Abstract
Alcoholic liver disease (ALD) is characterized by hepatocyte damage, inflammatory cell activation and increased intestinal permeability leading to the clinical manifestations of alcoholic hepatitis. Selected members of the family of microRNAs are affected by alcohol, resulting in an abnormal miRNA profile in the liver and circulation in ALD. Increasing evidence suggests that mRNAs that regulate inflammation, lipid metabolism and promote cancer are affected by excessive alcohol administration in mouse models of ALD. This communication highlights recent findings in miRNA expression and functions as they relate to the pathogenesis of ALD. The cell-specific distribution of miRNAs, as well as the significance of circulating extracellular miRNAs, is discussed as potential biomarkers. Finally, the prospects of miRNA-based therapies are evaluated in ALD.
Keywords: microRNA, miR-122, miR-155, Kupffer cell, gut permeability
Biogenesis and Function of MicroRNAs
MicroRNAs (miRNAs) are small noncoding RNAs that act at the posttranscriptional level to regulate expression of their respective target mRNAs and encoded proteins.1–3 MicroRNAs are transcribed from DNA as pri-miRNAs, transported from the nucleus to the cytosol as pre-miRNAs where they are processed to mature miRNAs, usually into 22 nucleic acid sequences. MicroRNAs act in the RISC (RNA-induced silencing complex) and the significance of their intracellular localization is under active investigation.4 Owing to the usually large number of target genes of individual miRNAs, miRNAs fine-tune all important biological processes in the liver including regeneration, metabolism, immunity, bile secretion, fibrosis, and hepatocellular cancer.5–8
In addition to intracellular localization, miRNAs are also found in body fluids including serum, plasma, urine, and saliva.9–11 Increasing evidence suggests that miRNAs are present in the protein fraction as well as in exosomes in the circulation.12 The high stability of miRNAs in the circulation makes them attractive for biomarker discovery in liver diseases.13 In this review, we summarize the central components of current knowledge on the role of microRNAs in alcoholic liver disease (ALD) (▶Table 1, ▶Fig. 1).
Table 1.
Distribution and function of microRNAs
| Distribution and function of microRNAs |
|---|
| Distribution |
| Nucleus |
| Cytoplasm |
| Extracellular |
| Body fluids (saliva, urine, bile) |
| Serum, plasma |
| Exosomes |
| Functions |
| Intracellular: Posttranscriptional regulation |
| Extracellular |
| Cell-to-cell communication |
| Interorgan comminations |
Fig. 1.
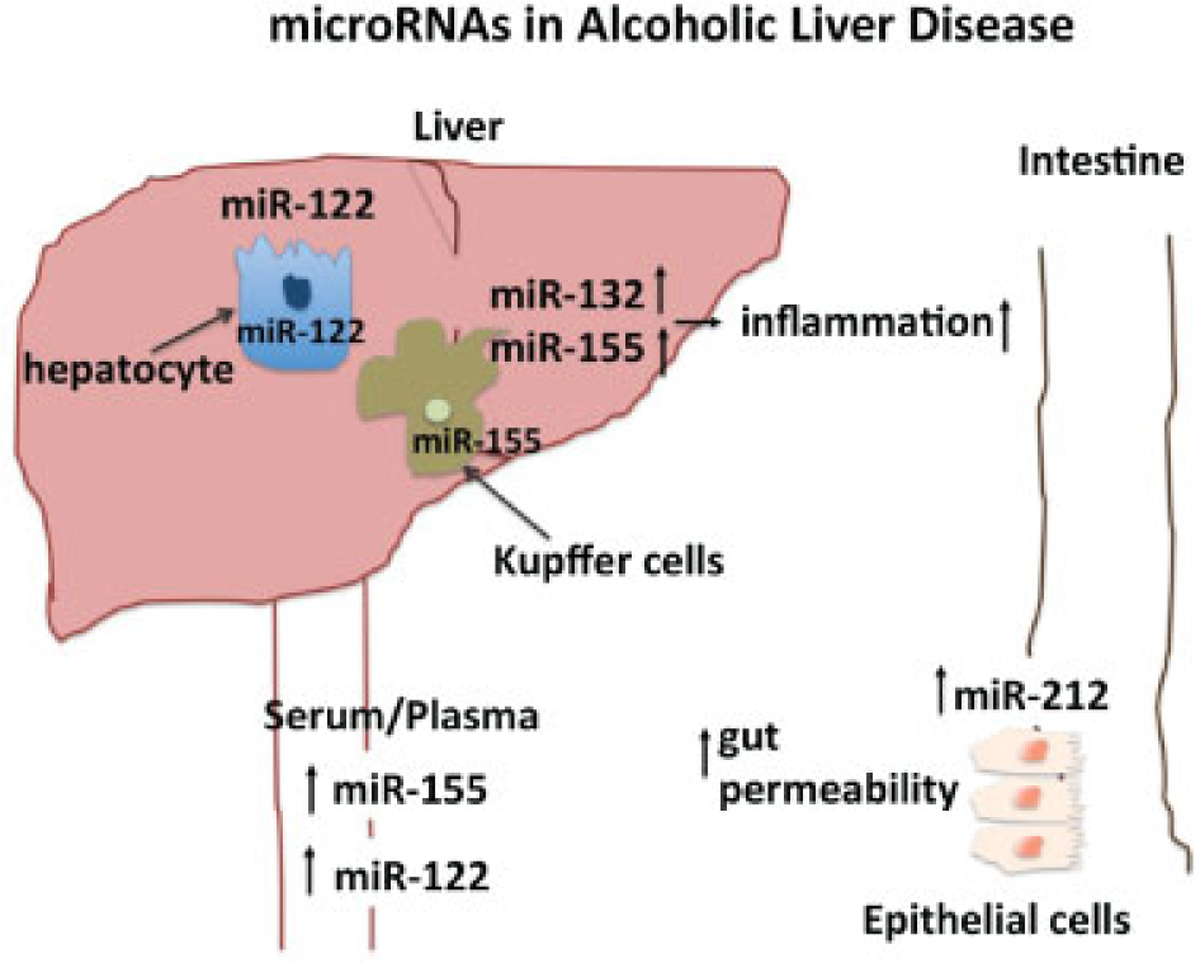
The diverse roles microRNAs (miRNAs) in the pathogenesis of alcoholic liver disease (ALD) in regulating steatosis, inflammation, and gut permeability. Ethanol and its metabolites induce miRNA dysregulation in a variety of organ systems. In the liver, miRNAs in hepatocytes regulate steatosis, while those in Kupffer cells yield a proinflammatory state, perpetuating liver injury. Additionally, alcohol’s effect on the intestines leads to increasing the permissiveness of the gut to inflammatory mediators such as lipopolysaccharide. The resulting endotoxemia contributes to hepatic damage and to the development of ALD.
Pathogenic Features of Alcoholic Liver Disease and Alcoholic Hepatitis
