Abstract
Accumulating evidence has demonstrated the crucial roles of long non-coding RNAs (lncRNAs) in various human cancers, including non-small cell lung cancer (NSCLC). However, to the best of our knowledge, the role of the lncRNA cancer susceptibility candidate 7 (CASC7) in NSCLC has not been clearly determined. The aim of the present study was to investigate the involvement of CASC7 in NSCLC. Marked downregulation of CASC7 was observed in NSCLC tissues and cell lines, and this downregulation of CASC7 was closely associated with distant metastasis, lymph node involvement and poor overall survival in NSCLC patients. Furthermore, overexpression of CASC7 significantly suppressed the proliferation, invasion and migration of the NSCLC cells A549 and H358, and promoted cell apoptosis in vitro. In addition, CASC7 was shown to act as a competing endogenous RNA by sponging miR-92a, which was proven to be an oncogenic miRNA in our previous study. The expression of miR-92a was upregulated in NSCLC tissues and cell lines, and was found to be inversely associated with CASC7 expression in NSCLC tissues. It was also demonstrated that CASC7 upregulated the expression of the tumor suppressor gene phosphatase and tensin homolog (a well-known target of miR-92a) by sequestration of miR-92a. Moreover, the tumor-suppressive effects of CASC7 were partly reversed by miR-92a overexpression in NSCLC cells. Collectively, the results of the present study indicated that CASC7 may act as a tumor-suppressive lncRNA that inhibits NSCLC progression by sponging miR-92a. These findings may improve our understanding of the potential mechanisms through which gain of CASC7 expression represses NSCLC progression.
Keywords: non-small cell lung cancer, long non-coding RNA, CASC7, miR-92a, phosphatase and tensin homolog axis, competing endogenous RNA
Introduction
Non-small cell lung cancer (NSCLC) is the most frequent type of lung cancer, comprising at least 80% of all lung cancer diagnoses (1). NSCLC is primarily divided into two subtypes, namely squamous cell carcinoma and adenocarcinoma, which are derived from the epithelial cells lining the larger and peripheral small airways, respectively (2). Despite the advances in NSCLC treatment, the prognosis of NSCLC patients remains poor, mainly due to tumor metastasis (3,4). Thus, exploring the mechanisms underlying the development of NSCLC is crucial.
Long non-coding RNAs (lncRNAs) are a class of RNA molecules>200 nucleotides in length, which have no protein-coding capacity (5). Recently, lncRNAs have been demonstrated to play key roles in various biological processes, including cell proliferation, migration and invasion (6-8). A large number of lncRNAs have been reported to be involved in the development of NSCLC, such as CRNDE, AWPPH and BANCR (9-11). Cancer susceptibility candidate 7 (CASC7) is a ~9.3-kb lncRNA, the function of which is largely unknown (12). Certain studies have demonstrated the involvement of CASC7 in severe asthma and spinal cord ischemia-reperfusion injury (13,14). In addition, CASC7 was reported to act as a tumor suppressor in colorectal cancer (CRC) and glioma tissues (15,16), but the precise mechanism of action of this cancer-related lncRNA remains unclear. Moreover, there are currently no reports on the role or mechanism of action of CASC7 in hepatocellular carcinoma (HCC).
LncRNAs have been shown to act as competing endogenous RNAs (ceRNAs) by sponging specific miRNAs, thereby preventing targeted transcripts of these miRNAs from being degraded (17). An example of this type of regulation is the lncRNA MNX1-AS1, which was shown to interact with miR-218-5p and regulate the expression of COMMD8 in HCC cells (18). Similarly, lncRNA NEAT1 acts as a ceRNA to regulate the expression of oncogene SOX2 through sponging miR-132 in glioma cells (19). In NSCLC, lncRNA 1308 has been identified as an oncogenic lncRNA that acts through regulating miR-124 (20). Notably, lncRNA CASC7 has been reported to inhibit growth and invasion of CRC cells through upregulating ING3 expression via sequestration of miR-21 (16). In addition, CASC7 was previously reported to target miR-21 in airway smooth muscle cells (13). Therefore, it was hypothesized that CASC7 may affect the development of NSCLC via its ceRNA role, which has not been previously investigated.
In the present study, the expression level and clinical significance of CASC7 in NSCLC were investigated. Furthermore, the regulatory role and underlying mechanism of action of CASC7 in the proliferation and invasion of NSCLC cells were examined, in order to determine whether CASC7 may serve as a new potential therapeutic target in NSCLC.
Materials and methods
Patients and samples
NSCLC tissues (n=80) and matched normal adjacent tissues (n=80) were obtained from patients who had undergone surgical resection at the Department of Thoracic Surgery, Changhai Hospital, between January 2016 and December 2017. The samples were snap-frozen and stored at -80°C prior to RNA extraction. The clinicopathological characteristics are summarized in Table I. Written informed consent was obtained from each patient. The protocol of the present study was approved by the Ethics Committee of Changhai Hospital (approval no. 2016-00113).
Table I.
Correlation between lncRNA CASC7 and the clinicopathological characteristics of patients with non-small cell lung cancer.
| Characteristics | All cases (n=80) | CASC7 expression
|
P-value | |
|---|---|---|---|---|
| High (n=31) | Low (n=49) | |||
| Sex | 0.7787 | |||
| Male | 48 | 18 | 30 | |
| Female | 32 | 13 | 19 | |
| Age (years) | 0.9097 | |||
| ≥60 | 51 | 20 | 31 | |
| <60 | 29 | 11 | 18 | |
| Tumor size (cm) | 0.0758 | |||
| ≥5 | 46 | 14 | 32 | |
| <5 | 34 | 17 | 17 | |
| Smoking | 0.7336 | |||
| No | 25 | 9 | 16 | |
| Yes | 55 | 22 | 33 | |
| Differentiation degree | 0.1096 | |||
| Moderate | 30 | 15 | 15 | |
| Poor | 50 | 16 | 34 | |
| Clinical stage | 0.0188 | |||
| I-II | 31 | 17 | 14 | |
| III-IV | 49 | 14 | 35 | |
| Distant metastasis | 0.0002b | |||
| No | 36 | 22 | 14 | |
| Yes | 44 | 9 | 35 | |
| Lymph node involvement | 0.0439a | |||
| No | 21 | 12 | 9 | |
| Yes | 59 | 19 | 40 | |
P<0.05,
P<0.01.
Reverse transcription-quantitative PCR (RT-qPCR) analysis
Total RNA was isolated from NSCLC tissues and cell lines using TRIzol reagent (TaKaRa Bio, Inc.). The RT of miR-92a was performed using the miScript II RT kit (Invitrogen; Thermo Fisher Scientific, Inc.), and the RT of lncRNA CASC7 and phosphatase and tensin homolog (PTEN) was performed using the SuperScript III First-Strand Synthesis System kit (Invitrogen; Thermo Fisher Scientific, Inc.). RT-qPCR assays were carried out using SYBR Premix Ex Taq II reagent (TaKaRa Bio, Inc.) on the 7900 HT Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The reaction mixtures were denatured at 95°C for 3 min, followed by 40 two-step cycles of 95°C for 10 sec and at 60°C for 30 sec. The primer sequences were as follows: CASC7 forward, 5′-GCT GCC AGG AGA AGG CAA GGA TC-3′ and reverse, 5′-AGG GTT AGA GCA GCC TTC GGA CT-3′; PTEN forward, 5′-TGG AAA GGG ACG AAC TGG TG-3′ and reverse, 5′-CAT AGC GCC TCT GAC TGG GA-3′; GAPDH forward, 5′-GAA GAT GGT GAT GGG ATT TC-3′ and reverse, 5′-AAC GCT TCA CGA ATT TGC GT-3′; miR-92a forward, 5′-CAC CTA TAT TGC ACT TGT CC-3′ and reverse, 5′-TGC GTG TCG TGG AGT C-3′; and U6 forward, 5′-CTC GCT TCG GCA GCA CA-3′ and reverse, 5′-GTC ATA CTC CTG CTT GCT GAT-3′. GAPDH was used as the internal reference for CASC7 and PTEN and U6 was used as reference for miR-92a. The relative gene expression was calculated using the 2−ΔΔCq method (21).
