Abstract
Family with sequence similarity 83 member A (FAM83A) has been recently observed to be upregulated in various types of cancer and hypothesized to be serve as an oncogene. The present study aimed to determine the functional roles and the underlying molecular mechanism of FAM83A in cervical cancer. The results demonstrated that although FAM83A expression was increased in cervical cancer compared with normal tissues, the expression levels of FAM83A were decreased in patients with advanced FIGO stage, deep stromal invasion, poor differentiation and/or lymph node metastasis and negatively associated with short survival time of patients with cervical cancer. FAM83A knockdown promoted cell proliferative, migratory and invasive abilities of CaSki and HeLa cells. A mouse xenograft model demonstrated that FAM83A knockdown promoted tumor growth in vivo. Mechanistically, RNA sequencing results revealed that knockdown of FAM83A increased the transcription of genes mainly associated with oncogenesis-associated pathways. In addition, FAM83A knockdown increased the protein levels of α1, α3, α5, β4 and β5 integrins in vitro and in vivo, and the expression of FAM83A was also negatively associated with the levels of these proteins in human cervical cancer tissue samples. In conclusion, the results of the present study suggested that FAM83A may exert a tumor-suppressive role in cervical cancer by suppressing the expression of integrins, which may offer new insight into the biological basis of cervical cancer.
Keywords: FAM83A, cervical cancer, cell proliferation, invasion, integrins
Introduction
Cervical cancer is the fourth most common cancer type among women worldwide, with 570,000 new cases and 311,000 deaths estimated in 2018; however, the incidence and mortality rates have decreased over the last few decades due to effective integration of human papillomavirus (HPV)-based testing through screening and HPV vaccination programs in a number of developed countries (1). Persistent infection with HPV is a primary cause of cervical cancer and its precursor lesions, and HPV testing is used for primary screening (2). HPV infection is common in sexually active women, with an estimated incidence rate of 74-80%, but cervical cancer is comparatively rare, as 90% of HPV infections clear within two years (3,4). Currently, it is not possible to predict which women who test positive for HPV may develop cervical cancer. Therefore, improved understanding of the underlying molecular mechanisms may facilitate the development of novel diagnostic markers and effective therapeutic targets for HPV-associated cancer.
Our previous study performed RNA sequencing to analyze the transcriptomes in healthy cervical tissues without HPV infection and cervical cancer tissues with high-risk HPV (HR-HPV) infection, and identified family with sequence similarity 83 member A (FAM83A) as one of the most upregulated genes in cervical cancer compared with healthy tissues (5). FAM83A is one of the eight-member FAM83 family of proteins; this 434-amino acid protein shares a conserved N-terminal domain of unknown function, termed the DUF1669 domain, with other FAM83 members and also contains serine-rich and proline-rich domains (6,7). FAM83A was originally identified as a novel tumor-specific gene BJ-TSA-9, located on chromosome 8q24, and was demonstrated to be highly expressed in lung cancer using bioinformatics (8). Additionally, FAM83A upregulation was identified in multiple human tumor types, including breast (6,9,10), pancreatic (11) and ovarian (12) cancer, as well as lung adenocarcinoma (13). Using a 3D laminin-rich extracellular matrix culture system, Lee et al (6) revealed that FAM83A was able to confer resistance to epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) via its putative interactions with proto-oncogene c-RAF and PI3K p85 in breast cancer. FAM83A depletion in breast cancer cells leads to suppressed proliferation and invasiveness in vitro as well as to suppressed tumor growth in vivo (14). In addition, analysis of a published breast cancer gene expression dataset reported that patients with breast cancer with high expression of FAM83A exhibited a poor clinical prognosis (15).
Based on the aforementioned studies, FAM83A is considered to be a candidate oncogene. However, the cellular mechanism of action of FAM83A in human cancer is not fully understood. The present study aimed to identify the functional roles and mechanisms of FAM83A in cervical cancer cells. The results of the present study may provide a potential target for cervical cancer therapeutics.
Materials and methods
Human samples
A total of 153 human cervical samples without preoperative treatment were obtained by surgical resection from patients with a mean age of 45 (range 28-66) years at the Department of Gynecologic Oncology at the Women's Hospital, Zhejiang University between January 2012 and December 2013. The Ethics Committee of the Women's Hospital approved the collection and use of human samples. All patients signed informed consent. Diagnosis of all samples was confirmed independently by two pathologists before further analysis.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
RNA extraction and RT-qPCR was performed as previously described (16). Briefly, total RNA was extracted from tissues or cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA synthesis was performed using a PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's instructions. The samples were analyzed on an ABI/ViiA7 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) using a SYBR® Premix Ex Taq kit (Takara Biotechnology Co., Ltd.). The thermocycling conditions were as follows: Denaturation at 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec, and the melting curve stage at 95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec. Gene quantification was performed using the 2−∆∆Cq method (16). GAPDH was used as an internal control. The primers used are presented in Table SI.
Immunohistochemistry (IHC)
IHC was performed on paraffin-embedded human cervical tissue sections and cervical tumor xenografts with the use of the primary antibodies. Briefly, each 4 μm-section was deparaffinized in xylene and rehydrated with 100, 85 and 75% ethanol in deionized water. The slides were placed in antigen retrieval solution, boiled at 100°C for 15 min and subsequently incubated with primary antibodies including anti-FAM83A (1:800; cat. no. bs-16014R; BIOSS), integrin α1 (1:100; cat. no. 22146-I-Ap; ProteinTech Group, Inc.), integrin α3 (1:1,000; cat. no. 66070-I-Ig; ProteinTech Group, Inc.), integrin α5 (1:100; cat. no. ET1701-58; Hangzhou HuaAn Biotechnology Co., Ltd.), integrin β4 (1:50; cat. no. ET1703-52; Hangzhou HuaAn Biotechnology Co., Ltd.) and integrin β5 (1:400; cat. no. bs-23987R; BIOSS) at 4°C overnight, followed by incubation with Dako Envision Peroxidase (Dako; Agilent Technologies, Inc.) for 1 h at room temperature. The antibody staining was visualized with DAB (Dako; Agilent Technologies, Inc.). All section slides were counterstained in hematoxylin. The IHC results were evaluated and scored based on the intensity of staining and the proportion of stained positive cells in a blinded manner by a technician (17). The frequency of positive cells were defined as follows: <5%, 0 points; 5-30%, 1 point; 31-70%, 2 points; >71%, 3 points. The staining intensity was scored as follows: No staining, 0 points; light brown staining, 1 point; brown staining, 2 points; dark brown staining, 3 points. The staining result was determined by multiplying the score for the frequency of positive cells with the score for staining intensity. The expression of FAM83A were graded as 'none' (score, 0), 'weak' (score, 1-3), 'moderate' (score, 4-6) or 'strong' (score, 7-9). 'Strong' staining was defined as 'high expression', whereas 'weak' or 'moderate' staining was considered 'low expression'.
Cell lines
Human cervical cancer cell lines HeLa and CaSki were purchased from the American Type Cell Culture and cultured in DMEM (Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). Cells were cultured at 37°C with 5% CO2. Both cell lines were negative for mycoplasma.
Small interfering (si)RNA transfection
FAM83A siRNA pool (si-FAM83A; cat. no. M-015040-00-0005) and non-targeting siRNA pool (si-NS; cat. no. D-001206-13-05) were purchased from GE Healthcare Dharmacon, Inc. HeLa and CaSki cells (3×105) were plated in 6-well plates and transfected with 50 μM si-NS or si-FAM83A using the DharmaFECT 1 transfection reagent (cat. no. T-2001-03; GE Healthcare Dharmacon, Inc) and incubated at 37°C in a humidified CO2 incubator for 24 h according to the manufacturer's instructions. The RNA was extracted at 48 h post-transfection, and protein was obtained at 72 h post-transfection. The transfection efficiency was confirmed by RT-qPCR and western blotting.
