Abstract
Background
CircRNAs have been found to play crucial roles in multiple tumor including non‐small cell lung cancer (NSCLC). Here, we researched the correlation between circRNA_103762 and chemotherapy resistance.
Methods
RT‐PCR assay was performed to detect circRNA_103762 and DNA damage inducible transcript 3 (CHOP) expression. CCK8 assay was performed to examine cell proliferation and IC50 of different drug. Migration and invasion assay was used to detect cell migration and invasion.
Results
In our study, circRNA_103762 expression was upregulated in NSCLC tissues and cell. Knockdown of circRNA_103762 can inhibited cell proliferation, migration and invasion in NSCLC. In addition, downregulation of circRNA_103762 promoted CHOP expression and inhibited multidrug resistance (MDR) in NSCLC.
Conclusion
Together, we demonstrated that circRNA_103762 is upregulated in NSCLC and functions as an oncogene in NSCLC, and circRNA_103762 enhanced MDR by inhibited CHOP expression in NSCLC cells. These results will help us understand the MDR of NSCLC, providing better effective therapy strategies for patients.
Keywords: CHOP, circRNA_103762, MDR, NSCLC
1. INTRODUCTION
Lung cancer remains the leading cause of cancer‐related deaths worldwide.1 Although many drugs have been developed and performed to suppress tumor progression, chemotherapy often fails as the tumor cells acquire multidrug resistance (MDR).2, 3 Thus, it is to find the molecular mechanisms of MDR in NSCLC that helps establish new therapeutic strategy.
Recently, it is reported non‐coding RNAs was associated with tumor, such as, miRNA,4 lncRNA,5, 6 and circRNA.7, 8 CircRNA, a class of non‐coding RNAs, covalently closed continuous loop without a 5'cap or a 3'Poly A tail.9, 10 Previous researches pointed out that circRNAs played an important role in tumor. For example, RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer.11 A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo‐YAP signaling.12 CircRNA_0023642 promotes migration and invasion of gastric cancer cells by regulating EMT.13 In addition, some studies pointed out that circRNAs are associated with MDR. For example, circular RNA‐MTO1 suppresses breast cancer cell viability and reverses monastrol resistance through regulating the TRAF4/Eg5 axis.14 Notably, it is reported circRNA also played a significant role in NSCLC.15, 16 However, the chemoresistance regulatory mechanisms of circRNA in NSCLC remain unclear. Thus, it is important to identify MDR‐associated circRNA and as biomarkers or therapeutic targets.
In this study, we firstly reported expression of circRNA_103762 in NSCLC tissues and cell lines and the function of circRNA_103762 in NSCLC. In addition, we also found that circRNA_103762 promoted chemotherapy resistance in NSCLC by targeting CHOP.
2. MATERIALS AND METHODS
2.1. Subjects and sample collection
Sixty tissue samples were recruited from NSCLC patients. All samples were collected between December 2017 and May 2019. The study was approved by the ethics committee of Southern Medical University Cancer Institute and Hospital. Informed consent was written from all patients involved in this study. All methods were performed in accordance with Health Insurance Portability and Accountability Act (HIPAA).
2.2. Cell culture
Human normal lung cell line Beas‐2B and NSCLC cell lines A549, NCI‐H1299, H1650, and H358 were purchased from the Cell Bank of Type Culture Collection. All of these cell lines were cultured in DMEM medium containing 10% fetal bovine serum (HyClone) and were maintained at 37°C in an atmosphere of 5% CO2.
2.3. Bioinformatics analysis
The microarray data of circRNA profiles in NSCLC samples and normal lung samples were from NCBI GEO Datasets (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE112214). After applying log 2 transformation, GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE112214) was performed to analyze normalized microarray data.
2.4. CCK8 assay
The NSCLC cell proliferation was detected performing CCK8 assay following the manufacturer's instructions. Briefly, at every 24 hours interval, 10 μL of CCK8 (Dojindo) was added to each well and incubated for 2 hours. The solution was examined spectrophotometrically at 450 nm.