In situ hybridization (ISH)
ISH detection for CASC7 was performed on NSCLC and normal tissues using a commercial ISH Detection kit (cat. no. AR0149; Boster Biological Technology, Ltd.). ISH staining was evaluated by two pathologists in a blinded manner.
Cell lines and cell culture
The A549, H358 and H2170 NSCLC cell lines and 293T cells, were obtained from ATCC. Normal human bronchial epithelial cells (16HBE) were also obtained from ATCC. All cells were maintained in DMEM supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) in a 5% CO2 incubator at 37°C.
Cell transfection
The miR-92a mimics (5′-UAU UGC ACU UGU CCC GGC CUG U-3′), mimics negative control (NC; 5′-UUC UCC GAA CGU GUC ACG UTT-3′), miR-92a inhibitor (5′-ACA GGC CGG GAC AAG UGC AAU A-3′) and inhibitor NC (5′-CAG UAC UUU UGU GUA GUA CAA-3′) were obtained from RiBoBio. The CASC7-overexpressing vector pcDNA-CASC7 and pcDNA vector were constructed by Qiagen, Inc. In addition, CASC7 siRNA (si-CASC7) and corresponding negative control siRNA (si-Scramble) were purchased from RiboBio. The sequences were as follows: si-CASC7, 5′-TGG AAC ACA TGG TCC AGC ACT TTA A-3′; and si-Scramble, 5′-TGG ACA CTG GTG ACC TCA CTA ATA A-3′.
After A540 and H358 cells in 6-well plates had grown to ~80% confluence, miR-92a mimics (20 nmol/l), miR-92a inhibitor (20 nmol/l), si-CASC7 (30 nM) or 2 µg pcDNA-CASC7 were transfected into cells at 37°C for 24 h using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Cell proliferation
The antiproliferative effect of CASC7 on NSCLC cells was evaluated by using MTT assay. At the end of transfection, 20 µl MTT solution (Sigma-Aldrich; Merck KGaA) was added to each well, and A540 and H358 cells (1×105/well) were cultured for 2 h. Subsequently, the absor-bance of the samples at 450 nm was detected by an iMark microplate reader (Bio-Rad Laboratories, Inc.).
Immunofluorescence
Following transfection, the cells were fixed in absolute ethyl alcohol for 30 min at room temperature. After washing twice with PBS, the fixed cells were stained with primary antibody against cleaved caspase-3 (1:100; cat. no. ab49822; Abcam) for 1 h at room temperature. Subsequently, secondary antibody conjugated with FITC (1:200, cat. no. ab116639; Abcam) was added for 2 h in the dark, and fluorescence images were captured and analyzed using an inverted fluorescence microscope (EVOS FL; AMF-4306, Thermo Fisher Scientific, Inc.; magnification, 100×).
Cell apoptosis
The apoptosis of A549 and H358 cells was examined using flow cytometry. Following transfection for 48 h, cells were collected and the apoptotic cells were identified using an Annexin V-FITC Apoptosis Detection kit (Abcam) according to the manufacturer's protocol. The fluorescence signals were collected by a FACScan flow cytometer (Beckman Coulter, Inc.) and then analyzed by FlowJo 8.7.1 software (FlowJo LLC).
Cell invasion
Transwell chambers (24-well, 8-µm pore polyethylene terephthalate membrane; BD Biosciences) coated with Matrigel (BD Biosciences) were used for the invasion assay. Briefly, A549 and H358 cell suspension containing 8×104 cells were added in the top chamber with DMEM, while the lower chamber contained DMEM supplemented with 20% FBS. After 24 h of incubation, the cells were stained with 0.1% crystal violet solution for 10 min at room temperature and photographed with a CKX41 inverted microscope (Olympus Corporation) at a magnification of ×200.
Wound healing assay
A540 and H358 cells (2×106/well) were seeded in 6-well plates coated with 50 µg/ml poly-D-lysine (Sigma-Aldrich; Merck KGaA) overnight at 37°C to allow cells to attach. When the cell reached ~90%, the cells were starved in serum-free medium for 24 h. Then, the cell monolayers were scratched using a 200-µl pipette tip and images from each well were captured at 0 and 48 h with a CKX41 inverted microscope (Olympus Corporation) at a magnification of ×200.
Dual-luciferase reporter assay
A total of 100 ng pGL3-CASC7 wild-type (wt) or pGL3-CASC7 mutant (mut) plasmid (Shanghai Jima Industrial Co., Ltd.) were co-transfected in cells with miR-92a mimics, together with 20 ng Renilla luciferase vector (Promega Corporation) as an internal normalization control in 24-well plates (2×105/well) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 24 h post-transfection, the luciferase activities were analyzed using the Dual-Luciferase Reporter Assay system (Promega Corporation) according to the manufacturer's protocol. Transfections were performed in duplicate and repeated three times.
Western blot analysis
Western blotting was performed to detect the expression of proteins, as described previously (22). Briefly, protein was extracted from cells using RIPA lysis buffer (Beyotime Institute of Biotechnology) with proteinase inhibitor. The concentrations of total cellular protein were determined using a BCA assay kit (Beyotime Institute of Biotechnology). Proteins (40 µg) were separated by 10% SDS-PAGE and transferred onto a PVDF membrane (EMD Millipore). The membrane was then blocked with 5% skimmed milk at 4°C overnight and probed with antibodies against PTEN (cat. no. 32199; 1:1,000), proliferating cell nuclear antigen (PCNA; cat. no. 92552; 1:1,000), E-cadherin (cat. no. 194982; 1:1,000), N-cadherin (cat. no. 202030; 1:1,000), fibronectin (cat. no. 32419; 1:1,000), vimentin (cat. no. 92547; 1:1,000) and β-actin (cat. no. 179467; 1:2,000) overnight at 4°C, followed by horseradish peroxidase-conjugated goat anti-rabbit IgG (cat. no. 205718; 1:10,000) for 1 h at room temperature. All antibodies were purchased from Abcam. The protein bands were developed using an ECL kit (GE Healthcare) and blot bands were quantified with ImageJ software (version 1.46; Rawak Software, Inc.).
Statistical analysis
Statistical analysis was performed by SPSS 18.0 (SPSS, Inc.). All data are presented as mean ± standard deviation. The overall survival was analyzed using the Kaplan-Meier method and log-rank test. Correlations between clinical characteristics and CASC7 expression were evaluated using the χ2 test. The correlations between CASC7 and miR-92a levels, and between CASC7 and PTEN levels, were evaluated using Pearson's coefficient. Comparisons among data were performed by one-way ANOVA followed by Tukey's post hoc test. P<0.05 was considered to indicate statistically significant differences.
