The lesion bypass pathway, translesion synthesis (TLS), exists in essentially all organisms and is considered a pathway for postreplicative gap repair and, at the same time, for lesion tolerance. As with the saying “a trip is not over until you get back home,” studying TLS only at the site of the lesion is not enough to understand the whole process of TLS. Recently, a genetic study uncovered that polymerase V (Pol V), a poorly expressed Escherichia coli TLS polymerase, is not only involved in the TLS step per se but also participates in the gap-filling reaction over several hundred nucleotides.
KEYWORDS: β-clamp, Pol IV, Pol V, RecA, translesion synthesis, evolution, gap-filling synthesis, postreplicative gap repair, replicative polymerase, untargeted mutagenesis
SUMMARY
The lesion bypass pathway, translesion synthesis (TLS), exists in essentially all organisms and is considered a pathway for postreplicative gap repair and, at the same time, for lesion tolerance. As with the saying “a trip is not over until you get back home,” studying TLS only at the site of the lesion is not enough to understand the whole process of TLS. Recently, a genetic study uncovered that polymerase V (Pol V), a poorly expressed Escherichia coli TLS polymerase, is not only involved in the TLS step per se but also participates in the gap-filling reaction over several hundred nucleotides. The same study revealed that in contrast, Pol IV, another highly expressed TLS polymerase, essentially stays away from the gap-filling reaction. These observations imply fundamentally different ways these polymerases are recruited to DNA in cells. While access of Pol IV appears to be governed by mass action, efficient recruitment of Pol V involves a chaperone-like action of the RecA filament. We present a model of Pol V activation: the 3′ tip of the RecA filament initially stabilizes Pol V to allow stable complex formation with a sliding β-clamp, followed by the capture of the terminal RecA monomer by Pol V, thus forming a functional Pol V complex. This activation process likely determines higher accessibility of Pol V than of Pol IV to normal DNA. Finally, we discuss the biological significance of TLS polymerases during gap-filling reactions: error-prone gap-filling synthesis may contribute as a driving force for genetic diversity, adaptive mutation, and evolution.
INTRODUCTION
Cells have evolved robust and versatile defense mechanisms to cope with various types of DNA damage derived from endogenous and exogenous sources. Endogenous DNA damage stems from normal metabolic processes. For instance, reactive oxygen species (ROS), derived from hydrogen peroxide (H2O2) and superoxide (O2–), are produced via respiration of mitochondria in eukaryotes (1). In the case of Escherichia coli, ROS are generated via aerobic metabolism under normal aerobic conditions (2, 3). It has also been reported that sublethal levels of bactericidal antibiotics stimulate ROS generation in cells (4). Through whole-genome sequencing approaches in various E. coli mutant strains, it has been suggested that oxidative DNA damage induced by ROS is the most potent endogenous source contributing to spontaneous mutagenesis (5). The intrinsic instability of DNA per se, such as that represented by an abasic lesion (apurinic/apyrimidinic [AP] site), is also a source of DNA damage (6). In addition, AP sites also appear via enzymatic action, such as uracil DNA glycosylase acting at uracil formed upon spontaneous deamination of cytosine (7). The steady-state number of lesions caused by endogenous sources in human DNA is estimated to be over 40,000 per cell (with lesions at the AP site being the most prevalent, estimated at ∼30,000) (8). Taken together, not surprisingly, the level of endogenous DNA damage is largely affected by various environmental and cellular parameters (e.g., temperature, pH, aerobic condition, cellular metabolism, availability of DNA repair enzymes, etc.).
With respect to exogenous sources of DNA damage, they are derived from numerous chemicals (e.g., polycyclic aromatic hydrocarbons [PAHs] formed during incomplete combustion in cigarette smoke or car exhausts [9] and alkylating agents during chemotherapy [10]) and environmental sources (e.g., UV light via sunshine [11]). Most DNA damage is effectively repaired through canonical DNA repair pathways, such as nucleotide excision repair (NER) and base excision repair (BER) (12). Inactivation of the canonical excision repair pathways makes cells highly sensitive to DNA damage (12, 13). However, even in the presence of efficient DNA repair systems, a subfraction of DNA damage escapes repair and has an increased likelihood of encountering the replicative DNA polymerase (Pol) on the leading strand (Fig. 1, step a) or the lagging strand (Fig. 1, step a′), at the fork. In the case of replication-blocking lesions, the replication fork skips over the DNA damage and reinitiates downstream in the leading strand (Fig. 1, step b) or skips over the DNA damage without the need of reinitiation in the lagging strand (Fig. 1, step b′). Thus, irrespective of the location of a lesion on either the leading or the lagging strand, the fork commonly leaves a single-stranded DNA (ssDNA) gap behind (see details in the next section) (14, 15). As a consequence, the unrepaired lesion is now located in the context of ssDNA (Fig. 1, step b or b′), where it becomes irreparable by regular DNA repair systems that function only on double-stranded DNA (dsDNA) (16).
FIG 1.
Schematic overview of postreplicative gap repair. (a) The fork moves from left to right. The red triangle represents a replication-blocking lesion in the leading strand. The green arrow represents the direction of the leading-strand Pol. (a′) Same as shown in panel a except that the replication-blocking lesion is in the lagging strand. The green arrow represents the direction of the lagging-strand Pol. (b and b′) In the case of the leading-strand lesion, the fork skips over the lesion and downstream repriming leaves an ssDNA gap. In the case of the lagging-strand lesion, the fork skips over the lesion and an ssDNA gap spontaneously appears due to the preexisting upstream Okazaki fragment. The green arrow represents the direction of Pol during gap-filling synthesis. Subsequently, the gap is repaired either via TLS (c to e) or HDGR (f to h). Independently of the initial location of the lesion, the structure of the gap is the same and its downstream processing therefore is not distinguished. (c) The pink line represents a short DNA patch, mediated by a TLS Pol, across and beyond the lesion. (d) A DNA Pol fills in the remaining gap, followed by ligation. (e) At certain steps (c and d), the lesion will be repaired by regular repair systems. (f) The lesion, located in ssDNA, is relocated to dsDNA via sister chromatid exchange. (g) A DNA Pol fills in the newly appeared gap. (h) Resolution of the Holliday junction and ligation. At any time (steps f to h), when the lesion is relocated to dsDNA, it will be repaired by regular repair systems. (i) Schematic illustration of the classical right-hand structure of DNA Pol.
In order to cope with DNA damage located in ssDNA, cells have evolved two so-called DNA damage tolerance strategies that take place in the context of postreplicative gap repair (Fig. 1). The most straightforward damage tolerance strategy involves specialized DNA Pols that directly bypass DNA lesions, a process called translesion synthesis (TLS) (Fig. 1, step c) (17–19). Whereas some types of DNA lesions are bypassed by the replicative Pols themselves (e.g., 8-oxo-dG) (20), in this review we define TLS as an event requiring the action of a specialized Pol (i.e., TLS Pol). TLS is potentially highly mutagenic when DNA lesions are noninstructive {e.g., AP site; TT pyrimidine-pyrimidone (6-4) photoproduct [TT (6-4)]}. Therefore, the resulting nascent strand often contains a mutation opposite the template lesion site (a process referred to as “targeted mutagenesis”). It should be noted that activation of TLS pathways in E. coli is unlikely to be induced by endogenous DNA damage that arises during normal cellular metabolism (3, 8) at a level that can be dealt with efficiently in the canonical DNA repair systems (5, 21). Thus, the TLS pathway is essentially activated by an excess amount of exogenous DNA-damaging agents that exceeds the capacity of canonical DNA repair systems (22, 23).
The second DNA damage tolerance strategy relying on template switching mechanisms, referred to as homology-dependent gap repair (HDGR) or homology-directed repair (HDR) (18, 24), aims at relocating the lesion into a double-stranded context by means of a homologous recombination intermediate with the complementary sister chromatid (Fig. 1, step f) (13, 16, 18, 25–27). Irrespective of the precise model, the main purpose of HDGR (which is also the case for TLS) is to prevent deleterious discontinuities in DNA strands and to reinstate the DNA damage from its irreparable ssDNA context to a repairable dsDNA context (Fig. 1, steps c to e or f to h). There is another template switching mechanism, usually referred to as fork regression or fork reversal, that allows a replication fork, arrested in the vicinity of a lesion or a secondary structure in the leading-strand template, to restart without formation of an ssDNA gap (18, 28).
Owing to the intrinsic low fidelity of TLS Pols, the process of TLS is highly mutagenic both at the lesion bypass step per se (targeted mutagenesis) and downstream from the lesion site on normal template DNA (untargeted mutagenesis) (Fig. 1, step b/b′ to c). As discussed later in this review, despite their high mutagenic load, TLS pathways are present in most species (Gram-negative and Gram-positive bacteria, lower and higher eukaryotes, etc.) (29–31), strongly suggesting that TLS pathways are likely to be beneficial for evolution in the long term and for prevention of persistent ssDNA gaps in the short term, rather than being an extra backup pathway. In E. coli, an emergency situation causing occurrence of persistent ssDNA gaps (e.g., an excess amount of DNA damage) leads to activation of the SOS response, which is an inducible protein network system, including the activation of the TLS pathway, in order to allow cells to survive (21–23). This SOS response is not a particular system in the Gram-negative bacterium E. coli (and its closely related bacteria); rather, it is broadly conserved among bacteria (21), including, for example, the Gram-negative bacterium Pseudomonas aeruginosa (a gammaproteobacterium distantly related to E. coli) (32) and the Gram-positive bacterium Bacillus subtilis (33).
The bypass of many different DNA lesions by different TLS Pols has been extensively studied in vitro (17, 34). However, the overall process of TLS in vivo has received little attention until recently (35, 36). The present article aims to discuss how the various Pols that are encoded and expressed at different levels in E. coli manage the overall process of postreplicative gap repair in vivo. We propose a TLS model involving an interplay between the error-prone TLS Pols and the high-fidelity Pols in the context of physiologically relevant conditions according to parameters such as genetically required factors and the expression level of individual Pols (17) and discuss its biological consequences.
WHAT HAPPENS WHEN A REPLICATION FORK ENCOUNTERS A LESION?
When an ongoing replication fork encounters a replication-blocking lesion in one of the template strands, it was initially expected that the fate of the fork would depend on whether the lesion is located in the leading- or lagging-strand template. For a lesion in the lagging strand, due to preexisting Okazaki fragments in the upstream region of the fork, the fork would go ahead, leaving a gap (i.e., an unfinished Okazaki fragment) (Fig. 1, step a′). Consequently, the lesion is processed in the context of postreplicative gap repair (Fig. 1, step b′) (18). With a leading-strand lesion, the fork may stall due to the concept of continuous leading-strand replication, as observed with reconstituted replication forks in vitro (37, 38). However, initial data from Okazaki et al. indicated that, in E. coli, normal replication is discontinuous in both leading and lagging strands (39). Discontinuous strand synthesis has also been demonstrated when E. coli is damaged by UV irradiation (14), suggesting that the fork can skip over UV-induced lesions during both leading- and lagging-strand synthesis.
The reason these observations had not been taken into account was the lack of biochemical evidence for leading-strand repriming. However, nearly 40 years after the classic in vivo observations, it was shown that repriming events relying on DnaG primase can occur during leading-strand synthesis in vitro, either in the presence of accessory factors related to de novo priming activities (PriA- or PriC-dependent pathway) (40) or in the absence of such factors (41). Since transcription is always implemented during DNA replication in vivo, an ongoing replication fork would occasionally encounter an ongoing transcription apparatus on the same template strand. When such a situation, i.e., where a leading-strand polymerase at a fork encounters an RNA polymerase with an RNA transcript on the same template strand (i.e., codirectional collision) in vitro, was mimicked, it was demonstrated that the fork removes the RNA polymerase while the RNA transcript is retained on the template strand. Subsequently, the leading-strand polymerase skips over the RNA transcript and utilizes it as a primer, leading to resumption of replication (42). In addition to the de novo repriming events described above, this event, RNA transcript-mediated replication resumption, may be an additional layer enabling cells to discontinuously synthesize the leading strand in vivo. In order to address whether the leading strand is discontinuously synthesized in E. coli, nascent genomic DNA was prepared from a temperature-sensitive ligase mutant under ligase-deficient conditions, which impair joining of nascent discontinuous strands, and the DNA size was analyzed. The results suggested that discontinuous synthesis of both leading and lagging strands likely operates in vivo (43, 44). In addition, based on a similar setup using the temperature-sensitive ligase mutant, it was recently proposed that leading-strand repriming is ∼70-fold less frequent than lagging-strand repriming, again suggesting the existence of discontinuous leading-strand synthesis in vivo (45).
These observations led to the suggestion that repriming of leading-strand synthesis can occur downstream from a replication-blocking lesion, leaving a gap behind the fork. As a consequence, the process of TLS became regarded as part of postreplicative gap repair for both leading- and lagging-strand lesions rather than a continuous process occurring at the fork (15), sometimes referred to as “TLS on the fly” (46). Indeed, the scenario of TLS being implemented behind the replication fork has been supported via single-molecule fluorescence assays in which the localization of a TLS Pol, Pol V, was shown to be distinct from the localization of the Pol III replisome following UV irradiation in vivo (47). Similarly, single-molecule fluorescence assays in vivo also indicated that Pol IV, another TLS Pol, principally colocalizes outside the replisome following exposure of DNA-damaging agents (e.g., 4-nitroquinoline N-oxide [4-NQO]), implying that Pol IV-mediated TLS also occurs predominantly behind, rather than at, the replication fork (48).
CHARACTERISTIC FEATURES OF DNA POLYMERASES IN E. COLI
E. coli possesses five DNA polymerases (Pol I to V), and their characteristic features are summarized in Table 1. DNA Pols are phylogenetically classified into families: in E. coli, Pol I, Pol II, and Pol III belong to A, B, and C families, respectively, while Pol IV and Pol V are Y family Pols (29). Although Pol I to III belong to different families, all three Pols possess 3′→5′ exonuclease activity to correct replication errors induced by themselves (i.e., proofreading function) (38). This proofreading function contributes to ∼100-fold-higher accuracy during DNA synthesis (49). Although these high-fidelity Pols (i.e., Pol I to III) have been shown to exhibit bypass properties for some lesions under certain experimental conditions (50–52), their contribution to TLS under physiological conditions is limited (largely unknown) except for a few examples (17). In contrast, the two remaining Y family Pols (i.e., Pol IV and V) have characteristic features of TLS Pols, namely, their lack of proofreading function and their capacity to accommodate bulky lesions given their spacious catalytic domain (29, 53). Such structurally characteristic features are described in “Structural Features of TLS Pols" (see below).
TABLE 1.
Features of DNA polymerases in E. colia
| Polymerase | Family | Proofreading function (3′→5′ exonuclease) | Error rate/base | Molecules/cell without SOS induction | Molecules/cell with SOS induction | Processivity (nt) without the β-clamp | Processivity (nt) with the β-clamp | Velocity (nt/s) |
|---|---|---|---|---|---|---|---|---|
| Pol I | A | Yes | 10–6 (176) | 400 (38) | Noninducible | 15–20 (113) | ≤700 (179, 180) | 13–15 (182, 183) |
| Pol II | B | Yes | 10–6 (177) | ≤50 (73) | ≤350 (73) | 5 (114) | ≥1,600 (114) | 20–30 (114) |
| Pol III | C | Yes | 10–6–10–7 (107) | 20 (112) | Noninducible | 10–15 (115) | ≥50,000 (181) | 730 (181) |
| Pol IV | Y | No | 10–3–10–5 (178) | 250 (74) | 2,500 (74) | 1 (94) | 300–400 (94) | 3–5 (94) |
| Pol V | Y | No | 10–3–10–5 (178) | Undetectable | 15–60 (75) | 1 (102) | 25 (65, 102) | 0.29 (65, 102) |
References are given in parentheses.
