Zinc deficiency in soils and crops is a global problem. We combine functional and phylogenetic analyses to unravel the role of rice F-bZIP homologs in the zinc deficiency response.
Keywords: Biofortification, F-bZIP, monocots, phylogenetic analysis, rice (Oryza sativa), zinc deficiency, ZIP transporters
Abstract
The F-bZIP transcription factors bZIP19 and bZIP23 are the central regulators of the zinc deficiency response in Arabidopsis, and phylogenetic analysis of F-bZIP homologs across land plants indicates that the regulatory mechanism of the zinc deficiency response may be conserved. Here, we identified the rice F-bZIP homologs and investigated their function. OsbZIP48 and OsbZIP50, but not OsbZIP49, complement the zinc deficiency-hypersensitive Arabidopsis bzip19bzip23 double mutant. Ectopic expression of OsbZIP50 in Arabidopsis significantly increases plant zinc accumulation under control zinc supply, suggesting an altered Zn sensing in OsbZIP50. In addition, we performed a phylogenetic analysis of F-bZIP homologs from representative monocot species that supports the branching of plant F-bZIPs into Group 1 and Group 2. Our results suggest that regulation of the zinc deficiency response in rice is conserved, with OsbZIP48 being a functional homolog of AtbZIP19 and AtbZIP23. A better understanding of the mechanisms behind the Zn deficiency response in rice and other important crops will contribute to develop plant-based strategies to address the problems of Zn deficiency in soils, crops, and cereal-based human diets.
Introduction
Zinc (Zn) is an essential micronutrient for all organisms because of its catalytic and structural roles in many proteins, with Zn-binding proteins estimated at ~10% of eukaryote proteomes (Andreini et al., 2006). Zn-deficient soils are widespread globally, particularly in large parts of Africa and Asia, affecting yield and nutritional quality of crops (Alloway, 2008). Rice (Oryza sativa L.) is one of the most important food crops worldwide, feeding nearly half of the world’s population, especially in Asia, as the staple food in many areas, but it does not provide enough essential mineral nutrients to match human requirements (Welch and Graham, 2004; Nakandalage et al., 2016). The risk of Zn deficiency is estimated to affect about one-third of the world’s human population, and it can lead to different degrees of growth retardation, immune dysfunction, and cognitive impairment (Prasad, 2009; Wessells and Brown, 2012). Improving Zn use efficiency and Zn accumulation in the edible parts of crops (Zn biofortification), in combination with agronomic strategies, constitute plant-based solutions to tackle these global problems (Cakmak, 2002; White and Broadley, 2009).
A better understanding of the molecular mechanisms of plant response to Zn deficiency can contribute to improve traits in crops such as Zn use efficiency, Zn accumulation, and adaptation to Zn-deficient soils. In the model plant Arabidopsis thaliana (Arabidopsis), the bZIP19 and bZIP23 transcription factors are the central regulators of the Zn deficiency response (Assunção et al., 2010). They are members of the F group of Arabidopsis basic-leucine zipper proteins (F-bZIPs), characterized by the presence of a region, at the N-terminus, rich in cysteine and histidine residues (Cys/His-rich motif). The third F-bZIP member is bZIP24, involved in regulation of salt stress tolerance (Jakoby et al., 2002; Yang et al., 2009). The bzip19bzip23 double mutant (bzip19/23) is hypersensitive to Zn deficiency, but has no visible phenotype under Zn sufficiency. The bZIP19 and bZIP23 transcription factors are localized to the nucleus and bind to a 10 bp Zinc Deficiency Response Element (ZDRE; RTGTCGACAY) that is present in the promoter region of their target genes (Assunção et al., 2010). Under Zn deficiency, bZIP19 and bZIP23, which are partially redundant, activate transcription of a small set of Zn homeostasis genes, involved in Zn transport and distribution. These include genes encoding ZIP (Zinc-regulated/Iron-regulated Protein) family Zn transporters involved in cellular Zn uptake, and genes encoding NAS (nicotianamine synthase) enzymes that produce the Zn chelator nicotianamine (NA) involved in Zn distribution (Guerinot, 2000; Assunção et al., 2010; Clemens et al., 2013; Inaba et al., 2015).
A phylogenetic analysis of F-bZIP homologs across land plants suggested conservation of the Zn deficiency response regulatory mechanism. It also showed that predicted orthologs of the AtbZIP19 and AtbZIP23 target ZIP genes, from representative land plant species, also contain ZDRE motifs in their promoters (Castro et al., 2017). This is supported by functional analysis of F-bZIP members from wheat (Triticum aestivum) and barley (Hordeum vulgare) (Evens et al., 2017; Nazri et al., 2017). In barley, seven F-bZIP homologs were identified, of which HvbZIP56 and HvbZIP62 complemented the Arabidopsis bzip19/23 mutant Zn deficiency phenotype. The complemented line with HvbZIP56 was also shown to rescue AtbZIP19/23 ZIP target gene regulation, and subcellular localization analysis of this protein indicated nuclear and cytosolic expression (Nazri et al., 2017). In wheat, 21 F-bZIP homologs, corresponding to seven groups with homeologs from A, B, and D genomes were identified, of which TabZIPF1-7DL and TabZIPF4-7AL rescue the bzip19/23 Zn deficiency phenotype. The TabZIPF1-7DL protein was also shown to bind to the ZDRE motif (Evens et al., 2017). In addition, ZDRE motifs were identified in the promoters of barley and wheat ZIP genes, overall supporting the conservation of the function of F-bZIPs in the Zn deficiency response regulatory network (Castro et al., 2017; Evens et al., 2017; Nazri et al., 2017). Also, in Brachypodium distachyon, the F-bZIP BdbZIP10 is suggested to be involved in regulation of Zn homeostasis (Martin et al., 2018). Considering the global importance of rice as a staple food crop, especially in areas with prevalence of Zn-deficient human diets, we performed the functional characterization of the rice F-bZIP homologs to uncover their role in the Zn deficiency response. We also performed a detailed phylogenetic analysis with F-bZIP homologs retrieved from representative monocot species. A better understanding of the mechanisms behind the Zn deficiency response in rice and in other important crops will contribute to effectively address Zn use efficiency and Zn biofortification in crops.
Materials and methods
Phylogenetic and synteny analysis
Phylogeny of monocot F-bZIP genes was determined as previously reported (Castro et al., 2017). Amino acid sequences of monocot F-bZIPs were obtained from the comparative database Plaza Monocots 4.5 (Van Bel et al., 2018) for the following species: Ananas comosus, Brachypodium distachyon, Elaeis guineensis, Hordeum vulgare, Musa acuminata, Oropetium thomaeum, Oryza brachyantha, Oryza sativa ssp. indica, Oryza sativa ssp. japonica, Phalaenopsis equestris, Phyllostachys edulis, Setaria italica, Sorghum bicolor, Spirodela polyrhiza, Triticum aestivum, Zea mays, Zostera marina, and Zoysia japonica ssp. nagirizaki. For the monocot sequences we opted to use the most recent nomenclature available in Plaza Monocots 4.5. A highly incomplete sequence, and indicated as an outlier by the database, was found for O. thomaeum (Oropetium_20150105_23063) and therefore it was excluded from the analysis. To outgroup and have a better evolutionary perspective of the F-bZIPs across land plants, other species were selected as being representative of major plant taxa, including a bryophyte (Physcomitrella pattens), a pteridophyte (Selaginella moellendorffii), a gymnosperm (Picea glauca), the basal angiosperm (Amborella trichopoda), the model angiosperm eudicot (Arabidopsis thaliana), and another three dicot species containing one Group 1 and one Group 2 F-bZIP (Prunus persica, Theobroma cacao, and Vitis vinifera) (Castro et al., 2017). Phylogenetic analysis was performed using CIPRES Science Gateway V3.3 (http://www.phylo.org) (Miller et al., 2011) and computed inputting 1000 bootstrap iterations, as previously described (Castro et al., 2017). The output tree was visualized in the SeaView Version 4 software (Gouy et al., 2010). To resolve syntenic relationships, we used the colinear gene pairs option at Plaza Monocots 4.5 (Van Bel et al., 2018) to identify colinear segments (multiplicons) associated with each F-bZIP gene present in a selected set of species (the above-mentioned monocots, and dicots A. thaliana and A. trichopoda). Information on multiplicon properties (#Archorpoints and Profile length) was retrieved for each pairwise gene comparison, and the ratio of #Archorpoints per Profile length was used as a measure of collinearity strength. The ratio was applied as the edge score for the construction of a network on Cytoscape (Shannon, 2003), using the yFiles_Organic layout and removing genes with no observed collinearity score. Edge thickness was set between the minimum (0.06) and maximum (0.75) observed scores.
