Abstract
Awns are bristle-like structures formed at the tip of the lemma on the florets of some cereal grasses. Wild-type wheat is awned, but awnletted and awnless variants have been selected and nowadays all forms are cultivated. In this study, we dissected the genetic control underlying variation of this characteristic feature by association mapping in a large panel of 1110 winter wheat cultivars of worldwide origin. We identified the B1 (Tipped 1) locus on chromosome 5A as the major determinant of awnlessness globally. Using a combination of fine-mapping and expression analysis, we identified a putative C2H2 zinc finger protein with an EAR domain, characteristic of transcriptional repressors, as a likely candidate for Tipped 1. This gene was found to be up-regulated in awnless B1 compared with awned b1 plants, indicating that misexpression of this transcriptional regulator may contribute to the reduction of awn length in B1 plants. Taken together, our study provides an entry point towards a better molecular understanding of the evolution of morphological features in cereals through selection and breeding.
Keywords: Association mapping, awns, B1, misexpression, Tipped 1, Triticum aestivum, wheat
We identified a candidate for the awnedness inhibitor Tipped 1, and showed that misexpression of this transcriptional repressor is the likely molecular cause underlying the evolution of awnlessness in wheat.
Introduction
Awns are stiff, bristle-like appendages characteristic of some grasses (Poaceae) such as wheat, barley, and rice, where they extend from the lemma of the florets (Fig. 1A). Wild-type wheat (Triticum aestivum L.) is awned, as are its progenitors and wild forms, while cultivated wheat incorporates awned, awnletted, and awnless forms. In the wild, awns aid in the dispersal of seeds to the site of germination, either by attaching seeds to passing animals, or by balancing the seed dispersal unit as it falls and propelling it into the ground (Harper et al., 1970; Elbaum et al., 2007). Awns can also provide protection from feeding damage by herbivorous animals, which conversely makes them an undesirable characteristic in forage crops (Christensen et al., 1977; Karren et al., 1994; Wallsten et al., 2008). Furthermore, awns have been reported to contribute to carbohydrate storage, water-use efficiency, and photosynthesis, the latter hypothesized to be of particular importance after leaf senescence due to drought or diseases (e.g. Weyhrich et al., 1995; Mozto and Giunta, 2002; Tambussi et al., 2007).
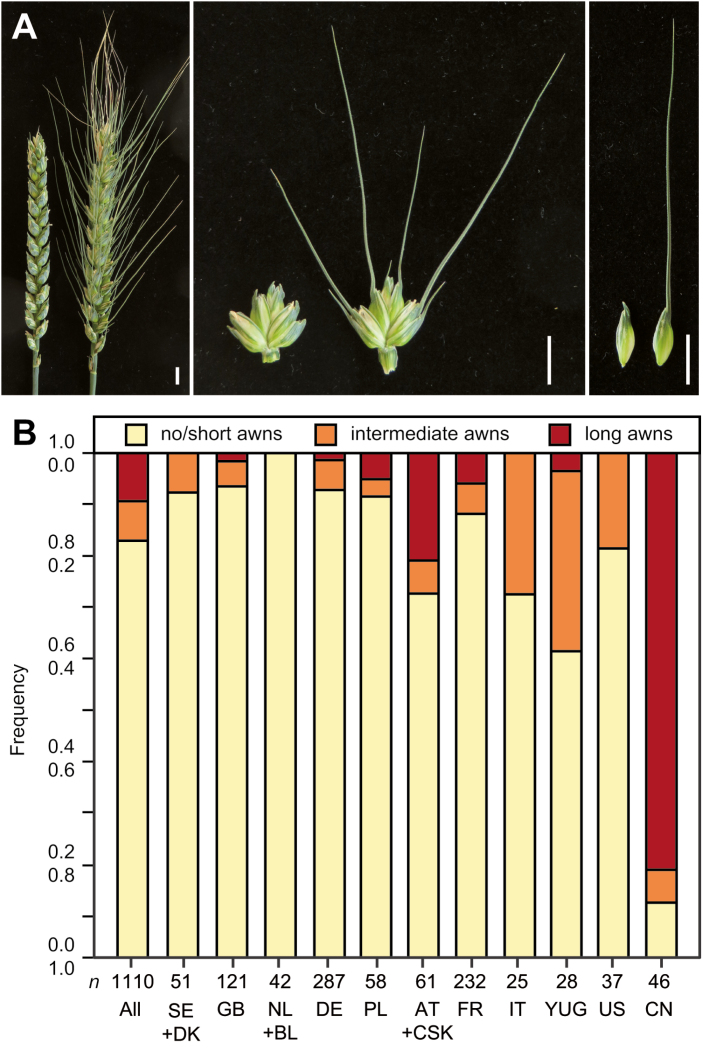
Fig. 1.
