Abstract
Accumulating evidence demonstrates that microRNAs (miRNAs or miRs) play important roles in the development and progression of human malignancies, including oral squamous cell carcinoma (OSCC); however, the unique roles of miRNAs are not yet fully understood in OSCC. The present study aimed to identify novel miRNAs associated with OSCC and to elucidate their functions. Based on a micro-array analysis, miR-144-3p was found to be one of the most significantly downregulated miRNAs in OSCC tissues. Its low expression was closely associated with tumor size, differentiation and lymph node metastasis. Functionally, miR-144-3p overexpression suppressed proliferation, promoted apoptosis, and suppressed the invasion and migration of OSCC cells. In addition, enhancer of zeste homolog 2 (EZH2), a well-known oncogene, was proven to be a direct target of miR-144-3p, and its protein expression was negatively regulated by miR-144-3p. Moreover, EZH2 expression was increased, and inversely correlated with the miR-144-3p level in OSCC tissues. Notably, EZH2 knockdown inhibited cell proliferation, promoted cell apoptosis, and suppressed the invasion and migration of OSCC cells, whereas EZH2 overexpression partially reversed the anticancer effects mediated by miR-144-3p overexpression. On the whole, the findings of the present study suggest that miR-144-3p functions as a tumor suppressor by targeting the EZH2 oncogene, and may thus be considered as a potential diagnostic and therapeutic target for OSCC.
Keywords: oral squamous cell carcinoma, miR-144-3p, enhancer of zeste homolog 2
Introduction
Oral squamous cell carcinoma (OSCC) is the sixth most prevalent type of human cancer, and approximately 275,000 cases are newly diagnosed, annually (1). Despite tremendous efforts being made in treatment modalities, the overall 5-year survival rates (<50%) remain poor due to the high recurrence and metastasis rate following surgery (2,3). Thus, there is an urgent need for the identification of key molecules which are responsible for and are involved in the progression of OSCC.
MicroRNAs (miRNAs or miRs) are short, non-coding RNAs that can bind to the 3'-untranslated region (3'-UTR) of target mRNAs to regulate gene expression, leading to the degradation of target mRNAs or translational inhibition of functional proteins (4). Emerging evidence indicates that miRNAs are extensively involved in the development of OSCC, functioning as oncogenes or tumor suppressors, depending on the target genes (5-7). For instance, Chou et al demonstrated that miR-486 overexpression led to growth inhibition and apoptosis induction by targeting discoidin domain receptor-1 (DDR1) in oral cancer cells (8). Another study revealed that miR-10a promoted tumor cell proliferation by regulating the glucose transporter 1 (GLUT1) oncogene in OSCC (9). Peng and Pang found that miR-140-5p overexpression suppressed the growth of OSCC tumor xenografts in mice by downregulating p21-activated kinase 4 (PAK4) (10). In addition, recent studies have reported that changes in miRNA profiles in cancer cells have the potential to serve as diagnostic markers for OSCC (11,12). These previous findings suggest that the manipulation of miRNAs may serve as a novel therapeutic approach for OSCC. However, to date, only a limited number of studies on the roles of miRNAs in OSCC have been conducted, at least to the best of our knowledge, and thus further extensive investigations are required.
In the present study, miRNA profiles were examined in tumor tissues from patients with OSCC using a microarray and miR-144-3p was found to be one of the most significantly downregulated miRNAs. Subsequently, gain-of-function experiments were performed to determine the roles of miR-144-3p. The findings suggest that miR-144-3p functions as a tumor suppressor by directly targeting enhancer of zeste homolog 2 (EZH2), and may thus be a novel target for the diagnosis and treatment of OSCC.
Materials and methods
Clinical specimens
The OSCC tissues and adjacent non-cancerous were collected from 50 patients with OSCC between May, 2017 and July, 2018 at the Clinical Research Center of Shaanxi Province for Dental and Maxillofacial Diseases, College of Stomatology, Xi'an Jiaotong University. These patients included 26 males and 24 females, and their age ranged from 22 to 72 years, with an average age of 44±6.9 years. The inclusion criteria were histologically established squamous cell carcinoma within the oral cavity that had primary surgical treatment with curative intent. Patients treated for OSCC prior to 2017, and those with recurrent tumors and distant metastasis at the time of diagnosis were excluded from the study. All patient characteristics are presented in Table I. The experimental protocol was approved by the Ethics Committee of the Xi'an Jiaotong University. Written informed consents for tissue donation were obtained from each patient for the research only.
Table I.
Association between miR-144-3p and clinicopathological features of patients with oral squamous cell carcinoma (OSCC).
| Clinical parameters | All cases (n=50) | miR-144-3p expression
|
P-value | |
|---|---|---|---|---|
| High (n=21) | Low (n=29) | |||
| Sex | 0.5356 | |||
| Male | 26 | 12 | 14 | |
| Female | 24 | 9 | 15 | |
| Age (years) | 0.1973 | |||
| ≥0 | 40 | 15 | 25 | |
| <50 | 10 | 6 | 4 | |
| Site | 0.3399 | |||
| Buccal mucosa | 27 | 13 | 14 | |
| Non-buccal mucosa | 23 | 8 | 15 | |
| Alcohol consumption | 0.7381 | |||
| Yes | 32 | 14 | 18 | |
| No | 18 | 7 | 11 | |
| Smoking habits | 0.1452 | |||
| Yes | 31 | 11 | 21 | |
| No | 19 | 10 | 8 | |
| Tumor size (cm) | 0.0025b | |||
| ≥2 | 22 | 4 | 18 | |
| <2 | 28 | 17 | 11 | |
| Differentiation | 0.021a | |||
| Well and moderate | 15 | 10 | 5 | |
| sPoor | 35 | 11 | 24 | |
| Lymph node metastasis | 0.044a | |||
| Present | 34 | 11 | 23 | |
| Absent | 16 | 10 | 6 | |
| cTNM stage | 0.2907 | |||
| I + II | 21 | 7 | 14 | |
| III + IV | 29 | 14 | 15 | |
P<0.05,
P<0.01.
miRNA microarray analysis
Total RNA was extracted from the OSCC tissues using the miRNeasy mini kit (Qiagen, Inc.). The samples were assessed using the miRCURY™ LNA Array (v.16.0). The procedure and imaging processes were as previously described (13).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the OSCC tissues and cells using TRIzol Reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The RNA was reverse transcribed into cDNA using the PrimeScript RT reagent kit (Takara Bio, Inc.) and the MicroRNA Reverse Transcription kit (Thermo Fisher Scientific, Inc.), respectively. miR-144a-3p and EZH2 expression levels were measured using a SYBR® PrimeScript™ RT-PCR kit (Takara Bio, Inc.) on a Light Cycler instrument (Bio-Rad Laboratories, Inc.). The primers of miR-144a-3p and EZH2 were as follows: miR-144-3p forward, 5′-GCC CCT ACA GTA TAG ATG ATG TA-3′ and reverse, 5′-GTG CAG GGT CCG AGG T-3′; U6 forward, 5′-GCT TCG GCA GCA CAT ATA CTA AAA T-3′ and reverse, 5′-CGC TTC ACG AAT TTG CGT GTC AT-3′; EZH2 forward, 5′-TTG TTG GCG GAA GCG TGT AAA ATC-3′ and reverse, 5′-TCC CTA GTC CCG CGC AAT GAG C-3′; GAPDH forward, 5′-GTG GTG AAG ACG CCA GT G GA-3′ and reverse, 5′-CGA GCC ACA TCG CTC AGA CA-3′. U6 and GAPDH were used as internal controls for detecting miR-144-3p and EZH2, respectively and fold changes were calculated using the 2-∆∆Cq method (14).