Plasmid construction and plant transformation
The pCaMV35S::bZIP-CFP-HA constructs for stable transformation of the Arabidopsis bzip19/23 double mutant were generated as follows: the full-length coding sequences (CDSs) of OsbZIP48 (LOC_Os06g50310), OsbZIP49 (LOC_Os01g58760), and OsbZIP50 (LOC_Os05g41540) were amplified from a rice (cv. Nipponbare) cDNA library using forward and reverse primers containing NotI and AscI restriction sites, respectively (Supplementary Table S1 at JXB online). The cloning into pEarleyGate-102 Gateway vector (Earley et al., 2006), carrying a Cauliflower mosaic virus (CaMV) 35S promoter, a C-terminal cyan fluorescent protein (CFP), and a HA-tag, the transformation into Agrobacterium tumefaciens, and production of the Arabidopsis bzip19/23 mutant were performed as described by Lilay et al. (2019). Transgenic plants were selected for Basta (phosphinothricin) resistance, and homozygous transgenic seeds (T3 generation) of three independent lines per construct were selected. The expression of each F-bZIP gene was confirmed in the respective lines by real-time quantitative reverse transcription–PCR (RT–qPCR). The lines were referred to as bzip19/23-OEOs48, bzip19/23-OEOs49, and bzip19/23-OEOs50.
Plant material and growth conditions
For hydroponic growth, rice seeds (cv. Nipponbare) were heat treated in paper bags for 5 d at 50 °C and then germinated on vermiculite with distilled water, at 28 °C, for 48 h in the dark, followed by 5 d with a 12 h light/dark cycle and 70% relative humidity. One-week-old seedlings were transferred to 5 liter pots with a lid containing four holes, one for every seedling held supported by rock-wool. A Yoshida nutrient solution (Almeida et al., 2016) buffered with 0.5 mM MES at pH 5.1 was used, with 0.15 µM ZnSO4 (control) or with no ZnSO4 added (–Zn). The plastic pots, for the control and –Zn treatments, were rinsed with 0.1 N HCl followed by five rinses with ultrapure water prior to use. The nutrient solutions were prepared with ultrapure water. Rice plants were grown for 12 weeks in either control or –Zn nutrient solution, which was aerated throughout the experiment, with four plants per pot and five pots per Zn treatment. The nutrient solutions were replaced once a week during the first 4 weeks, and twice in the weeks thereafter, with daily control and adjustment of pH. For the complementation analysis in Arabidopsis, the wild-type (accession Columbia, Col-0) and the bzip19 bzip23 double mutant (bzip19/23; in the Col-0 background, described by Assunção et al., 2010) were used in all experiments. The complementation analysis was part of a larger experimental set-up that simultaneously addressed complementation of bzip19/23 with Arabidopsis F-bZIPs (Lilay et al., 2018) and rice F-bZIPs (this study), thus data for the wild-type and bzip19/23, positive and negative control genotypes, respectively, are common in both studies. For agar-grown seedlings, sterilized seeds of the wild type, bzip19/23 mutant, and bzip19/23-OEOs48, bzip19/23-OEOs49, and bzip19/23-OEOs50 lines were sown on half-strength Murashige and Skoog (MS) medium containing 15 µM ZnSO4 (control) or no added Zn (–Zn) as described by Lilay et al. (2018). The lines were grown together in plates (~5 seedlings per genotype) with control or –Zn MS medium for 14 d. For each complementation line, three independently transformed transgenic lines were tested with at least four plates (replicates) per line and per Zn condition. For the analysis with hydroponically grown plants, sterilized seeds of the above-mentioned lines were germinated and grown for 8 weeks with a modified half-strength Hoagland nutrient solution with either 2 µM ZnSO4 (control) or 0.002 µM ZnSO4 (–Zn), with six plants per genotype, as described by Lilay et al. (2018). For the analysis with the soil-grown plants, 2-week-old seedlings of the above-mentioned lines, grown on control MS, were transplanted to a mixture of 80% peat–20% vermiculite, one seedling per 0.4 liter pot with six pots per line, and were grown for an additional 4 weeks. The hydroponics set-up and the MS plates with a 8/16 h light/dark cycle, and the pots with a 8/16 h and 16/8 h light/dark cycle were placed in a growth chamber, with 125 µmol m−2 s−1 white light, 22/20 °C light/dark temperature, and 70% relative humidity.
Yeast one-hybrid (Y1H)assay
Total RNA was isolated from rice seedlings, as described by Almeida et al. (2017). The cDNA expression library was synthesized according to HybriZAP-2.1 XR cDNA Synthesis and Library Construction Kits (Stratagene), as described by the manufacturer. The screening was performed according to the Matchmaker, One-Hybrid System (Clontech). The reporter vectors containing bait F and bait G were introduced into yeast strain PJ69-4A, as previously reported (Assunção et al., 2010). Yeast bait strains were transformed with 1 µg of the cDNA expression library. For each reporter vector, >1 million yeast colonies were screened in CM-His supplemented with 10–40 mM 3-aminotriazole (3-AT), as previously described (Ouwerkerk and Meijer, 2001). The identified clones were re-streaked on selective medium to confirm growth. Direct PCR on the yeast colonies was performed to amplify the cDNA insert, using specific primers for the library plasmid. The sequences were used to search for homology in the rice genome.
EMSA
The full-length CDSs of OsbZIP48 (LOC_Os06g50310) and OsbZIP49 (LOC_Os01g58760) were amplified from a rice (cv. Nipponbare) cDNA library using forward and reverse primers containing NotI and AscI restriction sites, respectively (Supplementary Table S1). The PCR products were purified with PureLink (Invitrogen) and cloned into the pENTR™/D-TOPO vector (Invitrogen) followed by in vitro site-directed recombination into the pSPUTK in vitro translation vector (Stratagene) using LR Clonase™ II Enzyme Mix (Invitrogen). All constructs were verified by restriction enzyme digestion analysis and DNA sequencing. The in vitro translation of OsbZIP48 and OsbZIP49, the oligonucleotide labeling, and the EMSA were performed as described by Assunção et al. (2010).
Real-time quantitative RT–PCR analysis
Roots and shoots from 4-week-old rice plants grown in control or –Zn hydroponic solution were separated and immediately frozen in liquid nitrogen in pools of two plants per pot and five pots per treatment. Roots and shoots were ground with liquid nitrogen using a mortar and pestle, and total RNA was extracted using the Direct-zol RNA Kit (Zymo Research). Fourteen-day-old Arabidopsis seedlings of the wild type, bzip19/23 mutant, and bzip19/23-OEOs48, bzip19/23-OEOs49, and bzip19/23-OEOs50 lines, grown in control or –Zn MS medium, were harvested and immediately frozen in liquid nitrogen in pools of five seedlings per line and per Zn treatment×3 different plates grown simultaneously and considered as biological replicates. This experiment was performed twice, with two independently transformed T3 homozygous lines per genotype. Seedlings were ground with liquid nitrogen in a microtube, with the help of polypropylene pestles, and total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). RNA quantity and integrity, and cDNA synthesis were assessed and performed as described by Lilay et al. (2018). Primers for RT–qPCR (Supplementary Table S2) were designed using NCBI Primer-BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast), and the primer amplification efficiency for each primer pair was between 1.9 and 2.1. RT–qPCR was performed with a LightCycler 96 Real-Time PCR System (Roche Diagnostics), using HOT FIREPol EvaGreen qPCR Mix (Solis BioDyne) in a 20 µl PCR mixture, as described by Lilay et al. (2018). Rice Eukaryotic elongation factor1-alpha (OseEF-1α, LOC_Os03g08020) and Arabidopsis Actin-2 (ACT2, At3g18780) were used as reference genes in rice and Arabidopsis gene expression analysis, respectively. Reactions were performed in 2–3 technical replicas per biological replicate and in three biological replicates per line and Zn treatment. The calculated cycle threshold (Ct) value for each gene was normalized to the reference gene calculated Ct value (OseEF-1α for rice and ACT2 for Arabidopsis). The relative transcript levels were expressed against the wild-type grown under control conditions, and calculated according to the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Subcellular localization analysis
Ten-day-old seedlings of the wild type, bzip19/23 mutant, and bzip19/23-OEOs48, bzip19/23-OEOs49, and bzip19/23-OEOs50 lines were grown in control or –Zn MS medium. For laser scanning confocal microscopy (LSCM) analysis, roots were transferred to microscope slides containing propidium iodide (PI) to stain root cell walls, and were visualized using a Leica TCS SP5 II laser scanning confocal inverted microscope (Leica Microsystems) with a HC PL APO CS ×63/1.30 Glycerine objective. Argon 458 nm and 514 nm laser lines were used for CFP and PI excitation, respectively. The emission settings were between 470 nm and 500 nm for CFP and 590 nm and 630 nm for PI. For each complementation line, three independently transformed T3 homozygous lines were tested, with observations of 2–3 seedlings per line and Zn condition.
Tissue element analysis
For rice plants, shoots and roots from 4-week-old hydroponically grown plants (five plants per Zn treatment), grown in control or –Zn nutrient solution, were harvested. For Arabidopsis plants, shoots and roots from 8-week-old hydroponically grown plants from the wild type, bzip19/23 mutant, and bzip19/23-OEOs48, bzip19/23-OEOs49, and bzip19/23-OEOs50 lines (four plants per line per Zn treatment), grown in control or –Zn nutrient solution, were harvested. For the bzip19/23 mutant and bzip19/23-OEOs49 lines grown at –Zn, six plants were harvested, following the same procedure, and were analyzed in pools of two. This experiment was performed twice, with two or three independently transformed T3 homozygous lines per genotype. Arabidopsis plants from bzip19/23-OEOs48, bzip19/23-OEOs49, and bzip19/23-OEOs50 were also grown on soil, and the shoots from four plants per line were harvested. Shoot and root harvest, tissue digestion, and multielemental analysis, using inductively-coupled plasma mass spectrometry (ICP-MS) (7900 ICP-MS, Agilent Technologies), were performed as described by Lilay et al. (2018).
