Abstract
Genome editing techniques are considered to be one of the most challenging yet efficient tools for assisting therapeutic approaches. Several studies have focused on the development of novel methods to improve the efficiency of gene editing, as well as minimise their off-target effects. Clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein (Cas9) is a tool that has revolutionised genome editing technologies. New applications of CRISPR/Cas9 in a broad range of diseases have demonstrated its efficiency and have been used in ex vivo models of somatic and pluripotent stem cells, as well as in in vivo animal models, and may eventually be used to correct defective genes. The focus of the present review was the recent applications of CRISPR/Cas9 and its contribution to the treatment of challenging human diseases, such as various types of cancer, neurodegenerative diseases and a broad spectrum of other disorders. CRISPR technology is a novel method for disease treatment, enhancing the effectiveness of drugs and improving the development of personalised medicine.
Keywords: clustered regularly interspaced short palindromic repeats, Cas9, gene editing, gene therapy, human diseases, neurodegenerative diseases, in vivo animal models, ex vivo models
1. Introduction
Since the discovery of the clustered regularly interspaced short palindromic repeats (CRISPR) system in 1987 (1), there have been significant advances in the field of gene therapy. The CRISPR system is an adaptive prokaryotic immune system, which serves as a bacterial defence mechanism against insertion of foreign genomic material and prevents the destructive impacts of mobile genetic elements delivered by phages and plasmids (2). CRISPR/Cas9 has been shown to boost the host immune system using the invading organisms' genetic material in order to protect the host from further invasion. The protective mechanism is completed with the acquisition of spacer sequences by CRISPR-associated spacer (Cas) proteins (3). Cas proteins are guided to the exogenous spacer sequences of foreign nucleic acids by CRISPR-associated RNA (crRNA) (4). The mechanism involves spacer identification and anchoring by Cas proteins, providing protection against further invasion. The existence of CRISPR was discovered in 1987 by Ishino et al (1), who cloned a portion of the CRISPR sequence together with the inhibitor of apoptosis gene (1). This discovery resulted in a novel method for gene-based therapeutics for the treatment of challenging disorders (5). Further analysis of prokaryotes, such as Archaeoglobus fulgidus, revealed other constituents of the CRISPR system, including non-messenger RNA sequencing, transcription of DNA repeats loci (target DNA sequences that are acquired and preserved in CRISPR loci) to small RNAs (Fig. 1) (6), and the Cas gene family (which is associated with CRISPR loci during immune processes) (7). Furthermore, identification of specific spacer sequences from the viral genome revealed how bacterial systems exhibited phenotypic resistance against the phage (7).
Figure 1.
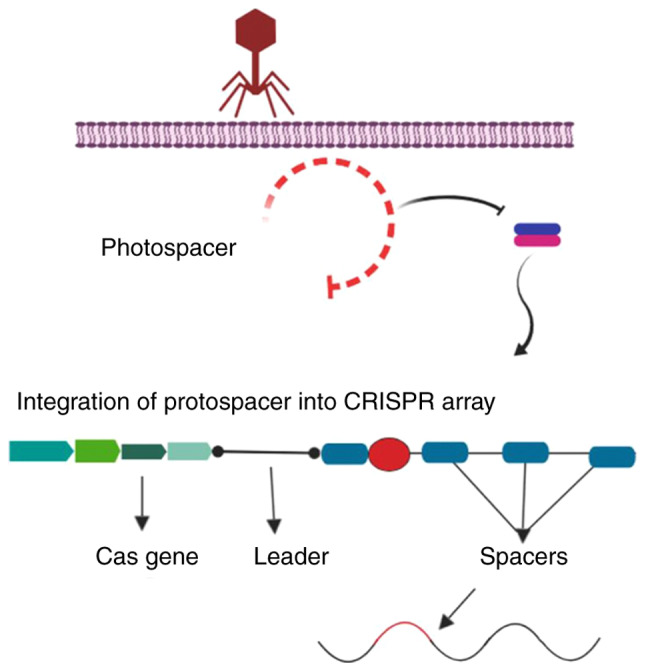
Schematic representation of the CRISPR immune system in the acquisition of foreign genetic material. The CRISPR system consists of a Cas operon containing Cas genes, and a CRISPR array that contains identical repeat sequences and spacers. In the case of viral or plasmid-based invasion, CRISPR acquires the protospacer sequence (red) of the viral DNA, which is achieved via a Cas1-Cas2 complex and integrated into the CRISPR array, which is further transcribed to pre-crRNA. CRISPR, clustered regularly interspaced short palindromic repeats.
The aim of the present review was to describe the challenges and achievements of CRISPR customisation, taking into consideration various factors that may affect therapeutic outcomes in vitro and in vivo.
2. DNA repair systems
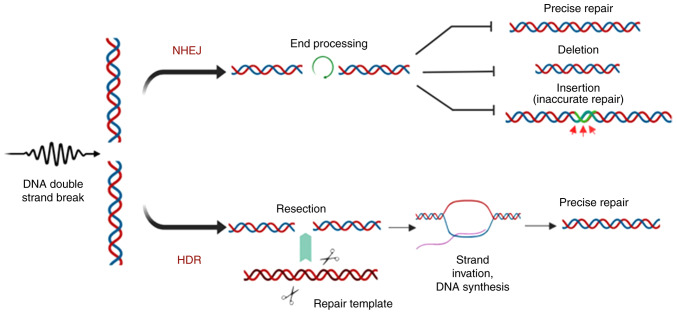
As a therapeutic method, CRISPR is used for both gene knockout and gene knock-in to correct a specific gene. Generation of double-strand breaks (DSBs) in the target gene results in the initiation of two endogenous DNA repair mechanisms: Non-homologous end joining (NHEJ) and homology-directed repair (HDR).
NHEJ joins a DSB without the use of a homologous DNA template (8,9), resulting in mutations, deletions and insertion of potentially nonsensical genetic material (Fig. 2) (10). Given the functional characteristics of NHEJ, it is considered an error-prone repair pathway, as it may result in the introduction of random indels and frameshifts in the sequence of a gene causing gene knockout, insertion or deletion of an amino acid, incomplete truncation and, ultimately, deactivation of the coded protein (Fig. 2). NHEJ is not limited to a specific cell cycle phase, whereas HDR occurs in the S or G phase and uses the sister chromatid as a template for homologous repair (11). HDR is a more precise DNA repair mechanism, the function of which has become a focus of attention along with site-specific gene editing tools in gene editing technology (9,12-16).
Figure 2.
DNA repair mechanisms used for gene editing. Formation of double-stranded breaks to initiate endogenous DNA repair by NHEJ, resulting in acci-dental insertions/deletions, or by HDR, which uses a template DNA strand for repair. NHEJ, non-homologous end joining; HDR, homology directed repair.
3. Overview of other gene editing tools
Various tools have been developed to perform knock-in and knockout in target genes to improve gene editing, and these have been used as therapeutic tools to treat certain diseases.
Zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs)
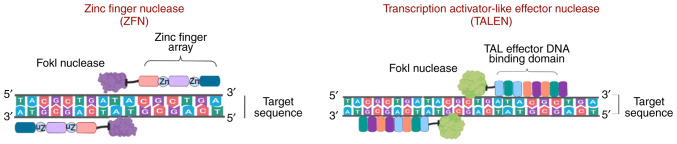
ZFNs are sequence-specific nucleases that are frequently found in eukaryotes and were first discovered as repetitive zinc-binding domains in Xenopus oocytes (17), which are currently known as zinc finger motifs. Analysis of the ZFN crystal structure revealed the presence of 30 cysteine and histidine residues (each in pair; Cys2-His2) bound to zinc ions that provide stability to their ββα structure (18). ZFNs also contain a non-specific Fok-1 restriction enzyme, which is bound to the DNA-binding domain of eukaryotic transcription factors known as zinc finger proteins (ZFPs) containing Cys2-His2 fingers. Each finger identifies almost three base pairs of DNA sequences of target DNA and assists the binding of the ZFN to a particular sequence (17,19). However, there are difficulties in assembling ZFP fingers to bind to specific extended DNA sequences. Maeder et al (20,21) designed and assembled ZPFs that could bind to a specific 200-bp DNA sequence. However, binding may occur at random sites of the genome and, thus, may complicate gene correction. Furthermore, frequent off-target effects in the number of loci is another concern (22,23). Researchers successfully addressed this issue by designing ZFN pairs where one ZFN binds to the forward strand and the other to the reverse strand. Each pair contains distinct heterodimer Fok-1 domains with opposite charges, assisting in the formation of DSBs (Fig. 3) (24-26).
Figure 3.
Comparison of ZFNs and TALENs, non-specific nucleases designed to cleave the genome at a specific site. ZFNs, zinc finger nucleases; TALENs, transcription activator-like effector nucleases.
Subsequently, another method for gene editing, termed TALENs, was developed, which is considered more efficient, and has more advanced potential gene therapy applications (27,28). TALENs are fusion proteins composed of TALE and Fok-1 nucleases. The proteins, 33-35 amino acids in length, contain repeat variable di-residues, which are central binding protein regions that may be customized (29,30). Compared with ZFNs, TALENs are more suitable for therapeutic use due to the 1:1 TALE-DNA binding affinity (31) and lower rates of cytotoxicity (32). However, TALENs delivery is dependent on vectors, and developing suitable vectors has proved challenging, as the size of the cDNA that encodes TALENs is larger than the cDNA which encodes ZFNs (Fig. 3) (33).
