Abstract
Background
Although plasma free hemoglobin (fHb) test is important for assessing intravascular hemolysis, it is still dependent on the gold standard Harboe method using manual and labor‐intensive spectrometric measurements at the wavelength of 380‐415‐450 nm. We established an automated fHb assay using a routine chemistry autoanalyzer that can be tuned to a wavelength of 380‐416‐450 nm.
Methods
The linearity, precision, accuracy, correlation, and sample carryover of fHb measurement using TBA200FRneo method and manual Harboe method were evaluated, respectively. fHb values measured by manual Harboe method were compared with those measured by our new automated TBA200FRneo method.
Results
fHb measurements were linear in the range of 0.05~38.75 µmol/L by TBA200FRneo and 0.05~9.69 µmol/L by manual Harboe method. Imprecision analysis (%CV) revealed 0.9~2.8% for TBA200FRneo method and 5.3~13.6% for the manual Harboe method. Comparison analysis showed 0.9986 of correlation coefficient (TBA200FRneo = 0.970 × Harboe + 0.12). In analytical accuracy analysis, the manual Harboe method revealed about 4 times higher average total error % (12.2%) than the TBA200FRneo automated method (2.8%). The sample carryover was −0.0016% in TBA200FRneo method and 0.0038% in Harboe method.
Conclusions
In the measurement of fHb, the automated TBA200FRneo method showed better performance than the conventional Harboe method. It is expected that the automated fHb assay using the routine chemistry analyzer can replace the gold standard Harboe method which is labor‐intensive and need an independent spectrophotometry equipment.
Keywords: automated chemistry analyzer, free hemoglobin, Harboe method, plasma hemoglobin, spectrophotometer
1. INTRODUCTION
The majority of the hemoglobin (Hb) is found inside the red blood cells, and not in the serum. Plasma free Hb (fHb) test is a blood test that measures the level of free Hb in the liquid part of the blood (the plasma), outside of the red blood cells. The determination of fHb is essential for diagnosing and monitoring intravascular hemolysis.1 The known methods for measuring Hb are chromogenic assays, spectrophotometric assays, immunonephelometric assays, and enzyme‐linked immunosorbent assays. Traditionally, the concentration of fHb is measured using chromogenic assays or spectrophotometry. Chromogenic assays were favored for sensitive detection of small quantities of Hb. However, the use benzidine, a chromogenic reducing substance, has been restricted because of its carcinogenicity.2, 3 Various spectrophotometric assays for measuring absorption at multiple wavelengths have been used for determining fHb levels to avoid carcinogenic chemicals.4, 5 However, their results could be influenced by the presence of bilirubin and turbidity.6 Direct spectrophotometric methods have been introduced to minimize the analytical interference of fHb; however, they are not sensitive enough and often require cumbersome calculations.4, 5, 7, 8 Among these spectrophotometric algorithms, the Harboe method is the most widely used and accepted as gold standard. These assays are not yet fully automated for diagnostic purposes in routine laboratories. Here, we focused on using spectrophotometric principles on an automated chemistry analyzer, and implemented to the TBA200FRneo chemistry autoanalyzer. The CAP survey program is a worldwide external quality assessment program, in which more than 2000 laboratories are participating. For fHb tests, only 92 laboratories participated in the 2018 CAP survey program, indicating that, although fHb analysis is a direct indicator of intravascular hemolysis, not many clinical laboratories perform fHb analysis because of the difficulties described above.
