Abstract
Insect courtship and mating depend on integration of olfactory, visual, and tactile cues. Compared to other insects, Bombyx mori, the domesticated silkworm, has relatively simple sexual behaviors as it cannot fly. Here by using CRISPR/Cas9 and electrophysiological techniques we found that courtship and mating behaviors are regulated in male silk moths by mutating genes in the sex determination cascade belonging to two conserved pathways. Loss of Bmdsx gene expression significantly reduced the peripheral perception of the major pheromone component bombykol by reducing expression of the product of the BmOR1 gene which completely blocked courtship in adult males. Interestingly, we found that mating behavior was regulated independently by another sexual differentiation gene, Bmfru. Loss of Bmfru completely blocked mating, but males displayed normal courtship behavior. Lack of Bmfru expression significantly reduced the perception of the minor pheromone component bombykal due to the down regulation of BmOR3 expression; further, functional analysis revealed that loss of the product of BmOR3 played a key role in terminating male mating behavior. Our results suggest that Bmdsx and Bmfru are at the base of the two primary pathways that regulate olfactory-based sexual behavior.
Author summary
The fundamental insect sexual behaviors, courtship and mating, result from successful integration of olfactory, vision, tactile and other complex innate behaviors. In the widely used insect model, Drosophila melanogaster, the sex determination cascade genes fruitless and doublesex are involved in the regulation of courtship and mating behaviors; however, little is known about the function of these sexual differentiation genes in regulating sex behaviors of Lepidoptera. Here we combine genetics and electrophysiology to investigate regulation pathway of sexual behaviors in the model lepidopteran insect, the domesticated silk moth, Bombyx mori. Our results support the presence of two genetic pathways in B. mori, named Bmdsx-BmOR1-bombykol and Bmfru-BmOR3-bombykal, which control distinct aspects of male sexual behavior that are modulated by olfaction. This is the first comprehensive report about the role of sex differentiation genes in the male sexual behavior in the silk moth.
Introduction
Sex determination pathways control the sexually dimorphic traits of males and females, including sexual differentiation and sexual behavior [1]. The genetic cascades of primary signalling that underlie sex determination in insects have high diversity among species. In the model insect Drosophila melanogaster, sex determination is controlled hierarchically by X:A, Sex-lethal (Sxl), transformer or transformer 2 (tra/tra2), doublesex (dsx), and fruitless (fru) [2, 3]. An X:A ratio of 1 promotes transcription of Sxl and results in feminization, while an X:A ratio of 0.5 results in Sxl suppression and male differentiation [4, 5]. Sxl proteins control the splicing of female tra mRNA which gives rise to functional proteins, while no functional Sxl proteins are produced in the male [6]. Tra and tra2 proteins control the splicing of dsx and fru which are located at the bottom of the sex determination pathway to maintain sexual development and behavior [7–9]. The sex determining genetic cascade based on tra/tra2 control of dsx and fru splicing is widely conserved in many Diptera, Coleoptera and Hymenoptera, while the domesticated Bombyx mori, lack tra/tra2 as regulators of dsx/fru [3]. In the silkworm, the sex determination cascade involves at least 4 distinct components: a female-enriched PIWI-interacting RNA (fem), a responding gene, BmMasc, a P-element somatic inhibitor (BmPSI)/ (BmImp), and Bmdsx [10–12]. The product of the W chromosome derived fem piRNA targets the downstream gene, BmMasc, to control Bmdsx sex-specific splicing; BmPSI and BmImp regulate Bmdsx splicing through binding CE1 sequences of Bmdsx pre-mRNA [10]. Although the upstream signal is not conserved, with X:A and fem as the primary signals in the fruit fly and silkworm, respectively, the role of dsx is conserved.
Insects have sex-specific splicing that generates a male- (dsxM) and female-specific (dsxF) dsx isoform. Previous reports have shown that the Bmdsx gene products (BmdsxM and BmdsxF) control sexually dimorphic traits such as formation of abdominal segments and external and internal genitalia [13–16]. Studies of D. melanogaster have shown that the development of numerous sexually dimorphic traits are controlled by Dmdsx, including DmdsxF controlled female-specific yolk gene transcription and female-specific spermathecae, DmdsxM controlled male-specific abdominal pigmentation and male-specific sex combs [17–20]. The DmdsxM gene products also regulate courtship behaviors, including licking, courtship song and copulatory behaviors [21]. The courtship behavior of D. melanogaster males consists of a series of discrete elements, including orientation toward the female, following the female, extending and vibrating one wing to produce a courtship song, licking the external genitalia, and attempting copulation [22]. DmdsxM is expressed in about 900 neurons of the central nervous system, the majority of which also express DmfruM [21]. DmfruM function is both necessary and sufficient for nearly all aspects of male courtship behavior, and it is expressed in a dispersed subset of approximately 2,000 neurons in the central and peripheral nervous systems [23–25]. Males lacking DmfruM appear to be normal externally yet are profoundly defective in most aspects of courtship behavior [22]. Moreover, DmdsxM is necessary and sufficient for DmfruM-independent courtship [26].
Although in recent years studies of neural and genetic mechanisms of sexual behavior in fruit flies indicate that DmdsxM and DmfruM are the master regulators of many sexually differentiated processes and behaviors [27, 28], how these master genes act to control neural development to build these complex behaviors by regulating downstream genes is still not fully understood [29]. Many studies have shown the fru gene to be conserved functionally with sex-specific splicing expression patterns among Diptera, including Anopheles gambiae, Ceratitis capitata, Aedes aegypti, Nasonia vitripennis, and Musca domestica, and a Blattodea, Blatella germanica [29–35]. However, the genetic regulatory mechanism of sexual behavior remains unclear in lepidopteran insects. Our previous study showed that loss of the BmdsxM blocks male sexual behavior. The defective expression of BmOR1 in male mutants of BmdsxM contributes to the failed courtship behavior of orientation and leads to subsequent rejection of males by females [13].
B. mori has a simple sex pheromone system: female silkmoths emit the sex pheromones bombykol [(E,Z)-10,12-hexadecadien-1-ol] and bombykal [(E,Z)-10,12-hexadecadienal] at a typical ratio of 11:1. The major pheromone bombykol triggers the full sexual behavior of the male moth [36, 37]. Two sex pheromone receptors, BmOR1 and BmOR3, have male specific expression and are specific for bombykol and bombykal, respectively [38]. The pheromone binding protein BmPBP1, which has a male-biased expression, is required for the selectivity of BmOR1 for bombykol [39]. Loss of function mutations in BmPBP1 or BmOR1 cause the disappearance of male sexual behavior [40, 41]. Previous studies showed that the sex determination gene Bmdsx controls the expression of BmPBP1 and of BmOR1 [42, 14]. These results suggest that Bmdsx promotes male sex behavior, activating specific receptors of the olfactory system.
In the current study, we used genetic and electrophysiological approaches to investigate the molecular regulatory mechanisms controlling sexual behaviors in the silkworm by analyzing the mating behaviors of silk moths with mutations in known sex determination factors. Using our previously reported non sex-specific mutants BmMasc, BmPSI, and Bmdsx [12, 13], and the Bmfru mutant described here, we found that the sex determination pathway influences the development of the olfactory system to regulate courtship and mating behaviors. The sex determination pathway regulates morphological development and also the response to bombykol. The sexually dimorphic antennal morphologies of the male mutants of BmMasc, BmPSI, and Bmdsx are affected, which result in abnormal responses to bombykol and loss of courtship behavior. In contrast, male mutants of Bmfru show normal courtship but defective mating behavior. Knockout of the Bmfru downstream gene, BmOR3, impairs the response to bombykal but not bombykol, with normal courtship but extended mating behavior. Our data provide in vivo evidence of the function of the sexual differentiation pathway in the regulation of olfactory-based sexual behavior of B. mori.
Results
Sex determination pathway mutants have altered courtship and mating behaviors
We used a binary transgenic CRISPR/Cas9 system to obtain a Bmfru mutant. The Bmfru gene had not been reported in the silkworm, so we searched the silkworm genome database and identified a locus that encodes a protein (NCBI Accession number: EU649701.1) with a conserved BTB domain found in the FRU proteins of D. melanogaster and M. domestica (S1A Fig). We designed a small guide RNA (sgRNA) to target exon 3 of the putative Bmfru gene (S1B Fig). Through germline transformation, we obtained a single U6-sgRNA transgenic line. To obtain heritable and homozygous mutants to assess preference behaviors, we performed a series of crossing strategies and PCR-based screening experiments (S1C Fig) as described previously [43]. The U6-sgRNA and nos-Cas9 parental transgenic lines were crossed with each other to obtain F1 founder moths, then ten random F1 female moths were crossed with wild-type males to obtain heterozygous mutants. F2 heterozygous mutant females were individually crossed with wild-type males to obtain independent lines of F3 heterozygous progeny, each potentially carrying a unique mutant allele. Individuals within F3 heterozygous lines were crossed with each other to obtain F4 homozygous mutants (S1C Fig) resulting in approximately 25% homozygous mutants in the F4 progeny. We characterized the mutations by PCR using gene-specific primer pairs. All of the homozygous mutants had four deleted base pairs that resulted in a premature termination codon (S1D Fig), confirming that the original CRISPR/Cas9-induced mutation had occurred.
