Abstract
Circular ribonucleic acids (circRNAs) serve a vital role in the pathological processes of a number of diseases. Previous microarray results of circRNA expression revealed that hsa_circ_0070933 and hsa_circ_0070934, two circRNAs associated with the La ribonucleoprotein 1B gene, were highly expressed in cutaneous squamous cell carcinoma (CSCC). The present study aimed to explore the specific role of these circRNAs in CSCC. Through reverse transcription-quantitative PCR, hsa_circ_0070933 and hsa_circ_0070934 expression levels in CSCC cell lines and a human keratino-cyte cell line were detected. Additionally, direct interactions between miR-1236-3p and HOXB7 or circ-0070934 were identified using RNA binding protein immunoprecipitation assays and dual-luciferase reporter assays. Cell Counting Kit-8, 5-ethynyl-2′-deoxyuridine, Transwell invasion and flow cytometry assays were used to assess the roles of miR-1236-3p or circ-0070934 in cell invasion, proliferation and apoptosis. Subsequently, in vivo tumor formation assays were used to verify the role of circ-0070934 in CSCC. The results demonstrated that the expression of circ-0070934 was stably upregulated in a number of CSCC cell lines compared with that in normal human keratinocytes. Knockdown of circ-0070934 inhibited the invasive and proliferative potential of CSCC cells and promoted apoptosis both in vivo and in vitro. In addition, circ-0070934 modulated HOXB7 expression through competitive binding with miR-1236-3p. In conclusion, the results of the present study demonstrated the effects of the circ-0070934/miR-1236-3p/HOXB7 regulatory axis on CSCC and provided a novel insight for the pathogenesis of CSCC.
Keywords: cutaneous squamous cell carcinoma, circular ribonucleic acids, competing endogenous RNAs, cell proliferation, cell invasion
Introduction
Nonmelanoma skin cancer (NMSC) is a common type of malignant neoplasm in Caucasian populations (1). NMSC accounts for more than one-third of all cancers in the US with an estimated incidence of >600,000 cases per year (2). Cutaneous squamous cell carcinoma (CSCC) is the second most frequent type of NMSC, accounting for ~20% of all NMSC cases (3). The long-term prognosis of patients with NMSC remains unsatisfactory despite promising progress in the diagnosis and treatment for CSCC over the past few decades. Thus, developing effective treatment methods for CSCC should be a priority for researchers. A number of genes including p16 and p53 have been confirmed to be involved in the pathogenesis of CSCC (4, 5). In addition, non-coding RNAs (ncRNAs) such as microRNA (miRNA/miR)-186 and long non-coding RNA (lncRNA) LINC00520 have also been verified to be involved in the pathology of CSCC (6,7). Despite these observations, there is still a need to explore the specific pathogenesis behind CSCC. Therefore, further research investigating the occurrence and development of CSCC in the aspect of epigenetics is pivotal to provide further guidance for clinicians.
In terms of structure, ncRNAs can be divided into circular (circ)RNAs and linear ncRNAs. The majority of research has focused on linear ncRNAs, especially miRNAs (8,9) and lncRNAs (10-12), whereas studies regarding circRNAs are rare, mainly since circRNAs were once considered by-products of incorrect slicing compared with the numerous classical linear ncRNAs (13). Andreeva and Cooper (13) reported in 2015 that circRNAs widely existed in animal and plant cell tissues and possessed a number of specific biological characteristics. This report has ensured that circRNAs gained attention from numerous researchers (14). Recent studies have indicated that circRNAs exert crucial effects in the pathological process of a number of diseases, such as a variety of tumor types, cardiovascular and digestive system diseases (15-18). For instance, Su et al (19) have revealed that the circRNA cTFRC regulates the expression of target genes through competitive binding with miR-107, which eventually participates in the pathogenesis of bladder cancer (19).
The results of circRNA expression microarray revealed that hsa_circ_0070933 and hsa_circ_0070934 (circ-0070934), two circRNAs associated with the La ribonucleoprotein 1B (LARP1B) gene, exhibited high expression levels in CSCC (20). Circ-0070934 is located at chr4:128995614-129012667, and its associated-gene symbol is LARP1B (http://www.circbase.org). The present study aimed to explore whether circ-0070934 may enhance the invasive and proliferative capacities of tumor cells and participate in the pathogenesis of CSCC.
In the present study, a series of experiments were performed to confirm whether circ-0070934 may function as a competing endogenous RNA (ceRNA) to modulate homeobox B7 (HOXB7) gene expression by collating miR-1236-3p in CSCC. Overall, the present study aimed to explore whether circ-0070934 may have a crucial role in CSCC pathogenesis and to provide a novel molecular target for the therapy of CSCC.
Materials and methods
Cell culture and transfection
Human embryonic cell line 293T, human keratinocyte cell line HaCaT and CSCC cell lines A431, HSC-5, SCC13 and SCL-1 were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences and were authenticated by STR profiling. All cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS; Sigma-Aldrich; Merck KGaA) in a humidified environment with 5% CO2 at 37°C. Cells with high viability were seeded in 6-well plates when they reached the logarithmic growth phase and were subjected to transfections with 5 µl circ-0070934 small interfering (si)RNAs, circ-0070934 short hairpin (sh)RNA vectors, circ-0070934 overexpression plasmids, miR-1236-3p inhibitors and miR-1236-3p mimics (synthesized by Shanghai GenePhama Co., Ltd.) and corresponding negative controls (NCs) using 5 µl Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) for 6 h at 37°C according to the manufacturer's instructions. The cells were collected 24 h post-transfection for further in vitro experiments. For in vivo studies, lentiviral particles carrying scrambled or circ-0070934 shRNA vectors (pLVX-shRNA 2-GFP-Puro; TSINGKE Biological Technology Co., Ltd.) were generated in 293T cells. A431 cells were then infected with the recombinant lentivirus, followed by selection with 2 µg/ml puromycin. Detailed sequences are presented in Table SI.