Alcoholic liver disease is caused by prolonged and excessive alcohol use in susceptible individuals. It is characterized by hepatic steatosis, steatohepatitis, and fibrosis/cirrhosis in advanced cases that are all biological processes potentially regulated by miRNAs.14 The clinical progression and manifestations of ALD parallel its pathogenesis. Changes from normal liver to steatosis, steatohepatitis, steatohepatitis with fibrosis, and cirrhosis represent progression of ALD. Through its direct effects, as well as its metabolic byproducts, acetaldehyde and reactive oxygen species (ROS), alcohol triggers mitochondrial and intracellular damage mechanisms.15 Even in the early phase of ALD, hepatocyte damage and apoptosis results in the release of cellular danger molecules that are sensed by immune cells in the liver, triggering inflammation. In addition to these endogenous danger signals, alcohol use results in increased levels of bacterial lipopolysaccharide (LPS) in the portal and systemic circulation due to increased intestinal permeability, leading to the activation of Kupffer cells.16–19 Excessive alcohol use adversely affects multiple organ systems including the gastrointestinal tracts and the liver.20,21 The gut–liver axis plays a critical role in the pathogenesis of ALD due to the alcohol-induced increase in gut permeability.22,23 Intake of excessive amounts of alcohol decreases the barrier function of the gut mucosa and results in increased levels of bacterial endotoxin in the portal circulation.24–27 MicroRNA-212 has been implicated in the alcohol-induced abnormal gut barrier function.28 Studies found that gut-derived endotoxin contributes to activation of Kupffer cells in the liver, leading to increased production of proinflammatory cytokines including TNFα, IL-1, IL-6, and other inflammatory mediators.17,29–31 It has also been demonstrated that TLR4 and its downstream signaling molecule, IRF3, are critical in alcohol-induced liver disease.15,32 Importantly, TLR4-mediated signaling is a major regulator of inflammation-associated microRNAs, and conversely, inflammation-associated microRNAs exert tight regulation on the mRNA levels of TLR4 and the majority of the signal transduction machinery downstream of TLR4.33,34 Thus, the components of the LPS/TLR4/inflammatory cascade are all closely regulated by miRNAs in ALD.13,35
Inflammation-Associated MicroRNAs Are Induced in Alcoholic Liver Disease
Inflammation-associated microRNAs (inflammiRs) are a group of miRNAs that regulate inflammation. These include miR-223, miRNA-155, miRNA-146a, miRNA-146b, miRNA-125, and miRNA-132, which play important roles in the regulation of inflammation by modulating expression of multiple components of TLR signaling and cytokine production, as well as being negative regulators of inflammation (▶Table 2).33,34 In the immediate early stage of the inflammatory processes, there is a rapid increase in the levels of miR-132, followed by increases in miR-155 and miR-146b. In the late stage of inflammation, miR-21 is induced and contributes to IL-10 production and is also involved in fibrosis.36 Although the kinetics of miRNA induction in the various stages of ALD awaits investigation, prolonged alcohol feeding in mice was shown to increase levels of miR-155 in the total liver, isolated hepatocytes, and in Kupffer cells.35,37
Table 2.