The purpose of this study was to apply the principles of direct spectrophotometric assays for measuring fHb in an automated routine chemistry analyzer without reagents. And we evaluated analytical performance of new method and compared with gold standard manual Harboe method.9
2. MATERIALS AND METHODS
2.1. Subjects
This study was conducted at the National Cancer Center, South Korea. Based on the Clinical and Laboratory Standards Institute (CLSI) guideline EP09‐A3, at least 40 samples were analyzed for comparison and bias estimation.10 Samples obtained from the laboratory for fHb measurement were used for linearity, comparison, and carryover analyses. Quality control data were used to access analytical imprecision. The protocol was approved by the Institutional Review Board (IRB) as a review exemption (NCC2017‐0154), as it was conducted for the purpose of diagnosis at a medical laboratory and for test method evaluation using residual human‐derived materials.11
2.2. Determination of plasma free Hb
For the measurement of fHb, 3 mL of whole blood was collected by venipuncture directly into a potassium‐EDTA tube. fHb measurement performed using the newly introduced method on the TBA200FRneo chemistry autoanalyzer (Toshiba Medical Systems). The TBA200FRneo was set up to measure the absorbance of fHb at 380‐416‐450 nm; the conditions had 1 nm (416 nm vs 415 nm) difference with those used for the gold standard Harboe method (380‐415‐450 nm).9 Set the TBA200FRneo autoanalyzer to measure the absorbance of the sample at 380 nm (A380 nm) for FreeHB1 order, the absorbance of the sample at 416 nm (A415 nm), for FreeHB2 order, and the absorbance of the sample at 450 nm (A450 nm) for FreeHB3 order. Place the sample in the sample cup and put it in the TBA200FRneo autoanalyzer and order FreeHB1, FreeHB2, and FreeHB3. Assay parameters are set according to the manufacturer's operation manual (No. 2B586‐285EN), as follows. In the configuration screen of the TBA200FRneo autoanalyzer, make 3 test orders named order FreeHB1, FreeHB2, and FreeHB3. Select bottle type as 100 mL, click reaction mode as endpoint photometer (“Photo” button), put sample volume as 35 μL and reagent (Na2CO3) volume as 300 μL, and set the reading time as “from 30 to 33 point (meaning from 135.0 to 148.5 s).” The wavelength for the reading is set to 380 nm, 416 nm, and 450 nm for each FreeHB1, FreeHB2, and FreeHB3 assay, respectively. Place the sample in the sample cup and put it in the TBA200FRneo autoanalyzer and order FreeHB1, FreeHB2, and FreeHB3. Wait for the three outputs. All the measurements are multiplied by 10 to correct dilution and finally calculate fHB using Harboe equation. We also determined fHb concentration by manual gold standard Harboe method: measuring absorbance on a Hitachi U‐2001 spectrophotometer (Hitachi High‐Tech Co) at 380‐415‐450 nm.9 fHb levels were calculated using the Harboe equation, fHb (mg/dL) = 83.6 (2 × A415 nm − A380 nm − A450 nm).9 The conversion factor for fHb measurement from mg/dL to µmol/L is 0.155.
2.3. Linearity analysis
The linearity of the assays was evaluated according to the CLSI EP6‐A.12 Pooled plasma samples were prepared at high (H, 38.75 µmol/L [250.0 mg/dL]) and low (L, 0.05 µmol/L [0.3 mg/dL]) concentrations. Five linearity materials, from level 1 to level 5, were prepared according to the CLSI guidelines, as follows: 1L, 0.75L + 0.25H, 0.50L + 0.50H, 0.25L + 0.75H, and 1H. These materials were tested in duplicate by the new method and the average values were analyzed for linearity. And same linearity analysis was performed by manual Harboe method, simultaneously.
2.4. Precision analysis
Precision was evaluated according to the CLSI document EP 05‐A313 using quality control material. The clinical decision level to determine hemolysis was 0.78 µmol/L (5 mg/dL), and the mean values of quality control materials for precision analysis were 0.81 µmol/L and 1.33 µmol/L (5.2 mg/dL and 8.6 mg/dL). Samples were tested in 2 runs per day (at 9:00 and 13:00) for 20 d, with 40 replicates per level. Repeatability (previously within‐run precision) and within‐laboratory imprecision (previously total precision) were evaluated and expressed as coefficient of variation (total CV %).
