Abstract
Placenta-derived exosomes play an important role in cellular communication both in the mother and the fetus. Their concentration and composition are altered in several pregnancy disorders, such as gestational diabetes mellitus (GDM). The isolation and characterization of placental exosomes from serum, plasma and tissues from patients with GDM have been previously described; however, to the best of our knowledge, to date, there is no study available on placental exosomes isolated from urine of patients with GDM. In the present study, placental exosomes were purified from urine the 1st, 2nd and 3rd trimester of gestation. Placental exosomes were characterized by transmission electron microscopy in cryogenic mode and by western blot analysis, confirming the presence of exosomal vesicles. The expression profile of five microRNAs (miR-516-5p, miR-517-3p, miR-518-5p, miR-222-3p and miR-16-5p) was determined by RT-qPCR. In healthy pregnant women, the expression of the miRNAs increased across gestation, apart from miR-516-5p, which was not expressed at the 2nd trimester. All the miRNAs examined were downregulated in patients with GDM at the 3rd trimester of gestation. The downregulated miRNAs affected several metabolic pathways closely associated with the pathophysiology of GDM. This provides further evidence of the regulatory role of miRNAs in the GDM. This also suggests that the of urinary exosomes may be an excellent source of biomarkers and therapeutic targets.
Keywords: placental exosomes, microRNAs, biomarkers, gestational diabetes
Introduction
Gestational diabetes mellitus (GDM) is defined as any degree of glucose intolerance identified for the first time during pregnancy. This condition begins when the function of pancreatic β cells is insufficient to overcome the insulin resistance that occurs during pregnancy (1). GDM is associated with pro-inflammatory processes, oxidative stress and endothelial cell dysfunction in the microvasculature of the placenta (2).
Although several studies have been conducted to identify biomarkers and to make a timely diagnosis of GDM (3,4), the oral glucose tolerance test (OGTT) is still the gold standard for the diagnosis of this condition. However, the cut-off values established by the World Health Organization (WHO) have been modified by several international consortiums, generating multiple criteria for the diagnosis and management of GDM (5). Moreover, the OGTT is considered by several authors to be invasive and to provide a late diagnosis during pregnancy (4,6).
Maternal-fetal cellular communication depends on factors secreted by the placenta. These factors can be markers of altered placental functions that are detectable in the maternal circulation from the early stages of pregnancy and, therefore, could have a predictive value in various pregnancy-related diseases (7). The human placenta expresses >500 microRNAs (miRNAs or miRs), almost exclusively expressed in this organ. For this reason, the study of placental miRNAs is essential for the understanding of the regulatory mechanisms of normal and complicated pregnancies (8).
Placental exosomes are released by exocytosis as part of mechanisms for communication and molecular regulation in normal pregnancy (9); however, their composition and concen-tration is a reflection of the cellular physiology, and important differences in these parameters have been identified in association with pathological states (10).
The chromosome 19 miRNA cluster (C19MC) located on chromosome 19q13.41, is the largest group of miRNAs identified to date. Some of the most important C19MC miRNAs are miR-516b-5p, miR-517-5p and miR-518a-3p (11), whose expression levels are modulated in response to stress, alterations in circulating glucose levels and other stimuli characteristic of pregnancy and are also contained in placental exosomes. The cellular targets of C19MC miRNAs carried as exosomal cargo have not been totally identified and may include non-trophoblastic placental cells, maternal organs, maternal blood system or fetal cells (12). The downregulation of several C19MC miRNAs has been observed in preeclampsia, gestational hypertension and fetal growth restriction (13). Furthermore, some miRNAs that are not coded by the C19CM, such as miR-16-5p and miR-222-3p, have also been associated with pregnancy complications, such as preeclampsia and GDM (14).
The expression of miRNAs contained in placental exosomes purified from serum and plasma of patients with GDM has been previously evaluated (15,16). However, to the best of our knowledge, there are no studies available to date on the characterization of miRNAs from urine exosomes of women with GDM. Urine is a biological fluid very useful for clinical applications due to its availability in large quantities, non-invasive collection and simple sample treatment. In the present study, the expression of miR-516-5p, miR-517-3p, miR-518-5p, miR-222-3p and miR-16-5p was evaluated in placental exosomes purified from urine in the 1st, 2nd and 3rd trimester of pregnancy with GDM and healthy controls, since the participation of these microRNAs in the pathophysiology of GDM is not yet fully understood.
Materials and methods
Study design and selection of participants
The present longitudinal study included 61 pregnant women who attended prenatal control between May, 2018 and April, 2019 at the Obstetrics Service of the Hospital Central 'Dr. Ignacio Morones Prieto', San Luis Potosí, Mexico. Patients with gestational hypertension, urinary infections, pre-existing diabetes mellitus, and chronic kidney disease were excluded.
The criteria used for the diagnosis of GDM were established by the Obstetrics Service of the Hospital Central 'Dr. Ignacio Morones Prieto' (Table SI) and following the WHO criteria (17). All participants were tested for oral glucose tolerance between weeks 24 and 28 of gestation (17).
Clinical and demographic data were obtained from the medical records of each participant in the first prenatal visit. The GDM group (n=27) was formed by patients who were diagnosed with GDM during the second trimester of pregnancy. The control group (n=34) was constituted by euglycemic women. The present study was approved by the Ethics Committee of the Hospital Central 'Dr. Ignacio Morones Prieto', with registration number 84-17 (Folio CONBIOET ICA-24-CEI-001-201604279). The study was conducted in accordance with the Declaration of Helsinki. The informed consent signature was obtained from each participant prior to the collection of the urine samples.
Sample collection and treatment
Three urine samples were collected per participant in a sterile vessel: The first during the 1st trimester (8-20 weeks), the second during the 2nd trimester (24-28 weeks) and finally in the 3rd trimester (32-39 weeks), as summarized in Table SII.
After collection, the urine samples were centrifuged at 300 × g for 15 min at room temperature to remove cells and cell debris. The samples were then centrifuged again at 1,200 × g for 10 min at 4°C and divided into 10 ml aliquots. A cocktail of protease inhibitors (PMSF, NaN3 and leupeptin) (18) was added to each aliquot and they were stored at -80°C until analysis.
Isolation of urinary exosomes
Composite samples (pools) were established prior to the isolation of exosomes and the amplification of miRNAs, mixing equivalent amounts of individual samples. A total of 5 pools containing 34 individual samples and 5 pools containing 27 individual samples were established from the healthy controls and GDM groups, respectively. Each pool was matched by age and body mass index (BMI).
Exosomes were purified from 10 ml of pooled urine samples using the Urine Exosome Purification and RNA Isolation Midi kit (Norgen Biotek Corp, Product # 58700) following the manufacturer's instructions. The Urine Exosome Purification kit isolates exosomes based on the isoelectric point of the proteins present in the microvesicles that are filtered through silicon carbide columns, which makes the method sensitive and specific (19). Purified exosomes were stored at -80°C for subsequent analysis.
Cryogenic transmission electron microscopy (cryo-TEM)
The morphology and size of the exosomes were examined by cryo-TEM. A volume of 3 to 5 µl of exosome samples suspended in phosphate-buffered saline was transferred to a Lacey carbon-coated grid. The grid was maintained at 85% humidity for 10 sec and then was immersed in liquid ethanol prepared without ice crystals at liquid nitrogen temperature (-196°C). Subsequenlty, the grid was mounted on a cryo-transfer sample holder at -175°C. The preparation was observed using a cryo-TEM Microscope JEM-2100 containing a LaB6 filament operated at different acceleration voltages (80,100,120,160 and 200 kV).