2.5. Migration assay and invasion assay
Transwell filters (8 µm pore size; Millipore) coated with or without Matrigel (BD Biosciences) were performed to evaluate the metastasis of NSCLC cells. Cells were resuspended with serum‐free medium and then seeded in the up chamber. A DMEM medium supplemented with 10% FBS was used to add in the bottom chamber. The cells were cultured for 48 hours. The cotton swabs were used to mechanically scrap the non‐invasive cells on the up chambers. Then, the migrated or invasive cells were stained with Hoechest 33342 and photographed using a light microscope as well as were counted the number.
2.6. Cell apoptosis
The NSCLC cells were treated according to standard protocols of cell apoptosis kit (BD Biosciences). For this, cells were collected and incubated with PI (BD Biosciences) and FITC (BD Biosciences). Later, cell apoptosis was examined by flow cytometry (C6; BD Biosciences).
2.7. Quantitative real‐time PCR
Total RNA was collected using TRIzol reagent (Invitrogen) in accordance with the manufacturer's protocol. The recoverAll™ Total Nucleic Acid Isolation Kit (Applied Biosystems) were performed to isolated total RNA in accordance with the manufacturer's protocol treated with RNase‐free DNase I (Qiagen) according to the manufacturer's instructions. For detection of gene expression, cDNA was synthesized by qPCR RT Kit (Toyobo) according to standard protocols. SYBR Green kit (Qiagen) was performed to qPCR. The GAPDH was used for internal control. Quantitative real‐time PCR was reacted to in triplicate in ABI StepOnePlus™ real‐time PCR system (Applied Biosystems). The primers were showed in Table 1.
Table 1.
The primer sequences included in this study
| Gene | Direction | Gene Direction primer sequence (5′‐3′) |
|---|---|---|
| circRNA_103762 | Forward | AAAACTATCTGGAATGCAGGAAGAGC |
| Reverse | ACTTACCGTTTCACATCGGCC | |
| CHOP | Forward | CTCCCAGAGCCCTCACTCTC |
| Reverse | TGCTTGAGCCGTTCATTCTC | |
| GAPDH | Forward | GAGTCAACGGATTTGGTCGT |
| Reverse | TTGATTTTGGAGGGATCTCG |
2.8. Western blot
Western blot analysis was treated according to standard protocols as previously study.13 The following primary antibodies directed against the antigens were used: CHOP (1:1000, mouse monoclonal; Cell Signaling). The second antibodies were from Cell Signaling. GAPDH was used as the control in the Western blots.
2.9. Statistical analysis
Student's t test was used for statistical analysis. All data were shown represent the mean ± SD. Differences with a P < .05 were considered significant.
3. RESULT
3.1. CircRNA_103762expression was increased in NSCLC tissues and cell lines
The microarray data in the GEO dataset (GSE112214) were performed to initially examine circRNAs expression, and normalized microarray data were analyzed by using GEO2R after applying log2 transformation. The microarray data showed circRNA_103762 expression was upregulated in NSCLC tissues compared with normal tissues (Figure 1A). To further explore the expression of circRNA_103762 in NSCLC, circRNA_103762 expression was detected by RT‐PCR assay. The results showed circRNA_103762 were increased in NSCLC tissues compared with adjacent normal tissues (Figure 1B). Notably, Kaplan‐Meier survival analysis showed higher circRNA_103762 expression in NSCLC patients was associated with lower survival rate (Figure 1C). The RT‐PCR also showed that circRNA_103762 expression was remarkably upregulated in NSCLC cell lines compared with normal lung cell line Beas‐2B (Figure 1D). Thus, these results revealed that circRNA_103762 expression was remarkably increased in NSCLC tissues and cell lines and negatively correlated with NSCLC survival, suggesting its dysregulation may promote to NSCLC progression.
Figure 1.