CRISPR
To design a customized CRISPR system, a single-guide (sg)RNA must be designed (34). Jinek et al (35) and Cong et al (5) used the CRISPR/Cas system as a genome editing tool and, since then, it has been extensively studied by researchers to identify other characteristics of the CIRSPR system, which may be used for eukaryotic genome editing and, thus, treatment of various diseases. Liang et al (36) used this system to correct a mutation in the haemoglobin subunit β (HBB) gene whilst working on the haemoglobin subunit δ (HBD) gene (which is homologous to the HBB gene) (36).
CRISPR classification
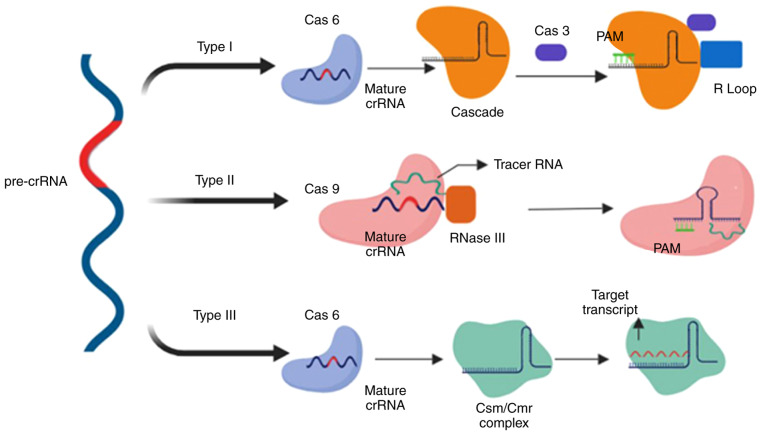
The CRISPR/Cas system is classified into two types, with each type comprising various subtypes (I-V) based on their flanking Cas genes and location of the target on foreign DNA (37), which have improved our under-standing of and ability to develop phage-resistant strains and the phylogenetic classification of bacteria. During phage/plasmid invasion, CRISPR/Cas functions in three phases: Adaptation, expression and interference. Each stage is associated with specific characteristics that result in antiplasmid or antiviral immunity (38). Adaptation consists of an integration process by which the invader-derived spacers (known as the spacer sequence) merge with the CRISPR array. In the next step, the CRISPR loci are transcribed into CRISPR-associated RNA (crRNA), which contains the spacer sequence. Subsequently, an endonuclease is produced and uses the spacer sequences as a guide to cleave the invader genome (Fig. 4) (39).
Figure 4.
Comparison of type I, II and III CRISPR systems in crRNA maturation and interference. Upon transcription of CRISPR following the acquisition stage, pre-crRNA undergoes a maturation stage, which is processed by Cas6 in type I and III. In type II, the maturation step is performed by Cas9 accompanied by tracer RNA and RNase III. The interference step varies notably between the different types. CRISPR, clustered regularly interspaced short palindromic repeats; crRNA, CRISPR-associated RNA; Cas, CRISPR-associated protein; pre-crRNA, precursor crRNA; PAM, protospacer adjacent motif.
The functional characteristics of the CRISPR/Cas system are defined by the properties of the Cas1 and Cas2 genes. Taxonomic studies initially classified the CRISPR/Cas system into three types based on the particular marker proteins: Cas3 (type I), Cas9 (type II) and Csm/Cmr (type III) (40). Subsequently, classes IV and V of the CRISPR editing system were added (41). The general classification of the CRISPR system is based on the genes that encode the functional proteins and factors (Cascade, Csm, Cmr complex or Cas9). Class 1 CRISPR systems functions consist of multi-subunit crRNA-effector complexes, and includes type I, III and putative type IV (41). Class 2 only consists of Cas9 as a single protein, which performs all the functions of the CRISPR system, a feature also observed in putative class V (40).
Type I CRISPR is defined by the significance of the Cascade complex followed by the Cas3 nuclease (42). Pre-crRNA, which is the product of a transcribed CRISPR array, is cleaved by Cas6e, resulting in the production of crRNA (43). crRNA, which is associated with Cascade, is responsible for locating the protospacer in the target DNA. Furthermore, another subunit of Cascade, Cas8, identifies and locates protospacer adjacent motif (PAM), which is a short sequence located near the target sequence (44). The PAM sequence is the crucial factor for type I Cascade-Cas immune defence mechanisms; its dysfunction results in an inability of crRNA to recognise the spacers in target DNA by Cascade proteins (45,46), and inhibits R-loop formation between crRNA and target DNA, a process which eventually results in viral evasion from CRISPR screening (47,48). In the presence of a fully functional CRISPR system, recognition leads to activation of the Cas3 nuclease, which creates nicks on the single-stranded DNA of the target (virus or plasmid), resulting in its degradation (Fig. 4) (49).
Unlike type I, type II relies on Cas9 as the sole Cas protein, two types of RNAs, RNase III and tracer RNA (13), and a PAM, which is located downstream of the protospacer sequence in target DNA and recognised by the Cas9 protein (50). A DSB is introduced by HNH nuclease domain cleaving one strand, and RuvC nuclease domain cleaving another (51). However, cleavage requires identification of a PAM sequence by Cas9, which results in dissociation of dsDNA and the formation of an R-loop between the crRNA and DNA. This structural change results in the binding of the tracer RNA to the cleavage target sequence (52,53).
Type III is unique to Cas6, whose endoribonuclease mechanism produces crRNA by cleaving pre-crRNA (5). Unlike previous models of the CRISPR system, this type introduces 8-nucleotide repeat sequences, known as crRNA tags, as a result of crRNA cleavage by Cas6. The crRNA tag, which is located downstream of the spacer sequence (54-56), undergoes maturation and is modified into six nucleotides (53). The size of the crRNA complex increases and forms Cas10-Csm in type III-A and Cas10-Cmr complex III-B systems (54). In contrast to type I and type II, type III targets both DNA and RNA, producing co-transcriptional crRNA-guided cleavage of the target DNA (57,58). The palm domain of Cas10 cleaves DNA strands (58) and Csm3 (type III-A), and Cmr4 (type III-B) cleave RNA transcripts (59,60). Another notable difference of type III is that the PAM is not necessarily essential for the system to initiate immune mechanisms (Fig. 4).
4. Overview of therapeutic approaches
The identification of the CRISPR system allowed for improvements to gene editing technologies, and may improve current therapeutic techniques in the field of medicine, genetics, embryology, bioinformatics and pathology. Compared with the other genome editing techniques, CRISPR/Cas9 is more cost-effective, easier to use and can be used to perform specific gene knockdown, or base insertions and substitutions with lower rates of mutations with the known mechanism of CRISPR system.
Neurodegenerative diseases
Neurodegenerative diseases are characterised by disruption of neuronal function or loss of neurons that cause progressive central nervous system dysfunction and, thus, are frequently difficult to treat (61). There are numerous pathological neurodegenerative disorders, including Alzheimer's disease (AD), which is a fatal form of progressive dementia, and Parkinson's disease, which is a progressive movement disorder. These neurodegenerative diseases are relatively common (62). Familial AD, which may be caused by point mutations or deletions in the genes encoding amyloid precursor protein (APP), presenilin (PSEN)1 or PSEN2 (62), accounts for <5% of AD cases. Sporadic AD, which develops due to environmental factors, such as ageing or injury following brain ischemia, accounts for >90% of AD cases (63). In vitro analyses of induced pluripotent stem cells (iPSCs) that have been treated with CRISPR/Cas9 provide future perspectives in the treatment of neurodegenerative diseases, such as AD (64). Several studies have used the CRISPR/Cas9 system to identify defective and upregulated genes in early- and late-onset AD. Analysis of chromosome 1q31-42 in early-onset familial AD revealed a point mutation in PSEN2 (65), which was found to be correlated with a significant increase in the β-amyloid peptide ratio (Aβ42-43/40) (66), a change in voltage-gated potassium channel expression (67), and an increase in the concentration of calcium in the endoplasmic reticulum of neurons (68), which leads to neurotoxicity, cognitive deterioration and short-term memory loss in patients with AD, primarily due to basal forebrain cholinergic neuron (BFCN) damage. When healthy BFCNs were transplanted into an AD mouse model, a significant improvement in learning ability was observed (69). These findings led to further analyses of BFCNs, which were established from in vitro development of iPSCs obtained from the fibroblasts of humans with a PSEN2 mutation (70). The PSEN2N141 iPSC-derived BFCNs, characterised by a lack of electrophysiological properties, were observed to exhibit an improvement in neural activity and amyloid ratio following correction of the PSEN2 gene using CRISPR/Cas9 (71). Other studies performing similar analyses confirmed the effect of the CRISPR/Cas9 genome editing system on cells, the damage of which was associated with AD.
