognostic value of TP53 mutations in circulating tumor DNA from breast cancer patients in China. Cancer Communications. 2020;40:260–269. 10.1002/cac2.12032
Contributor Information
Fei Ma, Email: drmafei@126.com.
Binghe Xu, Email: xubingheBM@163.com.
REFERENCES
- 1. Lane D. Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2(12):a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2(8):594‐604. [DOI] [PubMed] [Google Scholar]
- 3. Zhao P, Chen H, Wen D, Mou S, Zhang F, Zheng S. Personalized treatment based on mini patient‐derived xenografts and WES/RNA sequencing in a patient with metastatic duodenal adenocarcinoma. Cancer Commun. 2018;38(1). 10.1186/s40880-018-0323-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lefebvre C, Bachelot T, Filleron T, Pedrero M, Campone M, Soria JC, et al. Mutational Profile of Metastatic Breast Cancers: A Retrospective Analysis. PLoS Med. 2016;13(12):e1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Silwal‐Pandit L, Vollan HK, Chin SF, Rueda OM, McKinney S, Osako T, et al. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin Cancer Res. 2014;20(13):3569‐80. [DOI] [PubMed] [Google Scholar]
- 6. Silwal‐Pandit L, Langerod A, Borresen‐Dale AL. TP53 Mutations in Breast and Ovarian Cancer. Cold Spring Harb Perspect Med. 2017;7(1). 10.1101/cshperspect.a026252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dumay A, Feugeas JP, Wittmer E, Lehmann‐Che J, Bertheau P, Espie M, et al. Distinct tumor protein p53 mutants in breast cancer subgroups. Int J Cancer. 2013;132(5):1227‐31. [DOI] [PubMed] [Google Scholar]
- 8. Bertheau P, Lehmann‐Che J, Varna M, Dumay A, Poirot B, Porcher R, et al. p53 in breast cancer subtypes and new insights into response to chemotherapy. Breast. 2013;22(Suppl 2):S27‐9. [DOI] [PubMed] [Google Scholar]
- 9. Kim JY, Park K, Jung HH, Lee E, Cho EY, Lee KH, et al. Association between Mutation and Expression of TP53 as a Potential Prognostic Marker of Triple‐Negative Breast Cancer. Cancer Res Treat. 2016;48(4):1338‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res. 2006;12(4):1157‐67. [DOI] [PubMed] [Google Scholar]
- 11. Langerod A, Zhao H, Borgan O, Nesland JM, Bukholm IR, Ikdahl T, et al. TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer. Breast Cancer Res. 2007;9(3):R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Borresen AL, Andersen TI, Eyfjord JE, Cornelis RS, Thorlacius S, Borg A, et al. TP53 mutations and breast cancer prognosis: particularly poor survival rates for cases with mutations in the zinc‐binding domains. Genes Chromosomes Cancer. 1995;14(1):71‐5. [DOI] [PubMed] [Google Scholar]
- 13. Harris LN, Broadwater G, Lin NU, Miron A, Schnitt SJ, Cowan D, et al. Molecular subtypes of breast cancer in relation to paclitaxel response and outcomes in women with metastatic disease: results from CALGB 9342. Breast Cancer Res. 2006;8(6):R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gluck S, Ross JS, Royce M, McKenna EF, Jr. , Perou CM, Avisar E, et al. TP53 genomics predict higher clinical and pathologic tumor response in operable early‐stage breast cancer treated with docetaxel‐capecitabine +/‐ trastuzumab. Breast Cancer Res Treat. 2012;132(3):781‐91. [DOI] [PubMed] [Google Scholar]
- 15. Kostler WJ, Brodowicz T, Hudelist G, Rudas M, Horvat R, Steger GG, et al. The efficacy of trastuzumab in Her‐2/neu‐overexpressing metastatic breast cancer is independent of p53 status. J Cancer Res Clin Oncol. 2005;131(7):420‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Norberg T LJ, Ingana¨s M. Bergh J Comparison between p53 protein measurements using the luminometric immunoassay and immunohistochemistry with detection of p53 gene mutations using cDNA sequencing in human breast tumors. Int J Cancer 1998;79:376‐83. [DOI] [PubMed] [Google Scholar]