Identification of exosomal surface protein markers by western blot analysis
The identity of exosomes was assessed by western blot analysis, evaluating the presence of the surface proteins, CD9, CD63 and CD81. Additionally, the placental origin was evaluated by the presence of placental alkaline phosphatase (PLAP). Electrophoresis was performed under reducing conditions (12.5% SDS-PAGE), from 20 µg of protein, extracted using the Urine Exosome Purification and RNA Isolation Midi kit (Norgen Biotek Corp., product #58700). The protein concentration was determined using the bicinchoninic acid (BCA) method using the Pierce BCA Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.) and 20 µg of protein was loaded by lane. Western blot analysis was performed according to standard procedures using polyvinylidene difluoride membranes and an enhanced chemiluminescence system Optiblot ECL Substrate kit (ab133406, Abcam United Kingdom). The following antibodies were used: Anti-CD9 mouse monoclonal antibody (isotype IgG1; host species, mouse; this antibody reacts with human CD9 antigen, ab2215, Abcam United Kingdom, 1:1,000), anti-CD81 mouse monoclonal antibody (isotype IgG1; host species, mouse; this antibody reacts with human CD81 antigen, ab79559, Abcam United Kingdom, 1:1,000), anti-CD63 mouse monoclonal antibody, (isotype IgG1; host species, mouse; this antibody reacts with human CD63 antigen, ab8219, Abcam United Kingdom, 1:1,000) and anti-PLAP rabbit monoclonal antibody (isotype IgG; host species, rabbit; ab133602, Abcam United Kingdom, 1:500). The primary antibodies were incubated at room temperature for 2 h. The secondary antibodies used were as follows: Goat polyclonal secondary antibody to mouse IgG-H&L (HRP) (ab6789) and goat anti-rabbit IgG H&L(HRP) (ab6789), incubated overnight at 4°C. As a loading control, vesicles isolated from HepG2 [HEPG2] cells (ATCC® HB-8065™) were used. The stability of urinary exosomes was evaluated after 12 months of storage at -80°C by western blot analysis and cryo-TEM.
miRNA isolation and quantification
Total RNA was isolated from 400 µl of purified exosomes using the Urine Exosome Purification and RNA Isolation Midi kit (Norgen Biotek Corp.; cat. no. 58700) following the manufacturer's instructions. The RNA concentration was assessed using a NanoDrop 1000 spectrophotometer (NanoDrop Technologies Inc.). The cDNAs for the mature miRNAs (U6, miR-16-5p, miR-222-3p, miR-516b-5p, miR-517-5p and miR-518a-3p) were synthesized from 150 ng of total RNA using the Taq Man® microRNA test, (Applied Biosystems); TaqMan probes were used for each of the miRNAs to avoid unspecific amplification. The 5'-3' sequences of the primers are presented in Table SIII. The CFX96 touch thermal cycler was used (Bio-Rad Laboratories, Inc.).
All RT-qPCR reactions were performed in duplicate; Cq values were averaged and the 2−ΔΔCq method (20) was used to obtain the relative expression, where ΔΔCq was calculated by subtracting the ΔCq value from the mean of the control group (healthy pregnant women) with the ΔCq value of GMD women. miRNA levels were normalized with the use of the average of U6 snRNA Cq value as the housekeeping gene. The experimental conditions for reverse transcription and PCR are detailed in Tables SIV and SV, respectively.
miRNA target prediction
The evaluation for the interaction and target genes networks was performed using the miRTar-getLink program, which contains experimentally validated interactions in the latest version of miRTarBase 1 (version 6.0: September 15, 2015). The functional evaluation of the miRNAs was performed using DIANA-miRPath v3.0 (http://www.microrna.gr/miRPathv3). Additionally, using The Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/) bioinformatics tool, signaling pathways related to the study of miRNAs were evaluated, considering a value of P<0.05 as statistically significant.
Statistical analysis
Statistical analysis was performed with GraphPad Prism 5.0 software (GraphPad Software, Inc.). The data are expressed as the means ± SD or median ± IQR according data distribution, respectively. For the relative expression of the miRNAs, a Kruskal-Wallis ANOVA test and a non-parametric Dunn post-hoc test were performed. A value of P<0.05 was considered to indicate a statistically significant difference. The GDM predictive capacity was evaluated by means of receiver operating characteristic (ROC) curve analysis.
Results
Clinical characteristics of the population studied
The anthropometric, clinical and biochemical parameters of the pregnant women included in the present study are summarized in Table I. The mean age of the patients with GDM was 29.9±6.03 and 26.06±5.28 years for the healthy controls. The BMI was significantly higher in women that developed GDM (28.66±4.02 vs. 25.69±4.03, P=0.006; Table I).
Table I.
General characteristics of pregnant women included in the study.
| Clinical data | GDM (n=27) | Healthy (n=34) | P-value |
|---|---|---|---|
| Age (years) | 29.93±6.03 | 26.06±5.28 | 0.01 |
| BMI (kg/m2) | 28.66±4.02 | 25.69±4.03 | 0.006 |
| Glucose (mg/dl) | 89.60±14.15 | 78.00±8.48 | 0.0002 |
| Systolic blood pressure (mmHg) | 110.4±9.29 | 105.7±9.80 | 0.06 |
| Diastolic blood pressure (mmHg) | 69.44±8.59 | 68.74±9.96 | 0.77 |
| Urea (mg/dl) | 14.18±3.62 | 17.84±5.99 | 0.007 |
| Creatinine (mg/dl) | 0.55±0.08 | 0.57±0.12 | 0.35 |
| Hemoglobin (g/dl) | 13.45±1.23 | 12.68±1.28 | 0.021 |
| Leukocytes (×103/µl) | 8.64±2.06 | 8.40±1.77 | 0.64 |
GDM, gestational diabetes mellitus.
Isolation and characterization of placental exosomes from urine
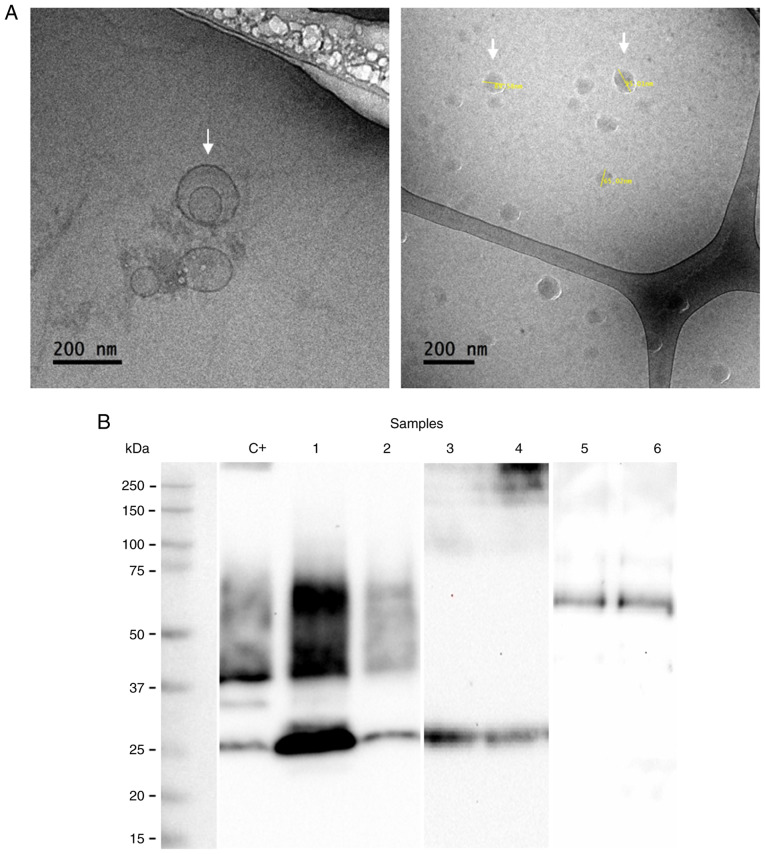
The presence of exosomes was demonstrated by morphological (cryo-TEM) and molecular characterization (western blot analysis) after 4 and 12 months of storage. The isolated exosomes exhibited a spherical shape with a size between 80 and 140 nm (Fig. 1A). The arrows in Fig. 1A indicate the presence of exosomes. The presence of CD63, CD9 and CD81 proteins associated with the exosome identity was corroborated by western blot analysis (Fig. 1B). In addition, the presence of PLAP indicates the placental origin of these vesicles (Fig. 1B).
Figure 1.
Characterization of urinary exosomes of placental origin in women with gestational diabetes mellitus. (A) Transmission electron microscopy in cryogenic mode. Arrows indicate the presence of exosomes and the size (B) Identification of exosomal markers by western blot. Loaded samples belong to urinary exosomes isolated from GDM patients. Loading control: C+ (vesicles isolated from HepG2 cells). Lanes 1 and 2, CD63 (30 kDa) and CD9 (25 kDa) markers; lanes 3 and 4, CD81 (25 kDa) marker; lanes 5 and 6, PLAP (52 kDa) marker.