Scientists from the New York Stem Cell Foundation Research Institute expanded their studies on PSEN1 and PSEN2 point mutations in familial AD (72). They demonstrated a correlation between the inflammasome (a component of an innate immune system in a myeloid cell) and PSEN2 mutations using CRISPR/Cas9 (71). A correction of the point mutation on the PSEN2 gene in iPSCs derived from AD patients was performed using CRISPR/Cas9 and template sgRNA, as well as single-stranded oligonucleotides (ssODNs) that could edit the sequence in exon 5 of the PSEN2 gene located on chromosome 1 (1q42.13) (73). In vitro gene editing using the CRISPR/Cas9 system reversed the effect of the mutated PSEN2 gene on the increase of amyloid plaques (Aβ42/40) in iPSC-derived BFCNs in the cerebrospinal fluid, which was directly associated with the onset of AD (71). The in vivo effect of CRISPR/Cas9 on the mutated APP gene shows the system's ability to reduce neurotoxicity in AD patients. The APPsw allele in the fibroblasts of human patients' with an APPsw mutation was transfected using CRISPR/Cas9 with a gRNA designed to specifically mutate the allele. There was a considerable decrease in heterozygous APP alleles (APPsw/WT) and in the β-amyloid ratio (73). Additionally, the S. pyogenes CRISPR system has been shown to target specific desired genes or alleles (e.g., the APPsw allele) without disruption or indel formation in the other alleles, such as APPWT (73). However, in vivo analysis in a Tg2576 mouse model (mice with an age-deficient cognitive ability that over-express mutant APP) using exosome-adeno-associated virus (AAV)1 with CRISPR/Cas9 and gRNA, showed indel formation and DNA frameshifts in the APPsw alleles (73). Although CRISPR/Cas9 may successfully alter the APPsw allele and reduce the β-amyloid ratio in brain cells, further investigations and clinical trials are required to improve the system and to reduce or ideally nullify indel formation.
Cancer therapy and drug discovery
Before the discovery of gene editing tools, several complications in cancer therapy could not be efficiently addressed, as drug resistance had not been effectively identified as a possible cause (74-76). CRISPR/Cas9 has facilitated the identification of genes associated with drug resistance by exposing drug-resistant cells to CRISPR/Cas9 gRNAs, each of which individually knock out a single gene at a time in each cell (77), and the results may be used to introduce alternative drugs with increased efficacy for improved outcomes (77).
For drug design, animal models and human cell lines are the optimal platforms for testing specific drug toxicity and efficacy prior to its use in humans. However, animal models, as well as in vitro analysis of human cell lines, may not provide representative and conclusive results regarding the effectiveness of a particular drug. CRISPR/Cas9 has provided a means to modify cells to allow them to more accurately represent human models for different types of cancer. ID8 (mouse ovarian surface epithelium) cells, which were obtained from ID8 mice with ovarian cancer with the TP53 and BRCA2 genes knocked out using CRISPR/Cas9, displayed characteristics of a high-grade serous carcinoma (78). High-grade serous ovarian carcinomas (HGSCs) exhibit reduced activity of TP53 and BRCA genes, loss of ability to form Rad51 foci (associated with DSB repair), and sensitivity to poly(ADP-ribose) polymerase inhibition (79). CRISPR/Cas9 has assisted in the development of more representative models of cancer that are likely to be increasingly used for evaluating drug safety and eliminating drug resistance in human diseases.
In addition to drug design, CRISPR/Cas9 has been proposed as a promising gene editing tool in cancer therapy. The dCas9 (mutated Cas9 without endonuclease activity, with added transcriptional activators on dCas9 or gRNA) is used to target specific genes by either activating or knocking them out, when combined with transcriptional activation or inhibition (80). Epigenome editing is another approach to cancer treatment, in which dCas9 is tethered to histone modifiers involved in DNA methylation to disturb processes associated with cancer progression, as DNA methylation is observed in the majority of cancers (81).
To determine the effects of the CRISPR/Cas9 system on different malignancies, such as leukaemia, K562 human myeloid leukaemia cells, which do not normally harbour a mutation in the isocitrate dehydrogenase (IDH2) gene, under-went a DSB and point mutation on the IDH2 R140Q locus following transfection with a plasmid which contained a sgRNA and CRISPR/Cas9. Following the mutation, gene repair was performed using another transfection with a pBS-SK+ vector with a CRISPR/Cas9 and fluorescent-template DNA followed by sgRNA to check gene correction rate on point-mutated cells (82). The results revealed high levels of H3K9me2, H3K27me2 and H3K4me3 expression, which indicated hyper-methylation of chromatin in mutated cells (83). In other studies, CRISPR/Cas9 was used as a tracking device to determine the effect of IL (interleukin)4-induced signal transducer and activator of transcription (Stat)6 activation on the elimination of leukaemia cells by using lentiviral vectors containing Cas9 and sgRNA for Stat6 (84). The results indicated that IL4 and its antileukemic effects were dependent on the ability of acute myeloid leukaemia cells to activate Stat6 (84), highlighting the potential of the CRISPR/Cas9 system as a therapeutic and diagnostic tool in various diseases. Several studies have been performed to assess the therapeutic potential of the CRISPR system for treating different types of cancer. 2CT-CRISPR assay is used to identify the genes causing resistance to immune cells (85). The test consists of two types of target cells and effector cells, such as melanoma cells and CD8+ T cells, respectively. The aim of the assay is to identify factors mediating the growth of melanoma cells following an immune system response, and to design efficient therapeutic methods to augment immunotherapy against cancer cells. These assays were performed in vivo on C57BL/6J mice (85). Other studies focused on modifying chimeric antigen receptor (CAR) T cells in order to target cancer cells, and showed that CRISPR/Cas9 was more effective compared with RNA interference, which is only partially effective (86). Thus, scientists improved the efficiency of the technique by using CRISPR/Cas9, TALENs and ZFN, which reduced the off-target effects (87-93). However, the possibility of recurrence following treatment is considered to be a notable disadvantage that should be taken into account when designing the most effective therapy to decrease the possibility of the need for repeated treatments (88). Recent studies have demonstrated that CAR T-cell therapy may a promising treatment for various diseases, including cancer. CRISPR is a system that may be used for improving CAR T cells. CAR permanent tonic signalling has been shown to reduce antitumor activity (89-93). Human CAR T cells modified with CRISPR/Cas9 gene editing, which contained 4-1BB and CD3z intracellular signalling domains, eliminated tumour cells by targeting the CD19+ B cells that are associated with increased tumorigenesis (89).
Duchenne muscular dystrophy (DMD)
DMD is an X-linked recessive muscular disorder caused by certain mutations of the DMD gene, which is located on chromosome 21. The mutation leads to a decrease in the levels of dystrophin, which is the protein responsible for normal muscular integrity (94). Men are more prone to this disease, as they carry one X chromo-some (95). Of all the mutations identified in the DMD gene, ~86% are deletions and are present in an exon (96). In an attempt to regenerate muscle tissues to replace damaged tissue, haematopoietic therapy using haematopoietic stem cells obtained using ex vivo expansion of myoblasts from satellite cells has been developed. However, this technique was not found to be beneficial for patients with DMD (97), as myoblasts lose their ability to engraft into muscle tissues (98). iPSCS and embryonic stem cells (ESCs) are capable of producing a vast quantity of skeletal myogenic progenitors, exhibit in vivo regenerative capacity, as well the ability to synthesize dystrophin (99,100); therefore, ESCs are the optimal cells for performing genome editing to reduce DMD symptoms (99). Different therapies have been developed to replace dystrophin deficiency, such as anti-inflammatory-based techniques, or to restore DMD gene expression, such as cell-based therapies (101). Genome editing techniques, including the ZFN, TALENs and CRISPR systems, are the most efficient of these techniques. In vitro, TALENs and CRISPR function by restoring dystrophin synthesis via gene knock-in (insertion of exon 44 in the DMD gene), and has been demonstrated to be effective, with minimal off-target effects. iPSCs that were collected from DMD fibroblasts of a specific patient with DMD with exon 44 missing were transfected with TALENs and CRISPR separately to insert exon 44 by creating indels in adjacent exons (101). In vitro removal of exons 45-55, instead of a single exon in the DMD gene in the patient's iPSC-derived cardiomyocytes and skeletal myotubes using sgRNA and CRISPR/Cas9 system, resulted in restoration of dystrophin protein synthesis and, consequently, creatine kinase levels, whose linkage causes muscle instability and disintegration (102). Such a deletion was observed to normalise miR32 (miRNA32) (102), which reduces dystrophin levels in muscular dystrophies, including Becker muscular dystrophy (103). These results indicate the potential benefits of larger deletions, which rectify the dysfunction of other factors affecting the function of dystrophin. In vivo, CRISPR/Cas9 has been used to correct mutations of the DMD gene, in mdx mice (mice with a point mutation in the DMD gene and lacking dystrophin expression). sgRNA vector plasmids were used to target exon 23 of the DMD gene and ssODN to activate HDR repair mechanisms and repair the lesions of the DMD gene. However, the results indicated a higher rate of NHEJ-based repair, which resulted in the formation of indels (12).
