Abstract
Unstructured clinical narratives are continuously being recorded as part of delivery of care in electronic health records, and dedicated tagging staff spend considerable effort manually assigning clinical codes for billing purposes. Despite these efforts, however, label availability and accuracy are both suboptimal. In this retrospective study, we aimed to automate the assignment of top-level International Classification of Diseases version 9 (ICD-9) codes to clinical records from human and veterinary data stores using minimal manual labor and feature curation. Automating top-level annotations could in turn enable rapid cohort identification, especially in a veterinary setting. To this end, we trained long short-term memory (LSTM) recurrent neural networks (RNNs) on 52,722 human and 89,591 veterinary records. We investigated the accuracy of both separate-domain and combined-domain models and probed model portability. We established relevant baseline classification performances by training Decision Trees (DT) and Random Forests (RF). We also investigated whether transforming the data using MetaMap Lite, a clinical natural language processing tool, affected classification performance. We showed that the LSTM-RNNs accurately classify veterinary and human text narratives into top-level categories with an average weighted macro F1 score of 0.74 and 0.68 respectively. In the “neoplasia” category, the model trained on veterinary data had a high validation accuracy in veterinary data and moderate accuracy in human data, with F1 scores of 0.91 and 0.70 respectively. Our LSTM method scored slightly higher than that of the DT and RF models. The use of LSTM-RNN models represents a scalable structure that could prove useful in cohort identification for comparative oncology studies. Digitization of human and veterinary health information will continue to be a reality, particularly in the form of unstructured narratives. Our approach is a step forward for these two domains to learn from and inform one another.
Introduction
Motivation
The increasingly worldwide adoption of electronic health records (EHRs) has populated clinical databases with millions of clinical narratives (descriptions of actual clinical practice). However, given the nature of the medical enterprise, a big portion of the data being recorded is in the form of unstructured free-text clinical notes. Cohorts of individuals with similar clinical characteristics require quality phenotype labels, oftentimes not readily available alongside clinical notes, to be studied adequately.
In place of such labeling, diagnostic codes are the most common surrogates to true phenotypes. In routine clinical practice, dedicated tagging staff read clinical narratives and assign these diagnostic codes to patients’ diagnoses from one or both of two coding systems: the International Classification of Diseases (ICD) and the Systematized Nomenclature of Medicine (SNOMED) [1]. However, this time-consuming, error-prone task leads to only 60–80% of the assigned codes reflecting actual patient diagnoses [2], misjudgment of severity of conditions, and/or omission of codes altogether. The relative inaccuracy of oncological medical coding [3–6] affects the quality of cancer registries [7] and cancer prevalence calculations [8–10], for example. Poorly-defined disease types and poorly-trained coding staff who overuse the “not otherwise specified” code when classifying text exacerbate the problem.
Challenges in clinical coding also exist in veterinary medicine in the United States, where neither clinicians nor medical coders regularly apply diagnosis codes to veterinary visits. There are few incentives for veterinary clinicians to annotate their records; a lack of 1) a substantial veterinary third-party payer system and 2) legislation enforcing higher standards of veterinary EHRs (the U.S. Health Information Technology for Economic and Clinical Health Act of 2009 sets standards for human EHRs) compound the problem. Billing codes are thus rarely applicable across veterinary institutions unless hospitals share the same management structure and records system; even then, hospital-specific modifications exist. Less than five academic veterinary centers of a total of thirty veterinary schools in the United States have dedicated medical coding staff to annotate records using SNOMED-CT-Vet [11], a veterinary extension of SNOMED constructed by the American Animal Hospital Association (AAHA) and maintained by the Veterinary Terminology Services Laboratory at the Virginia-Maryland Regional College of Veterinary Medicine [12].
The vast majority of veterinary clinical data is stored as free-text fields with very low rates of formal data curation, making data characterization a tall order. Further increasing variance in the data, veterinary patients come from many different environments, including hospitals [13], practices [14], zoos [15], wildlife reserves [16], army facilities [17], research facilities [18], breeders, dealers, exhibitors [19], livestock farms, and ranches [20].
It is thus important that a general method, agnostic of patient environment, is able to categorize EHRs for cohort identification solely based on free-text.
A primer on automatic text classification
Automatic text classification is an emerging field that uses a combination of tools such as human medical coding, rule-based systems queries [21], natural language processing (NLP), statistical analyses, data mining, and machine learning (ML) [22]. In a previous study [23], we have shown the feasibility of automatic annotation of veterinary clinical narratives across a broad range of diagnoses with minimal preprocessing, but further exploration is needed to probe what we can learn from human-veterinary comparisons. Automatically adding meaningful disease-related tags to human and veterinary clinical notes using the same machinery would be a huge step forward in that exploration and could facilitate cross-species findings downstream.
Said integration has the potential to improve both veterinary and human coding accuracy as well as comparative analyses across species. Comparative oncology, for example, has accelerated the development of novel human anti-cancer therapies through the study of companion animals [24], especially dogs [25–28]. The National Institute of Health recently funded a multi-center institution called the Arizona Cancer Evolution Center (ACE) that aims to integrate data from a broad array of species to understand the evolutionarily conserved basis for oncology. As this group utilizes animal clinical and pathology data to identify helpful traits like species-specific cancer resistance, they would greatly benefit from improved cohort discovery through automated record tagging.
15 out of 30 veterinary schools across the United States have formed partnerships with their respective medical schools in order to perform cross-species translational research within the Clinical and Translational Science Award One Health Alliance (COHA, [29]). Of these schools, only two have active programs to assign disease codes to their medical records. The data for the rest represents the very use case of automatic text classification.
Automatic medical text classification aims to reduce the human burden of handling unstructured clinical narratives. These computational NLP methods can be divided into two groups: a) semantic processing and subsequent ML; and b) deep learning.
Semantic processing and subsequent ML
Semantic processing methods range from simple dictionary-based keyword-matching techniques and/or direct database queries to tools capable of interpreting the semantics of human language through lemmatization (removal of inflectional word endings), part-of-speech tagging, parsing, sentence breaking, word segmentation, and entity recognition [30]. Building the underlying dictionaries and manually crafting the rules that capture these diverse lexical elements both require time and domain expertise.
There is a growing interest in medical concept classification for clinical text; as such, many domain-specific semantic NLP tools (with various objectives, frameworks, licensing conditions, source code availabilities, language supports, and learning curves) have been developed for the medical setting. Such tools include MedLEE [31], MPLUS [32], MetaMap [33], KMCI [34], SPIN [35], HITEX [36], MCVS [37], ONYX [38], MedEx [39], cTAKES [40], pyConTextNLP [41], Topaz [42], TextHunter [43], NOBLE [44], and CLAMP [45]. However, there is no single NLP tool that can handle the broad problem of general medical concept classification. Instead, each method solves specific problems and applies its unique set of constraints.
After clinical narratives are fed into the above semantic NLP tools, various clinical “concepts” or “terms” (e.g. conditions, diseases, age, body parts, periods of time, etc.) are extracted. These concepts can then be represented in a “term-document matrix,” which shows frequencies of the terms across documents. These frequencies can be used raw as features in a ML model, but more often than not, choices are made to transform these features to more meaningful spaces. This can be done via term frequency-inverse document frequency (tf-idf, which assigns weights to terms based on the frequency of the term in both the document of interest as well as the corpus at large), other vectorization techniques like Word2Vec [46], or manually curated rules.