RNA separation and reverse transcription-quantitative PCR (RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was utilized to extract total RNA from CSCC cells. The purity of the RNA was measured using a UV spectrophotometer. Subsequently, RNA was placed in a refrigerator at -80°C for later use. cDNAs were synthesized using the PrimeScript™ Reverse Transcription reagent kit (Takara Bio, Inc.) at 37°C for 15 min and 85°C for 5 sec. The expression levels of circRNAs, mRNAs and miRNAs were then measured using an ABI 7900HT PCR instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.) by initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec and extension at 72°C for 90 sec. The 2−ΔΔCq method (21) was used to calculate the relative gene expression normalized by GAPDH and U6. The primers used in the present study are listed in Table SI.
RNase R digestion
Total RNAs (5 µg) from A431 and SCL-1 cells were incubated at 37°C for 15 min, and RNase R (Epicentre; Illumina, Inc.) was used to remove linear RNAs at the ratio of 6 units: 1 µg. After RNase R treatment, the expression of circ-0070934 was detected using RT-qPCR.
Sanger sequencing
The amplified PCR product was inserted into the T vector (TSINGKE Biological Technology) for Sanger sequencing. After determination of the full length sequence, different primers were constructed by Invitrogen (Shanghai, China). Sanger sequencing was performed by Realgene (Nanjing, China).
Western blotting detection
Cells in each group were collected and mixed into 1 ml prepared lysis buffer (Beyotime Institute of Biotechnology) in each culture dish, followed by 5 min of lysis on ice. Lysate solutions were collected using RIPA lysis buffer (Beyotime Institute of Biotechnology) to extract total proteins. Protein concentrations were measured by bicinchoninic acid (BCA) assay (Beyotime Institute of Biotechnology). Protein samples (80 µg/lane) were subjected to 10% SDS-PAGE and transferred onto a PVDF membrane, and the membranes were blocked for 1 h in 5% skimmed milk at room temperature and incubated with anti-HOXB7 (1:1,000; cat. no. 12613-1-AP; ProteinTech Group, Inc.) and anti-GAPDH (1:1,000; cat. no. AG019; Beyotime Institute of Biotechnology) antibodies at 4°C overnight. The washing reagent was TBS containing 0.1% Tween-20 (Beyotime Institute of Biotechnology). The next day, the membranes were incubated with anti-rabbit horseradish peroxidase (HRP)-conjugated (cat. no. A0208) or anti-mouse HRP-conjugated (cat. no. A0216) secondary antibodies (1:1,000; Beyotime Institute of Biotechnology) at 37°C for 2 h. Protein bands were detected on X-ray film using an enhanced chemiluminescence detection system (Amersham; Cytiva).
Bioinformatics prediction
The potential targets of circ-0070934 were predicted through bioinformatics analyses using RegRNA (http://regrna2.mbc.nctu.edu.tw/) and circinteractome (https://circinteractome.nia.nih.gov/). The potential target genes of miR-1236-3p were searched and intersected using TargetScan (http://www.targetscan.org/vert_72/) and miRDB (http://mirdb.org/).
Dual-luciferase reporter assay
The 3′UTR sequences of circ-0070934 and homeobox B7 (HOXB7) were downloaded from the NCBI website (https://www.ncbi.nlm.nih.gov/), and circ-0070934 wild-type (WT) 3′UTR and HOXB7 WT 3′UTR sequences, as well as circ-0070934 mutant (MUT) 3′UTR and HOXB7 MUT 3′UTR sequences were constructed. Subsequently, 5×103 cells A431 and SCL-1 cells were seeded onto 96-well plates and co-transfected with 80 ng WT or MUT plasmids and 50 pmol/l miR-1236-3p mimics or NC using Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) for 6 h at 37°C according to the manufacturer's instructions. At 48 h post-transfection, fluorescence intensity was detected using the dual-luciferase reporter gene detection system and normalized to that of Renilla luciferase (Promega Corporation).
Transwell invasion assay
Transwell assays were used to determine the invasive abilities of CSCC cells. The upper chamber was pre-coated with Matrigel (BD Biosciences) at 37°C for 30 min, and 4×105 transfected A431 and SCL-1 cells were placed in 100 µl DMEM without serum, whereas 500 µl DMEM containing 10% FBS was placed in the bottom chamber. After 24-h incubation at 37°C, the cells on the upper surface of the membrane were wiped using cotton swabs, and the culture medium was removed. Formaldehyde was used at room temperature for 10 min to fix the cells, which were subsequently stained using 0.5% crystal violet at room temperature for 30 min. The number of invaded cells was counted in 10 randomly selected fields of view under a light microscope (×200 magnification).
Cell proliferation assays
After transfection, A431 and SCL-1 cells were cultured in 96-well plates (100 µl/well) at 1×106 cells/ml. After 24 h, 10 µl Cell Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology) solution was added and incubated with 5% CO2 at 37°C for 1 h. Finally, the culture medium was removed, and the absorbance at was measured at 450 nm using the TECAN infinite M200 multimode micro-plate reader (Tecan Group, Ltd.).
Cells seeded in 96-well plates with 5×103 cells/well were labeled with 50 µM medium containing 5-ethynyl-2′-deoxy-uridine (EdU; Guangzhou Ribobio Co., Ltd.) for 2 h, fixed with 4% paraformaldehyde and 0.5% Triton X-100 and incubated with anti-EdU working solution according to the manufacturer's instructions. Cell nuclei were dyed with DAPI (Beyotime Institute of Biotechnology). A total of five randomly selected fields of view in each well were captured using fluorescence microscopy (×200 magnification) to calculate EdU-positive cells. The experiments were performed in triplicate.
Flow cytometry assay
At 24 h post-transfection, 1×106 A431 and SCL-1 cells were cultured in 6-well plates. After 48 h, Annexin V-FITC/Propidium Iodide kit (BD Biosciences) was used to stain the treated cells according to the manufacturer's instructions. Next, the samples were detected with a BeamCyte flow cytometer (Changzhou Beam Diagnostics Automation Co. Ltd.) and analyzed early and late apoptosis with CytoSYS 1.0 software (Changzhou Beam Diagnostics Automation Co. Ltd.). All assays were repeated three times independently.