AIDS
The HIV-1 virus invades host immune cells through the CD4 receptor and interaction with CC chemokine receptor 5 (CCR5) and CXC chemokine receptor 4. Although developmental treatment for HIV-1 with combinational retroviral therapy has improved the quality of life of the patients, it fails to eradicate the HIV-1 virus from the body, resulting in high rates of morbidity and mortality (104). A 32-bp deletion in the CCR5 allele results in the deactivation of the CCR5 gene, which results in a high degree of resistance to HIV-1 infection (104,105). In 2000, a patient with acute myeloid leukaemia and HIV-1 infection underwent bone marrow transplantation (using allogeneic stem cells) from a donor carrying CCR5Δ32/Δ32 cells, which resulted in abrogation of HIV replication, and the HIV-1 virus was not detected in the body (106). Research has focused on the development of homozygous cells using gene editing technologies, such as zinc-finger nuclease (107-109). Human CD34+ hematopoietic stem/progenitor cells (HSPCs) from umbilical cord blood were transfected with ZFNs to knock out the CCR5 locus on chromosome 3 to establish a CCR5−/− clone (108-110). These cells were grafted into non-obese diabetic/severe combined immunodeficient/interleukin 2Rγnull (NOD/SCID/IL2Rγnull; NSG) mice, an ideal rodent model for examining HIV-1 infections (111) and haematopoiesis (112). The results of the experiment revealed HIV-1 replication control (110). Similar studies on CD34+ HSPCs using adenoviral vectors carrying CCR5-ZFN resulted in a more effective knockdown of CCR5−/− and fewer off-target effects compared with plasmid DNA ZFNs (109). These studies were extended to human patients with HIV infection who received CD4 T cells with dysfunctional CCR5 using ZFN60. The results indicated a high number of CD4 T cells. However, the rate of viral replication in cells with non-mutated CCR5 alleles (homozygous) was faster compared with cells with mutated CCR5 allele (heterozygous), which indicates that cells with homogenicity require knockout in both alleles to participate in the disease prognosis (107). Furthermore, using modified cells with ZFN does not result in permanent changes in vivo, as modified cells fail to control HIV-1 replication due to the presence of unmodified cells (113). Additionally, the adverse effects of adenoviral vectors must be considered (94). Wild-type (WT) iPSCs have been used to generate homozygous cells that harbour mutations in CCR5, termed CCR5-Δ32 (114). Generation of these cells was performed by transfecting WT iPSCs with a CRISPR/Cas9 plasmid with a specific sgRNA (115). The matured monocytes or macrophages from the modified iPSCs expressed resistance to HIV-1 infection (114-116). Another study on HIV-1-positive patients demonstrated that the presence of Cas9 and gRNA together in T-cells (specifically CD4+ T cells) that have been manipulated genetically by Cas9/gRNA confer resistance to HIV-1 infection (117). The same study experimented with excision of pro-viral HIV-1 DNA from T cells using Cas9/gRNA on the RSBN1 gene, without disrupting the normal function of the gene, which encodes histone demethylase, which is responsible for chromatin structure (117). Astrocyte cell lines were transfected with Cas9 protein with and without plasmids, and double fluorescent protein HIV-1 reporter RGH was utilised to determine the excision sites of pro-viral HIV-1 DNA using a gRNA; the results demonstrated there was a reduction in fluorescent protein in astrocytes with no alterations to their regular function and morphology (118).
Sickle cell disease (SCD)
In SCD, a recessive genetic disorder with a prevalence of 250,000 annually worldwide (119), a modified CRISPR/Cas9 system was used with guide strands that specifically target the HBB and CCR5 genes using the pX330 plasmid in human kidney cells (120). Off-target effects were found to be directly associated with the presence of adjacent acquisition motifs (AAMs) in the PAM sequence, which reduces or nullifies the cleavage of target genes via CRISPR/Cas9 (120). The rate of mutations due to interception of the correction of the genes by Cas9 was directly correlated with the distance between sgRNA and the PAM of the specific protospacer recognised by the particular sgRNA (121). Furthermore, to correct the mutations in patients with SCD, ribonucleoprotein (RNP) consisting of Cas9 protein and sgRNA trG10 (truncated sgRNA G10 that targets the first exon of the HBB gene) along with ssODNs was introduced into human HSPCs collected from blood samples of patients with SCD (121). The results demonstrated that the use of CRISP/CAS9 resulted in efficient gene correction with reduced off-target effects and with optimum activation of HDR, compared with previous studies that used ZFN mRNA electroporation with ssODNs (13). The various diseases that have been treated using the CRISPR/Cas9 system are listed in Table I.
Table I.
Strategies using the CRISPR/Cas9 system for the treatment of diseases.
| Disease | In vitro analysis | In vivo analysis | Result | Conclusion | Refs. |
|---|---|---|---|---|---|
| AIDS | CRISPR/Cas9 transfected WT iPSCs to generate homozygous CCR5−/− CC | NOD/SCID/IL2Rγnull; NSG mice were engrafted with R5−/−42 generated from CD34+ HSPCs by ZFN HIV+ patients were engrafted with CCR5−/− CD4 T cells by ZFN |
i) Resistance to HIV-1 infection In vivo animal model: Suppression of HIV-1 Replication iii) In vivo human model: HIV-1 replication could not be controlled |
CRISPR appears more promising than ZFN due to long-term effect and ability to mutate both CCR5 alleles | 108-112, 114-116 |
| Neurodegenerative diseases | i) Exon 5 of PSEN2 gene in iPSCs derived from AD patients were corrected using CRISPR/Cas9 | ii) Healthy PSEN2N141 iPSC-derived BFCNs underwent CRISPR/Cas9 gene correction and were transplanted into AD mice | i) Reversal in elevated amyloid plaques (Aβ42/40) ii) Neurological development | Reduction in AD neuropathological symptoms. Further analysis required for in vivo human therapy | 70-74 |
| DMD | i) Exon 44 knock-in in DMD gene in iPSCs of DMD patients using TALENs and CRISPR ii) 45-55 exon removal of DMD gene in iPSC-derived cardiomyocytes |
iii) Targeting exon 23 in DMD gene in mdx mice | ii) Normal function of miR32 iii) Gene correction, however causing indels due to more NHEJ repair compared to HDR |
Larger size deletion by CRISPR corrected errors on factors affecting dystrophin function. However, due to off-target effects, further analysis to modify CRISPR/Cas9 is necessary to reduce off-target effects | 101-103 |
| Haemophilia | i) iPSCs of HB patient were transfected with CRISPR/Cas9 and differentiated into Hepatocytes ii) Modified HDR cassette containing coding sequence of WT at cFIX mutation locus and modified codon at TALEN/Cas9 binding site |
iii) AAV8 using CRISPR-SaCas9 in hepatocytes of HB mice to restore F9 gene creating DSB and inserting cDNA to intron iv) Injection of in vitro generated hepatocytes into NOD/SCID mice |
ii) Enhance HDR activation leading to decrease in off0target Repair iii) Genotypic correction and phenotypic improvement iv) Secretion of human FIX in mice |
Improvements in reduction of off-target effects caused by CRISPR, which is promising for further analysis of CRISPR for treatment in humans | 122-125 |
| ASD | i) RNP-induced Cas9 on NPCs of Ai9 tdTomato mouse | ii) AAV9 with MCO was injected into Mecp2 KO mice iii) RNP-induced Cas9 in NPCs of Ai9 tdTomato mouse from hippocampus, striatum and cortex iv) CRISPR-Gold with Cas9 or Cpf1 on Thy1- YFP and Ai9 mice |
ii) Behavioural development i and iii) Significant genome editing iv) Reduction in XFS |
High dose of Mecp2 in liver cells causing liver metabolism dysfunction However, CRISPR-Gold showed minimal off-target effects and no effects on immune system98 | 129-134 |
| SCD | Using pX330 plasmid with CRISPR/Cas9 that contains truncated sgRNA G10 to target first exon in HBB gene in human HSPCs from SCD patients | - | Highly efficient gene correction and reduction in mortality | CRISPR showed fewer off-target effects and better HDR function compared with genome editing by ZFN | 120-123 |
CRISPR, clustered regularly interspaced short palindromic repeats; Cas9, CRISPR-associated protein; AIDS, acquired immune deficiency syndrome; HIV, human immunodeficiency virus; DMD, Duchenne muscular dystrophy; ASD, autism spectrum disorder; SCD, sickle cell disease; WT, wild-type; iPSCs, induced pluripotent stem cells; CCR5, CXC chemokine receptor 5; NOD/SCID/IL2Rγnull, non-obese diabetic/severe combined immunodeficient/interleukin 2Rγnull (NSG); PSEN2, presenilin 2; AD, Alzheimer's disease; ZFN, zinc-finger nucleases; BFCNs, basal forebrain cholinergic neurons; TALENs, transcription activator-like effector nucleases; NHEJ, non-homologous end joining; HDR, homology-directed repair; FIX, coagulation factor IX; cFIX, canine FIX; AAV, adeno-associated virus; DSB, double-strand break; RNP, ribonucleoprotein; Mecp2, methyl CpG binding protein 2 gene; MCO, brain isoform of Mecp2; HBB, haemoglobin subunit β; HSPCs, hematopoietic stem/progenitor cells.
Haemophilia
CRISPR/Cas9 using AAV vectors has been assessed for the development of novel therapeutic methods to treat X-linked genetic diseases (122), such as haemophilia, a challenging disease with a high mortality rate, which is characterised by mutation on the coagulation factor IX (FIX). To restore the function of the F9 gene in patients with haemophilia, an AAV8 vector system carrying codon-optimized SaCas9 cDNA and sgRNA was transfected into hepatocytes in an in vivo model of mice with haemophilia B to create DSBs in the exon near the F9 gene (exon 2-8), and insert cDNA into an intron of the gene (123). The results revealed a genotypic and phenotypic correction of haemophilia B mice by targeting hepatocytes without disrupting epithelial cells of the liver morphologically or phenotypically (123). Additional studies extended the therapeutic design using other vectors with capacity for larger constructs. A low dose of adenovirus (Adv) containing Cas9 and specific sgRNA containing a correct donor template (dsDNA) was transfected into hepatocytes of haemophilia B mice, which resulted in F9 gene correction with improved efficacy compared with other vectors (14). However, lack of restoration in coagulation factor was an unfortunate outcome, as the Adv resulted in an adverse immune response due to the presence of vector genome immunogenicity, and an insignificant rate of HDR. Thus, it was hypothesized that recombinant Adv may be more suitable (14). However, integration of cDNA into the host genome can result in genotoxicity by either activating potential oncogenes or damaging functional genes (124). Therefore, using iPSCs compared with vector may be more suitable. In vitro and in vivo CRISPR studies using human iPSCs from patients with haemophilia B reported interesting results. iPSC cell lines prepared from peripheral blood mononuclear cells from patients with haemophilia B were modified by inserting the complete F9 human cDNA using CRISPR/Cas9; these cells differentiated into hepatocytes, and were subsequently injected into NOD/SCID mice. Analysis of the mice following transplantation revealed secretion of human FIX (125), a promising result that may serve as the basis for future studies.