2.5. Accuracy analysis
To access the analytical accuracy of both methods, we made three levels of samples with 0.9% saline and lyophilized human hemoglobin powder (Sigma‐Aldrich). Expected level was 1.70, 1.40, 2.79 µmol/L (4.5, 9.0, and 18.0 mg/dL), respectively. And each level was measured in 2 runs during 3 different days in both methods: TBA200FRneo method and manual Harboe method. We accessed the mean of 6 measurements in each level. Using these data, we calculated recovery% (100 × [measured]/expected concentration), bias% (100 × [expected‐measured]/expected concentration), total CV%, and total error (absolute bias% + 1.96 × total CV%).
2.6. Comparison analysis
Method comparison was performed for 40 clinical samples (8 samples from healthy individuals, and 32 samples from hemolytic patients) covering clinically significant ranges of fHb, according to CLSI EP09‐A3.10 The fHb was evaluated in all the samples in both methods: TBA200FRneo method and manual Harboe method. All tests were conducted under the same conditions as routine clinical chemistry analysis. Calibration and quality control were performed in each method as routine clinical analysis.
2.7. Sample carryover analysis
Carryover test was performed using a minipool of the patient samples. A high‐concentration sample (20.15 µmol/L [130 mg/dL]) was tested 4 consecutive times (H1, H2, H3, and H4), and a low‐concentration sample (0.19 µmol/L [1.2 mg/dL]) was tested 4 consecutive times (L1, L2, L3, and L4) with both methods.14 The carryover was calculated as follows: carryover (%) = [L1 − (L3 + L4)/2 × 100/[(H2 + H3)/2 − (L3 + L4)/2].14 The acceptance criterion for carryover was set to less than 1.0%.15
2.8. Statistical analyses
Statistical analyses were performed using Microsoft Excel with R version 3.3.2 and CentOS Linux 7. The chi‐square test was used for statistical evaluation of correlation analysis. The differences between the results of the conventional method and our method were evaluated using Student's t test. Bland‐Altman plots were constructed.16 The level of significance for all statistical analyses was set to P < .05.
3. RESULTS
3.1. Linearity analysis
The results of the TBA200FRneo showed that the first‐order models were the best fits over the entire range from 0.05~38.75 µmol/L (0.3~250.0 mg/dL), with a maximum bias of 2.6%. The results of the manual Harboe method showed unacceptable linearity, because level 2 showed a 15.9% difference from the predicted first‐order line. According to the CLSI EP6‐A, an additional linearity test for the Harboe method was performed using plasma samples with level 1 (set as new L) and 2 (set as new H), which were sequentially mixed to make five‐level samples. The linearity of the CLSI EP6‐A was ideal for the first‐order model, with a maximum bias of −1.6%, between 0.05~9.69 µmol/L (62.5 mg/dL). The final linear ranges in each method are summarized in Table 1.
Table 1.
Summary of the performance evaluation of two methods for the measurement of plasma free hemoglobin
| TBA200FRneo method | Harboe method | |
|---|---|---|
| Linear range (µmol/L) | 0.05 ‐ 38.75 | 0.05 ‐ 9.69 |
| Imprecision at clinical decision level (%CV) | 2.7 | 13.6 |
| Carry over (%) | −0.0016 | 0.0038 |
Abbreviation: CV, coefficient of variation.
3.2. Precision analysis
The repeatability and within‐laboratory precision at near clinical decision level (level 1, 0.81 µmol/L [5.2 mg/dL]) and abnormal level (level 2, 1.33 µmol/L [8.6 mg/dL]) are shown in Table 2. Clinical decision level for determining intravascular hemolysis is 0.78 µmol/L (5.0 mg/dL). The CV% at each level was significantly lower for the TBA200FRneo method, indicating superior precision than the gold standard Harboe method.
Table 2.