Results
LncRNA CASC7 is downregulated in NSCLC tissues and cell lines
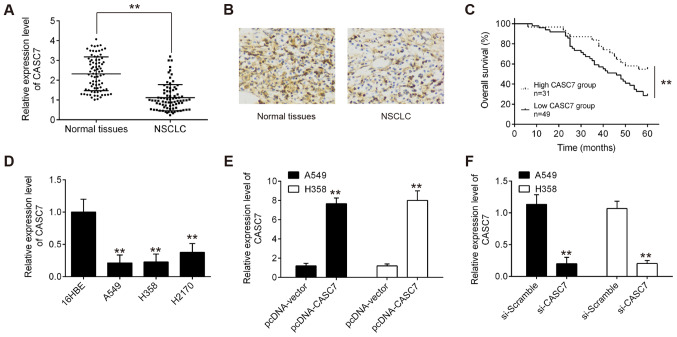
To investigate the roles of CASC7 in NSCLC, the expression levels of CASC7 were quantified in 80 pairs of NSCLC and matched normal adjacent tissues using RT-qPCR. As shown in Fig. 1A, CASC7 expression was significantly lower in tumor tissues compared with that in normal adjacent tissues. Furthermore, downregulation of CASC7 in NSCLC tissues was also observed by ISH assay (Fig. 1B). The correlations between the clinicopathological characteristics and CASC7 expression in 80 NSCLC patients are summarized in Table I. CASC7 expression was found to be negatively associated with distant metastasis and lymph node involvement. Next, Kaplan-Meier analysis demonstrated that patients with lower levels of CASC7 expression had poorer overall survival (OS) rates compared with those with higher levels of CASC7 expression (Fig. 1C). These data indicated that low CASC7 expression may play an important role in NSCLC progression.
Figure 1.
LncRNA CASC7 was found to be downregulated in NSCLC tissues and was correlated with poor prognosis in NSCLC patients. (A) CASC7 expression levels in 80 paired NSCLC and adjacent tissues by RT-qPCR; **P<0.01 vs. normal tissues. (B) Expression analysis of CASC7 in NSCLC and adjacent tissues from NSCLC patients using immunohistochemistry (magnification, ×400). (C) Kaplan-Meier analysis of overall survival in NSCLC patients with high and low CASC7 expression levels (n=31 and n=49, respectively); **P<0.01 vs. low CASC7 group. (D) RT-qPCR analysis of CASC7 expression levels in NSCLC cells (A549, H358 and H2170) and one normal human bronchial epithelial cell line (16HBE) that was used as a control. Data are presented as means ± standard deviation from three independent experiments; **P<0.01 vs. 16HBE cells. (E and F) RT-qPCR analysis of CASC7 expression levels in A549 and H358 cells following pcDNA-CASC7 or si-CASC7 transfection. Data are presented as means ± standard deviation from three independent experiments; **P<0.01 vs. pcDNA-vector or si-Scramble. NSCLC, non-small cell lung cancer; RT-qPCR, reverse transcription-quantitative PCR.
Overexpression of lncRNA CASC7 suppresses NSCLC cell proliferation in vitro
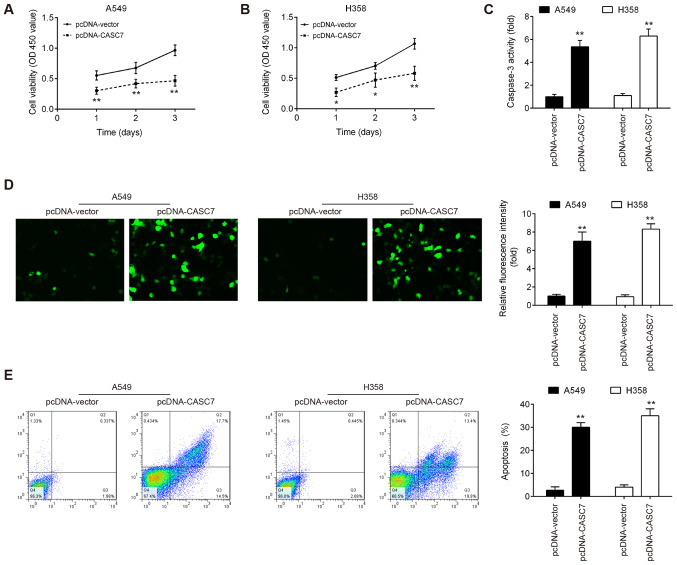
To further examine the biological role of CASC7 in NSCLC, the expression of CASC7 was first assessed in NSCLC cell lines. It was observed that the level of CASC7 was markedly decreased in NSCLC cell lines compared with that in normal human bronchial epithelial cells (16HBE) (Fig. 1D). As the expression of CASC7 was relatively lower in A549 (a widely used lung adenocarcinoma cell line) and H358 (a commonly used lung squamous cell carcinoma cell line) cells, CASC7 was overexpressed using pcDNA CASC7 or knocked down using si-CASC7 in these two cell lines. The pcDNA-CASC7 plasmids were added to both A549 and H358 cells, and it was observed that CASC7 was effectively upregulated compared with the pcDNA vector group (Fig. 1E). In addition, CASC7 expression was successfully reduced by si-CASC7 in A549 and H358 cells (Fig. 1F). The MTT assay revealed that overexpression of CASC7 signifi-cantly suppressed the proliferation of both A549 and H358 cells (Fig. 2A and B). Moreover, the activity and expression of caspase-3 in both NSCLC cell lines were markedly enhanced by lncRNA CASC7 overexpression (Fig. 2C and D). The effect of lncRNA CASC7 on cell apoptosis was also examined, and the results demonstrated that CASC7 overexpression promoted the apoptosis of A549 and H358 cells compared with the control group (Fig. 2E). Taken together, these results suggest that upregulation of CASC7 may exert suppressive effects on cell proliferation and promote apoptosis in NSCLC cells.
Figure 2.
Overexpression of lncRNA CASC7 suppressed NSCLC cell proliferation in vitro. (A and B) The effect of CASC7 overexpression on the proliferation of NSCLC cells was determined by Cell Counting Kit-8 assays. (C) The effect of CASC7 overexpression on the activity of caspase-3 was measured by a commercial kit. (D) The effect of CASC7 overexpression on the apoptosis-related cleaved caspase-3 protein was detected by immunofluorescence (magnification, ×200). (E) Cell apoptosis was measured by flow cytometry. Data are presented as means ± standard deviation from three independent experiments; *P<0.05, **P<0.01 vs. pcDNA-vector. NSCLC, non-small cell lung cancer.