Cells and cell culture
HSC-2, CAL-27 and SCC-4 cell lines were purchased from ATCC and were cultured in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) containing with 10% fetal bovine serum at 37°C in a 5% CO2 incubator. The normal human oral keratinocyte (NHOK) cells which served as control cells were obtained from the ScienCell Research Laboratories, Inc. (cat. no. 2610) and cultured as previously described (15).
Cell transfection
The miR-144-3p mimics and mimics negative control (mimics NC) were obtained from GenePharma. The EZH2 overexpression vector, pcDNA-EZH2, and pcDNA vector were constructed by GenePharma. In addition, EZH2 siRNA (si-EZH2) and si-Scramble were also purchased from GenePharma.
CAL-27 and SCC-4 cells (8.0×105/well) in a 6-well plate grown to approximately 80% confluence, and then respectively transfected with miR-144-3p mimics (20 nM), mimics NC (20 nM), si-EZH2 (50 nM), or 2 µg pcDNA-EZH2 using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). In addition, inhibition experiments were performed using the EZH2 inhibitor, GSK126 (a compound competes with S-adenosyl-methionine for binding to EZH2, thereby inhibiting histone methyltransferase activity without affecting EZH2 protein expression), as previously described (16). Briefly, the CAL-27 and SCC-4 cells were treated with 5 µM GSK126 (Shanghai HanXiang Life Technology Limited Corporation) or DMSO for 48 h and then collected for use in further experiments. The concentration of DMSO used was ≤1% to ensure the lack of cytotoxicity.
Cell proliferation assay
The anti-proliferative effect was measured using Cell Counting kit-8 (CCK-8) assay. Briefly, the CAL-27 and SCC-4 cells were prepared in 96-well plates containing a final volume of 100 µl/well. Following transfection with miR-144-3p mimics for 24 and 48 h, 10 µl CCK-8 solution was added to the cells followed by culture for a further 2 h. The absorbance value at 490 nm was read using a spectrophotometer (SpectraMax 190; Molecular Devices).
Cell apoptosis assay
The cells were harvested at 48 h post-transfection. After washing in ice-cold PBS, 100 µl binding buffer was added to the cells, followed by staining with 5 µl AnnexinV-FITC/PI (Roche Diagnostics GmbH) at room temperature. Following maintenance for 15 min in the dark, data were collected on an EPICS XL-MCL FACScan (BD Biosciences) and analyzed using FlowJo 8.7.1 software (FlowJo, LLC).
Cell invasion assays
For the invasion assay, a Transwell chamber (8 µm; BD Biosciences) coated with Matrigel was used. At 48 h post-transfection, 8×104 transfected cells in DMEM without serum were added to the upper chamber for incubation, while DMEM with 20% FBS was added to the lower chamber at 37°C for 24 h. The cells at the bottom chamber were then fixed with 4% paraformaldehyde for 30 min and stained with 0.5% crystal violet (Solarbio) for 30 min at room temperature. Subsequently, those cells that had invaded to the lower side of the membrane were counted as the number of cells under a 10/20X inverted microscope (Olympus Corp.) in 5 different fields and analyzed using ImageJ software (version 1.46; Rawak Software, Inc.).
Wound healing assay
The CAL-27 and SCC-4 cells were added in 6-well plates at a density of 2×105 per well and cultured until ~90% confluency was reached. Following transfection for 24 h, gaps were made using a 10 µl pipette tip. After washing with PBS to remove non-adherent cells, the scratches were photographed using a microscope (Carl Zeiss MicroImaging GmbH). The cells were then cultured with serum-free medium at 37°C for 24 h, and the scratches were photographed again. Distances were measured and analyzed using ImageJ software (version 1.46; Rawak Software, Inc.).
Luciferase assay
miRNA target prediction tools, including MiRanda (http://miranda.org.uk) and TargetScan Release 7.0 (http://targetscan.org/) were used to search for the putative targets of miR-144-3p. CAL-27 cells (8×104) were added to 24-well plates until ~90% confluency was reached, and the cells were then co-transfected with the luciferase reporter plasmids (wt-EZH2-UTR-pGL3 or mt-EZH2-UTR-pGL3) with or without miR-144-3p mimics using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h post-transfection, the activity of luciferase was measured using the dual-luciferase reporter assay system (Promega Corporation). Renilla luciferase was used for normalization. The experiments were independently performed in triplicate.
Western blot analysis
Western blot analysis was performed as previously described (17). Briefly, total protein was isolated and quantitated using BCA assay at 48 h post-transfection. The protein lysates (40 µg) were electrophoretically transferred onto PVDF membranes (EMD Millipore), followed blocking with 5% skim milk at 4°C overnight. The membranes were probed with primary antibodies against EZH2 (1:1,000; cat. no. 5246), E-cadherin (1:1,000; cat. no. 3195), N-cadherin (1:1,000; cat. no. 13116) and β-actin (1:1,000; cat. no. 4970), followed by incubation with secondary antibody (anti-rabbit IgG, 1:10,000; cat. no. 7074) for 1 h at room temperature. All antibodies were obtained from Cell Signaling Technology, Inc. Proteins bands were detected using an ECL detection system (GE Healthcare Life Sciences) and blot bands were quantified using ImageJ software (version 1.46; Rawak Software, Inc.).
Statistical analysis
SPSS 13.0 software package (SPSS, Inc.) was applied to analyze the data. All data are presented as the means ± SD. When only 2 groups were compared, the Student's t-test was used. One-way analysis of variance followed by Tukey's post hoc test was applied to compare differences between multiple groups. Categorical data were compared using the Pearson's Chi-squared test. P<0.05 was considered to indicate statistically significant differences. Spearman's correlation analysis was used for correlation analysis.