Statistical analysis
To compare lines or treatments, we used one-way ANOVA followed by Tukey’s post-hoc test, or Student’s t-test, as appropriate, calculated with IBM SPSS Statistics V22.0 software.
Results
Yeast-one-hybrid screening identifies rice F-bZIPs
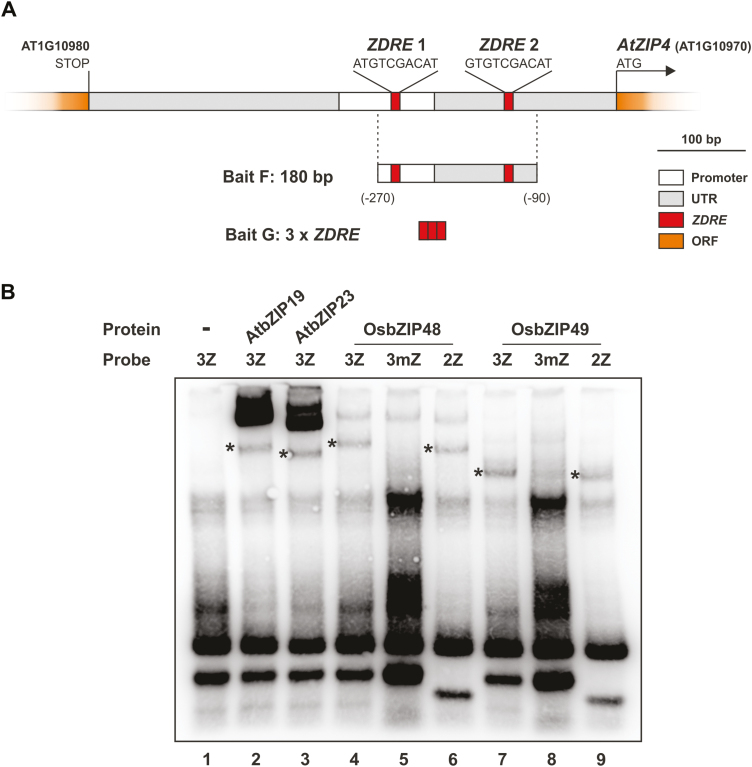
To identify rice transcription factors that bind to the ZDRE promoter element, we performed a Y1H screening. As bait we employed either a three tandem repeat of the ZDRE motif (RTGTCGACAY) (Fig. 1A, Bait F), or a 180 bp promoter fragment of the AtZIP4 Zn transporter gene (fragment G) that contains two ZDRE copies (Assunção et al., 2010) (Fig. 1A, Bait G). The corresponding yeast bait strains were screened with a rice cDNA expression library, resulting in the identification of LOC_Os06g50310 (one clone) and LOC_Os01g58760 (two clones) as binding to bait F, and LOC_Os06g50310 (two clones) as binding to bait G (Supplementary Table S3). These loci correspond to the rice F-bZIP members, OsbZIP48 and OsbZIP49, respectively.
Fig. 1.
Yeast-one-hybrid (Y1H) screening and EMSA analysis. (A) Schematic diagram of the bait fragments (F and G) used to construct the reporter vectors in the Y1H assay. (B) EMSA analysis with in vitro translated OsbZIP48, OsbZIP49, and, as reference, AtbZIP19 and AtbZIP23. Probe acronyms 2Z and 3Z correspond to two or three ZDRE (ATGTCGACAY) tandem repeats, respectively, and 3mZ corresponds to three tandem repeats of a ZDRE mutated version (ATGTAGACAY).
To validate the binding ability of OsbZIP48 and OsbZIP49 to the ZDRE motif, we carried out an in vitro binding assay using EMSA. Two and three tandem copies of ZDRE were used, and also three tandem copies of a modified ZDRE motif in which the TCGA core has been mutated to TAGA (RTGTAGACAY). The EMSA showed that both OsbZIP48 and OsbZIP49 can bind in vitro to three and two tandem copies of the ZDRE motif, but not to three tandem copies of the modified ZDRE version (Fig. 1B).
Annotated F-bZIP homologs in rice and phyl.ogenetic analysis of monocot F-bZIPs
Previously, we carried out a thorough phylogenetic characterization of F-bZIP homologs across land plants that identified the presence of three F-bZIP members in rice, namely LOC_Os06g50310, LOC_Os01g58760, and LOC_Os05g41540, which correspond to OsbZIP48, OsbZIP49, and OsbZIP50, respectively. OsbZIP48 was mapped to F-bZIP Group 1, together with Arabidopsis bZIP19 and bZIP23, whereas OsbZIP49 and OsbZIP50 were included in Group 2, together with Arabidopsis bZIP24 (Castro et al., 2017). The protein sequence alignments show a high amino acid sequence identity between the rice and Arabidopsis F-bZIP homologs (Supplementary Fig. S1A, B). Notably, the OsbZIP49 protein corresponds to a form truncated at the N-terminal end (Supplementary Fig. S1B).
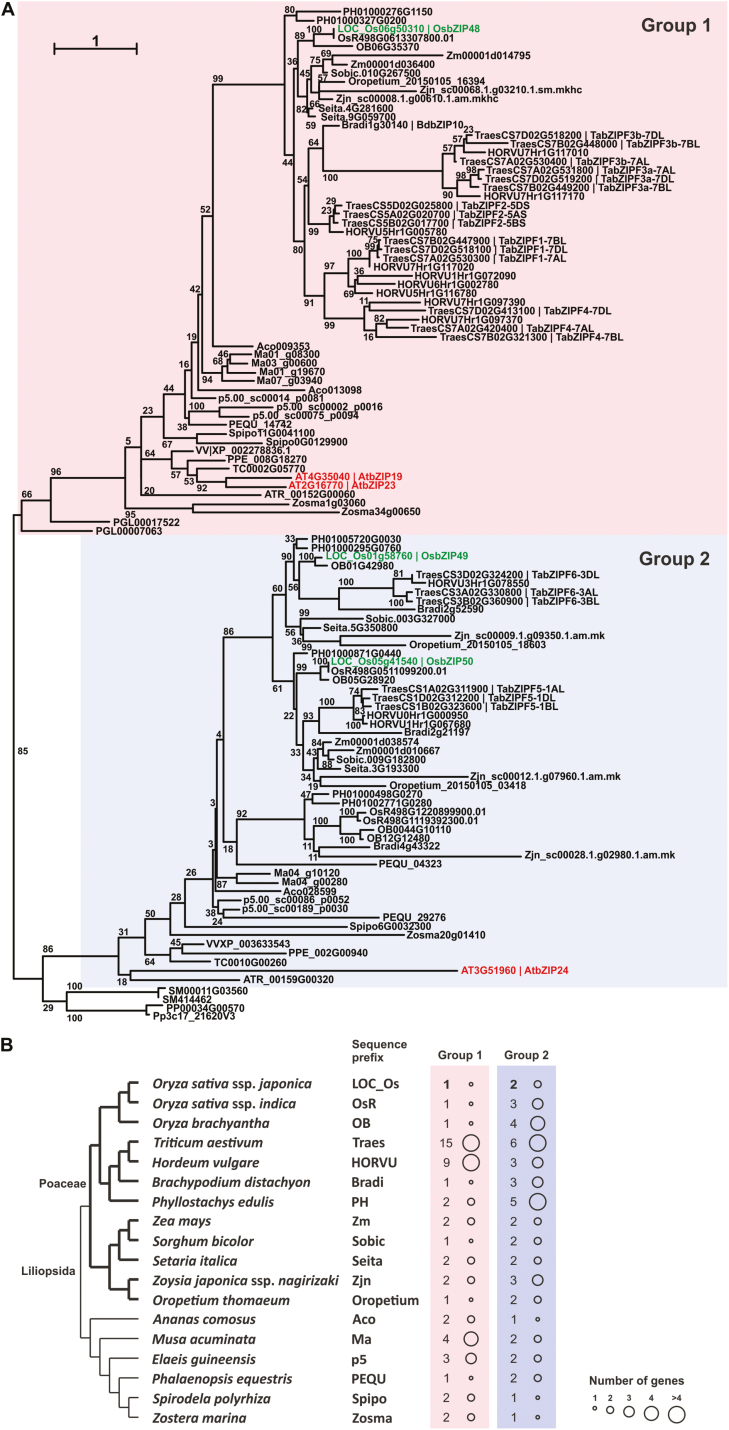
In our previous phylogenetic analysis (Castro et al., 2017), a monocot enrichment for Group 2 genes was suggested. Here, in order to obtain a more detailed evolutionary perspective of the monocot F-bZIP homologs, we increased resolution with the incorporation of additional genomes. Towards this end, a set of monocot species representative of the taxon’s phylogeny were selected. In total, 98 monocot F-bZIP sequences were identified (Supplementary Table S4), characterized by the presence of the bZIP domain and the F-bZIP characteristic Cys/His-rich motif, though truncated versions were observed. The full protein sequences were subsequently used for phylogenetic inference (Fig. 2A). Previously, we demonstrated that F-bZIP Group 1 and 2 emergence was associated with seed plant differentiation (Castro et al., 2017). Here, we once again observed that bryophyte and pteridophyte sequence positioning supports a single monophyletic origin for F-bZIP transcription factors, with differentiated branches forming Group 1 and Group 2 F-bZIPs. In this report, we introduced a collinearity analysis across F-bZIP members to further establish the robustness of Group 1 and Group 2 differentiation (Supplementary Fig. S2). This analysis serves as a survey to infer the evolution of gene family members, complementing phylogeny by looking at paralogs within their genomic context. Results showed no collinearity between Group 1 and 2 genes, but strong collinearity within group members, which supports the phylogenetic analysis in Fig. 2A.