Imprecision of two methods at low and high level of plasma free hemoglobin
| Method | Level | Repeatability | Within‐laboratory imprecision | ||
|---|---|---|---|---|---|
| SD (µmol/L) | CV (%) | SD (µmol/L) | CV (%) | ||
| TBA200FRneo method | Low (0.81 µmol/L) | 0.08 | 1.0 | 0.22 | 2.7 |
| High (1.33 µmol/L) | 0.12 | 0.9 | 0.37 | 2.8 | |
| Harboe method | Low (0.81 µmol/L) | 0.82 | 9.4 | 1.21 | 13.6 |
| High (1.33 µmol/L) | 1.52 | 9.7 | 1.94 | 12.4 | |
Repeatability (previously within‐run precision) and within‐laboratory imprecision (previously, total precision) are expressed as the coefficient of variation for each method. Clinical decision level for determining intravascular hemolysis is 0.78 µmol/L (5.0 mg/dL).
Abbreviations: CV, coefficient of variation; SD, standard deviation.
3.3. Accuracy analysis
Analytical accuracies are summarized in Table 3. Total error of level 1, 2, and 3 were 1.9%, 3.8%, and 2.8% in TBA200FRneo resulting in an average 2.8% of total error. In the manual Harboe method, the total error of level 1, 2, and 3 were 11.0%, 14.8%, and 10.9%, resulting in an average 12.2% of total error.
Table 3.
Results of accuracy analysis by the TBA200FRneo method and Harboe method
| TBA200FRneo method | Harboe method | |||||
|---|---|---|---|---|---|---|
| Level 1 | Level 2 | Level 3 | Level 1 | Level 2 | Level 3 | |
| Expected concentration (µmol/L) | 0.70 | 1.40 | 2.79 | 0.70 | 1.40 | 2.79 |
| Mean of measured concentration (µmol/L) | 0.70 | 1.39 | 2.77 | 0.68 | 1.30 | 2.70 |
| Bias (%) | 0.4 | −0.4 | −0.6 | −3.0 | −6.9 | −3.1 |
| Total CV (%) | 0.8 | 1.7 | 1.1 | 4.1 | 4.0 | 4.0 |
| Total error (%) | 1.9 | 3.8 | 2.8 | 11.0 | 14.8 | 10.9 |
| Recovery (%) | 100.4 | 99.6 | 99.4 | 97.0 | 93.1 | 96.9 |
Abbreviation: CV, coefficient of variation.
3.4. Comparison analysis
The comparison analysis between the two methods showed a correlation coefficient of 0.9989 (TBA200FRneo = 0.970 × Harboe + 0.12) in the range of 0.03~39.22 µmol/L (0.2 ~ 253.0 mg/dL). The mean/median values of healthy population (n = 8) were 0.54/0.56 µmol/L (range 0.03‐0.78 µmol/L) and those of hemolytic population (n = 32) were 5.44/1.76 µmol/L (range 0.79‐39.22 µmol/L). fHb values measured using TBA200FRneo and Harboe showed a mean difference of −1.5%. Graphical differences are shown in Figure 1.
Figure 1.
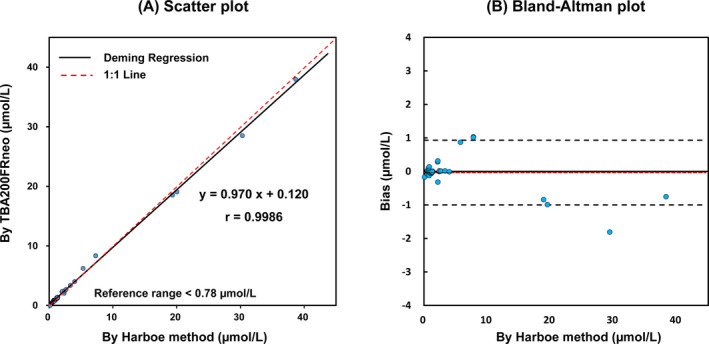
Comparison of gold standard Harboe method and new method using chemistry autoanalyzer, TBA200FRneo. A, Deming regression line is shown with 1:1 line; r = .9986. Plasma free hemoglobin in TBA200FRneo (µmol/L) = 0.970 × Harboe + 0.12. B, Bland‐Altman plots for plasma free hemoglobin measurements. Central dotted lines denote the average difference and ± 2SDs (black dot)
3.5. Sample carryover analysis
The results of carryover testing were −0.0016% for the TBA200FRneo method and 0.0038% for the Harboe method (Table 1). These results met the laboratory acceptance criteria, which was less than 1.0%.