Determination of localization by subcellular fractionation
The PARIS kit (Thermo Fisher Scientific, Inc.) was used to separate cytoplasmic and nuclear RNAs according to the manufacturer's protocol. Total RNA was separated from each fraction and determined using RT-qPCR, with U6 as a nuclear control marker and GAPDH as a cytoplasmic control marker.
RNA binding protein immunoprecipitation (RIP) assay
Magna RIP kit (EMD Millipore) was utilized to perform the RIP assay as previously described (22). The immunoprecipitation mixture was applied for RNA extraction and detection.
Mouse xenograft model
A total of 6 female BALB/c nude mice (age, 6 weeks; weight, 18-22 g) were purchased from the Model Animal Research Center of Nanjing University. Mice were housed in a sterile room under a 12-h light/dark cycle at ~23°C and 50% humidity, with ad libitum access to food and water. A total of 2×106 A431 cells transfected with hsa_circ_0070934 shRNA or NC were injected subcutaneously into the BALB/c nude mice. Tumor volumes were calculated every 4 days using the following formula: Tumor volume = (length × width2) / 2. At 4 weeks post-injection, the mice were anesthetized by intraperitoneal injection of sodium pentobarbital (40 mg/kg) and sacrificed by 10% formalin perfusion fixation of central nervous system; death was confirmed by complete stopping of the heartbeat and breathing, as well as disappearance of the foot withdrawal reflex. The tumor tissues were isolated and weighed. Then, the tumor tissues were analyzed using a TUNEL Apoptosis Detection kit (cat. no. C1086; Beyotime Institute of Biotechnology) according to the manufacturer's instructions. The study was approved by the Ethics Committee of The Affiliated Huaian No. 1 People's Hospital of Nanjing Medical University, and the experiments were performed following the National Institutes of Health guidelines on animal welfare.
Statistical analysis
SPSS 22.0 (IBM Corp.) and GraphPad Prism 6.0 (GraphPad Software, Inc.) were used for statistical processing. For normally distributed data with equal variance, the difference was evaluated by two-tailed Student's t-test (two group comparisons) or one-way ANOVA followed by the Bonferroni post hoc test (multigroup comparisons) as appropriate. For non-normally distributed data or data with unequal variances, the difference was evaluated by a nonparametric Mann-Whitney U test (two group comparisons) or the Kruskal-Wallis test followed by the Bonferroni post hoc test (multigroup comparisons). Data are presented as the mean ± SD. P<0.05 was considered to indicate a statistically signifi-cant difference.
Results
Expression of circ-0070934 in CSCC
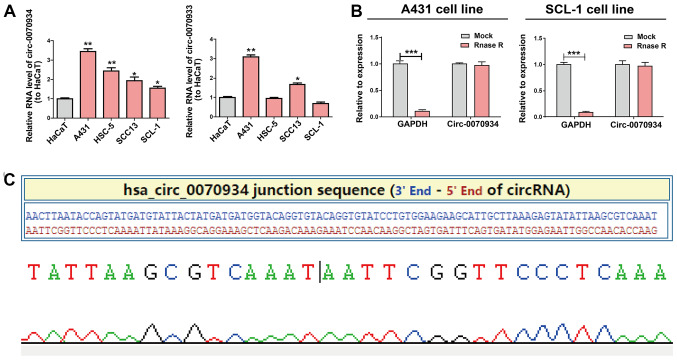
Based on the results of circRNA expression microarray (20), hsa_circ_0070933 and hsa_circ_0070934 were selected as the candidate circRNAs of interest. The expression patterns of these two circRNAs in CSCC cell lines and a human keratinocyte cell line (HaCaT) were examined using RT-qPCR. circ-0070934, but no circ-0070933, exhibited a stable high level in CSCC cell lines compared with those in HaCaT cells. (Fig. 1A). Thus, circ-0070934 was selected for subsequent experiments. RNase R was added to the total RNA samples to further determine the circular nature of circ-0070934, as RNase R dissolved linear RNAs that contained a free 3′ terminus, but did not affect circRNAs. The results revealed that circ-0070934 was indeed a circRNA with resistance to RNase R digestion (Fig. 1B). The circ-0070934 sequence was amplified using its primer and was identified to be identical to the sequence in circbase through Sanger sequencing (Fig. 1C).
Figure 1.
Expression of circ-0070934 in CSCC. (A) Expression levels of circ-0070934 in A431, SCL-1, HSC-5 and SCC13 CSCC cell lines and HaCaT cells were detected using reverse transcription-quantitative PCR. CSCC cell lines exhibited higher circ-0070934 levels compared with those in HaCaT cells. (B) Circ-0070934 is resistant to RNase R digestion. (C) The sequence of circ-0070934 in circBase was consistent with the results of Sanger sequencing. All experiments were conducted in triplicate. Data are reported as the mean ± SD. *P<0.05, **P<0.01 vs. HaCaT; ***P<0.001. SCSS, cutaneous squamous cell carcinoma; circ, circular RNA.