Overexpression of lncRNA CASC7 suppresses NSCLC cell invasion and migration in vitro
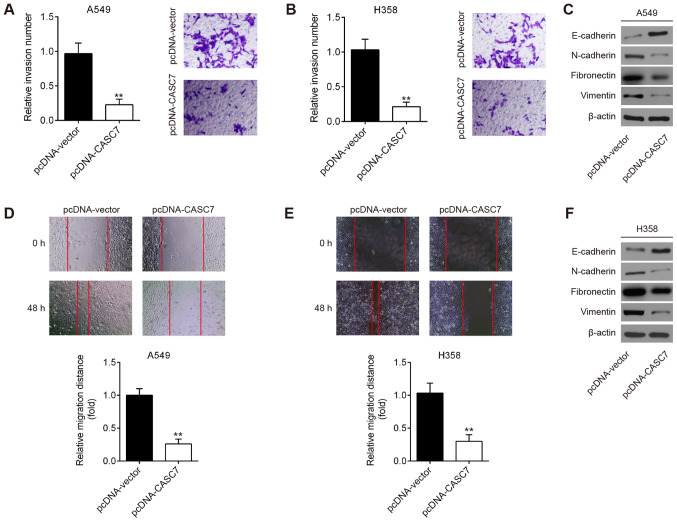
The effect of CASC7 on NSCLC cell invasion and migration was next assessed. Transwell and wound healing assays demonstrated that CASC7 overexpression suppressed the invasive and migratory capacities of A549 cells (Fig. 3A and D). Since epithelial-to-mesenchymal transition (EMT) is known to be a key pro-metastatic event, the expression of EMT markers was detected by western blotting. As shown in Fig. 3C, overexpression of CASC7 increased the expression of E-cadherin, whereas it decreased the expression of N-cadherin, fibronectin and vimentin, suggesting that CASC7 overexpression inhibits EMT in NSCLC cells. Similar results were observed in H358 cells (Fig. 3B, E and F). These data demonstrated that CASC7 overexpression exerted a significant suppressive effect on the invasion and migration of NSCLC cells in vitro.
Figure 3.
Overexpression of lncRNA CASC7 suppressed non-small cell lung cancer cell invasion and migration in vitro (A and B) The effect of CASC7 overexpression on the invasion of A549 and H358 cells was determined by Transwell assay with Matrigel coating (magnification, ×200). (C and F) The effect of CASC7 overexpression on the expression of epithelial-to-mesenchymal transition-related genes, including E-cadherin, N-cadherin, fibronectin and vimentin, was assessed by western blotting. (D and E) The effect of CASC7 overexpression on the migration of A549 and H358 cells was determined by wound healing assay (magnification, ×200). Data are presented as means ± standard deviation from three independent experiments; **P<0.01 vs. pcDNA-vector.
LncRNA CASC7 acts as a ceRNA for miR-92a in NSCLC cells
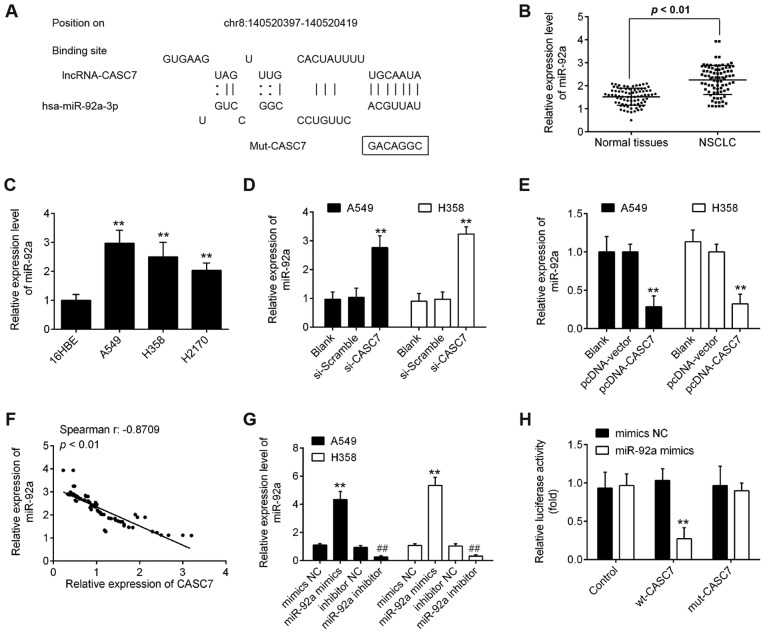
It is well-known that lncRNAs are likely to function as ceRNAs for special miRNAs, thus reversing the effects of miRNAs on the target genes (23,24). In the present study, starbase v2.0 (http://starbase.sysu.edu.cn/) was used to predict the potential targets of CASC7. As shown in Fig. 4A, miR-92a had a putative binding site with CASC7. miR-92a has been previously reported to be among the cancer-associated miRNAs (25-27). Additionally, our previous study demonstrated that miR-92a acts as an oncogene in the progression of NSCLC (28). Therefore, miR-92a was selected for further investigation. The expression levels of miR-92a were significantly upregulated in tumor tissues and NSCLC cell lines compared with those in adjacent normal tissues and 16HBE cells (Fig. 4B and C). Moreover, knockdown of CASC7 by si-CASC7 significantly increased miR-92a expression, while NSCLC cells transfected with pcDNA-CASC7 exhibited a marked inhibition of miR-92a expression (Fig. 4D and E). In addition, further correlation analysis revealed that the expression of CASC7 was inversely correlated with the expression of miR-92a in NSCLC tissues (Fig. 4F). In addition, the expression of miR-92a was detected by RT-qPCR 48 h after transfection of miR-92a mimics, miR-92a inhibitor, and their respective NCs. As shown in Fig. 4G, the expression of miR-92a was signifi-cantly increased following transfection of miR-92a mimics, whereas it was markedly decreased following transfection of miR-92a inhibitor, compared with their respective NCs.
Figure 4.
LncRNA CASC7 acts as a competing endogenous RNA for miR-92a in NSCLC cells. (A) Predicted miR-92a-binding sites on CASC7. (B) The miR-92a expression levels in 80 paired NSCLC and adjacent tissues were determined by RT-qPCR. P<0.01 vs. normal tissues. (C) RT-qPCR analysis of miR-92a expression levels in NSCLC cells (A549, H358 and H2170) and one normal human bronchial epithelial cell line (16HBE) that was used as a control. Data are presented as means ± standard deviation from three independent experiments; **P<0.01 vs. 16HBE cells. (D and E) The relative miR-92a expression in A549 and H358 cells transfected with si-CASC7 or pcDNA-CASC7 was analyzed by RT-qPCR. Data are presented as means ± standard deviation from three independent experiments; **P<0.01 vs. si-Scramble or pcDNA-vector. (F) The Spearman's rank test was used to analyze the association between CASC7 and miR-92a expression levels in NSCLC tissues (r:-0.8709, P<0.01). (G) RT-qPCR analysis of miR-92a expression levels in A549 and H358 cells following miR-92a mimics or miR-92a inhibitor transfection. (H) Luciferase reporter assay in 293T cells co-transfected with the reporter plasmid (or the corresponding mutant reporter) and the indicated miRNAs. miR-92a mimics significantly decreased the luciferase activity in wt-CASC7 but not in mut-CASC7. Data are presented as means ± standard deviation from three independent experiments, **P<0.01 vs. mimics NC, ##P<0.01 vs. inhibitor NC. NSCLC, non-small cell lung cancer; RT-qPCR, reverse transcription-quantitative PCR; wt, wild-type; mut, mutant.
Next, luciferase reporter assay was employed to validate the binding of miR-92a to lncRNA CASC7. As shown in Fig. 4H, overexpression of miR-92a reduced the luciferase activity of wt-CASC7, but did not affect the luciferase activity of mut-CASC7 in 293T cells. These data indicated that CASC7 interacts with miR-92a in NSCLC cells.