Cell proliferation, migration and invasion assays
To determine cell proliferation, 5,000 HeLa and CaSki cells were plated in triplicate in 96-well plates, and a Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc.) was performed at 24, 48, 72 and 96 h post-transfection. All experiments were performed in triplicate and repeated three times independently.
Migration assays were performed using Transwell filter chambers (Corning, Inc.). HeLa and CaSki cells were trans-fected with 50 nM siRNA in 12-well plates for 24 h, and cell viability was determined with a Trypan blue assay prior to the migration assay. A total of 1×105 cells with similar viability in 200 μl serum-free DMEM in each group were then seeded in the upper chamber of each Transwell, and 500 μl medium supplemented with 10% FBS was added to the lower chamber. After 16 h incubation for HeLa and 24 h for CaSki cells, the non-migrated cells were removed by cotton swabs, and cells that migrated through the membrane were fixed by 4% paraformaldehyde for 20 min at room temperature, stained with 0.1% crystal violet for 30 min at room temperature, and counted in five random visual fields per well under a light microscope (×100 magnification; Leica Microsystems GmbH).
Invasion assays were performed using Transwell filter chambers pre-coated with Matrigel for 30 min at 37°C. The detection was performed following incubation for 24 h of HeLa and 36 h for CaSki cells. All migration and invasion experiments were performed in duplicate and repeated independently three times.
Xenograft models
BALB/c female nude mice (n=12; age, 4 weeks) were purchased from Shanghai Laboratory Animal Center. All mice were housed in a dedicated SPF facility (temperature, 22±1°C; humidity, 50±10%; light/dark cycle, 12:12 h) with standard laboratory food and water, and used with the approval of the Animal Care and Use Committee of Zhejiang University. The mice were randomly divided in two groups in a blinded manner. Human cervical cancer HeLa cells transfected with scrambled short hairpin (sh)RNA (nc-shRNA) or FAM83A-shRNA were injected subcutaneously (1×107 cells/inoculum) into the right-side flank of each mouse (n=10). Tumor growth was detected with calipers every 4 days, and tumor volume was calculated using the following formula: Volume = (length × width2)/2. To further evaluate the metastatic potential of FAM83A, mice (n=2) were intraperitoneally transplanted with 1×107 tumor cells (HeLa/nc-shRNA and HeLa/FAM83A-shRNA) suspended in 200 μl PBS. Five weeks after transplantation, all mice were euthanized by cervical dislocation. Death was confirmed through close observation of the mice. The mice were assessed for any movement, including the respiratory movement of the chest, and checked for heartbeat by a stethoscope. Once the muscles of the mice stiffened and evidence of animal death was observed, the tumors were harvested and imaged.
Immunofluorescence staining
Experiments were performed on 4-μm paraffin-embedded tumor tissues mounted on slides. Paraffin was removed, and the sections were serially rehydrated by descending ethanol series as described above. Antigen retrieval was performed by heating the slides in EDTA buffer at 100°C for 15 min. Slides were blocked by incubating with 3% BSA (Wuhan Servicebio Technology Co., Ltd.) for 30 min at 37°C and then incubated with primary antibodies against FAM83A (1:500; BIOSS), integrin α1 (1:200; ProteinTech Group, Inc.), integrin α3 (1:1,000; ProteinTech Group, Inc.), integrin α5 (1:100; Hangzhou HuaAn Biotechnology Co., Ltd.), integrin β4 (1:100; ProteinTech Group, Inc.) and integrin β5 (1:100; BIOSS) at 4°C overnight, followed by the secondary antibodies Alexa Fluor 488-conjugated goat anti-mouse (1:400; cat. no. GB25303) or Cy-3-conjugated goat anti-rabbit (1:300; cat. no. GB21303; both from Wuhan Servicebio Technology Co., Ltd.) for 1 h at room temperature. The cell nuclei were stained with DAPI. Nikon Eclipse C1 fluorescence microscope with an integrated camera was used to visualize the fluorescence and acquire images from five representative fields of each sections (×400).
RNA sequencing (RNA-seq)
After transiently transfecting HeLa cells with si-FAM83A or si-NS for 48 h, total RNA of each group was extracted and subjected to poly(A) selection using Oligo(dT) beads (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RNA libraries were sequenced on the Illumina HiSeq/MiSeq platform at Novogene Co., Ltd. RNA-seq reads in fasta format were mapped to the human genome GRCh38 using TopHat (version 2.0.13) (18). DESeq was used for differential expression (DE) analysis (19). In DE tests, a gene was considered significantly differentially expressed when the adjusted P<0.05. Integrative Genomics Viewer (20) was used to visualize the expression of FAM83A in each group.
Western blot analysis
At 72 h post-transfection, HeLa and CaSki cells were lysed in RIPA buffer in presence of protease and phosphatase inhibitors (ThermoFisher Scientific, Inc.). The protein concentration was quantified using a BCA protein assay kit (Thermo Fisher Scientific, Inc.). Western blotting was performed following a standard procedure (21). The membranes were blocked with 5% skimmed milk for 1 h at room temperature and incubated with the primary antibodies overnight at 4°C, washed with TBS-Tween-20 and incubated with the secondary antibodies for 1 h at room temperature. The following antibodies were used: FAM83A (1:1,000; cat. no. SAB1103067; Sigma-Aldrich; Merck KGaA), integrin α1 (1:500; ProteinTech Group, Inc.), integrin α2 (1:1,000; cat. no. ET1611-57; Hangzhou HuaAn Biotechnology Co., Ltd.), integrin α3 (1:1,000; ProteinTech Group, Inc.), integrin α5 (1:1,000; Hangzhou HuaAn Biotechnology Co., Ltd.), integrin β4 (1:1,000; Hangzhou HuaAn Biotechnology Co., Ltd.), integrin β5 (1:1,000; BIOSS), phosphorylated (p)-PI3K (1:1,000; cat. no. 4228; Cell Signaling Technology, Inc.), p-AKT (1:1,000; cat. no. 4060; Cell Signaling Technology, Inc.), GAPDH (1:4,000; cat. no. sc-47724; Santa Cruz Biotechnology, Inc.), horseradish peroxidase (HRP)-conjugated anti-mouse IgG (1:1,000; Cell Signaling Technology, Inc.; cat. no. 7076) and anti-rabbit IgG (1:1,000; cat. no. 7074; Cell Signaling Technology, Inc.). Blots were visualized with SuperSignal West Pico Chemiluminescent Substrate (Pierce; Thermo Fisher Scientific, Inc.) and analyzed on a LAS-4000 Imaging System (Fuji Photo Film, Inc.). Band intensities were determined by Quantity One software v4.6.6 (Bio-Rad Laboratories, Inc.).
Statistical analysis
The data obtained using tissue samples are presented as the mean ± SEM; all other data are presented as the mean ± SD. The experiments were performed ≥3 times unless stated otherwise. Analyses were performed using GraphPad Prism 6.0 software (GraphPad Software, Inc.). Testing involving two groups in cervical tissues were performed using the nonparametric Mann-Whitney U test. Association between FAM83A expression and patient clinicopathological parameters were analyzed by the χ2 test and Fisher's exact probability test. Kaplan-Meier curves and the log-rank test were used to examine overall survival. For experiments in cell lines and xenograft models, two-tailed Student's t-test was used. P<0.05 was considered to indicate a statistically significant difference.