Fig. 2.
Phylogenetic analysis of the F-bZIP proteins in monocot plant species. (A) Phylogenetic tree representing 17 monocot species, the major taxa within monocots, plus representative species from other major plant taxa. The tree was constructed using maximum likehood and bootstrap values from 1000 replicates. Numbers on each branch represent the bootstrap percentages. (B) F-bZIP gene enrichment for each monocot plant species. Circle size represents the absolute number of expected genes present in the genome of each species.
In addition to the rice subspecies japonica F-bZIP members, we identified the F-bZIPs from subspecies indica, with OsR498G0613307800.01 mapped to Group 1 and OsR498G0511099200.01, OsR498G1119392300.01, and OsR498G1220899900.01 mapped to Group 2. Given the diversity of species employed in this phylogenetic analysis, it supports the previously observed enrichment of Group 2 F-bZIP monocots (Castro et al., 2017), in relation to Group 1, with the exception of wheat and barley (Fig. 2A), and indicates that the enrichment was confined to the Poaceae family, and was absent from the pineapple genome at the basis of the Poales order (Fig. 2A, B). In addition, Fig. 2B shows that all analyzed monocot species have at least one Group 1 F-bZIP member, in support of previous findings across land plant species (Castro et al., 2017).
Expression of rice F-bZIP and ZIP genes in response to Zn supply
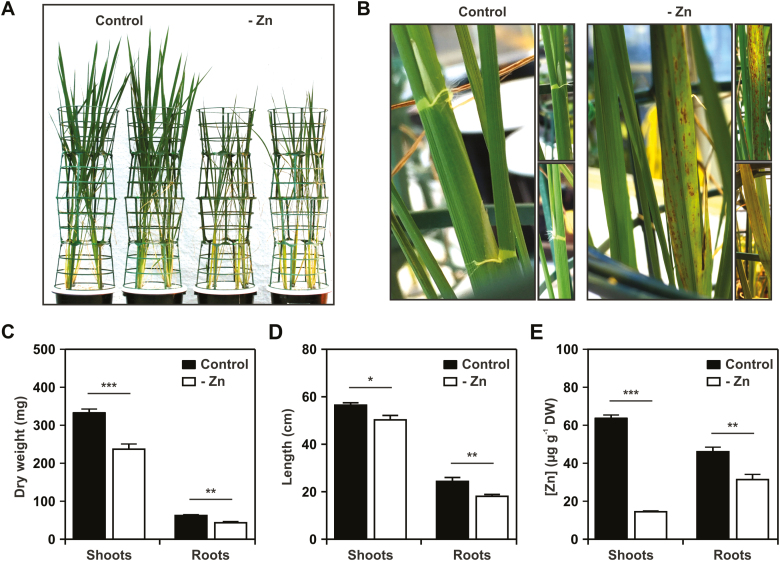
To further our understanding of F-bZIP function in monocot Zn homeostasis, we analyzed rice plants grown hydroponically with control or Zn-deficient conditions. In 12-week-old plants, there were visible phenotypic differences between control and Zn deficiency treatments, with the latter displaying reduced growth and development of leaf chlorosis and necrosis (Fig. 3A, B). Four-week-old plants already displayed a reduction in both shoot and root dry weight, and shoot and root length, under Zn deficiency (Fig. 3C, D). As expected, element analysis revealed a significant reduction in the concentration of Zn in shoots and roots of Zn deficiency-grown plants. This reduction was more pronounced in shoots than in roots (Fig. 3E).
Fig. 3.
Phenotypic analysis of hydroponically grown rice plants in response to Zn supply. (A) Twelve-week-old rice plants grown with control and Zn-deficient (–Zn) nutrient solution. (B) Detail showing chlorosis and necrosis development with –Zn. Shoot and root dry weight (C), shoot and root length (D), and shoot and root Zn concentration (E), in 4-week-old rice plants grown in control and –Zn nutrient solution. Data are presented as means ±SE (n≥5 plants). Statistically significant differences between control and –Zn were determined by Student t-test (*P<0.05, **P<0.01, ***P<0.001).
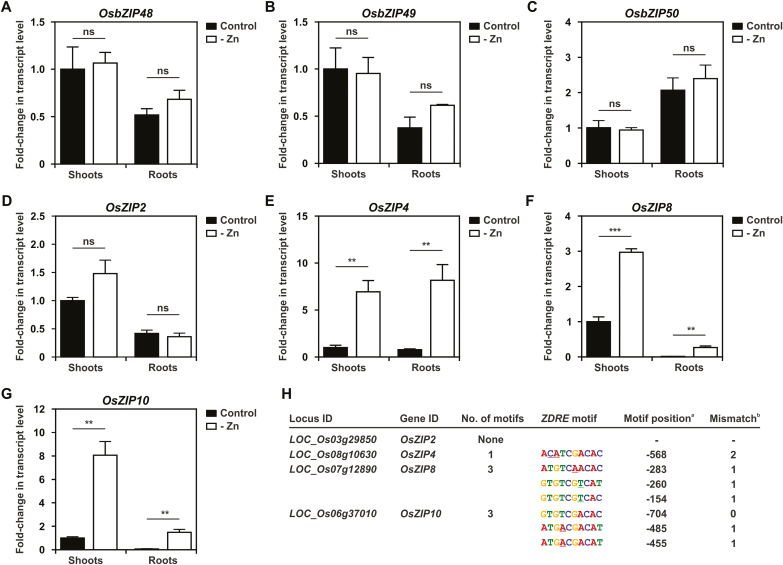
Next, we analyzed the transcript levels of the rice F-bZIP genes, OsbZIP48, OsbZIP49, and OsbZIP50. Our previous in silico analysis indicated that OsbZIP49 has a lower expression level than OsbZIP48 and OsbZIP50 (Castro et al., 2017). Here we show that OsbZIP48 and OsbZIP49 are expressed more in shoots than in roots, whereas OsbZIP50 has the opposite expression pattern (Fig. 4A–C). None of the F-bZIP genes showed variation in transcript levels between control and Zn deficiency in shoots or roots (Fig. 4A–C). We also determined the expression of four rice ZIP family member genes, OsZIP2, OsZIP4, OsZIP8, and OsZIP10. OsZIP2 expression was not significantly Zn deficiency responsive (Fig. 4D), whereas OsZIP4, OsZIP8, and OsZIP10 expression was significantly induced under Zn deficiency in both shoots and roots (Fig. 4E–G). We analyzed the promoter region of these rice ZIP genes to identify the presence of the ZDRE motif. In the OsZIP2 promoter, no ZDRE was found, whereas in OsZIP8 and OsZIP10, three ZDRE copies were identified in each promoter region, with one mismatch allowed. In the OsZIP4 promoter, a ZDRE-like motif, with two mismatches (ACATCGACAC), was detected (Fig. 4H).
Fig. 4.
Gene expression analysis of 4-week-old hydroponically grown rice plants in response to Zn supply. Shoot and root transcript levels of (A) OsbZIP48, (B) OsbZIP49, (C) OsbZIP50, (D) OsZIP2, (E) OsZIP4, (F) OsZIP8, and (G) OsZIP10, grown with control or –Zn nutrient solution. Bars represent mean fold change in the transcript level of three biological replicates ±SE. Statistically significant differences between control and –Zn conditions were determined by Student t-test (**P<0.01, ***P<0.001; ns indicates not significant). (H) Analysis of the number and position of detected ZDRE motifs in the promoter of each gene. aPosition of the first nucleotide of the ZDRE motif in relation to the start codon. bMismatches from the ZDRE consensus sequence (RTGTCGACAY).
