Abstract
The current researches have reported that circular RNA is an important regulatory factor in the progression of various human disease. However, the function and mechanism of most circular RNAs remain unknown in cancers including multiple myeloma. Our study has confirmed that hsa_circ_0007841 is up regulated in U266 doxorubicin resistant cells (U266R) and 8226 doxorubicin resistant cells (8226R) compared to U266 parent cells (U266P) and 8226 parent cells (8226P). Silence of hsa_circ_0007841 in U266R and 8226R could reduce the half-maximal inhibitory concentration which indicated reduction in chemoresistance. In doxorubicin resistant cells, the messenger RNA and protein level of ATP-binding cassette transporters G2 increased. Silence of hsa_circ_0007841 in drug resistant cells could decrease both the messenger RNA and protein levels of ATP-binding cassette transporters G2; reexpression of hsa_circ_0007841 could block the reduction. However, overexpression of hsa_circ_0007841 could effectively upregulate the ATP-binding cassette transporters G2 messenger RNA and protein level. Inhibition of ATP-binding cassette transporters G2 could block hsa_circ_0007841 overexpression induced chemoresistance in U266P and 8226P cells. What’s more, inhibition of ATP-binding cassette transporters G2 could reduce differences of half-maximal inhibitory concentration between parent cell lines and drug-resistant cell lines. Our data collectively suggest a new model in which hsa_circ_0007841 promotes acquired chemotherapy resistance by upregulating ATP-binding cassette transporters G2 providing a novel molecular basis of chemotherapy in multiple myeloma cancer.
Keywords: hsa_circ_0007841, ABCG2, chemoresistance, multiple myeloma, IC50
Introduction
Circular RNAs (circRNA) are a newly found class of noncoding RNAs with widespread expressions in human tissue.1,2 Circular RNAs can be divided into exonic circRNAs or intronic circRNAs dependent on whether they are generated from exons3 or introns.4,5 Circular RNAs are closed loops without 5′-3′ ends and poly a tail. Owing to the closed loops, circRNAs are resistant to exonuclease RNase R and stably exist in the cells.6
Both animal and human studies have suggested that circRNAs are evolutionarily conserved, indicating important roles of circRNAs in cellular physiology.7 One the most reported function of circRNAs is acting as endogenous miRNA sponges since some circRNAs have microRNA-binding sites allowing them to arrest microRNA.8,9 Abundance and relative stable structure make circRNAs to be effective sponges than linear messenger RNAs (mRNAs).10,11 CircRNAs could also regulate RNA-binding proteins activity by forming RNA–protein complex.12 Circular RNAs have been reported to regulate cancer development in various cancer including multiple myeloma (MM).13
Multiple myeloma is the second most common hematologic malignancy, which is characterized with uncontrolled proliferation of plasma cells within the bone marrow without initial symptoms.14 Multiple myeloma accounts 15% of hematological neoplasms with 4.5 to 6 cases per 100 000 people annually.15 Approximately 86 000 new MM cases occur worldwide every year.16
Despite the advent of novel agents such as thalidomide, bortezomib, lenalidomide, and autologous stem cell transplantation, MM still remains incurable with a 5-year survival rate of 40%.17 What’s more, most of the patients relapse or become resistance to chemotherapies, suggesting that drug resistance (DR) is responsible for the ineffective treatment of MM.
Despite the development of antimyeloma treatment, the acquisition of DR is still a major cause of MM relapse.18 Multiagent combination chemotherapy is used in MM treatment, such as VDT-PACE (doxorubicin, bortezomib, dexamethasone, thalidomide, cisplatin, cyclophosphamide, and etoposide).19,20 As one of the primary components of MM chemotherapy, resistance to doxorubicin (DOX) is quite common.21 The main mechanisms underlying DR is multifactorial processes including increase in drug efflux pumps proteins expression, decrease in drug uptake, changes in metabolism, repair of DNA damage, alterations of proliferation or apoptosis.22
Overexpression of drug efflux pumps proteins plays important roles in the DR of MM.23 P-glycoprotein (ABCB1), ATP-binding cassette transporters G2 (ABCG2) have been reported to be upregulated in response to many chemotherapeutic drugs which have been extensively studied in patients with MM. The upregulations of ABC transporters lead to the decrease in the intracellular concentrations of several drugs and limit their therapeutic activity in the treatment of MM. ATP-binding cassette transporters G2 transports diverse arrays of substrates. Initially, many of the ABCG2 substrates were reported to be chemotherapeutics such as mitoxantrone; tyrosine kinase inhibitors such as imatinib and gefitinib; flavopiridol, camptothecins, topotecan, irinotecan, SN-38, and so on. Some drug-selected cell lines that express ABCG2 increase resistance to anthracyclines (DOX, daunorubicin) and rhodamine 123. Other drugs also act as substrates of ABCG such as cimetidine, prazosin, statins, and zidovudine. Additionally, ABCG2 can also transport biological substrates such as estrone, 17β estradiol, porphyrins such as protoporphyrin IX; and the dietary carcinogen, 2-amino-1-methyl-6-phenylimidazo[4,5-b] pyridine.24
Previous study employed bioinformatics in high-throughput circRNA microarray and identified differentially expressed circRNAs in MM patient’s tissue.25 The hsa_circ_0007841 expressions were confirmed in the MM tissues of 86 patients, which closely associated with DR and poor prognosis. The hsa_circ_0007841 locates on chr3:127778944-127779504. The genomic length is 560 bp, and spliced sequence length is only 265 bp. It is transcripted by gene SEC61A1. Because of the limited researches on hsa_circ_0007841, more information and function of hsa_circ_0007841 need to be investigated in the future. In our study, we investigate the role of hsa_circ_0007841 in MM and the specific mechanism through biology experiments.