Results
FAM83A is upregulated in human cervical cancer tissues, but low expression is associated with a poor prognosis
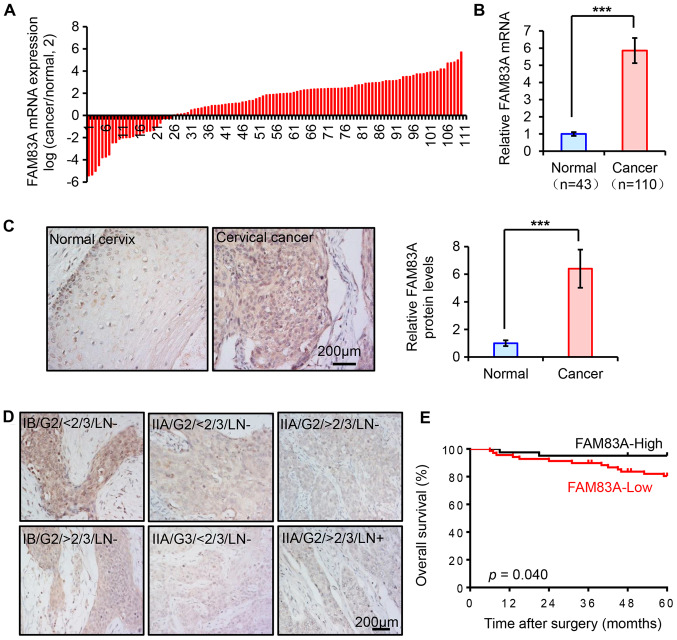
In our previous study, FAM83A mRNA was demonstrated to be upregulated in cervical cancer by RNA sequencing (5). In the present study, the expression pattern of FAM83A was assessed in another cohort of human cervical tissue samples from the Women's Hospital of Zhejiang University. The mRNA expression levels of FAM83A were quantified using RT-qPCR in 153 cervical specimens, including 43 healthy and 110 cancerous cervical tissues. Compared with the healthy cervix, FAM83A mRNA expression was significantly increased in cancer tissues (Fig. 1A and B).
Figure 1.
Expression of FAM83A in human cervical tissues. (A and B) Reverse transcription-quantitative PCR analysis of FAM83A mRNA levels in 43 normal cervixes and 110 cervical cancer tissues. GAPDH was used as the internal control. Data are presented as (A) log2 (cancer/normal) for each cervical cancer sample and (B) mean (2−ΔΔCT) ± SEM in the normal and cancer groups, respectively. (A) The numbers on the x axis indicate the patient number. (C and D) Representative immunohistochemical staining of FAM83A in normal cervixes and cervical cancer tissues with known clinicopathological parameters. (E) Overall survival curves of patients with cervical cancer with high or low FAM83A expression. ***P<0.001. FAM83A, family with sequence similarity 83 member A; IB, FIGO stage IB; IIA, FIGO stage IIA; G3, pathological grade 3; LN+, lymph node metastasis; >2/3, deep stromal invasion.
FAM83A IHC staining was performed in the aforementioned human cervical samples to determine whether the protein expression exhibited similar trends. The expression of FAM83A protein was weak in the healthy cervical epithelia, but significantly higher in cervical cancer tissues (Fig. 1C). As FAM83A is considered to be an oncogene, it was hypothesized that its upregulation may be associated with poor outcomes in patients with cervical cancer. However, in the present study, the upregulation of FAM83A protein expression was more likely to occur in patients with favorable clinical parameters, as the results demonstrated that FAM83A protein expression was inversely associated with advanced FIGO stage, deep stromal invasion, high pathological grade and lymph node metastasis, but not with other clinicopathological parameters including age, parametrium, margins, lymphovascular space invasion, tumor size or the presence of squamous cell carcinoma antigen in patients with cervical cancer (Fig. 1D; Table I). As demonstrated in Fig. 1D, the FAM83A protein was located both in the cytoplasm and the nucleus in cervical tissues, and differences in the localization were observed among groups with different FIGO stage, stomal invasion and lymph node metastasis . The intensity of FAM83A protein staining was similar in the cell nucleus and the cytoplasm of healthy cervical epithelium; in cervical cancer samples, high FAM83A expression was located both in nucleus and cytoplasm, but the localization was mainly in cytoplasm in patients with low FAM83A expression. Kaplan-Meier survival analysis results indicated that patients with low FAM83A protein expression exhibited a shorter overall survival time (Fig. 1E). Thus, FAM83A expression was frequently upregulated in human cervical cancer types, but negatively associated with poor prognostic parameters, suggesting the possibility that FAM83A may serve a suppressive role in cervical cancer.
Table I.
Association between FAM83A protein expression and clinicopathologic parameters of patients with cervical cancer.
| Characteristic | Total (n=110) | FAM83A protein expression
|
P-value | |
|---|---|---|---|---|
| Low | High | |||
| Age, years | 0.369 | |||
| ≤45 | 61 | 36 | 25 | |
| >45 | 49 | 33 | 16 | |
| Pelvic nodes | 0.003a | |||
| Negative | 92 | 52 | 40 | |
| Positive | 18 | 17 | 1 | |
| Parametrium | 1.000a | |||
| Negative | 99 | 62 | 37 | |
| Positive | 11 | 7 | 4 | |
| Margins | 1.000a | |||
| Negative | 107 | 67 | 40 | |
| Positive | 3 | 2 | 1 | |
| FIGO stageb | 0.046 | |||
| IA/IB | 76 | 43 | 33 | |
| IIA | 34 | 26 | 8 | |
| Pathological grade | 0.046a | |||
| 1/2 | 95 | 56 | 39 | |
| 3 | 15 | 13 | 2 | |
| Lymphovascular space invasion | 0.741 | |||
| Negative | 73 | 45 | 28 | |
| Positive | 37 | 24 | 13 | |
| Deep stromal invasion | 0.035 | |||
| <2/3 | 58 | 31 | 27 | |
| ≥2/3 | 52 | 38 | 14 | |
| Tumor size, cm | 0.554 | |||
| ≤4 | 99 | 63 | 36 | |
| >4 | 11 | 6 | 5 | |
| SCC-Ag, ng/ml | 0.245a | |||
| <4 | 96 | 58 | 38 | |
| ≥4 | 14 | 11 | 3 | |
P<0.05, Fisher's exact test;
FIGO 2014 (47). FIGO,
International Federation of Gynecology and Obstetrics; SCC-Ag, squamous cell carcinoma antigen; FAM83A, family with sequence similarity 83 member A.
Knockdown of FAM83A promotes aggressive phenotypes in cervical cancer cells
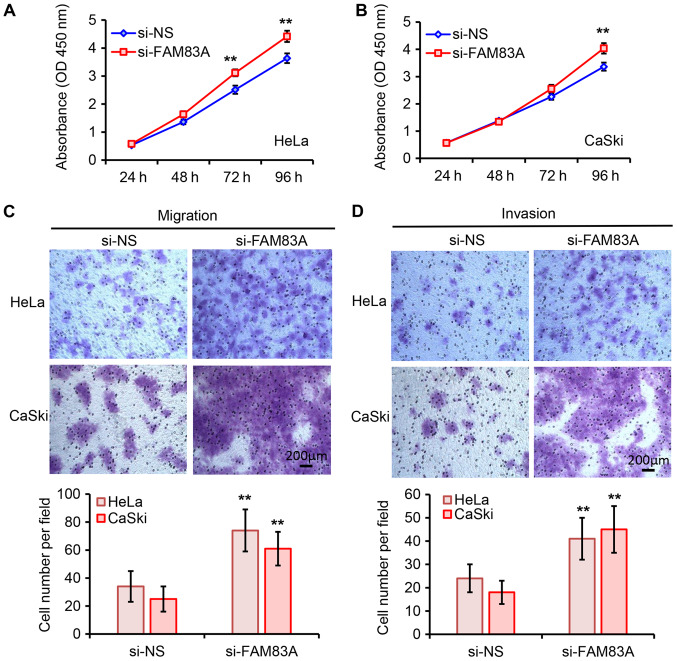
To further elucidate the biological roles of FAM83A in cervical cancer cells, FAM83A expression in HeLa and CaSki cells was silenced by transfection with si-FAM83A (Fig. S1). As demonstrated in Fig. 2A and B, knockdown of FAM83A significantly promoted HeLa and CaSki cell proliferation compared with that in the negative controls groups. The tumor cell migratory and invasive abilities were assessed using Transwell assays. Knockdown of FAM83A significantly increased the migratory and invasive abilities of HeLa and CaSki cells (Fig. 2C and D). Collectively, these results suggested that knockdown of FAM83A promoted cervical cancer cell proliferation and invasion in vitro.