microRNAs in liver diseases
| MicroRNA | Target genes |
|---|---|
| miR-155 | TNFα (positive regulation), SOCS-1, SHIP-1 |
| miR-132 | Neuroimmune microRNA |
| miR-122 | Lipid metabolism: HMCCoA, ApoE, MTTP, PCC1a |
| Fibrosis: P4HA1 | |
| Hepatocellular cancer: Igf1R, ADAM10, cyclin G1, KLF6 | |
| miR-21 | Fibrosis |
| miR-26a | Hepatocellular cancer |
Abbreviations: ADAM10, a disintegrin and metalloproteinase domain-containing protein 10; ApoE, apolipoprotein E; HMGCoA, 3-hydroxy-3-methylglutaryl-coenzymeA; Igf1R, insulin-like growth factor 1 receptor; KLF6, Kruppel-like factor 6; PGC1a, PPAR-gamma coactivator 1-alpha; SHIP-1, Src Homology-2 domain containing inositol 5-phosphate-1; SOCS, suppressor of cytokine signaling; TNFα, tumor necrosis factor alpha.
Kupffer Cell Activation and Sensitization to LPS Is Regulated by miRNA-155 in Alcoholic Liver Disease
Kupffer cells, resident liver macrophages, are important contributors to the inflammatory components of ALD. Previous studies showed that prolonged (5–7 days) exposure of human monocytes or RAW murine macrophages to alcohol results in a proinflammatory phenotype and sensitization to LPS.30,38 Interestingly, prolonged alcohol treatment of RAW macrophages resulted in increased miR-155 levels.37 Kupffer cells isolated from mice after a 4-week chronic alcohol feeding have increased sensitivity to LPS by producing significantly higher levels of TNFα compared with KCs from nonalcohol-exposed mice.30,37 We found that the increased TNFα production by KCs from chronic alcohol-fed mice had significantly higher levels of miR-155 compared with KCs from pairfed control mice. Chronic alcohol increased miR-155 in KCs via NF-kB induction.37
MicroRNA-132 and miR-212
MicroRNAs-132 and −212 are products of the same gene and share substantial homology.39 Independent studies discovered increases in miR-132 levels in the liver and brain and miR-212 increases in the intestine after chronic alcohol administration in mice.28,35,40,41 The complex roles of these microRNAs, both in the liver and in other organs, have yet to be evaluated in ALD. Increasing evidence suggests that alcohol-induced tissue damage, including ALD, is influenced not only by the direct effects of alcohol on the liver, but also by signals derived from other organs as a result of alcohol exposure. The best-studied evidence for this is the increased LPS in the portal circulation after excessive alcohol consumption. It remains to be seen whether alcohol-induced microRNAs generated in one organ could modify the function of another remote organ in alcohol-induced tissue damage.