Next, we analyzed courtship behavior of male mutants with a wild-type virgin female as the target and a mutant male (M-M) as the test at distances of 10 and 20 centimeters. The male sexual behavior of silk moth consists of the following steps: the male silk moth first recognizes the female by responding to a sex pheromone released by the female, the male moth then exhibits a programmed sequence of walking consisting of transitory bouts of straight-line walking, zig-zag turns and looping to climb toward the female. Then the male displays orientation, wing flapping (wing song), or turns around, and uses its forelegs to touch the female’s abdomen. After confirming the position of the female’s external gentitalia the male attempts to mate with the female using its claspers. Once the male mates with female, the male continues to flap its wings intermittently and copulates for several hours. The Bmfru mutant males did recognize wild-type females and displayed normal courtship behavior (10 cm: 91%, p = 0.034; 20 cm: 93%, p = 0.118). Nevertheless, despite many attempts at copulating, the Bmfru mutant males could not mate with the wild-type female (S1–S3 Movies). Thus, the courtship index of Bmfru mutant males was normal, whereas the mating index was zero (Fig 1). In contrast, BmMasc, BmPSI, and Bmdsx mutant males did not display any courtship behavior (i.e., no orientation, wing song, or turning around; Bmdsx behavior is illustrated in S4 and S5 Movies), so the courtship and mating indexes of these three mutants were zero (Fig 1).
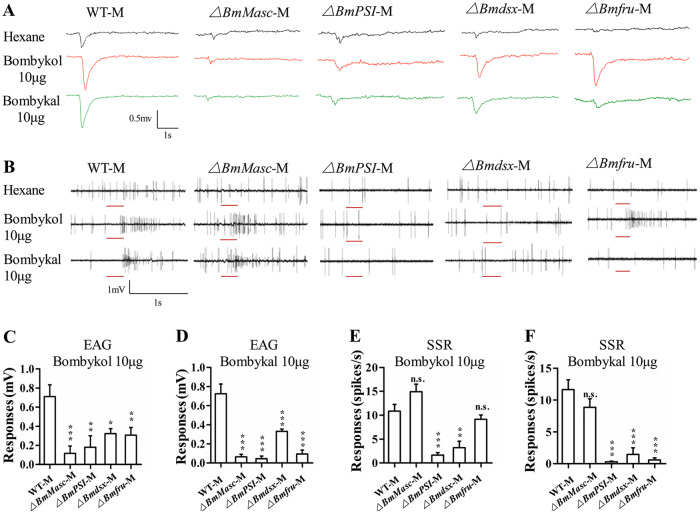
Fig 1. Loss of sex determination pathway genes impairs male courtship and mating behaviors.
(A) Diagram of the sex determination cascade in the silkworm. (B) Diagram of the behavioral test setup. The adult male (M-M) is placed at a distance of 10 cm or 20 cm from a wild-type adult female (WT-F, which releases pheromones) or from a wild-type male control, and behavior is monitored. (C and D) The courtship and mating behavior indexes for wild-type males (WT) and BmMasc, BmPSI, Bmdsx, and Bmfru mutant males. The results are expressed as percentage from 90 pairs tests with Fisher exact test. An index of 0% indicates the absence of courtship behavior or failed mating behavior.
Antennal structures are abnormal in BmMasc, BmPSI, and Bmdsx mutants
Since insect antennae play a key role in the olfactory-based chemical communication necessary for courtship behavior, we evaluated the morphology of antennal structures in the mutants. In wild-type B. mori, the length of male antenna (5.40±0.15) is longer than the female antenna (4.75±0.19), and the number of sensilla trichodiea in one SEM scan field on antennae of male (83.60±7.28) is much more than female (59.60±3.44) (Fig 2). The antennae were significantly shorter in males with mutations in BmMasc (18%, 4.45±0.09, p = 0.0001), and BmPSI (17%, 4.50±0.13, p = 0.0003), whereas antenna lengths were normal in Bmdsx (4.90±0.16, p = 0.065) and Bmfru mutant males (5.20±0.19, p = 0.7349) (Fig 2A and 2C). Using scanning electron microscopy (SEM), we observed that the numbers of sensilla trichoidea were significantly decreased in males with mutations in BmMasc (38%, 52.00±3.18, p = 0.0003), BmPSI (40%, 50.20±3.51, p = 0.0001), and Bmdsx (26%, 62.00±2.80, p = 0.0097) compared to wild-type males (83.60±7.28), whereas the Bmfru male mutants (96.00±4.14, p = 0.1894) had similar numbers of sensilla trichoidea to wild-type males (Fig 2B and 2D). The female mutant antennae had no significant change compared with wild-type females (Fig 2C and 2D). These results suggest that loss of BmMasc, BmPSI, or Bmdsx affects the development of antennal structures, which might underlie the observed dysfunctions in courtship and mating.
Fig 2. Silkworms with mutations in BmMasc, BmPSI, and Bmdsx have abnormal antennal structures.
(A) Gross morphology of antennae of wild-type and mutant males (M) and females (F). Scale bars: 1 mm. (B) SEM images of the sensilla trichoidea structures in the middle of the antennae of wild-type and mutant males. Scale bars: 50 μm. (C) Antennal lengths of wild-type and mutant adults. The results are expressed as the means ± SEM of 10 independent biological replicates. *** represent significant difference at the 0.001 level (one- way ANOVA), compared with the WT-F and WT-M; n.s. indicates that the difference is not statistically significant. (D) The number of sensilla trichoidea in one SEM scan field in wild-type and mutant adults. The results are expressed as the means ± SEM of 5 insects per group. ** and *** represents significant differences at the 0.01 and 0.001 level (one- way ANOVA) compared with the control; n.s. indicates that the difference is not statistically significant.
Electrophysiological analyses reveal abnormal responses to sex pheromone components upon loss of sex determination genes
The female silkmoth attracts males by emitting sex pheromones. The pheromone bombykol is critical to attracting the male moths, whereas the minor pheromone bombykal plays an antagonistic role in mating behavior [36, 44]. We tested the responsiveness of mutant males to bombykol and bombykal using two methods: Electroantennogram (EAG) and single sensillum recording (SSR). We used EAG to detect the responsiveness to bombykol or bombykal at the level of whole antennae and SSR to evaluate responses of individual long sensilla trichoidea as described previously [40]. Compared to wild-type males (0.71 ± 0.12), Bmdsx (0.32 ± 0.05, p = 0.0183) and Bmfru (0.31 ± 0.08, p = 0.0067) mutants, the BmMasc (0.12 ± 0.07, p = 0.0004) and BmPSI (0.18 ± 0.12, p = 0.0044) mutants showed significantly lower EAG responses to bombykol (Fig 3A and 3C). BmMasc (0.07 ± 0.03, p < 0.0001), BmPSI (0.05 ± 0.03, p < 0.0001), and Bmfru (0.09 ± 0.04, p < 0.0001) mutant males displayed significantly lower EAG responses to the minor pheromone bombykal than Bmdsx (0.33 ± 0.03, p = 0.0003) mutants; nevertheless, the response of the Bmdsx mutants was also significantly lower than that of the wild-type (0.72 ± 0.10) moths (Fig 3A and 3D). These findings suggested that loss of any of the functions exerted by these sex determination and sexual differentiation genes interrupts the neuronal response to pheromones at the level of the antennae.
Fig 3. Electrophysiological analyses reveal abnormalities in responses to pheromone components of male silkworms with mutations in sex determination genes.