Classification of DNA Lesions according to the Bypass Capacities of TLS Pols
The DNA double helix entails two grooves, termed major and minor, the ratio of widths between them being 7:4, respectively, in the regular B-form DNA (54). Base adducts in DNA can be located in either the major groove (e.g., 4-aminobiphenyl [55], N-2-acetylaminofluorene [56]) or the minor groove (e.g., benzo[a]pyrene [57], N-2-furfuryl [58]), essentially depending upon the position of the atom to which the adduct is bound (59–63). Interestingly, experimental evidence supports the notion that Pol IV and Pol V appear to be adapted for bypass of minor and major groove lesions (i.e., division of labor), respectively (17). It may seem awkward to refer to minor versus major groove location during TLS when the adduct is located at the ssDNA-to-dsDNA junction (Fig. 1, step b/b’). However, in the Pol active site, the conformation of the damaged nucleotide (at the ssDNA-to-dsDNA junction) may adopt, during the nucleotide insertion step, the same conformation as in dsDNA, allowing thus for Pol IV versus Pol V selectivity. Of note, the choice of a TLS polymerase at the insertion step is undoubtedly performed through stochastic, trial-and-error access of Pols (17), rather than in a scenario where a lesion would specifically recruit a Pol that is best suited for its bypass. Beyond major groove lesions, Pol V can bypass other types of replication-blocking lesions (e.g., AP site, cyclobutane pyrimidine dimer [CPD] [64], etc.) (17), lesions that may not be categorized into major or minor groove lesions. In this review, by definition, we will consider major and minor groove lesions as lesions that can be bypassed by Pol V and Pol IV, respectively.
The SOS Response
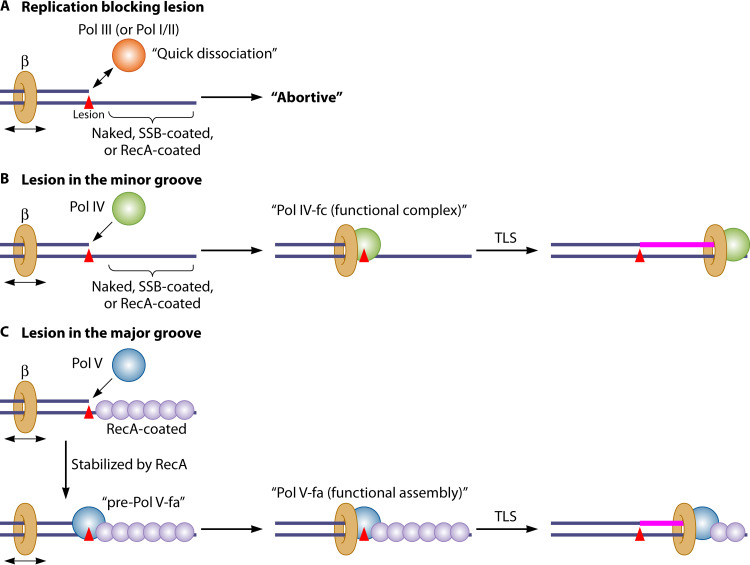
When a replicative Pol stalls at a replication-blocking lesion and then dissociates, a 3′ end of the nascent strand is typically located 1 nucleotide (nt) before the lesion site (65); upon dissociation, the stalled Pol leaves its processivity factor (the β-clamp) bound to the template DNA (65, 66). Downstream of the lesion, an exposed ssDNA gap first becomes coated by ssDNA binding (SSB) proteins, which are subsequently replaced by RecA proteins with the assistance of the mediator proteins RecFOR, leading to formation of a RecA nucleoprotein filament (termed RecA*) (16, 17, 67, 68). This RecA* stimulates autocleavage of the transcriptional repressor protein LexA (the core regulator of the SOS response), leading to upregulation of as many as 43 SOS genes, including those encoding DNA repair proteins and TLS Pols (69). LexA binding sites in the SOS genes show diverse sequence contexts modulating its binding affinity. Since LexA concentration decreases with time following induction of the SOS response, the individual SOS genes are expressed at different times during the ∼1-h period (22, 70, 71). In principle, the SOS response is turned on by the presence of persistent ssDNA gaps and turned off when these gaps are repaired. Thus, if gaps are repaired quickly, SOS genes turned on late fail to be induced. It should be noted that if ssDNA gaps persist, any Pols (or other types of DNA binding proteins like exonucleases and helicases [72]) can stochastically access vacant 3′ ends of nascent strands in gaps. If a high-fidelity Pol (i.e., Pol I to III) accesses the 3′ end in the vicinity of a lesion [e.g., TT (6-4)], it either quickly detaches as a consequence of nonproductive association/dissociation cycles (Fig. 2A) or causes degradation/resynthesis cycles upstream from the lesion site (65, 66).
FIG 2.
Stochastic access of DNA Pols on lesion-containing template DNA. Functional TLS requires a proper lesion-Pol combination. (A) We discuss only the case of a lesion that cannot be bypassed by high-fidelity Pols (i.e., Pol I to III). However, these Pols occasionally get involved in nonproductive degradation/resynthesis cycles at primer ends in the vicinity of lesions (65). (B) For a minor groove lesion, Pol IV following interaction with the β-clamp (i.e., Pol IV functional complex [Pol IV-fc]) bypasses the lesion in any stage of the SOS response. (C) For a major groove lesion, during the late SOS response, Pol V is stabilized by RecA, leading to the formation of a pre-Pol V-fa complex (Pol V with RecA*), followed by interaction with the β-clamp (Pol V functional assembly [Pol V-fa]), subsequently bypassing the lesion.
DNA Pols Induced by the SOS Response
As shown in Table 1, three Pols (i.e., Pol II, Pol IV, Pol V) are induced by the SOS response. Pol II and Pol IV are expressed under normal growth conditions, and their expressions are upregulated ∼7-fold for Pol II (73) and ∼10-fold for Pol IV (74) under the SOS response. In contrast, Pol V is virtually absent unless the SOS response is triggered (75). In the cases of Pol II and Pol IV, their transcriptional regulation is directly linked to their expression as Pols. On the other hand, expression of Pol V is not simply regulated at the transcriptional level. Pol V is encoded in the umuDC operon (UmuC, catalytic subunit; UmuD, auxiliary subunit). Under the SOS response, both proteins are expressed and can form a UmuD2C complex (UmuC with a UmuD dimer), although this complex has no polymerase activity. Instead, when the RecA nucleoprotein filament stimulates autocleavage of UmuD, converting it into the truncated form UmuD′ (lacking the N-terminal 24 amino acids of UmuD), UmuC interacts with the UmuD′2 homodimer, leading to the UmuD′2C complex that exhibits polymerase activity, termed Pol V (76, 77). Thus, expression of Pol V is regulated not only at the transcriptional level but also at the posttranslational level.
Structural Features of TLS Pols
Irrespective of sequence similarity (homology) of genes encoding Pols in any species, most Pols structurally exhibit a similar conformation, a so-called right-hand structure composed of palm, thumb, and finger domains (Fig. 1i) (78). Within this frame, when a Pol accesses a template/primer (T/P) that is composed of a long ssDNA (template) annealed with a short ssDNA (primer), one can envision an image of a right hand grabbing a chain. Although Y family TLS Pols also form a right-hand structure, they have an additional domain, called the little finger domain (or polymerase-associated domain), which is required for DNA binding (79, 80). The palm domain contains the catalytic domain for all Pols. The thumb and finger domains in TLS Pols are smaller than those of high-fidelity Pols, allowing TLS Pols to form more spacious catalytic domains (79, 80). Thus, whether the catalytic domain can physically accommodate a lesion (e.g., bulky adduct) becomes an important determinant for the lesion bypass ability of Pols. On the other hand, a more detailed structural analysis is required to analyze lesion bypass specificity of individual TLS Pols (78). Even if they belong to the same Y family, Pol IV and Pol V exhibit different lesion bypass specificities, as described in “Classification of DNA Lesions according to the Bypass Capacities of TLS Pols" (see above). In the case of Pol IV, structural data support the fact that Pol IV can accommodate minor groove lesions without interfering with its catalytic activity (81). In the case of Pol V, there is so far no structural data available; instead, its structural features have been studied through homology modeling in silico (53), and it has been proposed that Pol V may have structural similarities with human Pol η (Y family), which also has the ability to bypass CPD.
Should Pol II Be Regarded as a TLS Pol?
Background of Pol II.
Physiologically, the most important role of DNA polymerases is undoubtedly DNA replication in order to duplicate genomic DNA. In E. coli, Pol I and Pol III are assigned to this role while Pol II is dispensable (38). Although Pol II was identified in 1970, its physiological role is largely unknown (38). In 1990, Pol II was found to be regulated by the SOS response (82, 83). Since then, it has been speculated that Pol II may function as a TLS Pol. Until 1999, when both Pol IV and Pol V were identified (29), various studies have tried to discover lesion bypass activities in Pol I, Pol II, and Pol III (17, 38, 84). For instance, Pol I, Pol II, and Pol III all have the ability to bypass AP sites with comparable efficiencies in vitro (50). Currently, Pol II is often lumped into the category of TLS Pol, the main reason being that in addition to its SOS inducibility, Pol II has been demonstrated to be able to bypass an N-2-acetylaminofluorene adduct (AAF) in vivo as well as in vitro (85, 86). We discuss the molecular events that occur during AAF bypass by Pol II below.
AAF-dependent frameshift mutagenesis.
The AAF belongs to a large family of aromatic amides and preferentially binds to the C-8 position of guanine, forming AAF-dG (87). When binding to particular sequences such as the NarI sequence (3′-CCGAAFCGG), AAF represents a hot spot for –2 frameshift mutations in vivo (88). In the NarI context, the spontaneous –2 frameshift mutation frequency is below 10–8 in E. coli. In contrast, in the presence of an AAF-dG, the –2 frameshift mutation frequency reaches the 10–2 to 10–1 range but drops to the 10–4 to 10–3 range in the absence of Pol II (85). In addition, Pol II bypasses the AAF-containing NarI context via either the –1 or –2 frameshift intermediate with comparable efficiency in vitro (66). Thus, in vivo and in vitro, Pol II indirectly bypasses AAF via structured DNA such as frameshift intermediates rather than directly bypassing AAF per se, as demonstrated in the case of Pol V (88).
Insight into the definition of TLS Pol.
In the absence of Pol II, –2 frameshift mutagenesis in the AAF-containing NarI context is still detectable, and its frequency is much higher (104- to 105-fold) than that of spontaneous –2 frameshift mutagenesis in the NarI context in vivo (85). It may indicate that Pol III (and/or Pol I) has the ability to induce the –2 frameshift mutations in vivo. In the AAF-containing NarI context, indeed, Pol III efficiently inserts a nucleotide opposite AAF-dG and produces frameshift intermediates in vitro (65).
We suggest that the following three criteria have to be met in order for a Pol to qualify as a TLS Pol: (i) it can bypass a replication-blocking lesion; (ii) it can directly traverse a lesion irrespective of the DNA sequence context; and (iii) it can synthesize, in a single binding event, a productive DNA patch of 4 nt or more beyond a lesion site (65). When a Pol is able to elongate a slippage intermediate formed within a specific sequence context (e.g., AAF-dG in the NarI context by Pol II), we suggest that this event be qualified as a “lesion-skipping” event rather than a strictly defined TLS event. Indeed, this event is sequence specific and does not comply with criterion ii.
In the case of eukaryotes, TLS events often require two Pols: one Pol (inserter) inserts a nucleotide opposite a lesion, and another (extender) extends the nucleotide inserted by the inserter Pol (89). In this two-polymerase system, it is thought that the two required Pols are dynamically recruited in the vicinity of a lesion via assistance of multiple auxiliary proteins, leading to a seamless handoff from an inserter Pol to an extender Pol (90). Thus, to apply the above definition to the eukaryotic two-Pol system, it is a protein complex including multiple Pols that will be regarded as the TLS Pol.
What Is the Mechanism Underlying Functional Access of Pol IV to Template DNA?
If a lesion is located in the minor groove {e.g., [–ta]-benzo[a]pyrene-N2-dG ([–ta]-BaP-dG)}, Pol IV bypasses the lesion in vivo (57). Pol IV first accesses a 3′ end of nascent strand in the vicinity of the lesion and subsequently interacts with a freely sliding β-clamp, leading to the formation of a functional complex (Pol IV-fc) (Fig. 2B; Table 2). It should be stressed that the opposite scenario (i.e., the β-clamp recruits Pols) is unlikely, since the β-clamp is freely sliding on DNA (91). Interaction with the β-clamp is genetically essential for Pol IV-mediated TLS (92); moreover, it has been shown that in vitro under multiple-hit conditions (i.e., Pols freely access T/P during the reaction time), the processivity of Pol IV alone is only 1 nt (93, 94), indicating high instability of Pol IV on the T/P. These observations imply that only a minor fraction of transiently bound Pol IV will succeed in interacting with a freely sliding β-clamp. Thus, the rate-limiting factor for successful TLS by Pol IV is the formation of Pol IV-fc (Fig. 2B).
TABLE 2.
Nomenclature of protein complexes derived from Pol IV and Pol V
| Complexa | Components in the complexb |
|---|---|
| Pol IV-derived complex | |
| Pol IV-fc | 3 factors: T/P, Pol IV, β-clamp |
| Pol V-derived complex | |
| Pre-Pol V-fa | 3 factors: T/P, Pol V, RecA filament |
| Pol V-fa | 4 factors: T/P, Pol V, β-clamp, RecA filament |
| Pol V-fc | 4 factors: T/P, Pol V, β-clamp, a single RecA monomer |
| Pol V-dc | 2 factors: Pol V, a single RecA monomer |
fc, functional complex; fa, functional assembly; dc, dissociation complex.
, template/primer.
Intriguingly, under normal growth conditions, the basal expression level of Pol IV (∼250 molecules per cell) is the second most abundant among all Pols and is further induced to ∼2,500 molecules per cell under SOS induction (Table 1) (74). One may conclude that the high abundance of Pol IV counteracts its intrinsically low T/P binding affinity by increasing the probability of stochastic access, thus allowing the formation of Pol IV-fc required for the minor groove adduct bypass. This notion is supported by the observation that Pol IV-mediated TLS across [–ta]-BaP-dG adducts in vivo indeed occurs under non-SOS-induced conditions and is only moderately stimulated by SOS induction (S. Fujii, A. Isogawa, and R. Fuchs, unpublished data). In contrast, on a normal T/P, it is known that Pol IV-induced untargeted mutagenesis in vivo is below detection limits even under SOS-induced conditions, being detectable only upon artificially high overexpression levels (95, 96). These observations suggest that the number of Pol IV molecules required for stochastic access during TLS is below the number required for access on a normal T/P. Under physiological levels of expression, Pol IV cannot efficiently compete with high-fidelity Pols on a normal T/P, given its instability in the absence of interaction with the β-clamp (94). In contrast, when a postreplicative ssDNA gap contains a minor groove lesion, owing to the lack of elongation capacity by high-fidelity Pols, multiple attempts of access by Pol IV can lead to formation of a functional complex (Pol IV-fc) by interaction with a freely sliding β-clamp (Fig. 2B; Table 2). Formation of this functional complex is thus restricted to Pol IV-specific substrates, such as minor groove lesions.