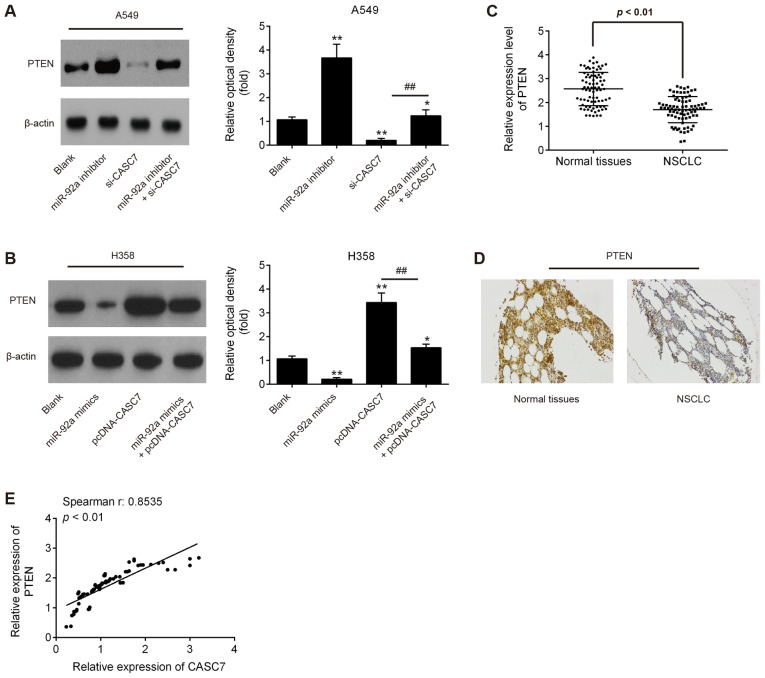
LncRNA CASC7 increases the expression of PTEN by acting as a ceRNA of miR-92a
PTEN is a well-known tumor suppressor, and plays an important role in cancer initiation and progression (29). Notably, several studies have demonstrated that miR-92a targets PTEN and suppresses its translation in different types of cancer cells (30-32). Given the association between miR-92a and CASC7, it was further investigated whether CASC7 acts a tumor suppressor through restoring the expression of PTEN via sequestration of miR-92a. Western blot assay demonstrated that the CASC7 knockdown by si-CASC7 led to a significant decrease of PTEN expression in A549 cells, whereas miR-92a knockdown reversed the inhibitory effect of CASC7 on the expression of PTEN (Fig. 5A). It was also observed that CASC7 overexpression induced a marked increase of PTEN expression in H358 cells, while miR-92a overexpression attenuated the promoting effect of CASC7 on PTEN protein levels (Fig. 5B). Furthermore, the levels of PTEN in NSCLC tissues were detected by RT-qPCR. As shown in Fig. 5C, the expression of PTEN was significantly decreased in NSCLC compared with that in adjacent tissues. In addition, using immunohistochemistry, a significantly lower expression of PTEN was observed in NSCLC tissues compared with that in adjacent tissues (Fig. 5D). Moreover, a positive correlation was detected between CASC7 and PTEN expression in NSCLC tissues (Fig. 5E). These data indicate that CASC7 may regulate the expression of PTEN by acting as a ceRNA of miR-92a.
Figure 5.
LncRNA CASC7 increased the expression of PTEN by acting as a competing endogenous RNA of miR-92a. A549 cells were co-transfected with si-CASC7 and miR-92a inhibitor, while H358 cells were co-transfected with miR-92a mimics and pcDNA-CASC7. Cells were then harvested for western blotting. (A and B) PTEN expression was measured by western blotting. Data are presented as means ± standard deviation from three independent experiments, *P<0.05, **P<0.01 vs. blank group; ##P<0.01 vs. si-CASC7 or pcDNA-CASC7. (C) PTEN expression levels in 80 paired NSCLC tissues and paired non-tumor tissues by RT-qPCR. P<0.01 vs. normal tissues. (D) Expression analysis of PTEN in NSCLC tissues and adjacent tissues using immunohistochemistry (magnification, ×200). (E) The Spearman's rank test was used to analyze the association between CASC7 and PTEN expression levels in NSCLC tissues (r:0.8535, P<0.01). NSCLC, non-small cell lung cancer; PTEN, phosphatase and tensin homolog; RT-qPCR, reverse transcription-quantitative PCR.
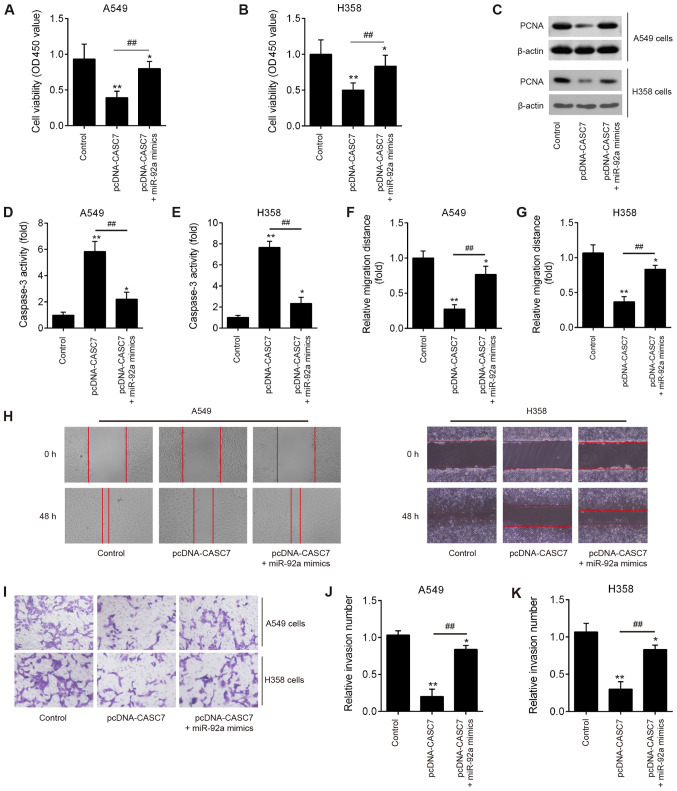
LncRNA CASC7 regulates miR-92a to suppress NSCLC cell growth and invasion
To further investigate whether miR-92a is involved in lncRNA CASC7-mediated inhibition of proliferation and invasion in NSCLC cells, pcDNA-CASC7 and miR-92a mimics were co-transfected into A549 and H358 cells. Compared with the control group, CASC7 overexpression significantly suppressed cell proliferation, whereas miR-92a overexpression partially reversed this inhibitory effect of CASC7 (Fig. 6A and B). In addition, the expression of a well-known marker of cell proliferation, PCNA, was significantly decreased in the pcDNA-CASC7 group compared with the control group in A549 and H358 cells, while this inhibitory effect was also reversed by overexpression of miR-92a (Fig. 6C). The activity of caspase-3 was also measured and found to be obviously increased by CASC7 overexpression compared with the control group, whereas upregulation of miR-92a attenuated this promoting effect (Fig. 6D and E). Wound healing and cell invasion experiments revealed that the decreased migration distance and reduced number of invading cells induced by CASC7 overexpression was partially abrogated by overexpression of miR-92a (Fig. 6F-K). These data indicated that CASC7 suppresses NSCLC cell growth and invasion by regulating miR-92a expression.