Materials and Methods
Cell Culture
THP-1, KM3, U266, and RPMI-8226 were obtained from ATCC cell bank (The Global Bioresource Center). The cells were cultured in the 1640 culture medium with 10% fetal bovine serum, 50 U/mL penicillin, and 50 g/mL streptomycin, The cells were incubated with 37 °C, 5% CO2 .The DOX resistant cell lines were obtained with culture of increase concentrations of DOX.
RNA Extraction
Total RNAs were extracted by TRIzol Reagent. We carried out qualitative real-time–polymerase chain reaction (qRT-PCR) with SYBR kit according to the manuscript instructions. Complementary DNAs were synthesized using the M-MLV Reverse Transcriptase Synthesis Kit (Promega) according to the protocol of the manufactory. Power SYBR Green PCR Master Mix Kit according to the manufacturer’s instructions were used for qRT-PCR.
Quantitative RT-PCR
RNA was reversely transcribed into complementary DNA by M-MLV Reverse Transcriptase Synthesis Kit. Real-time qRT-PCR was implemented by Power SYBR Green PCR Master Mix Kit. The total system is 10 µL. The mix was added into 384-well PCR plate, which was then placed in the real-time PCR instrument for PCR reaction. The PCR conditions: 95 °C, 5 minutes; 95 °C, 15 seconds, 40 cycles; 53 °C, 30 seconds; 72 °C, 35 seconds. β-actin was used as internal reference. 2−ΔΔCt methods were used to analyze results. The primers used for each target gene are shown as followings.
| Gene | Forward | Reverse |
|---|---|---|
| β-actin | 5′GTGGCCGAGGACTTTGATTG 3′ | 5′CCTGTAACAACGCATCTCATATT 3′ |
| circ_0007841 | 5′CTAACATCTGTGAAACCATCGT3′ | 5′TCATCACATACACGATAGACTG 3′ |
| ABCB1 | 5′TTGGCTGATGTTTGTGGGAA 3′ | 5′ CCAAAAATGAGTAGCACGCCT 3′ |
| ABCG2 | 5′CAGGTGGAGGCAAATCTTCGT 3′ | 5′ACCCTGTTAATCCGTTCGTTTT 3′ |
| ABCB11 | 5′TTGGCTGATGTTTGTGGGAAG3’ | 5′CCAAAAATGAGTAGCACGCCT 3′ |
| ABCC1 | 5′CTCTATCTCTCCCGACATGAC 3′ | 5′AGCAGACGATCCACAGCAAAA3′ |
| ABCC2 | 5′CCCTGCTGTTCGATATACCAAT3′ | 5′TCGAGAGAATCCAGAATAGGGA3′ |
| ABCC3 | 5′CACCAACTCAGTCAAACGTGC3′ | 5′GCAAGACCATGAAAGCGACTC3′ |
Cell Proliferation/Viability Assay and Half-Maximal Inhibitory Concentration Values
Briefly, cancer cells were suspended at the concentration of 1 × 105 cells/mL in cell culture and seeded in a 96-well plate; 1000 cells were seeded per well for growth assay. The cell growth was tested at day 0, day 2, day 4, and day 6. Cell viability was identified by the cell counting kit-8 (CCK8) according to the manufacturer’s instructions (Dojindo Molecular Technologies).