The results of TALENs have shown promise in haemophilia B genome editing. In vitro analysis of canine FIX (cFIX) using both CRISPR and TALENs, showed that TALENs resulted in fewer off-target effects as they act as dimmers, and this may explain the lower numbers of DSBs compared with CRISPR (124). HDR cassettes were designed, which contained 471 bps of the WT coding sequence at the cFIX mutation locus and altered codons at the TALEN/Cas9 binding sites, which were cloned into pscAAV-cFIXWT to reduce off-target effects. Using modified HDR cassettes improved HDR efficiency, thus reducing the off-target effects of gene editing techniques (124).
Autism spectrum disorder (ASD)
ASD is a primarily inherited neurodevelopmental condition that is characterised by difficulty in social interactions, with language and communication abnormalities, which may be identified in children during early development. The symptoms have been found to be genetically associated with fragile X, maternal 15q11-13 duplication (83) and 2q37 and 22q13.3 deletion (126). AAVs, specifically AAV9, improves the treatment of Rett syndrome, as AAV9 can effectively penetrate into brain cells (127). To enhance the ability of the vectors for gene delivery, a self-complementary AAV9 vector, along with a codon-optimized version of the major methyl CpG binding protein 2 (Mecp2) gene (a mutation in the Mecp2 gene causing Rett syndrome has been observed in almost 1% of ASD patients) (128). The function of this gene involves transcription regulation by activating and repressing neuron function, and its brain isoform, referred to as MCO, was injected intravenously into Mecp2 knockout mice and was shown to improve behavioural development (22). However, an increased level of Mecp2 in liver cells, as a result of an off-target effect, was observed when a high dose was used for treatment (129), which disrupted liver metabolism and function (130). Different types of AAV vectors, such as AAV8, were used to transfect enhanced green fluorescent protein in astrocytes, which are cells abundantly present in the mouse striatum. The results demonstrated a high rate of protein transport due to the high penetrative ability of AAV8 in brain cells (131), which may be used to design methods to transfer target genes to astrocytes in patients with ASD.
The CRISPR/Cas9 genome editing system has been used in neural and brain cells to deliver specific genes to target cells. RNP induced Cas9 with Simian vacuolating virus 40 nuclear localisation sequence in Ai9 tdTomato mouse neural progenitor cells in the hippocampus, striatum and cortex, and this demonstrated marked gene editing ability in vitro and in vivo, indicating successful neuron-specific targeting for future ASD treatment (132). CRISPR/Cas9 has been used for Huntington's disease to suppress mutant HTT (mHTT) gene, which is located on chromosome 4 and produces Huntington's disease protein when mutated. In vivo HTT gene targeting in HD140Q-KI mice with mHTT using an AVV vector containing the CRISPR/Cas9 system with four gRNAs resulted in a significant reduction of mHTT expression and, thus, a reduction in the expression of Huntington's disease protein in HD140Q-KI mice (133). Therefore, it may be possible to design an efficient gene editing tool to treat ASD or minimise the severity of the symptoms.
One of the most successful studies on Thy1-YFP and Ai9 mice with X-fragile syndrome resulted in a significant reduction in repetitive symptoms of X-fragile syndrome (XFS) by using CRISPR-Gold (CRISPR designed with gold nanoparticles) to deliver Cas9 or Cpf1 to the striatum via local intracranial injection. CRISPR system side effects included minimal off-target effects and no impact on the immune system, unlike AAVs. Additionally, the metabotropic glutamate receptor subtype 5 gene was selected as an editing target (134,135), since its signalling was found to be overactivated in XFS and other ASD syndromes (136,137). The study reported minimal side effects, indicating successful Cas9 or Cpf1 delivery and gene editing. The same study used CRSPR-Gold effect for gene editing of other cell types, such as glial cells, dysfunction of which is observed in numerous neurological and brain disorders (137,138). Such accomplishments have provided novel insights into CRISPR genome editing technology, and may be used to treat other rare diseases.
5. Factors affecting the use of the CRISPR/Cas9 system
Although the abilities of CRISPR/Cas9 system are clearly established and have been used in various applications, there are concerns regarding off-target mutations, which may limit its future perspectives. Data from several studies indicate that the off-target effects of the CRISPR/Cas9 system are among the most important consequences of this method, regardless of the cell type and target genes (5,14,16,33,36,43). Hybrid R-loop formation between sgRNA and the target DNA may result in double-stranded cleavage of DNA due to RNA-guided nucleases, the recognition of PAM sequences and the presence of adjacent AAMs (139). Additionally, it was demonstrated that such activity results in an increased degree and a high volume of off-target effects by CRISPR/Cas9 during gene treatment, specifically due to dsDNA break and NHEJ function (140). Various techniques and protocols have been designed to optimise the low specificity of CRISPR/Cas9 and to promote HDR-based repair over NHEJ, in order to reduce the mutation rate. Exposure of mini circle-iPSCs to cold shock or low temperatures after treatment with CRISPR/Cas system resulted in increased HDR function and, thus, reduced off-target effects. However, the rate of indel formation was not significantly affected (15). Another study designed to reduce the off-target effects investigated changing the ratio of sgRNA to Cas9 protein, and demonstrated that a higher ratio of sgRNA to Cas9 resulted in reduced incidence of off-target effects (139).
Selection of bacteria for harvesting Cas9 markedly affects the performance of CRISPR/Cas9. For example, several studies investigated the impact of the CRISPR/Cas9 system using three different species of bacteria; Streptococcus pyogenes Cas9 (SpCas9), S. thermophilus Cas9 (St1Cas9) and SaCas9 (139,141). The analysis of human cells transfected with Cas9 plasmids from bacteria exhibited increased activity, as well as reduced mutation rates, compared with SpCas9 and SaCas9 (124). In addition to the findings mentioned above, the base sequence of the AAM upstream of PAM plays a key role in sgRNA binding with protospacers on the target DNA (142). sgRNAs with a higher ratio of guanine and a lower ratio of adenine are more stable in binding with target DNA compared with sgRNAs with a higher ratio of cytosine (143).
Other challenges include plasmids with low specificity and random integration into the target DNA, which creates tracking obstacles (139).
6. Discussion
Limitations of previous genome editing tools led scientists to develop the CRISPR system, which has reduced the undesired effects whilst increasing efficiency compared with previous methods. CRISPR was designed to reduce off-target effects caused by mutations as a result of DNA breaks, thus resulting in a reduction of unwanted errors. CpfI endonuclease was introduced into the CRISPR system (Class II) to overcome the challenges mentioned above. CfpI is a single RNA-guided endonuclease that does not require tracer RNA, and studies using Cpf1 of Francisella novicida bacterium showed inactivation of RuvC-like domain avoiding dsDNA cleavage (16). In addition to the findings mentioned above, Cpf1 creates 5' overhangs, which can efficiently add a DNA sequence during genome editing via a non-HDR system, in contrast to Cas9, which forms blunt end cuts on target DNA (16).
Furthermore, other studies reported fewer or no off-target effects using Cpf1 compared with Cas9 by analysing on-target activity of Acidaminococcus sp. BV3L6 Cpf1 and Lachnospiraceae bacterium ND2006 Cpf1 in human cells, compared with SpCas9 (144). The effect of R-loops and their stability on the occurrence rates of an off-target impact are other factors to be considered in designing the most efficient CRISPR system and potential target sequences in gene editing techniques (145). However, other studies reported that TALENs was more efficient compared with the CRISPR system (29,30,32,33).
CRPSR has helped overcome the challenges of disease therapies by locating target genes that are the causes of drug resistance (88). Techniques, such as 2CT-CRISPR and dCas9, have increased the efficacy of drug therapy. Several factors, such as vector/plasmid and CRISPR selection based on size, have exerted a notable effect on the efficacy and delivery of CRISPR endonucleases to target genes (37,45). Therefore, selecting the most suitable vector with good penetration and a low rate of host immune activation for CRISPR delivery is required to carefully address the treatment of various diseases, such as AIDS, haemophilia, ASD and SCD.
Scientists at the University of Washington used vectors with CRISPR/Cas9 components from either Streptococcus pyogenes or Staphylococcus aureus to treat DMD (146). The results demonstrated that the expression of dystrophin using dual vectors exhibited increased efficiency compared with a single vector. Furthermore, they achieved a higher yield with SpCas9 compared with SaCas9, as demonstrated by the reduction in the off-target effects in both DMD and SCD (146,147). In addition, certain factors, such as size and PAM sequence recognition, further improve the efficiency of SpCas9 compared with SaCas9 for CRISPR/Cas9 therapy (147). The impact of the CRISPR/Cas9 system on other diseases has been shown to involve off-target effects and, eventually, the formation of indels due to the involvement of NHEJ-based repair, which is associated with an increased risk of mutations (148).