- 17. Fountzilas G, Giannoulatou E, Alexopoulou Z, Zagouri F, Timotheadou E, Papadopoulou K, et al. TP53 mutations and protein immunopositivity may predict for poor outcome but also for trastuzumab benefit in patients with early breast cancer treated in the adjuvant setting. Oncotarget. 2016;7(22):32731‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nong J, Gong Y, Guan Y, Yi X, Yi Y, Chang L, et al. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat Commun. 2018;9(1):3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma F, Guan Y, Yi Z, Chang L, Li Q, Chen S, et al. Assessing tumor heterogeneity using ctDNA to predict and monitor therapeutic response in metastatic breast cancer. Int J Cancer. 2019. 10.1002/ijc.32536. [DOI] [PubMed] [Google Scholar]
- 21. Yi Z, Ma F, Liu B, Guan X, Li L, Li C, et al. Everolimus in hormone receptor‐positive metastatic breast cancer: PIK3CA mutation H1047R was a potential efficacy biomarker in a retrospective study. BMC Cancer. 2019;19(1):442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466(7308):869‐73. [DOI] [PubMed] [Google Scholar]
- 23. Borresen‐Dale AL. TP53 and breast cancer. Hum Mutat. 2003;21(3):292‐300. [DOI] [PubMed] [Google Scholar]
- 24. Petitjean A, Achatz MI, Borresen‐Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26(15):2157‐65. [DOI] [PubMed] [Google Scholar]
- 25. Shahbandi A, Nguyen HD, Jackson JG. TP53 Mutations and Outcomes in Breast Cancer: Reading beyond the Headlines. Trends Cancer. 2020;6(2):98‐110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aas T, Borresen AL, Geisler S, Smith‐Sorensen B, Johnsen H, Varhaug JE, et al. Specific P53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nat Med. 1996;2(7):811‐4. [DOI] [PubMed] [Google Scholar]
- 27. Geisler S, Lonning PE, Aas T, Johnsen H, Fluge O, Haugen DF, et al. Influence of TP53 gene alterations and c‐erbB‐2 expression on the response to treatment with doxorubicin in locally advanced breast cancer. Cancer Res. 2001;61(6):2505‐12. [PubMed] [Google Scholar]
- 28. Kandioler‐Eckersberger D, Ludwig C, Rudas M, Kappel S, Janschek E, Wenzel C, et al. TP53 mutation and p53 overexpression for prediction of response to neoadjuvant treatment in breast cancer patients. Clin Cancer Res. 2000;6(1):50‐6. [PubMed] [Google Scholar]
- 29. Daisy Carvajal CT, Yang Hong, Binh T. Vu, David C. Heimbrook, and Lyubomir T. Vassilev. Activation of p53 by MDM2 antagonists can protect proliferating cells from mitotic inhibitors. Cancer Res. 2005;65(5):1918‐24. [DOI] [PubMed] [Google Scholar]
- 30. Di Leo A1 TM. Desmedt C, Paesmans M, Cardoso F, Durbecq V, Chan S, Perren T, Aapro M, Sotiriou C, Piccart MJ, Larsimont D, Isola J; TAX 303 translational study team. p‐53 gene mutations as a predictive marker in a population of advanced breast cancer patients randomly treated with doxorubicin or docetaxel in the context of a phase III clinical trial. Ann Oncol. 2007;18:997‐1003. [DOI] [PubMed] [Google Scholar]
- 31. Bonnefoi H, Piccart M, Bogaerts J, Mauriac L, Fumoleau P, Brain E, et al. TP53 status for prediction of sensitivity to taxane versus non‐taxane neoadjuvant chemotherapy in breast cancer (EORTC 10994/BIG 1‐00): a randomised phase 3 trial. Lancet Oncol. 2011;12(6):527‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The data used to support the findings of this study are available from the corresponding authors upon request.