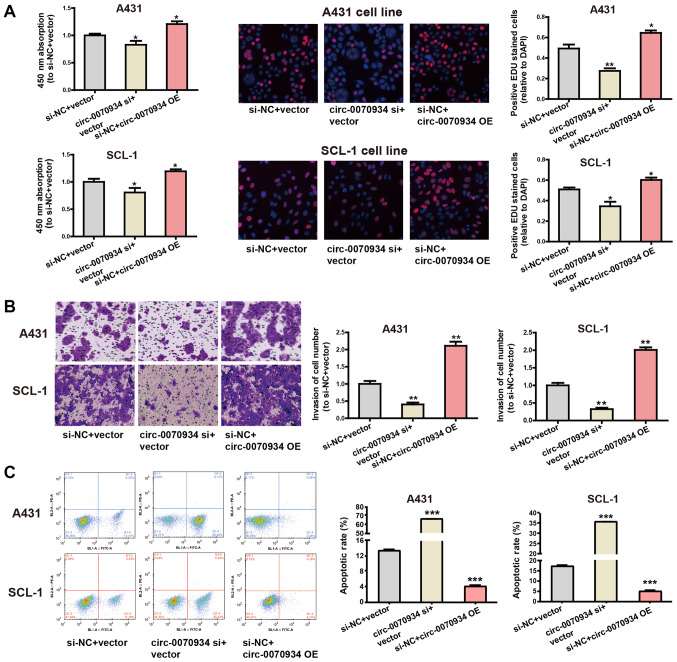
Overexpression of circ-0070934 stimulates the invasion and proliferation of CSCC cells and inhibits apoptosis
The expression of circ-0070934 was silenced using targeted siRNAs, and circ-0070934 overexpression plasmids were used for ectopic expression. After the circ-0070934 overexpression plasmids or siRNAs were transfected into CSCC cells, the transfection efficiency was determined by RT-qPCR (Fig. S1A). According to CCK-8 and EdU assays, knockdown of circ-0070934 decreased the rate of cell proliferation, whereas circ-0070934 overexpression enhanced the proliferative capacity of CSCC cells compared with that of the control cells (Fig. 2A). In addition, Transwell assay results demonstrated that knockdown of circ-0070934 inhibited the invasive capability of CSCC cells, whereas circ-0070934 overexpression promoted the invasive capability of CSCC cells compared with that of the control cells (Fig. 2B). Flow cytometric analysis of apoptosis revealed that knockdown of circ-0070934 promoted CSCC cell apop-tosis, which was suppressed by circ-0070934 overexpression compared with that in the control cells (Fig. 2C). Overall, these results indicated that elevated circ-0070934 enhanced the proliferative and invasive capabilities of CSCC cells, as well as inhibited apoptosis.
Figure 2.
Cytobiological changes of A431 and SCL-1 cells transfected with circ-0070934 siRNA or overexpression plasmids. (A) Knockdown of circ-0070934 suppressed cell proliferation, whereas circ-0070934 overexpression exerted the opposite effect. Cell Counting Kit-8 assays were performed to determine cell proliferation (left). Typical images of proliferating cells in EdU assays are presented (middle). The proliferating cells were quantified (right). (B) Transwell assays demonstrated that the knockdown of circ-0070934 inhibited cell invasion, whereas circ-0070934 overexpression had the opposite effect. (C) Flow cytometry results indicated that knockdown of circ-0070934 increased apoptosis compared with that in the control group, whereas circ-0070934 overexpression has an adverse effect. All experiments were conducted in triplicate. Data are reported as the mean ± SD. *P<0.05, **P<0.01, ***P<0.001 vs. siNC + vector. Circ, circular RNA; si, small interfering RNA; OE, overexpression vector.
Circ-0070934 serves as a sponge for miR-1236-3p
To further analyze the exact mechanisms of action through which circ-0070934 acts in CSCC, the subcellular localization of circ-0070934 was determined using separation experiments in the cytoplasm and the nucleus. The results demonstrated that circ-0070934 was primarily located within the cytoplasm of CSCC cells (Fig. 3A). This observation suggested that circ-0070934 may function through post-translational modifications. Considering that circRNAs can serve as miRNA sponge (23), the potential targets of circ-0070934 were predicted through bioinformatics analyses (RegRNA, circinteractome), and miR-1236-3p was indicated to be the potential complementary miRNA that can bind to circ-0070934 (Fig. 3B). To determine the association between circ-0070934 and the predicted miRNA, miR-1236-3p expression levels in CSCC cell lines and HaCaT cells were measured; the results of RT-qPCR demonstrated that compared with that in HaCaT, miR-1236-3p expression in the CSCC cell lines was downregulated (Fig. 3C). In addition, the results of the RIP binding assay demonstrated that the levels of circ-0070934 and miR-1236-3p were higher in the anti-Ago2 group than that in the anti-normal IgG group, which indicated that circ-0070934 and miR-1236-3p were in the same RNA-induced silencing complex (Fig. 3D). Subsequently, plasmids containing the MUT or WT circ-0070934 sequences were constructed (Fig. 3E). Dual-luciferase reporter gene assays demonstrated that cells co-transfected with circ-0070934 WT and miR-1236-3p mimics exhibited reduced luciferase activity compared with that in the control groups (Fig. 3F). However, the relative luciferase activity was not notably different between the cells co-transfected with circ-0070934 MUT and miR-1236-3p mimics and those transfected with the NC (Fig. 3F). These results indicated that circ-0070934 may serve as a collator for miR-1236-3p.
Figure 3.
circ-0070934 serves as a collator for miR-1236-3p. (A) Circ-0070934 is primarily located in the cytoplasm. (B) A Venn diagram of the results of the bioinformatics analysis. (C) miR-1236-3p expression is lower in CSCC cell lines compared with that in HaCaT cells. (D) RIP assay demonstrated that circ-0070934 and miR-1236-3p were enriched in products pulled down by anti-AGO2. (E) The binding sequence of WT circ-0070934 and miR-1236-3p was predicted, and the MUT circ-0070934 sequence was constructed. (F) Dual-luciferase reporter gene experiments were conducted to examine the binding between miR-1236-3p and circ-0070934 in CSCC cells. All experiments were conducted in triplicate. Data are reported as the mean ± SD. *P<0.05, **P<0.01, ***P<0.001 vs. HaCaT or as indicated. CSCC, cutaneous squamous cell carcinoma; miR, microRNA; WT, wild-type; MUT, mutant.
Circ-0070934 regulates the miR-1236-3p target gene HOXB7
The potential target genes of miR-1236-3p were searched and intersected by bioinformatics analysis (TargetScan, miRDB). It was identified that the binding site between HOXB7 and miR-1236-3p was highly consistent with that of circ-0070934 and miR-1236-3p. In order to further examine the interactions between miR-1236-3p and HOXB7, pGL3-HOXB7-WT and pGL3-HOXB7-MUT plasmids were constructed (Fig. 4A), which were then used to transfect CSCC cells. Compared with the control group, cells co-transfected with pGL3-HOXB7-WT and miR-1236-3p mimics exhibited significantly decreased luciferase activity; however, luciferase activity did not change in cells co-transfected with pGL3-HOXB7-MUT and miR-1236-3p mimics (Fig. 4B). Subsequently, the mRNA levels of HOXB7 in the CSCC cell lines was examined. Compared with HaCaT, the CSCC cell lines exhibited higher HOXB7 expression levels (Fig. 4C). Similarly, the expression levels of the HOXB7 protein in A431 and SCL-1 cells were also increased compared with those in HaCaT cells (Fig. 4D). These results suggested that HOXB7 was the target gene of miR-1236-3p.