Complementation analysis in the Arabidopsis bzip19/23 background
The Arabidopsis bzip19/23 double mutant is characterized by a Zn deficiency-hypersensitive phenotype (Assunção et al., 2010). Here, the complementation lines of each rice F-bZIP member (OsbZIP48, OsbZIP49, and OsbZIP50) in the bzip19/23 background were analyzed. To generate these heterologous complementation lines, we stably transformed the bzip19/23 double mutant with each of the rice F-bZIP gene cDNAs under control of the constitutive CaMV 35S promoter and containing a C-terminal CFP-HA fusion (i.e. bzip19/23-OEOs48, bzip19/23-OEOs49, and bzip19/23-OEOs50). The transcript level of the expressed rice gene in each line was verified, and a western blot with anti-HA confirmed the expected protein molecular weight for each rice bZIP-CFP-HA expressed protein in the heterologous complementation lines (Supplementary Fig. S3).
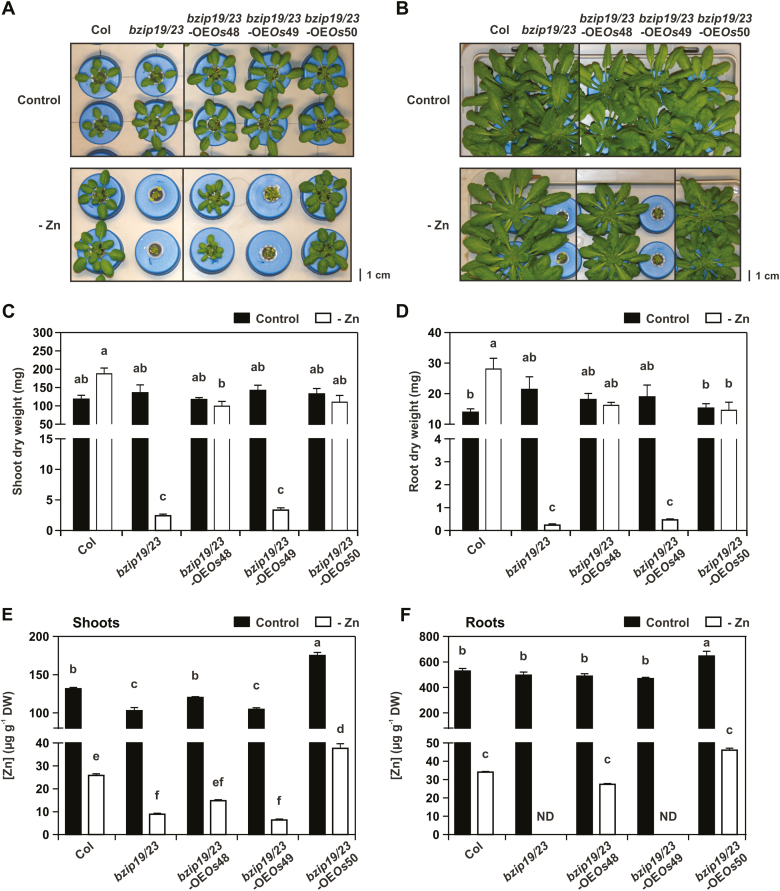
To test the complementation of the bzip19/23 mutant, the bzip19/23-OEOs48, bzip19/23-OEOs49, and bzip19/23-OEOs50 lines were grown in hydroponics with control or Zn-deficient conditions. The analysis showed that bzip19/23-OEOs48 and bzip19/23-OEOs50 lines complemented the bzip19/23 mutant phenotype under Zn deficiency, conferring phenotypes comparable with that of the wild-type. The bzip19/23-OEOs49 line, on the other hand, showed severe growth impairment under Zn deficiency, comparable with the bzip19/23 mutant (Fig. 5A). The plants were allowed to grow for 8 weeks, during which the Zn deficiency-hypersensitive phenotype of the bzip19/23 mutant and bzip19/23-OEOs49 lines became more evident (Fig. 5B). This was in agreement with the shoot and root tissue dry weight data (Fig. 5C,D). The lines were also grown with control or Zn-deficient MS medium, with bzip19/23-OEOs48 and bzip19/23-OEOs50 seedlings complementing the bzip19/23 mutant, whereas the performance of bzip19/23-OEOs49 seedlings was slightly better than that of the bzip19/23 mutant line (Supplementary Fig. S4).
Fig. 5.
Complementation study with Arabidopsis bzip19/23-OEOs48, bzip19/23-OEOs49, and bzip19/23-OEOs50 lines, the wild type (Col), and the bzip19/23 double mutant, grown in hydroponics with control or Zn-deficient (–Zn) nutrient solution. (A) Phenotype of 4-week-old plants and (B) 8-week-old plants. (C) Shoot and (D) root dry weight, and (E) shoot and (F) root Zn concentration, of 8-week-old plants grown under control (filled bar) or –Zn (open bar) conditions. Data are represented as means ±SE (n=4 plants for all treatments, except bzip19/23 and bzip19/23-OEOs49 at –Zn with n=6). Different letters indicate significant differences (P<0.05) after one-way ANOVA followed by Tukey’s post-hoc test.
The analysis of Zn concentration in the 8-week-old hydroponically grown plants showed that under control conditions, the shoot and root Zn concentration of the bzip19/23-OEOs48 line was not significantly different from that of the wild type, whereas the shoot concentration of the bzip19/23 mutant and bzip19/23-OEOs49 lines was significantly lower. Interestingly, the shoot Zn concentration of the bzip19/23-OEOs50 line was significantly higher, with an increase to ~1.3 that of the wild type (Fig. 5E, F). Under Zn deficiency, the roots harvested from the bzip19/23 mutant and bzip19/23-OEOs49 plants were extremely small (<0.5 mg DW), in line with the severity of the Zn deficiency phenotype (Fig. 5B), and were not included in the analysis. The Zn concentration in shoots of the bzip19/23 mutant and bzip19/23-OEOs49 lines was significantly lower than in the wild type, and the bzip19/23-OEOs48 and bzip19/23-OEOs50 lines, with the latter having a significantly higher Zn concentration than the wild type (Fig. 5E, F; Supplementary Fig. S7, S8).
Subcellular localization in response to Zn supply
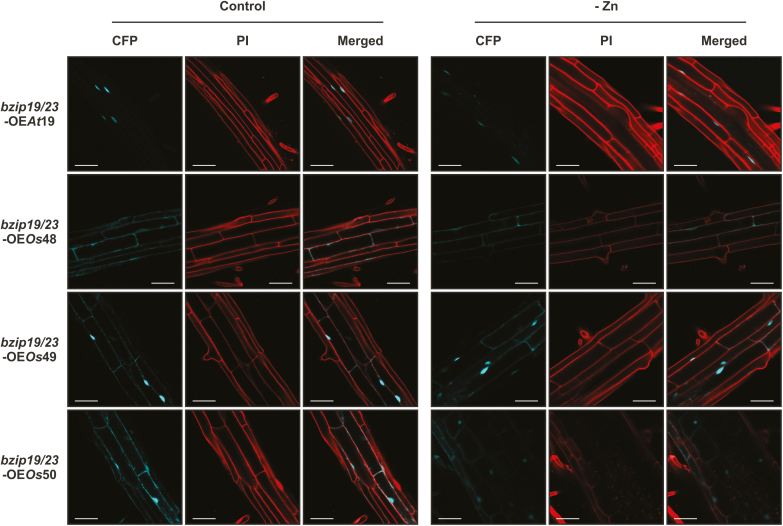
To investigate the subcellular localization of the rice F-bZIP proteins and address whether it is affected by cellular Zn status, we analyzed seedlings of the bzip19/23-OEOs48, bzip19/23-OEOs49, and bzip19/23-OEOs50 lines, grown with control or Zn-deficient medium. The fluorescence of the C-terminal CFP fluorophore was visualized with CLSM. The fluorescent signal for the three rice F-bZIP protein fusions (OsbZIP48–CFP, OsbZIP49–CFP, and OsbZIP50–CFP) localized in the cell nucleus and also in the cytosol (Fig. 6). The analysis also revealed that the subcellular localization of the three rice F-bZIPs was identical between seedlings grown under control conditions and Zn deficiency (Fig. 6). These results were consistent between the three independently transformed lines analyzed for each complementation line. In addition, we performed a localization analysis using transient expression in Nicotiana benthamiana leaves with each rice F-bZIP cDNA under control of the constitutive CaMV 35S promoter and containing an N-terminal green fluorescent protein (GFP) fluorophore. We found similar results for the three F-bZIP proteins (Supplementary Fig. S5).
Fig. 6.
Subcellular localization analysis of OsbZIP48–CFP, OsbZIP49–CFP, and OsbZIP50–CFP fusion proteins in roots of 10-day-old seedlings of Arabidopsis bzip19/23-OEOs48, bzip19/23-OEOs49, and bzip19/23-OEOs50 lines, respectively, grown with control or Zn-deficient (–Zn) MS medium. The nuclear-localized AtbZIP19–CFP is used for comparison. Emissions of CFP and PI were visualized with CLSM. Two to three seedlings of three independently transformed T3 homozygous lines of bzip19/23-OEOs48, bzip19/23-OEOs49, and bzip19/23-OEOs50 lines were analyzed. Scale bars=50 μm.