The importance of miR-212 has been highlighted in alcohol-induced gut permeability. Alcohol has increased the levels of miR-212 in caco-2 cells and this has been correlated with a decrease in the expression of tight junction proteins including occludens and zona occludens-1 (ZO-1).28
Liver Steatosis and miRNA-122 in Hepatocytes in Alcoholic Liver Disease
In hepatocytes, miR-122 is unique among hepatic microRNAs, representing approximately 70% of its miRnome.42–45 In hepatocytes, miRNA-122 directly and indirectly regulates a complex web of genes involved in lipid synthesis and export, as well as cholesterol homeostasis.44,46–49 Germline knockout (KO) and antisense oligonucleotides (ASO), or adeno-associated virus- (AAV) mediated knockdown (KD) models in mice have been shown to reduce serum cholesterol (high-density lipoprotein and low-density lipoprotein) and decrease serum triglycerides (TGs) through inhibition of critical regulatory steps involved in lipid metabolism such as HMG-CoA reductase, cytochrome P450 7a1, PGC1a, and ApoE.44,46,50 However, though both germline and liver-specific KO models demonstrated beneficial trends in serum lipid profiles, corroborating previous KD studies, both KO models were shown to have advanced steatohepatitis, contrary to previous ASO-mediated KD studies.44,46–48 In these KO mice, microsomal triglyceride transfer protein (MTTP) was found to be indirectly reduced, preventing the assembly and export of lipopro teins from the liver, thus leading to steatosis.47,48 It is key to note that while these pathways are significantly affected by modulation of miR-122 in vivo, a direct target or central mechanism has remained elusive. Studies have suggested that AMP-activated protein kinase (AMPK) and the PPAR family of proteins, both widely identified as master regulators of cellular metabolism, may act as primary effectors of this microRNA.46,51
Alcohol and Hepatitis C Infection
The clinical course and outcomes of the combination of prolonged alcohol use and chronic hepatitis C virus (HCV) infection result in accelerated progression to fibrosis and cirrhosis.52–54 Multiple pathogenic mechanisms have been identified as contributors to the combined liver damage by alcohol plus HCV, including effects on hepatocytes, HCV replication, and host immune defense.55,56 Importantly, miR-122 is a key host factor in HCV replication by providing a binding site for the promoter-region of the HCV virus, stabilizing the vector genome, and promoting viral replication.57 Because of the key role of miR-122 as host factor in HCV replication, it is not unexpected that miR-122 inhibition results in attenuation of HCV replication.58 Inhibition of miR-122 using Miravirsen, has resulted in attractive inhibition of HCV viral load in a recently published study, and currently Miravirsen is in phase 2 clinical trials.59,60 Using in vitro approaches, it has recently been shown that alcohol administration to hepatoma cells increases miR-122 levels via NF-kB activation and that miR-122-mediated upregulation of cyclinG1 also contributes to the alcohol-induced increase in HCV replication.61 Furthermore, alcohol increases GW182 and heat shock protein-90 in hepatoma cells and these proteins provide a stabilization platform for miR-122, thereby increasing HCV replication.62 Together, these data suggest that microRNA modulation by alcohol has an important role in the modulation of HCV infection and potentially other liver diseases.
Fibrosis in Alcoholic Liver Disease and the Role of MicroRNAs
miR-122 expression was significantly reduced in transactivated HSCs and in the livers of mice treated with CCl4. In vitro, an overexpression of miR-122 in LX2 cells inhibited the proliferation. It was found that P4HA1 is a primary target of miR-122. Additionally, overexpression led to decreased collagen maturation and ECM production.63 Ethanol has been shown to repress miRNA 21 in neural stem cells and neural progenitor cells that alter cellular development and maturation. Additionally, miR-21 influences PTEN/PI3K signaling, which ultimately contributes to the fibrotic response.64
Hepatocellular Cancer, MicroRNAs, and Alcoholic Liver Disease
Cancer cells have changes in their miRNA expression profile and also secrete miRNAs that may provide specific signatures. In hepatocellular cancer (HCC), decreases in miRNA-26a and miR-122 have been found in hepatocytes.65 In contrast, miR-221 is increased in HCC.