Figure 6.
LncRNA CASC7 regulates miR-92a expression to suppress NSCLC cell growth and invasion. A549 and H358 cells were co-transfected with pcDNA-CASC7 and miR-92a mimics, and were then harvested for subsequent experiments. (A and B) Cell viability was determined by Cell Counting Kit-8 assay. (C) The protein expression of PCNA was detected by western blotting. (D and E) The activity of caspase-3 was measured by a commercial kit. (F-H) The migration of A549 and H358 cells was determined by wound healing assay. (I-K) The invasion of A549 and H358 cells was determined by Transwell assay with Matrigel coating. Data are presented as means ± standard deviation from three independent experiments, *P<0.05, **P<0.01 vs. control group; ##P<0.01 vs. pcDNA-CASC7. NSCLC, non-small cell lung cancer; PCNA, proliferating cell nuclear antigen.
Discussion
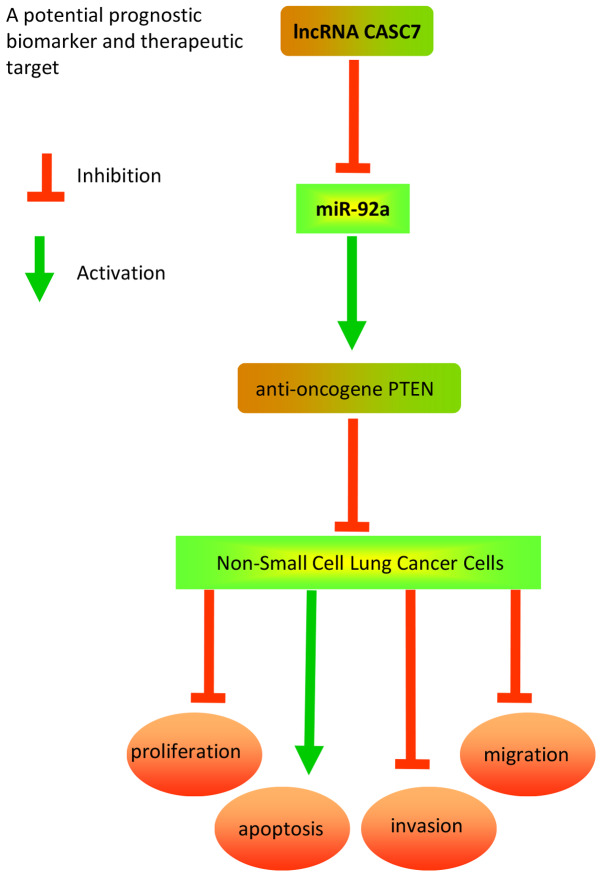
The present study demonstrated that CASC7 was down-regulated in NSCLC tissues and cell lines, and its low expression was closely correlated with poor prognosis and shorter survival in NSCLC patients. Furthermore, CASC7 overexpression sponged miR-92a to suppress cancer cell proliferation, invasion and migration via upregulating the expression of the anti-oncogene PTEN. Collectively, the results of the present study may provide new insight into the role of the CASC7/miR-92a/PTEN axis in NSCLC, which may prove to be a promising molecular target for NSCLC treatment. A schematic presentation summarizing the mechanism of action of CASC7 in NSCLC is presented in Fig. 7.
Figure 7.
Schematic diagram of the signaling pathway through which CASC7 inhibits the proliferation, invasion and migration of NSCLC cells. Overexpression of CASC7 effectively inhibits growth and invasion of NSCLC cells through upregulating the expression of the PTEN tumor suppressor gene via sequestration of miR-92a. NSCLC, non-small cell lung cancer; PTEN, phosphatase and tensin homolog.
As a newly identified lncRNA, there is very little research on the role of CASC7 in cancer. In glioma, CASC7 was found to be downregulated and exerted its tumor-suppressive effects via inactivation of the Wnt/β-catenin pathway (15). In CRC, CASC7 overexpression inhibited proliferation, migration and invasion, and promoted apoptosis in CRC cells, suggesting that CASC7 may be considered as a novel diagnostic marker of CRC (16). However, to the best of our knowledge, no data on the expression and role of CASC7 in NSCLC have been reported to date. In the present study, CASC7 was found to be significantly decreased in NSCLC tissues and cell lines, and its low expression was correlated with the clinicopathological characteristics of NSCLC patients, particularly distant metastasis and lymph node involvement. Furthermore, the high expression of CASC7 in the NSCLC cell lines A549 and H358 was found to suppress cancer cell proliferation, invasion and migration, and promote apoptosis. Collectively, these data indicate that CASC7 plays a key role in suppressing NSCLC development and progression; however, the underlying mechanism remains elusive.
Recently, ceRNA regulation has been implicated in lung carcinogenesis. For example, Dong et al demonstrated that lncRNA GAS5 acted as a tumor suppressor via the miR-205/PTEN axis in NSCLC (33). Jin et al reported that the lncRNA SNHG20 suppressed NSCLC growth in in vivo through suppressing miR-154 and enhancing ZEB2 and RUNX2 expression (34). Zhang et al demonstrated that the lncRNA FENDRR was downregulated in NSCLC tissues, whereas FENDRR overexpression inhibited the malignant phenotypes of NSCLC cell by competitively binding to miR-761 (35). Of note, it has been reported that CASC7 may act as a ceRNA to sponge miR-21 and regulate the expression of ING3 in CRC (16). Therefore, it was inferred that lncRNA CASC7 may act as a ceRNA, participating in NSCLC development. The present study, using bioinformatics analysis, identified miR-92a as a potential CASC7-binding miRNA, which had been previously demonstrated to be an oncogene in several cancers, such as HCC (36), CRC (37) and gastric cancer (38). Interestingly, our previous study had demonstrated that the expression of miR-92a was markedly upregulated in NSCLC, and overexpression of miR-92a promoted tumor progression through activating the PTEN/PI3K/AKT signaling pathway (28). As expected, the results of the present study demonstrated that CASC7 directly targeted miR-92a and negatively regulated the expression of miR-92a in vitro. Furthermore, the expression of CASC7 was found to be inversely correlated with the expression of miR-92a in NSCLC tissues. Importantly, CASC7 inhibited cell proliferation, invasion and migration through suppressing miR-92a expression in NSCLC cells. Collectively, these data suggest that the CASC7/miR-92a axis plays a key role in NSCLC growth and development. Of note, the overexpression of miR-92a could only partially reverse the promoting effects of CASC7 on PTEN expression, as well as its inhibitory effects on the proliferation, invasion and migration of NSCLC cells. All these results suggest that the tumor-suppressive effects of CASC7 may also be mediated by other targets, and future research should focus on the identification of other targets of CASC7.
It is well-known that the PTEN gene plays a tumor-suppressive role in several human cancers, including NSCLC (39,40). For example, Perumal et al demonstrated that PTEN inactivation induced migration and invasion by regulation of β-catenin and Snail/Slug in NSCLC cells (41). Liu et al reported that PTEN promoted G0/G1 arrest and apoptosis in NSCLC cells by regulating the expression levels of Skp2 (42). It has also been reported that miR-92a exerts its oncogenic effects by directly targeting PTEN in several types of cancer, including NSCLC (28,30,43,44). Given that both lncRNA CASC7 and PTEN interact with miR-92a, lncRNA CASC7 may regulate PTEN expression by competitively binding to miR-92a. In the present study, it was observed that CASC7 upregulated the expression of PTEN in A549 and H358 cells by inhibiting the expression of miR-92. Furthermore, PTEN expression was found to be markedly downregulated, and was positively correlated with CASC7 expression levels in NSCLC tissues. Taken together, these data strongly suggest that there is a ceRNA functional association among CASC7, PTEN and miR-92a in NSCLC.