Figure 4.
Circ-0070934 regulates HOXB7 by sequestering miR-1236-3p. (A) The binding sequence of WT HOXB7 and miR-1236-3p was predicted, and the MUT HOXB7 sequence was constructed. (B) Dual-luciferase reporter gene experiments demonstrated that HOXB7 interacted with miR-1236-3p. (C) The expression levels of HOXB7 were upregulated in cutaneous squamous cell carcinoma cell lines compared with those in HaCaT cells. (D) Western blotting analysis revealed that compared with those in HaCaT cells, the expression levels of the HOXB7 protein were significantly elevated in A431 and SCL-1 cells. All experiments were conducted in triplicate. Data are reported as the mean ± SD. *P<0.05, **P<0.01 vs. HaCaT or as indicated. Circ, circular RNA; HOXB7, homeobox B7; miR, microRNA; WT, wild-type; MUT, mutant.
Following from these observations, the present study explored whether circ-0070934 was able to bind miR-1236-3p to mediate the expression of HOXB7. In cells transfected with the circ-0070934 siRNA, the protein and mRNA expression levels of HOXB7 were decreased compared with those in cells transfected with the NC (Fig. 5A and B). However, co-transfection with the miR-1236-3p inhibitor prevented the circ-0070934 siRNA-induced downregulation of HOXB7 (Fig. 5A and B). In cells treated with the miR-1236-3p mimics, the mRNA and protein expression levels of HOXB7 were reduced, but this reduction was reversed by co-transfection with circ-0070934 overex-pression plasmids (Fig. 5C and D). Overall, these results demonstrated that circ-0070934 upregulated HOXB7 expression by suppressing miR-1236-3p.
Figure 5.
The circ-0070934/miR-1236-3p regulatory axis is crucial for the expression of HOXB7. (A and B) After CSCC cells were transfected with circ-0070934 siRNAs with or without miR-1236-3p inhibitors, the mRNA and protein expression levels of HOXB7 mRNA and protein were determined, with GAPDH used as a control. (C and D) After CSCC cells were transfected with miR-1236-3p mimics with or without circ-0070934 overexpression plasmids, the mRNA and protein expression levels of HOXB7 were detected. All experiments were conducted in triplicate. Data are reported as the mean ± SD. **P<0.01. CSCC, cutaneous squamous cell carcinoma; circ, circular RNA; miR, microRNA; HOX7B, homeobox 7B; siRNA, small interfering RNA; NC, negative control; inh, inhibitor; OE, overexpression plasmid.
The circ-0070934/miR-1236-3p regulatory axis is pivotal for CSCC cell function
The role of miR-1236-3p in CSCC cell function was analyzed. The effects of transfections with the miRNA mimics or inhibitors on miR-1236-3p expression levels in CSCC cells was determined by RT-qPCR (Fig. S1B). Cell proliferative and invasive capabilities were impeded by miR-1236-3p mimics, whereas the overexpression of circ-0070934 exerted the opposite effect (Fig. 6A-C). In addition, the miR-1236-3p mimics promoted CSCC cell apop-tosis, whereas overexpression of circ-0070934 suppressed apoptosis (Fig. 6D). These results further identified the regulatory role of the circ-0070934/miR-1236-3p/HOXB7 axis on CSCC cell function.
Figure 6.
The circ-0070934/miR-1236-3p regulatory axis is pivotal for cutaneous squamous cell carcinoma cell function. (A and B) Cell Counting Kit-8 and EdU assays were performed to determine the changes in the proliferative capability of cells transfected with miR-1236-3p mimics. This change was reversed by co-transfection with circ-0070934 overexpression plasmids. (C) Transwell assays were conducted to determine the changes in the invasive capability of cells transfected with miR-1236-3p mimics; co-transfection with circ-0070934 overexpression plasmids reversed this change. (D) Flow cytometry was used to assess the apoptotic rates of cells transfected with miR-1236-3p mimics; the effect was reversed by co-transfection with circ-0070934 overexpression plasmids. All experiments were conducted in triplicate. Results are reported as the mean ± SD. *P<0.05, **P<0.01, ***P<0.001. Circ, circular RNA; miR, microRNA; NC, negative control; OE, overexpression plasmid.
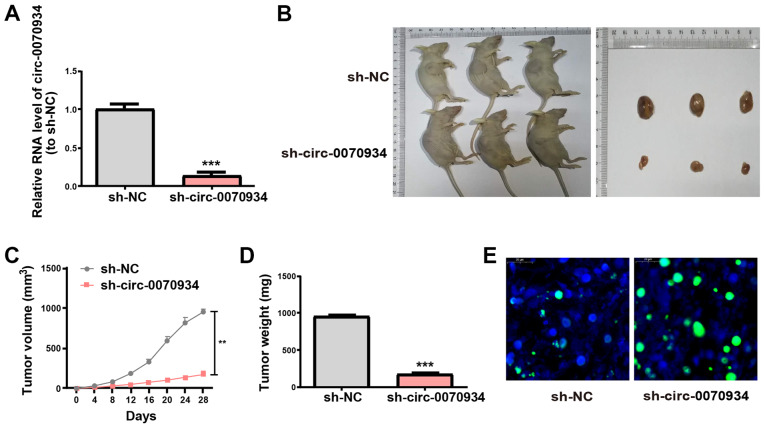
Knockdown of circ-0070934 in tumors inhibits CSCC growth
To investigate the role of circ-0070934 in CSCC tumor growth in vivo, A431 cells transfected with scrambled or circ-0070934 shRNAs were subcutaneously injected into nude mice. After the scrambled or circ-0070934 shRNAs were transfected into A431 cells, the transfection efficiency was detected (Fig. 7A). The results of the xenograft assay demonstrated that knockdown of circ-0070934 decreased the tumor volume and weight after four weeks after tumor inoculation (Fig. 7B-D). In addition, TUNEL assays demonstrated that xenograft tumors from the circ-0070934 shRNA group appeared to exhibit higher levels of apoptosis compared with those from the sh-NC group (Fig. 7E). These results were consistent with the in vitro results of the present study.