Predictive ML models (like Decision Trees [DTs], Random Forests [RFs], and Support Vector Machines [SVMs] [47]) that operate on the raw or transformed term-document matrix use this training data (input features and “ground-truth” labels) to make accurate predictions or decisions on unseen test data without explicit instructions on how to do so. They have been shown to achieve high classification accuracy in human [48, 49] and veterinary [50] free-text narratives for diseases well-represented in training datasets (e.g. diabetes, influenza, and diarrhea). However, these models generally do not classify under-represented diseases or conditions well, and opportunities for both public data generation and methodological innovation lie in this space [50].
Deep learning
Deep learning (DL) methods eliminate the need of feature engineering, harmonization, or rule creation. They learn hierarchical feature representations from raw data in an end-to-end fashion, requiring significantly less domain expertise than traditional ML approaches [51].
DL is quickly emerging in the literature as a viable alternative method to traditional ML for the classification of clinical narratives [47]. The technique can help in the recognition of a limited number of categories from biomedical text [52, 53]; identify psychiatric conditions of patients based on short clinical histories [54]; and accurately classify whether or not radiology reports indicate pulmonary embolism [55, 56] whilst outperforming baseline non-DL-based methods (e.g. RFs or DTs). Previous studies have shown the possibility of using DL to label clinical narratives with medical subspecialties [57] (e.g. cardiology or neurology) or medical conditions [58] (e.g. advanced cancer or chronic pain), outperforming concept-extraction based methods. Furthermore, the use of DL to analyze clinical narratives has also facilitated the prediction of relevant patient attributes, such as in-hospital mortality, 30-day unplanned readmission, prolonged length of stay, and final discharge diagnosis [59].
Traditional NLP methods boast interpretability and flexibility but come at the steep cost of data quality control, formatting, normalization, domain knowledge, and time needed to generate meaningful heuristics (which oftentimes are not even generalizable to other datasets). Automatic text classification using DL is thus a logical choice to bypass these steps, classifying medical narratives from EHRs by solely leveraging big data. We expect that our efforts could facilitate rapid triaging of documents and cohort identification for biosurveillance.
Materials and methods
Ethics statement
This research was reviewed and approved by Stanford’s Institutional Review Board (IRB), which provided a non-human subject determination under eProtocol 46979. Consent was not required.
Study design
This retrospective cross-sectional chart review study uses medical records collected routinely as part of clinical care from two clinical settings: the veterinary teaching hospital at Colorado State University (CSU) and the Medical Information Mart for Intensive Care (MIMIC-III) from the Beth Israel Deaconess Medical Center in Boston, Massachusetts [60]. Both datasets were divided in two smaller datasets—training datasets containing 70% of the original datasets (used to build TensorFlow [61] DL models), and validation datasets containing 30% of the original datasets.
The goal of our model was to predict top-level ICD version 9 (ICD-9) codes, which we considered our “ground-truth” labels. These codes are organized in a hierarchical fashion, with the top levels representing the grossest possible descriptors of clinical diseases or conditions (e.g., “neoplasia”). The MIMIC-III dataset provides ICD-9 codes for all its patients as-is. However, veterinary codes from the CSU were coded using SNOMED-CT, and thus needed to be converted to their closest equivalent top-level ICD-9 codes. Mapping between SNOMED-CT and ICD-9 codes was a challenging task but promoted semantic interoperability between our two domains. Table 1 shows our mapping between ICD (versions 9 and 10) codes and their counterparts in SNOMED-CT (including the Veterinary extension, SNOMED-CT-Vet). This mapping, which then allowed us to generate “ground-truth” labels for the veterinary data,was manually curated by two board-certified veterinarians trained in clinical coding (co-authors AZM and RLP). We also wanted to investigate the effect of using MetaMap [33], a NLP tool that extracts clinically-relevant terms, on our clinical narratives. Specifically, we explored whether or not training our models on these “MetaMapped” free-texts (that only contain extracted UMLS terms) would improve accuracy.
Table 1. Top-level coding mapping between ICD-9, ICD-10, and SNOMED-CT.
| Top-level category | Description | ICD-9 | ICD-10 | SNOMED-CT |
|---|---|---|---|---|
| 1 | Infectious and parasitic diseases | 001-139 | A00-B99 | 105714009, 68843000, 78885002, 344431000009103, 338591000009108, 40733004, 17322007 |
| 2 | Neoplasms | 140-239 | C00-D49 | 723976005, 399981008 |
| 3 | Endocrine, nutritional and metabolic diseases, and immunity disorders | 240-279 | E00-E90 | 85828009, 414029004, 473010000, 75934005, 363246002, 2492009, 414916001, 363247006, 420134006, 362969004 |
| 4 | Diseases of blood and blood-forming organs | 280-289 | D50-D89 | 271737000, 414022008, 414026006, 362970003, 11888009, 212373009, 262938004, 405538007 |
| 5 | Mental disorders | 290-319 | F00-F99 | 74732009 |
| 6 | Diseases of the nervous system | 320-359 | G00-G99 | 118940003, 313891000009106 |
| 7 | Diseases of sense organs | 360-389 | H00-H59, H60-H95 | 50611000119105, 87118001, 362966006, 128127008, 85972008 |
| 8 | Diseases of the circulatory system | 390-459 | I00-I99 | 49601007 |
| 9 | Diseases of the respiratory system | 460-519 | J00-J99 | 50043002 |
| 10 | Diseases of the digestive system | 520-579 | K00-K93 | 370514003, 422400008, 53619000 |
| 11 | Diseases of the genitourinary system | 580-629 | N00-N99 | 42030000 |
| 12 | Complications of pregnancy, childbirth, and the puerperium | 630-679 | O00-O99 | 362972006, 173300003, 362973001 |
| 13 | Diseases of the skin and subcutaneous tissue | 680-709 | L00-L99 | 404177007, 414032001, 128598002 |
| 14 | Diseases of the musculoskeletal system and connective tissue | 710-739 | M00-M99 | 105969002, 928000 |
| 15 | Congenital anomalies | 740-759 | Q00-Q99 | 111941005, 32895009, 66091009 |
| 16 | Certain conditions originating in the perinatal period | 760-779 | P00-P96 | 414025005 |
| 17 | Injury and poisoning | 800-899 | S00-T98 | 85983004, 75478009, 77434001, 417163006 |
Mapping of top-level categories was manually curated by two board-certified veterinarians trained in clinical coding.