Results
miR-144-3p expression is downregulated in OSCC tissues and cell lines
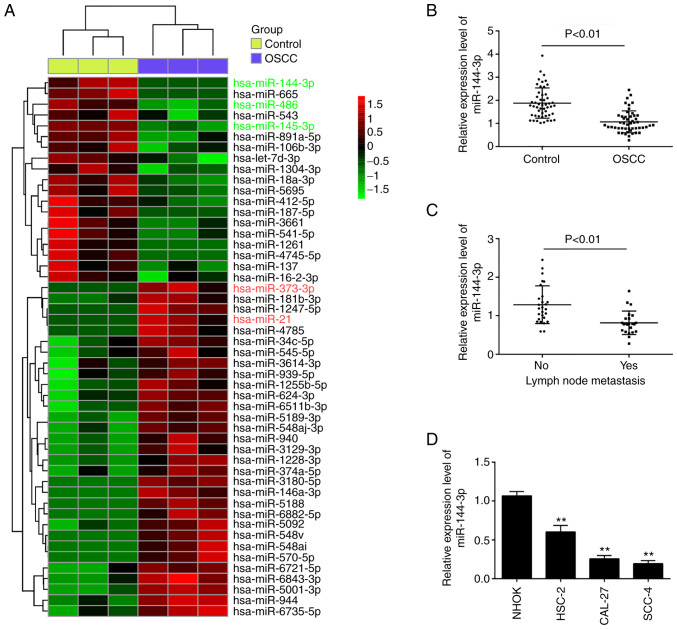
To investigate the role of miRNA expression in OSCC, the profiles of miRNAs were compared between OSCC tumors and adjacent non-cancerous tissues using microarray analysis. Following normalization of the raw data, 50 significant differentially expressed miRNAs (expression of 31 miRNAs was increased and that of 19 miRNAs was decreased) were observed in the OSCC group compared with the control group (Fig. 1A). Of these aberrant miRNAs, the expression levels of miR-486 and miR-145-3p were decreased, while those of miR-373-3p and miR-21 were increased; these findings are consistent with those of previous studies (5,12,17), indicating the reliability of the microarray in the present study. Among the downregulated miRNAs, miR-144-3p exhibited the lowest expression levels in the OSCC group compared with the control group, and some studies have demonstrated the potent suppressive roles of miR-144-3p in various types of human cancer (18,19). Therefore, this miRNA was selected for analysis in further experiments.
Figure 1.
miR-144-3p is downregulated in OSCC tissues and cell lines. (A) Heatmap of miRNA profiles representing the significantly up/downregulated miRNAs. The color code in the heat maps is linear, with green as the lowest and red as the highest. (B) Expression of miR-144-3p was validated by RT-qPCR analysis in OSCC tissues and matched tumor-adjacent tissues (n=50). P<0.01 vs. control group. (C) Expression of miR-144-3p was validated by RT-qPCR analysis in OSCC tissues with lymph node metastasis. P<0.01 vs. non-metastasis group. (D) Expression of miR-144-3p was measured in 4 OSCC cell lines (HSC-2, CAL-27 and SCC-4), and the HOK normal oral mucosa cell line, used as a control, by RT-qPCR analysis. Data are presented as the means ± SD of 3 independent experiments. **P<0.01 vs. NHOK cells. OSCC, oral squamous cell carcinoma.
To validate the microarray results, the expression of miR-144-3p was compared between tumor and adjacent non-cancerous tissues (n=50) by RT-qPCR. It was found that the miR-143-3p levels in the OSCC tissues were much lower than those in adjacent non-cancerous tissues (Fig. 1B). The levels of miR-144-3p were also detected in OSCC tissues from patients with lymph node metastasis. It was observed that miR-144-3p expression was notably decreased in tumor tissues with lymph node metastasis, suggesting that miR-144-3p level was closely associated with a higher incidence of metastasis (Fig. 1C). In addition, the levels of miR-144-3p in OSCC cells were measured. Consistent with the results obtained with the clinical samples, miR-143-3p expression was significantly downregulated in OSCC cells compared with that in the normal human oral keratinocyte (NHOK) cells (Fig. 1D).
Subsequently, the clinicopathological significance of miR-144-3p downregulation in OSCC was analyzed (Table I). It was found that a low expression of miR-144-3p was closely associated with tumor size, differentiation and lymph node metastasis. However, other clinicopathological parameters, such as sex, age, site, alcohol consumption, smoking habits and TNM stage exhibited no significant association with miR-144-3p expression. These data indicate that miR-144-3p may be a potential biomarker for the diagnosis of OSCC.
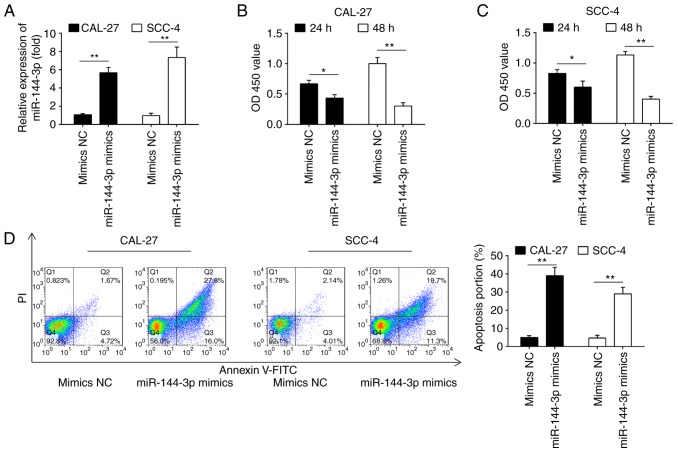
Overexpression of miR-144-3p suppresses cell proliferation and induces cell apoptosis
The frequent downregulation of miR-144-3p in OSCC cell lines and OSCC tissues suggests that miR-144-3p plays a role in OSCC carcinogenesis. To prove this hypothesis, the effects of the ectopic expression of miR-144-3p on cell growth were investigated in 2 OSCC cell lines (CAL-27 and SCC-4). The rationale of using these 2 cell lines is that both cell lines exhibited the lowest expression levels of miR-144-3p among the 3 cell lines examined (Fig. 1D). As shown in Fig. 2A, miR-144-3p expression was notably increased in the CAL-27 and SCC-4 cells following transfection with miR-144-3p mimics, which confirmed the transfection efficiency. CCK-8 assay revealed that miR-144-3p upregulation markedly suppressed the proliferation of the CAL-27 and SCC-4 cells compared with the cells transfected with the mimics control (Fig. 2B and C). Cell apoptosis analysis was then performed to further examine the effects of miR-144a-3p on cell proliferation. It was observed that miR-144-3p upregulation significantly increased cell apop-tosis compared to the mimics control (Fig. 2D). Thus, it was concluded that miR-144-3p may suppress cell proliferation by promoting cell apoptosis.
Figure 2.
Overexpression of miR-144-3p suppresses cell proliferation and promotes cell apoptosis. CAL-27 and SCC-4 cells were transfected with the 20 nM miR-144-3p mimics or mimics-NC for 48 h, and the cells were used for analysis. (A) Transfection efficiency was assessed by RT-qPCR analysis. Cell proliferation was measured using a Cell Counting kit-8 assay in (B) CAL-27 and (C) SCC-4 cells at the indicated time points. (D) Apoptosis was detected by flow cytometry. Data are presented as the means ± SD of 3 independent experiments. *P<0.05, **P<0.01 vs. mimics NC.