Awnedness in wheat. (A) Examples of an awnless and an awned wheat spike, floret, and lemma. The scale bar represents 1 cm. (B) Frequency of awnedness in the panel of 1110 winter wheat cultivars, scored as no/short awns (awns 0–4 mm), intermediate long awns (awns 5–25 mm), and long awns (awns>25 mm), dependent on the cultivars’ country of origin. AT, Austria; BE, Belgium; CN, China; CSK, former Czechoslovakia; DE, Germany; DK, Denmark; FR, France; GB, Great Britain; IT, Italy; NL, The Netherlands; PL, Poland; SE, Sweden; US, USA; YUG, former Yugoslavia, Serbia, Croatia.
Consequently, awned wheats should be the preferred type, but in many global wheat production areas awnless types predominate. It is often stated that awned wheats are better suited to warmer, drier environments while awnless wheats perform better in cooler, temperate regions (Teich, 1982; Rebetzke et al., 2016). However, despite a considerable body of existing studies, the debate around the benefits and costs of awns for grain yield is ongoing and their effects probably depend on environmental conditions. Rebetzke et al. (2016) reported an overall equivalent yield of awned–awnletted sister near isogenic lines, but also showed that awns modify spike morphology, for example increasing grain size but decreasing fertile spikelet number. Based on this, Guo and Schnurbusch (2016) summarized the influence of wheat awns on grain yield as a balance between the costs of awn formation and the benefits of their various functions. The variation in awn length across wheat cultivars may therefore be due to some of their properties and the adaptation of wheat to the different target environments, but may also be a simple consequence of breeders’ preferences. Notably, short or non-existent awns also facilitate harvesting, handling, and storage of grains.
From a developmental genetic perspective, awns provide an interesting model to study the molecular mechanisms underlying the formation of morphological characteristics in grasses, but also the evolution of traits through domestication and breeding. In wheat, three major inhibitors of awn development are known: Tipped 1 (B1), Tipped 2 (B2), and Hooded (Hd), which are located on chromosomes 5A, 6B, and 4A, respectively (Watkins and Ellerton, 1940; McIntosh et al., 2013). Genotypes homozygous for the wild-type b1 b2 hd alleles are fully awned, and as the awn length-reducing alleles act dominantly they have been designated B1, B2, and Hd. The two types of tip-awned wheats were designated Tipped 1 and Tipped 2, which differ in the distribution of awn length along the spikes. Tipped 1 produces very short awns at the base and center of the spike, but longer ones towards the apex, while for Tipped 2 the awn tips are of nearly equal length along the spike, with slightly longer awns, if they present at all, at the center (Watkins and Ellerton, 1940). Hooded also reduces awn length, but leads to curved and deformed awns, often with membranous lateral outgrowth. Importantly, each of these alleles alone will only reduce awn length, while awnletted or awnless phenotypes result from combinations of at least two of these three inhibitors. B1 or B2 with Hd results in very short awns, B1 with B2 in very short or no awns, and the triple combination B1, B2, and Hd appears to be required for complete and stable awnlessness (Yoshioka et al., 2017). B1 has been fine-mapped to a 7.5 cM interval (Mackay et al., 2014), and only recently was a putative candidate gene reported that may underlie the locus (DeWitt et al., 2020; Huang et al., 2020).