Pivotal Role of RecA for Assembly of Functional Pol V
If a lesion is located in the major groove (e.g., AAF), Pol V bypasses the lesion in vivo (85). For Pol V-mediated TLS, interaction with RecA is essential, as demonstrated both genetically (97–99) and biochemically (76, 100). Interaction of Pol V with the β-clamp is also genetically essential for Pol V-mediated TLS (92), as previously mentioned for Pol IV. It should be noted that in the context of a T/P, both the direction of the extension of Pol and growth of the RecA filament appear to be 5′→3′ relative to their respective strand (primer strand for Pol and template strand for RecA), thus leading to contact between a Pol and the 3′ tip of the RecA filament (see Fig. 5, step a).
FIG 5.
Pol V-mediated TLS: overall view of the complete postreplicative gap repair process. This scheme is likely to describe the mechanism involved in vivo, as depicted in Fig. 3C. (a) Before TLS, a 3′ end of the primer is located in the vicinity of a lesion. The template ssDNA is subject to binding of RecA proteins, leading to RecA nucleoprotein filament (RecA*) formation. It should be noted that the 3′ tip of the RecA filament is relative to the template strand, whereas the 3′ end extended by Pols is in the primer strand, putting them in opposite, converging directions. (b) When Pol V with a RecA monomer (i.e., Pol V-dc) dissociates following Pol V-mediated TLS, a short ssDNA gap appears between the 3′ end of the primer and the 3′ tip of RecA*. (c) This short gap prevents proper reaccess of a Pol V molecule to both the 3′ end of the primer and the 3′ tip of RecA*, simultaneously. Thereby, functional access of one of the high-fidelity Pols dominates, leading to synthesis of a short error-free patch. We propose that the short error-free patch may be produced by multiple successive accesses of high-fidelity Pols, and dissociation of the high-fidelity Pols eventually yields a primer that is exclusively compatible with the functional access of Pol V. (d) Pol V synthesizes a short DNA patch in an error-prone manner. A short gap again appears following dissociation of Pol V-dc. (e) The remaining overall ssDNA gap is filled in through repeated cycles of steps c and d.
Subsequently, both genetic requirements (i.e., interactions with both the β-clamp and RecA) were also demonstrated to be essential factors in vitro (101). When Pol V accesses a lesion-containing T/P in the vicinity of a RecA-coated ssDNA gap, the following scenario is likely to occur (Fig. 2C). The interaction of Pol V with the T/P is, as already noted for Pol IV, highly unstable (65, 101, 102); following RecA nucleoprotein filament (i.e., RecA*) formation, the RecA molecule located at a 3′ tip of RecA* stabilizes a Pol V molecule transiently bound to the T/P, leading to formation of a complex referred to as pre-Pol V- functional assembly (pre-Pol V-fa; i.e., Pol V with RecA*) (Table 2). The resulting increase in residency time allows Pol V to encounter and to associate with a freely sliding β-clamp. Consequently, a functional assembly of Pol V with RecA* and the β-clamp (Pol V-fa) is reached, allowing occurrence of productive TLS events (Fig. 2C; Table 2) (65, 101, 102).
In the RecA filament on ssDNA, each RecA monomer binds 3 nt (16); consequently, the length of the ssDNA gap between a 3′ end of primer and a 3′ tip of RecA* is 0, 1, or 2 nt long. Potentially, only one of these gap sizes is the proper length to form a pre-Pol V-fa (Table 2). To allow formation of a correct pre-Pol V-fa complex, adjustment of the gap size may occur owing to intrinsic dynamic features of RecA* (103) or as a result of the dynamic state of the 3′ end of primer that is subject to degradation/resynthesis cycles by high-fidelity Pols as mentioned previously (65).
Characteristic Features of Pol IV- and Pol V-Mediated TLS Pathways
The major difference between Pol IV and Pol V in terms of their respective activation procedures for TLS is the unique requirement of RecA for Pol V. Under SOS-induced conditions, the estimated number of Pol V (15 to 60 molecules per cell) (75) is ∼66-fold lower than that of Pol IV (∼2,500 molecules per cell) (Table 1) (74). Pol IV and Pol V implement different strategies to compensate for their low T/P affinity. For Pol IV, its high abundance compensates for its low affinity to the T/P, while Pol V qualitatively benefits from additional stabilization through a specific interaction with RecA. During SOS induction, in contrast to dinB encoding Pol IV that is induced early (69), the umuDC genes are induced late (104), while the active UmuD′2C (i.e., Pol V) appears only ∼50 min after SOS induction due to additional delays caused by UmuD processing into UmuD' and intrinsic UmuC and UmuD instability (23, 105, 106). In the case of a major groove lesion like AAF-dG, given that Pol V induction is delayed to a late time point during SOS induction, one can assume that all other Pols have randomly accessed the 3′ end of the primer in a nonproductive way (Fig. 2A). Thus, depending upon the type of lesion to be bypassed (e.g., minor or major groove lesion), the timing of TLS occurrence largely differs between Pol IV- and Pol V-mediated pathways. Since bypass across [–ta]-BaP-dG in vivo does not require SOS induction, Pol IV-mediated TLS can occur at an earlier time than can TLS mediated by Pol V, thus reducing the time harmful ssDNA is exposed. This notion is also supported by single-molecule fluorescence assays in vivo (48).
UNTARGETED MUTAGENESIS DURING COMPLETION OF THE WHOLE PROCESS OF POL V-MEDIATED TLS
Genetic instability during lesion bypass would be minimal if error-prone TLS Pols dissociated from template DNA as soon as the TLS patch reached the length (Fig. 1c, pink line) sufficient for extension by high-fidelity Pols (e.g., Pol III) (Fig. 3A) (65). We have previously shown that a 5-nt-long TLS patch (i.e., one distorted base pair at a lesion site plus four correct base pairs) is efficiently extended by Pol III (and similarly by Pol II) (65). In such a case, the occurrence of untargeted mutations would be limited to a short TLS patch (Fig. 3B). Unexpectedly, for Pol V-mediated TLS in vivo, untargeted mutagenesis is spread over essentially the whole postreplicative gap and even in the upstream region (Fig. 3C) (36). Our work revealed that the error frequency of Pol V-induced untargeted mutagenesis during gap-filling synthesis lies in the range of 10–5 per base. If a gap size is several hundred nucleotides, then cumulative untargeted mutations reach a 10–3 to 10–2 frequency for every TLS event. The data clearly indicate that in vivo, access of Pol V to undamaged template DNA is not strictly limited to a minimal TLS patch. In addition, in contrast to replication errors (107), untargeted mutations due to Pol V are refractory to mismatch repair (MMR) correction (36). Indeed, efficient MMR requires association with the replication apparatus (108, 109). Thus, the observed lack of efficient MMR correction is likely due to the absence of the replication fork context. Such a phenomenon has also been observed in Saccharomyces cerevisiae, where untargeted mutations induced by Pol ζ during gap-filling synthesis are refractory to MMR (35, 110). Taken together, in addition to the intrinsic low fidelity of TLS Pols, the ineffectiveness of MMR during postreplicative gap repair exacerbates the genetic instability caused by untargeted mutagenesis.
FIG 3.
TLS elicits untargeted mutagenesis during postreplicative gap repair. (A) Schematic representation of template DNA undergoing TLS-mediated postreplicative gap repair. (B) Expected pattern of untargeted mutations in the case in which the error-prone TLS Pol is restricted to the lesion bypass event per se. (C) Schematic representation of the untargeted mutation pattern as observed in vivo during Pol V-mediated TLS (36).
ALTERNATE ACCESSES OF LOW- AND HIGH-FIDELITY POLS DURING POSTREPLICATIVE GAP REPAIR
During the overall process of TLS in vivo, Pol V-mediated untargeted mutagenesis was revealed to extend over several hundred nucleotides around a lesion site (Fig. 3C) (36). The intensity of untargeted mutations downstream from the lesion site globally decreases with the distance from the lesion site. Surprisingly, the decrease is not uniform but exhibits an oscillating (wavy) pattern with periodicity (a few dozen nucleotides of a low-mutation stretch and around 50 nt of a high-mutation stretch), suggesting alternate accesses of high- and low-fidelity Pols, respectively (36). This in vivo observation is seemingly explainable in terms of stochastic access of any Pol to a vacant T/P. This notion (i.e., stochastic access) was demonstrated by means of single-molecule imaging in vitro: access of E. coli’s high- and low-fidelity Pols (Pol II, Pol III, Pol IV) to a T/P occurs stochastically depending on the Pols’ concentration (111).
In principle, any Pol can stochastically access vacant 3′ ends in nascent strands irrespective of whether or not the binding event will be productive, i.e., leads to DNA synthesis. The probability for stochastic access of a given Pol to a vacant T/P will depend upon the relative number of Pol molecules per cell (111). When Pol V functions, cells are under SOS-induced conditions. Under such conditions, the estimated numbers of molecules per cell are as follows: Pol I, ∼400 (38); Pol II, ≤350 (≤50 without SOS induction) (73); Pol III, ∼20 (112); Pol IV, ∼2,500 (∼250 without SOS induction) (74); Pol V, ∼38 (the mean of 15 to 60) (75) (Table 1). As the total number of high-fidelity Pols (i.e., high-fidelity Pol I to III) is ≤770, the relative proportions of high-fidelity Pols, Pol IV, and Pol V are approximately 23%, 76%, and 1.1%, respectively. Despite the high relative abundance of Pol IV (76%), its contribution to untargeted mutagenesis during gap-filling synthesis is essentially negligible (36). This observation reflects the above-mentioned feature of Pol IV, namely, its inefficient functional access to a normal T/P. Thereby, it was concluded that Pol V is the main contributor to untargeted mutagenesis (36)
How Are the Error-Free Stretches Produced?
Within the oscillating pattern of untargeted mutations, the apparent length of the valleys (i.e., error-free stretches) is relatively short (e.g., a few dozen nucleotides) (Fig. 3C) (36). The length is obviously much shorter than the processivity of any high-fidelity Pol (Pol I to III) in the presence of the β-clamp (Table 1). On the other hand, the processivities of high-fidelity Pols in the absence of the β-clamp are short (Table 1): Pol I, 15 to 20 nt (113); Pol II, ∼5 nt (114); Pol III, 10 to 15 nt (115). We suggest that when high-fidelity Pols access 3′ ends of primers, most Pols begin elongation and dissociate before spontaneously encountering a diffusing sliding β-clamp (Fig. 4A). When the relative number of molecules per cell for the individual high-fidelity Pols is taken into account, the apparent average processivity of the high-fidelity Pol (composed of Pol I to III) in the absence of the β-clamp becomes ∼12 nt. This size is seemingly a little small compared with the apparent length of the observed error-free stretches (a few dozen nucleotides). This may indicate that most, if not all, single error-free stretches are produced by a few accesses of high-fidelity Pols. It is noteworthy to mention that under SOS-induced conditions, any ssDNA is converted into RecA nucleoprotein filament (i.e., RecA*). High-fidelity Pols (Pol II, Pol III) can efficiently utilize such a RecA-coated T/P as well as an SSB-coated T/P in vitro (65).
FIG 4.
Stochastic access of DNA Pols on normal template DNA. (A) High-fidelity Pols can elongate primers independently of the status of the template DNA, such as naked, SSB coated, or RecA coated (65). When a high-fidelity Pol occasionally encounters a freely sliding β-clamp, the resulting complex exhibits increased residency time, leading to synthesis of a long DNA patch. (B) Pol IV quickly dissociates from template DNA. The vast majority of these transiently bound Pol IV molecules will not encounter a freely sliding β-clamp. Therefore, Pol IV binding events are either abortive or lead to 1-nt incorporation. (C) Pol V physically interacts with RecA on template DNA, leading to the formation of pre-Pol V-fa (i.e., Pol V with RecA*). The substantial increase in residency time of Pol V on template DNA allows Pol V to encounter a freely sliding β-clamp. Pol V interacting with both RecA and the β-clamp (i.e., Pol V-fa) synthesizes a short DNA patch in an error-prone manner. When Pol V together with a RecA monomer (Pol V-dc) dissociates, it leaves behind a short ssDNA gap between the 3′ end of the primer and the 3′ tip of RecA*.
Molecular Dynamics of Pol V during Production of Error-Prone Stretches
The apparent length of the “hills” (i.e., error-prone stretches) in the oscillating pattern is around 50 nt (Fig. 3C) (36). In contrast to Pol IV on a normal T/P (i.e., quick dissociation or 1-nt elongation) (Fig. 4B), Pol V becomes stabilized by interacting with the 3′ tip of RecA* (i.e., pre-Pol V-fa), thus providing a long enough residency time to interact with a freely sliding β-clamp (i.e., pre-Pol V-fa is converted to Pol V-fa) on a normal T/P (Fig. 4C; Table 2) (65, 102), as discussed above in the context of TLS (Fig. 2C) (65, 101, 102). While the first hill, in the pattern of Pol V-mediated untargeted mutagenesis, inevitably starts at the replication-blocking lesion, the boundaries of subsequent hills are less clear, as they depend upon the length of the intervening valleys. Consequently, downstream hills become broader and fuzzier. Pol V-fa begins to elongate a normal T/P with an average processivity of ∼25 nt (maximum elongation is up to around 100 nt), as determined in vitro (65, 102), a size that is compatible with the apparent length of the observed hills (36). Thereafter, Pol V likely dissociates from the T/P as a subcomplex composed of Pol V and a single RecA monomer ( Pol V-dissociation complex [Pol V-dc]) (Table 2) (36, 116). Upon dissociation of Pol V and its associated RecA monomer (Pol V-dc), a short ssDNA gap is generated between the vacant 3′ end of the primer and the new 3′ tip of RecA* (Fig. 4C).
What Is the Underlying Mechanism of the Alternate Accesses of Pol V and High-Fidelity Pols?
In addition to the untargeted mutation pattern indicating the alternate accesses of low- and high-fidelity Pols described above, intriguingly, the overall decline of untargeted mutations with distance from the lesion site is conspicuously moderate (36), given the numerical superiority of high-fidelity Pols over Pol V (relative ratio, ∼20:1). As a minor pathway, if a high-fidelity Pol occasionally encounters a freely sliding β-clamp leading to a long stretch of synthesis in an error-free manner (Fig. 4A), it would contribute to the overall decline of mutations.
What may be the mechanism that allows Pol V to dominate over the high-fidelity Pols in spite of its numerical inferiority? If the vacant T/P is formed upon dissociation of Pol V, the short ssDNA gap that is formed may be geometrically unsuitable for functional access of a free Pol V molecule which requires concomitant interaction with the T/P and RecA* (Fig. 5, step b). Under such circumstances, high-fidelity Pols may preferentially access the vacant 3′ end and synthesize, without the β-clamp, a relatively short error-free patch (Fig. 5, step c). During synthesis of such a short error-free patch by high-fidelity Pols, RecA molecules are dislodged from the 3′ tip of RecA* (65, 67), forming new 3′ tips of RecA*. Following dissociation of a high-fidelity Pol, the ssDNA gap size between a 3′ end of primer and a 3′ tip of RecA* is either 0, 1, 2, or 3 nt long (e.g., 3 nt is a case of concurrent dissociation of a Pol and a RecA monomer). Since functional access of Pol V is seemingly compatible with only one of these gap sizes, the next Pol following dissociation of a high-fidelity Pol may again frequently be a high-fidelity Pol rather than Pol V, leading to formation of a single error-free patch by multiple high-fidelity Pols. If the gap size becomes compatible with simultaneous binding of Pol V to the T/P and the 3′ tip of RecA*, loading of Pol V may be favored despite its lower number of molecules per cell. This notion seems to be supported by the observation that in vitro, when RecA* is stabilized by ATP-γ-S (a poorly hydrolyzable ATP analog), preferential access of Pol V over Pol III is observed (117). Subsequently, Pol V locked onto the T/P (i.e., pre-Pol V-fa) associates with a freely sliding β-clamp, and the resulting functional assembly of Pol V (i.e., Pol V-fa) synthesizes an error-prone patch (Fig. 5, step d; Table 2), restarting a new cycle (Fig. 5, step e).