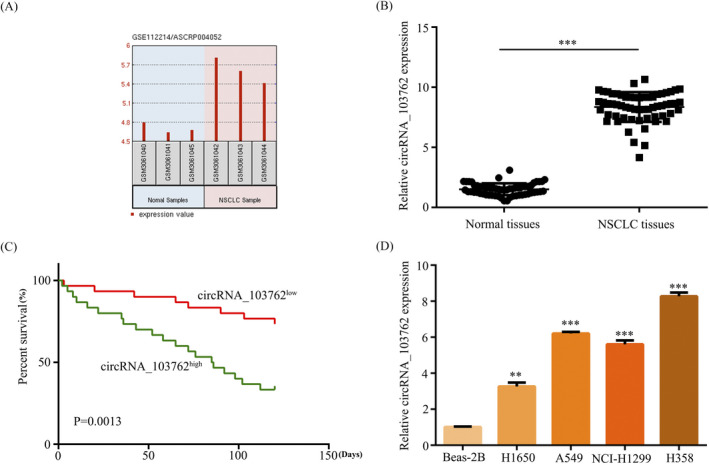
CircRNA_103762 expression was increased in NSCLC tissues and cell lines. A, GEO dataset (GSE112214) revealed that circRNA_103762 was remarkably upregulated in NSCLC tissues compared with normal tissues. B, Relative expression of circRNA_103762 was examined by qRT‐PCR in NSCLC tissues. C, The Kaplan‐Meier survival analysis revealed that overexpression of circRNA_103762 group has a worse overall survival compared with the low expression of circRNA_103762 group. D, Relative expression of circRNA_103762 was examined by qRT‐PCR in NSCLC cell line and Beas‐2B. The data shown represent the mean ± SD (n = 3). *P < .05, **P < .01, ***P < .001
3.2. CircRNA_103762 downregulation suppressed cell Proliferation, migration, and invasion in NSCLC
To further examine the role of circRNA_103762 in NSCLC, si‐circRNA_103762 specifically targeted at circRNA_103762 junction site were constructed and transfected into the H358 cell lines. The RT‐PCR showed circRNA_103762 expression was downregulated in H358/si‐circRNA_103762 cell compared with H358/si‐NC cell (Figure 2A). The CCK8 assay revealed that downregulation of circRNA_103762 inhibited the H358 cells proliferation (Figure 2B). In addition, the migration and invasion assay revealed that downregulation of circRNA_103762 inhibited migration (Figure 2C) and invasion (Figure 2D) in H358 cell. These results pointed out that circRNA_103762 acts as a tumor promoter in NSCLC.
Figure 2.
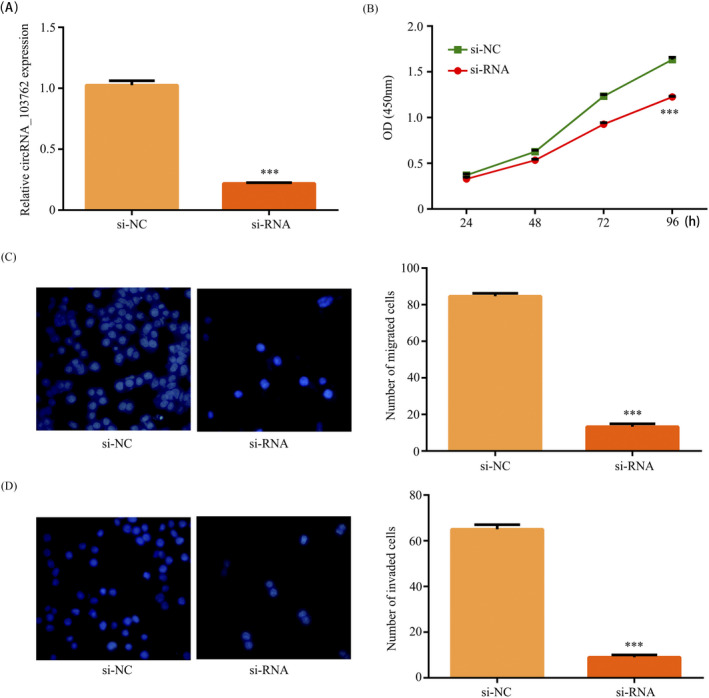
CircRNA_103762 downregulation suppressed cell Proliferation, migration, and invasion in NSCLC. A, Relative expression of circRNA_103762 in H358 cells transfected with si‐circRNA_103762 or si‐NC was detected by RT‐PCR. B, CCK8 assay was used to detected H358 cells proliferation. C, Migration assay was used to detected cell migration. D, Invasion assay was performed to examined cell invasion. The data shown represent the mean ± SD (n = 3). *P < .05, **P < .01, ***P < .001. All siRNA is si‐circRNA_103762
3.3. Drug resistance is associated with increased circRNA_103762 expression in H358 cell