Figure 7.
Knockdown of circ-0070934 in tumors inhibits cutaneous squamous cell carcinoma growth. (A) After A431 cells are treated with scrambled or circ-0070934 shRNAs, the mRNA levels of circ-0070934 were detected by reverse transcription-quantitative PCR. (B) Representative images of xenograft tumors in nude mice (n=3 mice/group). (C) Tumor volumes were monitored every 4 days for 4 weeks. (D) The weights of excised xenograft tumors at the end of the experiment. (E) Apoptosis levels in xenograft tumors of the sh-NC or sh-circ-0070934 groups were detected using TUNEL assay. Data are reported as the mean ± SD. **P<0.01, ***P<0.001 vs. sh-NC. Circ, circular RNA; shRNA, short hairpin RNA; NC, negative control.
Discussion
Previous studies have demonstrated that genes have various methods of transcriptional regulation. For example, miRNA, as an important regulatory factor, is a short-chain RNA with a length of ~22 nt that can serve a negative regulatory role on target gene expression by blocking the degradation and translation of the target gene (24). Numerous ncRNAs also share binding sites with miRNAs, appearing to collate miRNAs within cells, thus preventing miRNAs from inhibiting their target genes and subsequently allowing for the upregulation of the expression levels of the target gene; this mechanism of action is termed the ceRNA mechanism (25). Numerous recent studies have reported that circRNAs can regulate the invasion and proliferation of tumor cells in a similar manner to that of ceRNAs (26-29); however, limited attention has been paid to the mechanism of circRNA action in CSCC as ceRNAs. Thus, the present study aimed to investigate whether circ-0070934 may be used as a ceRNA to affect CSCC cell proliferation and invasion.
The results of the present study demonstrated that circ-0070934 expression levels in various CSCC cell lines were higher compared with those in the normal human kerati-nocyte cell line HaCaT. This result is consistent with a previous study, which has demonstrated that circ-0070934 expression is notably increased in CSCC samples compared with that in the corresponding control samples (20); however, the effect of circ-0070934 on CSCC cell function has yet to be confirmed. Additionally, knockdown of circ-0070934 expression inhibited the invasion and proliferation of CSCC cells and stimulated apoptosis compared with that in the negative control group. This suggested that circ-0070934 may be a pivotal factor, positively modulating CSCC cell proliferation and serving as an oncogene. Therefore, investigating the mechanism of action behind the effects of circ-0070934 on CSCC cell proliferation may be valuable for evaluating the occurrence, development and metastasis of CSCC.
Through separation of RNAs in the nucleus and the cytoplasm, the present study verified that the subcellular localization of circ-0070934 was primarily in the cytoplasm, indicating that circ-0070934 may act as a ceRNA. RIP assays demonstrated that circ-0070934 and miR-1236-3p were in the same RNA-induced silencing complex. And subsequent dual-luciferase reporter assays demonstrated that circ-0070934 was a molecular collator that upregulated HOXB7 expression through competitively binding miR-1236-3p. Previous studies have demonstrated that miR-1236-3p exerts an inhibitory effect on numerous tumor types such as lung adenocarcinoma, gastric and bladder cancer (30-32). Wang et al (33) have demonstrated that miR-1236-3p targets ZEB1 in high-grade serous ovarian carcinoma, reducing the migratory and invasive abilities of the cells (33). The results of the present study demonstrated that miR-1236-3p was downregulated in CSCC cell lines, and that the miR-1236-3p mimics inhibited CSCC cell invasion and proliferation as well as promoted apoptosis. In addition, HOXB7 was expressed at a higher level in CSCC cell lines compared with that in the normal control. Transfection with circ-0070934 siRNAs reduced the expression of HOXB7, which was consistent with the inhibitory effect of the miR-1236-3p mimics on HOXB7 expression levels. Of note, the invasive and proliferative capabilities of CSCC cells were inhibited by the miR-1236-3p mimics compared with those in the NC group, and this inhibition was reversed by overexpressing circ-0070934. In addition, the miR-1236-3p mimics increased the apoptotic rates of CSCC cells, whereas co-transfection with circ-0070934 overexpression plasmids rescued apoptosis. These results suggested that the circ-0070934/miR-1236-3p/HOXB7 regulatory axis may be involved in the occurrence and development of CSCC by modulating the invasive and proliferative abilities of CSCC cells and regulating their apoptosis.
HOX genes encode the transcription factor family that is crucial for growth regulation and differentiation in the process of embryonic development and maintaining adult tissue homeostasis (34) These genes serve roles in tumor cell migration, proliferation, invasion and apoptosis, and are often abnormally regulated in cancer (35,36). A total of 39 HOX genes are classified into chromosomal clusters (A, B, C and D) in humans, with each ~100 kb long, and they are located on chromosomes 7, 17, 2 and 12, respectively (37). The transcription factor HOXB7, a member of class I HOX genes, exerts a pivotal effect on tumorigenesis in several types of cancer, including gastric (38), pancreatic (39), lung (40), oral squamous (41) and breast (42) cancers. Tu et al (43) have reported that HOXB7 upregulation is associated with poor prognosis in patients with gastric cancer. Gao and Chen (44) have demonstrated that specific HOXB7 knockdown impedes CSCC cell migration and invasion and triggers apoptosis through the Wnt/β-catenin signaling pathway. The results of the present study demonstrated that overexpression of circ-0070934 led to an increase in the expression levels of HOXB7, a target of miR-1236-3p, which may result in abnormal proliferation, invasion and apoptosis of CSCC cells. However, the present study had a number of limitations. Firstly, tissue samples from patients with CSCC are required to further explore the clinical value of circ-0070934. Secondly, in situ hybridization fluorescence would be valuable to verify the association between circ-0070934 and miR-1236-3p in future studies. Additionally, whether there are other target genes or miRNAs which can interact with circ-0070934 needs to be explored.