Expression analysis of bZIP19/23 target genes in the heterologous complementation lines
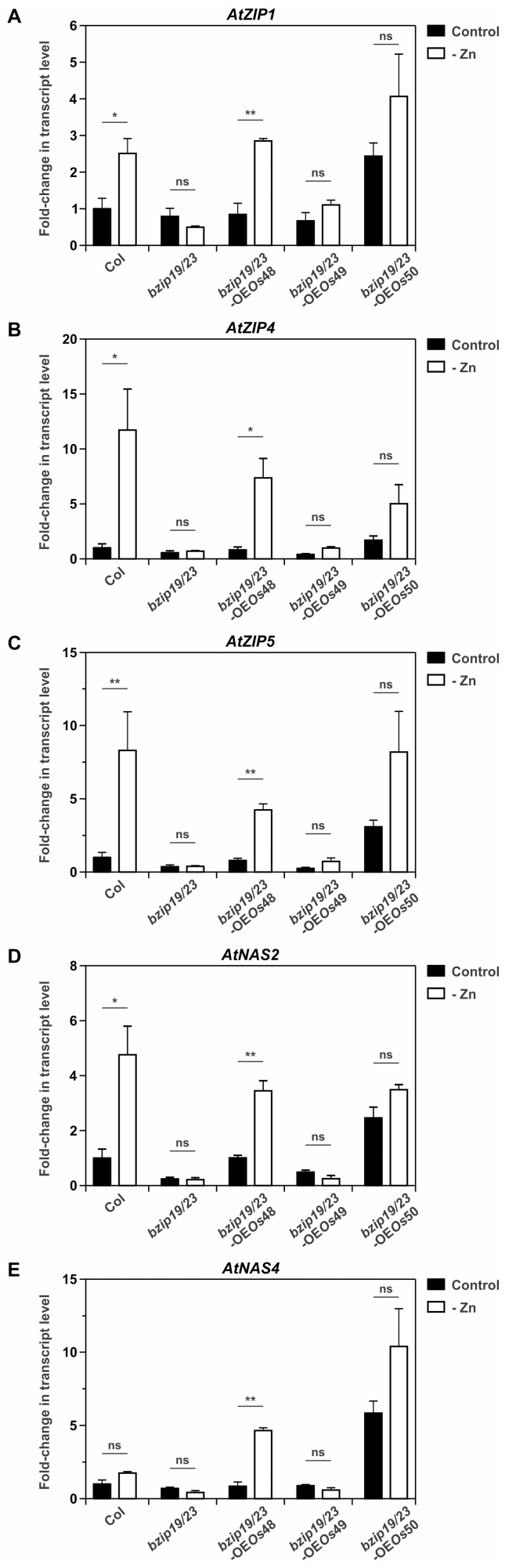
To further analyze the rice F-bZIP complementation lines, we investigated the expression profiles of a subset of the Arabidopsis bZIP19 and bZIP23 target genes, namely ZIP1, ZIP4, ZIP5, NAS2, and NAS4 (Assunção et al., 2010) in seedlings of bzip19/23-OEOs48, bzip19/23-OEOs49, and bzip19/23-OEOs50 lines grown under control conditions or Zn deficiency (Fig. 7; Supplementary Fig. S6). The transcript level profiles were overall comparable between the bzip19/23-OEOs48 line and wild type, showing a significant induction of gene expression under Zn deficiency, indicating that the Group 1 OsbZIP48 protein was capable of complementing the Arabidopsis bzip19/23 mutant (Fig. 7A–E). With regard to Group 2 rice F-bZIPs, the bzip19/23-OEOs49 line showed a transcript level profile that was overall comparable with that of the bzip19/23 mutant—displaying gene expression levels under both Zn conditions that collectively support the absence of functional complementation (Fig. 7A–E). Conversely, under Zn deficiency, the expression pattern in the bzip19/23-OEOs50 line was overall comparable with that of the wild type and bzip19/23-OEOs48, in agreement with the complementation analysis. Most significantly, under control conditions, bzip19/23-OEOs50 displayed a high transcript level for all genes analyzed, surpassing not only the null mutant levels but also those of the wild type (Fig. 7A–E).
Fig. 7.
Transcript level profiles of bZIP19 and bZIP23 target genes in 14-day-old seedlings of wild-type Arabidopsis (Col), the bzip19/23 double mutant, and bzip19/23-OEOs48, bzip19/23-OEOs49, and bzip19/23-OEOs50 lines, grown with control (filled bars) or Zn-deficient (–Zn, open bars) MS medium. Real-time quantitative RT–PCR was used to determine the transcript levels of (A) ZIP1, (B) ZIP4, (C) ZIP5, (D) NAS2, and (E) NAS4. Bars represent mean fold change in transcript level of three biological replicates ±SE. Data correspond to one independently transformed T3 homozygous line of bzip19/23-OEOs48, bzip19/23-OEOs49, and bzip19/23-OEOs50 lines. Data on the second independently transformed lines are shown in Supplementary Fig S6. Statistically significant differences between control and –Zn were determined by Student t-test (*P<0.05, **P<0.01; ns, indicates not significant).
Element analysis in soil-grown plants
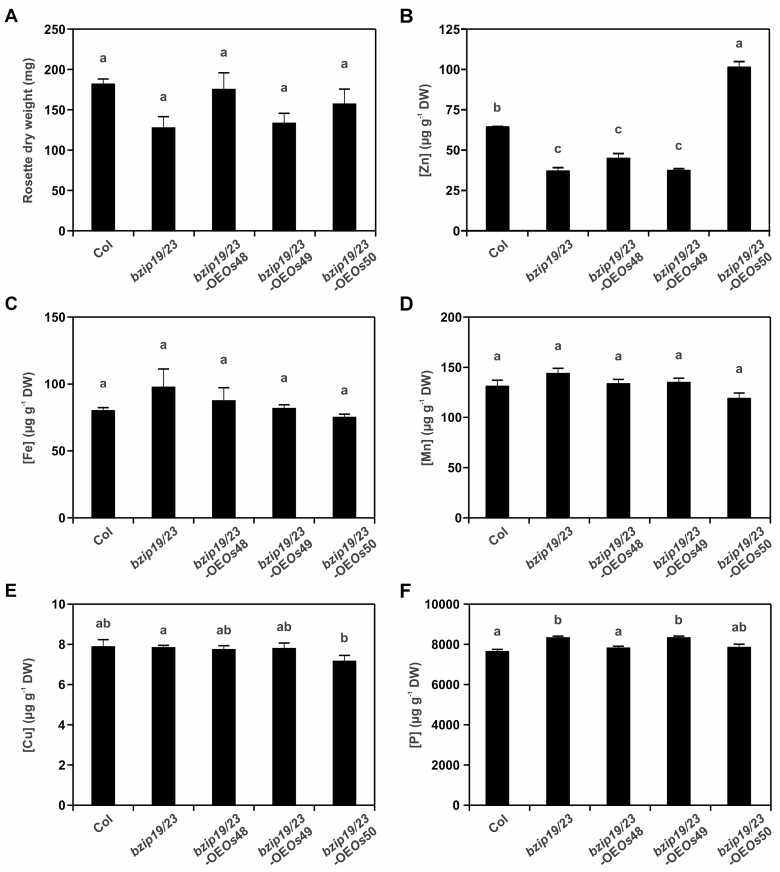
Finally, we also analyzed the shoot Zn concentration of soil-grown plants, which displayed a similar pattern when compared with the hydroponically grown plants under control conditions (Fig. 8A). Here, the bzip19/23-OEOs50 line showed a significantly higher Zn concentration, with an increase to ~1.5 that of the wild type (Fig. 8A). In addition, the analysis of other elements, namely Fe, Mn, Cu, and P, showed that their concentrations in shoots did not differ strongly between the wild type, the mutant, and the complementation lines (Fig. 8B–F; Supplementary Fig. S8).
Fig. 8.
Element analysis of shoots of 6-week-old plants of the Arabidopsis wild type (Col), bzip19/23 double mutant, and bzip19/23-OEOs48, bzip19/23-OEOs49, and bzip19/23-OEOs50 lines, grown on soil. Data correspond to one independently transformed T3 homozygous lines. Data on second and third independently transformed lines are shown in Supplementary Figs S7 and S8. Bars represent (A) shoot dry weight, (B) Zn concentration, (C) Fe concentration, (D) Mn concentration, (E) Cu concentration, and (F) P concentration. Data are represented as means ±SE (n=4 plants). Different letters indicate significant differences (P<0.05) after one-way ANOVA followed by Tukey’s post-hoc test.
Discussion
Phylogenetic analysis of monocot F-bZIPs
In this study, we identified and functionally characterized F-bZIP members of rice ssp. japonica, namely LOC_Os06g50310, LOC_Os01g58760, and LOC_Os05g41540, addressing their role in the Zn deficiency response. The classification of the bZIP family in Arabidopsis comprises 13 groups (designated A–M) (Jakoby et al., 2002; Dröge-Laser et al., 2018) and this classification has been extended to all major lineages of green plants in an evolutionary analysis (Corrêa et al., 2008). In this classification, the above-mentioned rice F-bZIPs correspond to OsbZIP48, OsbZIP49, and OsbZIP50, respectively (Corrêa et al., 2008). A different classification for rice bZIP family members comprises 11 groups (designated I–XI), in which the same rice F-bZIPs correspond to OsbZIP53, OsbZIP44, and OsbZIP7, respectively (Nijhawan et al., 2008). We adopted the former classification system.