To detect whether circRNA_103762 is involved in MDR, we established a cisplatin‐resistant lung cancer cell line (H358/CDDP). The CCK8 assay showed that IC50 values of different drugs were increased in H358/CDDP cell compared with H358 cell (Figure 3A). In addition, circRNA_103762 expression was upregulated in H358/CDDP cell (Figure 3B). To further examine the role of circRNA_103762 in NSCLC, si‐circRNA_103762 specifically targeted at circRNA_103762 junction site were constructed and transfected into the H358/CDDP cell. The RT‐PCR showed circRNA_103762 expression was downregulated in H358/CDDP/si‐circRNA_103762 cell compared with H358/CDDP/si‐NC cell (Figure 3C). The CCK8 assay showed that IC50 values of different drugs were decreased in H358/CDDP/ si‐circRNA_103762 cell and H358/si‐circRNA_103762 cell (Figure 3D). Thus, these results revealed that upregulation of circRNA_103762 is associated with MDR.
Figure 3.
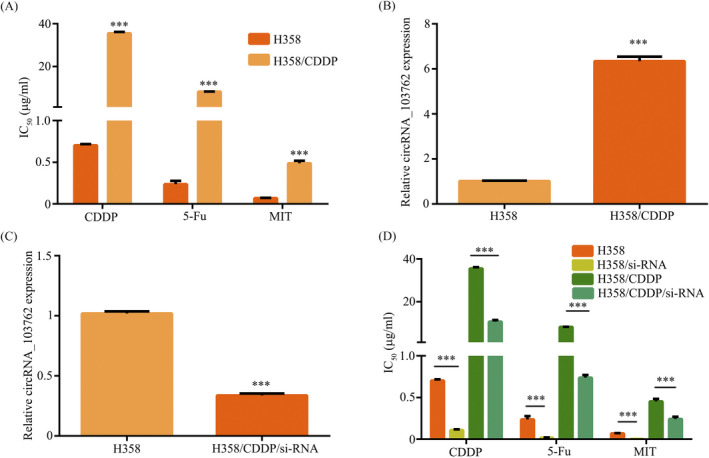
Drug resistance is associated with increased circRNA_103762 expression in H358 cell. A, The IC50 of different drugs on H358 and H358/CDDP cells. B, The circRNA_103762 expression was detected by RT‐PCR in H358 and H358/CDDP cells. C, Relative expression of circRNA_103762 in H358/CDDP cells transfected with si‐circRNA_103762 or si‐NC was detected by RT‐PCR. D, The IC50 of different drugs on H358, H358/si‐circRNA_103762, H358/CDDP and H358/CDDP/si‐circRNA_103762 cells. The data shown represent the mean ± SD (n = 3). *P < .05, **P < .01, ***P < .001. All siRNA is si‐circRNA_103762
3.4. CircRNA_103762 enhanced MDR by inhibited CHOP expression in NSCLC cells
Early reports pointed out that CHOP is related to tumor and plays a significant role in ER stress.17, 18, 19 CHOP expression can be induced by chemotherapeutic drugs and is associated with MDR.20 RT‐PCR and Western blot assay showed that the CHOP expression was increased in H358/si‐circRNA_103762 cell compared with control group (Figure 4A,4) and the CHOP expression was decreased in H358/CDDP cell compared with H358 cell (Figure 4C,D). To further explore whether circRNA_103762 enhanced MDR by inhibited CHOP expression in NSCLC cells, we established a knocking down CHOP cell, namely H358/si‐circRNA_103762/si‐CHOP cell and H358/CDDP/ si‐circRNA_103762/si‐CHOP cell. The RT‐PCR and Western blot showed the CHOP expression was decreased in