Figure 2.
Effects of FAM83A knockdown on cervical cancer cell proliferation, migration and invasion. (A and B) The effects of FAM83A knockdown on (A) HeLa and (B) CaSki cell proliferation were analyzed by Cell Counting Kit-8 assay at the indicated timepoints. (C and D) The effects of FAM83A knockdown on HeLa and CaSki cell (C) migration and (D) invasion were measured, and the representative images of migrated and invaded cells are presented. Magnification, ×100. **P<0.01 vs. si-NS. FAM83A, family with sequence similarity 83 member A; si, small interfering RNA; si-NS, scrambled control; OD, optical density.
Knockdown of FAM83A promotes tumor growth and invasion in vivo
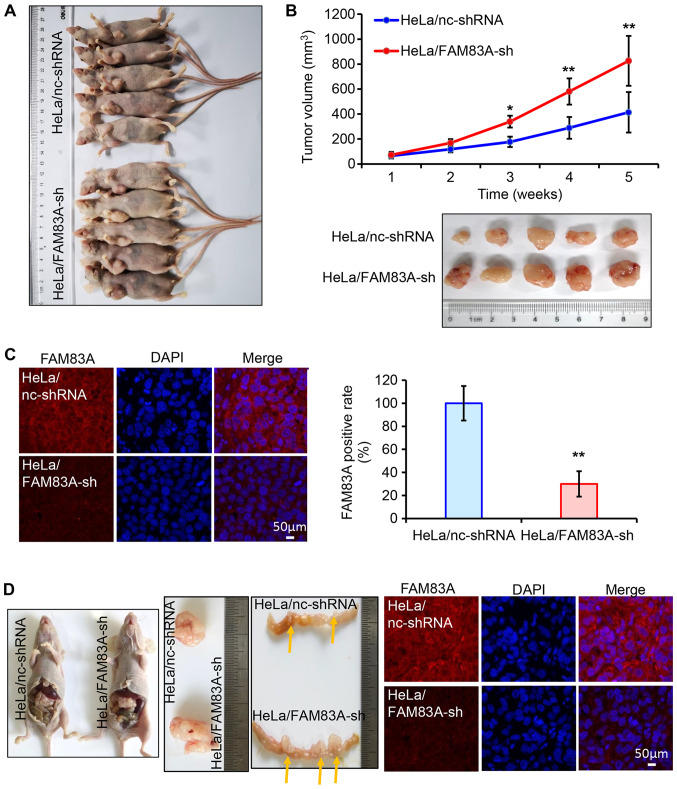
Subsequently, the role of FAM83A knockdown in cervical tumor growth and invasion was evaluated in vivo. BALB/c nude mice (n=10) were randomly injected subcutaneously with HeLa cells stably expressing either control or FAM83A shRNA (each n=5). Of note, HeLa/FAM83A-sh tumors exhibited more progressive growth compared with that of HeLa/nc-shRNA tumors, as the volumes of the tumors generated from HeLa/FAM83A-sh cells were higher compared with those of tumors from HeLa/nc-shRNA cells (Fig. 3A and B). Immunofluorescence staining in tumors dissected from the mice identified that tumors transfected with FAM83A shRNA exhibited lower FAM83A protein expression compared with those from the control group (Fig. 3C).
Figure 3.
FAM83A knockdown promotes tumor growth and invasion in nude mice. (A-C) HeLa/FAM83A-sh and HeLa/nc-shRNA cells were subcutaneously injected into nude mice (n=10). (A) Xenograft tumor models demonstrated that tumors originating from HeLa/FAM83A-sh cells were larger compared with those originating from the HeLa/nc-shRNA cells. (B) Tumor growth curves of each group and images of tumors from nude mice. (C) Immunofluorescence analysis of FAM83A protein expression (red) in xenograft tumors expressing control or FAM83A shRNA. Nuclear DNA was stained with DAPI (blue). (D) Xenograft tumor models were induced by intraperitoneal injection of HeLa/FAM83A-sh or HeLa/nc-shRNA cells. Local tumors from the abdominal cavity and colon invasive tumors in each group are presented. Yellow arrow indicates the tumor position. Immunofluorescence analysis was used to determine the expression of FAM83A protein expression (red) in the local tumors. *P<0.05, **P<0.01 vs. HeLa/nc-shRNA. sh/shRNA, short hairpin RNA; FAM83A, family with sequence similarity 83 member A.
The invasive potential of FAM83A was determined in vivo. HeLa/FAM83A-sh cells and HeLa/nc-shRNA cells were intraperitoneally transplanted into nude mice (n=2). The volume of the abdominal HeLa/FAM83A-sh tumor was larger compared with that of the HeLa/nc-shRNA tumor (Fig. 3D). In addition, HeLa/FAM83A-sh cells significantly increased the formation of tumor nodules in the colon compared with that in the HeLa/nc-shRNA group (Fig. 3D). The expression of FAM83A in these tumors was also confirmed by immunofluorescence. These results demonstrated that knockdown of FAM83A potentiated cervical tumor growth and invasion.
RNA-seq demonstrates that FAM83A knockdown alters key oncogenic signaling pathways
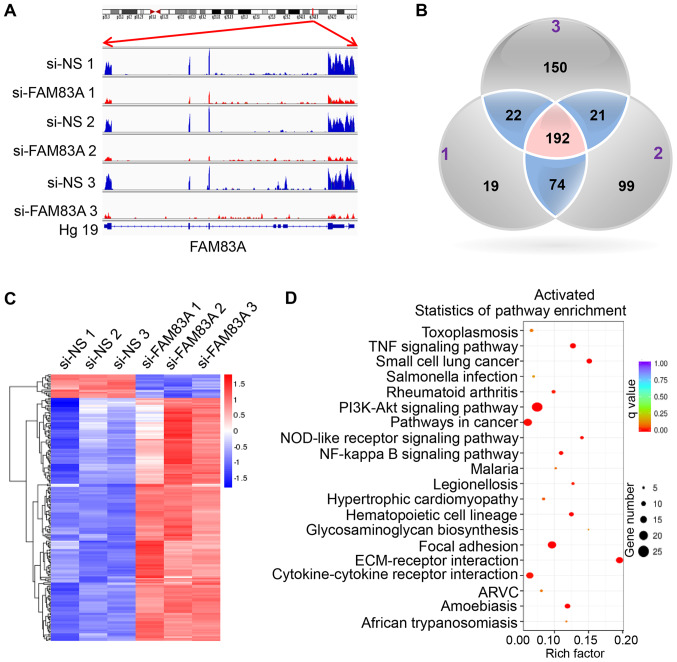
To identify the underlying mechanisms behind the observed effects of FAM83A, the present study investigated the signaling pathways regulated by FAM83A. RNA-seq was conducted in HeLa cells transiently transfected with si-FAM83A and the negative control groups (Table SII). The expression of FAM83A was significantly inhibited in HeLa cells transfected with FAM83A siRNA (Fig. 4A).