Our previous phylogenetic analysis of F-bZIP homologs across land plants suggested a monophyletic origin prior to seed plant evolution, where branching out of two clades (Group 1 and Group 2) was observed (Castro et al., 2017). Here, our phylogenetic and synteny analysis of monocot F-bZIP homologs supports the previously described emergence of F-bZIP Groups 1 and 2 associated with seed plant differentiation (Fig. 2; Supplementary Fig. S2). Rice bZIP48 belongs to Group 1, together with Arabidopsis bZIP19 and bZIP23 proteins, the central regulators of the Zn deficiency response (Assunção et al., 2010). The wheat TraesCS7D02G518100/TabZIPF1-7DL that complements the Arabidopsis bzip19/23 mutant (Evens et al., 2017) is also in Group 1. Rice bZIP49 and bZIP50 belong to Group 2, together with Arabidopsis bZIP24, involved in salt stress regulation, and with no significant role in the Zn deficiency response (Yang et al., 2009; Lilay et al., 2018). The analysis also indicates an enrichment of Group 2 F-bZIP genes in monocots, in relation to Group 1, that contrasts with the observed pattern in eudicots, where several species lost Group 2 F-bZIP homologs (Castro et al., 2017). Previously, we suggested that Group 1 F-bZIPs conserve a function in the Zn deficiency response regulation, whereas Group 2 F-bZIPs appear more prone to gene loss or expansion events that might lead to neo- or subfunctionalization (Castro et al., 2017).
Role of rice F-bZIPs in the zinc deficiency response
The role of the three rice F-bZIPs in the Zn deficiency response was investigated. The expression profiles of OsbZIP48, OsbZIP49, and OsbZIP50 in rice roots and shoots indicate that these genes are not responsive to Zn deficiency (Fig. 4), suggesting that a Zn-dependent activity of the transcription factors is probably not regulated at the transcriptional level. This is in line with results obtained for the Arabidopsis F-bZIP transcription factors (Lilay et al., 2018). In wheat and barley, some of the identified F-bZIP homologs showed gene expression induction under Zn deficiency (Evens et al., 2017; Nazri et al., 2017) and, for wheat, it was suggested that Zn deficiency induction of some F-bZIP genes would allow a sustained adaptive response under prolonged deficiency (Evens et al., 2017). Interestingly, among the wheat and barley F-bZIP homologs, those reported to complement the Arabidopsis bzip19/23 mutant, or to show in vitro binding to the ZDRE motif (i.e. TraesCS7D02G518100/TabZIPF1-7DL, HvbZIP56, and HvbZIP62), displayed only a small transcript induction, if any, upon Zn deficiency (Evens et al., 2017; Nazri et al., 2017).
The heterologous expression of the three rice F-bZIPs in Arabidopsis and N. benthamiana indicated a nuclear and also cytosolic localization (Fig. 6; Supplementary Fig. S5), in line with the results obtained for the barley HvbZIP56 expressed in Arabidopsis (Nazri et al., 2017), whereas AtbZIP19 and AtbZIP23, also with a 35S promoter, were shown to be nuclear localized (Inaba et al., 2015; Lilay et al., 2018). Our results additionally indicate that the cellular Zn status is not involved in the subcellular targeting of rice F-bZIP proteins, comparable with what was reported for Arabidopsis F-bZIPs (Lilay et al., 2018), suggesting that a Zn-dependent modulation of the activity of these transcription factors involves other mechanisms.
Several members of the rice ZIP family were characterized and shown to be involved in Zn uptake and transport (Bashir et al., 2012). We analyzed the expression of four rice OsZIP genes as Zn deficiency markers for our hydroponically grown rice plants, and verified the presence of ZDRE motifs in their promoter region (Fig. 4). Previously, the analysis of predicted orthologs of the Arabidopsis bZIP19/23 target ZIP genes in land plant species found ZDRE motifs in the promoters and identified a conserved substitution in the motif (i.e. RTGWCGACAY) (Castro et al., 2017). Here, we identified three promoter ZDRE motifs (RTGWCGACAY) in the Zn deficiency-induced OsZIP10, and three ZDRE variants, with one mismatch, in the Zn deficiency-induced OsZIP8 (Lee et al., 2010). For the strongly Zn deficiency-induced OsZIP4, functionally characterized as a Zn transporter (Ishimaru et al., 2005), no ZDRE motif was found but a ZDRE variant, with two mismatches, was detected, which deserves further analysis. Conversely, in the non-Zn deficiency-induced OsZIP2, similarly to its close homolog AtZIP2 (Assunção et al., 2010; Ishimaru et al., 2011), no motif was found in the promoter region. Overall, our data indicate an association between Zn deficiency-induced rice ZIP genes and the presence of promoter ZDRE motifs, as previously reported for ZIP genes from wheat, barley, and other land plant species (Castro et al., 2017; Evens et al., 2017; Nazri et al., 2017), supporting the conservation of the Zn deficiency response regulatory mechanism.
Our findings additionally show that OsbZIP48 and OsbZIP49 bind not only to the ZDRE motif in vitro but also to a ZDRE-containing AtZIP4 promoter fragment in a Y1H assay. The yeast screening did not identify OsbZIP50 possibly because, as we show, OsbZIP50 is expressed more in roots than in shoots, and the rice cDNA library was enriched for shoot transcripts. OsbZIP48 and OsbZIP49 showed the opposite expression pattern—more expressed in shoots than in roots. This was also observed between Arabidopsis bZIP19 and bZIP23 expression patterns and might be relevant for the individual role of each transcription factor in the Zn deficiency response (Lilay et al., 2018).
OsbZIP48 is a functional homolog of AtbZIP19 and AtbZIP23
The role of the three rice F-bZIPs in the Zn deficiency response was further investigated by complementation analysis in the Arabidopsis bzip19/23 double mutant background, namely bzip19/23-OEOs48, bzip19/23-OEOs49, and bzip19/23-OEOs50 lines (Figs 5, 7, 8). Our observations indicate that Group 1 OsbZIP48 is a functional homolog of AtbZIP19 and AtbZIP23. The bzip19/23-OEOs48 line complemented the bzip19/23 mutant, restoring Zn concentration levels in the plant similar to the wild type. In addition, the analysis of AtbZIP19/23 target gene expression showed Zn deficiency induction in the bzip19/23-OEOs48 line, similarly to the wild type, and to the Arabidopsis bZIP19 and bZIP23 complementation lines (bzip19/23-OE19 and bzip19/23-OE23) (Lilay et al., 2018). A wild-type expression pattern of Zn deficiency-responsive AtZIP target genes was also reported in the bzip19/23 complementation lines overexpressing barley HvbZIP56 and HvbZIP62 (Nazri et al., 2017). According to our results, a Zn-dependent regulation of rice F-bZIPs is unlikely to occur at the transcriptional level, and cellular Zn status does not seem to interfere with their subcellular targeting. Furthermore, the expression profile of the target genes in the bzip19/23-OEOs48 line suggests that cellular Zn status can repress or limit the activity of OsbZIP48. This is consistent with previous findings from Arabidopsis bZIP19 and bZIP23, supporting that Zn deficiency sensing is required for the activity of these transcription factors (Lilay et al., 2018).
The bzip19/23-OEOs49 line did not complement the bzip19/23 mutant and displayed its severe Zn deficiency phenotype, with the Zn shoot concentration in line with the reported critical deficiency level below 15–20 µg g−1 DW (Marschner, 1995). This was consistent with a similar pattern of target gene expression between bzip19/23-OEOs49 and the bzip19/23 mutant. Group 2 OsbZIP49 is a protein lacking much of the N-terminal region typically found in F-bZIPs which very probably disturbs its function, explaining the lack of complementation. The conservation of the bZIP domain, with the basic region and leucine-zipper domain, involved in DNA binding and directing dimerization, respectively (Vinson et al., 2006), is consistent with our EMSA and Y1H results indicating binding of OsbZIP49 to the ZDRE motif.
The Group 2 OsbZIP50 complemented the bzip19/23 mutant in the bzip19/23-OEOs50 line, which is interesting considering that the Group 2 Arabidopsis bZIP24 has not been associated with the regulation of Zn deficiency response. The most remarkable difference between the bzip19/23-OEOs50 line and the wild-type and bzip19/23-OEOs48 lines is the increased shoot Zn concentration found in bzip19/23-OEOs50 plants grown under Zn-sufficient conditions while the concentration of other elements was not significantly affected. Moreover, the expression of the target genes was consistently higher in the bzip19/23-OEOs50 line, under control conditions, than in the wild type or bzip19/23-OEOs48 lines. This suggests that the enhanced shoot Zn concentration in bzip19/23-OEOs50 is caused by higher expression of, at least, the analyzed genes. The Arabidopsis bZIP19 and bZIP23 target genes are transcriptionally activated upon Zn deficiency, with Zn sufficiency limiting or repressing their activation (Lilay et al., 2018). Our results suggest that the effect of Zn sufficiency in repressing target gene activation by OsbZIP50 is less efficient than in AtbZIP19, AtbZIP23, and OsbZIP48. The increased expression of AtbZIP19/23 target genes and the enhanced shoot Zn concentration in the bzip19/23-OEOs50 line under Zn sufficiency provide a novel perspective suggesting that the Zn-dependent activity of the F-bZIP transcription factors could be modulated to improve plant Zn acquisition.