In this study, we phenotyped awns in a large panel of 1110 winter wheat cultivars of worldwide origin. We identified one major quantitative trait locus (QTL) on chromosome 5A, corresponding to Tipped 1 (B1), explaining 75% of the genotypic variation. The two alleles at this locus separated awned from awnletted or awnless cultivars. Fine-mapping and expression analysis led to the identification of a putative zinc finger transcription factor, containing an EAR motif characteristic of transcriptional repressors, as the candidate underlying B1. Although no polymorphism was found in the gene itself, a stronger expression signal was detected in B1 genotypes with reduced awn length compared with fully awned wild-type b1 plants. Collectively, our results substantiate the findings from the two recent publications (DeWitt et al., 2020; Huang et al., 2020) and suggest misexpression of B1 to be a likely molecular mechanism underlying the reduction of awn length by Tipped 1 in wheat.
Materials and methods
Plant material and experimental design
This study is based on a panel of 1110 soft winter wheat (T. aestivum L.) cultivars that has been described in detail previously ( Würschum et al., 2015, 2017; Boeven et al., 2016). In brief, it includes cultivars from worldwide origin released during the past decades, but with a focus on cultivars from Europe. Principal coordinate and phylogenetic analyses based on genome-wide markers revealed no major population structure (Boeven et al., 2016). The test locations were Hohenheim [HOH, 48°42'54.4''N, 9°11' 22.6''E, 400 m above sea level (asl)] where 460 genotypes were evaluated in 2012, and in 2013 the entire panel was evaluated in Hohenheim, at Ihinger Hof (IHO, 48°44'42.6''N, 8°55'30.8''E, 493 m asl), and in Eckartsweier (EWE, 48°3'18.3''N, 7°52'16.8''E, 141 m asl). The experiment was conducted in a partially replicated design with a replication rate of 1.25 per location (Williams et al., 2011). Entries were sown in observation plots of two rows and 1.25 m length.
Awns were scored on a scale from 0 to 2, where 0 denotes no or very short awns with a length between 0 mm and 4 mm, 1 denotes awns with a length between 5 mm and 25 mm, and 2 indicates awns with a length >25 mm. Phenotypic data were analyzed as described previously (Würschum et al., 2017). In brief, best linear unbiased estimates (BLUEs) were estimated across all four environments, assuming fixed effects for the genotype. Heritability (h2) was estimated following the approach suggested by Piepho and Möhring (2007; formula [19]). All statistical analyses were performed using the statistical software R (R Development Core Team, 2014) and ASReml-R 3.0 (Gilmour et al., 2009).
Genotypic analysis and association mapping
All lines were genotyped by genotyping-by-sequencing (GBS) at Diversity Arrays Technology (Yarralumla, Australia) using the Wheat GBS 1.0 assay (DArTseq), resulting in silico DArT markers that show the presence or absence of DArT clones and single nucleotide polymorphism (SNP) markers present in some DArT clones. Markers with a minor allele frequency <0.05 were removed, resulting in a total of 23 720 markers for which a map position was available (Li et al., 2015). The CloneIDs of the silico DArT markers were given a ‘D’ prefix and those of the SNP markers a ‘S’ prefix. Marker imputation was done with LinkImpute (Money et al., 2015). The physical positions of the markers were determined by BLASTing the sequences of the DArT clones against the wheat genome (IWGSC RefSeq v1.0).
For association mapping, an additive genetic model was chosen and mapping was done with a mixed model incorporating a kinship matrix as described previously (Würschum et al., 2017). To control for multiple testing, a Bonferroni-corrected threshold of P<0.01 was applied. The total proportion of genotypic variance (pG) explained by the detected QTL was calculated by fitting the significantly associated markers in the order of the strength of their association simultaneously in a linear model. The ratio pG=R2adj / h2, where R2adj refers to the adjusted R2 from the linear model and h2 to the heritability of the trait, yielded the proportion of genotypic variance (Utz et al., 2000). The pG values of individual QTL were accordingly derived from the sums of squares of the QTL (SSQTL) in this linear model. The allele substitution (α) effects were derived as the regression coefficient from models with only the marker under consideration.