The Region Upstream of the Lesion Displays a Dynamic State
Unexpectedly, during the process of Pol V-mediated TLS in vivo, untargeted mutagenesis was also observed in the region upstream from the lesion site, albeit with somewhat reduced intensity compared to the downstream region (Fig. 3C) (36). These events likely result from a scenario in which exonucleases (with or without helicases) degrade the 3′ end of the nascent strand in the vicinity of a lesion or of a nonextendable short TLS patch produced by Pol V, thus expanding the single-stranded region upstream from the lesion site, followed by RecA* formation. Subsequently, during gap filling of the region upstream of the lesion, Pol V-fa is formed and induces untargeted mutations. This observation implies that until the late SOS response, the region upstream from the lesion is in a dynamic dsDNA ⇄ ssDNA equilibrium state. During that period, high-fidelity Pols are essentially responsible for the filling-in reaction, while the accuracy of the filling-in reaction will be lower than that of nascent strands at a replication fork due to lack of efficient MMR correction. When Pol V becomes available in late SOS response, the subfraction of these 3′ ends located upstream from the lesion will be extended in an error-prone way by Pol V. Thus, even before Pol V bypasses a lesion, genome integrity is already compromised.
HOW DOES POL V IMPLEMENT ELONGATION ON RecA-COATED ssDNA?
Pol V Interacts with a Dynamically Formed RecA Filament
When Pol V is expressed, all ssDNA regions are already converted into RecA-coated ssDNA (RecA*). Globally, the apparent growth of the RecA filament has been shown to occur in the 5′→3′ direction on ssDNA (118). However, the RecA filament formation is a highly dynamic process, and individual RecA monomers associate at and dissociate from both ends of the filament at different rates, leading to a net apparent growth in the 5′→3′ direction (Fig. 6) (103).
FIG 6.
Dynamics of RecA* formation. The directionality of RecA filament formation was deduced from the following biochemical parameters (103). The dissociation rates (koff) of RecA monomers in RecA* at the 5′ and 3′ ends are essentially the same (5′ end, ∼0.12 s−1; 3′ end, ∼0.16 s−1), and their association rates (kon) are as follows: 5′ end, ∼0.11 s−1 at 100 nM RecA; 3′ end, ∼0.18 s−1 at 8 nM RecA. This leads to the dissociation constants (Kd) at the 5′ end of ∼100 nM and at the 3′ end of ∼8 nM (103). Thus, whereas both ends of RecA* are in a highly dynamic state, the significant difference of Kd (i.e., the large difference in kon values dependent upon the concentration of RecA) leads to the apparent 5′→3′ directionality of RecA* formation.
Through the isolation of various RecA mutants in vivo, it has been suggested that, genetically, the 3′ tip of RecA* is essential for UmuD′2C-induced mutagenesis (i.e., targeted mutagenesis via Pol V), and this RecA function has been referred to as the third role of RecA (75, 97–99). Thereafter, it has also been shown that Pol V requires direct contact at the 3′ tip of RecA* for activation of Pol V in vitro (119) and that the ultimate RecA monomer becomes stably associated with Pol V, forming a complex composed of Pol V and a RecA monomer (116). It should be stressed that the stability of RecA bound to Pol V is much higher than the stability of the 3′-terminal RecA monomer in RecA*; indeed, 50% of the RecA monomer bound to Pol V dissociates in ∼30 min (116), while at the 3′ end of RecA*, the RecA monomer dissociates with a half-life (t1/2) of ∼4.3 s (= ln2/koff [where koff, the dissociation rate, is ∼0.16 s−1]).
Interaction of RecA with Pol V.
Genetically, certain RecA mutants (e.g., recA730) stimulate SOS mutator activities through constitutive SOS induction (99). The molecular basis of this phenotype is likely to rely on its higher affinities for binding to ssDNA than of wild-type (wt) RecA (120). In addition to the enhancement of the Pol V expression level in recA730 through constitutive SOS induction, the possibility that the RecA730 monomer may directly modulate Pol V’s activity has been raised and tested (121). Indeed, a RecA730 monomer has been biochemically shown to enhance Pol V activity in comparison to a wt RecA monomer. In addition, a truncated form of RecA730 (RecA730-ΔC17) exhibits a higher ssDNA binding affinity than does RecA730 (122) and further enhances Pol V’s activity in comparison to RecA730 in vitro (121). Taken together, these data indicate that Pol V’s catalytic activity depends upon its interaction with a RecA monomer and is modulated by the specific ssDNA binding affinity of the RecA variant. Along the same lines, it has also been demonstrated that a surface of RecA monomer exposed at the 3′-terminal in RecA* is important for activation of Pol V in vitro (123). In the same study, it was proposed that based on molecular modeling in silico, a RecA monomer bound on UmuC (catalytic subunit of Pol V) could exhibit multiple conformational states during Pol V activation.
It should be noted that the use of RecA as an auxiliary subunit may not be restricted to E. coli (and its closely related bacteria). Based on homology modeling in silico, various bacteria, including Gram-positive bacteria, seemingly possess Pol V-like systems regulated by direct interaction with RecA (31).
Biochemical Features of RecA* Formation
The first step in RecA* formation is a nucleation step whereby ∼5 RecA monomers associate on a short stretch of ssDNA (∼20 nt), followed by an extension phase to form RecA* (103, 124, 125). In other words, the requirement of the nucleation step indicates that individual RecA monomers bound to ssDNA are intrinsically very unstable (e.g., very high koff of RecA) (103, 124–127). In vitro, the efficiency of RecA* formation largely depends upon the nature of the nucleotide analogue that is used as a cofactor (128, 129). The affinity of RecA for ssDNA decreases in the order ATP > no cofactor > ADP, and only ATP efficiently supports representative RecA functions, such as coprotease and strand exchange reactions, in the context of RecA*. Thereby, as a model nucleotide cofactor, ATP has been frequently used to understand the dynamics of RecA* in vitro (126, 128, 130). Cellular concentrations of ATP and ADP in E. coli are estimated to be 3 mM and 0.25 mM, respectively (131). During RecA* formation in vitro, the different cofactors induce distinct RecA* filaments. Indeed, RecA-ATP (or RecA-ATP-γ-S) and RecA-ADP (or RecA with no cofactor) induce filaments on ssDNA referred to as extended and compressed, respectively (132–134). During de novo filament formation, RecA occupies 3 nt in the extended filament (helical pitch, ∼95 Å) (16), while it occupies 5 nt in the compressed filament (helical pitch, ∼75 Å) (135).
Dynamics of RecA monomers at the RecA* terminus.
Activation of Pol V is triggered locally by the contact between Pol V and a 3′ tip of RecA* on the T/P. As described above, the RecA monomer that is associated with Pol V largely influences its activation and elongation mode. Before discussing the role of RecA associated with Pol V, we describe some fundamental RecA monomer features below.
Conversion of a RecA filament from an extended to a compressed filament was reversibly observed on the same ssDNA molecule in response to changing the nucleotide cofactor, implying that the conversion can elastically occur without varying the nucleotide occupation of RecA (133, 136). RecA possesses a DNA-dependent ATPase activity, and its turnover number (kcat) for ATP hydrolysis has been estimated at ∼0.3 to 0.5 s−1 in the presence of ssDNA (137). Based on relatively similar ranges of koff (5′ end, ∼0.12 s−1; 3′ end, ∼0.16 s−1) (103) and kcat of RecA, dissociation of RecA monomers from the ends of RecA* could be linked to ATP hydrolysis, accompanied with a conformational change of RecA (i.e., RecA-ATP → RecA-ADP) (103, 127). On the other hand, as single RecA monomers (i.e., not in RecA*) are unstable when bound to ssDNA, their dissociation is most likely not linked to ATP hydrolysis.
The RecA Fluttering Model
Background of the proposed model.
When we proposed, in 2004, a model as to how Pol V implements elongation on RecA*, we thought that Pol V-fa per se (Fig. 2C and 4C; Table 2) would perform elongation accompanied by a RecA filament sliding process with the concomitant dissociation of RecA monomers from the 5′ tip of RecA*, as the 3′ tip of RecA* is capped by Pol V (101). While sliding on ssDNA was demonstrated for ssDNA binding (SSB) proteins, sliding of RecA* on ssDNA appears to be unlikely (68, 138). In addition, as it was reported that Pol V can appropriate a single RecA molecule from a 3′ tip of RecA* in vitro (116), we no longer needed to posit that the RecA monomer at the 3′ tip of RecA* was irremovable.
How is Pol V activated on RecA-coated ssDNA?
Since the extended form of the RecA filament is an active form supporting RecA functions (e.g., recombination) (139), we consider the RecA-ATP filament exclusively below. At the outset, Pol V accesses RecA-coated ssDNA, leading to the formation of pre-Pol V-fa, a prerequisite for subsequent conversion into Pol V-fa via interaction with the β-clamp (Fig. 7A; Table 2). As discussed above, the RecA monomer at the 3′ tip of RecA* has greater affinity for Pol V than toward the next RecA monomer in RecA*. Therefore, in the context of Pol V-fa, when the RecA monomer at the 3′ tip of RecA* dissociates from the RecA*, it remains stably bound to Pol V. We refer to the transition of the RecA monomer from the 3′ tip of RecA* to Pol V as “RecA capture.” We propose the following scenario to describe the RecA capture event. If a RecA monomer spontaneously dissociates from the 3′ tip of RecA* (i.e., dissociation of the RecA-ATP monomer), it will readily regain its original binding site (i.e., a 3-nt ssDNA gap) due to its high local concentration, becoming again a part of RecA*, while Pol V remains as Pol V-fa. On the other hand, if dissociation of the terminal RecA monomer is accompanied by ATP hydrolysis (formation of RecA-ADP), it will be unable to regain access to its original binding site, as it would occupy 5 nt instead of the 3 nt left upon its dissociation. We thus propose that the RecA capture event is an irreversible process triggered by ATP hydrolysis at the 3′ tip of RecA*.
FIG 7.
RecA fluttering model. RecA molecules are numbered starting from the 3′ end of RecA*. ATP and ADP forms of RecA are shown in light purple and yellow, respectively. If Pol V can carry out elongation (i.e., there is enough free ssDNA space), we refer to it as “movable.” If not, we call it “immovable.” (A) Pre-Pol V-fa (i.e., Pol V with RecA*) is converted into Pol V-fa following interaction with the β-clamp. The 3′ end of the RecA molecule detaches from the filament while remaining tightly bound to Pol V. During that process, referred to as RecA capture, RecA-ATP is converted into RecA-ADP and Pol V becomes a fully functional complex (Pol V-fc, i.e., Pol V with a RecA monomer and the β-clamp). During the RecA capture event, a 3-nt gap forms and RecA molecule 2 becomes the new 3′ tip of RecA*. The transition between Pol V-fa and Pol V-fc is irreversible due to ATP-to-ADP conversion. Moreover, due to steric hindrance, both Pol V-fa and Pol V-fc complexes are immovable. It should be noted that if RecA capture occurs at the pre-Pol V-fa state, it leads to a nonproductive event, since Pol V with a RecA monomer (i.e., Pol V-dc) dissociates immediately. (B) Upon dissociation of RecA molecule 2, the ensuing 6-nt gap allows Pol V-fc to enter its movable state. Thus, in the presence of the β-clamp, “activation” of Pol V occurs on the T/P substrate through dynamic cooperation with RecA*. Elongation proceeds via either the discontinuous or continuous mode. (C) Discontinuous elongation mode. RecA molecule 1 in Pol V-fc repeatedly binds to and detaches from template DNA (RecA fluttering). Since access of RecA-ADP spatially requires 5 nt on ssDNA (135), Pol V-fc synthesizes 1 nt of the 6-nt gap and then pauses due to steric hindrance until RecA molecule 3 dissociates, generating an 8-nt gap. Pol V-fc can synthesize 3 nt until reaching a 5-nt gap, which again blocks further synthesis. Elongation by Pol V-fc proceeds in an inchworm-like motion (i.e., multiple cycles of 3 nt except for 1-nt incorporation at the first cycle). It should be noted that if ATP gets spontaneously bound to the RecA monomer in Pol V-fc, it can be converted into Pol V-fa during elongation, and Pol V-fa would then need to be reactivated via the RecA capture process. (D) Continuous elongation mode. When the 3′ tip of RecA* dissociates before encountering the moving Pol V-fc complex, smooth elongation (without stalling) is maintained until Pol V dissociates. (E) Based on the processivity of Pol V-fc on a normal T/P (i.e., ∼25 nt) (65, 102), ∼9 RecA molecules have been released from the 3′ end of the filament during elongation. When Pol V-dc dissociates from the T/P, Pol V-fc is either in an immovable state, leaving a 5-nt gap, or in a movable state, leaving a 5- to 8-nt gap.
In vitro, under single-hit conditions (e.g., experimental conditions involving a trap that prevents a Pol from rebinding its T/P once dissociated), Pol V with RecA* (i.e., pre-Pol V-fa) lacks measurable processivity on a normal T/P (Table 2) (102). Thus, in the event that RecA capture occurs in the pre-Pol V-fa state, Pol V with a RecA monomer would immediately dissociate from the T/P (i.e., in the form of Pol V-dc), resulting in a nonproductive event (Fig. 7A). On the other hand, in the context of Pol V-fa, owing to the additional stability conferred by the interaction with the β-clamp, the event of RecA capture leads to a formation referred to as a Pol V functional complex (Pol V-fc; i.e., Pol V with a RecA monomer and the β-clamp) (Fig. 7A; Table 2). We propose that although a 3-nt ssDNA gap appears accompanied by the RecA capture event, it could not support Pol V-mediated DNA synthesis due to steric hindrance between the RecA-ADP monomer on Pol V and a newly formed 3′ tip of RecA* (Fig. 7A and B, RecA molecule 2). We refer to this sterically blocked form of Pol V as its “immovable state.” The transition of Pol V-fa to Pol V-fc is irreversible (Fig. 7A; Table 2). Consequently, DNA synthesis by Pol V-fc can occur only upon dissociation of the next 3′ tip of RecA* (koff, ∼0.16 s−1), thus generating a 6-nt gap and turning Pol V from an immovable to a movable state (Fig. 7B).
What is RecA fluttering?
During elongation, the RecA monomer that is part of Pol V-fc likely behaves as either a 5′ tip of RecA* or a free RecA monomer. In either case, given the low binding affinity of a single RecA monomer to ssDNA, we propose that the RecA monomer in Pol V-fc performs rapid association and dissociation cycles on ssDNA, referred to as RecA fluttering (Fig. 7C and D). As described in “Interaction of RecA with Pol V" (see above), the RecA monomer associated with Pol V actively influences its catalytic activity (121), and multiple conformational states of the RecA monomer bound to UmuC during activation of Pol V have been proposed (123). These reports seemingly fit well with the concept of RecA fluttering. We propose that RecA fluttering serves to properly position Pol V with the β-clamp on the T/P at the nucleotide insertion step during elongation. Indeed, in vitro, under multiple-hit conditions, and in the absence of RecA, Pol V alone has the capacity to produce much longer elongation products than Pol V with the β-clamp in certain natural DNA sequence contexts (102). This observation implies that the β-clamp impairs proper positioning of Pol V on the T/P at such difficult-to-replicate DNA sequences or at lesion-containing template DNA (102). It is noteworthy that the RecA-ADP form may be advantageous for Pol V-fc elongation owing to its lower ssDNA binding affinity. Otherwise, due to the lack of sliding capacity of RecA on ssDNA (68, 138), a RecA monomer with higher ssDNA binding affinity such as RecA-ATP may modestly behave as an obstacle during Pol V-fc elongation.