Differential expression of placental exosomal miRNAs in the urine of patients with GDM and healthy pregnant women
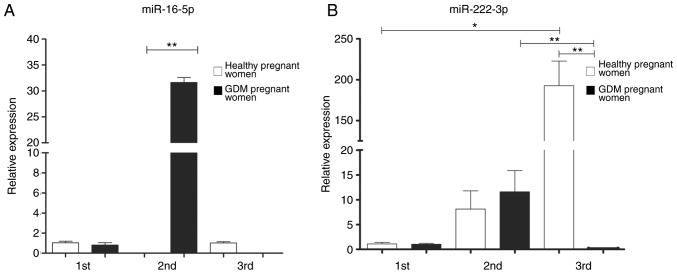
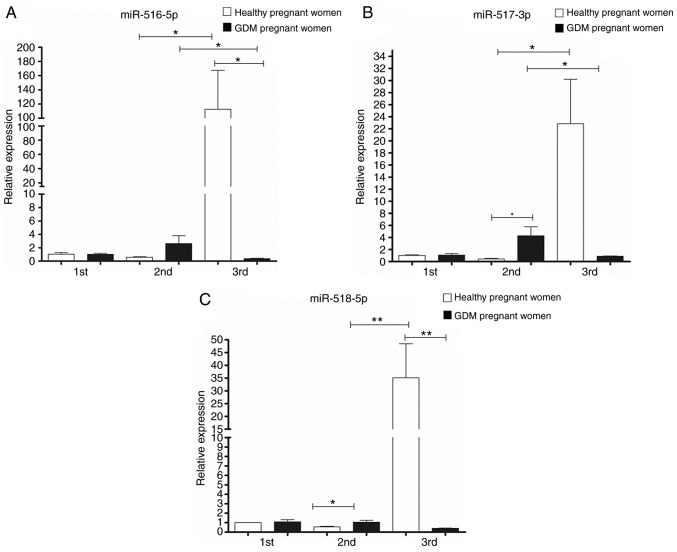
The relative expression of each miRNA was determined by RT-qPCR, using the 2−ΔΔCq method and U6 as a constitutive expression gene for normalization. In the 1st trimester of pregnancy, non-significant differences were observed in the expression of the five analyzed miRNAs. However, statistically significant differences were found between the study groups in the 2nd and 3rd trimester of pregnancy, for the miRNAs coded by C19MC, as well as for miR-222-3p (Figs. 2 and 3).
Figure 2.
Expression of (A) miR-16-5p and (B) miR-222-3p in exosomes of healthy pregnant women and with GDM through pregnancy. A non-parametric Kruskall-Wallis statistical test was used with a Dunn's post hoc test was performed. *P<0.05, **P<0.01.
Figure 3.
Expression of (A) miR-516-5p, (B) miR-517-3p and (C) miR-518-5p in urinary exosomes of placental origin of healthy pregnant women and gestational diabetics during the three trimesters of gestation. A non-parametric Kruskall-Wallis statistical test was used with a Dunn's post hoc test was performed. *P<0.05, **P<0.01.
The expression of miR-222-3p in the placental exosomes from healthy women increased significantly as the pregnancy progressed, as shown in Fig. 2; however, the expression of this miRNA in the pregnant women with GDM significantly decreased during the 3rd trimester in comparison to the healthy controls (Fig. 2).
Regarding the expression of 'trophomiRs' (miR-516-5p, miR-517-3p and miR-518-5p), the expression profile observed in samples from healthy pregnant women exhibited an increase in their expression as the pregnancy progressed, from the 1st to the 3rd trimester. In the case of the GDM group, the expression of these trophomiRs increased from the 1st to the 2nd trimester, with miR-517-3p and miR-518-5p exhibiting statistically significant differences (upregulated) compared to the healthy pregnant women. In the GDM group, a marked decrease was observed in the 3rd trimester and the expression levels of the three miRNAs were significantly downregulated compared to the healthy pregnant group (Fig. 3).
Since the expression levels of miR-16-5p, miR-517-3p and miR-518-5p in patients with GDM were differed markedly from those of the healthy controls in the 2nd trimester, the diagnostic potential of these miRNAs was further investigated. A ROC curve constructed based on exosomal miRNAs levels in the early 2nd trimester, revealed that the area under the curve (AUC) was 1 (CI 1-1) for miR-16-5p, miR-517-3p and miR-518-5p, indicating the high diagnostic accuracy of these miRNAs for the differentiation between patients with GDM and healthy women. The performance of these miRNAs to predict GDM is shown in Table II.
Table II.
ROC analysis of the microRNAs.
| MicroRNA | AUC | IC 95% | Standard error | P-value |
|---|---|---|---|---|
| miR-16-5p | 1.00 | 1.00-1.00 | 0 | 0.009 |
| miR-222-3p | 0.688 | 0.2893-1.086 | 0.203 | 0.387 |
| miR-516b-5p | 0.938 | 0.7615-1.113 | 0.090 | 0.043 |
| miR-517-5p | 1.00 | 1.00-1.00 | 0 | 0.034 |
| miR-518-3p | 1.00 | 1.00-1.00 | 0 | 0.021 |
Evaluation of interaction networks of common target genes between the studied miRNAs
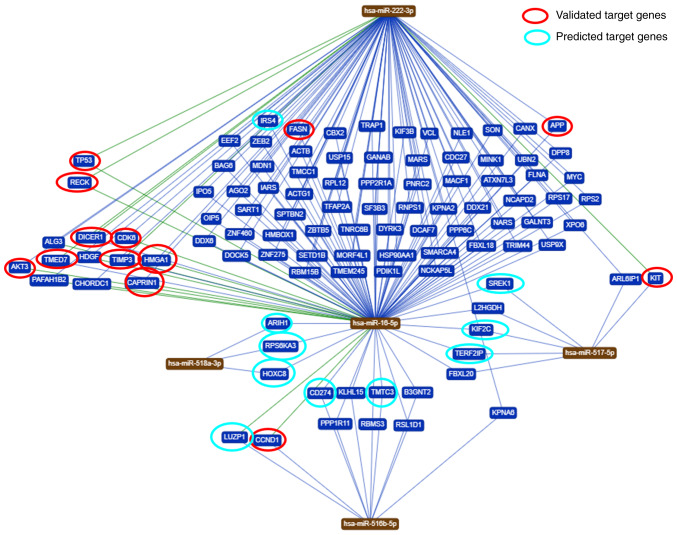
To identify genes regulated by the five miRNAs analyzed in the present study, a bioinformatics analysis was performed. Based on the bioinformatic analysis, it was demonstrated that these miRNAs are predicted to regulate different target genes. In total, 102 genes were shared among the five miRNAs evaluated in the present study (Fig. 4). An in-depth analysis of the potential functions of the five miRNAs under study (https://ccb-web.cs.unisaarland.de/mirtargetlink/), revealed that some of the target genes predicted by the bioinformatics analysis have been previously experimentally validated or predicted (Table III).
Figure 4.
Common targets between the five studied microRNAs. The evaluation for the interaction and target genes networks was performed using the miRTargetLink program. Validated (red) and predicted (green) target genes are highlighted.
Table III.
MicroRNAs validated and/or predicted target genes.