7. Conclusion
The effects of CRISPR/Cas9 on humans requires further investigation, as human iPSCs and mouse iPSCs vary in response based on the specific type of CRISPR system used. Furthermore, the issue of indels as a result of NHEJ-based repair must be reduced in order to reduce the off-target effects caused by indels. Therefore, studies are focusing on designing a CRISPR/Cas system for gene editing with a lower risk of mutations utilizing HDR.
Numerous studies have demonstrated the use of CRISPR, ZFNs and TALENS as powerful gene editing tools. Although ZFNs and TALENS represent important advances in gene editing, their capacity is currently limited for effective use. The discovery of CRISPR, which exhibits higher efficiency and fewer off-target effects, has provided opportunities for scientists to use this technique widely and develop CRISPR-based gene therapy. However, the issue of off-target effects must be addressed and, thus, should be the focus of future studies, with the aim of further developing this technology for use in human gene therapy.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
MA drafted the paper and designed the manuscript structure. NK drafted the manuscript and arranged the data of the studies. Both authors have read and approved the final version of the manuscript for publication.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
All the authors declare that they have no competing interests.
References
- 1.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–5433. doi: 10.1128/JB.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 3.Karginov FV, Hannon GJ. The CRISPR system: Small RNA-guided defense in bacteria and archaea. Mol Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrangou R, Marraffini LA. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 7.Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Hüttenhofer A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci USA. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst) 2005;4:639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Davis AJ, Chen DJ. DNA double strand break repair via non-homologous end-joining. Transl Cancer Res. 2013;2:130–143. doi: 10.3978/j.issn.2218-676X.2013.04.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bibikova M, Golic M, Golic KG, Carroll GD. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoban MD, Cost GJ, Mendel MC, Romero Z, Kaufman ML, Joglekar AV, Ho M, Lumaquin D, Gray D, Lill GR, et al. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood. 2015;125:2597–2604. doi: 10.1182/blood-2014-12-615948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan Y, Ma Y, Li Q, Sun Z, Ma L, Wu L, Wang L, Zeng L, Shao Y, Chen Y, et al. CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse. EMBO Mol Med. 2016;8:477–488. doi: 10.15252/emmm.201506039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Q, Mintier G, Ma-Edmonds M, Storton D, Wang X, Xiao X, Kienzle B, Zhao D, Feder JN. 'Cold shock' increases the frequency of homology directed repair gene editing in induced pluripotent stem cells. Sci Rep. 2018;8:2080. doi: 10.1038/s41598-018-20358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: Crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 19.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 20.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid 'open-source' engineering of custom-ized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maeder ML, Thibodeau-Beganny S, Sander JD, Voytas DF, Joung JK. Oligomerized pool engineering (OPEN): An 'open-source' protocol for making customized zinc-finger arrays. Nat Protoc. 2009;4:1471–1501. doi: 10.1038/nprot.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, Nowrouzi A, Bartholomae CC, Wang J, Friedman G, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 24.Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 25.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 26.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 27.Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: Finding plant genes for disease and defense. Curr Opin Plant Biol. 2010;13:394–401. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 28.Scholze H, Boch J. TAL effectors are remote controls for gene activation. Curr Opin Microbiol. 2011;14:47–53. doi: 10.1016/j.mib.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease archi-tecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 31.Zhu H, Lau CH, Goh SL, Liang Q, Chen C, Du S, Phang RZ, Tay FC, Tan WK, Li Z, et al. Baculoviral transduction facilitates TALEN-mediated targeted transgene integration and Cre/LoxP cassette exchange in human-induced pluripotent stem cells. Nucleic Acids Res. 2013;41:e180. doi: 10.1093/nar/gkt721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mussolino C, Alzubi J, Fine EJ, Morbitzer R, Cradick TJ, Lahaye T, Bao G, Cathomen T. TALENs facilitate targeted genome editing in human cells with high specificity and low cytotoxicity. Nucleic Acids Res. 2014;42:6762–6773. doi: 10.1093/nar/gku305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta RM, Musunuru K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J Clin Invest. 2014;124:4154–4161. doi: 10.1172/JCI72992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonu-clease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, Lv J, Xie X, Chen Y, Li Y, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6:363–372. doi: 10.1007/s13238-015-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shabbir MA, Hao H, Shabbir MZ, Hussain HI, Iqbal Z, Ahmed S, Sattar A, Iqbal M, Li J, Yuan Z. Survival and evolution of CRISPR-Cas system in prokaryotes and its applications. Front Immunol. 2016;7:375. doi: 10.3389/fimmu.2016.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR/Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, et al. An updated evolutionary classification of CRISPR/Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, Horvath P, Siksnys V. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westra ER, van Erp PB, Künne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V, Seidel R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci USA. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA. 2011;108:10098–11103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hochstrasser ML, Taylor DW, Bhat P, Guegler CK, Sternberg SH, Nogales E, Doudna JA. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci USA. 2014;111:6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR/Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen F, Ding X, Feng Y, Seebeck T, Jiang Y, Davis GD. Targeted activation of diverse CRISPR-Cas systems for mammalian genome editing via proximal CRISPR targeting. Nat Commun. 2017;8:14958. doi: 10.1038/ncomms14958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hatoum-Aslan A, Maniv I, Marraffini LA. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc Natl Acad Sci USA. 2011;108:21218–21222. doi: 10.1073/pnas.1112832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng W, Feng M, Feng X, Liang YX, She Q. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 2015;43:406–417. doi: 10.1093/nar/gku1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, Marraffini LA. Co-transcriptional DNA and RNA Cleavage during Type III CRISPR/Cas Immunity. Cell. 2015;161:1164–1174. doi: 10.1016/j.cell.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Staals RH, Zhu Y, Taylor DW, Kornfeld JE, Sharma K, Barendregt A, Koehorst JJ, Vlot M, Neupane N, Varossieau K, et al. RNA targeting by the type III-A CRISPR/Cas Csm complex of thermus thermophilus. Mol Cell. 2014;56:518–530. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamulaitis G, Kazlauskiene M, Manakova E, Venclovas Č, Nwokeoji AO, Dickman MJ, Horvath P, Siksnys V. Programmable RNA shredding by the type III-A CRISPR-Cas system of streptococcus thermophilus. Mol Cell. 2014;56:506–517. doi: 10.1016/j.molcel.2014.09.027. [DOI] [PubMed] [Google Scholar]
- 61.Cicero CE, Mostile G, Vasta R, Rapisarda V, Signorelli SS, Ferrante M, Zappia M, Nicoletti A. Metals and neurodegenerative diseases. A systematic review. Environ Res. 2017;159:82–94. doi: 10.1016/j.envres.2017.07.048. [DOI] [PubMed] [Google Scholar]
- 62.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: The AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 63.Armstrong RA. What causes Alzheimer's disease? Folia Neuropathol. 2013;51:169–188. doi: 10.5114/fn.2013.37702. [DOI] [PubMed] [Google Scholar]
- 64.Heidenreich M, Zhang F. Applications of CRISPR/Cas systems in neuroscience. Nat Rev Neurosci. 2016;17:36–44. doi: 10.1038/nrn.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu CE, Jondro PD, Schmidt SD, Wang K, et al. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 66.Tomita T, Maruyama K, Saido TC, Kume H, Shinozaki K, Tokuhiro S, Capell A, Walter J, Grünberg J, Haass C, et al. The presenilin 2 mutation (N141I) linked to familial Alzheimer disease (Volga German families) increases the secretion of amyloid beta protein ending at the 42nd (or 43rd) residue. Proc Natl Acad Sci USA. 1997;94:2025–2030. doi: 10.1073/pnas.94.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sachse CC, Kim YH, Agsten M, Huth T, Alzheimer C, Kovacs DM, Kim DY. BACE1 and presenilin/γ-secretase regulate proteolytic processing of KCNE1 and 2, auxiliary subunits of voltage-gated potassium channels. FASEB J. 2013;27:2458–2467. doi: 10.1096/fj.12-214056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stutzmann GE, Caccamo A, LaFerla FM, Parker I. Dysregulated IP3 signaling in cortical neurons of knock-in mice expressing an Alzheimer's-linked mutation in presenilin1 results in exaggerated Ca2+ signals and altered membrane excitability. J Neurosci. 2004;24:508–513. doi: 10.1523/JNEUROSCI.4386-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yue W, Li Y, Zhang T, Jiang M, Qian Y, Zhang M, Sheng N, Feng S, Tang K, Yu X, et al. ESC-derived basal forebrain cholinergic neurons ameliorate the cognitive symptoms associated with Alzheimer's disease in mouse models. Stem Cell Reports. 2015;5:776–790. doi: 10.1016/j.stemcr.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paull D, Sevilla A, Zhou H, Hahn AK, Kim H, Napolitano C, Tsankov A, Shang L, Krumholz K, Jagadeesan P, et al. Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells. Nat Methods. 2015;12:885–892. doi: 10.1038/nmeth.3507. [DOI] [PubMed] [Google Scholar]