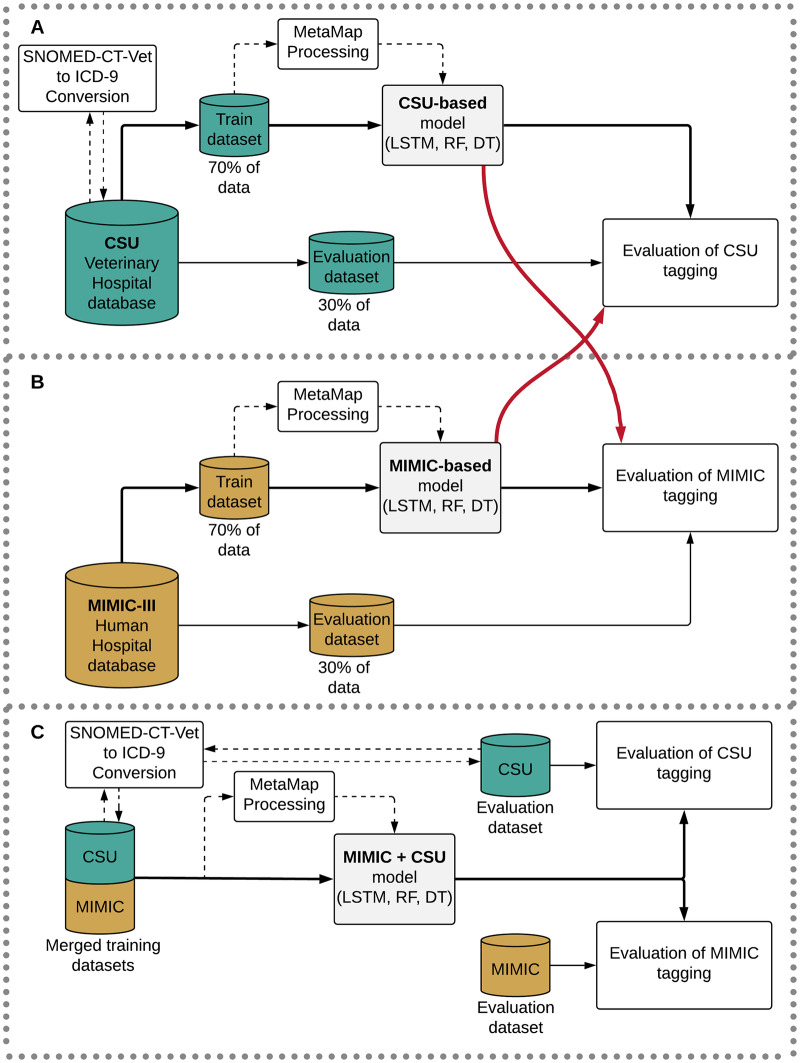
Finally, we measured the validation accuracy of the models, calculating the F1 score of our model and relevant non-DL baselines in each top-level disease category. We also explored the possibility of out-of-domain generalization, testing the MIMIC-trained model on the CSU validation data and vice versa (and ran separate tests for “MetaMapped” versions of each dataset, as well). Finally, we investigated the effect of merging the MIMIC and CSU training datasets to test the efficacy of data augmentation. Fig 1 shows a diagram of our study design. Our code to run all models can be found in a public repository (https://github.com/rivas-lab/FasTag).
Fig 1. Diagram of the training and evaluation design.
Relevant acronyms: MIMIC: Medical Information Mart for Intensive Care; CSU: Colorado State University; MetaMap, a tool for recognizing medical concepts in text; LSTM: long-short term memory recurrent neural network classifier; RF: Random Forest classifier; DT: Decision Tree classifier.
Data
Veterinary medical hospital at Colorado State University (CSU)
The CSU is a tertiary care referral teaching hospital with inpatient and outpatient facilities, serving all specialties of veterinary medicine. After consultation, veterinarians enter patient information into a custom-built veterinary EHR, including structured fields such as entry and discharge dates, patient signalment (species, breed, age, sex, etc.), and SNOMED-CT-Vet codes. There are also options to input free-text clinical narratives with various sections including history, assessment, diagnosis, prognosis, and medications. These records are subsequently coded; the final diagnostic codes represent single or multiple specific diagnoses or post-coordinated expressions (a combination of two or more concepts). For our study, we used the free-text clinical narratives (data input) and the SNOMED-CT-Vet codes (labels), which we subsequently converted to ICD-9 top-level codes as per Table 1.
Medical Information Mart for Intensive Care (MIMIC-III)
The Beth Israel Deaconess Medical Center is a tertiary care teaching hospital at Harvard Medical School in Boston, Massachusetts. The MIMIC-III database, a publicly available dataset which we utilize in this study, contains information on patients admitted to the critical care unit at the hospital [60]. These records are coded for billing purposes and have complete diagnoses per patient (the database is publicly available, and thus represents the best possible medical coding annotation scenario for a hospital). We were interested in the free-text hospital discharge summaries (data input) and the corresponding ICD-9 codes (labels) for the patients in this database. Free-text fields in MIMIC-III have been de-identified to protect privacy.
Comparing the sources
The CSU dataset contains medical records from 33,124 patients and 89,591 hospital visits between February 2007 and July 2017. Patients encompassed seven mammalian species, including dogs (Canis Lupus, 80.8%), cats (Felis Silvestris, 11.4%), horses (Equus Caballus, 6.5%), cattle (Bos Taurus, 0.7%), pigs (Sus Scrofa, 0.3%), goats (Capra hircus, 0.2%), sheep (Ovis Aries, 0.1%), and other unspecified mammals (0.1%). In contrast, the MIMIC-III database contains medical records from 38,597 distinct human adult patients (aged 16 years or above) and 7,870 neonates admitted between 2001 and 2008, encompassing 52,722 unique hospital admissions to the critical care unit between 2001 and 2012. Table 2 summarizes the category breakdowns of both databases. For this analysis, only those patients with a diagnosis in their record were considered.
Table 2. Database statistics of patients, records, and species (records with diagnosis).
| CSU | MIMIC | |
|---|---|---|
| Data | ||
| Medical Records | 89,591 | 52,722 |
| Patients | 33,124 | 41,126 |
| Hospital Visits | 89,591 | 49,785 |
| Species | ||
| Humans (Homo Sapiens) | n.a. | 52,722 |
| Dogs (Canis Lupus) | 72,420 | n.a. |
| Cats (Felis Silvestris) | 10,205 | n.a. |
| Horses (Equus Caballus) | 5,819 | n.a. |
| Other mammals | 1,147 | n.a. |
| Category | ||
| Infectious | 11,454 | 10,074 |
| Neoplasia | 36,108 | 6,223 |
| Endo-Immune | 17,295 | 24,762 |
| Blood | 10,171 | 13,481 |
| Mental | 511 | 10,989 |
| Nervous | 7,488 | 9,168 |
| Sense organs | 15,085 | 2,688 |
| Circulatory | 8,733 | 30,054 |
| Respiratory | 11,322 | 17,667 |
| Digestive | 22,776 | 14,646 |
| Genitourinary | 8,892 | 14,932 |
| Pregnancy | 136 | 133 |
| Skin | 21,147 | 4,241 |
| Musculoskeletal | 22,921 | 6,739 |
| Congenital | 3,347 | 2,334 |
| Perinatal | 54 | 3,661 |
| Injury | 9,873 | 16,121 |
The mappings in Table 1 were used to generate the categories and numbers presented here in Table 2. The seventeen categories represent the text classification labels.