| Target Official symbol | Official full name | Sequence accession ID (Gene) | MicroRNA associated with regulation | Predicted/validated | Gene function |
|---|---|---|---|---|---|
| FASN | Fatty acid synthase | NM_004104.5 | miR-16-5p | Validated (RT-qPCR) | Regulate components of the insulin/PI3K-AKT (21). |
| AKT3 (miRTarBase ID MIRT003329) | AKT serine/threonine kinase 3 | NM_005465.7 | miR-16-5p | Validated (luciferase reporter assay) | Involved in a wide variety of biological processes including cell proliferation, differentiation, apoptosis, tumorigenesis, as well as glycogen synthesis and glucose uptake. Key regulators of endothelial cell functions and angiogenesis (22). |
| CAPRIN1 (miRTarBase ID MIRT005086) | Cell cycle associated protein 1 | NM_005898.5 | miR-16-5p | Validated (luciferase reporter assay/RT-qPCR/western blot analysis) | Involved in cell proliferation (23). |
| HMGA1 (miRTarBase ID MIRT000270) | High mobility group AT-hook 1 | NM_002131.4 | miR-16-5p | Validated (luciferase reporter assay/RT-qPCR/western blot analysis) | Involved in cell proliferation and in the development of human malignancies (23,24). |
| CDK6 (miRTarBase ID MIRT000939) | Cyclin-dependent kinase 6 | NM_001259.8 | miR-16-5p | Validated (RT-qPCR/flow cytometry/luciferase reporter assay/western blot analysis) | Gene related to the cell cycle. Control of the G1/S transition (25). |
| TP53 (miRTarBase ID MIRT005764) | Tumor protein p53 | NM_000546.6 | miR-16-5p and miR-222-3p | Validated (western blot anal ysis/luciferase reporter assay) | Encodes a tumor suppressor protein that contains transcriptional activation, DNA binding, and oligomerization domains. The encoded protein responds to various cellular stresses to regulate the expression of target genes, leading to cell cycle arrest, apoptosis, senescence, DNA repair, or changes in metabolisms (26,27). |
| RECK (miRTarBase ID MIRT031373) | Reversion inducing cysteine rich protein with kazal motifs | NM_021111.3 | miR-16-5p and miR-222-3p | Validated (luciferase reporter assay/RT-qPCR/western blot analysis) | Associated with cellular apoptosis. Suppresses tumor invasion and metastasis by downregulating the activity of matrix metalloproteinase 9 (MMP9) (28). |
| APP (miRTarBase ID MIRT031838) | Amyloid-beta precursor protein | NM_000484.4 | miR-16-5p | Validated (ELISA/immunopre cipitation/luciferase reporter assay/RT-qPCR/western blot analysis) | Regulator Aβ, Tau, inflammation and oxidative stress in Alzheimer's disease (29). |
| CCND1 (miRTarBase ID MIRT001225) | Cyclin D1 | NM_053056.3 | miR-16-5p | Validated (luciferase reporter assay/RT-qPCR/western blot analysis/flow cytometry) | Well-known regulator of cell cycle progression (30-32). |
| IRS4 | Insulin receptor substrate 4 | NM_003604.2 | miR-16-5p | Predicted | Involved in cell growth and glucose homeostasis (33). |
| DICER1 (miRTarBase ID MIRT006377) | Dicer 1, ribonuclease III | NM_030621.4 | miR-222-3p | Validated (luciferase reporter assay) | It plays a crucial role in the biogenesis of small regulatory RNAs, such as microRNAs (miRNAs) and small interfering RNAs (siRNAs). Dicer has also been found to participate in chromosomal fragmentation during apoptosis or in inflammatory processes (34). |
| TMED7 (miRTarBase ID MIRT002334) | Transmembrane p24 trafficking protein 7 | NM_181836.6 | miR-222-3p | Validated (western blot analysis; RT-qPCR) | Involved in vesicular protein trafficking between endosomal structures in resting cells and the Golgi apparatus, therefore, it is functionally similar to the Rab family (35,36). |
| TIMP3 (miRTarBase ID MIRT003451) | TIMP metallopeptidase inhibitor 3 | NM_000362.5 | miR-222-3p | Validated (flow cytometry/immunohistochemistry/luciferase reporter assay/RT-qPCR/western blot analysis/immunohistochemistry/in situ hybridization/ELISA) | The proteins encoded by this gene family are inhibitors of the matrix metalloproteinases, a group of peptidases involved in degradation of the extracellular matrix (ECM). Expression of this gene is induced in response to mito genic stimulation (37,38). |
| KIT (miRTarBase ID MIRT001779) | KIT proto-oncogene, receptor tyrosine kinase | NM_000222.3 | miR-222-3p | Validated (luciferase reporter assay/RT-qPCR/microarray/western blot analysis) | Related to cell proliferation and melanogenesis (39,40). Involved in erythropoiesis, erythroleukemic cell growth, and TRAIL apoptotic signal in lung cancer cells (41). Oncogene in many types of cancer, involved in the pathogenesis of PTC (42). Promotes expansion of the first erythroblasts, linked to cancer therapy (43). Regulation of gastrointestinal stromal tumors (GIST) (44). |
| ARIH1 (miRTarBase ID MIRT549302) | Ariadne RBR E3 ubiquitin-protein ligase 1 | NM_005744.5 | miR-518a-3p | Predicted [PAR-CLIP (45)] | Associated with cellular proliferation and modification of proteins in nuclear bodies (46). Potent mediator of DNA damage-induced translation arrest that protects stem and cancer cells against genotoxic stress (47). |
| RPS6KA3 (miRTarBase ID MIRT554630) | Ribosomal protein S6 kinase A3 | NM_004586.3 | miR-518a-3p | Predicted [PAR-CLIP (48)] | Associated with cellular processes such as cell growth, motility, survival, and proliferation (49). Related to Coffin-Lowry syndrome (50). |
| HOXC8 (miRTarBase ID MIRT562161) | Homeobox C8 | NM_022658.4 | miR-518a-3p | Predicted [PAR-CLIP (45)] | Associated to cellular processes such as cell growth, motility, survival, and proliferation in cancer cells (51-53). |
| LUZP1 (miRTarBase ID MIRT444756) | Leucine zipper protein 1 | NM_033631.4 | miR-516b-5p | Predicted [PAR-CLIP (54)] | Involved in exosomal processes, ventricular septum development (55,56). |
| CD274 (miRTarBase ID MIRT445289) | CD274 molecule | NM_001267706.1 | miR-516b-5p | Predicted [PAR-CLIP (54)] | Expressed by T cells, B cells, and monocytes is a potent regulator of immune responses (57). Programmed death ligand 1 (PD-L1, CD274) has been reported to be expressed abnormally in many cancers (58). |
| TMTC3 (miRTarBase ID MIRT445289) | Transmembrane O-mannosyltransferase targeting cadherins 3 | NM_181783.4 | miR-516b-5p | Predicted [PAR-CLIP (59)] | Role in proteolysis and stress response in the endoplasmic reticulum. It can play a direct role in the physiology of immune cells in long-term graft function (60). |
| KIF2C (miRTarBase ID MIRT455884) | Kinesin family member 2C | NM_001297655.2 | miR-517-5p | Predicted [PAR-CLIP (59)] | Modulator in microtubule depolymerization, bipolar spindle formation, and chromosome segregation, has been reported to take roles in cancer biology (61,62). |
| TERF2 (miRTarBase ID MIRT490157) | TERF2 interacting protein | NM_018975.4 | miR-517-5p | Predicted [PAR-CLIP (48)] | Folding human telomeres into loops to prevent unwanted DNA repair and final binding of chromosomes (63). |
| SREK1 (miRTarBase ID MIRT611178) | splicing regulatory glutamic acid and lysine rich protein 1 | NM_001323533.2 | miR-517-5p | Predicted [PAR-CLIP (64)] | Nucleic acid binding and nucleotide binding (55,56). |
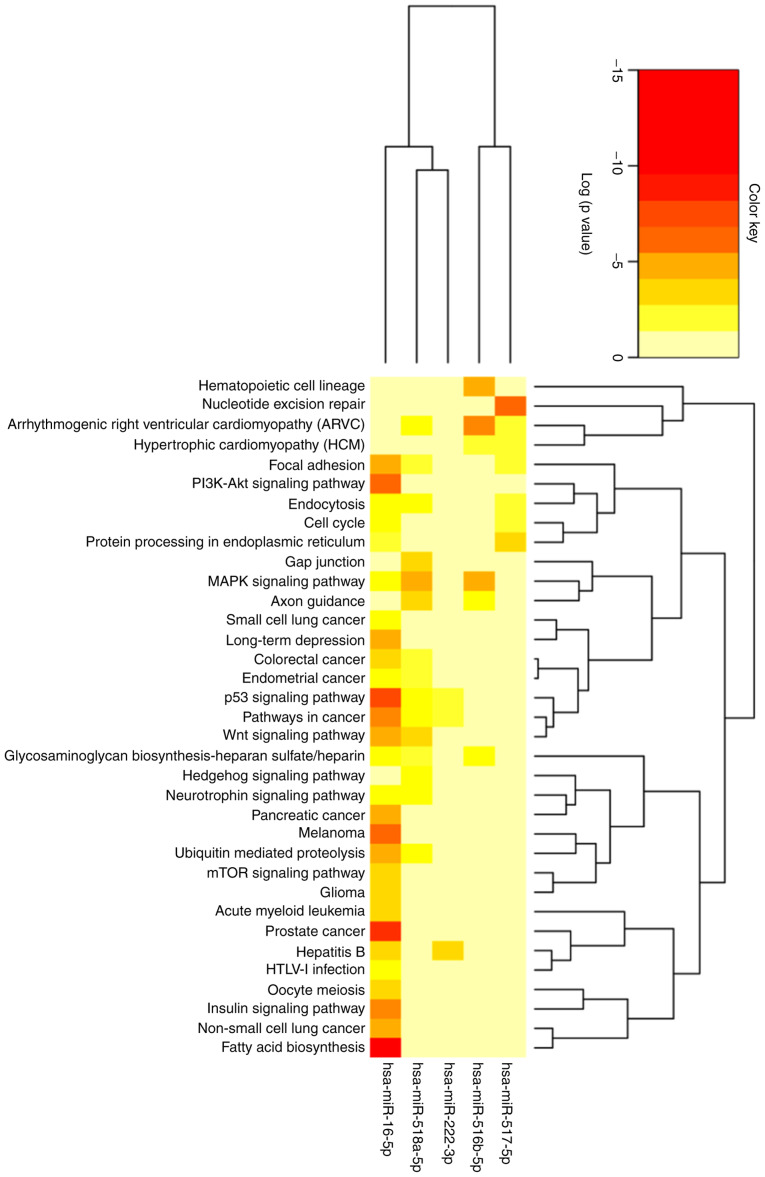
The 102 genes identified were related to 62 pathways predicted to be involved in different metabolic, molecular and cell regulation processes. In total, 32 routes presented a value of P<0.05, indicating that these particular pathways are regulated by at least one of the studied miRNAs. Some of the most significative pathways are the biosynthesis of fatty acids, the signaling pathway of PI3K-Akt and the signaling of insulin, as shown in Fig. 5 and in further detail in Table SVI.