(A) Representative EAGs of wild-type and mutant BmMasc, BmPSI, Bmdsx, and Bmfru male moths in response to hexane (upper panel), 10 μg bombykol (middle panel), and 10 μg bombykal (lower panel). (B) Representative single sensillum recording (SSR) from wild-type and mutant BmMasc, BmPSI, Bmdsx, and Bmfru males in response to hexane (upper panel), 10 μg bombykol (middle panel), and 10 μg bombykal (lower panel). The stimulus was applied for 300 ms, indicated by a red line under the trace. (C and D) Mean responses of male antennae to C) 10 μg of bombykol and D) 10 μg bombykal. The statistical significance between WT (n = 10) and BmMasc (n = 7), BmPSI (n = 5), Bmdsx (n = 8), and Bmfru (n = 11) mutant responses was analyzed with one-way ANOVA. Data are shown as means ± SEM; *, **, and *** represent significant differences at the 0.05, 0.01, and 0.001 levels, respectively, compared with the WT-M. (E and F) Mean responses of neurons in male sensillum trichodea to E) 10 μg of bombykol and F) 10 μg bombykal. The statistical significance between WT (n = 30) and BmMasc (n = 50), BmPSI (n = 44), Bmdsx (n = 30), and Bmfru (n = 76) mutants was analyzed with one-way ANOVA. Data are shown as means ± SEM; ** and *** indicates p < 0.01 and p < 0.001, compared with the WT-M, and n.s. indicates no significance.
Next, we examined the responses of neurons within the long sensillum trichodea of the four mutants and wild-type males using SSR. From the spike traces of wild-type silkworms, two neurons (expressing BmOR1 or BmOR3) were distinguished in the long sensillum trichodea. A larger amplitude was induced by the BmOR1 neuron responding to bombykol, and a smaller one was evoked by the BmOR3 neuron responding to bombykal (Fig 3B). The neuronal responses of the BmPSI (bombykol: 1.64 ± 0.53, p < 0.0001 and bombykal: 0.30 ± 0.11, p < 0.0001) and Bmdsx (bombykol: 3.20 ± 1.35, p value = 0.0014 and bombykal: 1.47 ± 1.03, p < 0.0001) mutants to both bombykol and bombykal were significantly lower than wild-type (bombykol: 10.83 ± 1.43 and bombykal: 11.63 ± 1.54) whereas the neuronal responses of the BmMasc mutants to bombykol (14.90 ± 1.60, p = 0.1037) and bombykal (8.86 ± 1.33, p = 0.1374) were similar to wild-type males (Fig 3B, 3E and 3F). Although the single sensillum trichodea in the BmMasc mutants responded normally to bombykol and bombykal, it is possible that a decrease in the number of sensillum trichodea may contribute to the failure of courtship behavior of these mutants. The Bmfru mutants responded normally to bombykol (9.13 ± 0.92, p = 0.7342) but did not respond to bombykal (0.61 ± 0.31, p < 0.0001) (Fig 3B, 3E and 3F). This suggested that Bmfru mutants display normal courtship behavior because they have a normal response to bombykol.
Expression of male-biased olfactory system genes is altered in mutants
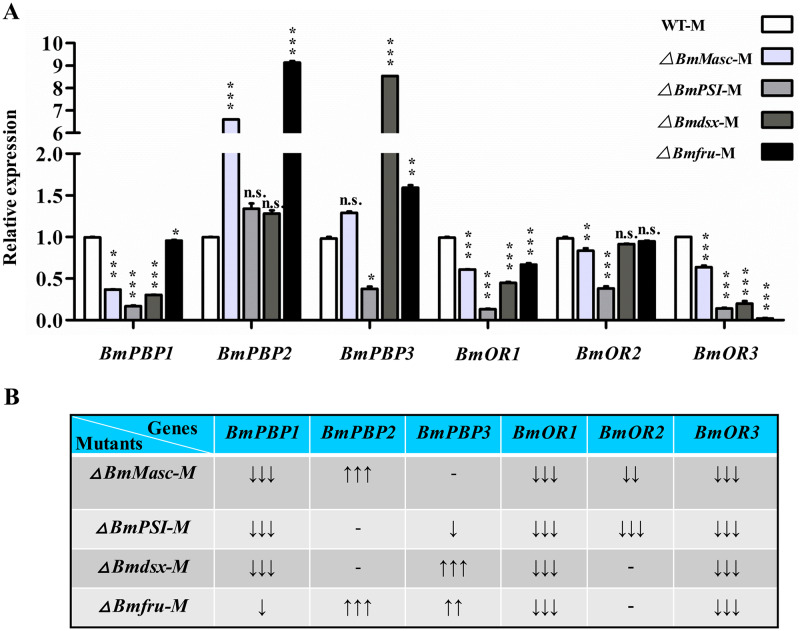
Olfaction plays an important role in insect behaviors such as mate recognition. To test whether expression of olfactory system genes in the antennae were affected by loss of sex determination pathway genes, we quantified expression of genes that encode factors necessary for mate recognition by males, notably, pheromone binding proteins BmPBP1, BmPBP2, and BmPBP3 and odorant receptors BmOR1, BmOR2, and BmOR3 [45]. BmPBP1 binds to bombykol, and its dysfunction causes the failure of male courtship behavior [41]. Although BmPBP1 was expressed at wild-type levels in male mutants in Bmfru, it was significantly decreased in the males mutant in BmMasc, BmPSI, and Bmdsx. Compared to wild-type males, BmPBP2 and BmPBP3 RNA expression levels were significantly higher in male mutants in BmMasc, Bmdsx, and Bmfru but lower in BmPSI mutants (Fig 4). Levels of OR1 and OR3 were significantly lower in all mutants (Fig 4). Mutations in Bmdsx reduce significantly the expression not only of PBP1 and BmOR1, but also BmOR3, and increase the expression of BmPBP3. No significant data are available for BmOR2 in Bmdsx mutant. Furthermore, mutations in Bmfru reduce significantly the expression of BmOR1 and much more of BmOR3, and increase the expression of BmPBP2, and possibly also of BmPBP3. No significant data are available for BmOR2. So the expression of BmOR3 is decreased in males by both Bmdsx and Bmfru, with Bmfru having an higher effect. BmOR1 higher expression in males is promoted mainly by Bmdsx but also by Bmfru (Fig 4). BmOR2 expression was normal except in the BmMasc and BmPSI mutant where it was expressed at levels lower than those observed in wild-type males (Fig 4). These expression patterns suggested that BmPSI is required in males for normal expression of 5 out of the 6 olfactory genes tested.
Fig 4. Expression change of olfactory sensory system genes in sex determination gene mutants.
(A) Relative mRNA expression levels of BmPBP1, BmPBP2, BmPBP3, BmOR1, BmOR2, and BmOR3 in WT and mutant males. Three individual biological replicates were performed with real-time quantitative PCR (qPCR). Plotted are means ± SEM. *, ** and *** represent significant differences at the 0.05, 0.01 and 0.001 levels with one- way ANOVA, comparing each gene was with the corresponding WT-M; n.s. represents not significant. (B) Summary of the expression change of olfactory system genes in each mutant. ‘-’ represents no significant change compare to WT, ‘↓’, ‘↓↓’, ‘↓↓↓’ represent significant decreased (*, **, ***) compare to WT, ‘↑↑’, ‘↑↑↑’ represent significant increased (**, ***) compare to WT.
The normal response to sex pheromones by a single sensillum of the BmMasc mutant as shown by SSR may be due to the expression of BmOR1 and BmOR3 at about 60% of wild-type levels. Interestingly, expression of BmOR3 was only 2% of wild-type levels in the Bmfru mutant, whereas the BmOR1 gene was expressed at about 70% of levels in the wild-type males and BmPBP1 expression was normal. Such relatively low expression of BmOR1 and BmPBP1 likely allowed the normal response to bombykol resulting in the normal courtship behavior of these mutant males. Altogether, these results suggest that sex determination pathway genes have an important role in establishing the sexually dimorphic expression of olfactory system genes, whereby Bmdsx primarily contributes to the expression of BmPBP1 and BmOR1 while Bmfru primarily contributes to the expression of BmOR3.
Loss of BmOR3 does not alter courtship behavior but does extend mating time
To further analyze the olfactory system-related genes in the antennae of Bmfru mutant males, we compared antennal transcriptomes of the adult Bmfru mutant male silk moths to those of wild-type males using RNA-seq. We identified 273 differentially expressed genes, 176 of which were down-regulated and 97 of which were up-regulated (S2A Fig). We found that olfactory system-related genes including BmPBP1, BmPBP2, BmPBP3, BmOR1, and BmOR3 were differentially expressed (S2B Fig) as shown by RT-qPCR. In the Bmfru mutant antennae, BmOR1 expression was 0.99-fold lower and BmOR3 was 5.32-fold lower than in the wild-type antennae.