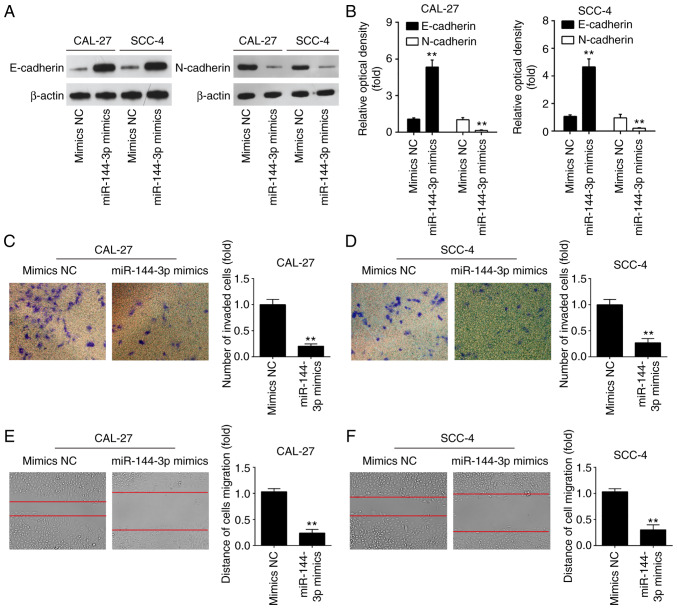
Overexpression of miR-144-3p inhibited cell invasion and migration
miR-144-3p has previously been found to regulate the epithelial-mesenchymal transition (EMT) process in gastric cancer (20). Therefore, it was hypothesized that the tumor suppressive effects of miR-144-3p may involve EMT. To verify this hypothesis, the effects of miR-144-3p on the expression levels of EMT markers were examined at the protein level in OSCC cells. As was expected, miR-144-3p mimics markedly upregulated the protein level of the epithe-lial marker, E-cadherin, and downregulated the expression of the mesenchymal marker, N-cadherin, in the CAL-27 and SCC-4 cells, suggesting that miR-144-3p suppressed the EMT process (Fig. 3A and B). To further examine the effects of miR-144-3p in the migratory and invasive abilities of OSCC cells, Transwell assay and wound healing assay were conducted. miR-144-3p upregulation suppressed the invasion and migration of CAL-27 and SCC-4 cells compared to the mimics NC group (Fig. 3C-F). The above-mentioned data indicate that miR-144-3p suppresses the migration and invasion of OSCC cells.
Figure 3.
Overexpression of miR-144-3p suppresses cell invasion and migration. CAL-27 and SCC-4 cells were transfected with the 20 nM miR-144-3p mimics or mimics-NC for 48 h, and the cells were used for analysis. (A and B) The expression levels of E-cadherin and N-cadherin were measured by western blot analysis. (C and D) Cell invasion was determined by Transwell assay. (E and F) Cell migration was detected by wound healing assay. Data are presented as the means ± SD of 3 independent experiments. **P<0.01 vs. mimics NC.
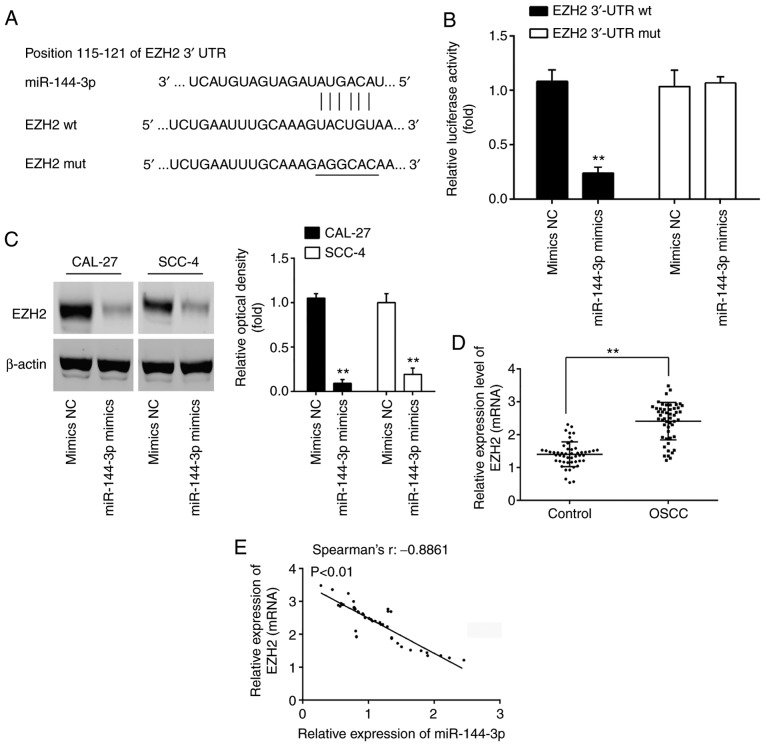
EZH2 is a direct target of miR-144-3p in OSCC cells
To better understand the mechanisms through which miR-144-3p exerts its tumor suppressive effects, the present study sought to identify the mRNA targets of miR-144-3p. Thus, TargetScan 7.0 (http://targetscan.org/) and miRanda (http://miranda.org.uk) were used to search for the predict targets of miR-144-3p, particularly for those that were associated with cell growth or invasion. Bioinformatics analysis predicted that EZH2 may be a potential target of miR-144-3p, since the EZH2 3'UTR possesses a miR-144-3p binding site (Fig. 4A). In order to confirm the interaction between miR-144-3p and EZH2, a luciferase assay was performed. It was found that miR-144-3p mimics decreased the relative luciferase activity of EZH2 3'-UTR wt. However, no significant differences were observed in the luciferase activity of EZH2-3'UTR mut reporter by transfection with miR-144-3p mimics (Fig. 4B). Western blot analysis was then performed to explore the association between miR-144-3p and EZH2 protein expression. Compared to the mimics NC group, miR-144-3p overexpression significantly decreased the expression of EZH2 in both the CAL-27 and SCC-4 cells (Fig. 4C). In addition, the expression levels of EZH2 in 50 pairs of OSCC clinical samples were measured by RT-qPCR. In accordance with the findings of previous studies (21,22), EZH2 was highly expressed in the OSCC group compared with the control group (Fig. 4D). Furthermore, Spearman's correlation analysis revealed a significant inverse correlation between the EZH2 and miR-144-3p expression levels in the tumor tissues (Fig. 4E). These data suggest that miR-144-3p suppresses the expression of EZH2 oncogene in OSCC.
Figure 4.
EZH2 is a direct target of miR-144-3p in OSCC cells. (A) Putative binding sites of miR-144-3p and EZH2. (B) Luciferase assay of CAL-27 cells co-transfected with EZH2 3'-UTR wt or EZH2 3'-UTR mut and 20 nM miR-144-3p mimics and mimics NC, as indicated (n=3). Data are presented as the means ± SD of 3 independent experiments. **P<0.01 vs. mimics NC. (C) Protein expression of EZH2 following transfection with 20 nM miR-144-3p mimics was measured by western blot analysis. **P<0.01 vs. mimics NC. (D) Expression of EZH2 was measured by RT-qPCR analysis in OSCC tissues and matched tumor-adjacent tissues (n=50). **P<0.01 vs. control group. (E) Spearman's rank correlation analysis revealed a negative correlation between the expression of EZH2 and miR-144-3p (r=-0.8861, P<0.01). OSCC, oral squamous cell carcinoma; EZH2, enhancer of zeste homolog 2.