H358/si‐circRNA_103762/si‐CHOP cell (Figure 4E,F) and H358/CDDP/ si‐circRNA_103762/si‐CHOP cell (Figure 4G,H). in addition, The CCK8 assay showed that IC50 values of different drugs were increased in H358/si‐circRNA_103762/si‐CHOP cell (Figure 4I) and H358/CDDP/ si‐circRNA_103762/si‐CHOP cell (Figure 4J). Thus, circRNA_103762 enhanced MDR by inhibited CHOP expression in NSCLC cells.
Figure 4.
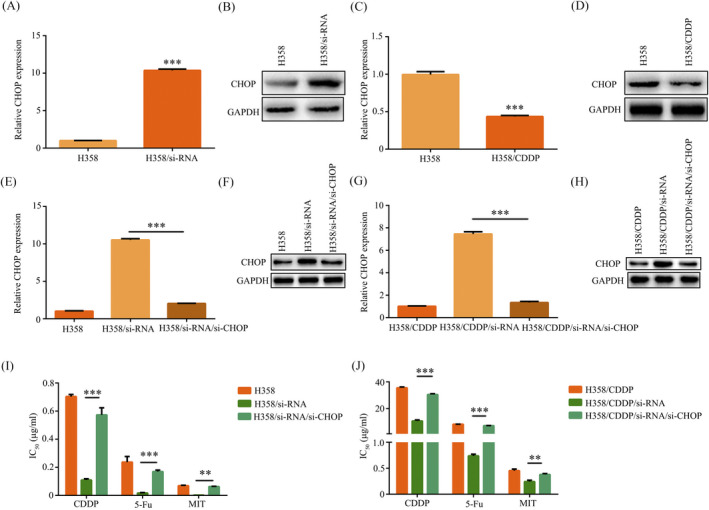
CircRNA_103762 enhanced multidrug resistance by inhibited CHOP expression in NSCLC cells. The CHOP expression was detected by RT‐PCR (A) and Western blot (B) in H358 and H358/si‐circRNA_103762. The CHOP expression was detected by RT‐PCR (C) and Western blot (D) in H358 and H358/CDDP. The CHOP expression was detected by RT‐PCR (E) and Western blot (F) in H358, H358/si‐circRNA_103762 and H358/siRNA/si‐CHOP. The CHOP expression was detected by RT‐PCR (G) and Western blot (H) in H358, H358/CDDP/si‐circRNA_103762 and H358/CDDP/si‐circRNA_103762/si‐CHOP. I, The IC50 of different drugs on H358, H358/si‐circRNA_103762 and H358/si‐circRNA_103762/si‐CHOP. J, The IC50 of different drugs on H358/CDDP, H358/CDDP/si‐circRNA_103762 and H358/CDDP/si‐circRNA_103762/si‐CHOP. The data shown represent the mean ± SD (n = 3). *P < .05, **P < .01, ***P < .001. All siRNA is si‐circRNA_103762
4. DISCUSSION
Recently, several researches have pointed out that circRNAs plays a significant roles in progression and metastasis in tumor. For example, circular RNA hsa_circ_0055538 regulates the malignant biological behavior of oral squamous cell carcinoma through the p53/Bcl‐2/caspase signaling pathway.21 Emerging roles of circRNA_001569 targeting miR‐145 in the proliferation and invasion of colorectal cancer.22 Circular RNA circ‐ABCB10 promotes breast cancer proliferation and progression through sponging miR‐1271.23 In addition, circRNA can affect function of tumor by sponging miRNA. For example, circRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR‐630.24 Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR‐143.25 Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via miR‐1275/FOXK1 axis.26 In this study, circRNA_103762 in NSCLC was firstly reported upregulation and found to be a tumor promoter.