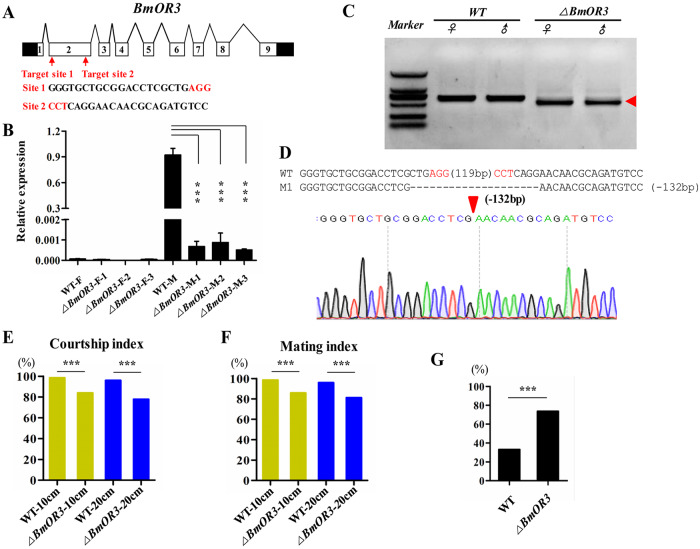
To further investigate the function of BmOR3, we used a binary transgenic CRISPR/Cas9 system to target exon 2 of BmOR3 (Fig 5A). We established three independent U6-sgRNA parental transgenic lines and then made crosses within each line to obtain F1 founder moths. Quantitative real-time RT-PCR showed a decreased in BmOR3 mRNA levels of F1 founder individuals by over 99% in each of the three mutant lines compared with wild type moths (Fig 5A and 5B). Characterization of the somatic mutations by PCR using gene-specific primer pairs indicated that mutants had deletions at the target site caused by non-homologous end joining-induced indels (Fig 5C and 5D). These results suggested that F1 individuals carried truncated proteins of BmOR3, so we used the F1 founder moths to analyze adult behavior.
Fig 5. Loss of BmOR3 expression extends mating time.
(A) Structure of the BmOR3 gene with nine exons indicated by boxes (black boxes, 5’- and 3’-UTRs; white boxes, coding exons). Target sites 1 and 2 are binding sites for sgRNAs. (B) The mean transcript levels (± SEM) of BmOR3 are down-regulated significantly compared to wild-type levels in the three BmOR3 mutant male (M) and female (F) lines. At least five males with mixed antenna were examined for each line. *** indicates p < 0.001 compared with the relevant control using one-way ANOVA. (C) Somatic mutations were induced in the F1 founder animals following crosses of nos-Cas9 with U6-sgRNA strains. PCR analyses with primers to amplify a region of 600 bp revealed deletion mutations in the G0 mutants. The red arrowhead indicates the deleted region. (D) Deletion mutation in the heterozygous offspring after crossing nos-Cas9 and U6-BmOR3sgRNA transgenic silkworm lines. The targeting sequence is shown in black, and the PAM sequence is in red. The deletion size in nucleotides is indicated above the red arrow at the site of the deletion. (E and F) Courtship and mating behavior indexes of BmOR3 mutant and WT males. Data are shown as percentage from 150 pairs tests with chi-squared test. ***p < 0.001. (G) Percentage of autosegregated WT and mutant males *** indicates significant difference at the 0.001 level with chi-squared test.
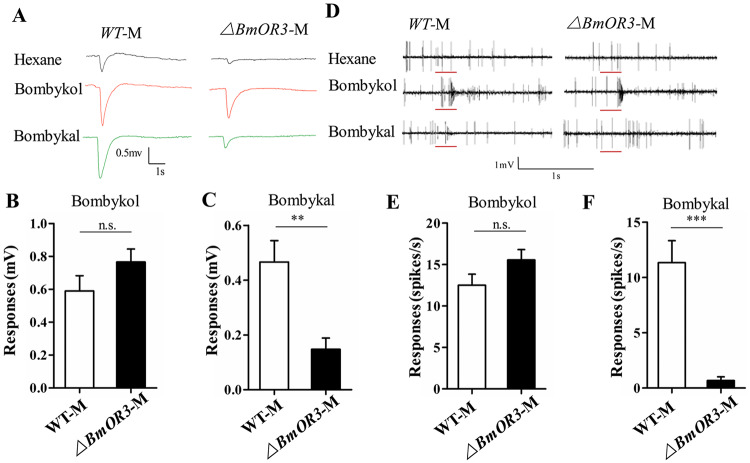
The BmOR3 mutant males displayed normal courtship behavior, including orientation, wing song, and turning, and the courtship and mating indexes were only slightly below normal (Fig 5E and 5F). As noted previously, the normal duration of copulation is several hours. Although almost all wild-type males had autosegregated after 12 hours of mating, most of the BmOR3 mutant males had not (Fig 5G). Electrophysiological analyses revealed that the BmOR3 mutants responded normally to bombykol but had lost responsiveness to bombykal (Fig 6); this was also the case for the Bmfru mutants. These findings indicated that BmOR3 is not necessary for courtship.
Fig 6. BmOR3 mutant male silk moths have nearly normal electrophysiological responses to bombykol but not bombykal.
(A) Representative EAGs from wild-type and BmOR3 mutant male moths in response to hexane (upper panel), 10 μg bombykol (middle panel), and 10 μg bombykal (lower panel). (B and C) Mean responses of WT (n = 9) and BmOR3 mutant (n = 9) male antennae to B) 10 μg of bombykol and C) 10 μg of bombykal. Data are means ± SEM; n.s. indicates no significant difference and ** represents a significant difference at the 0.01 level as determined by Student’s t-test. (D) Representative single sensillum recording (SSR) of wild-type and BmOR3 mutant male moths in response to hexane (upper panel), 10 μg bombykol (middle panel), and 10 μg bombykal (lower panel). The stimulus was applied for 300 ms as indicated with a red line under the trace. (E and F) Mean (± SEM) responses of neurons in male sensillum trichodea to E) 10 μg of bombykol or F) 10 μg of bombykal in WT (n = 26) and BmOR3 mutant (n = 40) male moths. *** indicates p < 0.001 and n.s. indicates no significant difference as determined by Student’s t-test.
Discussion
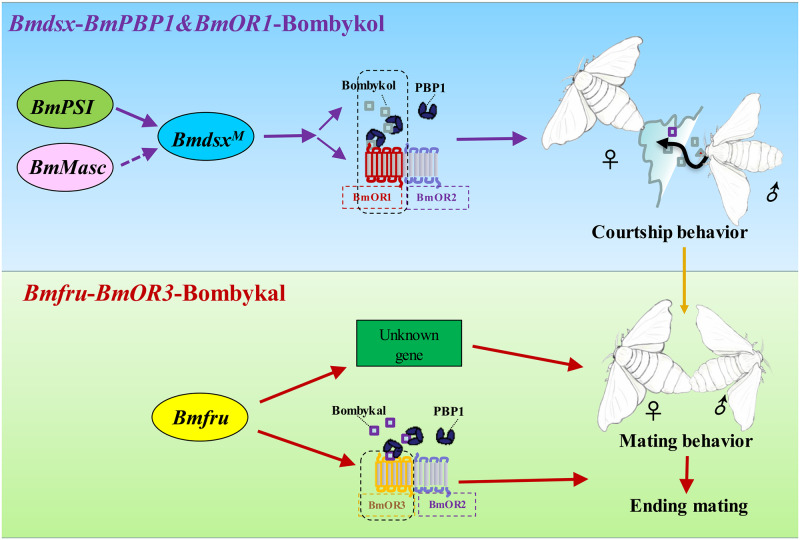
Previous studies have shown that BmOR1 and BmOR3 encode sex pheromone receptors in the silkworm [37, 39, 40]. Although the mechanism of sex determination in the silkworm has also been studied in recent years [13, 46, 47], it has not revealed a connection between these two major pathways which control the sexual behavior and morphology of the insect. Here, we provide genetic evidence for a functional interplay between these sex pheromone receptors and genes in the sex determination pathway. Our results support the notion that the Bmdsx-BmOR1-bombykol pathway regulates courtship behavior and that Bmfru-BmOR3-bombykal regulates mating (Fig 7).
Fig 7. Proposed genetic regulation pathway of sexual behavior in B. mori.
Sex determination pathway factors, olfactory sensory factors, and sex pheromones influence courtship and mating behavior. The Bmdsx-BmOR1/3-Bombykol and Bmfru-BmOR3-Bombykal cascades are the two primary pathways involved in olfactory-based sexual behavior. Disruption of the sex pathway gene Bmdsx blocks the expression of both BmOR1 and BmOR3, whereas disruption of Bmfru mainly inhibits expression of BmOR3. Thus, mutation of Bmdsx leads to an abnormal response to bombykol and bombykal and inhibits courtship and mating by male moths, but mutation of Bmfru disrupts termination of mating due to an abnormal response to bombykal.