To the best of our knowledge, the present study is the first to demonstrate that the expression of CASC7 is low in NSCLC tissues and cell lines, and that this low expression is closely associated with distant metastasis and lymph node involvement. There is currently no treatment regulating the expression of CASC7 in NSCLC. Thus, in the future, more clinical samples must be collected in order to determine whether chemotherapeutic drugs affect the expression levels of CASC7 in NSCLC.
In conclusion, we herein demonstrated that CASC7 acts as a tumor suppressor by upregulating PTEN by acting as a ceRNA of miR-92a in NSCLC cells. These findings suggest that CASC7 may be a promising target for the treatment of NSCLC.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China Grants (grant no. 81802288).
Availability of data and materials
All the datasets generated and/or analyzed during the present study are included in this published article.
Authors' contributions
LC, XL, YZ and JZ performed the experiments, contributed to data analysis and wrote the manuscript. LC, XL, YZ and JZ analyzed the data. CL and LY conceptualized the study design, contributed to data analysis and experimental materials. All authors have read and approved the final version of this manuscript.
Ethics approval and consent to participate
All individuals provided informed consent regarding the use of their specimens for the purposes of clinical research. The present study was approved by the Ethics Committee of Changhai Hospital (approval no. 2016-00113).
Patient consent for publication
Not applicable.
Competing interests
All the authors declare that they have no competing interests.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Chen G, Yi XH. Pathology and genetics of disease and tumours of the lung, pleura in China. Zhonghua Bing Li Xue Za Zhi. 2005;34:490–493. In Chinese. [PubMed] [Google Scholar]
- 3.Ramalingam SS, Owonikoko TK, Khuri FR. Lung cancer: New biological insights and recent therapeutic advances. CA Cancer J Clin. 2011;61:91–112. doi: 10.3322/caac.20102. [DOI] [PubMed] [Google Scholar]
- 4.Mostafa AA, Morris DG. Immunotherapy for lung cancer: Has it finally arrived? Front Oncol. 2014;4:288. doi: 10.3389/fonc.2014.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Wang R, Ma Z, Feng L, Yang Y, Tan C, Shi Q, Lian M, He S, Ma H, Fang J. LncRNA MIR31HG targets HIF1A and P21 to facilitate head and neck cancer cell proliferation and tumori-genesis by promoting cell-cycle progression. Mol Cancer. 2018;17:162. doi: 10.1186/s12943-018-0916-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng C, Liu S, Lu S, Yu X, Lai J, Wu Y, Chen S, Wang L, Yu Z, Luo G, Li Y. The c-Myc-regulated lncRNA NEAT1 and paraspeckles modulate imatinib-induced apoptosis in CML cells. Mol Cancer. 2018;17:130. doi: 10.1186/s12943-018-0884-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang G, Sun J, Zhao H, Li H. Long non-coding RNA (lncRNA) growth arrest specific 5 (GAS5) suppresses esophageal squamous cell carcinoma cell proliferation and migration by inactivating phosphatidylinositol 3-kinase (PI3K)/AKT/Mammalian target of rapamycin (mTOR) signaling pathway. Med Sci Monit. 2018;24:7689–7696. doi: 10.12659/MSM.910867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan YF, Yu ZP, Cui XY. lncRNA colorectal neoplasia differentially expressed (CRNDE) promotes proliferation and inhibits apoptosis in non-small cell lung cancer cells by regulating the miR-641/CDK6 axis. Med Sci Monit. 2019;25:2745–2755. doi: 10.12659/MSM.913420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song Z, Du J, Zhou L, Sun B. lncRNA AWPPH promotes proliferation and inhibits apoptosis of non-small cell lung cancer cells by activating the Wnt/β-catenin signaling pathway. Mol Med Rep. 2019;19:4425–4432. doi: 10.3892/mmr.2019.10089. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Li Y, Han L, Zhang P, Sun S. SUMO1P3 is associated clinical progression and facilitates cell migration and invasion through regulating miR-136 in non-small cell lung cancer. Biomed Pharmacother. 2019;113:108686. doi: 10.1016/j.biopha.2019.108686. [DOI] [PubMed] [Google Scholar]
- 12.van Heesch S, van Iterson M, Jacobi J, Boymans S, Essers PB, de Bruijn E, Hao W, MacInnes AW, Cuppen E, Simonis M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014;15:R6. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu JH, Li C, Zhang CH, Zhang ZH. LncRNA-CASC7 enhances corticosteroid sensitivity via inhibiting the PI3K/AKT signaling pathway by targeting miR-21 in severe asthma. Pulmonology. 2020;26:18–26. doi: 10.1016/j.pulmoe.2019.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Pan L, Jiang A, Yin M. Hydrogen sulfide upregulated lncRNA CasC7 to reduce neuronal cell apoptosis in spinal cord ischemia-reperfusion injury rat. Biomed Pharmacother. 2018;98:856–862. doi: 10.1016/j.biopha.2017.12.079. [DOI] [PubMed] [Google Scholar]
- 15.Gong X, Liao X, Huang M. LncRNA CASC7 inhibits the progression of glioma via regulating Wnt/β-catenin signaling pathway. Pathol Res Pract. 2019;215:564–570. doi: 10.1016/j.prp.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Fu C, Xu Q, Wei X. Long non-coding RNA CASC7 inhibits the proliferation and migration of colon cancer cells via inhibiting microRNA-21. Biomed Pharmacother. 2017;95:1644–1653. doi: 10.1016/j.biopha.2017.09.052. [DOI] [PubMed] [Google Scholar]
- 17.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji D, Wang Y, Sun B, Yang J, Luo X. Long non-coding RNAMNX1-AS1 promotes hepatocellular carcinoma proliferation and invasion through targeting miR-218-5p/COMMD8 axis. Biochem Biophys Res Commun. 2019;513:669–674. doi: 10.1016/j.bbrc.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 19.Zhou K, Zhang C, Yao H, Zhang X, Zhou Y, Che Y, Huang Y. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol Cancer. 2018;17:105. doi: 10.1186/s12943-018-0849-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Guo X, Li Q, Ran P, Xiang X, Yuan Y, Dong T, Zhu B, Wang L, Li F, et al. Long non-coding RNA1308 promotes cell invasion by regulating the miR-124/ADAM 15 axis in non-small-cell lung cancer cells. Cancer Manag Res. 2018;10:6599–6609. doi: 10.2147/CMAR.S187973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.E C, Li C, Li H, Yang J. Silencing of a novel lncRNA LOC105369748 suppresses the progression of hepatocel-lular carcinoma by sponging miR-5095 from MBD2. J Cell Physiol. 2019;234:18504–18512. doi: 10.1002/jcp.28486. [DOI] [PubMed] [Google Scholar]