In conclusion, the present study revealed the significance of circ-0070934 modulation of HOXB7 expression levels by acting as a ceRNA and collating miR-1236-3p in the pathogenesis of CSCC. Thus, further studies investigating circRNAs may be clinically valuable for diagnosing and treating CSCC and other diseases.
Supplementary Data
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
DWZ and CRZ designed the experiments. DWZ and HYW performed the experiments. CRZ and DDW wrote the manuscript. All authors discussed the results and revised the manuscript. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of The Affiliated Huaian No. 1 People's Hospital of Nanjing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Apalla Z, Lallas A, Sotiriou E, Lazaridou E, Ioannides D. Epidemiological trends in skin cancer. Dermatol Pract Concept. 2017;7:1–6. doi: 10.5826/dpc.0702a01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leiter U, Eigentler T, Garbe C. Epidemiology of skin cancer. Adv Exp Med Biol. 2014;810:120–140. doi: 10.1007/978-1-4939-0437-2_7. [DOI] [PubMed] [Google Scholar]
- 3.Newlands C, Currie R, Memon A, Whitaker S, Woolford T. Non-melanoma skin cancer: United Kingdom National Multidisciplinary Guidelines. J Laryngol Otol. 2016;130(Suppl 2):S125–S132. doi: 10.1017/S0022215116000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satgunaseelan L, Chia N, Suh H, Virk S, Ashford B, Lum T, Ranson M, Clark J, Gupta R. p16 expression in cutaneous squamous cell carcinoma of the head and neck is not associated with integration of high risk HPV DNA or prognosis. Pathology. 2017;49:494–498. doi: 10.1016/j.pathol.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Takahara M, Xie L, Takeuchi S, Tu Y, Nakahara T, Uchi H, Moroi Y, Furue M. Levels of the EMT-related protein Snail/Slug are not correlated with p53/p63 in cutaneous squamous cell carcinoma. J Cutan Pathol. 2013;40:651–656. doi: 10.1111/cup.12142. [DOI] [PubMed] [Google Scholar]
- 6.Hu X, Liu Y, Ai P, He S, Liu L, Chen C, Tan Y, Wang T. MicroRNA-186 promotes cell proliferation and inhibits cell apoptosis in cutaneous squamous cell carcinoma by targeting RETREG1. Exp Ther Med. 2019;17:1930–1938. doi: 10.3892/etm.2019.7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mei XL, Zhong S. Long noncoding RNA LINC00520 prevents the progression of cutaneous squamous cell carcinoma through the inactivation of the PI3K/Akt signaling pathway by downregulating EGFR. Chin Med J (Engl) 2019;132:454–465. doi: 10.1097/CM9.0000000000000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions - beyond repression of gene expression. Nat Rev Genet. 2014;15:599–612. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 9.Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov. 2016;6:235–246. doi: 10.1158/2159-8290.CD-15-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hubé F, Ulveling D, Sureau A, Forveille S, Francastel C. Short intron-derived ncRNAs. Nucleic Acids Res. 2017;45:4768–4781. doi: 10.1093/nar/gkw1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leisegang MS, Fork C, Josipovic I, Richter FM, Preussner J, Hu J, Miller MJ, Epah J, Hofmann P, Günther S, et al. Long Noncoding RNA MANTIS Facilitates Endothelial Angiogenic Function. Circulation. 2017;136:65–79. doi: 10.1161/CIRCULATIONAHA.116.026991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang Y, Xiao Z, Siverly AN, Lawhon SE, Ton BN, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018;50:1705–1715. doi: 10.1038/s41588-018-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu LP, He YJ, Hou JC, Chen X, Zhou SY, Yang SJ, Li J, Zhang HD, Hu JH, Zhong SL, et al. The role of circRNAs in cancers. Biosci Rep. 2017;37:BSR20170750. doi: 10.1042/BSR20170750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreeva K, Cooper NG. MicroRNAs in the Neural Retina. Int J Genomics. 2014;2014:165897. doi: 10.1155/2014/165897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patop IL, Kadener S. circRNAs in Cancer. Curr Opin Genet Dev. 2018;48:121–127. doi: 10.1016/j.gde.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greene J, Baird AM, Brady L, Lim M, Gray SG, McDermott R, Finn SP. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front Mol Biosci. 2017;4:38. doi: 10.3389/fmolb.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan X, Weng X, Zhao Y, Chen W, Gan T, Xu D. Circular RNAs in Cardiovascular Disease: An Overview. BioMed Res Int. 2017;2017:5135781. doi: 10.1155/2017/5135781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheng JQ, Liu L, Wang MR, Li PY. Circular RNAs in digestive system cancer: Potential biomarkers and therapeutic targets. Am J Cancer Res. 2018;8:1142–1156. [PMC free article] [PubMed] [Google Scholar]
- 19.Su H, Tao T, Yang Z, Kang X, Zhang X, Kang D, Wu S, Li C. Circular RNA cTFRC acts as the sponge of MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer. 2019;18:27. doi: 10.1186/s12943-019-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sand M, Bechara FG, Gambichler T, Sand D, Bromba M, Hahn SA, Stockfleth E, Hessam S. Circular RNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci. 2016;83:210–218. doi: 10.1016/j.jdermsci.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 21.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 22.Bi W, Huang J, Nie C, Liu B, He G, Han J, Pang R, Ding Z, Xu J, Zhang J. CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of β-catenin pathway. J Exp Clin Cancer Res. 2018;37:275. doi: 10.1186/s13046-018-0936-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su Y, Xu C, Liu Y, Hu Y, Wu H. Circular RNA hsa_circ_0001649 inhibits hepatocellular carcinoma progression via multiple miRNAs sponge. Aging (Albany NY) 2019;11:3362–3375. doi: 10.18632/aging.101988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Towler BP, Jones CI, Newbury SF. Mechanisms of regulation of mature miRNAs. Biochem Soc Trans. 2015;43:1208–1214. doi: 10.1042/BST20150157. [DOI] [PubMed] [Google Scholar]