Exposure to bombykol, the major female pheromone, was sufficient to induce pheromone source orientation behavior in male moths, as was the artificial activation of BmOR1 expressing olfactory receptor neurons [48, 49]. Previous studies have shown that BmOR1 and BmOR3 are specifically expressed in adult males [50]. This sexually dimorphic pattern of gene expression suggested that BmOR1 and BmOR3 are regulated by Bmdsx and Bmfru. TALEN-mediated knock out of BmOR1 indicated that BmOR1 is the sex pheromone receptor that mediates the pheromone response in male silkmoths [40]. The minor component, bombykal, was thought to negatively modulate the initiation of orientation behavior [44]. However, our results support a conclusion that BmOR3 has little effect on the initiation of orientation behavior. In our study, BmOR3 mutants which did not respond to bombykal had normal courtship but had extended mating behavior. Additionally, Bmfru mutants, which expressed almost no BmOR3, had normal courtship behavior but did not mate. This suggests that the BmOR3-bombykal interaction does not play a key role in the recognition of females by male moths and that Bmfru might act through other pathways to control mating behavior. Daimon et al. showed that the wing fluttering response of B. mori males to bombykol is strongly inhibited by bombykal, thereby indicating that bombykal acts as a behavioral antagonist [44]. All of these observations suggest that bombykal may play role in terminating copulation behavior.
In dipteran insects, fru is located downstream of tra in the sex determination pathway, and dsx is involved in the regulation of sexual behavior [34, 51]. No gene homologous to tra has been found in the silkworm genome, indicating that the sex determination pathway in B. mori is different from dipterans. Additionally, Bmfru is not like Dmfru, which has sex-specific splicing. Bmfru was found to have male-baised expression in the brain and testis [52]. This suggests that the regulatory mechanisms involving fru gene expression are different between fruitfly and silkworm. In Drosophila, a PSI–U1 snRNP interaction regulates male mating behavior, and one of its direct targets is Dmfru [53]. This suggests that DmPSI regulates mating behavior through Dmfru and other genes. By contrast, in the silkworm, BmPSI and BmMasc are upstream of the sex determination factor Bmdsx [11, 13]. Knockout of BmPSI caused the failure of courtship. In addition to BmOR2, the genes encoding BmOR1, BmOR3, BmPBP1 and BmPBP, were downregulated in the BmPSI mutant males produced in our study. This suggested that BmPSI might exert pleiotropic effects on olfactory development. On the other hand, BmOR2 was expressed normally in BmMasc mutant males, and changes in expression of other olfactory system genes were more moderate in BmMasc mutant males than in BmPSI mutant males. Moreover, the BmMasc mutant responded to pheromones normally as shown by SSR, even though the signal from whole antennae was decreased as shown using EAG. These results suggest that olfactory neurons are functionally normal but may be decreased in number in the BmMasc mutant.
In conclusion, by using a comprehensive set of knockout mutations in genes Bmdsx, BmPSI, BmMasc, Bmfru, and BmOR3, we showed that the sex determination cascade in B. mori contributes to the establishment of olfactory system-based sexual behaviors. We found that the sex determination genes control behavior by regulating expression of genes encoding olfactory receptors. BmMasc and BmPSI act as upstream signals and Bmdsx acts as the performer located at the bottom of the cascade to control courtship behaviors by regulating BmOR1, as well as BmOR2 (BmPSI) and both copies of BmOR3. The Bmfru gene product contributes to courtship behavior by regulating BmOR3, BmOR1 (slight effect) and other unknown pathways. Future work should focus on how these sex determination genes affect neuronal modulations that influence sexual behaviors.
Materials and methods
Silkworm strain
Silkworms of the Nistari genetic background (a multivoltine, non-diapausing strain) were used in all experiments. Larvae were reared on fresh mulberry leaves under standard conditions. BmMasc, BmPSI and Bmdsx mutants are described in our previous reports. The parental transgenic U6-sgRNA and nos-Cas9 lines were reared separately. Crossing these two lines produce heteroallelic mutations in somatic or germ cells of F1 individuals. F1 individuals carrying heteroallelic mutations were used in this study. The detection of genomic mutations and measurement of mRNA levels among these three F1 lines were reported previously [12, 13].
Plasmid construction and germline transformation
To target the Bmfru and BmOR3 genes, plasmids pBac[IE1-DsRed2-U6-sgRNAs] (U6-sgRNA) were constructed to express sgRNA under the control of the silkworm U6 promoter and the DsRed fluorescence marker gene, under control of an IE1 promoter. The sgRNA targeting sequences were selected by manually searching genomic regions for sequences that matched the 5′-GG-N18-NGG-3′ rule [54]. sgRNA sequences were checked bioinformatically for potential off-target binding to the relevant silkworm genomic sequence using CRISPRdirect (http://crispr.dbcls.jp/) [55]. All sgRNA and oligonucleotide primer sequences for plasmid construction are listed in S1 Table. Plasmid construction was performed as described previously [13]. Each U6-sgRNA plasmid was mixed with a piggyBac helper plasmid and microinjected separately into fertilized eggs at the pre-blastoderm stage. G0 adults were mated to WT moths, and the resulting G1 progeny were scored for the presence of the DsRed marker gene product using fluorescence microscopy (Nikon AZ100).
Genotyping analysis
For BmOR3 mutant, animal from U6-sgRNA transgenic line was mated with the nos-Cas9 line to obtain mutated F1 animals. For Bmfru mutant, animal from female founder animal was mated with male WT to obtain mutated F2 animals. Genomic DNA of mutated animals was extracted at the larval stage using standard SDS lysis-phenol treatment after incubation with proteinase K, followed by RNase treatment and ethanol precipitation. The resulting individual DNA samples from mutant animals were separated by sex using PCR amplification with primers specific to the W chromosome (S1 Table). Mutation events were detected by PCR amplification using gene-specific primers that bound upstream or downstream from each target (S1 Table). Amplified products were visualized by agarose gel electrophoresis. Amplicons were sub-cloned into the pJET-1.2 vector (Fermentas), and six positive clones of each line (one Bmfru and three BmOR3 lines) were selected and sequenced using Illumina NextSeq 500 platform (Sunnybio, Shanghai).
Photography and scanning electron microscopy (SEM)
Antennae of mutant and wild-type animals were dissected, photographed under a light microscope (Nikon, Tokyo, Japan) using a digital camera (Nikon DS-Ri1, Japan), and lengths measured on the images.
The dissected antennae were fixed overnight in a solution of 90 ml 70% ethanol, 5 ml acetic acid, and 5 ml 37% methyl aldehyde, dehydrated in a series of 70%, 80%, 90%, and 100% ethanol baths for 5 min each, and dried (CO2 for 6 h in a Critical Point Dryer). The dissected materials were coated with gold by JFC-1600 sputter (JEOL, Rigaku, Japan), and the middle parts of the antennae were observed by SEM using a JSM-6360LV microscope (JEOL, Rigaku, Japan).
Analysis of courtship and mating behavior
BmMasc, BmPSI, Bmdsx, and BmOR3 mutant males were from heterozygous F1 individuals. Bmfru mutant males could be distinguished by a behavior test monitoring in the presence of WT virgin females. In the absence of being able to fly, silkworm sexual behavior is dependent on walking distance. Therefore, we set a test field to measure movement within a radius of 10 cm or 20 cm as reported in a previous study [43] which was sufficient for male silk moths to recognize females. Once the male mates with the female, it continues to flap its wings and can remain copulated for several hours. Experientially, continuous mating for 30 minutes ensures normal sperm transfer and reproduction. So we defined and evaluated a courtship index by the following steps: (1) male moves toward the female and successfully displays orientation behaviors; (2) wing song; and (3) reorientation and tipping the abdomen. Courtship index was recorded as 1 when male moth displayed these three steps. The mating index was evaluated by measuring whether the male copulated with the female continuously for 30 minutes. Here, we set up 30 or 50 pairs as a group to measure the courtship or mating index, 3 independent biological replicates for each mutants or WT. The behavioral assays were performed at 25 °C and 60% relative humidity under normal ambient light.
Electroantennogram (EAG) recordings
Five to eleven antennas each taken from more than five virgin male moths 1–2 days after eclosion were used for EAG recordings. EAG values were recorded by using a method similar to one previously reported [56]. The antenna was cut off at the base from the head, and a few terminal segments of the antenna were excised to achieve better contact. The cut ends of the antenna were connected with two recording glass electrodes filled with 0.1 M KCl. Five to eleven individuals of each genotype were evaluated. The recordings were performed under an Olympus SZ61 microscope.