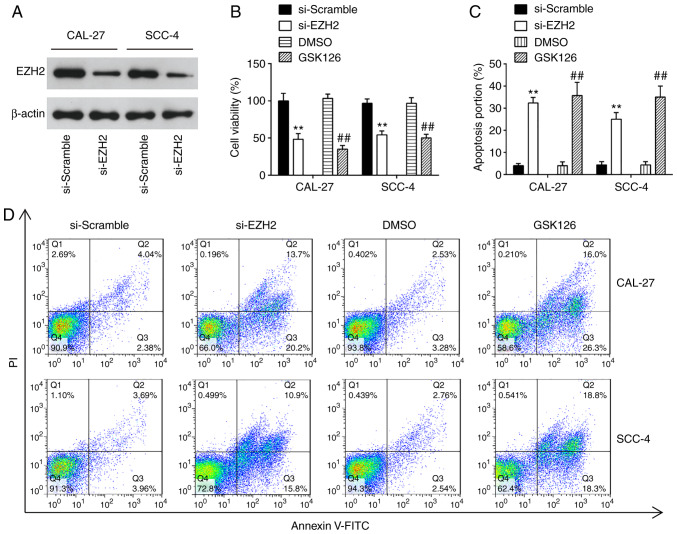
Knockdown of EZH2 inhibits cell proliferation, promotes cell apoptosis and suppresses cell invasion and migration
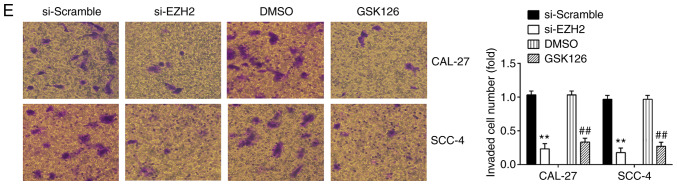
EZH2 is a well-known oncogene in several human cancers, including prostate cancer (23), bladder cancer (24) and renal cell carcinomas (25). In the present study, to examine the effects of EZH2 on OSCC cells, cell proliferation, apoptosis, invasion and migration were examined in the CAL-27 and SCC-4 cells following transfection with si-NC or si-EZH2 for 48 h. Western blot analysis revealed that EZH2 expression was significantly decreased in the CAL-27 and SCC-4 cells transfected with si-EZH2 for 48 h compared with the si-Scramble group (Fig. 5A). The results of CCK-8 assay revealed that the knock-down of EZH2 4 significantly suppressed the proliferation of OSCC cells, compared with the si-Scramble group (Fig. 5B). Additionally, EZH2 inhibition markedly promoted cell apoptosis compared with the si-Scramble group (Fig. 5C and D). Furthermore, Transwell assay suggested that the invasive abilities of the CAL-27 and SCC-4 cells were suppressed by the knockdown of EZH2 (Fig. 5E). Consequently, EZH2 silencing exerted similar effects to miR-144-3p overexpression on OSCC cells.
Figure 5.
Knockdown of EZH2 suppresses cell proliferation, promotes cell apoptosis, and inhibits cell invasion and migration. CAL-27 and SCC-4 cells were transfected with either 50 nM si-EZH2 or 5 µM GSK126 for 48 h, and the cells were used for analysis. (A) Transfection efficiency was assessed by RT-qPCR analysis. (B) Cell proliferation was measured using a Cell Counting kit-8 assay in CAL-27 and SCC-4 cells at the indicated time points. (C and D) Apoptosis was detected by flow cytometry. (E) Cell invasion was determined by Transwell assay. Data are presented as the means ± SD of 3 independent experiments. **P<0.01 vs. si-Scramble; ##P<0.01 vs. DMSO. EZH2, enhancer of zeste homolog 2.
To further confirm the inhibitory effect of EZH2 knock-down on OSCC cells, the CAL-27 and SCC-4 cells were treated with 5 µM GSK126, an Ezh2 inhibitor, for 48 h. The results revealed that GSK126 treatment suppressed the proliferation, promoted the apoptosis and inhibited the invasion and migration of CAL-27 and SCC-4 cells; these findings were similar to those observed with si-EZH2 on the cellular functions of OSCC cells. Taken together, these findings suggest the feasibility of EZH2 for use in clinical trials for the control of OSCC.
miR-144-3p regulates the biological behavior of OSCC cells by targeting EZH2
To determine whether the EZH2 oncogene is responsible for the inhibitory effects of miR-144-3p in OSCC cells, pcDNA-EZH2 combined with miR-144-3p mimics or mimics NC were added to the CAL-27 and SCC-4 cells. Western blot analysis revealed that EZH2 expression was upregulated by transfection with pcDNA-EZH2 plasmid in the CAL-27 and SCC-4 cells (Fig. 6A). Subsequently, CCK-8 assay demonstrated that the concomitant overexpression of miR-144-3p mimics and pcDNA-EZH2 abrogated this inhibitory effect of miR-144-3p mimics on cell proliferation (Fig. 6B). Consistently, the pro-apoptotic effect by miR-144-3p was markedly abolished by pcDNA-EZH2 (Fig. 6C). Furthermore, the overexpression of EZH2 partially reversed the inhibitory effects of miR-144-3p on the cell invasion and migration (Fig. 6D and E). These results suggest that EZH2 is a functional mediator of miR-144-3p in OSCC cells.
Figure 6.
Overexpression of EZH2 abrogates the anti-tumor effects of miR-144-3p mimics. CAL-27 and SCC-4 cells were co-transfected with 20 nM miR-144-3p mimics, mimics NC, 2 µg pcDNA-EZH2 and pcDNA-vector for 48 h, and the cells were used for further analysis. (A) Protein levels of EZH2 were detected by western blot analysis. (B) Cell proliferation was measured using a Cell Counting kit-8 assay. (C) Apoptosis was detected by flow cytometry. (D) Cell invasion was determined by Transwell assay. (E) Cell migration was detected by wound healing assay. Data are presented as the means ± SD of 3 independent experiments. *P<0.05 and **P<0.01 vs. control group; ##P<0.01 vs. miR-144-3p mimics group. EZH2, enhancer of zeste homolog 2.
Discussion
In the present study, it was demonstrated that miR-144-3p was expressed at low level in OSCC tissues and cells. Clinical association analysis indicated that the low expression of miR-144-3p was associated with tumor size, differentiation and lymph node metastasis. Moreover, miR-144-3p inhibited the growth and invasive ability of OSCC cells by suppressing the EZH2 oncogene. This suggests that miR-144-3p has the potential to function as a novel therapeutic target for OSCC.