Expression analysis
To study the expression of the three candidate genes, TraesCS5A01G542600, TraesCS5A01G542700, and TraesCS5A01G542800, spikes of awned and awnless genotypes were sampled in the field at the stage when the second node of the stem is visible (BBCH stage 32). RNA was extracted with the Qiagen RNeasy® Plant Kit (QIAGEN GmbH, Hilden, Germany) and reverse transcribed into cDNA with the M-MuLV reverse transcriptase from Genaxxon bioscience (Ulm, Germany). The sequences of the three genes were BLASTed against the wheat genome to identify genes with sequence similarity, which for all three identified homoeologous genes on chromosomes 4B and 4D. The obtained sequences were aligned to generate gene-specific primers for the quantitative PCR (qPCR).
For TraesCS5A01G542600, the primers were 5'-TTGTGATCGGGCAGACGTTT-3' as forward primer and 5'-GGCTCCTGCACCAGTTTGT-3' as reverse primer, and the PCR was run at an annealing temperature of 63 °C. For TraesCS5A01G542700, the primers were 5'- GGCTGGAGAAGCTCCTTGTG-3' as forward primer and 5'- GTTGAGATGGCCCTTGTATACAGTC-3' as reverse primer, and the PCR was run at an annealing temperature of 60 °C. For TraesCS5A01G542800, the primers were 5'-CCACCAGAACGCTCACAAGCT-3' as forward primer and 5'-ACGATATCCTGCTCGCCAAGC-3' as reverse primer, and the PCR was run at an annealing temperature of 65 °C. As the control gene, we used Ta2291 (ADP-ribosylation factor) (Paolacci et al., 2009). qPCR was performed on a Roche LightCycler® 480 II with the Genaxxon bioscience GreenMasterMix and two (TraesCS5A01G542600, TraesCS5A01G542700) or four (TraesCS5A01G542800) technical replications per genotype.
Results
Variation of awnedness in wheat
We investigated awnedness in a panel of 1110 winter wheat cultivars of global origin grown in field trials at four environments (Fig. 1A). Awnedness was classified into three categories: no or short awns (awns 0–4 mm), intermediate long awns (5–25 mm), and long awns (>25 mm). We observed a significant genotypic variance and a comparably low genotype-by-environment interaction variance, indicating that awnedness is mainly determined by the genotype (see Supplementary Table S1 at JXB online). Consequently, the heritability was high at 0.94. Most of the cultivars of this panel had no or short awns (82.6%), some had intermediate long awns (7.8%), and 9.6% had long awns (Fig. 1B). This can mainly be attributed to the composition of the panel, with a majority of the cultivars originating in Europe, where the awnless types are predominant. For example, of the British, French, and German cultivars, only 1.7, 6.0, and 1.4%, respectively, had long awns. The highest frequency (82.6%) of long awns was found in the Chinese lines. In general, there appeared to be a higher frequency of the intermediate or long-awned genotypes in the countries of lower latitude, which might indicate an adaptive advantage under the warmer and drier conditions prevalent in these regions. Alternatively, this might be due to breeders’ preferences or the composition of this panel. Nevertheless, the available variation presented a good entry point to study the genetic control underlying this trait.