Figure 5.
Heatmap showing the signaling pathways involved in regulation by the five studied microRNAs. Using the bioinformatics tool, The Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/), signaling pathways related to the study of microRNAs were evaluated, considering a value of P<0.05 to indicate statistically significant differences (the color scale represents statistical significance).
Discussion
To date, there are several methods wich can be used to examine miRNA profiling, such as RT-qPCR, microarrays and direct sequencing. Each method has its advantages and limitations; however, RT-qPCR seems to have better sensitivity and is the only platform capable of generating absolute quantification. In the present study, as well as in several studies (65), RT-qPCR techniques were used to validate candidate biomarkers identified through a literature search or based on previous findings (66-69).
The relative expression of miR-516-5p, miR-517-3p, miR-518-5p, miR-222-3p and miR-16-5p in urinary placental exosomes from patients with GDM were evaluated in the present study, at different stages of pregnancy. To the best of our knowledge, there is no previous study available in the literature to date characterizing these miRNAs expressed in urinary placental exosomes.
It was found that the miRNAs expression levels differed across pregnancy. Accordingly, it has been reported that in the maternal plasma, a substantial amount of placental-associated miRNAs are differently expressed during the 1st, 2nd and 3rd trimester of gestation, probably to meet the different regulatory demands of pregnancy (70).
In the present study, women with GDM exhibited additional risk factors, such as being older and overweight, compared to healthy pregnant women; both conditions are related to high adiposity, which favors a pro-inflammatory response by producing adipokines. Both the production of adipokines and the high levels of glucose in circulation are considered factors that induce the overexpression of miR-16-5p (14). In the case of miR-16-5p, it was found that in the 2nd trimester, its expression was only detectable in exosomes purified from patients with GDM, while in healthy controls, it was undetectable. miR-16-5p is a modulator of the PI3K/Akt signaling pathway, through the regulation of genes, such as Pi3Kr1 and Pi3kr3, mTOR and Mapk3, among others. In the present study, bioinformatics analysis revealed that the signaling pathways of PI3K/Akt (P<0.0001), Wnt (P<0.0001), insulin (P=0.004) and mTOR (P=0.016) were significantly regulated by miR-16-5p (Table SVI). The overexpression of these cell signaling pathways has been associated with diabetes mellitus and GDM (21). In addition, genes encoding for proteins 1 and 2 of the insulin receptor substrate (IRS1/IRS2) are targets of miR-16-5p (21,71). Therefore, the upregulation of miR-16-5p in patients with GDM at the 2nd trimester, will result in a negative regulation of IRS1 and IRS2, which could lead to abnormal Wnt/β-catenin signaling and, eventually, in diabetes (72). However, following the diagnosis of GDM, patients were subjected to pharmacological treatment (metformin, insulin, or a combination of both), diet and exercise. It is suggested that treatment with metformin may influence the pattern of expression observed in the 3rd trimester of pregnancy. Ortega et al identified that the circulating expression of miR-140-5p (-16%, P=0.004) and miR-222 (-47%, P=0.03) decreased significantly following metformin treatment in patients with type 2 diabetes (T2D) (73). Furthermore, Demirsoy et al demonstrated that a total of 13 miRNAs were found significantly downregulated following the treatment of patients with T2D with metformin (P<0.05) (74). The effect of the treatment, together with the complex and dynamic expression of miRNAs, which vary according to the requirements in the regulation of specific stages of development, may explain the differential expression of miRNAs across gestation.
A similar profile was observed for miR-222-3p, with the only difference that this microRNA was also expressed in the second trimester of healthy pregnancy. On the other hand, the role of miR-222-3p in the development of T2D has been previously reported and has been associated with the pathogenesis of GDM (75). Zhao et al reported that miR-222-3p expression was significantly decreased in women with GDM compared to controls at 17 weeks of gestation in serum samples (76). Similarly, Pheiffer et al reported a decrease in the expression of this miRNA in the serum of women with GDM compared to the controls at 26 weeks of gestation (77). Furthermore, the expresion of this miRNA increases in the adipose tissue of women with GDM, and negatively correlates with the levels of glucose transporter 4 (GLUT4) and estrogen receptor (ER) (78). It is important to consider that, since expression miR-222-3p is dependent on tissue and insulin sensibility, the expression between adipose tissue and placenta could be differential. Recently, the authors reported that in the urine metabolome profile of women with GDM in the 3rd trimester of pregnancy, 14 metabolites related with the steroid hormone biosynthesis and tryptophan metabolism were significantly elevated [i.e., L-tryptophan, urobilinogen, ceramide (d18:0/23:0), 21-deoxycortisol]. The upregulation of these pathways could trigger insulin resistance and may respond to oxidative stress and inflammation during GDM (79). The majority of the genes involved in the insulin resistance pathway are targets of miR-16-5p and miR-222-3p (8,80). These miRNAs, contained in exosomes, can reach different tissues and organs, thereby affecting metabolic processes in patients with GDM.
In the present study, the amplification of miRNAs coded by the C19MC (miR-516b-5p, miR-517-5p and miR-518a-3p) is an indirect evidence of the placental origin of the exosomes that were purified, which agrees with the detection of the exosomal marker, PLAP, by western blot analysis. The expression profile of this particular set of miRNAs was characterized by an increase across gestation in healthy pregnant and no statistically significant differences were found between both study groups in the 1st trimester. However, in women with GDM the expression was differential, and it was characterized by an increase in the 2nd trimester followed by a reduction in the 3rd trimester of pregnancy.
To date, there are only a few studies available on the altered expression of C19MC miRNAs in pregnancy disorders (13,81,82). None of these studies however, have evaluated the expression of these miRNAs in urinary exosomes derived from the placenta and, for this reason, the results of the expression levels obtained in the present study cannot bed compared directly with those of other reports in the literature. Hromadnikova et al reported that 1st trimester circulating plasma exosomes possess the identical C19MC miRNA expression profile as placental tissues derived from patients with gestational hypertension, preeclampsia and fetal growth restriction following labor (83). Further studies are required to elucidate whether the downregulation of C19MC miRNAs in placental exosomes is associated with the pathophysiology of GDM or if it is a compensatory mechanism.
The bioinformatics prediction carried out in the present study through KEGG revealed that miR-16-5p, miR-222-3p, miR-516b-5p, miR-517-5p and miR-518a-3p shared common target genes. The fact that redundant regulation through several miRNAs is required to modulate a physiological process constitutes a security molecular process. Some of the target genes shared by the 5 miRNAs selected in the present study (irs4, galnt, reck, alg3, akt3, timp3, kit, l2hgdh, ki2fc, rap1, fb12, hoxc8 and pd-l1) are expressed in a variety of tissues, including placenta and adipose tissue. Exosomes potentially reach these tissues, reflecting placental regulation as part of maternal-fetal communication. Future functional studies are needed to elucidate the effects of microRNAs expression on maternal and fetal tissues and also to follow offspring during extrauterine life.
The present study supports the use of placenta-derived exosomes as part of biomarker screening strategies to identify women at risk of later development of pregnancy-related complications. The placental exosomes released to the systemic circulation and collected in the urine represent a non-invasive source of signaling molecules, including miRNAs, whose abnormal expression profile due to defective placental functions, reflects the expression profile of the trophoblast cells and, consequently, provides more precise information about pregnancy disorders. In the 1st trimester of gestation, at least for the selected set of miRNAs under study, we were not able to find significant differences in the levels of expression in GDM or healthy patients. At the 2nd trimester, 3 miRNAs (mir-16-5p, miR-517-3p and miR-518-5p, exhibited a good diagnostic value. However, since the sample size in the present study was small, these results need to be interpreted with caution and further studies, including an external validation cohort with a larger size number are required.
The scenery for validating exosomal miRNAs for their intended use in clinical applications is still complex. To date, the most promising markers of gestational diabetes are: miR-29a, miR-222, miR-16-5p, miR-17-5p and miR-20a-5p (14,71,76,78,84). In future miRNAs expression studies during pregnancy, several aspects should be taken into account: Gestational age (since the expression of miRNAs changes through pregnancy), the type of sample studied, the sex of the offspring and the route of delivery. Therefore, these variables must be established between study groups. Further analyses need to be conducted to explore a larger population to validate these findings and to evaluate their diagnostic predictive value at the 1st trimester of gestation. Despite this limitation, the results revealed herein demonstrate alterations in the expression of miRNAs across gestation and a differential expression profile within urinary exosomes as a result of pathophysiological alterations in women with GDM. In general, miRNAs that regulate the innate and specific immune system are expressed throughout pregnancy. By contrast, miRNAs that promote cell differentiation and control cell cycle progression are mainly expressed in advanced gestation.