The greatest body of literature has characterized the significance of miR-122 in the development of HCC. Genome-wide expression profiling has shown decreased miR-122 expression and increased miR-122 predicted target genes in tumor tissues.50 This decrease of miR-122 expression in HCC has been found to be associated with worsening prognosis and metastasis by many studies.66–70 Additional studies have shown the involvement of miR-122 in cellular proliferation, hepatobiliary differentiation, apoptosis, and tumor invasion. Currently, several target genes of miR-122 have been identified to be involved in hepatocarcinogenesis, such as A disintegrin and metalloproteinase domain-containing protein 10 (ADAM10), Kruppel-like-factor 6 (KLF6), insulin-like growth factor 1 receptor (Igf1R), cyclin G1, and Wnt1.68,71,72
Circulating MicroRNAs in Alcoholic Liver Disease
MicroRNAs are present in various body fluids including blood, saliva, urine, bile, etc. This makes them attractive for biomarker discovery particularly because of their stability and easy detection. MicroRNAs are present in different compartments of the serum and plasma including the plasma protein fraction in association with Ago-2 or in extracellular vesicles, including exosomes that are small (20–150 nm) membrane-coated vesicles or in macrovesicles (size 150–1000 nm). The exact biological function of circulating microRNAs is under active investigation. Current knowledge suggests that they are involved in cell-to-cell communication and likely serve as markers of cellular damage, stress and disease. The latter provides potential for evaluation of miRNA “signatures” of tissue damage and injury in disease conditions including liver disease.
Much investigation focused on miRNA-122 as a maker of liver damage, and studies consistently demonstrate that circulating miRNA-122 levels are increased in various types of liver disease including alcoholic, non-alcoholic fatty liver disease, viral hepatitis (HCV and HBV), and drug (acetaminophen) induced liver injury.73–75 In most of these studies, correlation between serum ALT and miRNA-122 was linear suggesting that serum/plasma miR-122 is a good marker of liver injury.
MicroRNAs that regulate inflammation are also found in the circulation in ALD. In a recent study, we showed that in mouse models of liver diseases, inflammation-associated miRNAs are also present in the circulation. For example, miR-155 levels were increased both in ALD as well as in TLR-induced, but not in drug-induced, hepatocyte-damage models.12
A major limitation in current diagnostic tools is the lack of disease-specific biomarkers in alcoholic liver injury, and the lack of specific and/or non-invasive markers that could differentiate between liver injury and inflammation. It remains to be seen whether microRNAs (single or a panel of microRNA) will have the potential to provide such diagnostic tools as biomarkers.
MicroRNAs as Therapeutic Targets in Alcoholic Liver Disease
Therapeutic strategies targeting miRNAs can involve miRNA replacement or inhibition. To date, there have been no reports on miRNA-driven therapy in ALD.76 However, we showed that inhibition of miRNA-155 in alcohol-treated Kupffer cells and macrophages attenuates LPS-induced TNFα production.37 In HCC models, restoration of the reduced levels of miR-26a yielded an attenuation in the development of HCC.65 Further studies may reveal miRNA targets for therapy of ALD.
Future Perspectives
Understanding the function and role of microRNAs in ALD has many potential aspects that can be translated to improved care and potential treatment of patients with ALD.
First, miRNAs may serve as biomarkers to distinguish patients who have severe or moderate disease and/or aid physicians in predicting clinical outcomes. Although it is unlikely that a particular circulating miRNA would become a marker of disease, it is conceivable that a specific microRNA “signature” would indicate association with ALD and/or its various clinical stages.
Second, beyond the potential biomarker, circulating microRNAs (ex-miRNA) may have a yet to be defined biological function in cell-to-cell communication. The biodistribution and function of extracellular miRNAs is a novel research area. In a recent study, we showed that injection of a MicroRNA mimic into miR-155 deficient mice resulted in its rapid distribution and clearance of miRNA in the circulation.77 We also found that exmiRNA-155 enters hepatocytes in miR-155-deficient mice, suggesting that miRs may have a potential to modulate functions of cells and/or organs outside of their origin.
Finally, miRNAs are potential therapeutic targets in ALD. For example, increased miR-155 expression in Kupffer cells that contributes to LPS sensitivity and increased TNFα production could be a feasible target to attenuate TNFα production in ALD. Given that the extent of TNFα increase is a predictor of mortality in acute alcoholic hepatitis, it is tempting to speculate that attenuation of TNF by targeting miRNAs would result in reduced, but not eliminated, TNF levels. Previous clinical trials using anti-TNFα antibodies that block TNFα resulted in increased infections; thus, moderate reduction of TNF levels though miRNA inhibition might be an attractive approach in this patient population. Because miRNAs affect only posttranslational processes, anti-miRNA therapies result only in partial changes in their target gene products. A technical barrier to the theoretical approach of inhibition of miR-155 in KCs is cell-specific delivery of the miRNA inhibitor.
Acknowledgments
Some of the scientific content in this review article was supported by NIH grants AA020744, AA017729, AA021907, and AA022283. The authors thank Merin Mac-Donald for assistance with the preparation of the manuscript.