Although circRNA has been found to be involved in tumorigenesis,14, 27 circRNA is still rarely studied in MDR. in this study, we found that circRNA_103762 expression was upregulated in H358/CDDP cell. In addition, the expression of circRNA_103762 was increased in NSCLC patients after cisplatin chemotherapy compared with before cisplatin chemotherapy. We further confirmed that downregulation of circRNA_103762 expression can reduce IC50 values of different drugs. Therefore, circRNA_103762 can enhance MDR in NSCLC.
Early reports pointed out that CHOP is related to tumor and plays a significant role in ER stress.17, 18, 19 CHOP expression can be induced by chemotherapeutic drugs and is associated with MDR.20 In this study, we discovered that circRNA_103762 knockdown promoted CHOP expression in H358/si‐circRNA_103762 cell and H358/CDDP/ si‐circRNA_103762 cell. Besides, CHOP knockdown would rescue IC50 values of different drugs of circRNA_103762 silenced NSCLC cell lines. Thus, circRNA_103762 enhanced MDR by inhibited CHOP expression in NSCLC cells.
In conclusion, we demonstrated that circRNA_103762 is upregulated in NSCLC and functions as an oncogene in NSCLC, and circRNA_103762 enhanced MDR by inhibited CHOP expression in NSCLC cells. These results will help us understand the MDR of NSCLC, providing better effective therapy strategies for patients.
Xiao G, Huang W, Zhan Y, Li J, Tong W. CircRNA_103762 promotes multidrug resistance in NSCLC by targeting DNA damage inducible transcript 3 (CHOP). J Clin Lab Anal. 2020;34:e23252 10.1002/jcla.23252
Guanhua Xiao and Wenqi Hwang contribute equally to this article.
Contributor Information
Jing Li, Email: jingli163@hotmail.com.
Wancheng Tong, Email: WanchengTong@hotmail.com.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7‐34. [DOI] [PubMed] [Google Scholar]
- 2. Zhang JF, Li M, Miao JY, Zhao BX. Biological activities of novel pyrazolyl hydroxamic acid derivatives against human lung cancer cell line A549. Eur J Med Chem. 2014;83:516‐525. [DOI] [PubMed] [Google Scholar]
- 3. Tan CS, Gilligan D, Pacey S. Treatment approaches for EGFR‐inhibitor‐resistant patients with non‐small‐cell lung cancer. Lancet Oncol. 2015;16:e447‐e459. [DOI] [PubMed] [Google Scholar]
- 4. Wu D, Liu J, Chen J, He H, Ma H, Lv X. miR‐449a suppresses tumor growth, migration, and invasion in non‐small cell lung cancer by targeting a HMGB1‐mediated NF‐kappaB signaling pathway. Oncol Res. 2019;27:227‐235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang R, Feng N, Wang Y, et al. SNPs in LncRNA genes are associated with non‐small cell lung cancer in a Chinese population. J Clin Lab Anal. 2019;33:e22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lou B, Wei D, Zhou X, Chen H. Long non‐coding RNA KDM5B anti‐sense RNA 1 enhances tumor progression in non‐small cell lung cancer. J Clin Lab Anal. 2020;34:e22897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang H, Shen Y, Li Z, et al. The biogenesis and biological functions of circular RNAs and their molecular diagnostic values in cancers. J Clin Lab Anal. 2020;34:e23049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li Z, Ruan Y, Zhang H, Shen Y, Li T, Xiao B. Tumor‐suppressive circular RNAs: mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110:3630‐3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141‐157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1‐7. [DOI] [PubMed] [Google Scholar]