Figure 4.
Downstream transcriptional targets of FAM83A in HeLa cells. RNA sequencing analysis was performed on HeLa cells transfected with si-NS or si-FAM83A. (A) Read coverage maps demonstrating the RNA expression levels of FAM83A in the si-FAM83A and si-NS groups. Samples si-NS 1-3 were three independent repeats of si-NS-treated HeLa cells; samples si-FAM83A 1-3 were three independent repeats of HeLa cells transfected with si-FAM83A. Distribution of FAM83A reads was visualized along with the FAM83A gene transcript using the Integrative Genomics Viewer. (B) Venn diagram depicting the overlapping genes differentially expressed following FAM83A knockdown in HeLa cells. The numbers (1-3) in the Venn diagram indicate the three independent replicates. (C) A heatmap of the 192 differentially expressed genes from the RNA-seq experiment following FAM83A knockdown. Red represents upregulation; blue represents downregulation. (D) The top 20 most significant Kyoto Encyclopedia of Genes and Genomes pathways activated in response to FAM83A inhibition. FAM83A, family with sequence similarity 83 member A; si, small interfering RNA; si-NS, scrambled control; Hg19, human genome version 19.
In total, 192 differentially expressed genes were identified in si-FAM83A-transfected cells compared with the control groups, with 175 upregulated and 17 downregulated genes (Fig. 4B and C; Table SIII). KEGG pathway analysis of the 192 differentially expressed genes identified a significant activation of key oncogenesis-associated pathways following FAM83A knockdown, including the 'PI3K/AKT signaling pathway', 'TNF signaling pathway', 'focal adhesion' and 'ECM-receptor interaction' (Fig. 4D), while the 'proteoglycans in cancer' and 'microRNAs in cancer' pathways were suppressed (Fig. S2). To confirm the KEGG pathway analysis results, the activated PI3K/AKT pathways was validated in vitro. The results demonstrated that knockdown of FAM83A significantly increased the expression of p-PI3K and p-AKT proteins in HeLa and CaSki cells (Fig. S3).
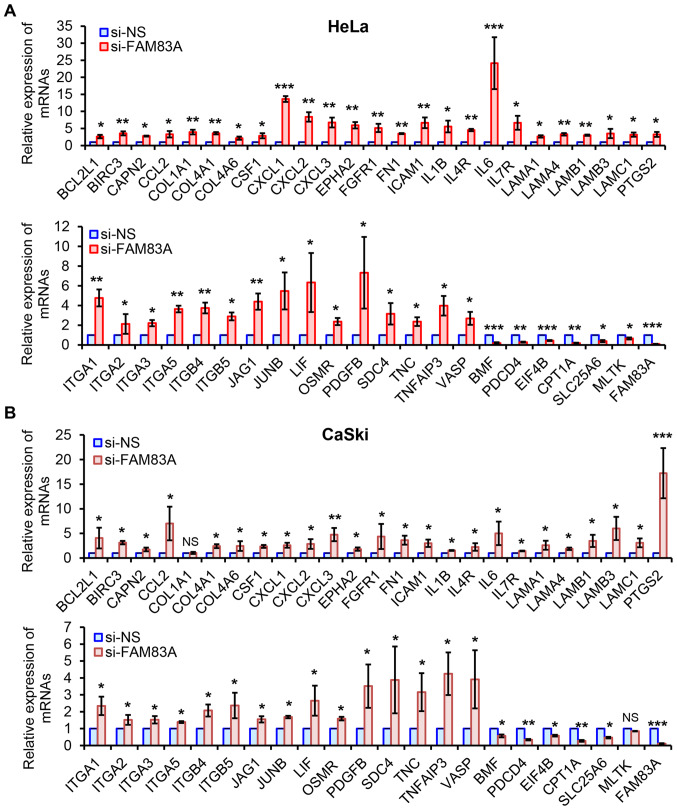
A total of 46 candidate genes, including 40 genes from the top four activated pathways and six from the main suppressed pathways in both HeLa and CaSki cells, were analyzed by RT-qPCR. In line with the RNA-seq data, all 46 genes were similarly regulated following FAM83A inhibition in HeLa cells (Fig. 5A). Although each expression pattern of the 46 genes was different compared with those in HeLa cells, similar expression patterns of 44 candidates were observed in CaSki cells following FAM83A knockdown, with the exception of collagen type 1 α1 and mitogen-activated protein kinase kinase kinase 20 (Fig. 5B). Therefore, FAM83A inhibition may activate multiple downstream genes mainly involved in oncogenesis-related pathways.
Figure 5.
Experimental validation of RNA-seq data in HeLa and CaSki cells. (A and B) Quantitative PCR validation of selected 46 genes and FAM83A in (A) HeLa and (B) CaSki cells with or without FAM83A knockdown. The relative mRNA levels of individual gene transcripts are presented as fold-change of FAM83A knockdown group compared with the si-NS group. The experiments were performed in triplicate. NS, not significant; *P<0.05, **P<0.01, ***P<0.001 vs. si-NS. FAM83A, family with sequence similarity 83 member A; si, small interfering RNA; si-NS, scrambled control.
FAM83A knockdown activates integrins in vitro and in vivo
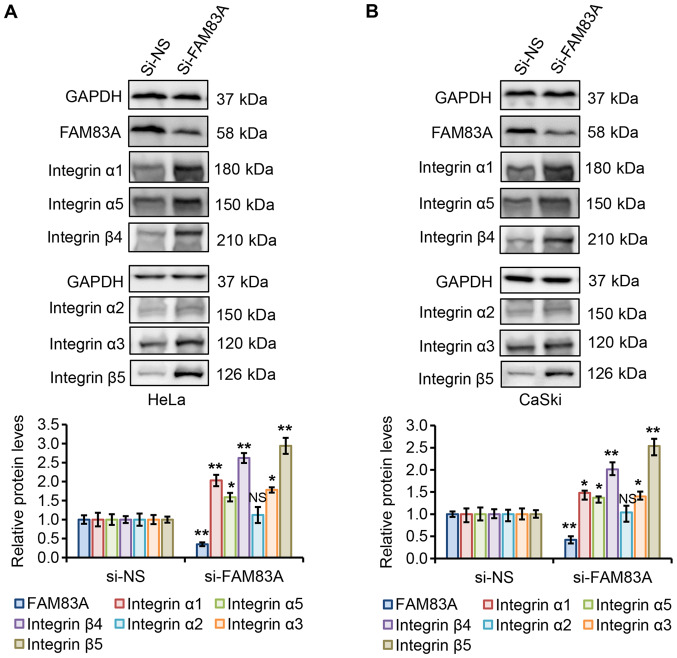
The main cell adhesion receptors, known as integrins, are essential for migration and invasion and have been implicated in almost every step of cancer progression, from primary tumor to metastasis (22,23). Dysregulated integrin-mediated adhesion to the extracellular matrix (ECM) and signaling is a precursor in the majority of invasive tumors (24-26). To validate these candidate integrins, additional in vitro and in vivo experiments were performed. The effects of FAM83A knockdown on the protein expression levels of integrins α1, α2, α3, α5, β4 and β5 in HeLa and CaSki cells were determined. As presented in Fig. 6A and B, FAM83A knockdown significantly increased the integrin α1, α3, α5, β4 and β5 protein expression levels in HeLa and CaSki cells compared with that in the corresponding si-NS groups, whereas integrin α2 protein expression was not significantly affected.