Abbreviations
- AAV
adeno-associated virus
- ADAM10
a disintegrin and metalloproteinase domain-containing protein 10
- ALD
alcoholic liver disease
- AMPK
AMP-activated protein kinase
- ApoE
apolipoprotein E
- ASO
antisense oligonucleotides
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- HCC
hepatocellular cancer
- HSC
hepatic stellate cell
- Igf1R
insulin-like growth factor 1 receptor
- IL-1
interleukin-1
- IL-6
interleukin-6
- IRF3
interferon regulatory factor
- KC
Kupffer cells
- KD
knockdown
- KLF6
Kruppel-like factor 6
- KO
knockout
- LPS
lipopolysaccharide
- miRNA and miR
microRNA
- MTTP
microsomal triglyceride transfer protein
- NF-kB
nuclear factor kappa beta
- RISC
RNA-induced silencing complex
- ROS
reactive oxygen species
- TG
triglyceride
- TLR
toll-like receptor
- TNFα
tumor necrosis factor alpha
- ZO-1
zona occludens-1
References
- 1.Ambros V The functions of animal microRNAs. Nature 2004; 431(7006):350–355 [DOI] [PubMed] [Google Scholar]
- 2.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science 2005;309(5740):1519–1524 [DOI] [PubMed] [Google Scholar]
- 3.Barringhaus KG, Zamore PD. MicroRNAs: regulating a change of heart. Circulation 2009;119(16):2217–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim DH, Saetrom P, Snøve O Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A 2008;105(42):16230–16235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136(2):215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet 2005;37(5):495–500 [DOI] [PubMed] [Google Scholar]
- 7.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell 2006;11(4): 441–450 [DOI] [PubMed] [Google Scholar]
- 8.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol 2009;21(3):452–460 [DOI] [PubMed] [Google Scholar]
- 9.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem 2010;56(11):1733–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105(30):10513–10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shigehara K, Yokomuro S, Ishibashi O, et al. Real-time PCR-based analysis of the human bile microRNAome identifies miR-9 as a potential diagnostic biomarker for biliary tract cancer. PLoS ONE 2011;6(8):e23584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bala S, Petrasek J, Mundkur S, et al. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology 2012;56(5):1946–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol 2013;10(9):542–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Shea RS, Dasarathy S, McCullough AJ; Practice Guideline Committee of the American Association for the Study of Liver Diseases; Practice Parameters Committee of the American College of Gastroenterology. Alcoholic liver disease. Hepatology 2010;51(1):307–328 [DOI] [PubMed] [Google Scholar]
- 15.Petrasek J, Iracheta-Vellve A, Csak T, et al. STING-IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc Natl Acad Sci U S A 2013;110(41): 16544–16549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adachi Y, Moore LE, Bradford BU, Gao W, Thurman RG. Antibiotics prevent liver injury in rats following long-term exposure to ethanol. Gastroenterology 1995;108(1):218–224 [DOI] [PubMed] [Google Scholar]
- 17.Mandrekar P, Szabo G. Signalling pathways in alcohol-induced liver inflammation. J Hepatol 2009;50(6):1258–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao B, Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology 2011;141(5):1572–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao B, Seki E, Brenner DA, et al. Innate immunity in alcoholic liver disease. Am J Physiol Gastrointest Liver Physiol 2011;300(4): G516–G525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massey VL, Arteel GE. Acute alcohol-induced liver injury. Front Phys 2012;3:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szabo G, Petrasek J, Bala S. Innate immunity and alcoholic liver disease. Dig Dis 2012;30(Suppl 1):55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szabo G, Bala S. Alcoholic liver disease and the gut-liver axis. World J Gastroenterol 2010;16(11):1321–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szabo G, Bala S, Petrasek J, Gattu A. Gut-liver axis and sensing microbes. Dig Dis 2010;28(6):737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keshavarzian A, Farhadi A, Forsyth CB, et al. Evidence that chronic alcohol exposure promotes intestinal oxidative stress, intestinal hyperpermeability and endotoxemia prior to development of alcoholic steatohepatitis in rats. J Hepatol 2009; 50(3):538–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Endotoxemia Rao R. and gut barrier dysfunction in alcoholic liver disease. Hepatology 2009;50(2):638–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bode C, Bode JC. Alcohol’s role in gastrointestinal tract disorders. Alcohol Health Res World 1997;21(1):76–83 [PMC free article] [PubMed] [Google Scholar]