Fifth role of RecA.
While the first and second roles of RecA correspond to upregulation of UmuDC expression via stimulation of LexA autocleavage and posttranslational cleavage of UmuD to UmuD′, respectively (17), we previously suggested that the so-called third role of RecA (97–99) corresponds to the stabilization of Pol V on the T/P via pre-Pol V-fa (i.e., Pol V with RecA*) formation (Table 2) (17). As discussed above, the pre-Pol V-fa complex facilitates the interaction of Pol V with a freely sliding β-clamp, leading to Pol V-fa formation (Fig. 4C). Additionally, another role of RecA allows smooth elongation of Pol V on difficult-to-replicate sequences (e.g., secondary structure) by stretching such sequences that otherwise would lead to strong pause sites (fourth role of RecA) (17, 102). Indeed, it has been demonstrated that RecA* suppresses –2 frameshift mutagenesis in vitro, by likely stretching out the slipped –2 frameshift intermediate (66). In contrast to all roles (i.e., first, second, third, and fourth) that involve RecA in the context of RecA*, its role in Pol V-fc implicates RecA as a monomer. Here, we propose that the RecA fluttering mode, in which the RecA monomer actively functions for proper positioning of Pol V bound to the β-clamp on the T/P during elongation, represents the fifth role of RecA.
Two elongation modes of Pol V-fc.
We propose that within the context of a movable Pol V-fc complex, the RecA fluttering mode allows Pol V to sense whether DNA synthesis is possible when the gap is ≥6 nt long (movable state) or impossible if the gap is only 3 nt (immovable state) (Fig. 7B). Subsequently, elongation on RecA-coated ssDNA is operated via two, nonexclusive elongation modes (discontinuous and continuous) depending upon the position of the 3′ tip of RecA* with respect to Pol V-fc. In the discontinuous mode, as a RecA-ADP monomer requires 5 nt for binding to ssDNA, elongation of Pol V-fc on a 6-nt gap would be sterically blocked after only 1-nt incorporation, leaving a 5-nt gap (Fig. 7C). In the subsequent cycle, relying on dissociation of a RecA monomer from the 3′ tip of RecA*, Pol V-fc encounters an 8-nt gap and succeeds in incorporating 3 nt, leaving a 5-nt gap in each cycle (Fig. 7C). During elongation, if the RecA-ADP in Pol V-fc is spontaneously converted to RecA-ATP, Pol V-fc could be converted into Pol V-fa via contact of the RecA-ATP with a 3′ tip of RecA* (Table 2); in such a case, Pol V-fa can be reactivated through the process of RecA capture as described above (Fig. 7A). In the continuous mode, when not encountering a blockade (i.e., a 3′ tip of RecA*), Pol V-fc can continue the elongation process uninterruptedly (Fig. 7D). We accordingly propose that a DNA patch synthesized by Pol V-fc could be produced by either mode or by a combination of both modes. When the Pol V-fc consumes its residency time on the T/P, it dissociates as Pol V-dc (Pol V with a RecA monomer), being a nonproductive byproduct in a physiological context (Fig. 7E). Based on the average processivity of Pol V-fc (i.e., ∼25 nt), in the case of the discontinuous mode, one can estimate that a total of 9 cycles have been performed (Fig. 8, steps d to f).
FIG 8.
Overall view of the RecA fluttering model. (a) When Pol V accesses a vacant 3′ end of a primer, it becomes stabilized by RecA*, leading to pre-Pol V-fa (Pol V with RecA*) formation. (b and c) pre-Pol V-fa is first converted into Pol V-fa by interacting with the β-clamp and then to Pol V-fc by capturing the terminal RecA molecule from the filament. ATP hydrolysis is associated with the RecA capture event. (d) When the new 3′ tip of RecA* dissociates, it generates a 6-nt gap, and Pol V-fc can start elongation. We propose that during elongation, Pol V-fc exhibits a dynamic state during which its associated RecA molecule alternately binds to and detaches from the DNA substrate, a motion we refer to as RecA fluttering. RecA fluttering, which we also refer to as the fifth role of RecA, ensures proper positioning of Pol V with the β-clamp during DNA synthesis. (e) Discontinuous elongation. Pol V-fc fills in 1 nt of the 6-nt gap. (f) In the next cycle, Pol V-fc fills in the first 3 nt of the 8-nt gap. Elongation of Pol V-fc is discontinuous until dissociation. (g) Continuous elongation. Pol V-fc continues elongation without stalling as long as it is not sterically hindered by the 3′ tip of RecA*. (h) When Pol V-fc exhausts its residency time, Pol V-dc (Pol V with a RecA monomer) dissociates from the T/P, leaving behind the β-clamp and a 5- to 8-nt gap on the T/P.
The apparent koff of Pol V-dc is estimated to be ∼0.012 s−1 as derived from the residency time of Pol V-fc, ∼86 s (= processivity/velocity, ∼25 nt/∼0.29 nt s−1) (65, 102). When Pol V-dc dissociates, it concomitantly creates a 5- to 8-nt gap depending on whether dissociation occurs at the immovable or the movable state (Fig. 7E). The kinetics of nucleotide cofactor exchange from RecA-ADP to RecA-ATP (e.g., ADP → no cofactor → ATP) as indirectly monitored by ATPase activity seemingly reaches a plateau value at ∼10 min (126). The residency time of Pol V-fc (∼86 s) may thus indicate that during elongation, most RecA monomers in Pol V-fc are in the ADP (or no cofactor) state or even after dissociation (Pol V-dc). In addition, a nucleotide cofactor such as ATP-γ-S was observed to remain stably bound to RecA associated with Pol V over 1 h (116).
The overall view of the RecA fluttering model is delineated in Fig. 8. It is noteworthy that while the average processivity of Pol V-fc is ∼25 nt, the distribution of processive synthesis is relatively broad, up to around 100 nt in vitro (65, 102). This observation may indicate that processive synthesis entails a combination of discontinuous and continuous elongation modes. In addition, during postreplicative gap repair in vivo, the average size distribution of Pol V-induced error-prone patches is not strictly confined to ∼25-nt long patches but is slightly larger (i.e., ∼50-nt patches) (Fig. 3C) (36). This observation may also indicate that these error-prone patches are generated via a combination of discontinuous and continuous elongation modes in vivo.
Extremely low velocity of Pol V-fc.
Intriguingly, on a normal T/P, the velocity of Pol V-fc is over 10-fold lower than that of Pol IV-fc (∼0.29 versus 3 to 5 nt s−1) (Tables 1 and 2) (65, 94, 102). This significant difference likely reflects the unique feature of Pol V during elongation, i.e., its functional association with RecA. Residency times of RecA monomers at the 5′ and 3′ ends of RecA*, deduced from their (koff)–1 values, are estimated to be ∼8.3 s and ∼6.3 s, respectively (103). Thus, the residency time of Pol V-fc (∼86 s) is 1 order of magnitude higher than the residency times of RecA monomers at the ends of RecA*. This suggests that within the residency time of Pol V-fc, multiple RecA monomers (∼14) are committed to dissociate from the 3′ ends of RecA*. Intriguingly, Pol V-fc requires ∼10 s to fill the 3 nt appearing upon dissociation of a single RecA monomer, a duration similar to the residency times of a single RecA monomer at the ends of RecA*. Taken together, there appears to be fine-tuning between the velocity of Pol V-fc and the dynamic state of RecA at the 3′ end of RecA*.
DISTINCT MODES OF ACTION DURING DNA SYNTHESIS FOR POL IV AND POL V
In contrast to Pol IV-fc (velocity, 3 to 5 nt s−1; processivity, 300 to 400 nt) (94), Pol V-fc exhibits lower velocity (∼0.29 nt s−1) and lower processivity (∼25 nt) on a normal T/P (Tables 1 and 2) (65, 102). Interestingly, the residency times of Pol IV-fc and Pol V-fc on a T/P exhibit similar durations, ∼97 s (average, 60 to 133 s [= processivity/velocity, i.e., 300 to 400 nt/3 to 5 nt s−1]) and ∼86 s (∼25 nt/∼0.29 nt s−1), respectively. Thus, despite significant differences between Pol IV and Pol V in terms of velocity and processivity on a normal T/P, their affinities for a T/P are similar, implying that the residency times of both Pol IV and Pol V may largely depend upon their interaction with the β-clamp (65, 94, 102).
Pol IV and Pol V exhibit distinct lesion bypass features, not only in terms of their substrate specificities (minor versus major groove lesions, respectively) but also in the way their processivities are affected by lesions. The processivity of Pol V-fc (∼25 nt on a normal T/P) is only moderately reduced by the presence of a lesion (e.g., reduced to ∼18 nt in TLS across an AAF-dG) (65, 102). In contrast, in vitro, the processivity of Pol IV-fc during [–ta]-BaP-dG bypass is intriguingly short (around 5 nt) compared to its processivity on a normal T/P (300 to 400 nt) (Tables 1 and 2) (94), suggesting that most of Pol IV’s residency time is consumed by the lesion bypass step (S. Fujii, A. Isogawa, and R. Fuchs, unpublished data). A similar phenomenon also occurs for Pol II during the bypass of an AAF-dG lesion in the NarI sequence context, as described in “AAF-dependent frameshift mutagenesis" (see above), namely, its processivity on a –2 frameshift intermediate is only 3 nt, while it is >1,600 nt on a normal T/P (Table 1) (66, 114). It thus indicates that Pol IV and Pol V largely differ in their modes of action during lesion bypass. Indeed, the fact that Pol V-fc can deal with a T/P almost as efficiently whether or not a lesion is present can be attributed to its fine positioning by virtue of a RecA monomer in Pol V-fc (i.e., the fifth role of RecA). This notion is also supported by the observation that under multiple-hit conditions in vitro, Pol IV (or Pol II) alone can carry out robust DNA synthesis on both a normal and a lesion-containing T/P (N-2-furfuryl-dG for Pol IV; AAF-dG for Pol II) (66, 140); in contrast, under multiple-hit conditions in vitro, Pol V alone can perform DNA synthesis on a normal T/P but not on a lesion-containing T/P (101).
IMPACT OF TLS AND PERSPECTIVES
In E. coli, TLS Pols modestly contribute to survival but are essential for induced mutagenesis (17), strongly suggesting that in addition to preventing persistent ssDNA gaps, they represent a major driving force for genetic diversity in both short-term (e.g., adaptive mutations) and long-term (e.g., evolution) ranges (17, 141, 142). The biological significance of genetic diversity in the short-term range is illustrated by, for instance, the appearance of antibiotic-resistant bacteria in clinics or mutations in the immunoglobulin gene locus (i.e., somatic hypermutation) in higher eukaryotes. The latter is driven by TLS Pol-mediated targeted and/or untargeted mutagenesis restricted to a localized region in the genome (143). The former is presumed to result from antibiotic-induced stress that leads to SOS induction, causing genome-wide mutagenesis mediated by TLS Pols (144).
Contribution of TLS Pols to Stress-Induced Mutagenesis
Environmental changes leading to slow growth or growth arrest (e.g., stationary phase, nutrient deprivation) result in accumulation of RpoS protein (a sigma factor, σs), the master regulator of the general stress response in E. coli (145). RpoS binds to RNA Pol and alters its preference for promoters. The RpoS directly or indirectly regulates up to 10% of the genome (∼500 genes) (146). This RpoS response is essential for the stress-induced mutagenesis described below (142, 147). As part of the RpoS regulon, Pol IV is upregulated ∼2-fold independent of its upregulation by the SOS response (148). In addition, it has been reported that for full expression of Pol II under the SOS response, the RpoS response is also required (149).
Molecular mechanisms of mutagenesis have been studied extensively to apprehend the origin of genetic diversity. Major attention has been paid to the mechanisms of mutagenesis in nondividing cells, as these events are critical for gene diversification in adaptation and evolution (142, 147, 150). To understand the contribution of TLS Pols to genetic diversity, it is important to assess how and where TLS Pols access DNA. In E. coli, a stress-induced mutagenesis assay, for adaptive or stationary-phase mutagenesis, has been widely used (151). This system involves a Lac– to Lac+ reversion assay via –1 frameshift mutagenesis. Briefly, the genetic requirements of the assay are as follows: the target lac allele must be located on an F′ conjugative plasmid; multiple copies of dinB are needed (i.e., Pol IV) (the chromosomal single copy of dinB does not support the phenotype); a conjugational factor, TraI (nickase and helicase activities), is required for F′ conjugation; and recombinational factors such as RecA are needed (96, 150, 152). These requirements may imply that the overexpressed Pol IV can access a T/P produced as a RecA-mediated recombination intermediate that resulted from F′ conjugation (142, 150). Involvement of TLS Pols during stress-induced mutagenesis has also been reported in other bacteria, Bacillus subtilis (153) and Pseudomonas putida (154). Interestingly, it has been suggested that the Mfd protein (a transcription-coupled repair [TCR] factor) might be associated with the stress-induced mutagenesis pathway in both Bacillus subtilis (155) and Pseudomonas putida (156). This may indicate that under these conditions, the working places of TLS Pols are biased toward actively transcribing gene loci.
With respect to antibiotic-resistant bacteria, a representative model study might be the case of ciprofloxacin (clinically used antimicrobial agent targeting topoisomerases)-resistant E. coli cells. Ciprofloxacin induces DNA double-strand breaks (DSBs) by interfering with topoisomerases (157). Since antibiotic resistance against ciprofloxacin largely relies on the presence of all SOS-induced Pols (i.e., Pol II, IV, and V), it has been proposed that the Pols participate in the repair process of ciprofloxacin-induced DSBs, consequently contributing to antibiotic resistance via error-prone synthesis mediated by Pol IV and Pol V (158). It has been recently reported that during such events, the intensity of the RpoS response in individual cells is critically important (159). Similar DSB-induced TLS Pol-dependent mechanisms seem to occur in Salmonella enterica (160) and perhaps in Pseudomonas aeruginosa (161).
Physiological Roles of TLS Pols beyond the Canonical TLS Pathway
A recent finding that Pol V participates in gap-filling synthesis associated with the process of TLS (36) raises the possibility that TLS Pols participate in any gap-filling synthesis irrespective of the origin of the ssDNA gap. For instance, during late SOS response, RecA-dependent recombinational repair is activated (Fig. 1, step f) (13, 69, 162). During that process, an ssDNA gap, in all points similar to the gap formed following TLS patch formation, appears (Fig. 1, steps c and d versus steps f and g). In support of this notion, it has been suggested that TLS Pols potentially participate in homologous recombination in vivo (163). This seems to be in good agreement with the participation of TLS Pols during the ciprofloxacin-induced DSB repair as described above (144, 158). Under metabolic stress conditions, the participation of TLS Pols (i.e., Pol IV and V) in recombinational repair has also been genetically indicated (164). In other examples, although the molecular mechanisms are largely unknown, Pol IV may participate in NER and recombinational repair (165) and may be recruited at transcription stalling sites mediated by interaction with NusA (a transcription elongation factor) (166). In addition, it has been reported that TLS Pols can genetically participate in NER in both log and stationary phases (167), and TLS Pols confer a competitive fitness advantage during stationary phase (168). The working place of TLS Pols appears thus not to be restricted to the sole context of lesion bypass. Consequently, the name “specialized Pols” instead of “TLS Pols” may be more appropriate to reflect their diverse functions, and one may thus categorize Pol I and III as replicative Pols and the remaining Pol II, IV, and V as specialized Pols.