4. DISCUSSION
fHb test is an important emergency test to assess hemolysis within the vasculature. During intravascular hemolysis, Hb molecules in RBCs are released into the plasma. The fHb is then captured by haptoglobins in circulation. The Hb‐haptoglobin complex is rapidly eliminated by the liver, leading to a reduction in plasma haptoglobin.17 Dimers of alpha‐beta globin that are not bound by haptoglobin are small enough (molecular weight, 34 000 Da) to be filtered by the glomerulus, leading to hemoglobinuria.18 During the physiological process after hemolysis, fHb is the most suitable clinical marker for rapidly detecting intravascular hemolysis.
fHb assay is not readily available in clinical laboratories, despite its excellent clinical utility. This is because of the difficulty of performing the manual Harboe method. This conventional gold standard method requires independent spectrophotometry, which is not commonly used in clinical laboratory recently and needs a skilled technician. The technician must also calculate the Hb concentration by measuring the absorbance at three wavelengths; the plasma samples must be placed in the cuvette manually for each test. Sending the samples to external laboratories where the test is available might extend the turnaround time to days, and it would become ineffective as an emergency test. Laboratory automation is irreversible trends.19, 20 The automation of the Harboe method means adding one more test item of fHb to the already operating chemistry autoanalyzer system. This implementation would, therefore, be an attractive alternative for labor‐intensive manual spectrometric measuring fHb in a clinical laboratory.
We focused on using the principles of spectrophotometry on the automated chemistry analyzer. We applied the direct spectrophotometric assay for measuring fHb to the automated routine chemistry analyzer without any reagents. This assay could be applied to any automated analyzer adopting spectrophotometric principle. In this study, fHb was analyzed using a triple wave method (380, 416, and 450 nm) on the TBA200FRneo chemistry autoanalyzer, similar to the Harboe method. The absorbance at 416 nm, which represents oxyhemoglobin, was corrected for the interference of bilirubin and turbidity by subtracting the absorbance at 380 and 450 nm, similar to the Harboe method using 415 nm.6, 9 Similar efforts to apply manual spectrometric assays to automated equipment have already been attempted in the indocyanine green test.21, 22 We obtained similar results when we established an indocyanine green test on the same automated chemistry analyzer.22
Overall, new automated method showed better performance than the conventional Harboe method. Linearity of TBA200FRneo method was acceptable in the range of 0.05~38.75 µmol/L, which was broader than the range of the Harboe method. Possible causes of narrow linear assay range of manual Harboe method include: the aging of the light source, differences in cuvette, or the decreased sensitivity of detector inside the spectrophotometer.23 The TBA200FRneo method showed acceptable imprecision, with less than 1.0% repeatability and less than 2.8% total precision, even in the low level. The precision results of the TBA200FRneo method were about fourfold to 10‐fold superior than those of the Harboe method. In analytical accuracy, the manual Harboe method revealed about 4 times higher average total error % (12.2%) than the TBA200FRneo automated method (2.8%). The correlation coefficient between the two methods was excellent as 0.9989. Although the slope was 0.970, the mean percent difference was within 10%. Furthermore, the TBA200FRneo method showed minimal carryover in fHb analysis.