Several studies have demonstrated that miRNAs play essential roles in controlling multiple steps of OSCC occurrence and development, including proliferation, apoptosis, invasion and metastasis. For example, miR-188 overexpression has been reported to inhibit OSCC cell growth and invasion by targeting SIX1 (26). Cui et al revealed that miR-378-3p/5p suppressed OSCC metastasis by inhibiting kallikrein-related peptidase 4 (KLK4) expression (6). Ding et al demonstrated that miR-145 overexpression suppressed the growth of OSCC xenograft tumors in vivo (27). Therefore, the identification of novel oncogenic or tumor suppressive miRNA involved in OSCC progression is beneficial for the discovery of novel therapeutic targets for OSCC. In the present study, using a microarray, a number of miRNAs were found to be aberrantly expressed in OSCC tissues; in particular, miR-144-3p expression displayed the most downregulated changes, which was supported by a previous study (28). Notably, low expression of miR-144-3p was observed in lymph node metastasis tissues, suggesting the association with higher incidence of lymph node metastasis. Of note, the low expression of miR-144-3p was associated with the tumor size, differentiation and lymph node metastasis. Thus, it is worthy to further detect the serum miR-144-3p level in OSCC patients to clarify its significance as a diagnostic biomarker.
It is worth mentioning that miR-144-3p is mainly considered as a tumor suppressor in many solid tumors (29). For example, miR-144-3p has been demonstrated to function as a tumor suppressor in gastric cancer by activating enhancer-binding protein 4 (AP4) regulation (30). Wu et al illustrated that miR-144-3p targets MAPK6 to suppress cervical cancer cell growth of cervical cancer (19). Furthermore, miR-144-5p, another isoform of miR-144, directly targets SDC3 in renal cell carcinoma (RCC) to inhibit the cancer cell growth (31). Additionally, the study by Chen et al demonstrated that the low expression of miR-144-3p contributes to the prediction in lung cancer patients (32). These previous studies support the important roles of miR-144-3p in different types of cancer. Although there is evidence to indicate the suppressive role of miR-144-3p in OSCC (28,33), the unique role of miR-144-3p and its exact mechanisms in OSCC remain largely unclear. In the present study, it was found that miR-143-3p upregulation significantly suppressed OSCC cell proliferation, induced apoptosis, and effectively suppressed the migration and invasion of OSCC cells by gain-of-function experiments, which is consistent with the findings of a previous study (28). These data indicate that miR-144-3p functions as a tumor suppressor in OSCC progression.
EZH2 belongs to the polycomb-group (PcG) family, and affects embryonic stem cell pluripotency and self-renewal (34,35). Additionally, increasing evidences have revealed that EZH2 was overexpressed in multiple tumors, such as hepatocellular carcinoma, colorectal cancer, and lung cancer (36). Furthermore, EZH2 has been indicated to function as an oncogene by decreasing the activity of histone methyltransferase and silencing the downstream anti-oncogene (37,38). For example, EZH2 expression is associated with the clinical characteristics of patients with colorectal cancer (CC), and the inhibition of EZH2 reduces the proliferation and invasion of CC cells (39). Furthermore, EZH2 has also been identified as an oncogene in breast and prostate cancer (40). Moreover, the use of inhibitors of EZH2 for cancer treatment has been an active area (41,42), and have moved to clinical trials (ClinicalTrials.gov Identifier: NCT02395601 and NCT02082977). Moreover, a number of researchers have reported that EZH2 is well regulated by miRNAs. For instance, Sun et al found that EZH2 was a direct target gene of miR-4465 in non-small cell lung cancer (43). Fan et al discovered miR-217 can regulate EZH2 in nasopharyngeal carcinoma (44). Herein, it was confirmed that EZH2 was directly targeted by miR-144-3p. It was also found that the EZH2 expression level was increased, and inversely correlated with the miR-144-3p levels in OSCC tissues. Finally, it was demonstrated that EZH2 inhibition exhibited a similar role with miR-144-3p mimics, whereas the overexpression of EZH2 abolished the tumor suppressive effect of miR-144-3p mimics on OSCC cells, suggesting that the EZH2 oncogene mediates the role of miR-144-3p in OSCC.
Although the results of recent genomic studies have enhanced the understanding of the role of EZH2 in cancer development, the detailed mechanisms underlying EZH2 function are still complex in distinct cancer types. Generally, EZH2 is the enzymatic subunit of polycomb repressive complex 2 (PRC2), which methylates lysine 27 of histone H3 (H3K27) to promote transcriptional silencing (45). Given its role as a transcriptional regulator, several efforts have been dedicated to the identification of downstream targets or pathways that are driven by EZH2. Researchers have revealed that EZH2 contributes to carcinogenesis by functioning as an oncogene through the silencing of E-cadherin and DNA-damage repair pathways (46,47). The ectopic expression of EZH2 also promotes cancers driven by the loss of SMARCB1 in malignant rhabdoid tumor (48). On the contrary, EZH2 has been shown to interact with PCNA-associated factor (PAF) to the β-catenin complex, and promoting transcriptional activation Wnt target genes in colon cancer cells (49). In addition, the phosphorylation of EZH2 by AKT functions as a co-activator for critical transcription factors, such as androgen receptor (AR) in prostate cancer cells (50). Therefore, EZH2 functions as a double-facet molecule in regulation of gene expression via repression or activation mechanism, depending on the different cellular contexts. In the present study, it was proven that miR-144-3p functions as a tumor suppressor by suppressing EZH2. However, the detailed mechanisms underlying the inhibition of cell viability, and cell invasion and migration by the knockdown of EZH2 warrant further investigation in the future.
However, there are still some limitations to the present study. Due to the limitation in experimental conditions and funds, further research is required in the future to investigate the expression levels of miR-144-3p in more clinical samples. Furthermore, the present study investigated the cellular function of miR-144-3p and its underlying mechanisms in OSCC; however, in vivo studies and clinical trial data are required to validate the preliminary in vitro results obtained in the present study. Therefore, the function of miR-144-3p in OSCC warrants further investigation in vivo.