MetaMap
Our hypothesis was that there would be a plethora of extraneous information, domain- and setting-specific misspellings, abbreviations, and jargon in the clinical narratives in both human and veterinary settings. In order to potentially resolve some of these issues, we used MetaMap Lite [62], a NLP tool which leverages the Unified Medical Language System (UMLS) Metathesaurus to identify SNOMED [63] or ICD-9 [64] codes from clinical narratives. MetaMap’s algorithm includes five steps: 1) parsing of text into simple noun phrases; 2) variant generation of phrases to include all derivations of words (i.e. synonyms, acronyms, meaningful spelling variants, combinations, etc.); 3) candidate retrieval of all UMLS strings that contains at least one variant from the previous step; 4) evaluation and ranking of each candidate, mapping between matched term and the Metathesaurus concept using metrics of centrality, variation, coverage, and cohesiveness; 5) construction of complete mappings to include those mappings that are involved in disjointed parts of the phrase (e.g. ‘ocular’ and ‘complication’ can together be mapped to a single term, ‘ocular complication’). MetaMap incorporates the use of ConText [65], an algorithm for the identification of negation in clinical narratives.
We proceeded to make a “MetaMapped” version of each training and validation dataset to see whether extracting and inputting only clinical terms from the narratives into the models would increase their accuracy. For additional information and examples of how we used and evaluated MetaMap, please refer to S1 Text and S1–S3 Tables.
Model architecture
We chose a long short-term memory (LSTM) recurrent neural network (RNN) architecture (which is able to handle variable-length sequences while using previous inputs to inform current time steps) for this multi-label text classification task [66]. The LSTM shares parameters across time steps as it unrolls, which allows it to handle sequences of variable length. In this case, these sequences are a series of word “embeddings” (created by mapping specific words to corresponding numeric vectors) from clinical narratives. Words are represented densely (rather than sparsely, as in Bag-of-Words or tf-idf models) using the Global Vectors for Word Representation (GloVe) [67] word embeddings. These embeddings learn a vector space representation of words such that words with similar contexts appear in a similar vector space and also capture global statistical features of the training corpus.
LSTMs have proven to be flexible enough to be used in many different tasks, such as machine translation, image captioning, medication prescription, and forecasting disease diagnosis using structured data [66]. The RNN can efficiently capture sequential information and theoretically model long-range dependencies, but empirical evidence has shown this is difficult to do in practice [68]. The sequential nature of text lends itself well to LSTMs, which have memory cells that can maintain information for over multiple time steps (words) and consist of a set of gates that control when information enters and exits memory, making them an ideal candidate architecture.
We first trained FasTag, the LSTM model, over a variety of hyperparameters on MIMIC data and calculated the model’s validation accuracy over all combinations of them, finding the set of [learning rate = 0.001, dropout rate = 0.5, batch size = 256, training epochs = 100, hidden layer size = 200, LSTM layers = 1] to be the optimal setting. We proceeded to use this hyperparameter set for all FasTag models trained, assuming that this set would be amenable to the task at hand regardless of training dataset. We then proceeded to train a set of six models on three datasets (MIMIC, CSU, and MIMIC+CSU) where each dataset had a version that was processed with MetaMap and a version that was not.
Evaluation
We aimed to characterize the performance of FasTag in both absolute and relative senses by establishing its empirical classification accuracy and the accuracy of non-DL alternatives. Several ML classifiers have similarly aimed to classify clinical narratives [47, 69]. We selected two of these classifiers (DTs and RFs) as relevant non-DL baseline comparator methods. DTs are ML models constructed around a branching boolean logic [70]. Each node in the tree can take a decision that leads to other nodes in a tree structure; there are no cycles allowed. The RF classifier is an ensemble of multiple DTs created by randomly selecting samples of the training data. The final prediction is done via a consensus voting mechanism of the trees in the forest.
We featurized the narratives using tf-idf, a statistic that reflects word importance in the context of other documents in a corpus and a standard ML modeling strategy for representing text, to convert the narratives into a text-document matrix [47]. The hyperparameters of both baseline models (DT and RF), like for FasTag, were tuned on the validation set.
For all models we trained (FasTag, DT, and RF), we used the same validation set evaluation metrics previously reported for MetaMap [62]: a) precision, defined as the proportion of documents which were assigned the correct category; b) recall, defined as the proportion of documents from a given category that were correctly identified; and c) F1 score, defined as the harmonic average of precision and recall. Formulas for these metrics are provided below:
| (1) |
| (2) |
| (3) |
Our task is framed as a multi-label classification problem, where each approach predicts multiple top-level ICD-9 categories for each observation using a single model. In order to combine all class-specific F1 scores, we averaged the F1 score for each label, weighting the labels by their supports (the number of true instances for each label, to account for label imbalance).
Domain adaptation
The portability of trained algorithms on independent domains has previously been used as a metric of model robustness in systems that leverage NLP and ML [71]. We evaluated the ability of our trained FasTag LSTM models to be used in a cross-species context. We utilized the MIMIC-trained model to classify the medical records in the CSU database and vice versa, assessing performance as before. We also assessed the classifier trained on the combined training set.
Results
We investigated the application of FasTag to free-text unstructured clinical narratives on two cohorts: veterinary medical records from CSU, and human medical records in the MIMIC-III database.
We trained FasTag (as well as DT/RF baselines) on the human, veterinary, and merged (human and veterinary) datasets and tested each on their own domain as well as the other domains. We built FasTag using the Python programming language (version 2.7), TensorFlow [61] (version 1.9), and the scikit-learn library (version 0.19.2) [72]; we built the baselines using Python and scikit-learn as well. The training was performed on an Amazon® Deep Learning AMI, a cloud-based platform running the Ubuntu operating system with pre-installed CUDA dependencies. FasTag’s training procedure was epoch-based; that is, our data was split into “batches” of size 256, and we calculated cross-entropy loss and updated the model using the Adam optimizer after each of these batches were input into the model. An “epoch” is said to have finished every time the entire dataset has passed through the model in batches. As is standard with most epoch-based model-training procedures, we trained FasTag until our validation loss increased between epochs three consecutive times. For the DT and RF baselines, we performed validation-set model selection across a grid of hyperparameters, including: information criterion; max features; max depth (of tree[s]); number of estimators; and tf-idf vector normalization type (L1 or L2). Average weighted macro F1 scores for models across all categories are shown in Table 3; a full list of F1 scores by category can be found in S4 Table. The “neoplasia” category results, which we found notable, are shown in Table 4.
Table 3. Average F1 scores using various training and validation dataset combinations for all categories.
| Configuration | Model evaluation (Weighted F1 score) | ||||
|---|---|---|---|---|---|
| Training | Validation | MetaMap | DT | RF | LSTM |
| MIMIC | MIMIC | No | 0.60 | 0.64 | 0.65 |
| Yes | 0.60 | 0.63 | 0.70 | ||
| CSU | CSU | No | 0.55 | 0.61 | 0.72 |
| Yes | 0.54 | 0.60 | 0.75 | ||
| MIMIC | CSU | No | 0.22 | 0.24 | 0.28 |
| Yes | 0.23 | 0.20 | 0.31 | ||
| CSU | MIMIC | No | 0.31 | 0.20 | 0.23 |
| Yes | 0.28 | 0.19 | 0.36 | ||
| MIMIC + CSU | CSU | No | 0.57 | 0.62 | 0.67 |
| Yes | 0.57 | 0.62 | 0.76 | ||
| MIMIC + CSU | MIMIC | No | 0.60 | 0.63 | 0.58 |
| Yes | 0.60 | 0.63 | 0.60 | ||
| MIMIC + CSU | MIMIC + CSU | No | 0.59 | 0.64 | 0.68 |
| Yes | 0.59 | 0.63 | 0.71 | ||
| Average | 0.489 | 0.506 | 0.571 | ||
Evaluation metrics for Decision Tree (DT), Random Forest (RF), and the FasTag Long Short Term Memory (LSTM) Recurrent Neural Network on validation datasets with and without MetaMap term extraction. Bolded and underlined numbers represent the best scores for the specific configuration of training data, validation data, and MetaMap toggle.