Pheromone components (bombykol and bombykal, 96% purity, purchased from Nimrod Inc., Changzhou, China) were diluted in hexane (98% purity, Sigma-Aldrich Co., St. Louis, MO, USA) and a dose of 10 μg was used for each trial as previously described [43]. Hexane was used as the control. Briefly, a filter paper strip (2.5 × 0.9 cm) was wet with 10 μl test solution and allowed to dry for 3 min, then the paper strip was inserted into a Pasteur pipette placed perpendicularly through a hole in a metal-lined tube with a humid airflow of 0.5 L/min.
Signals were amplified 10-fold (10 s, starting 1s before stimulation) by a high impedance pre-amplifier (IDAC-2 USB System, Syntech, Kirchzarten, Germany) then sent to a computer via an analog-digital converter. Off-line analysis was carried out by EAGpro 2.0 software (Syntech, The Netherlands). Relative EAG responses for each compound were calculated by subtracting EAG response for the blank from the EAG response to the test compound.
Single sensillum recordings (SSR)
To perform single sensillum recordings, a 1–2 day-old virgin male silk moth was placed in a remodeled 1-ml plastic pipette with the protruding head fixed by dental wax. The exposed antenna was attached to a cover-slip with double-faced adhesive tape. A recording tungsten wire electrode was inserted into the sensilla and reference tungsten wire electrodes were inserted into the compound eyes. Data were obtained for 3–6 individuals for each genotype and 10–20 sensilla for each individual were examined. The recordings were performed under a LEICA Z16 APO microscope at 920 × magnification.
Pheromone components were prepared as described in the “Electroantennogram (EAG) recordings” section. Humid air flow was set at 1.4 L/min, and a stimulus air pulse for 300 ms was controlled by a Syntech Stimulus controller (CS-55, Syntech, Kirchzarten, Germany). Signals were amplified 10-fold (10 s, starting 1 s before stimulation) by a high impedance pre-amplifier (IDAC-4 USB System, Syntech, Kirchzarten, Germany) then output to a computer via an analog-digital converter. Off-line analysis was carried out using AUTOSPIKE, v. 3.9, software (Syntech, Kirchzarten, Germany). The filter setting was 300 Hz at low cutoff and 2 kHz at high cutoff. The responses were measured by counting the number of action potentials within 1 s after stimulation. The number of olfactory sensory neurons housed in a single sensillum was determined based on the differences in spike amplitudes.
Quantitative RT-PCR
Total RNA was extracted from 10 silkworm antennae from 5 males per genotype using Trizol reagent (Invitrogen) and treated with RNase-free DNAse I (Ambion). cDNAs were synthesized using the Omniscript Reverse transcriptase kit (Qiagen) in a 20 μl reaction mixture containing 1 μg total RNA. Quantitative real-time RT-PCR (RT-qPCR) assays were performed using SYBR Green Realtime PCR Master Mix (Thermo Fisher Scientific) on an Eppendorf Real-time PCR System MasterCycler RealPlex instrument. RT-qPCR reactions were carried out with gene-specific primers (S1 Table). A 10-fold serial dilution of pooled cDNA was used as the template for standard curves. Quantitative mRNA measurements were performed in three independent biological replicates, and data were normalized to the amount of Bmrp49 mRNA [12].
RNA-seq protocol and data analysis
Illumina sequencing as perfomered as our previous study [57], total RNA was isolated from 10 Bmfru mutant and WT antennas using TRIzol (Invitrogen, Carlsbad, CA, USA), and the residual DNA was removed with RNase-free DNase I (New England BioLabs, Ipswich, MA, USA) for 30 min at 37 °C. For RNA-seq, library construction and sequencing using an Illumina HiSeq 2000 were conducted by BGI Genomic Services (Shenzhen, China), briefly described as follows. The mRNA was enriched using oligo (dT) magnetic beads samples were mixed with a fragmentation buffer, and the mRNA reduced to short fragments (~200 bp). The first strand of the cDNA was synthesized using random hexamer primers,buffer, dNTPs, RNase H, and DNA polymerase I were added to synthesize the second strand, and the double-stranded cDNA was purified with magnetic beads followed by performing end repair and 3’-end single nucleotide adenine addition. Finally, sequencing adaptors were ligated to the fragments which were enriched by PCR amplification, and an Agilent 2100 Bioanaylzer and an ABI Step One Plus Real-Time PCR System were used to quantify libraries. The library products were sequenced using an Illumina HiSeq 2000 (BGI Biotech Co. Ltd.). The raw sequencing data were qualified, filtered, and mapped to the reference silkworm genome database (http://silkworm.genomics.org.cn/) using tophat/bowtie2. The UniGene abundances were measured in fragments per kb of exon per million fragments mapped (FPKM). The differentially expressed genes were annotated functionally using Gene Ontology and Kyoto Encyclopedia of Genes and Genomes annotations.
Statistical analysis
Behavioral and RT-qPCR data were analyzed in GraphPad Prism 6, and electrophysiological data were analyzed in Spike 3.9 and EAGpro 2.0. Experimental data were analyzed with the Fisher exact test, chi-squared test (SPSS 20.0), ANOVA (one-way ANOVA, Dunnett’s multiple comparisons test) or Student’s t-test (Graphpad 6.0). At least three independent replicates were used for each treatment and means ± SEM were plotted. Detailed statistical information relating to each experiment is provided in the relevant Method Details or figure legends.
Supporting information
(A) The BmFRU protein, which contains the BTB domain conserved in dipteran insects, D. melanogaster and M. domestica. (B) Schematic representation of the exon/intron boundaries of the Bmfru gene. Exons are shown as boxes. Untranslated regions are shown as black boxes and coding regions as open boxes. Thin lines represent the introns and numbers are the lengths in kilobase pairs (kb). Target site locations are noted and PAM sequences are shown in red. (C) Crossing scheme to produce homozygous mutations. The binary transgenic CRISPR/Cas9 system in this study contains two lines, one of which contains the full Cas9 ORF driven by the nanos (nos) promoter, and the other contains a U6 promoter-driven sgRNA. These two lines also encode the reporter genes EGFP and DsRed2, respectively. The two transgenic lines were crossed to produce founder animals that express both Cas9 and Bmfru sgRNAs. The founder female silkworms were backcrossed with wild-types to obtain heterozygous offspring (F2, Fru+/-). F2 heterozygous mutant females were individually crossed with wild-type males to obtain distinct F3 heterozygous lines. F3 moths heterozygous for the mutations were sib-mated to generate independent lines of homozygous animals (F4, Fru-/-). (D) Homozygous mutations confirmed by sequence analysis. The targeting sequence is shown in blue and the PAM sequence in red. The deleted base pairs (bp), ATGC, are indicated by the broken line.
(TIF)
(A) Plot of significantly differentially expressed genes in 10 mixed Bmfru mutant male antennas compared to 10 mixed WT adult male antennas. False discovery rate (FDR) was used to determine the threshold of p values in multiple tests. We use FDR ≤ 0.001 and the absolute value of log2Ratio ≥ 1 as thresholds to determine significant differences in gene expression. Yellow represents up-regulated genes, blue represents down-regulated genes, and gray represents genes without significant differences. (B) Olfactory sensory system genes with changes significant at p<0.05.
(TIF)
(MP4)
(MP4)
(MP4)
(MP4)
(MP4)
(DOCX)
(XLSX)
The workbook contains two sheets. Sheet 1 shows data for antennal length. Sheet 2 presents data for the number of sensilla trichoidea in a single SEM scan field.
(XLSX)
The workbook contains six sheets. Each sheet contains raw data for a separateEAG experiment.
(XLSX)
The workbook contains six sheets. Each sheet contains raw data for a separate SSR experiment.
(XLSX)
Q-PCR data showing the mRNA level for different mutations compared to WT.
(XLSX)
Acknowledgments
We thank Jiqin Li and Xiaoyan Gao for SEM technique support. We thank all members of the Huang lab and Prof. Anjiang Tan for technical assistance and helpful discussions. We would like to thank Dr. Jacqueline Wyatt and Prof. Marian Goldsmith for proof reading the manuscript. We thank three anonymous reviewers for their constructive comments.
Data Availability
The transcriptome data are available from the Dryad database (DOI: https://doi.org/10.5061/dryad.3xsj3txbw).