Six thousand cells were seeded per well for drug viability assay. The cells were cultured overnight and then treated with DOX at different concentrations. After 72 hours’ treatment, 10-µL CCK8 was added and cultured for another 2 hours, then the absorbance of 450 nm was measured by Synergy H4 Hybred Reader (BioTek Instruments). Relative growth inhibition and the half-maximal inhibitory concentration (IC50) values were analyzed through the GraphPad Prism 6.0 software (GraphPad Software, Inc).
Western Blot
NETN 150 lysis buffer was prepared (0.5% NP-40, 20 mM Tris [pH 8.0], 150 mM NaCl, 6 mM EDTA). Cells were collected by trypsinization and lysated by NETN buffer. Proteins were quantified by Bradford kit; 20 µg protein was loaded and separated through sodium dodecyl sulphate–polyacrylamide gel electrophoresis and transferred to the NC membrane (Axygen). Immobilon Western Chemiluminescent HRP Substrate kit (Millipore Corporation) was used for detection.
Statistical Analysis
We use SPSS 20.0 software for statistical analysis. The data were shown as mean ± standard deviation. We used Mann-Whitney test for pair wise comparison, and Kruskal-Wallis H test for multiple comparison.26 P < .05 indicated significant difference. *P < .05, **P < .01, and ***P < .001.
Results
Hsa_Circ_0007841 Was Upregulated in MM DR Cells
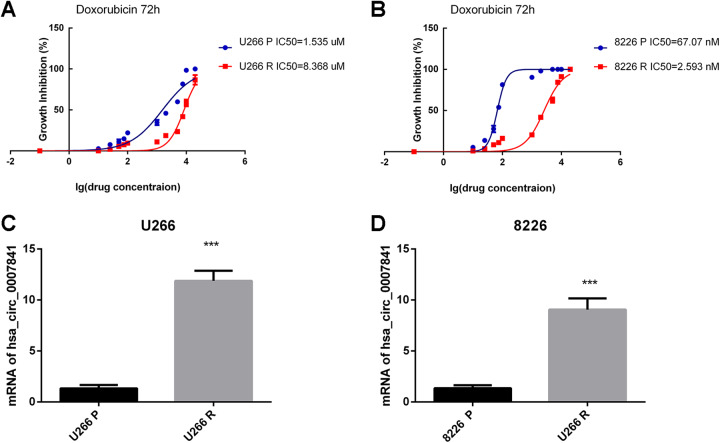
As previous study showed,25 hsa_circ_0007841 was significantly upregulated in bortezomib-resistant cell lines. In our study, we obtained DOX resistant MM cell lines by DOX treatment with increasing concentrations. To identify the DR of MM cells, we carried out the IC50 assays. The results showed that the U266 resistant cell line (U266R) had higher IC50 than U266 parent cell line (U266P; Figure 1A); and the 8226 resistant cell line (8226R) had higher IC50 than 8226 parent cell line (8226P; Figure 1B). As many drugs could be the substrate of ABCG2, we carried out the experiments of IC50s of gefitinib. The supplemental results showed that MM resistant cells are also resistant to another ABCG2 substrate drugs gefitinib (Supplemental Figure 1).
Figure 1.
Hsa_circ_0007841 was upregulated in MM chemotherapy resistance cells. A, IC50s of doxorubicin in U266P and U266R cells at 72 hours. B, IC50s of doxorubicin in 8226P and 8226R cells at 72 hours. C, Real-time PCR was carried out in U266P and U266R cells to identify hsa_circ_0007841 expression level. D, Real-time PCR was carried out in 8226 and 8226R cells to identify hsa_circ_0007841 expression level. IC50, half-maximal inhibitory concentration; MM, multiple myeloma; PCR, polymerase chain reaction.
We tested the expression level of hsa_circ_0007841 in both parent and drug resistant cell lines. The results showed that hsa_circ_0007841 was upregulated in both U266R and 8226R cells lines suggesting its important role in MM chemoresistance (Figure 1C and D).