Figure 6.
FAM83A knockdown activates integrins α1, α3, α5, β4 and β5 in cervical cancer cells. (A and B) Western blot analysis of FAM83A and integrins α1, α2, α3, α5, β4 and β5 in (A) HeLa and (B) CaSki cells with or without FAM83A knockdown. GAPDH protein level was used as the loading control. NS, not significant; *P<0.05, **P<0.01 vs. si-NS. FAM83A, family with sequence similarity 83 member A; si, small interfering RNA; si-NS, scrambled control.
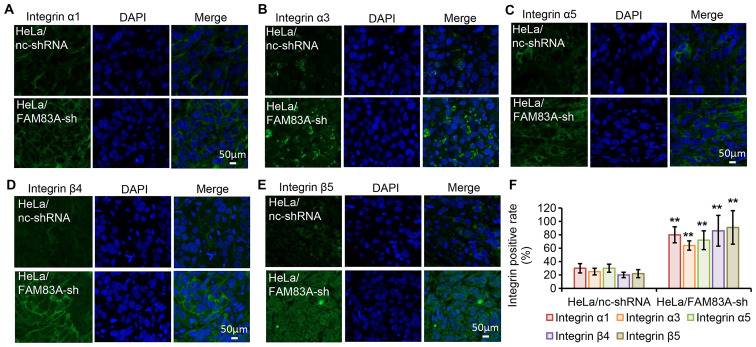
To evaluate the effect in vivo, the protein expression levels of integrins α1, α3, α5, β4 and β5 were investigated in tumor xenografts harvested from nude mice by immunofluorescence staining (Fig. 7A-F). Consistent with the in vitro findings, tumors expressing FAM83A shRNA exhibited increased protein expression levels of the five tested integrins. These results further suggest that integrins α1, α3, α5, β4 and β5 may be involved in the tumor-promoting effects of FAM83A knockdown.
Figure 7.
FAM83A knockdown activates integrins α1, α3, α5, β4 and β5 in mouse xenograft models. (A-F) Immunofluorescence analysis of integrin α1, α3, α5, β4 and β5 protein expression (green) in HeLa xenograft tumors generated using cells transfected with nc-shRNA or FAM83A-sh. Nuclear DNA was stained with DAPI (blue). **P<0.01 vs. HeLa/nc-shRNA. sh/shRNA, short hairpin RNA; FAM83A, family with sequence similarity 83 member A; nc, negative control.
Clinical association among FAM83A and integrins α1, α3, α5, β4 and β5 in human cervical cancer tissues
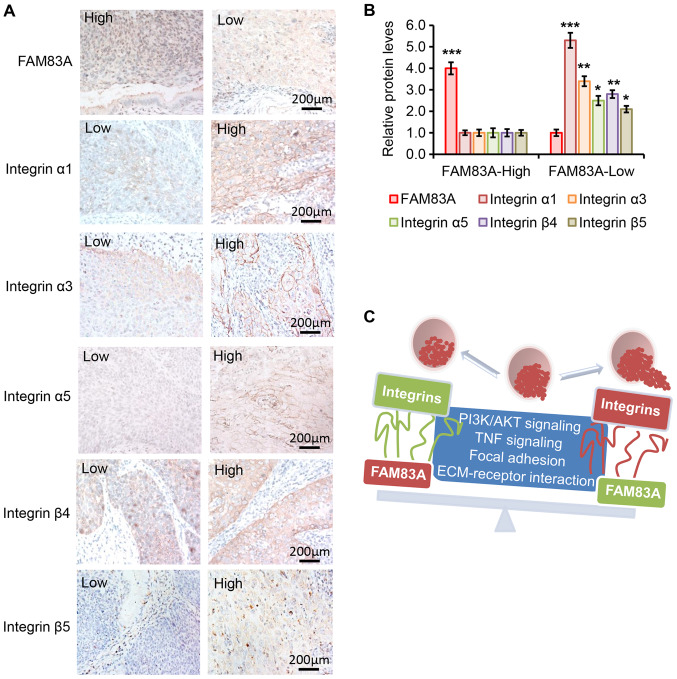
The clinical relevance of the associations among FAM83A and integrins α1, α3, α5, β4 and β5 were determined in clinical samples from 20 patients with cervical cancer (10 with high and 10 with low FAM83A expression) by IHC. As presented in Fig. 8A and B, the protein expression levels of integrins α1, α3, α5, β4 and β5 were negatively associated with FAM83A protein expression level. For example, the protein expression of integrin α1 was higher in the cervical cancer tissue samples with low FAM83A expression compared with that in samples with high FAM83A expression, whereas the expression of integrin α1 was lower in cancer tissues with high FAM83A expression. In addition, the protein expression levels of integrin α3, α5, β4 and β5 were higher in cervical cancer samples with low FAM83A expression compared with those in tissues with high FAM83A expression. Collectively, the results suggested an association between FAM83A and certain integrin subtypes in human cervical cancer samples.
Figure 8.
Protein levels of FAM83A and integrins α1, α3, α5, β4 and β5 in human cervical cancer specimens. (A) Representative images of the staining of FAM83A and integrins α1, α3, α5, β4 and β5 in tissues from patients with cervical cancer in the high and low FAM83A expression groups. 'High' and 'low' in each image indicate each protein level separately. (B) Relative protein expressions of FAM83A and integrins in cervical cancer tissues from patients in the low and high FAM83A expression groups determined by immunohistochemistry analysis (n=10 patients per group). (C) Schematic diagram demonstrating that upregulation of FAM83A inhibits the PI3K/AKT and TNF signaling, focal adhesion and ECM-receptor interaction oncogenic pathways, followed by inhibition of certain integrins to attenuate cervical cancer progression. FAM83A, family with sequence similarity 83 member A; TNF, tumor necrosis factor; ECM, extracellular matrix.
Discussion
FAM83A belongs to a recently discovered oncogenic FAM83 family and has been demonstrated to be significantly upregulated in a variety of types of cancer, such as breast and lung cancer (6,10,12,14,27,28). Thus, FAM83A is presumed to be a novel oncogene. However, the results of the present study suggested that FAM83A upregulation may serve a tumor-suppressing role in cervical cancer in vitro and in vivo, at least in part by suppressing integrins. It was speculated that the contrasting results identified in different types of cancer may indicate the context dependency of FAM83A functions.
The present clinical observations in human cervical samples have suggested complex roles for FAM83A. FAM83A mRNA expression was profiled across all human tumor and paired healthy tissues from TCGA and GTEx projects using GEPIA (29). As presented in Fig. S4, the mRNA expression of FMA83A was upregulated in bladder urothelial carcinoma, cervical squamous cell carcinoma, endocervical adenocarcinoma, head and neck squamous cell carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, ovarian serous cystadenocarcinoma and pancreatic adenocarcinoma, but suppressed in esophageal carcinoma, acute myeloid leukemia and skin cutaneous melanoma. The mRNA expression of FAM83A was not altered in other types of tumors, indicating the tissue specificity of FAM83A expression. The present results demonstrated that FAM83A expression was upregulated in human cervical cancer compared with the normal cervix. Chen et al (11) have reported that elevated FAM83A expression was attributed to the genomic amplification of chromosome 8q24.13 and c-MYC gene amplification. FAM83A is located near the HPV integration hot spots (30,31), which may affect the expression of FAM83A. In the present study, FAM83A upregulation was identified to be negatively associated with advanced FIGO stage, deep stromal invasion, poor differentiation, lymph node metastasis and poor prognosis. Although FAM83A was upregulated in cervical cancer tissues, it is likely that FAM83A serves tumor-suppressive role. The results of the present study suggested that FAM83A may be used as a potential biomarker for cervical cancer progression.