- 11. Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL, Yang Y. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res. 2018;37:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zheng X, Chen L, Zhou Y, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo‐YAP signaling. Mol Cancer. 2019;18:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou LH, Yang YC, Zhang RY, Wang P, Pang MH, Liang LQ. CircRNA_0023642 promotes migration and invasion of gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci. 2018;22:2297‐2303. [DOI] [PubMed] [Google Scholar]
- 14. Liu Y, Dong Y, Zhao L, Su L, Luo J. Circular RNAMTO1 suppresses breast cancer cell viability and reverses monastrol resistance through regulating the TRAF4/Eg5 axis. Int J Oncol. 2018;53:1752‐1762. [DOI] [PubMed] [Google Scholar]
- 15. Qi Y, Zhang B, Wang J, Yao M. Upregulation of circular RNA hsa_circ_0007534 predicts unfavorable prognosis for NSCLC and exerts oncogenic properties in vitro and in vivo. Gene. 2018;676:79‐85. [DOI] [PubMed] [Google Scholar]
- 16. Zhao F, Han Y, Liu Z, Zhao Z, Li Z, Jia K. circFADS2 regulates lung cancer cells proliferation and invasion via acting as a sponge of miR‐498. Biosci Rep. 2018;38:BSR20180570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Simard JC, Durocher I, Girard D. Silver nanoparticles induce irremediable endoplasmic reticulum stress leading to unfolded protein response dependent apoptosis in breast cancer cells. Apoptosis. 2016;21:1279‐1290. [DOI] [PubMed] [Google Scholar]
- 18. Chiu TL, Su CC. Tanshinone IIA increases protein expression levels of PERK, ATF6, IRE1alpha, CHOP, caspase3 and caspase12 in pancreatic cancer BxPC3 cellderived xenograft tumors. Mol Med Rep. 2017;15:3259‐3263. [DOI] [PubMed] [Google Scholar]
- 19. Dilshara MG, Jayasooriya RG, Park SR, Choi YH, Choi IW, Kim GY. Caffeic acid phenethyl ester enhances TRAIL‐mediated apoptosis via CHOP‐induced death receptor 5 upregulation in hepatocarcinoma Hep3B cells. Mol Cell Biochem. 2016;418:13‐20. [DOI] [PubMed] [Google Scholar]
- 20. Tan W, Liao Y, Qiu Y, et al. miRNA 146a promotes chemotherapy resistance in lung cancer cells by targeting DNA damage inducible transcript 3 (CHOP). Cancer Lett. 2018;428:55‐68. [DOI] [PubMed] [Google Scholar]
- 21. Su W, Sun S, Wang F, Shen Y, Yang H. Circular RNA hsa_circ_0055538 regulates the malignant biological behavior of oral squamous cell carcinoma through the p53/Bcl‐2/caspase signaling pathway. J Transl Med. 2019;17:76. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22. Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR‐145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680‐26691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ‐ABCB10 promotes breast cancer proliferation and progression through sponging miR‐1271. Am J Cancer Res. 2017;7:1566‐1576. [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Y, Liu H, Li W, et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR‐630. Aging (Albany NY). 2017;9:1585‐1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tang YY, Zhao P, Zou TN, et al. Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR‐143. DNA Cell Biol. 2017;36:901‐908. [DOI] [PubMed] [Google Scholar]
- 26. Ma X, Yang X, Bao W, et al. Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via miR‐1275/FOXK1 axis. Biochem Biophys Res Commun. 2018;498:1009‐1015. [DOI] [PubMed] [Google Scholar]
- 27. Gao D, Zhang X, Liu B, et al. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics. 2017;9:1175‐1188. [DOI] [PubMed] [Google Scholar]