Table 4. F1 scores using various training and validation dataset combinations for the “neoplasia” category.
| Configuration | Model evaluation (Weighted F1 score) | ||||
|---|---|---|---|---|---|
| Training | Validation | MetaMap | DT | RF | LSTM |
| MIMIC | MIMIC | No | 0.39 | 0.45 | 0.66 |
| Yes | 0.4 | 0.45 | 0.76 | ||
| CSU | CSU | No | 0.81 | 0.86 | 0.91 |
| Yes | 0.8 | 0.86 | 0.91 | ||
| MIMIC | CSU | No | 0.3 | 0.53 | 0.69 |
| Yes | 0.45 | 0.37 | 0.75 | ||
| CSU | MIMIC | No | 0.46 | 0.58 | 0.70 |
| Yes | 0.5 | 0.58 | 0.54 | ||
| MIMIC + CSU | CSU | No | 0.74 | 0.8 | 0.87 |
| Yes | 0.74 | 0.8 | 0.87 | ||
| MIMIC + CSU | MIMIC | No | 0.4 | 0.47 | 0.67 |
| Yes | 0.42 | 0.45 | 0.72 | ||
| MIMIC + CSU | MIMIC + CSU | No | 0.81 | 0.86 | 0.85 |
| Yes | 0.81 | 0.86 | 0.90 | ||
| Average | 0.574 | 0.637 | 0.771 | ||
Evaluation metrics for the “neoplasia” category Decision Tree (DT), Random Forest (RF), and the FasTag Long Short Term Memory (LSTM) Recurrent Neural Network on validation datasets with and without MetaMap term extraction. Bolded and underlined numbers represent the best scores for the specific configuration of training data, validation data, and MetaMap toggle.
Discussion
Applying DL to unstructured free-text clinical narratives in electronic health records offers a relatively simple, low-effort means to bypass the traditional bottlenecks in medical coding. Circumventing the need for data harmonization was very important for the datasets, which contained a plethora of domain- and setting-specific misspellings, abbreviations, and jargon (these issues would have greatly impacted the performance of standard ML models, and indeed, these were the cause of misclassifications by FasTag, as well). MetaMap was useful in this regard given its ability to parse clinical data, but much work is still needed to improve recognition of terms in veterinary and human domains (as evidenced by only low-to-moderate gains, and in some cases, losses, in performance in “MetaMapped” datasets [S4 Table]).
There is moderate evidence of domain adaptation (where a model trained on MIMIC data is useful in the CSU validation set, or vice versa) in the “neoplasia” category, with F1 scores of 0.69-0.70 (Table 4). This process involved training a model on the data in one database and testing on the data in the other without fine-tuning. It is evident that the high classification accuracy (F1 score = 0.91) obtained by the CSU model in the neoplasia category is decreased when testing the same model on the MIMIC data. One possible explanation is the difference in clinical settings; CSU is a tertiary care veterinary hospital specializing in oncological care, and the clinical narratives that arise in a critical care unit like in the MIMIC dataset do not necessarily compare. Moreover, the records were not coded in the same way, the clinicians did not receive the same training, and the documents apply to different species altogether (see S3 Table for an example of an example narrative unique to veterinary care). Despite these differences, however, our LSTM model was general enough to be able to accurately classify medical narratives at the top level of depth independently in both datasets. The achieved cross-domain accuracy is thus nonetheless encouraging. Given enough training data and similar-enough clinical narratives, one could conceivably imagine a general model that is highly effective across domains.
Models performed usually better on their respective validation datasets in those categories with more training samples. For example, the CSU-trained model (25,276 samples) had significantly better performance in the “neoplasia” category than the MIMIC-trained model (4,356 samples), while the MIMIC-trained model (21,038 samples) had better performance in the diseases of the circulatory system category than the CSU-trained model (6,133 samples). The corollary to this is that the biggest impediment to model performance within a category was the lack of training data. Unlike in the genetics community, where there exist hundreds of thousands of research samples available to researchers through DUAs [73], there is definitely a dearth of de-identified clinical text narratives alongside quality labels like in MIMIC-III. Along the same vein, when training on a mixed dataset of MIMIC and CSU data, we observed that the performance of the resultant classifier was significantly better on CSU than MIMIC validation data across various top-level categories (S4 Table). We hypothesize that a combination of the inherent differences in data across the two domains and the larger number of CSU records in the training set led to this performance gap. We additionally hypothesize that mixing training data from more similar data sources, in contrast, would result in strictly better performance outcomes on test data from both sources.
Insights gained through this work on generalizing across clinical and veterinary domains could be informative in training models attempting to generalize across different clinical institutions but within the same clinical domain. Linguistic variation within a clinical domain is due to factors like geography, clinical specialty, and patient population, among others. This variation manifests across many characteristics such as syntax [74–76], semantics [77], and workflow procedures [78]. The common practices to address domain heterogeneity are to re-train models from scratch [78] or to utilize domain adaptation techniques like distribution mapping [79] for the task of interest. Overall, we hypothesize that adapting models across clinical institutions will bear better results than when adapting them across clinical domains (like we have attempted in this work).
The usefulness of even top-level characterizations in the veterinary setting cannot be understated; usually, a veterinarian must read the full, unstructured text in order to get any information about the patient they are treating. Rapid selection of documents with specific types of clinical narratives (such as oncological cases, which our model performed well on) could lead to better cohort studies for comparative research. The repeated use of a series of such LSTM models for subsequent, increasingly-specific classifications thus represents a scalable, hierarchical tagging structure that could prove extremely useful in stratifying patients by specific diseases, severities, and protocols.
Conclusion
In this era of increasing deployment of EHRs, it is important to provide tools that facilitate cohort identification. Our deep learning approach, FasTag, was able to automatically classify medical narratives with minimal human preprocessing. In a future with enough training data, it is possible to foresee a scenario in which these models can accurately tag every clinical concept, regardless of data input. The expansion of veterinary data availability and the subsequently enormous potential of domain adaptation like we saw in the neoplasia category could prove to be exciting chapters in reducing bottlenecks in public health research at large; it is thus of critical importance to continue studying novel sources of data that can rapidly be used to augment classification models.
A reliable addition to existing rule-based and natural language processing strategies, deep learning is a promising tool for accelerating public health research.
Supporting information
(DOCX)
Comparison of reviewer and MetaMap term extractions for 19 records. True Positive (TP) = MetaMap correctly matched term to code; False Negative (FN) = MetaMap did not extract a term the experts did; False Positive (FP) = MetaMap matched a term not identified by the expert; Total = Total number of terms matched in that document.