As FAM83A overexpression could drive HMEC transformation and confer resistance to EGFR-TKIs, and knockdown of FAM83A suppressed growth and tumorigenicity in breast cancer (6,14,27), it was initially hypothesized that FAM83A may also serve similar functions in cervical cancer. However, in the present study, knockdown of FAM83A promoted cell proliferation, migration, invasion and tumorigenicity in vitro and in vivo. By examining how FAM83A knockdown promoted tumor growth and invasion, the present study identified 192 altered genes that associated FAM83A with four key signaling pathways using RNA-seq. It was demonstrated that FAM83A knockdown resulted in the activation of the PI3K/AKT and TNF signaling pathways, focal adhesion and ECM-receptor interaction. Opposite roles were reported by Lee et al (6), who demonstrated that FAM83A activated the PI3K/AKT pathways in breast cancer. Although the results of the present study were different from the reported data, both studies observed that FAM83A exerted its roles via the same pathway. The differences in the results may have occurred due to the different function of FAM83A in the different types of cancer. To assess the present findings, 46 genes from the top pathways in HeLa and CaSki cells were further validated, and it was demonstrated that FAM83A knockdown upregulated the expression of these pathway-related genes compared with that in the si-NS-transfected cells. It is possible that FAM83A, despite being generally considered to be an oncogene, may exert tumor-suppressive activities in cervical cancer. Similar results have been observed with other genes; for instance, polo-like kinase 1 (Plk1) has been considered to be a classical oncogene in a number of tumor types (32). However, de Cárcer et al (33) revealed that Plk1 overexpression exerted a tumor suppressor role, and the increased levels of Plk1 expression was associated with improved prognosis in patients with breast cancer. In addition, similar observations have been made for viral E7-induced expression of tumor-suppressive p16Ink4a (34). High-level of p16Ink4a expression is the clinical biomarker for high-risk HPV-associated lesions and cancers (35). Thus, one reasonable assumption may be that the HPV viral oncoproteins may also contribute to the altered roles of FMA83A in cervical cancer.
The results of the present study also suggested that FAM83A knockdown upregulated a set of integrin subunits. Integrins are members of a diverse family of glycoproteins that form heterodimeric receptors for ECM molecules (36). In total, 24 transmembrane heterodimers are generated from a combination of its 18 α and eight β subunits (36). Integrins are essential for cell migration, invasion and tumor metastasis, not only because they directly alter adhesion to the ECM, but also as they respond to intracellular cues that control pro-survival signaling pathways (37). Aberrant integrin expression is frequently detected in various types of cancer (24-26). For example, most β1 integrins are necessary for mammary tumorigenesis (38,39). In addition, integrin α5β1 upregulation contributes to poor tumor prognosis and progression in lung cancer (40). Integrin αvβ3 is significantly upregulated at the invasive front of melanoma cells and angiogenic blood vessels (41). The laminin-binding integrin α6β4 regulates the mesenchymal-to-epithelial transition oncogenic signaling, activates PI3K and correlates with the progression to invasive carcinoma, such as breast carcinoma, thyroid cancer and gastric carcinoma (42-45). The present study demonstrated that FAM83A knockdown upregulated integrins α1, α3, α5, β4 and β5 in cervical cancer cells and animal models. Thus, the results of the present study provided evidence that cervical cancer samples with low FAM83A expression exhibited high protein expression of integrins α1, α3, α5, β4 and β5. In addition, it was indicated that FAM83A acted as a tumor suppressor and exerted its suppressive role at least partially via the inhibition of integrins α1, α3, α5, β4 and β5. In addition, laminins and collagens have been reported to be closely associated with integrins to promote tumor progression (40,46). The results of the present study demonstrated FAM83A knockdown also significantly increased the expression of several members of the laminin and collagen family, indicating a potential association among integrins, collagens and laminins.
In conclusion, the present study identified novel potential roles of FAM83A in cervical cancer. Understanding the exact role of FAM83A in cervical cancer and the molecular mechanisms by which FAM83A knockdown activates integrins may offer new insight into the biological basis of cervical cancer and lay the groundwork for future studies.
Supplementary Data
Acknowledgments
Not applicable.
Funding
This study was supported by the National Natural Scientific Foundation of China (grant no. 81702552 to JX), the National Key Research and Development Program of China (grant no. 2016YFC1302900 to WL) and the Fundamental Research Funds for the Central Universities (grant no. 2019QNA7035 to JX).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JX and WL conceived the project, designed the study, developed the methodology and analyzed the data. JX performed experiments. JX wrote the manuscript. WL revised the manuscript and supervised the study. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
Ethics approval was obtained from the Ethics Committee of Women's Hospital, Zhejiang University School of Medicine (approval no. IRB-20190070). Written informed consent was obtained from each patient. The animal study was approved by the Animal Care and Use Committee of Zhejiang University (approval no. ZJU20170031).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Muñoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 3.Hutchinson DJ, Klein KC. Human papillomavirus disease and vaccines. Am J Health Syst Pharm. 2008;65:2105–2112. doi: 10.2146/ajhp070627. [DOI] [PubMed] [Google Scholar]
- 4.Markowitz LE, Dunne EF, Saraiya M, Lawson HW, Chesson H, Unger ER. Centers for Disease Control and Prevention (CDC); Advisory Committee on Immunization Practices (ACIP): Quadrivalent human papillomavirus vaccine: Recommendations of the advisory committee on immunization practices (ACIP) MMWR Recomm Rep. 2007;56:1–24. [PubMed] [Google Scholar]
- 5.Xu J, Liu H, Yang Y, Wang X, Liu P, Li Y, Meyers C, Banerjee NS, Wang HK, Cam M, et al. Genome-wide profiling of cervical RNA-binding proteins identifies human papillomavirus regulation of RNASEH2A expression by viral E7 and E2F1. MBio. 2019;10 doi: 10.1128/mBio.02687-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SY, Meier R, Furuta S, Lenburg ME, Kenny PA, Xu R, Bissell MJ. FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice. J Clin Invest. 2012;122:3211–3220. doi: 10.1172/JCI60498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fulcher LJ, Bozatzi P, Tachie-Menson T, Wu KZL, Cummins TD, Bufton JC, Pinkas DM, Dunbar K, Shrestha S, Wood NT, et al. The DUF1669 domain of FAM83 family proteins anchor casein kinase 1 isoforms. Sci Signal. 2018;11:eaao2341. doi: 10.1126/scisignal.aao2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Dong X, Yin Y, Su Y, Xu Q, Zhang Y, Pang X, Zhang Y, Chen W. BJ-TSA-9, a novel human tumor-specific gene, has potential as a biomarker of lung cancer. Neoplasia. 2005;7:1073–1080. doi: 10.1593/neo.05406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel CA, Jackson MW. HER2-positive breast cancer cells expressing elevated FAM83A are sensitive to FAM83A loss. PLoS One. 2017;12:e0176778. doi: 10.1371/journal.pone.0176778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant S. FAM83A and FAM83B: Candidate oncogenes and TKI resistance mediators. J Clin Invest. 