Future questions that need to be addressed in the context of physiological roles of specialized Pols pertain to the cross talk among DNA transactions (replication, transcription, repair pathways, etc.). Until now, these DNA transactions have been frequently viewed as individual, well-defined pathways, worked out at the genetic and biochemical levels, each on its own, separate from the others. These pathways may now appear as intellectual and artificial constructions that do not reflect the complexity occurring within cells, where distinct DNA lesions and stress-induced conditions are processed by different pathways simultaneously; as a consequence, these pathways will interfere and clash with each other, generating unforeseen intermediates that might become substrates for specialized Pols and possibly leading to cytotoxicity and/or mutations.
Untargeted Mutagenesis during Postreplicative Gap Repair as a Beneficial Source of Diversity in Evolution
Classically, genome-wide mutagenesis has been extensively studied through the analysis of mutator mutants (e.g., MMR-defective strains) to unravel contributions of the inactivated pathways to genome integrity (169). As with genome-wide mutators, Pol V can induce “targeted mutagenesis” at randomly located lesions generated by exposure to mutagens. However, as discussed throughout this review, Pol V also implements “untargeted mutagenesis” in restricted regions adjacent to the lesions (i.e., local hypermutagenesis) (Fig. 3C) (36), resembling somatic hypermutation in immunoglobulin gene loci in higher eukaryotes. Such localized transient hypermutability might contribute to the occurrence of local high-frequency mutations, clustered mutations, and mutation showers (170, 171) and may be beneficial to evolution (172). In the case of higher eukaryotes, such mechanisms would contribute to phenomena such as immunoglobulin diversification and acceleration of tumorigenesis during chemotherapy.
As mentioned above, the site of action of TLS Pols may be directed to local regions associated with recombinational repair, NER, actively transcribed genes, and so on. Furthermore, the action of TLS Pols is not restricted to periods of growth but also can occur during stationary phase. In addition, since Pol V-induced untargeted mutations are inefficiently repaired by MMR (36) (leading to a ≥104-fold increase compared to regular DNA replication errors [169, 173]), one daughter cell will acquire novel genetic information while the other daughter cell will retain the original information. It is noteworthy that Pol IV frequently induces –1 frameshift mutations at monotonous base runs (95) while Pol V essentially induces base substitutions (36). Taken together, such features may be physiologically advantageous for generating genetic diversity over short and long time periods.
Why Have Many Different Pol V Models Been Proposed over the Years?
Since Pol V was identified, various and sometimes conflicting models to explain its biochemical features in vitro have been proposed (17, 77, 88). We suggest that the subtlety of Pol V’s biochemistry stems from two distinct roles that RecA imparts to Pol V: (i) the ATP-bound form of RecA in RecA* facilitates the interaction of Pol V with a freely sliding β-clamp (third role of RecA) and (ii) the ADP-bound form of a RecA monomer (fifth role of RecA) allows the proper positioning of Pol V bound to the β-clamp during DNA synthesis. Complete physiological reconstitution of Pol V’s mode of action is thus made difficult by the stepwise involvement of RecA-ATP first, followed by RecA-ADP, in addition to the indispensable role of the β-clamp on Pol V’s mode of action. As a consequence, the biochemical properties of Pol V acutely depend upon the experimental conditions, such as the nature of the nucleotide cofactor, as well as the geometry of the T/P, leading to conflicting results and contributing to the enigmatic features of Pol V in vitro (17, 77, 88). With the integration of most of the published data, the RecA fluttering model is likely to become a paradigm that reconciles many perplexing and long-standing disputes in the field (Fig. 7 and 8).
SOS-Dependent Untargeted Mutagenic Roads
Last but certainly not least, Pol IV is essentially inefficient in gap-filling synthesis on normal template DNA as described in “What Is the Mechanism Underlying Functional Access of Pol IV to Template DNA?” (see above). Therefore, in contrast to postreplicative gap repair associated with Pol V-mediated TLS (Fig. 3C), the mutation spectrum associated with Pol IV-mediated TLS is expected to entail a minimal untargeted mutagenesis load, as shown in Fig. 3B.
Below we describe two situations where, in addition to Pol IV, Pol V becomes involved in completion of the whole process of minor groove lesion bypass. First, some minor groove lesions were found to require both Pol IV and Pol V for their bypass (84, 85). In the case of benzo[a]pyrene (BaP), following metabolic activation in vivo, the ultimate metabolite (diol-epoxide) reacts primarily at the N2 position of dG (174), forming [+ta]-BaP-dG and its stereoisomer [–ta]-BaP-dG, which represent the major and a minor adduct, respectively (175). As mentioned above, the [–ta]-BaP-dG adduct is bypassed by Pol IV under normal growth conditions in vivo. In contrast, bypass of [+ta]-BaP-dG requires both Pol IV and Pol V in vivo (57, 85). Thus, depending upon the precise chemistry and conformation of the minor groove adduct, successful bypass requires collaboration between Pol IV and Pol V. In this situation where Pol V is required, TLS necessarily occurs in the late SOS response and entails error-prone Pol V-mediated untargeted mutagenesis.
Second, when the number of minor groove lesions is high, i.e., exhausting the capacity of rapid Pol IV-mediated bypass, cells enter into the late SOS response, where Pol V is expressed and can functionally access the RecA-coated ssDNA gap. Thus, depending upon the timing of Pol IV-mediated lesion bypass completion, the untargeted mutation pattern gradually shifts from largely error-free (Fig. 3B) to error-prone (Fig. 3C) owing to the participation of Pol V.
Finally, considering TLS in vivo from the sole perspective of lesion bypass accompanied by a targeted mutation event reflects a viewpoint described by the saying “you can’t see the forest for the trees.” As mentioned throughout this review, on the basis of the overall context of postreplicative gap repair, our vision of TLS pathways should be thoroughly revisited.
ACKNOWLEDGMENTS
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
We declare no competing interests.
Conceptualization of the article and writing of the original draft was done by S.F.; writing, reviewing, and editing the final version was done by S.F. and R.P.F.
REFERENCES
- 1.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. 2003. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J 17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 2.Park S, You X, Imlay JA. 2005. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx− mutants of Escherichia coli. Proc Natl Acad Sci U S A 102:9317–9322. doi: 10.1073/pnas.0502051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imlay JA. 2013. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohanski MA, DePristo MA, Collins JJ. 2010. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell 37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster PL, Lee H, Popodi E, Townes JP, Tang H. 2015. Determinants of spontaneous mutation in the bacterium Escherichia coli as revealed by whole-genome sequencing. Proc Natl Acad Sci U S A 112:E5990–E5999. doi: 10.1073/pnas.1512136112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindahl T, Andersson A. 1972. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry 11:3618–3623. doi: 10.1021/bi00769a019. [DOI] [PubMed] [Google Scholar]
- 7.Lindahl T. 1993. Instability and decay of the primary structure of DNA. Nature 362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura J, Mutlu E, Sharma V, Collins L, Bodnar W, Yu R, Lai Y, Moeller B, Lu K, Swenberg J. 2014. The endogenous exposome. DNA Repair (Amst) 19:3–13. doi: 10.1016/j.dnarep.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips DH. 1983. Fifty years of benzo(a)pyrene. Nature 303:468–472. doi: 10.1038/303468a0. [DOI] [PubMed] [Google Scholar]
- 10.Cheung-Ong K, Giaever G, Nislow C. 2013. DNA-damaging agents in cancer chemotherapy: serendipity and chemical biology. Chem Biol 20:648–659. doi: 10.1016/j.chembiol.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 11.LeClerc JE, Borden A, Lawrence CW. 1991. The thymine-thymine pyrimidine-pyrimidone(6-4) ultraviolet light photoproduct is highly mutagenic and specifically induces 3′ thymine-to-cytosine transitions in Escherichia coli. Proc Natl Acad Sci U S A 88:9685–9689. doi: 10.1073/pnas.88.21.9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedberg EC, Walker GC, Siede W. 1995. DNA repair and mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- 13.Courcelle J, Hanawalt PC. 2003. RecA-dependent recovery of arrested DNA replication forks. Annu Rev Genet 37:611–646. doi: 10.1146/annurev.genet.37.110801.142616. [DOI] [PubMed] [Google Scholar]
- 14.Rupp WD, Howard-Flanders P. 1968. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol 31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 15.Lehmann AR, Fuchs RP. 2006. Gaps and forks in DNA replication: rediscovering old models. DNA Repair (Amst) 5:1495–1498. doi: 10.1016/j.dnarep.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Kuzminov A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev 63:751–813. doi: 10.1128/MMBR.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs RP, Fujii S. 2013. Translesion DNA synthesis and mutagenesis in prokaryotes. Cold Spring Harb Perspect Biol 5:a012682. doi: 10.1101/cshperspect.a012682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marians KJ. 2018. Lesion bypass and the reactivation of stalled replication forks. Annu Rev Biochem 87:217–238. doi: 10.1146/annurev-biochem-062917-011921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henrikus SS, van Oijen AM, Robinson A. 2018. Specialised DNA polymerases in Escherichia coli: roles within multiple pathways. Curr Genet 64:1189–1196. doi: 10.1007/s00294-018-0840-x. [DOI] [PubMed] [Google Scholar]
- 20.Hsu GW, Ober M, Carell T, Beese LS. 2004. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature 431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 21.Baharoglu Z, Mazel D. 2014. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol Rev 38:1126–1145. doi: 10.1111/1574-6976.12077. [DOI] [PubMed] [Google Scholar]
- 22.Michel B. 2005. After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol 3:e255. doi: 10.1371/journal.pbio.0030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maslowska KH, Makiela Dzbenska K, Fijalkowska IJ. 2019. The SOS system: a complex and tightly regulated response to DNA damage. Environ Mol Mutagen 60:368–384. doi: 10.1002/em.22267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujii S, Isogawa A, Fuchs RP. 2018. Chronological switch from translesion synthesis to homology-dependent gap repair in vivo. Toxicol Res 34:297–302. doi: 10.5487/TR.2018.34.4.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rupp WD, Wilde CE, Reno DL, Howard-Flanders P. 1971. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J Mol Biol 61:25–44. doi: 10.1016/0022-2836(71)90204-x. [DOI] [PubMed] [Google Scholar]
- 26.Bichara M, Pinet I, Lambert IB, Fuchs R. 2007. RecA-mediated excision repair: a novel mechanism for repairing DNA lesions at sites of arrested DNA synthesis. Mol Microbiol 65:218–229. doi: 10.1111/j.1365-2958.2007.05790.x. [DOI] [PubMed] [Google Scholar]
- 27.Bichara M, Fuchs RPP, Cordonnier A, Lambert IB. 2009. Preferential post-replication repair of DNA lesions situated on the leading strand of plasmids in Escherichia coli. Mol Microbiol 71:305–314. doi: 10.1111/j.1365-2958.2008.06527.x. [DOI] [PubMed] [Google Scholar]
- 28.Michel B, Sinha AK, Leach D. 2018. Replication fork breakage and restart in Escherichia coli. Microbiol Mol Biol Rev 82:e00013-18. doi: 10.1128/MMBR.00013-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. 2001. The Y-family of DNA polymerases. Mol Cell 8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 30.Goodman MF. 2002. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem 71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 31.Timinskas K, Venclovas Č. 2019. New insights into the structures and interactions of bacterial Y-family DNA polymerases. Nucleic Acids Res 47:4393–4405. doi: 10.1093/nar/gkz198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cirz RT, O'Neill BM, Hammond JA, Head SR, Romesberg FE. 2006. Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J Bacteriol 188:7101–7110. doi: 10.1128/JB.00807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Au N, Kuester-Schoeck E, Mandava V, Bothwell LE, Canny SP, Chachu K, Colavito SA, Fuller SN, Groban ES, Hensley LA, O'Brien TC, Shah A, Tierney JT, Tomm LL, O'Gara TM, Goranov AI, Grossman AD, Lovett CM. 2005. Genetic composition of the Bacillus subtilis SOS system. J Bacteriol 187:7655–7666. doi: 10.1128/JB.187.22.7655-7666.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao L, Washington M. 2017. Translesion synthesis: insights into the selection and switching of DNA polymerases. Genes 8:24–25. doi: 10.3390/genes8010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kochenova OV, Daee DL, Mertz TM, Shcherbakova PV. 2015. DNA polymerase ζ-dependent lesion bypass in Saccharomyces cerevisiae is accompanied by error-prone copying of long stretches of adjacent DNA. PLoS Genet 11:e1005110. doi: 10.1371/journal.pgen.1005110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isogawa A, Ong JL, Potapov V, Fuchs RP, Fujii S. 2018. Pol V-mediated translesion synthesis elicits localized untargeted mutagenesis during post-replicative gap repair. Cell Rep 24:1290–1300. doi: 10.1016/j.celrep.2018.06.120. [DOI] [PubMed] [Google Scholar]
- 37.Wu CA, Zechner EL, Marians KJ. 1992. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. I. Multiple effectors act to modulate Okazaki fragment size. J Biol Chem 267:4030–4044. [PubMed] [Google Scholar]
- 38.Kornberg A, Baker T. 1992. DNA replication, 2nd ed W.H. Freeman, New York, NY. [Google Scholar]
- 39.Okazaki R, Okazaki T, Sakabe K, Sugimoto K, Sugino A. 1968. Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proc Natl Acad Sci U S A 59:598–605. doi: 10.1073/pnas.59.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heller RC, Marians KJ. 2006. Replication fork reactivation downstream of a blocked nascent leading strand. Nature 439:557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 41.Yeeles JTP, Marians KJ. 2011. The Escherichia coli replisome is inherently DNA damage tolerant. Science 334:235–238. doi: 10.1126/science.1209111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pomerantz RT, O'Donnell M. 2008. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature 456:762–766. doi: 10.1038/nature07527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amado L, Kuzminov A. 2006. The replication intermediates in Escherichia coli are not the product of DNA processing or uracil excision. J Biol Chem 281:22635–22646. doi: 10.1074/jbc.M602320200. [DOI] [PubMed] [Google Scholar]