Hsa_circ_0007841 Enhances the DR of MM Cells
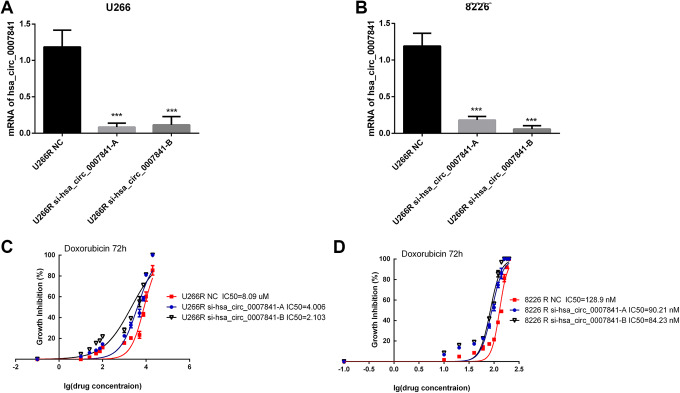
As the report and our results showed that hsa_circ_0007841 was upregulated in MM DR cells,25 we wanted to investigate the function and mechanism of hsa_circ_0007841 in chemoresistance. In U266R and 8226R cells, we silenced hsa_circ_0007841 (Figure 2A and B) and tested the DR by IC50 assays (Figure 2C and D). Silence of hsa_circ_0007841 decreased the IC50s of both U266R and 8226R cells (Figure 2C and D). The results confirmed that hsa_circ_0007841 could enhance the DR of MM cells.
Figure 2.
Hsa_circ_0007841 contributes to doxorubicin resistance in multiple myeloma.A, hsa_circ_0007841 siRNA-A and siRNA-B were transfected in U266R cells. Real-time PCR was carried out to identify the effect of gene silence. B, hsa_circ_0007841 siRNA-A and siRNA-B were transfected in 8226R cells. Real-time PCR was carried out to identify the effect of gene silence. C, IC50s of doxorubicin in U266R NC and U266R si-hsa_circ_0007841-A/B cells at 72 hours. D, IC50s of doxorubicin in U8226R NC and U8226R si-hsa_circ_0007841-A/B cells at 72 hours. IC50, half-maximal inhibitory concentration; PCR, polymerase chain reaction; siRNA, small interfering RNA.
ABCG2 Was Upregulated in MM DR Cells
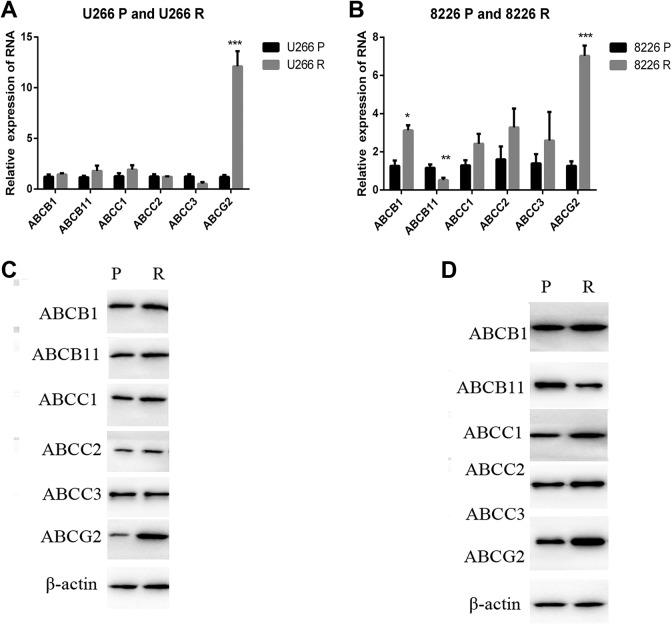
As researches have reported that ABC transporters play important roles in cancer chemotherapy resistance27 we carried out the real-time PCR to identify the relative expression of several important ABC transporters in both MM parent and drug resistant cells (Figure 3A and B). We identified the expression of ABCB1, ABCB11, ABCC1, ABCC2, ABCC3, and ABCG2. The results showed that ABCG2 was significantly upregulated in both U266R and 8226R cells (Figure 3A and B) .Western blot identified the increase in ABC transporters protein level. The results totally confirmed the upregulation of ABCG2 in MM resistant cells, suggesting that ABCG2 played important roles in MM DR. But whether hsa_circ_0007841 leads to the DR through the regulation of ABCG2 remains unclear.
Figure 3.
ABCG2 is upregulated in MM doxorubicin resistant cells. A, The mRNA level of indicated ABC transporters were identified in U266P and U266R cells by real-time PCR. B, The mRNA level of indicated ABC transporters was identified in 8226P and 8226R cells by real-time PCR. C, The protein level of indicated ABC transporters was identified in U266P and U266R cells by real-time PCR. D, The protein level of indicated ABC transporters was identified in 8226 and 8226R cells by real-time PCR. ABCG2, ATP-binding cassette transporters G2; MM, multiple myeloma; mRNA, messenger RNA; PCR, polymerase chain reaction.