The limitations of this study are as follows: the difference between the measured values at 415 nm and 416 nm, and the no validation for the interference. For differences in the measurement at 415 nm and 416 nm spectra, we compared the measurements of 20 different concentrations of the samples at 415‐380‐450 nm setting and 416‐380‐450 nm setting using same spectrophotometer. In this comparison experiment, the correlation coefficient (r) was .986 and mean bias was +0.15% in the range of 0.5~20.0 µmol/L. Since TBA200FRneo method basically uses the same spectrometry principles as the Harboe method, it is expected to exhibit interference effects similar to those of the Harboe method. This part needs further evaluation. This assay could be affected by hemolysis due to not internal factors such as improper sample management. Because it only measures the final products of hemolysis and cannot distinguish in vivo hemolysis from ex vivo hemolysis due to improper sample management. This is the same with gold standard Harboe method. To determine in vivo clinical hemolysis, other laboratory findings (such as positive direct anti‐globulin result, decrease of serum haptoglobin level, increase of serum indirect bilirubin, and presence of hemoglobinuria) should be concerned.
In summary, we implemented the measurement of fHb for performance on a routine chemistry autoanalyzer that can be set to a wavelength of 416‐380‐450 nm; the setting is 1 nm different from gold standard Harboe method using 415‐380‐450 nm. We demonstrated that the fHb assay using an automatic chemistry analyzer showed acceptable linearity over a clinically relevant range, precision, and good correlation with the conventional Harboe method. It is expected that the automated fHb test using the routine chemistry autoanalyzer can replace the conventional Harboe method using spectrophotometer, without preparation of additional equipment or reagent.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest. Only the authors are responsible for the development of the content and writing of the paper.
AUTHOR CONTRIBUTIONS
Hee‐Jung Chung involved in conceptualization, formal analysis, methodology, project administration, software, supervision, visualization, writing original draft, and writing—review and editing. Jae‐Woo Chung involved in data curation, investigation, resources, software, and validation. Joowon Yi is involved in data curation. Mina Hur involved in writing—review and editing. Tae Hwan Lee involved in data curation and investigation. Sang‐Hyun Hwang involved in funding acquisition and conceptualization. Yoon Kyung Song involved in project administration, investigation, and resources. Do Hoon Lee involved in funding acquisition, conceptualization, funding acquisition.
ACKNOWLEDGMENT
This study was supported by a grant from the National Cancer Center,South Korea (NCC 1510100) and Korean Association of External Quality Assessment Service (KEQAS), South Korea (2020‐04).
Chung H‐J, Chung J‐W, Yi J, et al. Automation of Harboe method for the measurement of plasma free hemoglobin. J Clin Lab Anal. 2020;34:e23242 10.1002/jcla.23242
Hee‐Jung Chung and Jae‐Woo Chung contributed equally to this work.
Contributor Information
Hee‐Jung Chung, Email: dhlee@ncc.re.kr.
Do Hoon Lee, Email: dhlee@ncc.re.kr.
REFERENCES
- 1. Rother RP, Bell L, Hillmen P, et al. The clinical sequelae of intravascular hemolysis and extracellular plasma hemoglobin: a novel mechanism of human disease. JAMA. 2005;293(13):1653‐1662. [DOI] [PubMed] [Google Scholar]
- 2. Pillier‐Loriette C, Wingerter P, Gerota J, et al. Determination of plasma hemoglobin in fresh plasma for therapeutic use by the tmb (3,3′,5,5′‐tetramethylbenzidine) method. Rev Fr Transfus Immunohematol. 1983;26(3):299‐312. [DOI] [PubMed] [Google Scholar]