- 44.Amado L, Kuzminov A. 2013. Low-molecular-weight DNA replication intermediates in Escherichia coli: mechanism of formation and strand specificity. J Mol Biol 425:4177–4191. doi: 10.1016/j.jmb.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cronan GE, Kouzminova EA, Kuzminov A. 2019. Near-continuously synthesized leading strands in Escherichia coli are broken by ribonucleotide excision. Proc Natl Acad Sci U S A 116:1251–1260. doi: 10.1073/pnas.1814512116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hedglin M, Benkovic SJ. 2017. Eukaryotic translesion DNA synthesis on the leading and lagging strands: unique detours around the same obstacle. Chem Rev 117:7857–7877. doi: 10.1021/acs.chemrev.7b00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson A, McDonald JP, Caldas VEA, Patel M, Wood EA, Punter CM, Ghodke H, Cox MM, Woodgate R, Goodman MF, van Oijen AM. 2015. Regulation of mutagenic DNA polymerase V activation in space and time. PLoS Genet 11:e1005482. doi: 10.1371/journal.pgen.1005482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henrikus SS, Wood EA, McDonald JP, Cox MM, Woodgate R, Goodman MF, van Oijen AM, Robinson A. 2018. DNA polymerase IV primarily operates outside of DNA replication forks in Escherichia coli. PLoS Genet 14:e1007161. doi: 10.1371/journal.pgen.1007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kunkel TA, Bebenek K. 2000. DNA replication fidelity. Annu Rev Biochem 69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 50.Paz-Elizur T, Takeshita M, Goodman M, O'Donnell M, Livneh Z. 1996. Mechanism of translesion DNA synthesis by DNA polymerase II. Comparison to DNA polymerases I and III core. J Biol Chem 271:24662–24669. doi: 10.1074/jbc.271.40.24662. [DOI] [PubMed] [Google Scholar]
- 51.Paz-Elizur T, Takeshita M, Livneh Z. 1997. Mechanism of bypass synthesis through an abasic site analog by DNA polymerase I. Biochemistry 36:1766–1773. doi: 10.1021/bi9621324. [DOI] [PubMed] [Google Scholar]
- 52.Nevin P, Gabbai CC, Marians KJ. 2017. Replisome-mediated translesion synthesis by a cellular replicase. J Biol Chem 292:13833–13842. doi: 10.1074/jbc.M117.800441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee CH, Chandani S, Loechler EL. 2006. Homology modeling of four Y-family, lesion-bypass DNA polymerases: the case that E. coli Pol IV and human Pol κ are orthologs, and E. coli Pol V and human Pol η are orthologs. J Mol Graph Model 25:87–102. doi: 10.1016/j.jmgm.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 54.Wing R, Drew H, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. 1980. Crystal structure analysis of a complete turn of B-DNA. Nature 287:755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- 55.Broyde S, Hingerty BE, Srinivasan AR. 1985. Influence of the carcinogen 4-aminobiphenyl on DNA conformation. Carcinogenesis 6:719–725. doi: 10.1093/carcin/6.5.719. [DOI] [PubMed] [Google Scholar]
- 56.Patnaik S, Cho BP. 2010. Structures of 2-acetylaminofluorene modified DNA revisited: insight into conformational heterogeneity. Chem Res Toxicol 23:1650–1652. doi: 10.1021/tx100341u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seo KY, Nagalingam A, Miri S, Yin J, Chandani S, Kolbanovskiy A, Shastry A, Loechler EL. 2006. Mirror image stereoisomers of the major benzo[a]pyrene N2-dG adduct are bypassed by different lesion-bypass DNA polymerases in E. coli. DNA Repair (Amst) 5:515–522. doi: 10.1016/j.dnarep.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 58.Ghodke PP, Gore KR, Harikrishna S, Samanta B, Kottur J, Nair DT, Pradeepkumar PI. 2016. The N2-furfuryl-deoxyguanosine adduct does not alter the structure of B-DNA. J Org Chem 81:502–511. doi: 10.1021/acs.joc.5b02341. [DOI] [PubMed] [Google Scholar]
- 59.Brown K, Hingerty BE, Guenther EA, Krishnan VV, Broyde S, Turteltaub KW, Cosman M. 2001. Solution structure of the 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine C8-deoxyguanosine adduct in duplex DNA. Proc Natl Acad Sci U S A 98:8507–8512. doi: 10.1073/pnas.151251898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O'Handley SF, Sanford DG, Xu R, Lester CC, Hingerty BE, Broyde S, Krugh TR. 1993. Structural characterization of an N-acetyl-2-aminofluorene (AAF) modified DNA oligomer by NMR, energy minimization, and molecular dynamics. Biochemistry 32:2481–2497. doi: 10.1021/bi00061a005. [DOI] [PubMed] [Google Scholar]
- 61.Zaliznyak T, Bonala R, Johnson F, de Los Santos C. 2006. Structure and stability of duplex DNA containing the 3-(deoxyguanosin-N2-yl)-2-acetylaminofluorene (dG(N2)-AAF) lesion: a bulky adduct that persists in cellular DNA. Chem Res Toxicol 19:745–752. doi: 10.1021/tx060002i. [DOI] [PubMed] [Google Scholar]
- 62.Broyde S, Wang L, Zhang L, Rechkoblit O, Geacintov NE, Patel DJ. 2008. DNA adduct structure-function relationships: comparing solution with polymerase structures. Chem Res Toxicol 21:45–52. doi: 10.1021/tx700193x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stavros KM, Hawkins EK, Rizzo CJ, Stone MP. 2014. Base-displaced intercalation of the 2-amino-3-methylimidazo[4,5-f]quinolone N2-dG adduct in the NarI DNA recognition sequence. Nucleic Acids Res 42:3450–3463. doi: 10.1093/nar/gkt1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park H, Zhang K, Ren Y, Nadji S, Sinha N, Taylor J-S, Kang C. 2002. Crystal structure of a DNA decamer containing a cis-syn thymine dimer. Proc Natl Acad Sci U S A 99:15965–15970. doi: 10.1073/pnas.242422699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fujii S, Fuchs RP. 2004. Defining the position of the switches between replicative and bypass DNA polymerases. EMBO J 23:4342–4352. doi: 10.1038/sj.emboj.7600438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fujii S, Fuchs RP. 2007. Interplay among replicative and specialized DNA polymerases determines failure or success of translesion synthesis pathways. J Mol Biol 372:883–893. doi: 10.1016/j.jmb.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 67.Fujii S, Isogawa A, Fuchs RP. 2006. RecFOR proteins are essential for Pol V-mediated translesion synthesis and mutagenesis. EMBO J 25:5754–5763. doi: 10.1038/sj.emboj.7601474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bell JC, Plank JL, Dombrowski CC, Kowalczykowski SC. 2012. Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature 491:274–278. doi: 10.1038/nature11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Friedman N, Vardi S, Ronen M, Alon U, Stavans J. 2005. Precise temporal modulation in the response of the SOS DNA repair network in individual bacteria. PLoS Biol 3:e238. doi: 10.1371/journal.pbio.0030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Culyba MJ, Kubiak JM, Mo CY, Goulian M, Kohli RM. 2018. Non-equilibrium repressor binding kinetics link DNA damage dose to transcriptional timing within the SOS gene network. PLoS Genet 14:e1007405. doi: 10.1371/journal.pgen.1007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Courcelle J, Hanawalt PC. 1999. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol Gen Genet 262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- 73.Qiu Z, Goodman MF. 1997. The Escherichia coli polB locus is identical to dinA, the structural gene for DNA polymerase II. Characterization of Pol II purified from a polB mutant. J Biol Chem 272:8611–8617. doi: 10.1074/jbc.272.13.8611. [DOI] [PubMed] [Google Scholar]
- 74.Kim SR, Matsui K, Yamada M, Gruz P, Nohmi T. 2001. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol Genet Genomics 266:207–215. doi: 10.1007/s004380100541. [DOI] [PubMed] [Google Scholar]
- 75.Sommer S, Boudsocq F, Devoret R, Bailone A. 1998. Specific RecA amino acid changes affect RecA-UmuD'C interaction. Mol Microbiol 28:281–291. doi: 10.1046/j.1365-2958.1998.00803.x. [DOI] [PubMed] [Google Scholar]
- 76.Tang M, Shen X, Frank EG, O'Donnell M, Woodgate R, Goodman MF. 1999. UmuD'(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci U S A 96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fuchs RP, Fujii S, Wagner J. 2004. Properties and functions of Escherichia coli: Pol IV and Pol V. Adv Protein Chem 69:229–264. doi: 10.1016/S0065-3233(04)69008-5. [DOI] [PubMed] [Google Scholar]
- 78.Yang W, Gao Y. 2018. Translesion and repair DNA polymerases: diverse structure and mechanism. Annu Rev Biochem 87:239–261. doi: 10.1146/annurev-biochem-062917-012405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK. 2001. Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis. Mol Cell 8:417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 80.Ling H, Boudsocq F, Woodgate R, Yang W. 2001. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 81.Kottur J, Sharma A, Gore KR, Narayanan N, Samanta B, Pradeepkumar PI, Nair DT. 2015. Unique structural features in DNA polymerase IV enable efficient bypass of the N2 adduct induced by the nitrofurazone antibiotic. Structure 23:56–67. doi: 10.1016/j.str.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 82.Bonner CA, Hays S, McEntee K, Goodman MF. 1990. DNA polymerase II is encoded by the DNA damage-inducible dinA gene of Escherichia coli. Proc Natl Acad Sci U S A 87:7663–7667. doi: 10.1073/pnas.87.19.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iwasaki H, Nakata A, Walker GC, Shinagawa H. 1990. The Escherichia coli polB gene, which encodes DNA polymerase II, is regulated by the SOS system. J Bacteriol 172:6268–6273. doi: 10.1128/jb.172.11.6268-6273.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nohmi T. 2006. Environmental stress and lesion-bypass DNA polymerases. Annu Rev Microbiol 60:231–253. doi: 10.1146/annurev.micro.60.080805.142238. [DOI] [PubMed] [Google Scholar]
- 85.Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP. 2000. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J 19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Becherel OJ, Fuchs RP. 2001. Mechanism of DNA polymerase II-mediated frameshift mutagenesis. Proc Natl Acad Sci U S A 98:8566–8571. doi: 10.1073/pnas.141113398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kriek E, Miller JA, Juhl U, Miller EC. 1967. 8-(N-2-fluorenylacetamido)guanosine, an arylamidation reaction product of guanosine and the carcinogen N-acetoxy-N-2-fluorenylacetamide in neutral solution. Biochemistry 6:177–182. doi: 10.1021/bi00853a029. [DOI] [PubMed] [Google Scholar]
- 88.Fuchs RPP, Fujii S. 2007. Translesion synthesis in Escherichia coli: lessons from the NarI mutation hot spot. DNA Repair (Amst) 6:1032–1041. doi: 10.1016/j.dnarep.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 89.Prakash S, Johnson RE, Prakash L. 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem 74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 90.Vaisman A, Woodgate R. 2017. Translesion DNA polymerases in eukaryotes: what makes them tick? Crit Rev Biochem Mol Biol 52:274–303. doi: 10.1080/10409238.2017.1291576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kong XP, Onrust R, O'Donnell M, Kuriyan J. 1992. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell 69:425–437. doi: 10.1016/0092-8674(92)90445-I. [DOI] [PubMed] [Google Scholar]
- 92.Becherel OJ, Fuchs RPP, Wagner J. 2002. Pivotal role of the beta-clamp in translesion DNA synthesis and mutagenesis in E. coli cells. DNA Repair (Amst) 1:703–708. doi: 10.1016/S1568-7864(02)00106-4. [DOI] [PubMed] [Google Scholar]
- 93.Wagner J, Gruz P, Kim SR, Yamada M, Matsui K, Fuchs RP, Nohmi T. 1999. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol Cell 4:281–286. doi: 10.1016/S1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- 94.Wagner J, Fujii S, Gruz P, Nohmi T, Fuchs RP. 2000. The beta clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep 1:484–488. doi: 10.1093/embo-reports/kvd109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim SR, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc Natl Acad Sci U S A 94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Slechta ES, Bunny KL, Kugelberg E, Kofoid E, Andersson DI, Roth JR. 2003. Adaptive mutation: general mutagenesis is not a programmed response to stress but results from rare coamplification of dinB with lac. Proc Natl Acad Sci U S A 100:12847–12852. doi: 10.1073/pnas.1735464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Blanco M, Herrera G, Collado P, Rebollo JE, Botella LM. 1982. Influence of RecA protein on induced mutagenesis. Biochimie 64:633–636. doi: 10.1016/s0300-9084(82)80102-8. [DOI] [PubMed] [Google Scholar]
- 98.Dutreix M, Moreau PL, Bailone A, Galibert F, Battista JR, Walker GC, Devoret R. 1989. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriol 171:2415–2423. doi: 10.1128/jb.171.5.2415-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sweasy JB, Witkin EM, Sinha N, Roegner-Maniscalco V. 1990. RecA protein of Escherichia coli has a third essential role in SOS mutator activity. J Bacteriol 172:3030–3036. doi: 10.1128/jb.172.6.3030-3036.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. 1999. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD', RecA, and SSB and is specialized for translesion replication. J Biol Chem 274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 101.Fujii S, Gasser V, Fuchs RP. 2004. The biochemical requirements of DNA polymerase V-mediated translesion synthesis revisited. J Mol Biol 341:405–417. doi: 10.1016/j.jmb.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 102.Fujii S, Fuchs RP. 2009. Biochemical basis for the essential genetic requirements of RecA and the beta-clamp in Pol V activation. Proc Natl Acad Sci U S A 106:14825–14830. doi: 10.1073/pnas.0905855106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T. 2006. Real-time observation of RecA filament dynamics with single monomer resolution. Cell 126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 104.Crowley DJ, Courcelle J. 2002. Answering the call: coping with DNA damage at the most inopportune time. J Biomed Biotechnol 2:66–74. doi: 10.1155/S1110724302202016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rangarajan S, Woodgate R, Goodman MF. 1999. A phenotype for enigmatic DNA polymerase II: a pivotal role for pol II in replication restart in UV-irradiated Escherichia coli. Proc Natl Acad Sci U S A 96:9224–9229. doi: 10.1073/pnas.96.16.9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pham P, Rangarajan S, Woodgate R, Goodman MF. 2001. Roles of DNA polymerases V and II in SOS-induced error-prone and error-free repair in Escherichia coli. Proc Natl Acad Sci U S A 98:8350–8354. doi: 10.1073/pnas.111007198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fujii S, Akiyama M, Aoki K, Sugaya Y, Higuchi K, Hiraoka M, Miki Y, Saitoh N, Yoshiyama K, Ihara K, Seki M, Ohtsubo E, Maki H. 1999. DNA replication errors produced by the replicative apparatus of Escherichia coli. J Mol Biol 289:835–850. doi: 10.1006/jmbi.1999.2802. [DOI] [PubMed] [Google Scholar]
- 108.Simmons LA, Davies BW, Grossman AD, Walker GC. 2008. β Clamp directs localization of mismatch repair in Bacillus subtilis. Mol Cell 29:291–301. doi: 10.1016/j.molcel.2007.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hasan AMM, Leach D. 2015. Chromosomal directionality of DNA mismatch repair in Escherichia coli. Proc Natl Acad Sci U S A 112:9388–9393. doi: 10.1073/pnas.1505370112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lehner K, Jinks-Robertson S. 2009. The mismatch repair system promotes DNA polymerase zeta-dependent translesion synthesis in yeast. Proc Natl Acad Sci U S A 106:5749–5754. doi: 10.1073/pnas.0812715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhao G, Gleave ES, Lamers MH. 2017. Single-molecule studies contrast ordered DNA replication with stochastic translesion synthesis. Elife 6:e32177. doi: 10.7554/eLife.32177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McHenry C, Kornberg A. 1977. DNA polymerase III holoenzyme of Escherichia coli. Purification and resolution into subunits. J Biol Chem 252:6478–6484. [PubMed] [Google Scholar]