Silence of hsa_circ_0007841 Downregulated ABCG2
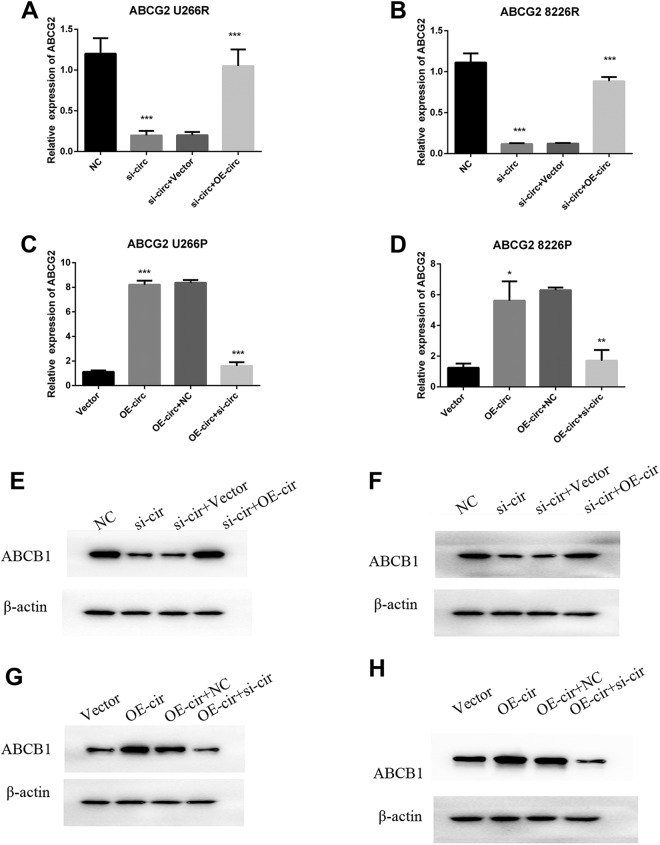
To investigate whether ABCG2 is the downstream of hsa_circ_0007841, we tested the influence of hsa_circ_0007841 on ABCG2 expression by overexpression or silence of it. In U266R and 8226R cells with high level of hsa_circ_0007841, we silenced it by transfection of small interfering RNAs. The figure showed that silence of hsa_circ_0007841 could reduce the mRNA (Figure 4A and B) and protein level of ABCG2 (Figure 4E and F); reexpression of hsa_circ_0007841 in si-hsa_circ_0007841 (si-circ) rescued the ABCG2 mRNA (Figure 4A and B) and protein level (Figure 4E and F). On the other hand, overexpression of hsa_circ_0007841 could upregulate ABCG2 mRNA (Figure 4C and D) and protein level (Figure 4E and F) in U266P and 8226P cells. In addition, silence of hsa_circ_0007841 could specifically block the overexpression induced ABCG2 upregulation. All the results above showed that hsa_circ_0007841 promoted the expression of ABCG2; silence of hsa_circ_0007841 decreased the expression of ABCG2.
Figure 4.
Hsa_circ_0007841 promotes the ABCG2 upregulation. A, hsa_circ_0007841 was silenced in U299R cells and then transfected with hsa_circ_0007841 overexpression plasmids. Real-time PCR was carried out to identify the mRNA level of ABCG2. B, hsa_circ_0007841 was silenced in 8226R cells and then transfected with hsa_circ_0007841 overexpression plasmids. Real-time PCR was carried out to identify the mRNA level of ABCG2. C, hsa_circ_0007841 plasmids and vector were transfected in U266P cells, then the hsa_circ_0007841 was silenced in overexpression cells. Real-time PCR was carried out to identify the mRNA level of ABCG2. D, hsa_circ_0007841 plasmids and vector were transfected in 8226P cells, then the hsa_circ_0007841 was silenced in overexpression cells. Real-time PCR was carried out to identify the mRNA level of ABCG2. E, hsa_circ_0007841 was silenced in U299R cells and then transfected with hsa_circ_0007841 overexpression plasmids. Western blot was carried out to identify the protein level of ABCG2. F, hsa_circ_0007841 was silenced in 8226R cells and then transfected with hsa_circ_0007841 overexpression plasmids. Western blot was carried out to identify the protein level of ABCG2. G, hsa_circ_0007841 plasmids and vector were transfected in U266P cells. Western blot was carried out to identify the protein level of ABCG2. H, hsa_circ_0007841 plasmids and vector were transfected in 8226P cells. Western blot was carried out to identify the protein level of ABCG2. ABCG2, ATP-binding cassette transporters G2; mRNA, messenger RNA; PCR, polymerase chain reaction.