- 71.Ortiz-Virumbrales M, Moreno CL, Kruglikov I, Marazuela P, Sproul A, Jacob S, Zimmer M, Paull D, Zhang B, Schadt EE, et al. CRISPR/Cas9-Correctable mutation-related molecular and phys-iological phenotypes in iPSC-derived Alzheimer's PSEN2N141I neurons. Acta Neuropathol Commun. 2017;5:77. doi: 10.1186/s40478-017-0475-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu C, Zhang L, Liu H, Cheng K. Delivery strategies of the CRISPR/Cas9 gene-editing system for therapeutic applications. J Control Release. 2017;266:17–26. doi: 10.1016/j.jconrel.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.György B, Lööv C, Zaborowski MP, Takeda S, Kleinstiver BP, Commins C, Kastanenka K, Mu D, Volak A, Giedraitis V, et al. CRISPR/Cas9 mediated disruption of the swedish app allele as a therapeutic approach for early-onset Alzheimer's disease. Mol Ther Nucleic Acids. 2018;11:429–440. doi: 10.1016/j.omtn.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mirzaei HR, Sahebkar A, Salehi R, Nahand JS, Karimi E, Jaafari MR, Mirzaei H, Sahebkar A, Salehi R. Boron neutron capture therapy: Moving toward targeted cancer therapy. J Cancer Res Ther. 2016;12:520–525. doi: 10.4103/0973-1482.176167. [DOI] [PubMed] [Google Scholar]
- 75.Pourhanifeh MH, Mohammadi R, Noruzi S, Hosseini SA, Fanoudi S, Mohamadi Y, Hashemzehi M, Asemi Z, Mirzaei HR, Salarinia R, Mirzaei H. The role of fibromodulin in cancer pathogenesis: Implications for diagnosis and therapy. Cancer Cell Int. 2019;19:157. doi: 10.1186/s12935-019-0870-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shafabakhsh R, Pourhanifeh MH, Mirzaei HR, Sahebkar A, Asemi Z, Mirzaei H. Targeting regulatory T cells by curcumin: A potential for cancer immunotherapy. Pharmacol Res. 2019;147:104353. doi: 10.1016/j.phrs.2019.104353. [DOI] [PubMed] [Google Scholar]
- 77.Scott A. How CRISPR is transforming drug discovery. Nature. 2018;555:S10–S11. doi: 10.1038/d41586-018-02477-1. [DOI] [PubMed] [Google Scholar]
- 78.Walton J, Blagih J, Ennis D, Leung E, Dowson S, Farquharson M, Tookman LA, Orange C, Athineos D, Mason S, et al. CRISPR/Cas9-mediated Trp53 and Brca2 knockout to generate improved murine models of ovarian high-grade serous carcinoma. Cancer Res. 2016;76:6118–6129. doi: 10.1158/0008-5472.CAN-16-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cruz C, Castroviejo-Bermejo M, Gutiérrez-Enríquez S, Llop-Guevara A, Ibrahim YH, Gris-Oliver A, Bonache S, Morancho B, Bruna A, Rueda OM, et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann Oncol. 2018;29:1203–1210. doi: 10.1093/annonc/mdy099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klann TS, Black JB, Chellappan M, Safi A, Song L, Hilton IB, Crawford GE, Reddy TE, Gersbach CA. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat Biotechnol. 2017;35:561–568. doi: 10.1038/nbt.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brabetz O, Alla V, Angenendt L, Schliemann C, Berdel WE, Arteaga MF, Mikesch JH. RNA-guided CRISPR/Cas9 system-mediated engineering of acute myeloid leukemia mutations. Mol Ther Nucleic Acids. 2017;6:243–248. doi: 10.1016/j.omtn.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu J, Zwaigenbaum L, Szatmari P, Scherer SW. Molecular cytogenetics of autism. Curr Genomics. 2004;5:347–364. doi: 10.2174/1389202043349246. [DOI] [Google Scholar]
- 84.Peña-Martínez P, Eriksson M, Ramakrishnan R, Chapellier M, Högberg C, Orsmark-Pietras C, Richter J, Andersson A, Fioretos T, Järås M. Interleukin 4 induces apoptosis of acute myeloid leukemia cells in a Stat6-dependent manner. Leukemia. 2018;32:588–596. doi: 10.1038/leu.2017.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, Gartner JJ, Jia L, Steinberg SM, Yamamoto TN, et al. Identification of essential genes for cancer immunotherapy. Nature. 2017;548:537–542. doi: 10.1038/nature23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mirzaei H, Yazdi F, Salehi R, Mirzaei HR. SiRNA and epigenetic aberrations in ovarian cancer. J Canc Res Ther. 2016;12:498–508. doi: 10.4103/0973-1482.153661. [DOI] [PubMed] [Google Scholar]
- 87.Mirzaei HR, Pourghadamyari H, Rahmati M, Mohammadi A, Nahand JS, Rezaei A, Mirzaei H, Hadjati J. Gene-knocked out chimeric antigen receptor (CAR) T cells: Tuning up for the next generation cancer immunotherapy. Cancer Lett. 2018;423:95–104. doi: 10.1016/j.canlet.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 88.Dai WJ, Zhu LY, Yan ZY, Xu Y, Wang QL, Lu XJ. CRISPR-Cas9 for in vivo gene therapy: Promise and hurdles. Mol Ther Nucleic Acids. 2016;5:e349. doi: 10.1038/mtna.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mirzaei HR, Jamali A, Jafarzadeh L, Masoumi E, Alishah K, Fallah Mehrjardi K, Emami SAH, Noorbakhsh F, Till BG, Hadjati J. Construction and functional characterization of a fully human anti-CD19 chimeric antigen receptor (huCAR)-expressing primary human T cells. J Cell Physiol. 2019;234:9207–9215. doi: 10.1002/jcp.27599. [DOI] [PubMed] [Google Scholar]
- 90.Gomes-Silva D, Mukherjee M, Srinivasan M, Krenciute G, Dakhova O, Zheng Y, Cabral JMS, Rooney CM, Orange JS, Brenner MK, Mamonkin M. Tonic 4-1BB costimulation in chimeric antigen receptors impedes T Cell survival and is vector-dependent. Cell Rep. 2017;21:17–26. doi: 10.1016/j.celrep.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mirzaei HR, Mirzaei H, Lee SY, Hadjati J, Till BG. Prospects for chimeric antigen receptor (CAR) γδ T cells: A potential game changer for adoptive T cell cancer immunotherapy. Cancer Lett. 2016;380:413–423. doi: 10.1016/j.canlet.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mirzaei HR, Mirzaei H, Namdar A, Rahmati M, Till BG, Hadjati J. Predictive and therapeutic biomarkers in chimeric antigen receptor T-cell therapy: A clinical perspective. J Cell Physiol. 2019;234:5827–5841. doi: 10.1002/jcp.27519. [DOI] [PubMed] [Google Scholar]
- 93.Mirzaei HR, Rodriguez A, Shepphird J, Brown CE, Badie B. Corrigendum: Chimeric antigen receptors T cell therapy in solid tumor: Challenges and clinical applications. Front Immunol. 2019;10:780. doi: 10.3389/fimmu.2019.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mah JK. Current and emerging treatment strategies for Duchenne muscular dystrophy. Neuropsychiatr Dis Treat. 2016;12:1795–1807. doi: 10.2147/NDT.S93873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee SH, Lee JH, Lee KA, Choi YC. Clinical and genetic characterization of female dystrophinopathy. J Clin Neurol. 2015;11:248–251. doi: 10.3988/jcn.2015.11.3.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bladen CL, Salgado D, Monges S, Foncuberta ME, Kekou K, Kosma K, Dawkins H, Lamont L, Roy AJ, Chamova T, et al. The TREAT-NMD DMD Global Database: Analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum Mutat. 2015;36:395–402. doi: 10.1002/humu.22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mendell JR, Kissel J, Amato AA, King W, Signore L, Prior TW, Sahenk Z, Benson S, McAndrew PE, Rice R, et al. Myoblast transfer in the treatment of Duchenne's muscular dystrophy. N Engl J Med. 1995;333:832–838. doi: 10.1056/NEJM199509283331303. [DOI] [PubMed] [Google Scholar]
- 98.Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, Partridge T, Buckingham M. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- 99.Darabi R, Arpke RW, Irion S, Dimos JT, Grskovic M, Kyba M, Perlingeiro RC. Human ES- and iPS-derived myogenic progenitors restore DYSTROPHIN and improve contractility upon transplantation in dystrophic mice. Cell Stem Cell. 2012;10:610–619. doi: 10.1016/j.stem.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shimizu-Motohashi Y, Miyatake S, Komaki H, Takeda S, Aoki Y. Recent advances in innovative therapeutic approaches for Duchenne muscular dystrophy: From discovery to clinical trials. Am J Transl Res. 2016;8:2471–2489. [PMC free article] [PubMed] [Google Scholar]
- 101.Li HL, Fujimoto N, Sasakawa N, Shirai S, Ohkame T, Sakuma T, Tanaka M, Amano N, Watanabe A, Sakurai H, et al. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015;4:143–154. doi: 10.1016/j.stemcr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Young CS, Hicks MR, Ermolova NV, Nakano H, Jan M, Younesi S, Karumbayaram S, Kumagai-Cresse C, Wang D, Zack JA, et al. A single CRISPR/Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell Stem Cell. 2016;18:533–540. doi: 10.1016/j.stem.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cacchiarelli D, Incitti T, Martone J, Cesana M, Cazzella V, Santini T, Sthandier O, Bozzoni I. miR-31 modulates dystro-phin expression: New implications for Duchenne muscular dystrophy therapy. EMBO Rep. 2011;12:136–114. doi: 10.1038/embor.2010.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marmor M, Sheppard HW, Donnell D, Bozeman S, Celum C, Buchbinder S, Koblin B. Homozygous and heterozygous CCR5-Delta32 genotypes are associated with resistance to HIV infection. J Acquir Immune Defic Syndr. 2001;27:472–481. doi: 10.1097/00042560-200108150-00009. [DOI] [PubMed] [Google Scholar]