2012;122:3048–3051. doi: 10.1172/JCI64412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S, Huang J, Liu Z, Liang Q, Zhang N, Jin Y. FAM83A is amplified and promotes cancer stem cell-like traits and chemore-sistance in pancreatic cancer. Oncogenesis. 2017;6:e300. doi: 10.1038/oncsis.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snijders AM, Lee SY, Hang B, Hao W, Bissell MJ, Mao JH. FAM83 family oncogenes are broadly involved in human cancers: An integrative multi-omics approach. Mol Oncol. 2017;11:167–179. doi: 10.1002/1878-0261.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Xiao X, Ji X, Liu B, Amos CI. RNA-seq analysis of lung adenocarcinomas reveals different gene expression profiles between smoking and nonsmoking patients. Tumour Biol. 2015;36:8993–9003. doi: 10.1007/s13277-015-3576-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cipriano R, Miskimen KL, Bryson BL, Foy CR, Bartel CA, Jackson MW. Conserved oncogenic behavior of the FAM83 family regulates MAPK signaling in human cancer. Mol Cancer Res. 2014;12:1156–1165. doi: 10.1158/1541-7786.MCR-13-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pawitan Y, Bjöhle J, Amler L, Borg AL, Egyhazi S, Hall P, Han X, Holmberg L, Huang F, Klaar S, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: Derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Q, Wang X, Yu Z, Wu X, Chen X, Li J, Zhu Z, Liu B, Su L. Transducin (β)-like 1 X-linked receptor 1 promotes gastric cancer progression via the ERK1/2 pathway. Oncogene. 2017;36:1873–1886. doi: 10.1038/onc.2016.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang Y, Yu H, Liang X, Xu J, Cai X. Chk1-induced CCNB1 overexpression promotes cell proliferation and tumor growth in human colorectal cancer. Cancer Biol Ther. 2014;15:1268–1279. doi: 10.4161/cbt.29691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winograd-Katz SE, Fässler R, Geiger B, Legate KR. The integrin adhesome: From genes and proteins to human disease. Nat Rev Mol Cell Biol. 2014;15:273–288. doi: 10.1038/nrm3769. [DOI] [PubMed] [Google Scholar]
- 23.Hamidi H, Ivaska J. Every step of the way: Integrins in cancer progression and metastasis. Nat Rev Cancer. 2018;18:533–548. doi: 10.1038/s41568-018-0038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seguin L, Desgrosellier JS, Weis SM, Cheresh DA. Integrins and cancer: Regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol. 2015;25:234–240. doi: 10.1016/j.tcb.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raab-Westphal S, Marshall JF, Goodman SL. Integrins as therapeutic targets: Successes and cancers. Cancers (Basel) 2017;9:110. doi: 10.3390/cancers9090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamidi H, Pietilä M, Ivaska J. The complexity of integrins in cancer and new scopes for therapeutic targeting. Br J Cancer. 2016;115:1017–1023. doi: 10.1038/bjc.2016.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cipriano R, Graham J, Miskimen KL, Bryson BL, Bruntz RC, Scott SA, Brown HA, Stark GR, Jackson MW. FAM83B mediates EGFR- and RAS-driven oncogenic transformation. J Clin Invest. 2012;122:3197–3210. doi: 10.1172/JCI60517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartel CA, Parameswaran N, Cipriano R, Jackson MW. FAM83 proteins: Fostering new interactions to drive oncogenic signaling and therapeutic resistance. Oncotarget. 2016;7:52597–52612. doi: 10.18632/oncotarget.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, Cherniack AD, Ambrogio L, Cibulskis K, Bertelsen B, et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506:371–375. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Z, Zhu D, Wang W, Li W, Jia W, Zeng X, Ding W, Yu L, Wang X, Wang L, et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat Genet. 2015;47:158–163. doi: 10.1038/ng.3178. [DOI] [PubMed] [Google Scholar]
- 32.Ito Y, Miyoshi E, Sasaki N, Kakudo K, Yoshida H, Tomoda C, Uruno T, Takamura Y, Miya A, Kobayashi K, et al. Polo-like kinase 1 overexpression is an early event in the progression of papillary carcinoma. Br J Cancer. 2004;90:414–418. doi: 10.1038/sj.bjc.6601540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Cárcer G, Venkateswaran SV, Salgueiro L, El Bakkali A, Somogyi K, Rowald K, Montañés P, Sanclemente M, Escobar B, de Martino A, et al. Plk1 overexpression induces chromosomal instability and suppresses tumor development. Nat Commun. 2018;9:3012. doi: 10.1038/s41467-018-05429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munger K, Gwin TK, McLaughlin-Drubin ME. p16 in HPV-associated cancers. Oncotarget. 2013;4:1864–1865. doi: 10.18632/oncotarget.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malhone C, Longatto-Filho A. Cervical, ovarian and endometrial tumor markers: Potential clinical value. Semin Ultrasound CT MR. 2019;40:350–357. doi: 10.1053/j.sult.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 36.van der Flier A, Sonnenberg A. Function and interactions of integrins. Cell Tissue Res. 2001;305:285–298. doi: 10.1007/s004410100417. [DOI] [PubMed] [Google Scholar]
- 37.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 38.Cagnet S, Faraldo MM, Kreft M, Sonnenberg A, Raymond K, Glukhova MA. Signaling events mediated by α3β1 integrin are essential for mammary tumorigenesis. Oncogene. 2014;33:4286–4295. doi: 10.1038/onc.2013.391. [DOI] [PubMed] [Google Scholar]
- 39.White DE, Kurpios NA, Zuo D, Hassell JA, Blaess S, Mueller U, Muller WJ. Targeted disruption of beta1-integrin in a transgenic mouse model of human breast cancer reveals an essential role in mammary tumor induction. Cancer Cell. 2004;6:159–170. doi: 10.1016/j.ccr.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 40.Roman J, Ritzenthaler JD, Roser-Page S, Sun X, Han S. alpha5beta1-integrin expression is essential for tumor progression in experimental lung cancer. Am J Respir Cell Mol Biol. 2010;43:684–691. doi: 10.1165/rcmb.2009-0375OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brooks PC, Clark RA, Cheresh DA. Requirement of vascular integrin alpha v beta 3 for angiogenesis. Science. 1994;264:569–571. doi: 10.1126/science.7512751. [DOI] [PubMed] [Google Scholar]
- 42.Trusolino L, Bertotti A, Comoglio PM. A signaling adapter function for alpha6beta4 integrin in the control of HGF-dependent invasive growth. Cell. 2001;107:643–654. doi: 10.1016/S0092-8674(01)00567-0. [DOI] [PubMed] [Google Scholar]
- 43.Serini G, Trusolino L, Saggiorato E, Cremona O, De Rossi M, Angeli A, Orlandi F, Marchisio PC. Changes in integrin and E-cadherin expression in neoplastic versus normal thyroid tissue. J Natl Cancer Inst. 1996;88:442–449. doi: 10.1093/jnci/88.7.442. [DOI] [PubMed] [Google Scholar]
- 44.Khwaja A, Rodriguez-Viciana P, Wennström S, Warne PH, Downward J. Matrix adhesion and Ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/Akt cellular survival pathway. EMBO J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaw LM, Rabinovitz I, Wang HH, Toker A, Mercurio AM. Activation of phosphoinositide 3-OH kinase by the alpha6beta4 integrin promotes carcinoma invasion. Cell. 1997;91:949–960. doi: 10.1016/S0092-8674(00)80486-9. [DOI] [PubMed] [Google Scholar]
- 46.Rathinam R, Alahari SK. Important role of integrins in the cancer biology. Cancer Metastasis Rev. 2010;29:223–237. doi: 10.1007/s10555-010-9211-x. [DOI] [PubMed] [Google Scholar]
- 47.FIGO Committee on Gynecologic Oncology FIGO staging for carcinoma of the vulva, cervix, and corpus uteri. Int J Gynaecol Obstet. 2014;125:97–98. doi: 10.1016/j.ijgo.2014.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.