ATP-Binding Cassette Transporters G2 Is Responsible for hsa_Circ_0007841 Induced Chemotherapy Resistance
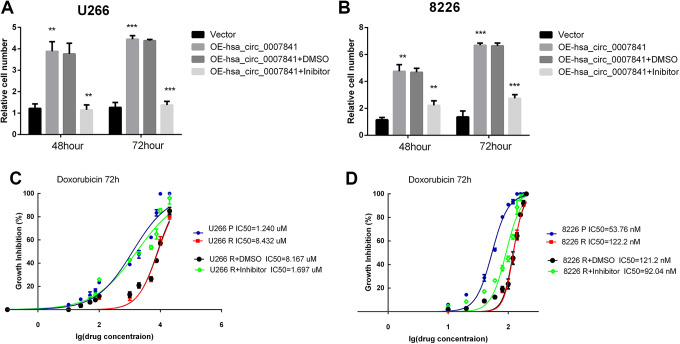
As the results showed that hsa_circ_0007841 was responsible for ABCG2 expression, we wanted to confirm whether hsa_circ_0007841 induced ABCG2 was the main cause for the DR. We carried out the CCK8 assay and IC50 assays to confirm the ABCG2 function in MM acquired chemoresistance. We used ABCG2 inhibitor RG14620 to inhibit the ABCG2 activation. In U266P and 8226P cells, ABCG2 inhibitor could effectively block the hsa_circ_0007841 induced DR (Figure 5A and B). What’s more, inhibition of ABCG2 could decrease the IC50s of U266R and 8226R cells (Figure 5C and D).Overall, ABCG2 is responsible for hsa_circ_0007841 induced DR.
Figure 5.
Inhibition of ABCG2 block the hsa_circ_0007841 induced drug resistance. A, hsa_circ_0007841 overexpression plasmids were transfected in U226P cells and then treated with ABCG2 inhibitor RG14620. The live cell numbers were tested by CC88 at 48 hours and 72 hours. B, hsa_circ_0007841 overexpression plasmids were transfected in 8226P cells and then treated with ABCG2 inhibitor RG14620. The live cell numbers were tested by CC88 at 48 hours and 72 hours. C, U266P and U266 R cells were treated with different concentration of doxorubicin with or without RG14620. Cell viability was tested at 72 hours and IC50 was calculated. D, 8226P and 8226R cells were treated with different concentration of doxorubicin with or without RG14620. Cell viability was tested at 72 hours and IC50 was calculated. ABCG2, ATP-binding cassette transporters G2; IC50, half-maximal inhibitory concentration.
Discussion
The morbidity of MM has increased year by year duo to the aging population partly.28,29 New therapies have improved the prognosis of patients with MM in recent years. Despite development in treatment, the acquisition of DR is the major limitation of MM therapy failure.20,30 Increased level of drug transporter protein, such as ABCG2, ABCB1, and other multidrug resistant proteins, is a major reason for chemotherapy resistance in MM.31 Precision medicine therapy depends on biomarkers, which could predict the efficacy of drug and reduce disease recurrence. Our study focused on the function of biomarker and its target in MM.
In previous study, bioinformatics methods with high through sequence tissues were carried out in patients with MM cancer. The study results showed that hsa_circ_0007841 was upregulated in MM drug resistant cell lines and closely associated with poor survival in patients.25
In our study, we found that ABCG2 expression was upregulated in drug resistant MM cells. The membrane-associated protein encoded by ABCG2 is one important members of ABC transporters superfamily. ABC genes are divided into 7 distinct subfamilies (ABCA, ABCB, ABCC, ABCD, ABCE, ABCF, and ABCG). ATP-binding cassette transporters G2 is a member of the G subfamily. ABCB1, ABCC1, and ABCG2 are the most important ABC transporters.32 ABC family proteins could transport various molecules across membranes. Human ABCG2 are also known as breast cancer resistance protein which functions as a homodimer.33 ATP-binding cassette transporters G2 is capable of transporting a wide range of traditional chemotherapy agents such as topotecan, methotrexate, SN-38, and mitoxantrone out of cancer cells.34 What’s more, recent studies suggested that ABCG2 could also transport many molecular-targeted agents, such as imatinib, sunitinib, vemurafenib, and sorafenib.35-37 In our study, we found that ABCG2 could regulate DOX resistance in MM cell lines. Inhibition of ABCG2 could effectively increase DOX sensitivity in MM cells.