- 113.Matson SW, Bambara RA. 1981. Short deoxyribonucleic acid repair patch length in Escherichia coli is determined by the processive mechanism of deoxyribonucleic acid polymerase I. J Bacteriol 146:275–284. doi: 10.1128/JB.146.1.275-284.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bonner CA, Stukenberg PT, Rajagopalan M, Eritja R, O'Donnell M, McEntee K, Echols H, Goodman MF. 1992. Processive DNA synthesis by DNA polymerase II mediated by DNA polymerase III accessory proteins. J Biol Chem 267:11431–11438. [PubMed] [Google Scholar]
- 115.Fay PJ, Johanson KO, McHenry CS, Bambara RA. 1981. Size classes of products synthesized processively by DNA polymerase III and DNA polymerase III holoenzyme of Escherichia coli. J Biol Chem 256:976–983. [PubMed] [Google Scholar]
- 116.Jiang Q, Karata K, Woodgate R, Cox MM, Goodman MF. 2009. The active form of DNA polymerase V is UmuD'(2)C-RecA-ATP. Nature 460:359–363. doi: 10.1038/nature08178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pham P, Bertram JG, O'Donnell M, Woodgate R, Goodman MF. 2001. A model for SOS-lesion-targeted mutations in Escherichia coli. Nature 409:366–370. doi: 10.1038/35053116. [DOI] [PubMed] [Google Scholar]
- 118.Register JC, Griffith J. 1985. The direction of RecA protein assembly onto single strand DNA is the same as the direction of strand assimilation during strand exchange. J Biol Chem 260:12308–12312. [PubMed] [Google Scholar]
- 119.Schlacher K, Cox MM, Woodgate R, Goodman MF. 2006. RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature 442:883–887. doi: 10.1038/nature05042. [DOI] [PubMed] [Google Scholar]
- 120.Lavery PE, Kowalczykowski SC. 1992. Biochemical basis of the constitutive repressor cleavage activity of recA730 protein. A comparison to recA441 and recA803 proteins. J Biol Chem 267:20648–20658. [PubMed] [Google Scholar]
- 121.Schlacher K, Leslie K, Wyman C, Woodgate R, Cox MM, Goodman MF. 2005. DNA polymerase V and RecA protein, a minimal mutasome. Mol Cell 17:561–572. doi: 10.1016/j.molcel.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 122.Eggler AL, Lusetti SL, Cox MM. 2003. The C terminus of the Escherichia coli RecA protein modulates the DNA binding competition with single-stranded DNA-binding protein. J Biol Chem 278:16389–16396. doi: 10.1074/jbc.M212920200. [DOI] [PubMed] [Google Scholar]
- 123.Gruber AJ, Erdem AL, Sabat G, Karata K, Jaszczur MM, Vo DD, Olsen TM, Woodgate R, Goodman MF, Cox MM. 2015. A RecA protein surface required for activation of DNA polymerase V. PLoS Genet 11:e1005066. doi: 10.1371/journal.pgen.1005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. 2006. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature 443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 125.Lu C-H, Chang T-T, Cho C-C, Lin H-C, Li H-W. 2017. Stable nuclei of nucleoprotein filament and high ssDNA binding affinity contribute to enhanced RecA E38K recombinase activity. Sci Rep 7:14964. doi: 10.1038/s41598-017-15088-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Menetski JP, Kowalczykowski SC. 1985. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J Mol Biol 181:281–295. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- 127.Arenson TA, Tsodikov OV, Cox MM. 1999. Quantitative analysis of the kinetics of end-dependent disassembly of RecA filaments from ssDNA. J Mol Biol 288:391–401. doi: 10.1006/jmbi.1999.2705. [DOI] [PubMed] [Google Scholar]
- 128.Ellouze C, Selmane T, Kim HK, Tuite E, Nordén B, Mortensen K, Takahashi M. 1999. Difference between active and inactive nucleotide cofactors in the effect on the DNA binding and the helical structure of RecA filament dissociation of RecA-DNA complex by inactive nucleotides. Eur J Biochem 262:88–94. doi: 10.1046/j.1432-1327.1999.00357.x. [DOI] [PubMed] [Google Scholar]
- 129.Wigle TJ, Lee AM, Singleton SF. 2006. Conformationally selective binding of nucleotide analogues to Escherichia coli RecA: a ligand-based analysis of the RecA ATP binding site. Biochemistry 45:4502–4513. doi: 10.1021/bi052298h. [DOI] [PubMed] [Google Scholar]
- 130.Roca AI, Singleton SF. 2003. Direct evaluation of a mechanism for activation of the RecA nucleoprotein filament. J Am Chem Soc 125:15366–15375. doi: 10.1021/ja0270165. [DOI] [PubMed] [Google Scholar]
- 131.Buckstein MH, He J, Rubin H. 2008. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol 190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cox JM, Tsodikov OV, Cox MM. 2005. Organized unidirectional waves of ATP hydrolysis within a RecA filament. PLoS Biol 3:e52. doi: 10.1371/journal.pbio.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.van Loenhout MTJ, van der Heijden T, Kanaar R, Wyman C, Dekker C. 2009. Dynamics of RecA filaments on single-stranded DNA. Nucleic Acids Res 37:4089–4099. doi: 10.1093/nar/gkp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Boyer B, Danilowicz C, Prentiss M, Prévost C. 2019. Weaving DNA strands: structural insight on ATP hydrolysis in RecA-induced homologous recombination. Nucleic Acids Res 47:7798–7808. doi: 10.1093/nar/gkz667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yu X, Egelman EH. 1992. Structural data suggest that the active and inactive forms of the RecA filament are not simply interconvertible. J Mol Biol 227:334–346. doi: 10.1016/0022-2836(92)90702-L. [DOI] [PubMed] [Google Scholar]
- 136.Nishinaka T, Doi Y, Hara R, Yashima E. 2007. Elastic behavior of RecA-DNA helical filaments. J Mol Biol 370:837–845. doi: 10.1016/j.jmb.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 137.Brenner SL, Mitchell RS, Morrical SW, Neuendorf SK, Schutte BC, Cox MM. 1987. recA protein-promoted ATP hydrolysis occurs throughout recA nucleoprotein filaments. J Biol Chem 262:4011–4016. [PubMed] [Google Scholar]
- 138.Roy R, Kozlov AG, Lohman TM, Ha T. 2009. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature 461:1092–1097. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kowalczykowski SC. 2015. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb Perspect Biol 7:a016410. doi: 10.1101/cshperspect.a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jarosz DF, Cohen SE, Delaney JC, Essigmann JM, Walker GC. 2009. A DinB variant reveals diverse physiological consequences of incomplete TLS extension by a Y-family DNA polymerase. Proc Natl Acad Sci U S A 106:21137–21142. doi: 10.1073/pnas.0907257106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Roth JR, Kugelberg E, Reams AB, Kofoid E, Andersson DI. 2006. Origin of mutations under selection: the adaptive mutation controversy. Annu Rev Microbiol 60:477–501. doi: 10.1146/annurev.micro.60.080805.142045. [DOI] [PubMed] [Google Scholar]
- 142.Galhardo RS, Hastings PJ, Rosenberg SM. 2007. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol 42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Methot SP, Di Noia JM. 2017. Molecular mechanisms of somatic hypermutation and class switch recombination. Adv Immunol 133:37–87. doi: 10.1016/bs.ai.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 144.Cirz RT, Romesberg FE. 2007. Controlling mutation: intervening in evolution as a therapeutic strategy. Crit Rev Biochem Mol Biol 42:341–354. doi: 10.1080/10409230701597741. [DOI] [PubMed] [Google Scholar]
- 145.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: S-dependent genes, promoters, and sigma factor selectivity. J Bacteriol 187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fitzgerald DM, Hastings PJ, Rosenberg SM. 2017. Stress-induced mutagenesis: implications in cancer and drug resistance. Annu Rev Cancer Biol 1:119–140. doi: 10.1146/annurev-cancerbio-050216-121919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Layton JC, Foster PL. 2003. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol Microbiol 50:549–561. doi: 10.1046/j.1365-2958.2003.03704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dapa T, Fleurier S, Bredeche M-F, Matic I. 2017. The SOS and RpoS regulons contribute to bacterial cell robustness to genotoxic stress by synergistically regulating DNA polymerase Pol II. Genetics 206:1349–1360. doi: 10.1534/genetics.116.199471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Maisnier-Patin S, Roth JR. 2015. The origin of mutants under selection: how natural selection mimics mutagenesis (adaptive mutation). Cold Spring Harb Perspect Biol 7:a018176. doi: 10.1101/cshperspect.a018176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cairns J, Foster PL. 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128:695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McKenzie GJ, Lee PL, Lombardo MJ, Hastings PJ, Rosenberg SM. 2001. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol Cell 7:571–579. doi: 10.1016/S1097-2765(01)00204-0. [DOI] [PubMed] [Google Scholar]
- 153.Sung HM, Yeamans G, Ross CA, Yasbin RE. 2003. Roles of YqjH and YqjW, homologs of the Escherichia coli UmuC/DinB or Y superfamily of DNA polymerases, in stationary-phase mutagenesis and UV-induced mutagenesis of Bacillus subtilis. J Bacteriol 185:2153–2160. doi: 10.1128/jb.185.7.2153-2160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tegova R, Tover A, Tarassova K, Tark M, Kivisaar M. 2004. Involvement of error-prone DNA polymerase IV in stationary-phase mutagenesis in Pseudomonas putida. J Bacteriol 186:2735–2744. doi: 10.1128/jb.186.9.2735-2744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gómez-Marroquín M, Martin HA, Pepper A, Girard ME, Kidman AA, Vallin C, Yasbin RE, Pedraza-Reyes M, Robleto EA. 2016. Stationary-phase mutagenesis in stressed Bacillus subtilis cells operates by Mfd-dependent mutagenic pathways. Genes 7:33. doi: 10.3390/genes7070033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ukkivi K, Kivisaar M. 2018. Involvement of transcription-coupled repair factor Mfd and DNA helicase UvrD in mutational processes in Pseudomonas putida. DNA Repair (Amst) 72:18–27. doi: 10.1016/j.dnarep.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 157.Drlica K. 1999. Mechanism of fluoroquinolone action. Curr Opin Microbiol 2:504–508. doi: 10.1016/s1369-5274(99)00008-9. [DOI] [PubMed] [Google Scholar]
- 158.Cirz RT, Chin JK, Andes DR, de Crécy-Lagard V, Craig WA, Romesberg FE. 2005. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol 3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pribis JP, García-Villada L, Zhai Y, Lewin-Epstein O, Wang AZ, Liu J, Xia J, Mei Q, Fitzgerald DM, Bos J, Austin RH, Herman C, Bates D, Hadany L, Hastings PJ, Rosenberg SM. 2019. Gamblers: an antibiotic-induced evolvable cell subpopulation differentiated by reactive-oxygen- induced general stress response. Mol Cell 74:785–800. doi: 10.1016/j.molcel.2019.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Prieto AI, Ramos-Morales F, Casadesús J. 2006. Repair of DNA damage induced by bile salts in Salmonella enterica. Genetics 174:575–584. doi: 10.1534/genetics.106.060889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Boles BR, Singh PK. 2008. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad Sci U S A 105:12503–12508. doi: 10.1073/pnas.0801499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Boudsocq F, Campbell M, Devoret R, Bailone A. 1997. Quantitation of the inhibition of Hfr × F− recombination by the mutagenesis complex UmuD'C. J Mol Biol 270:201–211. doi: 10.1006/jmbi.1997.1098. [DOI] [PubMed] [Google Scholar]
- 163.Delmas S, Matic I. 2006. Interplay between replication and recombination in Escherichia coli: impact of the alternative DNA polymerases. Proc Natl Acad Sci U S A 103:4564–4569. doi: 10.1073/pnas.0509012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Moore JM, Correa R, Rosenberg SM, Hastings PJ. 2017. Persistent damaged bases in DNA allow mutagenic break repair in Escherichia coli. PLoS Genet 13:e1006733. doi: 10.1371/journal.pgen.1006733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Williams AB, Hetrick KM, Foster PL. 2010. Interplay of DNA repair, homologous recombination, and DNA polymerases in resistance to the DNA damaging agent 4-nitroquinoline-1-oxide in Escherichia coli. DNA Repair (Amst) 9:1090–1097. doi: 10.1016/j.dnarep.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cohen SE, Lewis CA, Mooney RA, Kohanski MA, Collins JJ, Landick R, Walker GC. 2010. Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli. Proc Natl Acad Sci U S A 107:15517–15522. doi: 10.1073/pnas.1005203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Janel-Bintz R, Napolitano RL, Isogawa A, Fujii S, Fuchs RP. 2017. Processing closely spaced lesions during nucleotide excision repair triggers mutagenesis in E. coli. PLoS Genet 13:e1006881. doi: 10.1371/journal.pgen.1006881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yeiser B, Pepper ED, Goodman MF, Finkel SE. 2002. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc Natl Acad Sci U S A 99:8737–8741. doi: 10.1073/pnas.092269199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jayaraman R. 2009. Mutators and hypermutability in bacteria: the Escherichia coli paradigm. J Genet 88:379–391. doi: 10.1007/s12041-009-0058-2. [DOI] [PubMed] [Google Scholar]
- 170.Drake JW, Bebenek A, Kissling GE, Peddada S. 2005. Clusters of mutations from transient hypermutability. Proc Natl Acad Sci U S A 102:12849–12854. doi: 10.1073/pnas.0503009102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Drake JW. 2007. Mutations in clusters and showers. Proc Natl Acad Sci U S A 104:8203–8204. doi: 10.1073/pnas.0703089104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Martincorena I, Luscombe NM. 2013. Non-random mutation: the evolution of targeted hypermutation and hypomutation. Bioessays 35:123–130. doi: 10.1002/bies.201200150. [DOI] [PubMed] [Google Scholar]
- 173.Fijalkowska IJ, Schaaper RM, Jonczyk P. 2012. DNA replication fidelity in Escherichia coli: a multi-DNA polymerase affair. FEMS Microbiol Rev 36:1105–1121. doi: 10.1111/j.1574-6976.2012.00338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Conney AH. 1982. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res 42:4875–4917. [PubMed] [Google Scholar]
- 175.Cheng SC, Hilton BD, Roman JM, Dipple A. 1989. DNA adducts from carcinogenic and noncarcinogenic enantiomers of benzo[a]pyrene dihydrodiol epoxide. Chem Res Toxicol 2:334–340. doi: 10.1021/tx00011a011. [DOI] [PubMed] [Google Scholar]
- 176.Bebenek K, Joyce CM, Fitzgerald MP, Kunkel TA. 1990. The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J Biol Chem 265:13878–13887. [PubMed] [Google Scholar]
- 177.Cai H, Yu H, McEntee K, Kunkel TA, Goodman MF. 1995. Purification and properties of wild-type and exonuclease-deficient DNA polymerase II from Escherichia coli. J Biol Chem 270:15327–15335. doi: 10.1074/jbc.270.25.15327. [DOI] [PubMed] [Google Scholar]
- 178.Tang M, Pham P, Shen X, Taylor JS, O'Donnell M, Woodgate R, Goodman MF. 2000. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- 179.Camps M, Loeb LA. 2004. When pol I goes into high gear: processive DNA synthesis by pol I in the cell. Cell Cycle 3:114–118. doi: 10.4161/cc.3.2.651. [DOI] [PubMed] [Google Scholar]
- 180.López de Saro FJ, O'Donnell M. 2001. Interaction of the beta sliding clamp with MutS, ligase, and DNA polymerase I. Proc Natl Acad Sci U S A 98:8376–8380. doi: 10.1073/pnas.121009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mok M, Marians KJ. 1987. The Escherichia coli preprimosome and DNA B helicase can form replication forks that move at the same rate. J Biol Chem 262:16644–16654. [PubMed] [Google Scholar]
- 182.Dahlberg ME, Benkovic SJ. 1991. Kinetic mechanism of DNA polymerase I (Klenow fragment): identification of a second conformational change and evaluation of the internal equilibrium constant. Biochemistry 30:4835–4843. doi: 10.1021/bi00234a002. [DOI] [PubMed] [Google Scholar]
- 183.Singh K, Srivastava A, Patel SS, Modak MJ. 2007. Participation of the fingers subdomain of Escherichia coli DNA polymerase I in the strand displacement synthesis of DNA. J Biol Chem 282:10594–10604. doi: 10.1074/jbc.M611242200. [DOI] [PubMed] [Google Scholar]