Funding Statement
YH lab was supported by grants from the National Science Foundation of China (31802005, 31530072 and 31420103918), Strategic Priority Research Program of Chinese Academy of Sciences (XDB11010500), the National Postdoctoral Program for Innovative Talents (BX201700268). The authors gratefully acknowledge the support of the SA-SIBS scholarship program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Williams TM, Carroll SB. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat Rev Genet 2009; 10:797–804. 10.1038/nrg2687 [DOI] [PubMed] [Google Scholar]
- 2.Murray SM, Yang SY, Van Doren M. Germ cell sex determination: a collaboration between soma and germline. Curr Opin Cell Biol 2010; 22:722–729. 10.1016/j.ceb.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gempe T, Beye M. Function and evolution of sex determination mechanisms, genes and pathways in insects. Bioessays 2011; 33: 52–60. 10.1002/bies.201000043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graham PL, Yanowitz JL, Penn JK, Deshpande G, Schedl P. The translation initiation factor eIF4E regulates the sex-specific expression of the master switch gene Sxl in Drosophila melanogaster. PLoS Genet 2011; 7: e1002185 10.1371/journal.pgen.1002185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hashiyama K, Hayashi Y, Kobayashi S. Drosophila Sex lethal gene initiates female development in germline progenitors. Science 2011; 333: 885–888. 10.1126/science.1208146 [DOI] [PubMed] [Google Scholar]
- 6.Siera SG, Cline TW. Sexual back talk with evolutionary implications: stimulation of the Drosophila sex-determination gene sex-lethal by its target transformer. Genetics 2008; 180: 1963–1981. 10.1534/genetics.108.093898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valcárcel J, Singh R, Zamore PD, Green MR. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature 1993; 362: 171–175. 10.1038/362171a0 [DOI] [PubMed] [Google Scholar]
- 8.Mattox W, Baker BS. Autoregulation of the splicing of transcripts from the transformer-2 gene of Drosophila. Genes Dev 1991; 5: 786–796. 10.1101/gad.5.5.786 [DOI] [PubMed] [Google Scholar]
- 9.Heinrichs V, Ryner LC, Baker BS. Regulation of sex-specific selection of fruitless 5’ splice sites by transformer and transformer-2. Mol Cell Biol 1998; 18: 450–458. 10.1128/mcb.18.1.450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiuchi T, Koga H, Kawamoto M, Shoji K, Sakai H, Arai Y, et al. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 2014; 509: 633–636. 10.1038/nature13315 [DOI] [PubMed] [Google Scholar]
- 11.Sakai H, Sumitani M, Chikami Y, Yahata K, Uchino K, Kiuchi T, et al. Transgenic Expression of the piRNA-Resistant Masculinizer Gene Induces Female-Specific Lethality and Partial Female-to-Male Sex Reversal in the Silkworm, Bombyx mori. PLoS Genet 2016; 12: e1006203 10.1371/journal.pgen.1006203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Chen S, Zeng B, James AA, Tan A, Huang Y. Bombyx mori P-element Somatic Inhibitor (BmPSI) is a key auxiliary factor for silkworm male sex determination. PLoS Genet 2017; 13: e1006576 10.1371/journal.pgen.1006576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J, Zhan S, Chen S, Zeng B, Li Z, James AA, et al. Sexually dimorphic traits in the silkworm, Bombyx mori, are regulated by doublesex. Insect Biochem Mol Biol 2017; 80: 42–51. 10.1016/j.ibmb.2016.11.005 [DOI] [PubMed] [Google Scholar]
- 14.Suzuki MG, Funaguma S, Kanda T, Tamura T, Shimada T. Role of the male BmDSX protein in the sexual differentiation of Bombyx mori. Evol Dev 2005; 7: 58–68. 10.1111/j.1525-142X.2005.05007.x [DOI] [PubMed] [Google Scholar]
- 15.Xu J, Wang Y, Li Z, Ling L, Zeng B, James AA, et al. Transcription activator-like effector nuclease (TALEN)-mediated female-specific sterility in the silkworm, Bombyx mori. Insect Mol Biol 2014; 23: 800–807. 10.1111/imb.12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan J, Xu H, Ma S, Guo H, Wang F, Zhang L, et al. Ectopic expression of the male BmDSX affects formation of the chitin plate in female Bombyx mori. Mol Reprod Dev 2014; 81: 240–247. 10.1002/mrd.22290 [DOI] [PubMed] [Google Scholar]
- 17.Burtis KC, Coschigano KT, Baker B., Wensink PC. The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J 1991; 10: 2577–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams TM, Selegue JE, Werner T, Gompel N, Kopp A, Carroll SB. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell 2008; 134: 610–623. 10.1016/j.cell.2008.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka K, Barmina O, Sanders LE, Arbeitman MN, Kopp A. Evolution of sex-specific traits through changes in HOX-dependent doublesex expression. PLoS Biol 2011; 9: e1001131 10.1371/journal.pbio.1001131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo SD, Baker BS. Constraints on the evolution of a doublesex target gene arising from doublesex’s pleiotropic deployment. Proc Natl Acad Sci U S A 2015; 112: E852–861. 10.1073/pnas.1501192112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat Neurosci 2010; 13: 458–466. 10.1038/nn.2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Philipsborn AC, Jörchel S, Tirian L, Demir E, Morita T, Stern DL, Dickson BJ. Cellular and behavioral functions of fruitless isoforms in Drosophila courtship. Curr Biol 2014; 24: 242–251. 10.1016/j.cub.2013.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demir E, Dickson BJ. fruitless splicing specifies male courtship behavior in Drosophila. Cell 2005; 121: 785–794. 10.1016/j.cell.2005.04.027 [DOI] [PubMed] [Google Scholar]
- 24.Cachero S, Ostrovsky AD, Yu JY, Dickson BJ, Jefferis GS. Sexual dimorphism in the fly brain. Curr Biol 2010; 20: 1589–1601. 10.1016/j.cub.2010.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manoli DS, Foss M, Villella A, Taylor BJ, Hall JC, Baker BS. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature 2005; 436: 395–400. 10.1038/nature03859 [DOI] [PubMed] [Google Scholar]
- 26.Pan Y, Baker BS. Genetic identification and separation of innate and experience-dependent courtship behaviors in Drosophila. Cell 2014; 156: 236–248. 10.1016/j.cell.2013.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellendersen BE, von Philipsborn AC. Neuronal modulation of D. melanogaster sexual behaviour. Curr Opin Insect Sci 2017; 24: 21–28. 10.1016/j.cois.2017.08.005 [DOI] [PubMed] [Google Scholar]
- 28.Auer TO, Benton R. Sexual circuitry in Drosophila. Curr Opin Neurobiol 2016; 38: 18–26. 10.1016/j.conb.2016.01.004 [DOI] [PubMed] [Google Scholar]
- 29.Nojima T, Neville MC, Goodwin SF. Fruitless isoforms and target genes specify the sexually dimorphic nervous system underlying Drosophila reproductive behavior. Fly (Austin) 2014; 8: 95–100. 10.4161/fly.29132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gailey DA, Billeter JC, Liu JH, Bauzon F, Allendorfer JB, Goodwin SF. Functional conservation of the fruitless male sex-determination gene across 250 Myr of insect evolution. Mol Biol Evol 2006; 23: 633–643. 10.1093/molbev/msj070 [DOI] [PubMed] [Google Scholar]
- 31.Salvemini M, Robertson M, Aronson B, Atkinson P, Polito LC, Saccone G. Ceratitis capitata transformer-2 gene is required to establish and maintain the autoregulation of Cctra, the master gene for female sex determination. Int J Dev Biol 2009; 53: 109–120. 10.1387/ijdb.082681ms [DOI] [PubMed] [Google Scholar]
- 32.Salvemini M, D’Amato R, Petrella V, Aceto S, Nimmo D, Neira M, Alphey L, Polito LC, Saccone G. The orthologue of the fruitfly sex behaviour gene fruitless in the mosquito Aedes aegypti: evolution of genomic organisation and alternative splicing. PLoS One 2013; 8: e48554 10.1371/journal.pone.0048554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertossa RC, van de Zande L, Beukeboom LW. The Fruitless gene in Nasonia displays complex sex-specific splicing and contains new zinc finger domains. Mol Biol Evol 2009; 26: 1557–1569. 10.1093/molbev/msp067 [DOI] [PubMed] [Google Scholar]