- 27.Thurman RG, Bradford BU, Iimuro Y, et al. Role of Kupffer cells, endotoxin and free radicals in hepatotoxicity due to prolonged alcohol consumption: studies in female and male rats. J Nutr 1997; 127(5, Suppl):903S–906S [DOI] [PubMed] [Google Scholar]
- 28.Tang Y, Banan A, Forsyth CB, et al. Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol Clin Exp Res 2008;32(2): 355–364 [DOI] [PubMed] [Google Scholar]
- 29.McClain CJ, Barve S, Deaciuc I, Kugelmas M, Hill D. Cytokines in alcoholic liver disease. Semin Liver Dis 1999;19(2):205–219 [DOI] [PubMed] [Google Scholar]
- 30.Kishore R, McMullen MR, Cocuzzi E, Nagy LE. Lipopolysaccharide-mediated signal transduction: stabilization of TNF-alpha mRNA contributes to increased lipopolysaccharide-stimulated TNF-alpha production by Kupffer cells after chronic ethanol feeding. Comp Hepatol 2004;3(Suppl 1):S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hines IN, Wheeler MD. Recent advances in alcoholic liver disease III. Role of the innate immune response in alcoholic hepatitis. Am J Physiol Gastrointest Liver Physiol 2004;287(2):G310–G314 [DOI] [PubMed] [Google Scholar]
- 32.Hritz I, Mandrekar P, Velayudham A, et al. The critical role of toll-like receptor (TLR) 4 in alcoholic liver disease is independent of the common TLR adapter MyD88. Hepatology 2008;48(4): 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol 2012;30:295–312 [DOI] [PubMed] [Google Scholar]
- 34.O’Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol 2011;11(3): 163–175 [DOI] [PubMed] [Google Scholar]
- 35.Bala S, Szabo G. MicroRNA signature in alcoholic liver disease. Int J Hepatol 2012;2012:498232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alam MM, O’Neill LA. MicroRNAs and the resolution phase of inflammation in macrophages. Eur J Immunol 2011;41(9): 2482–2485 [DOI] [PubMed] [Google Scholar]
- 37.Bala S, Marcos M, Kodys K, et al. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor alpha (TNFalpha) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem 2011;286(2):1436–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandrekar P, Bala S, Catalano D, Kodys K, Szabo G. The opposite effects of acute and chronic alcohol on lipopolysaccharide-induced inflammation are linked to IRAK-M in human monocytes. J Immunol 2009;183(2):1320–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wanet A, Tacheny A, Arnould T, Renard P. miR-212/132 expression and functions: within and beyond the neuronal compartment. Nucleic Acids Res 2012;40(11):4742–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lippai D, Bala S, Csak T, Kurt-Jones EA, Szabo G. Chronic alcohol-induced microRNA-155 contributes to neuroinflammation in a TLR4-dependent manner in mice. PLoS ONE 2013;8(8):e70945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lippai D, Bala S, Catalano D, Kodys K, Szabo G. MicroRNA-155 deficiency prevents alcohol-induced serum endotoxin increase and small bowel inflammation in mice. Alcohol Clin Exp Res 2014; 38(8):2217–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol 2002;12(9):735–739 [DOI] [PubMed] [Google Scholar]
- 43.Xu H, He JH, Xiao ZD, et al. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology 2010;52(4):1431–1442 [DOI] [PubMed] [Google Scholar]
- 44.Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005;438(7068):685–689 [DOI] [PubMed] [Google Scholar]
- 45.Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A. miR-122, a paradigm for the role of microRNAs in the liver. J Hepatol 2008;48(4):648–656 [DOI] [PubMed] [Google Scholar]
- 46.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 2006;3(2):87–98 [DOI] [PubMed] [Google Scholar]
- 47.Hsu SH, Wang B, Kota J, et al. Essential metabolic, anti-inflamma-tory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest 2012;122(8):2871–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai WC, Hsu SD, Hsu CS, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest 2012; 122(8):2884–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie J, Ameres SL, Friedline R, et al. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat Methods 2012;9(4):403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burchard J, Zhang C, Liu AM, et al. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol 2010;6:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elmén J, Lindow M, Schütz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008;452(7189):896–899 [DOI] [PubMed] [Google Scholar]