In conclusion, the findings of the present study demonstrate that miR-144-3p is frequently downregulated in OSCC and serves as a potential tumor suppressor in OSCC. The findings suggest that the miR-144-3p/EZH2 axis may be a promising target for the treatment of OSCC.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
LH and LL performed the experiments, contributed to data analysis and wrote the manuscript. LH and LL analyzed the data. LD conceptualized the study design, contributed to data analysis and experimental materials. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All individuals provided informed consent for the use of human specimens for clinical research. The present study was approved by the College of Stomatology, Xi'an Jiaotong University Ethics Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45:309–316. doi: 10.1016/j.oraloncology.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Sasahira T, Kurihara M, Bhawal UK, Ueda N, Shimomoto T, Yamamoto K, Kirita T, Kuniyasu H. Downregulation of miR-126 induces angiogenesis and lymphangiogenesis by activation of VEGF-A in oral cancer. Br J Cancer. 2012;107:700–706. doi: 10.1038/bjc.2012.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marsh D, Suchak K, Moutasim KA, Vallath S, Hopper C, Jerjes W, Upile T, Kalavrezos N, Violette SM, Weinreb PH, et al. Stromal features are predictive of disease mortality in oral cancer patients. J Pathol. 2011;223:470–481. doi: 10.1002/path.2830. [DOI] [PubMed] [Google Scholar]
- 4.Li T, Cho WC. MicroRNAs: Mechanisms, functions and progress Genomics Proteomics. Bioinformatics. 2012;10:237–238. doi: 10.1016/j.gpb.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang XJ, Jin Y, Song JL, Deng F. MiR-373 promotes proliferation and metastasis of oral squamous cell carcinoma by targeting SPOP. Eur Rev Med Pharmacol Sci. 2019;23:5270–5276. doi: 10.26355/eurrev_201906_18193. [DOI] [PubMed] [Google Scholar]
- 6.Cui Z, Sun S, Liu Q, Zhou X, Gao S, Peng P, Li Q. MicroRNA-378-3p/5p suppresses migration and invasion of oral squamous carcinoma cells by inhibiting KLK4 expression. Biochem Cell Biol. 2019;98:154–163. doi: 10.1139/bcb-2019-0017. [DOI] [PubMed] [Google Scholar]
- 7.Zhang C, Hao Y, Sun Y, Liu P. Quercetin suppresses the tumorigenesis of oral squamous cell carcinoma by regulating microRNA-22/WNT1/β-catenin axis. J Pharmacol Sci. 2019;140:128–136. doi: 10.1016/j.jphs.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Chou ST, Peng HY, Mo KC, Hsu YM, Wu GH, Hsiao JR, Lin SF, Wang HW, Shiah SG. MicroRNA-486-3p functions as a tumor suppressor in oral cancer by targeting DDR1. J Exp Clin Cancer Res. 2019;38:281. doi: 10.1186/s13046-019-1283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen YH, Song Y, Yu YL, Cheng W, Tong X. miRNA-10a promotes cancer cell proliferation in oral squamous cell carcinoma by upregulating GLUT1 and promoting glucose metabolism. Oncol Let. 2019;17:5441–5446. doi: 10.3892/ol.2019.10257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng M, Pang C. MicroRNA-140-5p inhibits the tumorigenesis of oral squamous cell carcinoma by targeting p21-activated kinase 4. Cell Biol Int. 2019 doi: 10.1002/cbin.11213. Ahead of print. [DOI] [PubMed] [Google Scholar]
- 11.Parpart S, Wang XW. microRNA regulation and its consequences in cancer. Curr Pathobiol Rep. 2013;1:71–79. doi: 10.1007/s40139-012-0002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao Y, Qu Y, Dang S, Yao B, Ji M. MiR-145 inhibits oral squamous cell carcinoma (OSCC) cell growth by targeting c-Myc and Cdk6. Cancer Cell Int. 2013;13:51. doi: 10.1186/1475-2867-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei D, Wang W, Shen B, Zhou Y, Yang X, Lu G, Yang J, Shao Y. MicroRNA199a-5p suppresses migration and invasion in oral squamous cell carcinoma through inhibiting the EMTrelated transcription factor SOX4. Int J Mol Med. 2019;44:185–195. doi: 10.3892/ijmm.2019.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Wei D, Shen B, Wang W, Zhou Y, Yang X, Lu G, Yang J, Shao Y. MicroRNA199a-5p functions as a tumor suppressor in oral squamous cell carcinoma via targeting the IKKβ/NF-κB signaling pathway. Int J Mol Med. 2019;43:1585–1596. doi: 10.3892/ijmm.2019.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang S, Wang Z, Zhou J, Huang J, Zhou L, Luo J, Wan Y, Long H, Zhu B. EZH2 Inhibitor GSK126 suppresses anti-tumor immunity by driving production of myeloid-derived suppressor cells. Cancer Res. 2019;79:2009–2020. doi: 10.1158/0008-5472.CAN-18-2395. [DOI] [PubMed] [Google Scholar]
- 17.Yan Y, Wang X, Veno MT, Bakholdt V, Sørensen JA, Krogdahl A, Sun Z, Gao S, Kjems J. Circulating miRNAs as biomarkers for oral squamous cell carcinoma recurrence in operated patients. Oncotarget. 2017;8:8206–8214. doi: 10.18632/oncotarget.14143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang W, Xu Z, Yu L, Che J, Zhang J, Yang J. MicroRNA-144-3p suppressed TGF-B1-induced lung cancer cell invasion and adhe-sion by regulating the Src-Akt-Erk pathway. Cell Biol Int. 2019 doi: 10.1002/cbin.11158. Ahead of print. [DOI] [PubMed] [Google Scholar]
- 19.Wu J, Zhao Y, Li F, Qiao B. MiR-144-3p: A novel tumor suppressor targeting MAPK6 in cervical cancer. J Physiol Biochem. 2019;75:143–152. doi: 10.1007/s13105-019-00681-9. [DOI] [PubMed] [Google Scholar]
- 20.Li B, Zhang S, Shen H, Li C. MicroRNA-144-3p suppresses gastric cancer progression by inhibiting epithelial-to-mesenchymal transition through targeting PBX3. Biochem Biophys Res Commun. 2017;484:241–247. doi: 10.1016/j.bbrc.2017.01.084. [DOI] [PubMed] [Google Scholar]
- 21.Zhao L, Yu Y, Wu J, Bai J, Zhao Y, Li C, Sun W, Wang X. Role of EZH2 in oral squamous cell carcinoma carcinogenesis. Gene. 2014;537:197–202. doi: 10.1016/j.gene.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Wan Q, Wang W, Mai L, Sha L, Mashrah M, Lin Z, Pan C. LncRNA ADAMTS9-AS2 promotes tongue squamous cell carcinoma proliferation, migration and EMT via the miR-600/EZH2 axis. Biomed Pharmacother. 2019;112:108719. doi: 10.1016/j.biopha.2019.108719. [DOI] [PubMed] [Google Scholar]