In conclusion, the expression profile of miRNAs carried as the molecular cargo of placental urinary exosomes is different between healthy and women with GDM; the differences are evident and quantifiable from the 2nd trimester, being even more evident towards the 3rd trimester. The alteration of miRNA expression modulates pathways related to the development of insulin resistance, pro-inflammatory response and metabolic homeostasis, which are implicated in the physiopathology of GDM. The expression of miR-16-5p in the 2nd trimester may be an important molecular mechanism involved in insulin resistance in women who develop GDM, although validation studies are required. C19MC miRNAs also exhibit a differential profile in patients with GDM in comparison to healthy controls, being upregulated in the second trimester in the former group followed by a reduction in the third trimester of pregnancy. The differential expression of miRNAs in various development steps is probably to meet the different regulatory demands of pregnancy. To the best of our knowledge, the present study is the first to report the expression of miRNAs in placental urinary exosomes, suggesting that urine is a potential biological fluid for the research of pathological conditions during pregnancy.
Supplementary Data
Acknowledgments
The authors would like to thank the participation of Medical students of the specialty of Gynecology at the Hospital Central 'Dr.Ignacio Morones Prieto' for their support in patient recruitment.
Funding
The present study was funded by CONACYT-Fondo Sectorial de Salud (grant no. 290239 to YLH).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YLH and MSB were involved in the study conceptualization. ASHVO, NVDF, EMM, DPPP and DAGL were involved in the study methodology. MSB was involved in software analysis. ASHVO and JAL were involved in data validation. YLH, MSB, EMM, DPPP and ASHVO were involved in formal analysis. DEN was involved in the analysis and interpretation of data. JCTO was involved in the investigative aspects of the study. YLH was responsible for the study resources. ASHVO was involved in the writing and preparation of the original draft of the manuscript. YLH, EMM and JAL were involved in the writing, reviewing and editing of the manuscript. DEN was involved in the critical revision of the manuscript. MSB and YLH supervised the study. YLH was involved in project administration and in funding acquisition. All authors have read and agreed to the published version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Hospital Central 'Dr. Ignacio Morones Prieto', with registration number 84-17 (Folio CONBIOETICA-24- CEI-001-201604279). The study was conducted in accordance with the Declaration of Helsinki. The informed consent signature was obtained from each participant before the collection of urine samples.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Basina M. Gestational diabetogenesis. J Women's Health Care. 2012;01:e106. doi: 10.4172/2167-0420.1000e106. [DOI] [Google Scholar]
- 2.Sáez T, de Vos P, Sobrevia L, Faas MM. Is there a role for exosomes in foetoplacental endothelial dysfunction in gestational diabetes mellitus? Placenta. 2018;61:48–54. doi: 10.1016/j.placenta.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Powe CE. Early pregnancy biochemical predictors of gestational diabetes mellitus. Curr Diab Rep. 2017;17:12. doi: 10.1007/s11892-017-0834-y. [DOI] [PubMed] [Google Scholar]
- 4.Rodrigo N, Glastras SJ. The emerging role of biomarkers in the diagnosis of gestational diabetes mellitus. J Clin Med. 2018;7:120. doi: 10.3390/jcm7060120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akgöl E, Abuşoğlu S, Gün FD, Ünlü A. Prevalence of gestational diabetes mellitus according to the different criterias. Turk J Obstet Gynecol. 2017;14:18–22. doi: 10.4274/tjod.38802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karcaaltincaba D, Calis P, Ocal N, Ozek A, Altug Inan M, Bayram M. Prevalence of gestational diabetes mellitus evaluated by universal screening with a 75-g,2-h oral glucose tolerance test and IADPSG criteria. Int J Gynecol Obstet. 2017;138:148–151. doi: 10.1002/ijgo.12205. [DOI] [PubMed] [Google Scholar]
- 7.Ilekis JV, Tsilou E, Fisher S, Abrahams VM, Soares MJ, Cross JC, Zamudio S, Illsley NP, Myatt L, Colvis C, et al. Placental origins of adverse pregnancy outcomes: Potential molecular targets: An executive workshop summary of the eunice kennedy shriver national institute of child health and human development. Am J Obstet Gynecol. 2016;215(1 Suppl):S1–S46. doi: 10.1016/j.ajog.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guarino E, Poggi CD, Grieco GE, Cenci V, Ceccarelli E, Crisci I, Sebastiani G, Dotta F. Circulating MicroRNAs as biomarkers of gestational diabetes mellitus: Updates and perspectives. Int J Endocrinol. 2018;2018:6380463. doi: 10.1155/2018/6380463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Cuffe JSM, Holland O, Salomon C, Rice GE, Perkins AV. Review: Placental derived biomarkers of pregnancy disorders. Placenta. 2017;54:104–110. doi: 10.1016/j.placenta.2017.01.119. [DOI] [PubMed] [Google Scholar]
- 11.Fu G, Brkić J, Hayder H, Peng C. MicroRNAs in human placental development and pregnancy complications. Int J Mol Sci. 2013;14:5519–5544. doi: 10.3390/ijms14035519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouyang Y, Mouillet JF, Coyne CB, Sadovsky Y. Review: Placenta-specific microRNAs in exosomes-good things come in nano-packages. Placenta. 2014;35(Suppl):S69–S73. doi: 10.1016/j.placenta.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hromadnikova I, Kotlabova K, Ondrackova M, Pirkova P, Kestlerova A, Novotna V, Hympanova L, Krofta L. Expression profile of C19MC microRNAs in placental tissue in pregnancy-related complications. DNA Cell Biol. 2015;34:437–457. doi: 10.1089/dna.2014.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y, Tian F, Li H, Zhou Y, Lu J, Ge Q. Profiling maternal plasma microRNA expression in early pregnancy to predict gestational diabetes mellitus. Int J Gynecol Obstet. 2015;130:49–53. doi: 10.1016/j.ijgo.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Salomon C, Torres MJ, Kobayashi M, Scholz-Romero K, Sobrevia L, Dobierzewska A, Illanes SE, Mitchell MD, Rice GE. A gestational profile of placental exosomes in maternal plasma and their effects on endothelial cell migration. PLoS One. 2014;9:e98667. doi: 10.1371/journal.pone.0098667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salomon C, Scholz-Romero K, Sarker S, Sweeney E, Kobayashi M, Correa P, Longo S, Duncombe G, Mitchell MD, Rice GE, Illanes SE. Gestational diabetes mellitus is associated with changes in the concentration and bioactivity of placenta-derived exosomes in maternal circulation across gestation. Diabetes. 2016;65:598–609. doi: 10.2337/db15-0966. [DOI] [PubMed] [Google Scholar]
- 17.Diagnostic Criteria and Classification of Hyperglycaemia First Detected in Pregnancy: A world health organization guideline. Diabetes Res Clin Pract. 2014;103:341–363. doi: 10.1016/j.diabres.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Gonzales PA, Zhou H, Pisitkun T, Wang NS, Star RA, Knepper MA, Yuen PS. Isolation and purification of exosomes in urine. Methods Mol Biol. 2010;641:89–99. doi: 10.1007/978-1-60761-711-2_6. [DOI] [PubMed] [Google Scholar]
- 19.Abdalla M. Comprehensive coverage of exosomes purification and exosomal RNA isolation from different types of liquid biopsies. https://www.exosome-rna.com/upcoming-webinar-comprehensive-coverage-of-exosome-purification-and-exosomal-rna-isolation-from-different-types-of-liquid-biopsies/. Accessed April 15, 2019.