We provide a detailed monocot F-bZIPs phylogenetic analysis that supports the branching of plant F-bZIPs into Group 1 and Group 2. Our results show that the rice F-bZIP transcription factors play a role in the Zn deficiency response, with Group 1 OsbZIP48 being a functional homolog of Arabidopsis bZIP19 and bZIP23, supporting a conservation of this regulatory mechanism in land plants. More knowledge on the regulators and mechanisms of the Zn deficiency response in crops will contribute to effectively address Zn use efficiency and Zn biofortification, and develop plant-based strategies to address the problems of Zn deficiency in soils and in human diets.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Multiple sequence alignment of F-bZIP members.
Fig. S2. Pairwise collinearity analysis between monocots F-bZIP genes.
Fig. S3. Transcript level profiles and western blot analysis.
Fig. S4. Complementation analysis.
Fig. S5. Subcellular localization analysis.
Fig. S6. Transcript level profiles of the bzip19/23-OEOs48, -OEOs49, and -OEOs50 second lines.
Fig. S7. Element analysis of the bzip19/23-OEOs48 and -OEOs49 second lines.
Fig. S8. Element analysis of the bzip19/23-OEOs50 second and third lines.
Table S1. Primers used for cloning.
Table S2. Primers used in real-time quantitative RT–PCR analysis.
Table S3. Clones sequenced in the Y1H screening.
Table S4. Monocot F-bZIPs sequences and IDs.
Acknowledgements
The research was funded by the Danish Council for Independent Research, DFF-YDUN-program (4093-00245B), co-funded by the Portuguese Foundation for Science and Technology, FCT-IF-program (IF/01641/2014; IF/01126/2012) [GHL, PHC, AC, NJMS, and AGLA], Research Unit-GREEN-IT (UID/Multi/04551/2013) [DMA], NORTE2020 (NORTE-01-0145-FEDER-000007) [HA], EU-COST Action FA0905 STSM [JGG], and the ZonMWZenith program (grant 93512008) [MGMA].
References
- Alloway BJ. 2008. Zinc in soils and crop nutrition. Brussels: International Zinc Association Publications. [Google Scholar]
- Almeida DM, Almadanim MC, Lourenço T, Abreu IA, Saibo NJ, Oliveira MM. 2016. Screening for abiotic stress tolerance in rice: salt, cold, and drought. Methods in Molecular Biology 1398, 155–182. [DOI] [PubMed] [Google Scholar]
- Almeida DM, Gregorio GB, Oliveira MM, Saibo NJ. 2017. Five novel transcription factors as potential regulators of OsNHX1 gene expression in a salt tolerant rice genotype. Plant Molecular Biology 93, 61–77. [DOI] [PubMed] [Google Scholar]
- Andreini C, Banci L, Bertini I, Rosato A. 2006. Zinc through the three domains of life. Journal of Proteome Research 5, 3173–3178. [DOI] [PubMed] [Google Scholar]
- Assunção AG, Herrero E, Lin YF, et al. . 2010. Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proceedings of the National Academy of Sciences, USA 107, 10296–10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir K, Ishimaru Y, Nishizawa NK. 2012. Molecular mechanisms of zinc uptake and translocation in rice. Plant and Soil 361, 189–201. [Google Scholar]
- Cakmak I. 2002. Plant nutrition research: priorities to meet human needs for food in sustainable ways. Plant and Soil 247, 3–24. [Google Scholar]
- Castro PH, Lilay GH, Muñoz-Mérida A, Schjoerring JK, Azevedo H, Assunção AGL. 2017. Phylogenetic analysis of F-bZIP transcription factors indicates conservation of the zinc deficiency response across land plants. Scientific Reports 7, 3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S, Deinlein U, Ahmadi H, Höreth S, Uraguchi S. 2013. Nicotianamine is a major player in plant Zn homeostasis. BioMetals 26, 623–632. [DOI] [PubMed] [Google Scholar]
- Corrêa LG, Riaño-Pachón DM, Schrago CG, dos Santos RV, Mueller-Roeber B, Vincentz M. 2008. The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS One 3, e2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dröge-Laser W, Snoek BL, Snel B, Weiste C. 2018. The Arabidopsis bZIP transcription factor family—an update. Current Opinion in Plant Biology 45, 36–49. [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. 2006. Gateway-compatible vectors for plant functional genomics and proteomics. The Plant Journal 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Evens NP, Buchner P, Williams LE, Hawkesford MJ. 2017. The role of ZIP transporters and group F bZIP transcription factors in the Zn-deficiency response of wheat (Triticum aestivum). The Plant Journal 92, 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution 27, 221–224. [DOI] [PubMed] [Google Scholar]
- Guerinot ML. 2000. The ZIP family of metal transporters. Biochimica et Biophysica Acta 1465, 190–198. [DOI] [PubMed] [Google Scholar]
- Inaba S, Kurata R, Kobayashi M, Yamagishi Y, Mori I, Ogata Y, Fukao Y. 2015. Identification of putative target genes of bZIP19, a transcription factor essential for Arabidopsis adaptation to Zn deficiency in roots. The Plant Journal 84, 323–334. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Bashir K, Nishizawa NK. 2011. Zn uptake and translocation in rice plants. Rice 4, 21–27. [Google Scholar]
- Ishimaru Y, Suzuki M, Kobayashi T, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. 2005. OsZIP4, a novel zinc-regulated zinc transporter in rice. Journal of Experimental Botany 56, 3207–3214. [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. 2002. bZIP transcription factors in Arabidopsis. Trends in Plant Science 7, 106–111. [DOI] [PubMed] [Google Scholar]
- Lee S, Kim SA, Lee J, Guerinot ML, An G. 2010. Zinc deficiency-inducible OsZIP8 encodes a plasma membrane-localized zinc transporter in rice. Molecules and Cells 29, 551–558. [DOI] [PubMed] [Google Scholar]
- Lilay GH, Castro PH, Campilho A, Assunção AGL. 2019. The Arabidopsis bZIP19 and bZIP23 activity requires zinc deficiency—insight on regulation from complementation lines. Frontiers in Plant Science 9, 1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Marschner H. 1995. Mineral nutrition of higher plants. San Diego: Academic Press. [Google Scholar]
- Martin RC, Vining K, Dombrowski JE. 2018. Genome-wide (ChIP-seq) identification of target genes regulated by BdbZIP10 during paraquat-induced oxidative stress. BMC Plant Biology 18, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2011. The CIPRES science gateway. Proceedings of the 2011 TeraGrid Conference on Extreme Digital Discovery - TG ’11. New York: ACM Press. [Google Scholar]
- Nakandalage N, Nicolas M, Norton RM, Hirotsu N, Milham PJ, Seneweera S. 2016. Improving rice zinc biofortification success rates through genetic and crop management approaches in a changing environment. Frontiers in Plant Science 7, 764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazri AZ, Griffin JHC, Peaston KA, Alexander-Webber DGA, Williams LE. 2017. F-group bZIPs in barley—a role in Zn deficiency. Plant, Cell & Environment 40, 2754–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhawan A, Jain M, Tyagi AK, Khurana JP. 2008. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiology 146, 333–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwerkerk PBF, Meijer AH. 2001. Yeast one-hybrid screening for DNA–protein interactions . Current Protocols in Molecular Biology 55, 12.12.1–12.12.12. [DOI] [PubMed] [Google Scholar]
- Prasad AS. 2009. Impact of the discovery of human zinc deficiency on health. Journal of the American College of Nutrition 28, 257–265. [DOI] [PubMed] [Google Scholar]
- Shannon P. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel M, Diels T, Vancaester E, Kreft L, Botzki A, Van de Peer Y, Coppens F, Vandepoele K. 2018. PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Research 46, D1190–D1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson C, Acharya A, Taparowsky EJ. 2006. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochimica et Biophysica Acta 1759, 4–12. [DOI] [PubMed] [Google Scholar]
- Welch RM, Graham RD. 2004. Breeding for micronutrients in staple food crops from a human nutrition perspective. Journal of Experimental Botany 55, 353–364. [DOI] [PubMed] [Google Scholar]
- Wessells KR, Brown KH. 2012. Estimating the global prevalence of zinc deficiency: results based on zinc availability in national food supplies and the prevalence of stunting. PLoS One 7, e50568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PJ, Broadley MR. 2009. Biofortification of crops with seven mineral elements often lacking in human diets—iron, zinc, copper, calcium, magnesium, selenium and iodine. New Phytologist 182, 49–84. [DOI] [PubMed] [Google Scholar]
- Yang O, Popova OV, Süthoff U, Lüking I, Dietz KJ, Golldack D. 2009. The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 436, 45–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.