- 3. Goyal MM, Basak A. Estimation of plasma haemoglobin by a modified kinetic method using o‐tolidine. Indian J Clin Biochem. 2009;24(1):36‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Soloni FG, Cunningham MT, Amazon K. Plasma hemoglobin determination by recording derivative spectrophotometry. Am J Clin Pathol. 1986;85(3):342‐347. [DOI] [PubMed] [Google Scholar]
- 5. Copeland BE, Dyer PJ, Pesce AJ. Hemoglobin determination in plasma or serum by first‐derivative recording spectrophotometry. Evaluation of the procedure of soloni, cunningham, and amazon. Am J Clin Pathol. 1989;92(5):619‐624. [DOI] [PubMed] [Google Scholar]
- 6. Noe DA, Weedn V, Bell WR. Direct spectrophotometry of serum hemoglobin: an allen correction compared with a three‐wavelength polychromatic analysis. Clin Chem. 1984;30(5):627‐630. [PubMed] [Google Scholar]
- 7. Fairbanks VF, Ziesmer SC, O'Brien PC. Methods for measuring plasma hemoglobin in micromolar concentration compared. Clin Chem. 1992;38(1):132‐140. [PubMed] [Google Scholar]
- 8. Wong SS, Schenkel OJ. Quantification of plasma hemoglobin in the presence of bilirubin with bilirubin oxidase. Ann Clin Lab Sci. 1995;25(3):247‐251. [PubMed] [Google Scholar]
- 9. Harboe M. A method for determination of hemoglobin in plasma by near‐ultraviolet spectrophotometry. Scand J Clin Lab Invest. 1959;11:66‐70. [DOI] [PubMed] [Google Scholar]
- 10. CLSI . Measurement Procedure Comparison and Bias Estimation Using Patient Samples; Approved Guideline‐Third Ed. CLSI document ep09‐a3. Wayne, PA: Clinical and Laboratory Standards Institute; 2013. [Google Scholar]
- 11. Korea enforcement decree of the bioethics and safety act. Article 36 (2). 2012. Database: National law information center [internet]. http://www.law.go.kr. Accessed January 28, 2020.
- 12. CLSI . Evaluation of the Linearity of Quantitative Measurement Procedures: A Statistical Approach; Approved Guideline‐First Ed. CLSI document ep06‐a. Wayne, PA: Clinical and Laboratory Standards Institute; 2003. [Google Scholar]
- 13. CLSI . Evaluation of Precision of Quantitative Measurement Procedures; Approved Guideline‐Third Ed. CLSI document ep05‐a3. Wayne, PA: Clinical and Laboratory Standards Institute; 2014. [Google Scholar]
- 14. Kaplan LA, Pesce AJ. Clinical Chemistry: Theory, Analysis, Correlation (5th edn). St. Louis, MO: Mosby; 2010. [Google Scholar]
- 15. Broughton PM. Carry‐over in automatic analysers. J Automat Chem. 1984;6(2):94‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dewitte K, Fierens C, Stockl D, et al. Application of the Bland–Altman plot for interpretation of method‐comparison studies: a critical investigation of its practice. Clin Chem. 2002;48(5):799‐801; author reply 801‐792. [PubMed] [Google Scholar]
- 17. Barcellini W, Fattizzo B. Clinical applications of hemolytic markers in the differential diagnosis and management of hemolytic anemia. Dis Markers. 2015; 2015:1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smith A, McCulloh RJ. Hemopexin and haptoglobin: allies against heme toxicity from hemoglobin not contenders. Front Physiol. 2015;6:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chung H‐J, Song YK, Hwang S‐H, et al. Experimental fusion of different versions of the total laboratory automation system and improvement of laboratory turnaround time. J Clin Lab Anal. 2018;32(5):e22400‐e22400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matthijs O, Sc M, Joris D, et al. Progress in automated urinalysis. Ann Lab Med. 2019;39(1):15‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park J‐H, Won EJ, Choi H‐J, et al. Performance evaluation of automated clinical chemistry analyzer for indocyanine green (icg) R15 test. Lab Med Online. 2016;6(3):140‐146. [Google Scholar]
- 22. Seong MW, Song SH, Oh JY, et al. Establishment of an indocyanine green test using an automatic chemistry analyzer. Clin Chem Lab Med. 2006;44(2):196‐198. [DOI] [PubMed] [Google Scholar]
- 23. Lee WC. Annual report of the Korean Association of External Quality Assessment Service on general chemistry (2018). J Lab Med Qual Assur. 2019;41(2):51‐64. [Google Scholar]