(XLSX)
(XLSX)
A 2-year old female dog patient with recurrent otitis and allergic dermatitis. Both the narrative (left) and the “MetaMapped” version (right) show that the treatment included prednisone (among other important clinical details). For the purposes of this manuscript, the pet and owner’s name were manually de-identified.
(XLSX)
Sheet 1: Long Short Term Memory (LSTM) Recurrent Neural Network (RNN) [FasTag]; Sheet 2: Decision Trees (DT); Sheet 3: Random Forests (RF).
(XLSX)
Training with CSU data (green), MIMIC data (yellow) or MIMIC+CSU (purple); validating on CSU data (Panel A) or MIMIC data (Panel B). The color of the category text is darkened (black) and the box made bigger if it surpasses the threshold of an F1 score of at least 0.70 (dotted horizontal line).
(TIF)
Acknowledgments
The authors wish to acknowledge Dr. Katie M. Kanagawa for her valuable support in editing this manuscript and Devin Johnson, DVM, MS, for her contribution to clinical coding and comparison with coding from the MetaMap tool.
Data Availability
The veterinary data presented here belongs to the Colorado State University (CSU), which may grant access to this data on a case-by-case basis to researchers who obtain the necessary Data Use Agreement (DUA) and IRB approvals. The CSU Research division, which maintains the data, can be contacted at cvmbs-research@colostate.edu. Inquiries about access to the data should be directed to Dr. Tim Hackett, Senior Associate Dean for Veterinary Health Systems, College of Veterinary Medicine and Biomedical Sciences, CSU (tim.hackett@colostate.edu). The human data presented here belongs to the Beth Israel Deaconess Medical Center in Boston, Massachusetts and can be accessed after signing a DUA with the MIT Lab for Computational Physiology at https://mimic.physionet.org/gettingstarted/access.
Funding Statement
M.A.R. is supported by Stanford University and a National Institute of Health center for Multi and Trans-ethnic Mapping of Mendelian and Complex Diseases grant (5U01 HG009080). This work was supported by National Human Genome Research Institute (NHGRI) of the National Institutes of Health (NIH) under awards R01HG010140. C.D.B. is a Chan Zuckerberg Biohub Investigator. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Moriyama IM, Loy RM, Robb-Smith AHT, Rosenberg HM, Hoyert DL. History of the statistical classification of diseases and causes of death; 2011. [Google Scholar]
- 2. Benesch C, Witter DM Jr, Wilder AL, Duncan PW, Samsa GP, Matchar DB. Inaccuracy of the International Classification of Diseases (ICD-9-CM) in identifying the diagnosis of ischemic cerebrovascular disease. Neurology. 1997;49(3):660–664. 10.1212/WNL.49.3.660 [DOI] [PubMed] [Google Scholar]
- 3. Abraha I, Serraino D, Giovannini G, Stracci F, Casucci P, Alessandrini G, et al. Validity of ICD-9-CM codes for breast, lung and colorectal cancers in three Italian administrative healthcare databases: a diagnostic accuracy study protocol: Table 1; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim SC, Gillet VG, Feldman S, Lii H, Toh S, Brown JS, et al. Validation of claims-based algorithms for identification of high-grade cervical dysplasia and cervical cancer. Pharmacoepidemiol Drug Saf. 2013;22(11):1239–1244. 10.1002/pds.3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moar KK, Rogers SN. Impact of coding errors on departmental income: an audit of coding of microvascular free tissue transfer cases using OPCS-4 in UK. Br J Oral Maxillofac Surg. 2012;50(1):85–87. 10.1016/j.bjoms.2011.01.005 [DOI] [PubMed] [Google Scholar]
- 6. Friedlin J, Overhage M, Al-Haddad MA, Waters JA, Aguilar-Saavedra JJR, Kesterson J, et al. Comparing methods for identifying pancreatic cancer patients using electronic data sources. AMIA Annu Symp Proc. 2010;2010:237–241. [PMC free article] [PubMed] [Google Scholar]
- 7. German RR, Wike JM, Bauer KR, Fleming ST, Trentham-Dietz A, Namiak M, et al. Quality of cancer registry data: findings from CDC-NPCR’s Breast and Prostate Cancer Data Quality and Patterns of Care Study. J Registry Manag. 2011;38(2):75–86. [PubMed] [Google Scholar]
- 8. Paviot BT, Gomez F, Olive F, Polazzi S, Remontet L, Bossard N, et al. Identifying prevalent cases of breast cancer in the French case-mix databases. Methods Inf Med. 2011;50(02):124–130. 10.3414/ME09-01-0064 [DOI] [PubMed] [Google Scholar]
- 9. Fisher BT, Harris T, Torp K, Seif AE, Shah A, Huang YSV, et al. Establishment of an 11-Year Cohort of 8733 Pediatric Patients Hospitalized at United States Free-standing Children’s Hospitals With De Novo Acute Lymphoblastic Leukemia From Health Care Administrative Data; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Polednak AP, Phillips C. Cancers coded as tongue not otherwise specified: relevance to surveillance of human papillomavirus-related cancers. J Registry Manag. 2014;41(4):190–195. [PubMed] [Google Scholar]
- 11. Maccabe AT, Crawford L, Heider LE, Hooper B, Mann CJ, Pappaioanou M. Association of American Veterinary Medical Colleges (AAVMC): 50 Years of History and Service. J Vet Med Educ. 2015;42(5):395–402. 10.3138/jvme.0615-089R [DOI] [PubMed] [Google Scholar]
- 12.Virginia-Maryland Regional College of Veterinary Medicine. Research Resources: Virginia-Maryland Regional College of Veterinary Medicine. Virginia Polytechnic Institute and State University; 1993.