Increasing reports have revealed connections between aberrant expression of circRNAs and cancer progression. CircRNAs could function as a microRNA sponge to regulate the microRNA level, which indirectly regulates the expression of microRNA target genes.38 In our study, we found that hsa_circ_0007841 could promote cancer chemotherapy resistance through the upregulation of ABCG2. It was the first time to investigate the function of hsa_circ_0007841 on MM cells. However, the specific mechanism has not been investigated. We will carry on the research to investigate how hsa_circ_0007841 regulated ABCG2. We want to find out the target microRNAs that are regulated by hsa_circ_0007841 and indirectly influence ABCG2 level.
In our study, we confirmed the important role of hsa_circ_0007841 in MM cancer chemotherapy resistance. Hsa_circ_0007841 could upregulate the expression of ABCG2, which is responsible for the DR. Silence of hsa_circ_0007841 or inhibition of ABCG2 could reverse the chemoresistance of MM cells. We provided a new mechanism for hsa_circ_0007841-induced cancer chemoresistance. Our study suggested that the combination of ABCG2 inhibitor and hsa_circ_00078416 inhibitor could be a promising therapy for MM cells.
Supplemental Material
supplemental_results for Hsa_Circ_0007841 Enhances Multiple Myeloma Chemotherapy Resistance Through Upregulating ABCG2 by Yan Song, Ning Hu, Xiaowei Song and Juhong Yang in Technology in Cancer Research & Treatment
Acknowledgments
Thanks to all the authors. Thanks to the good experimental environment provided by hospitals and laboratories.
Abbreviations
- ABCG2
ATP-binding cassette transporters G2
- CCK8
cell counting kit-8
- circRNA
circular RNAs
- DOX
doxorubicin
- DR
drug resistance
- IC50
half-maximal inhibitory concentration
- MM
multiple myeloma
- mRNA
messenger RNAs
- qRT-PCR
real-time–polymerase chain reaction
Authors’ Note: Our experiments did not involve any patient or animal experiments.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Juhong Yang  https://orcid.org/0000-0003-2216-0585
https://orcid.org/0000-0003-2216-0585
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Enuka Y, Lauriola M, Feldman ME, Chen AS, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2015;44(3):1370–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu YC, Li JR, Sun CH, et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2015;44(D1):D209–D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. [DOI] [PubMed] [Google Scholar]
- 5. Zheng LL, Li JH, Jie W, et al. deepBase v2.0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res. 2015;44(D1):D196–D202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fluss RA, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with Pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. [DOI] [PubMed] [Google Scholar]
- 7. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qu S, Yang X, Li X, Wang J, Li H. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. [DOI] [PubMed] [Google Scholar]
- 9. Malte KL, Harald S, George H, et al. The circular RNA ciRs-126 predicts survival in critically ill patients with acute kidney injury. Kidney Int Rep. 2018;3(5):1144–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li P, Xiao QY, Guan CL. The emerging landscape of circular RNA ciRS-7 in cancer (Review). Oncol Rep. 2015;33(6):2669–2674. [DOI] [PubMed] [Google Scholar]
- 11. Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther. 2018;187:31–44. [DOI] [PubMed] [Google Scholar]
- 12. Ding Y, Zhang Y. Research progress and research strategy of competitive endogenous RNA. Chemistry of Life. 2015;35(1):89–95. [Google Scholar]
- 13. Belousova EA, Filipenko ML, Kushlinskii NE. Circular RNA: New regulatory molecules. Bull Exp Biol Med. 2018;164(6):803–815. [DOI] [PubMed] [Google Scholar]
- 14. Richardson PG, Anderson KC, Hideshima T, Mitsiades C, Tonon G. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer. 2007;7(8):585–598. [DOI] [PubMed] [Google Scholar]
- 15. Palumbo A, Hajek R, Delforge M, et al. Continuous lenalidomide treatment for newly diagnosed multiple myeloma. N Engl J Med. 2012;366(19):1759–1769. [DOI] [PubMed] [Google Scholar]
- 16. Mcnally RJQ, Cartwright RA. Epidemiology of multiple myeloma. Hematology. 1998;3(1):1–15. [DOI] [PubMed] [Google Scholar]
- 17. Brenner H, Gondos A, Pulte D. Recent major improvement in long-term survival of younger patients with multiple myeloma. Blood. 2008;111(5):2521–2526. [DOI] [PubMed] [Google Scholar]