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Kwon DN, Chang BS, Kim JH. MicroRNA dysregulation in liver and pancreas of CMP-Neu5Ac hydroxylase null mice disrupts insulin/PI3K-AKT signaling. Biomed Res Int. 2014;2014:236385. doi: 10.1155/2014/236385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spinetti G, Fortunato O, Caporali A, Shantikumar S, Marchetti M, Meloni M, Descamps B, Floris I, Sangalli E, Vono R, et al. MicroRNA-15a and MicroRNA-16 Impair human circulating proangiogenic cell functions and are increased in the proangiogenic cells and serum of patients with critical limb ischemia. Circ Res. 2013;112:335–346. doi: 10.1161/CIRCRESAHA.111.300418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaddar T, Rouault JP, Chien WW, Chebel A, Gadoux M, Salles G, Ffrench M, Magaud JP. Two new miR-16 targets: Caprin-1 and HMGA1, proteins implicated in cell proliferation. Biol Cell. 2009;101:511–524. doi: 10.1042/BC20080213. [DOI] [PubMed] [Google Scholar]
- 24.Palmieri D, D'Angelo D, Valentino T, De Martino I, Ferraro A, Wierinckx A, Fedele M, Trouillas J, Fusco A. Downregulation of HMGA-targeting microRNAs has a critical role in human pituitary tumorigenesis. Oncogene. 2012;31:3857–3865. doi: 10.1038/onc.2011.557. [DOI] [PubMed] [Google Scholar]
- 25.Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. MiR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–5404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin Y, Stephen CW, Luciani MG, Fåhraeus R. p53 stability and activity is regulated by Mdm2-mediated induction of alter-native p53 translation products. Nat Cell Biol. 2002;4:462–467. doi: 10.1038/ncb801. [DOI] [PubMed] [Google Scholar]
- 27.Marcel V, Perrier S, Aoubala M, Ageorges S, Groves MJ, Diot A, Fernandes K, Tauro S, Bourdon JC. Δ160p53 is a novel N-terminal p53 isoform encoded by Δ133p53 transcript. FEBS Lett. 2010;584:4463–4468. doi: 10.1016/j.febslet.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y, Xia Y, Niu H, Chen Y. MiR-16 induced the suppression of cell apoptosis while promote proliferation in esophageal squamous cell carcinoma. Cell Physiol Biochem. 2014;33:1340–1348. doi: 10.1159/000358701. [DOI] [PubMed] [Google Scholar]
- 29.Parsi S, Smith PY, Goupil C, Dorval V, Hébert SS. Preclinical evaluation of miR-15/107 family members as multifactorial drug targets for Alzheimer's disease. Mol Ther Nucleic Acids. 2015;4:e256. doi: 10.1038/mtna.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L, Kappeler A, Brunner T, Vassella E. MiR-15a and miR-16 are implicated in cell cycle regulation in a Rb-Dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553–5559. doi: 10.1158/0008-5472.CAN-08-4277. [DOI] [PubMed] [Google Scholar]
- 31.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 32.Lerner M, Harada M, Lovén J, Castro J, Davis Z, Oscier D, Henriksson M, Sangfelt O, Grandér D, Corcoran MM. DLEU2, frequently deleted in malignancy, functions as a critical host gene of the cell cycle inhibitory microRNAs miR-15a and miR-16-1. Exp Cell Res. 2009;315:2941–2952. doi: 10.1016/j.yexcr.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Sebastiani G, Guarino E, Grieco GE, Formichi C, Poggi CD, Ceccarelli E, Dotta F. Circulating microRNA (miRNA) expression profiling in plasma of patients with gestational diabetes mellitus reveals upregulation of miRNA miR-330-3p. Front Endocrinol (Lausanne) 2017;8:345. doi: 10.3389/fendo.2017.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koralewska N, Ciechanowska K, Pokornowska M, Figlerowicz M, Kurzyńska-Kokorniak A. Human ribonuclease Dicer-structure and functions. Postepy Biochem. 2019;65:173–182. doi: 10.18388/pb.2019_267. In Polish. [DOI] [PubMed] [Google Scholar]
- 35.Doyle SL, Husebye H, Connolly DJ, Espevik T, O'Neill LA, McGettrick AF. The GOLD domain-containing protein TMED7 inhibits TLR4 signalling from the endosome upon LPS stimu-lation. Nat Commun. 2012;3:707. doi: 10.1038/ncomms1706. [DOI] [PubMed] [Google Scholar]
- 36.Füllekrug J, Suganuma T, Tang BL, Hong W, Storrie B, Nilsson T. Localization and recycling of gp27 (hp24gamma3): Complex formation with other p24 family members. Mol Biol Cell. 1999;10:1939–1955. doi: 10.1091/mbc.10.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wisniewska M, Goettig P, Maskos K, Belouski E, Winters D, Hecht R, Black R, Bode W. Structural determinants of the ADAM inhibition by TIMP-3: Crystal structure of the TACE-N-TIMP-3 complex. J Mol Biol. 2008;381:1307–1319. doi: 10.1016/j.jmb.2008.06.088. [DOI] [PubMed] [Google Scholar]
- 38.Kotronis K, Zafrakas M, Papasozomenou P, Timologou A, Miliaras D, Tarlatzis BC, Grimbizis G. Protein expression pattern of tissue inhibitor of metalloproteinase-3 (TIMP3) in endometriosis and normal endometrium. Gynecol Endocrinol. 2019;35:1103–1106. doi: 10.1080/09513590.2019.1625880. [DOI] [PubMed] [Google Scholar]
- 39.Felicetti F, Errico MC, Bottero L, Segnalini P, Stoppacciaro A, Biffoni M, Felli N, Mattia G, Petrini M, Colombo MP, et al. The Promyelocytic leukemia zinc Finger-MicroRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;68:2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 40.Felicetti F, Errico MC, Segnalini P, Mattia G, Carè A. MicroRNA-221 and -222 pathway controls melanoma progression. Expert Rev Anticancer Ther. 2008;8:1759–1765. doi: 10.1586/14737140.8.11.1759. [DOI] [PubMed] [Google Scholar]
- 41.Garofalo M, Quintavalle C, Di Leva G, Zanca C, Romano G, Taccioli C, Liu CG, Croce CM, Condorelli G. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008;27:3845–3855. doi: 10.1038/onc.2008.6. [DOI] [PubMed] [Google Scholar]
- 42.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu C G, Franssila K, Suster S, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gits CM, van Kuijk PF, Jonkers MB, Boersma AW, van Ijcken WF, Wozniak A, Sciot R, Rutkowski P, Schöffski P, Taguchi T, et al. MiR-17-92 and miR-221/222 cluster members target KIT and ETV1 in human gastrointestinal stromal tumours. Br J Cancer. 2013;109:1625–1635. doi: 10.1038/bjc.2013.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-Binding protein and MicroRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elmehdawi F, Wheway G, Szymanska K, Adams M, High AS, Johnson CA, Robinson PA. Human homolog of drosophila ariadne (HHARI) is a marker of cellular proliferation associated with nuclear bodies. Exp Cell Res. 2013;319:161–172. doi: 10.1016/j.yexcr.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 47.von Stechow L, Typas D, Carreras Puigvert J, Oort L, Siddappa R, Pines A, Vrieling H, van de Water B, Mullenders LH, Danen EH. The E3 Ubiquitin ligase ARIH1 protects against genotoxic stress by initiating a 4EHP-Mediated mRNA translation arrest. Mol Cell Biol. 2015;35:1254–1268. doi: 10.1128/MCB.01152-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011;8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 49.Kennedy SA, Jarboui MA, Srihari S, Raso C, Bryan K, Dernayka L, Charitou T, Bernal-Llinares M, Herrera-Montavez C, Krstic A, et al. Extensive rewiring of the EGFR network in colorectal cancer cells expressing transforming levels of KRASG13D. Nat Commun. 2020;11:499. doi: 10.1038/s41467-019-14224-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen N, Liu Y, Zhang K, Lyu Y, Gao M, Ma J, Xu L, Gai Z. Analysis of RPS6KA3 gene mutation in a Chinese pedigree affected with coffinlowry syndrome. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2019;36:798–800. doi: 10.3760/cma.j.issn.1003-9406.2019.08.011. In Chinese. [DOI] [PubMed] [Google Scholar]
- 51.Zhang J, Yang M, Li D, Zhu S, Zou J, Xu S, Wang Y, Shi J, Li Y. Homeobox C8 is a transcriptional repressor of E-cadherin gene expression in non-small cell lung cancer. Int J Biochem Cell Biol. 2019;114:105557. doi: 10.1016/j.biocel.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 52.Gong C, Zou J, Zhang M, Zhang J, Xu S, Zhu S, Yang M, Li D, Wang Y, Shi J, Li Y. Upregulation of MGP by HOXC8 promotes the proliferation, migration, and EMT processes of triple-negative breast cancer. Mol Carcinog. 2019;58:1863–1875. doi: 10.1002/mc.23079. [DOI] [PubMed] [Google Scholar]