- 13. Cummings KJ, Rodriguez-Rivera LD, Mitchell KJ, Hoelzer K, Wiedmann M, McDonough PL, et al. Salmonella enterica serovar Oranienburg outbreak in a veterinary medical teaching hospital with evidence of nosocomial and on-farm transmission. Vector Borne Zoonotic Dis. 2014;14(7):496–502. 10.1089/vbz.2013.1467 [DOI] [PubMed] [Google Scholar]
- 14. Krone LM, Brown CM, Lindenmayer JM. Survey of electronic veterinary medical record adoption and use by independent small animal veterinary medical practices in Massachusetts. J Am Vet Med Assoc. 2014;245(3):324–332. 10.2460/javma.245.3.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Witte CL, Lamberski N, Rideout BA, Fields V, Teare CS, Barrie M, et al. Development of a case definition for clinical feline herpesvirus infection in cheetahs (Acinonyx jubatus) housed in zoos. J Zoo Wildl Med. 2013;44(3):634–644. 10.1638/2012-0183R.1 [DOI] [PubMed] [Google Scholar]
- 16. Griffith JE, Higgins DP. Diagnosis, treatment and outcomes for koala chlamydiosis at a rehabilitation facility (1995–2005). Aust Vet J. 2012;90(11):457–463. 10.1111/j.1751-0813.2012.00963.x [DOI] [PubMed] [Google Scholar]
- 17. Poppe JL. The US Army Veterinary Service 2020: knowledge and integrity. US Army Med Dep J. 2013; p. 5–10. [PubMed] [Google Scholar]
- 18. Committee AMR, Field K, Bailey M, Foresman LL, Harris RL, Motzel SL, et al. Medical records for animals used in research, teaching, and testing: public statement from the American College of Laboratory Animal Medicine. ILAR J. 2007;48(1):37–41. 10.1093/ilar.48.1.37 [DOI] [PubMed] [Google Scholar]
- 19. Shalev M. USDA to require research facilities, dealers, and exhibitors to keep veterinary medical records. Lab Anim. 2003;32(6):16. [DOI] [PubMed] [Google Scholar]
- 20. Robinson TP, Wint GRW, Conchedda G, Van Boeckel TP, Ercoli V, Palamara E, et al. Mapping the global distribution of livestock. PLoS One. 2014;9(5):e96084 10.1371/journal.pone.0096084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gundlapalli AV, Redd D, Gibson BS, Carter M, Korhonen C, Nebeker J, et al. Maximizing clinical cohort size using free text queries; 2015. [DOI] [PubMed] [Google Scholar]
- 22. Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc. 2014;21(2):221–230. 10.1136/amiajnl-2013-001935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nie A, Zehnder A, Page RL, Zhang Y, Pineda AL, Rivas MA, et al. DeepTag: inferring diagnoses from veterinary clinical notes. NPJ Digit Med. 2018;1:60 10.1038/s41746-018-0067-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garden OA, Volk SW, Mason NJ, Perry JA. Companion animals in comparative oncology: One Medicine in action. Vet J. 2018;240:6–13. 10.1016/j.tvjl.2018.08.008 [DOI] [PubMed] [Google Scholar]
- 25. Saba C, Paoloni M, Mazcko C, Kisseberth W, Burton JH, Smith A, et al. A Comparative Oncology Study of Iniparib Defines Its Pharmacokinetic Profile and Biological Activity in a Naturally-Occurring Canine Cancer Model. PLoS One. 2016;11(2):e0149194 10.1371/journal.pone.0149194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. LeBlanc AK, Mazcko CN, Khanna C. Defining the Value of a Comparative Approach to Cancer Drug Development. Clin Cancer Res. 2016;22(9):2133–2138. 10.1158/1078-0432.CCR-15-2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burton JH, Mazcko C, LeBlanc A, Covey JM, Ji J, Kinders RJ, et al. NCI Comparative Oncology Program Testing of Non-Camptothecin Indenoisoquinoline Topoisomerase I Inhibitors in Naturally Occurring Canine Lymphoma. Clin Cancer Res. 2018;24(23):5830–5840. 10.1158/1078-0432.CCR-18-1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paoloni M, Webb C, Mazcko C, Cherba D, Hendricks W, Lana S, et al. Prospective molecular profiling of canine cancers provides a clinically relevant comparative model for evaluating personalized medicine (PMed) trials. PLoS One. 2014;9(3):e90028 10.1371/journal.pone.0090028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lustgarten JL, Zehnder A, Shipman W, Gancher E, Webb TL. Veterinary informatics: forging the future between veterinary medicine, human medicine, and One Health initiatives—a joint paper by the Association of Veterinary Informatics (AVI) and the CTSA One Health Alliance (COHA); 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc. 2011;18(5):544–551. 10.1136/amiajnl-2011-000464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1(2):161–174. 10.1136/jamia.1994.95236146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christensen L, Haug P, Fiszman M. MPLUS: a probabilistic medical language understanding system. In: Proceedings of the ACL-02 workshop on Natural language processing in the biomedical domain; 2002. p. 29–36.
- 33. Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001; p. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 34. Denny JC, Irani PR, Wehbe FH, Smithers JD, Spickard A 3rd. The KnowledgeMap project: development of a concept-based medical school curriculum database. AMIA Annu Symp Proc. 2003; p. 195–199. [PMC free article] [PubMed] [Google Scholar]
- 35. Liu K, Mitchell KJ, Chapman WW, Crowley RS. Automating tissue bank annotation from pathology reports—comparison to a gold standard expert annotation set. AMIA Annu Symp Proc. 2005; p. 460–464. [PMC free article] [PubMed] [Google Scholar]
- 36. Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy SN, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak. 2006;6:30 10.1186/1472-6947-6-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Elkin PL, Brown SH, Husser CS, Bauer BA, Wahner-Roedler D, Rosenbloom ST, et al. Evaluation of the content coverage of SNOMED CT: ability of SNOMED clinical terms to represent clinical problem lists. Mayo Clin Proc. 2006;81(6):741–748. 10.4065/81.6.741 [DOI] [PubMed] [Google Scholar]
- 38. Christensen LM, Harkema H, Haug PJ, Irwin JY, Chapman WW. ONYX; 2009. [PMC free article] [PubMed] [Google Scholar]
- 39. Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010;17(1):19–24. 10.1197/jamia.M3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17(5):507–513. 10.1136/jamia.2009.001560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chapman BE, Lee S, Kang HP, Chapman WW. Document-level classification of CT pulmonary angiography reports based on an extension of the ConText algorithm. J Biomed Inform. 2011;44(5):728–737. 10.1016/j.jbi.2011.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wagner M, Tsui F, Cooper G, Espino JU, Harkema H, Levander J, et al. Probabilistic, Decision-theoretic Disease Surveillance and Control. Online J Public Health Inform. 2011;3(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jackson MSc RG, Ball M, Patel R, Hayes RD, Dobson RJB, Stewart R. TextHunter–A User Friendly Tool for Extracting Generic Concepts from Free Text in Clinical Research. AMIA Annu Symp Proc. 2014;2014:729–738. [PMC free article] [PubMed] [Google Scholar]
- 44. Tseytlin E, Mitchell K, Legowski E, Corrigan J, Chavan G, Jacobson RS. NOBLE – Flexible concept recognition for large-scale biomedical natural language processing; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee HJ, Xu H, Wang J, Zhang Y, Moon S, Xu J, et al. UTHealth at SemEval-2016 task 12: an end-to-end system for temporal information extraction from clinical notes. In: Proceedings of the 10th International Workshop on Semantic Evaluation (SemEval-2016); 2016. p. 1292–1297.