- 18. Sonneveld P, Lokhorst HM, Vossebeld P. Drug resistance in multiple myeloma. Semin Hematol. 1997;47(4 suppl 5):34–39. [PubMed] [Google Scholar]
- 19. Barlogie B, Anaissie E, Bolejack V, et al. High CR and near-CR rate with bortezomib incorporated into up-front therapy of multiple myeloma with tandem transplants. Br J Haematol. 2006;24:321–329. [Google Scholar]
- 20. Rajkumar SV. Multiple myeloma: 2016 update on diagnosis, risk-stratification, and management. Am J Hematol. 2016;91(7):719–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pan YZ, Wang X, Bai H, Wang CB, Zhang Q, Xi R. Autophagy in drug resistance of the multiple myeloma cell line RPMI8226 to doxorubicin. Genet Mol Res. 2015;14(2):5621–5629. [DOI] [PubMed] [Google Scholar]
- 22. Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–627. [DOI] [PubMed] [Google Scholar]
- 23. Dalton WS, Grogan TM, Meltzer PS, et al. Drug-resistance in multiple myeloma and non-Hodgkin’s lymphoma: detection of p-glycoprotein and potential circumvention by addition of verapamil to chemotherapy. J Clin Oncol. 1989;7(4):415–424. [DOI] [PubMed] [Google Scholar]
- 24. Stacy AE, Jansson PJ, Richardson DR. Molecular pharmacology of ABCG2 and its role in chemoresistance. Mol Pharmacol. 2013;84(5):655–669. [DOI] [PubMed] [Google Scholar]
- 25. Gao M, Li C, Xiao H, et al. hsa_circ_0007841: a novel potential biomarker and drug resistance for multiple myeloma. Front Oncol. 2019;9:1261 doi:10.3389/fonc.2019.01261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jin W, Xu H, Zhang S, et al. Tumor-infiltrating NETs predict postsurgical survival in patients with pancreatic ductal adenocarcinoma. Ann Surg Oncol. 2019;26(2):635–643. doi:10.1245/s10434-018-6941-4 [DOI] [PubMed] [Google Scholar]
- 27. Haber M, Henderson MJ, Norris MD, Fletcher JI. ABC transporters in cancer: more than just drug efflux pumps. Nat Rev Cancer. 2010;10(2):147–156. [DOI] [PubMed] [Google Scholar]
- 28. Di Marzo L, Desantis V, Solimando AG, et al. Microenvironment drug resistance in multiple myeloma: emerging new players. Oncotarget. 2016;7(37):60698–60711. doi:10.18632/oncotarget.10849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nikesitch N, Ling SCW. Molecular mechanisms in multiple myeloma drug resistance. J Clin Pathol. 2016;69(2):97–101. doi:10.1136/jclinpath-2015-203414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith L, McCourt O, Henrich M, et al. Multiple myeloma and physical activity: a scoping review. BMJ Open. 2015;5(11):e9576.doi:10.1136/bmjopen-2015-009576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sonneveld P, Lokhorst HM, Vossebeld P. Drug resistance in multiple myeloma. Semin Hematol. 1997;34(4 suppl 5):34–39. [PubMed] [Google Scholar]
- 32. Jin W, Liao X, Lv Y, et al. MUC1 induces acquired chemoresistance by upregulating ABCB1 in EGFR-dependent manner. Cell Death Dis. 2017;8:e2980 doi:10.1038/cddis.2017.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Doyle LA, Yang W, Abruzzo LV, et al. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci U S A. 1998;95:15665–15670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Borst P, Elferink RO. Mammalian ABC transporters in health and disease. Annu Rev Biochem. 2003;71:537–592. [DOI] [PubMed] [Google Scholar]
- 35. Hegedus C, Laczka CO, Apáti A, et al. Interaction of nilotinib, dasatinib and bosutinib with ABCB1 and ABCG2: implications for altered anti-cancer effects and pharmacological properties. Br J Pharmacol. 2009;158(4):1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lagas JS, van Waterschoot RAB, van Tilburg VACJ, et al. Brain accumulation of dasatinib is restricted by P-glycoprotein (ABCB1) and breast cancer resistance protein (ABCG2) and can be enhanced by elacridar treatment. Clin Cancer Res. 2009;15(7):2344–2351. [DOI] [PubMed] [Google Scholar]
- 37. Wu CP, Sim H, Huang Y, et al. Overexpression of ATP-binding cassette transporter ABCG2 as a potential mechanism of acquired resistance to vemurafenib in BRAF(V600E) mutant cancer cells. Biochem Pharmacol. 2013;85(3):325–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Z, Qing X, Dongmei H, et al. Circular RNA: new star, new hope in cancer. BMC Cancer. 2018;18(1):834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplemental_results for Hsa_Circ_0007841 Enhances Multiple Myeloma Chemotherapy Resistance Through Upregulating ABCG2 by Yan Song, Ning Hu, Xiaowei Song and Juhong Yang in Technology in Cancer Research & Treatment