- 23.Jiang F, Qi W, Wang Y, Wang W, Fan L. lncRNA PEG10 promotes cell survival, invasion and migration by sponging miR-134 in human bladder cancer. Biomed Pharmacother. 2019;114:108814. doi: 10.1016/j.biopha.2019.108814. [DOI] [PubMed] [Google Scholar]
- 24.Qi H, Wen B, Wu Q, Cheng W, Lou J, Wei J, Huang J, Yao X, Weng G. Long noncoding RNA SNHG7 accelerates prostate cancer proliferation and cycle progression through cyclin D1 by sponging miR-503. Biomed Pharmacother. 2018;102:326–332. doi: 10.1016/j.biopha.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Yu H, Song H, Liu L, Hu S, Liao Y, Li G, Xiao X, Chen X, He S. MiR-92a modulates proliferation, apoptosis, migration and invasion of osteosarcoma cell lines by targeting Dickkopf-related protein 3. Biosci Rep. 2019;39:BSR20190410. doi: 10.1042/BSR20190410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun L, Jin X, Xie L, Xu G, Cui Y, Chen Z. Swainsonine represses glioma cell proliferation, migration and invasion by reduction of miR-92a expression. BMC Cancer. 2019;19:247. doi: 10.1186/s12885-019-5425-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Sun B, Zhang J, Liu M, Guan L. Alkannin inhibits proliferation, migration and invasion of hepatocellular carcinoma cells via regulation of miR-92a. Biomed Pharmacother. 2019;114:108782. doi: 10.1016/j.biopha.2019.108782. [DOI] [PubMed] [Google Scholar]
- 28.Lu C, Shan Z, Hong J, Yang L. MicroRNA-92a promotes epithelial-mesenchymal transition through activation of PTEN/PI3K/AKT signaling pathway in non-small cell lung cancer metastasis. Int J Oncol. 2017;51:235–244. doi: 10.3892/ijo.2017.3999. [DOI] [PubMed] [Google Scholar]
- 29.Fang L, Wang X, Sun Q, Papakonstantinou E, S'ng C, Tamm M, Stolz D, Roth M. IgE Downregulates PTEN through MicroRNA-21-5p and stimulates airway smooth muscle cell remodeling. Int J Mol Sci. 2019;20:E875. doi: 10.3390/ijms20040875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Cao H, Xu D, Zhu K. MicroRNA-92a promotes metastasis of nasopharyngeal carcinoma by targeting the PTEN/AKT pathway. Onco Targets Ther. 2016;9:3579–3588. doi: 10.2147/OTT.S105470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ke TW, Wei PL, Yeh KT, Chen WT, Cheng YW. MiR-92a promotes cell metastasis of colorectal cancer through PTEN-Mediated PI3K/AKT pathway. Ann Surg Oncol. 2015;22:2649–2655. doi: 10.1245/s10434-014-4305-2. [DOI] [PubMed] [Google Scholar]
- 32.Zhang G, Zhou H, Xiao H, Liu Z, Tian H, Zhou T. MicroRNA-92a functions as an oncogene in colorectal cancer by targeting PTEN. Dig Dis Sci. 2014;59:98–107. doi: 10.1007/s10620-013-2858-8. [DOI] [PubMed] [Google Scholar]
- 33.Dong L, Li G, Li Y, Zhu Z. Upregulation of long noncoding RNA GAS5 inhibits lung cancer cell proliferation and metastasis via miR-205/PTEN axis. Med Sci Monit. 2019;25:2311–2319. doi: 10.12659/MSM.912581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin L, Jiang X, He G, Shi J, Su F, Zhu H. SNHG20 knockdown suppresses proliferation, migration and invasion, and promotes apoptosis in non-small cell lung cancer through acting as a miR-154 sponge. Biomed Pharmacother. 2019;112:108648. doi: 10.1016/j.biopha.2019.108648. [DOI] [PubMed] [Google Scholar]
- 35.Zhang MY, Zhang ZL, Cui HX, Wang RK, Fu L. Long non-coding RNA FENDRR inhibits NSCLC cell growth and aggressiveness by sponging miR-761. Eur Rev Med Pharmacol Sci. 2018;22:8324–8332. doi: 10.26355/eurrev_201812_16530. [DOI] [PubMed] [Google Scholar]
- 36.Shigoka M, Tsuchida A, Matsudo T, Nagakawa Y, Saito H, Suzuki Y, Aoki T, Murakami Y, Toyoda H, Kumada T, et al. Deregulation of miR-92a expression is implicated in hepatocellular carcinoma development. Pathol Int. 2010;60:351–357. doi: 10.1111/j.1440-1827.2010.02526.x. [DOI] [PubMed] [Google Scholar]
- 37.Liu GH, Zhou ZG, Chen R, Wang MJ, Zhou B, Li Y, Sun XF. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol. 2013;34:2175–2181. doi: 10.1007/s13277-013-0753-8. [DOI] [PubMed] [Google Scholar]
- 38.Wu Q, Yang Z, Wang F, Hu S, Yang L, Shi Y, Fan D. MiR-19b/20a/92a regulates the self-renewal and proliferation of gastric cancer stem cells. J Cell Sci. 2013;126:P4220–P4229. doi: 10.1242/jcs.127944. [DOI] [PubMed] [Google Scholar]
- 39.Zhao YB, Zhao J, Zhang LJ, Shan RG, Sun ZZ, Wang K, Chen JQ, Mu JX. MicroRNA-370 protects against myocardial isch-emia/reperfusion injury in mice following sevoflurane anesthetic preconditioning through PLIN5-dependent PPAR signaling pathway. Biomed Pharmacother. 2019;113:108697. doi: 10.1016/j.biopha.2019.108697. [DOI] [PubMed] [Google Scholar]
- 40.Ding K, Wu Z, Wang N, Wang X, Wang Y, Qian P, Meng G, Tan S. MiR-26a performs converse roles in proliferation and metastasis of different gastric cancer cells via regulating of PTEN expression. Pathol Res Pract. 2017;213:467–475. doi: 10.1016/j.prp.2017.01.026. [DOI] [PubMed] [Google Scholar]
- 41.Perumal E, So Youn K, Sun S, Seung-Hyun J, Suji M, Jieying L, Yeun-Jun C. PTEN inactivation induces epithelial-mesenchymal transition and metastasis by intranuclear translocation of β-catenin and snail/slug in non-small cell lung carcinoma cells. Lung Cancer. 2019;130:25–34. doi: 10.1016/j.lungcan.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 42.Liu L, Huang L, He J, Cai S, Weng Y, Huang S, Ma S. PTEN inhibits non-small cell lung cancer cell growth by promoting G0/G1 arrest and cell apoptosis. Oncol Lett. 2019;17:1333–1340. doi: 10.3892/ol.2018.9719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qin LB, Li ZY, Li H, Fan XQ, Liu HG, Dong XM, Jia WY. Inhibitive effects of microRNA-34a on protecting against ischemia-reperfusion injury of vital organs in hemorrhagic shock pregnant mice. Eur Rev Med Pharmacol Sci. 2018;22:1812–1818. doi: 10.26355/eurrev_201803_14600. [DOI] [PubMed] [Google Scholar]
- 44.Xiao J, Yu W, Hu K, Li M, Chen J, Li Z. MiR-92a promotes tumor growth of osteosarcoma by targeting PTEN/AKT signaling pathway. Oncol Rep. 2017;37:2513–2521. doi: 10.3892/or.2017.5484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the datasets generated and/or analyzed during the present study are included in this published article.