- 46. Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed Representations of Words and Phrases and their Compositionality. In: Burges CJC, Bottou L, Welling M, Ghahramani Z, Weinberger KQ, editors. Advances in Neural Information Processing Systems 26. Curran Associates, Inc.; 2013. p. 3111–3119. [Google Scholar]
- 47. Wang Y, Sohn S, Liu S, Shen F, Wang L, Atkinson EJ, et al. A clinical text classification paradigm using weak supervision and deep representation. BMC Med Inform Decis Mak. 2019;19(1):1 10.1186/s12911-018-0723-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Koopman B, Karimi S, Nguyen A, McGuire R, Muscatello D, Kemp M, et al. Automatic classification of diseases from free-text death certificates for real-time surveillance. BMC Med Inform Decis Mak. 2015;15:53 10.1186/s12911-015-0174-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Berndorfer S, Henriksson A. Automated Diagnosis Coding with Combined Text Representations. Stud Health Technol Inform. 2017;235:201–205. [PubMed] [Google Scholar]
- 50. Anholt RM, Berezowski J, Jamal I, Ribble C, Stephen C. Mining free-text medical records for companion animal enteric syndrome surveillance. Prev Vet Med. 2014;113(4):417–422. 10.1016/j.prevetmed.2014.01.017 [DOI] [PubMed] [Google Scholar]
- 51. Goodfellow I, Bengio Y, Courville A. Deep Learning. MIT Press; 2016. [Google Scholar]
- 52. Agibetov A, Blagec K, Xu H, Samwald M. Fast and scalable neural embedding models for biomedical sentence classification; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Du Y, Pan Y, Wang C, Ji J. Biomedical semantic indexing by deep neural network with multi-task learning. BMC Bioinformatics. 2018;19(Suppl 20):502 10.1186/s12859-018-2534-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tran T, Kavuluru R. Predicting mental conditions based on “history of present illness” in psychiatric notes with deep neural networks. J Biomed Inform. 2017;75S:S138–S148. 10.1016/j.jbi.2017.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen MC, Ball RL, Yang L, Moradzadeh N, Chapman BE, Larson DB, et al. Deep Learning to Classify Radiology Free-Text Reports. Radiology. 2018;286(3):845–852. 10.1148/radiol.2017171115 [DOI] [PubMed] [Google Scholar]
- 56. Banerjee I, Ling Y, Chen MC, Hasan SA, Langlotz CP, Moradzadeh N, et al. Comparative effectiveness of convolutional neural network (CNN) and recurrent neural network (RNN) architectures for radiology text report classification. Artif Intell Med. 2019;97:79–88. 10.1016/j.artmed.2018.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Weng WH, Wagholikar KB, McCray AT, Szolovits P, Chueh HC. Medical subdomain classification of clinical notes using a machine learning-based natural language processing approach; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gehrmann S, Dernoncourt F, Li Y, Carlson ET, Wu JT, Welt J, et al. Comparing deep learning and concept extraction based methods for patient phenotyping from clinical narratives. PLoS One. 2018;13(2):e0192360 10.1371/journal.pone.0192360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rajkomar A, Oren E, Chen K, Dai AM, Hajaj N, Hardt M, et al. Scalable and accurate deep learning with electronic health records. NPJ Digit Med. 2018;1:18 10.1038/s41746-018-0029-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Johnson AEW, Pollard TJ, Shen L, Lehman LWH, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3:160035 10.1038/sdata.2016.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. arXiv. 2016;.
- 62. Demner-Fushman D, Rogers WJ, Aronson AR. MetaMap Lite: an evaluation of a new Java implementation of MetaMap; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Barros JM, Duggan J, Rebholz-Schuhmann D. Disease mentions in airport and hospital geolocations expose dominance of news events for disease concerns. J Biomed Semantics. 2018;9(1):18 10.1186/s13326-018-0186-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hanauer DA, Saeed M, Zheng K, Mei Q, Shedden K, Aronson AR, et al. Applying MetaMap to Medline for identifying novel associations in a large clinical dataset: a feasibility analysis; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Harkema H, Dowling JN, Thornblade T, Chapman WW. ConText: An algorithm for determining negation, experiencer, and temporal status from clinical reports; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pham T, Tran T, Phung D, Venkatesh S. DeepCare: A Deep Dynamic Memory Model for Predictive Medicine. arXiv. 2016;.
- 67. Pennington J, Socher R, Manning C. Glove: Global Vectors for Word Representation; 2014. [Google Scholar]
- 68.Pascanu R, Mikolov T, Bengio Y. On the difficulty of training Recurrent Neural Networks. arXiv. 2012;. [Google Scholar]
- 69. Segura-Bedmar I, Colón-Ruíz C, Tejedor-Alonso MÁ, Moro-Moro M. Predicting of anaphylaxis in big data EMR by exploring machine learning approaches; 2018. [DOI] [PubMed] [Google Scholar]
- 70. Yu Z, Bernstam E, Cohen T, Wallace BC, Johnson TR. Improving the utility of MeSH® terms using the TopicalMeSH representation. J Biomed Inform. 2016;61:77–86. 10.1016/j.jbi.2016.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ye, Ye Y, Wagner MM, Cooper GF, Ferraro JP, Su H, et al. A study of the transferability of influenza case detection systems between two large healthcare systems; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12(85):2825–2830. [Google Scholar]
- 73. Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–209. 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Friedman C. A broad-coverage natural language processing system. Proc AMIA Symp. 2000; p. 270–274. [PMC free article] [PubMed] [Google Scholar]
- 75. Stetson PD, Johnson SB, Scotch M, Hripcsak G. The sublanguage of cross-coverage. Proc AMIA Symp. 2002; p. 742–746. [PMC free article] [PubMed] [Google Scholar]
- 76. Friedman C, Kra P, Rzhetsky A. Two biomedical sublanguages: a description based on the theories of Zellig Harris; 2002. [DOI] [PubMed] [Google Scholar]
- 77. Wu Y, Denny JC, Trent Rosenbloom S, Miller RA, Giuse DA, Wang L, et al. A long journey to short abbreviations: developing an open-source framework for clinical abbreviation recognition and disambiguation (CARD); 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sohn S, Wang Y, Wi CI, Krusemark EA, Ryu E, Ali MH, et al. Clinical documentation variations and NLP system portability: a case study in asthma birth cohorts across institutions. J Am Med Inform Assoc. 2018;25(3):353–359. 10.1093/jamia/ocx138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhang Y, Tang B, Jiang M, Wang J, Xu H. Domain adaptation for semantic role labeling of clinical text. J Am Med Inform Assoc. 2015;22(5):967–979. 10.1093/jamia/ocu048 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Comparison of reviewer and MetaMap term extractions for 19 records. True Positive (TP) = MetaMap correctly matched term to code; False Negative (FN) = MetaMap did not extract a term the experts did; False Positive (FP) = MetaMap matched a term not identified by the expert; Total = Total number of terms matched in that document.
(XLSX)
(XLSX)
A 2-year old female dog patient with recurrent otitis and allergic dermatitis. Both the narrative (left) and the “MetaMapped” version (right) show that the treatment included prednisone (among other important clinical details). For the purposes of this manuscript, the pet and owner’s name were manually de-identified.
(XLSX)
Sheet 1: Long Short Term Memory (LSTM) Recurrent Neural Network (RNN) [FasTag]; Sheet 2: Decision Trees (DT); Sheet 3: Random Forests (RF).
(XLSX)
Training with CSU data (green), MIMIC data (yellow) or MIMIC+CSU (purple); validating on CSU data (Panel A) or MIMIC data (Panel B). The color of the category text is darkened (black) and the box made bigger if it surpasses the threshold of an F1 score of at least 0.70 (dotted horizontal line).
(TIF)
Data Availability Statement
The veterinary data presented here belongs to the Colorado State University (CSU), which may grant access to this data on a case-by-case basis to researchers who obtain the necessary Data Use Agreement (DUA) and IRB approvals. The CSU Research division, which maintains the data, can be contacted at cvmbs-research@colostate.edu. Inquiries about access to the data should be directed to Dr. Tim Hackett, Senior Associate Dean for Veterinary Health Systems, College of Veterinary Medicine and Biomedical Sciences, CSU (tim.hackett@colostate.edu). The human data presented here belongs to the Beth Israel Deaconess Medical Center in Boston, Massachusetts and can be accessed after signing a DUA with the MIT Lab for Computational Physiology at https://mimic.physionet.org/gettingstarted/access.