- 34.Meier N, Käppeli SC, Hediger Niessen M, Billeter JC, Goodwin SF, Bopp D. Genetic control of courtship behavior in the housefly: evidence for a conserved bifurcation of the sex-determining pathway. PLoS One 2013; 8: e62476 10.1371/journal.pone.0062476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clynen E, Ciudad L, Bellés X, Piulachs MD. Conservation of fruitless’ role as master regulator of male courtship behaviour from cockroaches to flies. Dev Genes Evol 2011; 221: 43–48. 10.1007/s00427-011-0352-x [DOI] [PubMed] [Google Scholar]
- 36.Kaissling KE, Kasang G, Bestmann H, Stransky W, Vostrowsky O. A new pheromone of the silkworm moth Bombyx mori. Naturwissenschaften 1978; 65: 382–384. [Google Scholar]
- 37.Sakurai T, Nakagawa T, Mitsuno H, Mori H, Endo Y, Tanoue S, et al. Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc Natl Acad Sci U S A 2004; 101: 16653–16658. 10.1073/pnas.0407596101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science 2005; 307: 1638–1642. 10.1126/science.1106267 [DOI] [PubMed] [Google Scholar]
- 39.Grosse-Wilde E, Svatos A, Krieger J. A pheromone-binding protein mediates the bombykol-induced activation of a pheromone receptor in vitro. Chem Senses 2006; 31: 547–555. 10.1093/chemse/bjj059 [DOI] [PubMed] [Google Scholar]
- 40.Sakurai T, Mitsuno H, Mikami A, Uchino K, Tabuchi M, Zhang F, et al. Targeted disruption of a single sex pheromone receptor gene completely abolishes in vivo pheromone response in the silkmoth. Sci Rep 2015; 5: 11001 10.1038/srep11001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shiota Y, Sakurai T, Daimon T, Mitsuno H, Fujii T, Matsuyama S, et al. In vivo functional characterisation of pheromone binding protein-1 in the silkmoth, Bombyx mori. Sci Rep 2018; 8: 13529 10.1038/s41598-018-31978-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki MG, Funaguma S, Kanda T, Tamura T, Shimada T. Analysis of the biological functions of a doublesex homologue in Bombyx mori. Dev Genes Evol 2003; 213: 345–354. 10.1007/s00427-003-0334-8 [DOI] [PubMed] [Google Scholar]
- 43.Liu Q, Liu W, Zeng B, Wang G, Hao D, Huang Y. Deletion of the Bombyx mori odorant receptor co-receptor (BmOrco) impairs olfactory sensitivity in silkworms. Insect Biochem Mol Biol 2017; 86: 58–67. 10.1016/j.ibmb.2017.05.007 [DOI] [PubMed] [Google Scholar]
- 44.Daimon T, Fujii T, Fujii T, Yokoyama T, Katsuma S, Shinoda T, et al. Reinvestigation of the sex pheromone of the wild silkmoth Bombyx mandarina: the effects of bombykal and bombykyl acetate. J Chem Ecol 2012; 38: 1031–1035. 10.1007/s10886-012-0164-0 [DOI] [PubMed] [Google Scholar]
- 45.Wanner KW, Anderson AR, Trowell SC, Theilmann DA, Robertson HM, Newcomb RD. Female-biased expression of odourant receptor genes in the adult antennae of the silkworm, Bombyx mori. Insect Mol Biol 2007; 16: 107–119. 10.1111/j.1365-2583.2007.00708.x [DOI] [PubMed] [Google Scholar]
- 46.Katsuma S, Kiuchi T, Kawamoto M, Fujimoto T, Sahara K. Unique sex determination system in the silkworm, Bombyx mori: current status and beyond. Proc Jpn Acad Ser B Phys Biol Sci 2018; 94: 205–216. 10.2183/pjab.94.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sawanth SK, Gopinath G, Sambrani N, Arunkumar KP. The autoregulatory loop: A common mechanism of regulation of key sex determining genes in insects. J Biosci 2016; 41: 283–294. 10.1007/s12038-016-9609-x [DOI] [PubMed] [Google Scholar]
- 48.Sakurai T, Mitsuno H, Haupt SS, Uchino K, Yokohari F, Nishioka T, et al. A single sex pheromone receptor determines chemical response specificity of sexual behavior in the silkmoth Bombyx mori. PLoS Genet 2011; 7: e1002115 10.1371/journal.pgen.1002115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tabuchi M, Sakurai T, Mitsuno H, Namiki S, Minegishi R, Shiotsuki T, et al. Pheromone responsiveness threshold depends on temporal integration by antennal lobe projection neurons. Proc Natl Acad Sci U S A 2013; 110: 15455–15460. 10.1073/pnas.1313707110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka K, Uda Y, Ono Y, Nakagawa T, Suwa M, Yamaoka R, et al. Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr Biol 2009; 19: 881–890. (Erratum in 2011, 21: 623.) 10.1016/j.cub.2009.04.035 [DOI] [PubMed] [Google Scholar]
- 51.Heinrichs V, Ryner LC, Baker BS. Regulation of sex-specific selection of fruitless 5’ splice sites by transformer and transformer-2. Mol Cell Biol 1998; 18: 450–458. 10.1128/mcb.18.1.450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohbayashi F. Structural and functional analyses on the Bombyx mori genes homologous to Drosophila doublesex and fruitless. Ph.D. thesis, The University of Tokyo; 2001.
- 53.Wang Q, Taliaferro JM, Klibaite U, Hilgers V, Shaevitz JW, Rio DC. The PSI-U1 snRNP interaction regulates male mating behavior in Drosophila. Proc Natl Acad Sci U S A 2016; 113: 5269–5274. 10.1073/pnas.1600936113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Li Z, Xu J, Zeng B, Ling L, You L, et al. The CRISPR/Cas System mediates efficient genome engineering in Bombyx mori. Cell Res 2013; 23: 1414–1416. 10.1038/cr.2013.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naito Y, Hino K, Bono H, Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 2015; 31: 1120–1123. 10.1093/bioinformatics/btu743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu GH, Xu J, Cui Z, Dong XT, Ye ZF, Niu DJ, et al. Functional characterization of SlitPBP3 in Spodoptera litura by CRISPR/Cas9 mediated genome editing. Insect Biochem Mol Biol 2016; 75: 1–9. 10.1016/j.ibmb.2016.05.006 [DOI] [PubMed] [Google Scholar]
- 57.Xu J, Yu Y, Chen K, Huang Y. Intersex regulates female external genital and imaginal disc development in the silkworm. Insect Biochem Mol Biol 2019; 108: 1–8. 10.1016/j.ibmb.2019.02.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) The BmFRU protein, which contains the BTB domain conserved in dipteran insects, D. melanogaster and M. domestica. (B) Schematic representation of the exon/intron boundaries of the Bmfru gene. Exons are shown as boxes. Untranslated regions are shown as black boxes and coding regions as open boxes. Thin lines represent the introns and numbers are the lengths in kilobase pairs (kb). Target site locations are noted and PAM sequences are shown in red. (C) Crossing scheme to produce homozygous mutations. The binary transgenic CRISPR/Cas9 system in this study contains two lines, one of which contains the full Cas9 ORF driven by the nanos (nos) promoter, and the other contains a U6 promoter-driven sgRNA. These two lines also encode the reporter genes EGFP and DsRed2, respectively. The two transgenic lines were crossed to produce founder animals that express both Cas9 and Bmfru sgRNAs. The founder female silkworms were backcrossed with wild-types to obtain heterozygous offspring (F2, Fru+/-). F2 heterozygous mutant females were individually crossed with wild-type males to obtain distinct F3 heterozygous lines. F3 moths heterozygous for the mutations were sib-mated to generate independent lines of homozygous animals (F4, Fru-/-). (D) Homozygous mutations confirmed by sequence analysis. The targeting sequence is shown in blue and the PAM sequence in red. The deleted base pairs (bp), ATGC, are indicated by the broken line.
(TIF)
(A) Plot of significantly differentially expressed genes in 10 mixed Bmfru mutant male antennas compared to 10 mixed WT adult male antennas. False discovery rate (FDR) was used to determine the threshold of p values in multiple tests. We use FDR ≤ 0.001 and the absolute value of log2Ratio ≥ 1 as thresholds to determine significant differences in gene expression. Yellow represents up-regulated genes, blue represents down-regulated genes, and gray represents genes without significant differences. (B) Olfactory sensory system genes with changes significant at p<0.05.
(TIF)
(MP4)
(MP4)
(MP4)
(MP4)
(MP4)
(DOCX)
(XLSX)
The workbook contains two sheets. Sheet 1 shows data for antennal length. Sheet 2 presents data for the number of sensilla trichoidea in a single SEM scan field.
(XLSX)
The workbook contains six sheets. Each sheet contains raw data for a separateEAG experiment.
(XLSX)
The workbook contains six sheets. Each sheet contains raw data for a separate SSR experiment.
(XLSX)
Q-PCR data showing the mRNA level for different mutations compared to WT.
(XLSX)
Data Availability Statement
The transcriptome data are available from the Dryad database (DOI: https://doi.org/10.5061/dryad.3xsj3txbw).