- 25.Conte F, Fiscon G, Chiara M, Colombo T, Farina L, Paci P. Role of the long non-coding RNA PVT1 in the dysregulation of the ceRNA-ceRNA network in human breast cancer. PLoS One. 2017;12:e0171661. doi: 10.1371/journal.pone.0171661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen G, Shi Y, Zhang Y, Sun J. CircRNA_100782 regulates pancreatic carcinoma proliferation through the IL6-STAT3 pathway. OncoTargets Ther. 2017;10:5783–5794. doi: 10.2147/OTT.S150678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Zhang S, Wu J, Cui J, Zhong L, Zeng L, Ge S. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36:4551–4561. doi: 10.1038/onc.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.He JH, Li YG, Han ZP, Zhou JB, Chen WM, Lv YB, He ML, Zuo JD, Zheng L. The CircRNA-ACAP2/Hsa-miR-21-5p/ Tiam1 Regulatory Feedback Circuit Affects the Proliferation, Migration, and Invasion of Colon Cancer SW480 Cells. Cell Physiol Biochem. 2018;49:1539–1550. doi: 10.1159/000493457. [DOI] [PubMed] [Google Scholar]
- 29.Ma HB, Yao YN, Yu JJ, Chen XX, Li HF. Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. Am J Transl Res. 2018;10:592–604. [PMC free article] [PubMed] [Google Scholar]
- 30.Bian T, Jiang D, Liu J, Yuan X, Feng J, Li Q, Zhang Q, Li X, Liu Y, Zhang J. miR-1236-3p suppresses the migration and invasion by targeting KLF8 in lung adenocarcinoma A549 cells. Biochem Biophys Res Commun. 2017;492:461–467. doi: 10.1016/j.bbrc.2017.08.074. [DOI] [PubMed] [Google Scholar]
- 31.An JX, Ma MH, Zhang CD, Shao S, Zhou NM, Dai DQ. miR-1236-3p inhibits invasion and metastasis in gastric cancer by targeting MTA2. Cancer Cell Int. 2018;18:66. doi: 10.1186/s12935-018-0560-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manthei U, Nickells MW, Barnes SH, Ballard LL, Cui WY, Atkinson JP. Identification of a C3b/iC3 binding protein of rabbit platelets and leukocytes. A CR1-like candidate for the immune adherence receptor. J Immunol. 1988;140:1228–1235. [PubMed] [Google Scholar]
- 33.Wang Y, Yan S, Liu X, Zhang W, Li Y, Dong R, Zhang Q, Yang Q, Yuan C, Shen K, et al. miR-1236-3p represses the cell migration and invasion abilities by targeting ZEB1 in high-grade serous ovarian carcinoma. Oncol Rep. 2014;31:1905–1910. doi: 10.3892/or.2014.3046. [DOI] [PubMed] [Google Scholar]
- 34.Miksiunas R, Mobasheri A, Bironaite D. Homeobox Genes and Homeodomain Proteins: New Insights into Cardiac Development, Degeneration and Regeneration. Adv Exp Med Biol. 2020;1212:155–178. doi: 10.1007/5584_2019_349. [DOI] [PubMed] [Google Scholar]
- 35.Carrera M, Bitu CC, de Oliveira CE, Cervigne NK, Graner E, Manninen A, Salo T, Coletta RD. HOXA10 controls proliferation, migration and invasion in oral squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8:3613–3623. [PMC free article] [PubMed] [Google Scholar]
- 36.Hur H, Lee JY, Yun HJ, Park BW, Kim MH. Analysis of HOX gene expression patterns in human breast cancer. Mol Biotechnol. 2014;56:64–71. doi: 10.1007/s12033-013-9682-4. [DOI] [PubMed] [Google Scholar]
- 37.Holland PW. Evolution of homeobox genes. Wiley Interdiscip Rev Dev Biol. 2013;2:31–45. doi: 10.1002/wdev.78. [DOI] [PubMed] [Google Scholar]
- 38.Joo MK, Park JJ, Yoo HS, Lee BJ, Chun HJ, Lee SW, Bak YT. The roles of HOXB7 in promoting migration, invasion, and anti-apoptosis in gastric cancer. J Gastroenterol Hepatol. 2016;31:1717–1726. doi: 10.1111/jgh.13330. [DOI] [PubMed] [Google Scholar]
- 39.Tsuboi M, Taniuchi K, Shimizu T, Saito M, Saibara T. The transcription factor HOXB7 regulates ERK kinase activity and thereby stimulates the motility and invasiveness of pancreatic cancer cells. J Biol Chem. 2017;292:17681–17702. doi: 10.1074/jbc.M116.772780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monterisi S, Lo Riso P, Russo K, Bertalot G, Vecchi M, Testa G, Di Fiore PP, Bianchi F. HOXB7 overexpression in lung cancer is a hallmark of acquired stem-like phenotype. Oncogene. 2018;37:3575–3588. doi: 10.1038/s41388-018-0229-9. [DOI] [PubMed] [Google Scholar]
- 41.Wang K, Jin J, Ma T, Zhai H. MiR-376c-3p regulates the proliferation, invasion, migration, cell cycle and apoptosis of human oral squamous cancer cells by suppressing HOXB7. Biomed Pharmacother. 2017;91:517–525. doi: 10.1016/j.biopha.2017.04.050. [DOI] [PubMed] [Google Scholar]
- 42.Heinonen H, Lepikhova T, Sahu B, Pehkonen H, Pihlajamaa P, Louhimo R, Gao P, Wei GH, Hautaniemi S, Jänne OA, et al. Identification of several potential chromatin binding sites of HOXB7 and its downstream target genes in breast cancer. Int J Cancer. 2015;137:2374–2383. doi: 10.1002/ijc.29616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tu W, Zhu X, Han Y, Wen Y, Qiu G, Zhou C. Overexpression of HOXB7 is associated with a poor prognosis in patients with gastric cancer. Oncol Lett. 2015;10:2967–2973. doi: 10.3892/ol.2015.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao D, Chen HQ. Specific knockdown of HOXB7 inhibits cutaneous squamous cell carcinoma cell migration and invasion while inducing apoptosis via the Wnt/beta-catenin signaling pathway. Am J Physiol Cell Physiol. 2018;315:C675–C86. doi: 10.1152/ajpcell.00291.2017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.