- 53.Fang X, Yan R. MiR-152 inhibits the proliferation and invasion of chordoma cells by targeting HOXC8. J Int Med Res. 2019;47:5185–5193. doi: 10.1177/0300060519870915. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Gottwein E, Corcoran DL, Mukherjee N, Skalsky RL, Hafner M, Nusbaum JD, Shamulailatpam P, Love CL, Dave SS, Tuschl T, et al. Viral MicroRNA targetome of KSHV-Infected primary effusion lymphoma cell lines. Cell Host Microbe. 2011;10:515–526. doi: 10.1016/j.chom.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.The Gene Ontology Consortium The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng X, Veverka V, Radhakrishnan A, Waters LC, Muskett FW, Morgan SH, Huo J, Yu C, Evans EJ, Leslie AJ, et al. Structure and interactions of the human programmed cell death 1 receptor. J Biol Chem. 2013;288:11771–11785. doi: 10.1074/jbc.M112.448126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He PX, Ma ZL, Han H, Zhang XY, Niu SH, Du LN, Zheng YC, Liu HM. Expression of programmed death ligand 1 (PD-L1) is associated with metastasis and differentiation in gastric cancer. Life Sci. 2020;242:117247. doi: 10.1016/j.lfs.2019.117247. [DOI] [PubMed] [Google Scholar]
- 59.Whisnant AW, Bogerd HP, Flores O, Ho P, Powers JG, Sharova N, Stevenson M, Chen CH, Cullen BR. In-Depth analysis of the interaction of HIV-1 with cellular microRNA biogenesis and effector mechanisms. MBio. 2013;4:e000193. doi: 10.1128/mBio.00193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Racapé M, Duong Van Huyen JP, Danger R, Giral M, Bleicher F, Foucher Y, Pallier A, Pilet P, Tafelmeyer P, Ashton-Chess J, et al. The involvement of SMILE/TMTC3 in endoplasmic reticulum stress response. PLoS One. 2011;6:e19321. doi: 10.1371/journal.pone.0019321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gan H, Lin L, Hu N, Yang Y, Gao Y, Pei Y, Chen K, Sun B. KIF2C exerts an oncogenic role in nonsmall cell lung cancer and is negatively regulated by miR-325-3p. Cell Biochem Funct. 2019;37:424–431. doi: 10.1002/cbf.3420. [DOI] [PubMed] [Google Scholar]
- 62.McHugh T, Zou J, Volkov VA, Aurélie Bertin A, Talapatra SK, Rappsilber J, Dogterom M, Welburn JPI. The depolymerase activity of MCAK shows a graded response to Aurora B kinase phosphorylation through allosteric regulation. J Cell Sci. 2019;132:jcs228353. doi: 10.1242/jcs.228353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nečasová I, Janoušková E, Klumpler T, Hofr C. Basic domain of telomere guardian TRF2 reduces D-loop unwinding whereas Rap1 restores it. Nucleic Acids Res. 2017;45:12170–12180. doi: 10.1093/nar/gkx812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karginov FV, Hannon GJ. Remodeling of Ago2-mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev. 2013;27:1624–1632. doi: 10.1101/gad.215939.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moldovan L, Batte KE, Trgovcich J, Wisler J, Marsh CB, Piper M. Methodological challenges in utilizing miRNAs as circulating biomarkers. Version 2 J Cell Mol Med. 2014;18:371–390. doi: 10.1111/jcmm.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Munaut C, Tebache L, Blacher S, Noël A, Nisolle M, Chantraine F. Dysregulated circulating miRNAs in preeclampsia. Biomed Rep. 2016;5:686–692. doi: 10.3892/br.2016.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li D, Li J. Association of miR-34a3p/5p, miR-1413p/5p, and miR-24 in decidual natural killer cells with unexplained recurrent spontaneous abortion. Med Sci Monit. 2016;22:922–929. doi: 10.12659/MSM.895459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song GY, Song WW, Han Y, Wang D, Na Q. Characterization of the role of microRNA-517a expression in low birth weight infants. J Dev Orig Health Dis. 2013;4:522–526. doi: 10.1017/S204017441300024X. [DOI] [PubMed] [Google Scholar]
- 69.Li J, Chen L, Tang Q, Wu W, Gu H, Liu L, Wu J, Jiang H, Ding H, Xia Y, et al. The role, mechanism and potentially novel biomarker of microRNA-17-92 cluster in macrosomia. Sci Rep. 2015;5:17212. doi: 10.1038/srep17212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cai M, Kolluru GK, Ahmed A. Small molecule, big prospects: MicroRNA in pregnancy and its complications. J Pregnancy. 2017;2017:6972732. doi: 10.1155/2017/6972732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao YL, Jia YJ, Xing BH, Shi DD, Dong XJ. Plasma microRNA-16-5p-17-5p and -20a-5p: Novel diagnostic biomarkers for gestational diabetes mellitus. J Obstet Gynaecol Res. 2017;43:974–981. doi: 10.1111/jog.13317. [DOI] [PubMed] [Google Scholar]
- 72.Geng Y, Ju Y, Ren F, Qiu Y, Tomita Y, Tomoeda M, Kishida M, Wang Y, Jin L, Su F, et al. Insulin receptor substrate 1/2 (IRS1/2) regulates Wnt/β-catenin signaling through blocking autophagic degradation of dishevelled2. J Biol Chem. 2014;289:11230–11241. doi: 10.1074/jbc.M113.544999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ortega FJ, Mercader JM, Moreno-Navarrete JM, Rovira O, Guerra E, Esteve E, Xifra G, Martínez C, Ricart W, Rieusset J, et al. Profiling of circulating MicroRNAs reveals common micrornas linked to type 2 diabetes that change with insulin sensitization. Diabetes Care. 2014;37:1375–1383. doi: 10.2337/dc13-1847. [DOI] [PubMed] [Google Scholar]
- 74.Demirsoy İH, Ertural DY, Balci Ş, Çınkır Ü, Sezer K, Tamer L, Aras N. Profiles of circulating MiRNAs following metformin treatment in patients with type 2 diabetes. J Med Biochem. 2018;37:499–506. doi: 10.2478/jomb-2018-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Collares CV, Evangelista AF, Xavier DJ, Rassi DM, Arns T, Foss-Freitas MC, Foss MC, Puthier D, Sakamoto-Hojo ET, Passos GA, et al. Identifying common and specific microRNAs expressed in peripheral blood mononuclear cell of type 1, type 2, and gestational diabetes mellitus patients. BMC Res Notes. 2013;6:491. doi: 10.1186/1756-0500-6-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao C, Dong J, Jiang T, Shi Z, Yu B, Zhu Y, Chen D, Xu J, Huo R, Dai J, et al. Early second-trimester serum MiRNA profiling predicts gestational diabetes mellitus. PLoS One. 2011;6:e23925. doi: 10.1371/journal.pone.0023925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pheiffer C, Dias S, Rheeder P, Adam S. Decreased expression of circulating miR-20a-5p in South African women with gestational diabetes mellitus. Mol Diagn Ther. 2018;22:345–352. doi: 10.1007/s40291-018-0325-0. [DOI] [PubMed] [Google Scholar]
- 78.Shi Z, Zhao C, Guo X, Ding H, Cui Y, Shen R, Liu J. Differential expression of microRNAs in omental adipose tissue from gestational diabetes mellitus subjects reveals miR-222 as a regulator of Erα expression in estrogen-induced insulin resistance. Endocrinology. 2014;155:1982–1990. doi: 10.1210/en.2013-2046. [DOI] [PubMed] [Google Scholar]
- 79.López-Hernández Y, Herrera-Van Oostdam A, Toro-Ortiz JC, López JA, Salgado-Bustamante M, Murgu M, Torres-Torres LM. Urinary metabolites altered during the third trimester in pregnancies complicated by gestational diabetes mellitus: Relationship with potential upcoming metabolic disorders. Int J Mol Sci. 2019;20:1186. doi: 10.3390/ijms20051186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ibarra A, Vega-Guedes B, Brito-Casillas Y, Wägner AM. Diabetes in pregnancy and MicroRNAs: Promises and limitations in their clinical application. Noncoding RNA. 2018;4:32. doi: 10.3390/ncrna4040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sadovsky Y, Mouillet JF, Ouyang Y, Bayer A, Coyne CB. The function of trophomirs and other micrornas in the human placenta. Cold Spring Harb Perspect Med. 2015;5:a023036. doi: 10.1101/cshperspect.a023036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hromadnikova I, Kotlabova K, Ivankova K, Krofta L. First trimester screening of circulating C19MC microRNAs and the evaluation of their potential to predict the onset of preeclampsia and IUGR. PLoS One. 2017;12:e0171756. doi: 10.1371/journal.pone.0171756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hromadnikova I, Dvorakova L, Kotlabova K, Krofta L. The prediction of gestational hypertension, preeclampsia and fetal growth restriction via the first trimester screening of plasma exosomal C19MC microRNAs. Int J Mol Sci. 2019;20:2972. doi: 10.3390/ijms20122972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Esteves JV, Enguita FJ, Machado UF. MicroRNAs-Mediated regulation of skeletal muscle GLUT4 expression and translocation in insulin resistance. J Diabetes Res. 2017;2017:7267910. doi: 10.1155/2017/7267910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.