- 52.Marcellin P, Asselah T, Boyer N. Fibrosis and disease progression in hepatitis C. Hepatology 2002;36(5, Suppl 1):S47–S56 [DOI] [PubMed] [Google Scholar]
- 53.Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med 2013;19(7): 837–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hajarizadeh B, Grebely J, Dore GJ. Epidemiology and natural history of HCV infection. Nat Rev Gastroenterol Hepatol 2013; 10(9):553–562 [DOI] [PubMed] [Google Scholar]
- 55.Szabo G, Wands JR, Eken A, et al. Alcohol and hepatitis C virus—interactions in immune dysfunctions and liver damage. Alcohol Clin Exp Res 2010;34(10):1675–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Szabo G, Aloman C, Polyak SJ, Weinman SA, Wands J, Zakhari S. Hepatitis C infection and alcohol use: a dangerous mix for the liver and antiviral immunity. Alcohol Clin Exp Res 2006;30(4): 709–719 [DOI] [PubMed] [Google Scholar]
- 57.Jopling CL, Schütz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 2008;4(1):77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jopling CL. Targeting microRNA-122 to treat hepatitis C virus infection. Viruses 2010;2(7):1382–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Janssen HL, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med 2013;368(18): 1685–1694 [DOI] [PubMed] [Google Scholar]
- 60.Lindow M, Kauppinen S. Discovering the first microRNA-targeted drug. J Cell Biol 2012;199(3):407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hou W, Bukong TN, Kodys K, Szabo G. Alcohol facilitates HCV RNA replication via up-regulation of miR-122 expression and inhibition of cyclin G1 in human hepatoma cells. Alcohol Clin Exp Res 2013; 37(4):599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bukong TN, Hou W, Kodys K, Szabo G. Ethanol facilitates hepatitis C virus replication via up-regulation of GW182 and heat shock protein 90 in human hepatoma cells. Hepatology 2013;57(1): 70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J, Ghazwani M, Zhang Y, et al. miR-122 regulates collagen production via targeting hepatic stellate cells and suppressing P4HA1 expression. J Hepatol 2013;58(3):522–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miranda RC, Pietrzykowski AZ, Tang Y, et al. MicroRNAs: master regulators of ethanol abuse and toxicity? Alcohol Clin Exp Res 2010;34(4):575–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kota J, Chivukula RR, O’Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009;137(6):1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene 2009;28(40):3526–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bai S, Nasser MW, Wang B, et al. MicroRNA-122 inhibits tumor-igenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem 2009;284(46): 32015–32027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeng C, Wang R, Li D, et al. A novel GSK-3 beta-C/EBP alpha-miR-122-insulin-like growth factor 1 receptor regulatory circuitry in human hepatocellular carcinoma. Hepatology 2010;52(5): 1702–1712 [DOI] [PubMed] [Google Scholar]
- 69.Mizuguchi Y, Mishima T, Yokomuro S, et al. Sequencing and bioinformatics-based analyses of the microRNA transcriptome in hepatitis B-related hepatocellular carcinoma. PLoS ONE 2011; 6(1):e15304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog 2013;52(4):297–303 [DOI] [PubMed] [Google Scholar]
- 71.Fornari F, Gramantieri L, Giovannini C, et al. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res 2009;69(14): 5761–5767 [DOI] [PubMed] [Google Scholar]
- 72.Xu J, Zhu X, Wu L, et al. MicroRNA-122 suppresses cell proliferation and induces cell apoptosis in hepatocellular carcinoma by directly targeting Wnt/β-catenin pathway. Liver Int 2012;32(5): 752–760 [DOI] [PubMed] [Google Scholar]
- 73.Zhang X, Zhang Z, Dai F, et al. Comparison of circulating, hepatocyte specific messenger RNA and microRNA as biomarkers for chronic hepatitis B and C. PLoS ONE 2014;9(3):e92112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ding X, Ding J, Ning J, et al. Circulating microRNA-122 as a potential biomarker for liver injury. Mol Med Rep 2012;5(6): 1428–1432 [DOI] [PubMed] [Google Scholar]
- 75.Elfimova N, Schlattjan M, Sowa JP, Dienes HP, Canbay A, Odenthal M. Circulating microRNAs: promising candidates serving as novel biomarkers of acute hepatitis. Front Phys 2012;3:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Szabo G, Sarnow P, Bala S. MicroRNA silencing and the development of novel therapies for liver disease. J Hepatol 2012;57(2): 462–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Momen-Heravi F, Bala S, Bukong T, Szabo G. Exosome-mediated delivery of functionally active miRNA-155 inhibitor to macrophages. Nanomedicine (Lond) 2014;10(7):1517–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]