- 105.Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, MacDonald ME, Stuhlmann H, Koup RA, Landau NR. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/S0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 106.Hütter G, Nowak D, Mossner M, Ganepola S, Müssig A, Allers K, Schneider T, Hofmann J, Kücherer C, Blau O, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 107.Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.DiGiusto DL, Cannon PM, Holmes MC, Li L, Rao A, Wang J, Lee G, Gregory PD, Kim KA, Hayward SB, et al. Preclinical development and qualification of ZFN-mediated CCR5 disruption in human hematopoietic stem/progenitor cells. Mol Ther Methods Clin Dev. 2016;3:16067. doi: 10.1038/mtm.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li L, Krymskaya L, Wang J, Henley J, Rao A, Cao LF, Tran C A, Torres-Coronado M, Gardner A, Gonzalez N, et al. Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther. 2013;21:1259–1269. doi: 10.1038/mt.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, Cannon PM. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kumar P, Ban HS, Kim SS, Wu H, Pearson T, Greiner DL, Laouar A, Yao J, Haridas V, Habiro K, et al. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ishikawa F, Yasukawa M, Lyons B, Yoshida S, Miyamoto T, Yoshimoto G, Watanabe T, Akashi K, Shultz LD, Harada M. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor {gamma} chain(null) mice. Blood. 2005;106:1565–1573. doi: 10.1182/blood-2005-02-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, Down J, Denaro M, Brady T, Westerman K, et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ye L, Wang J, Beyer AI, Teque F, Cradick TJ, Qi Z, Chang JC, Bao G, Muench MO, Yu J, et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc Natl Acad Sci USA. 2014;111:9591–9596. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR/Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hütter G. Stem cell transplantation in strategies for curing HIV/AIDS. AIDS Res Ther. 2016;13:31. doi: 10.1186/s12981-016-0114-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kaminski R, Chen Y, Fischer T, Tedaldi E, Napoli A, Zhang Y, Karn J, Hu W, Khalili K. Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Sci Rep. 2016;6:22555. doi: 10.1038/srep22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huang Z, Nair M. A CRISPR/Cas9 guidance RNA screen platform for HIV provirus disruption and HIV/AIDS gene therapy in astrocytes. Sci Rep. 2017;7:5955. doi: 10.1038/s41598-017-06269-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lervolino LG, Baldin PE, Picado SM, Calil KB, Viel AA, Campos LA. Prevalence of sickle cell disease and sickle cell trait in national neonatal screening studies. Rev Bras Hematol Hemoter. 2011;33:49–54. doi: 10.5581/1516-8484.20110015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.DeWitt MA, Magis W, Bray NL, Wang T, Berman JR, Urbinati F, Heo SJ, Mitros T, Muñoz DP, Boffelli D, et al. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci Transl Med. 2016;8:360ra134. doi: 10.1126/scitranslmed.aaf9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stephens CJ, Kashentseva E, Everett W, Kaliberova L, Curiel DT. Targeted in vivo knock-in of human alpha-1-anti-trypsin cDNA using adenoviral delivery of CRISPR/Cas9. Gene Ther. 2018;25:139–156. doi: 10.1038/s41434-018-0003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ohmori T, Nagao Y, Mizukami H, Sakata A, Muramatsu SI, Ozawa K, Tominaga SI, Hanazono Y, Nishimura S, Nureki O, Sakata Y. CRISPR/Cas9-mediated genome editing via post-natal administration of AAV vector cures haemophilia B mice. Sci Rep. 2017;7:4159. doi: 10.1038/s41598-017-04625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bergmann T, Ehrke-Schulz E, Gao J, Schiwon M, Schildgen V, David S, Schildgen O, Ehrhardt A. Designer nuclease-medi-ated gene correction via homology-directed repair in an in vitro model of canine hemophilia. B J Gene Med. 2018;20:e3020. doi: 10.1002/jgm.3020. [DOI] [PubMed] [Google Scholar]
- 125.Lyu C, Shen J, Wang R, Gu H, Zhang J, Xue F, Liu X, Liu W, Fu R, Zhang L, et al. Targeted genome engineering in human induced pluripotent stem cells from patients with hemophilia B using the CRISPR-Cas9 system. Stem Cell Res Ther. 2018;9:92. doi: 10.1186/s13287-018-0839-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jacquemont ML, Sanlaville D, Redon R, Raoul O, Cormier-Daire V, Lyonnet S, Amiel J, Le Merrer M, Heron D, de Blois MC, et al. Array-based comparative genomic hybridisation identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J Med Genet. 2006;43:843–849. doi: 10.1136/jmg.2006.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gray SJ, Matagne V, Bachaboina L, Yadav S, Ojeda SR, Samulski RJ. Preclinical differences of intravascular AAV9 delivery to neurons and glia: A comparative study of adult mice and nonhuman primates. Mol Ther. 2011;19:1058–1069. doi: 10.1038/mt.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 129.Matagne V, Ehinger Y, Saidi L, Borges-Correia A, Barkats M, Bartoli M, Villard L, Roux JC. A codon-optimized Mecp2 transgene corrects breathing deficits and improves survival in a mouse model of Rett syndrome. Neurobiol Dis. 2017;99:1–11. doi: 10.1016/j.nbd.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 130.Kyle SM, Saha PK, Brown HM, Chan LC, Justice MJ. MeCP2 co-ordinates liver lipid metabolism with the NCoR1/HDAC3 corepressor complex. Hum Mol Genet. 2016;25:3029–3041. doi: 10.1093/hmg/ddw156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pignataro D, Sucunza D, Vanrell L, Lopez-Franco E, Dopeso-Reyes IG, Vales A, Hommel M, Rico AJ, Lanciego JL, Gonzalez-Aseguinolaza G. Adeno-associated viral vectors serotype 8 for cell-specific delivery of therapeutic genes in the central nervous system. Front Neuroanat. 2017;11:12. doi: 10.3389/fnana.2017.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Staahl BT, Benekareddy M, Coulon-Bainier C, Banfal AA, Floor SN, Sabo JK, Urnes C, Munares GA, Ghosh A, Doudna JA. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat Biotechnol. 2017;35:431–434. doi: 10.1038/nbt.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yang S, Chang R, Yang H, Zhao T, Hong Y, Kong HE, Sun X, Qin Z, Jin P, Li S, Li XJ. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington's disease. J Clin Invest. 2017;127:2719–2724. doi: 10.1172/JCI92087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lee B, Lee K, Panda S, Gonzales-Rojas R, Chong A, Bugay V, Park HM, Brenner R, Murthy N, Lee HY. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat Biomed Eng. 2018;2:497–507. doi: 10.1038/s41551-018-0252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Silverman JL, Smith DG, Rizzo SJ, Karras MN, Turner SM, Tolu SS, Bryce DK, Smith DL, Fonseca K, Ring RH, Crawley JN. Negative allosteric modulation of the mGluR5 receptor reduces repetitive behaviors and rescues social deficits in mouse models of autism. Sci Transl Med. 2012;4:131ra51. doi: 10.1126/scitranslmed.3003501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tao J, Wu H, Coronado AA, de Laittre E, Osterweil EK, Zhang Y, Bear MF. Negative allosteric modulation of mGluR5 partially corrects pathophysiology in a mouse model of Rett syndrome. J Neurosci. 2016;36:11946–11958. doi: 10.1523/JNEUROSCI.0672-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Almad AA, Maragakis NJ. Glia: An emerging target for neurological disease therapy. Stem Cell Res Ter. 2012;3:37. doi: 10.1186/scrt128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Alagoz M, Kherad N, Gavaz M, Yuksel A. New genetic approaches for early diagnosis and treatment of autism spectrum disorders. Rev J Autism Dev Disoed. 2019;6:367–380. doi: 10.1007/s40489-019-00167-w. [DOI] [Google Scholar]
- 139.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Klein M, Eslami-Mossallam B, Arroyo DG, Depken M. Hybridization kinetics explains CRISPR/Cas Off-targeting rules. Cell Rep. 2018;22:1413–1423. doi: 10.1016/j.celrep.2018.01.045. [DOI] [PubMed] [Google Scholar]
- 141.Chari R, Mali P, Moosburner M, Church GM. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods. 2015;12:823–826. doi: 10.1038/nmeth.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Matsoukas IG. Commentary: CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria. Front Bioeng Biotechnol. 2017;5:57. doi: 10.3389/fbioe.2017.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ. CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, et al. Engineered CRISPR/Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rutkauskas M, Sinkunas T, Songailiene I, Tikhomirova MS, Siksnys V, Seidel R. Directional R-Loop formation by the CRISPR-Cas surveillance complex cascade provides efficient Off-target site rejection. Cell Rep. 2015;10:1534–1543. doi: 10.1016/j.celrep.2015.01.067. [DOI] [PubMed] [Google Scholar]
- 146.Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD, Hauschka SD, Chamberlain JR, Chamberlain JS. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun. 2017;8:14454. doi: 10.1038/ncomms14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Morsy SG, Tonne JM, Zhu Y, Lu B, Budzik K, Krempski JW, Ali SA, El-Feky MA, Ikeda Y. Divergent susceptibilities to AAV-SaCas9-gRNA vector-mediated genome-editing in a single-cell-derived cell population. BMC Res Notes. 2017;10:720. doi: 10.1186/s13104-017-3028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Divergent susceptibilities to AAV-SaCas9-gRNA vector-mediated genome-editing in a single-cell-derived cell population. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.