- 23.Labbe DP, Sweeney CJ, Brown M, Galbo P, Rosario S, Wadosky KM, Ku SY, Sjostrom M, Alshalalfa M, Erho N, et al. TOP2A and EZH2 provide early detection of an aggressive prostate cancer subgroup. Clin Cancer Res. 2017;23:7072–7083. doi: 10.1158/1078-0432.CCR-17-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu D, Li Y, Luo G, Xiao X, Tao D, Wu X, Wang M, Huang C, Wang L, Zeng L, Jiang G. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett. 2017;388:281–291. doi: 10.1016/j.canlet.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Chen Y, Geng H, Qi C, Liu Y, Yue D. Overexpression of YB1 and EZH2 are associated with cancer metastasis and poor prognosis in renal cell carcinomas. Tumour Biol. 2015;36:7159–7166. doi: 10.1007/s13277-015-3417-z. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Liu H. microRNA-188 is downregulated in oral squamous cell carcinoma and inhibits proliferation and invasion by targeting SIX1. Tumour Biol. 2016;37:4105–4113. doi: 10.1007/s13277-015-4246-9. [DOI] [PubMed] [Google Scholar]
- 27.Ding J, Sun D, Xie P. Elevated microRNA-145 inhibits the development of oral squamous cell carcinoma through inactivating ERK/MAPK signaling pathway by down-regulating HOXA1. Biosci Rep. 2019;39:BSR20182214. doi: 10.1042/BSR20182214. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Li X, Li Y, Jiang C, Chen L, Gan N. MicroRNA-144-3p inhibits tumorigenesis of oral squamous cell carcinoma by downregulating ERO1L. J Cancer. 2020;11:759–768. doi: 10.7150/jca.33267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu M, Lin Y, Zhou Y, Jin H, Hou B, Wu Z, Li Z, Jian Z, Sun J. MiR-144 suppresses cell proliferation, migration, and invasion in hepatocellular carcinoma by targeting SMAD4. Onco Targets Ther. 2016;9:4705–4714. doi: 10.2147/OTT.S88233. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Mushtaq F, Zhang J, Li J. miR-144 suppresses cell proliferation and invasion in gastric cancer through downregulation of activating enhancer-binding protein 4. Oncol Lett. 2019;17:5686–5692. doi: 10.3892/ol.2019.10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamada Y, Arai T, Kojima S, Sugawara H, Kato M, Okato A, Yamazaki K, Naya Y, Ichikawa T, Seki N. Regulation of antitumor miR-144-5p targets oncogenes: Direct regulation of syndecan-3 and its clinical significance. Cancer Sci. 2018;109:2919–2936. doi: 10.1111/cas.13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen N, Feng L, Lu K, Li P, Lv X, Wang X. STAT6 phosphor-ylation upregulates microRNA-155 expression and subsequently enhances the pathogenesis of chronic lymphocytic leukemia. Oncol Lett. 2019;18:95–100. doi: 10.3892/ol.2019.10294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pedersen NJ, Jensen DH, Lelkaitis G, Kiss K, Charabi BW, Ullum H, Specht L, Schmidt AY, Nielsen FC, Buchwald C. MicroRNA-based classifiers for diagnosis of oral cavity squamous cell carcinoma in tissue and plasma. Oral Oncol. 2018;83:46–52. doi: 10.1016/j.oraloncology.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 34.Gu W, Zhang E, Song L, Wang Z, Tu L, Tian F, Aikenmu K, Chu G, Zhao J. Long noncoding RNA HOXD-AS1 aggravates osteosarcoma carcinogenesis through epigenetically inhibiting p57 via EZH2. Biomed Pharmacother. 2018;106:890–895. doi: 10.1016/j.biopha.2018.06.173. [DOI] [PubMed] [Google Scholar]
- 35.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kondo Y. Targeting histone methyltransferase EZH2 as cancer treatment. J Biochem. 2014;156:249–257. doi: 10.1093/jb/mvu054. [DOI] [PubMed] [Google Scholar]
- 37.Matsukawa Y, Semba S, Kato H, Ito A, Yanagihara K, Yokozaki H. Expression of the enhancer of zeste homolog 2 is correlated with poor prognosis in human gastric cancer. Cancer Sci. 2006;97:484–491. doi: 10.1111/j.1349-7006.2006.00203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raman JD, Mongan NP, Tickoo SK, Boorjian SA, Scherr DS, Gudas LJ. Increased expression of the polycomb group gene, EZH2, in transitional cell carcinoma of the bladder. Clin Cancer Res. 2005;11:8570–8576. doi: 10.1158/1078-0432.CCR-05-1047. [DOI] [PubMed] [Google Scholar]
- 39.Song-Bing H, Hao Z, Jian Z, Guo-Qiang Z, Tuo H, Dai-Wei W, Wen G, Lin G, Yi Z, Xiao-Feng X, et al. Inhibition of EZH2 expression is associated with the proliferation, apoptosis and migration of SW620 colorectal cancer cells in vitro. Exp Biol Med. 2015;240:546–555. doi: 10.1177/1535370215573463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo KH, Hennighausen L. EZH2 methyltransferase and H3K27 methylation in breast cancer. Int J Biol Sci. 2012;8:59–65. doi: 10.7150/ijbs.8.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim W, Bird GH, Neff T, Guo G, Kerenyi MA, Walenssky LD, Orkin SH. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun J, Tian X, Lu SQ, Hu HB. MicroRNA-4465 suppresses tumor proliferation and metastasis in non-small cell lung cancer by directly targeting the oncogene EZH2. Biomed Pharmacother. 2017;96:1358–1362. doi: 10.1016/j.biopha.2017.11.070. [DOI] [PubMed] [Google Scholar]
- 44.Fan DC, Zhao YR, Qi H, Hou JX, Zhang TH. MiRNA-506 presents multiple tumor suppressor activities by targeting EZH2 in nasopharyngeal carcinoma. Auris Nasus Larynx. 2020 doi: 10.1016/j.anl.2019.12.007. Ahead of print. [DOI] [PubMed] [Google Scholar]
- 45.Gall Troselj K, Novak Kujundzic R, Ugarkovic D. Polycomb repressive complex's evolutionary conserved function: The role of EZH2 status and cellular background. Clin Epigenetics. 2016;8:55. doi: 10.1186/s13148-016-0226-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao K, He J, Wang YF, Jin SD, Fan Y, Fang N, Qian J, Xu TP, Guo RH. EZH2-mediated epigenetic suppression of EphB3 inhibits gastric cancer proliferation and metastasis by affecting E-cadherin and vimentin expression. Gene. 2019;686:118–124. doi: 10.1016/j.gene.2018.11.015. [DOI] [PubMed] [Google Scholar]
- 47.Ito T, Teo YV, Evans SA, Neretti N, Sedivy JM. Regulation of cellular senescence by polycomb chromatin modifiers through distinct DNA damage- and histone methylation-dependent path-ways. Cell Rep. 2018;22:3480–3492. doi: 10.1016/j.celrep.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knutson SK, Warholic NM, Wigle TJ, Klaus CR, Allain CJ, Raimondia A, Scott MP, Chesworth R, Moyer MP, Copeland RA, et al. Durable tumor regression in genetically altered malignant rhab tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci USA. 2013;110:7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jung HY, Jun S, Lee M, Kim HC, Wang X, Ji H, Park J., II PAF and EZH2 induce Wnt/β-catenin signaling hyperactivation. Mol Cell. 2013;52:193–205. doi: 10.1016/j.molcel.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Q, Wang G, Li Q, Jiang W, Kim JS, Wang R, Zhu S, Wang X, Yan L, Yi Y, et al. Polycomb group proteins EZH2 and EED directly regulate androgen receptor in advanced prostate cancer. Int J Cancer. 2019;145:415–426. doi: 10.1002/ijc.32118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.