Identification of Tipped 1 as the major QTL underlying awnedness
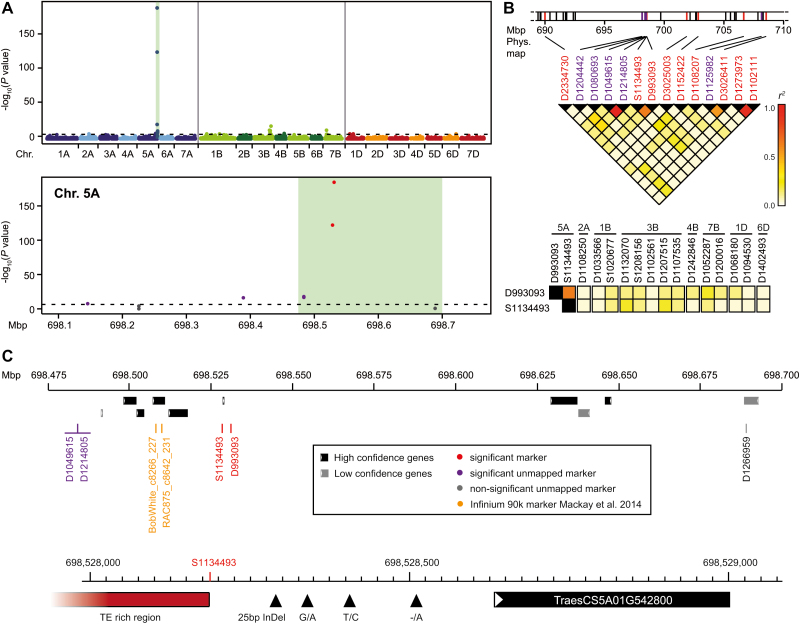
Genome-wide association mapping based on 23 720 mapped markers identified a major QTL at the end of the long arm of chromosome 5A (Fig. 2A, B). Additional putative QTL were found on several other chromosomes, most notably on 3B (Supplementary Table S2). In Chinese Spring, which is awnless and presumably B2 Hd, deletion of the short arm of chromosome 3B resulted in a partially awned phenotype, suggesting another awn development locus located on this chromosome (Ma et al., 2012). Intriguingly, the five markers on chromosome 3B are genetically mapped to this chromosome, but their physical positions were found to be on chromosome 5A, in the region of the major QTL. The linkage disequilibrium (LD) between these markers and those indicative of the major QTL on 5A was rather low, indicating that they present a separate locus and were not identified due to their LD with the major QTL (Fig. 2B). However, owing to the very strong association of the 5A QTL, the latter cannot be ruled out completely.
Fig. 2.
Fine-mapping of the major awnedness inhibitor locus B1 (Tipped 1). (A) Manhattan plots showing results from the genome-wide association mapping and subsequent narrowing down of the target region. (B) Linkage disequilibrium structure at the B1 (Tipped 1) locus on chromosome 5A and between the two most significantly associated markers of the Tipped 1 locus (D993093 and S1134493) and markers of the other putative QTL. (C) Fine-mapping of the Tipped 1 locus and identification of the candidate gene.
Regarding the proportion of explained genotypic variance, one QTL dominates in this panel: the QTL on chromosome 5A explaining 74.6% (Supplementary Table S2). Owing to its chromosomal localization, we concluded that this locus corresponds to the major awnedness inhibitor locus B1 (Tipped 1). It was identified by several significantly associated markers, of which two showed much stronger association signals than all the others (Fig. 2A). The proportion of explained genotypic variance can almost fully be captured by one marker, suggesting a single gene underlying this locus. Analysis of the LD among the significant markers indicated a rapid LD decay, which facilitates fine-mapping (Fig. 2B).
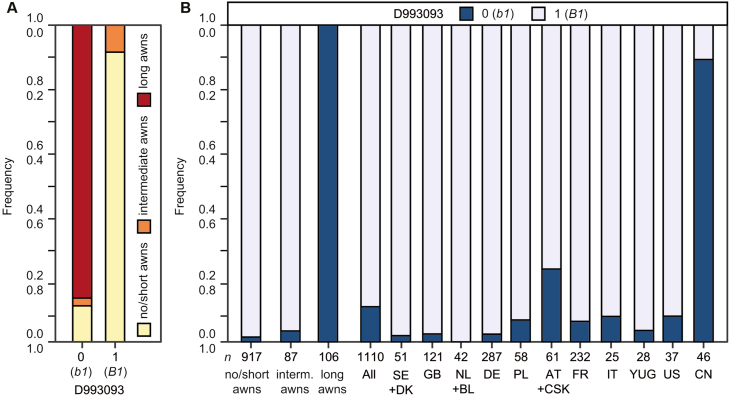
Using the most strongly associated marker D993093 (physical position at base pair 698 530 966) as a proxy for Tipped 1, we found that most of the wild-type b1 plants had long awns (86.2%), few had intermediate long awns (2.4%) possibly due to either B2 or Hd, and some had no or short awns (11.4%), possibly carrying B2 and Hd (Fig. 3). Of the B1 plants, 91.5% had no or short awns and the remainder had intermediate awns. Thus, all 106 cultivars with long awns carried the wild-type b1, while the vast majority of the genotypes with no, short, or intermediate long awns carried B1. The example of Chinese Spring illustrates that awnlessness can also be achieved through B2 and Hd, possibly in combination with other QTL, but does not necessarily require B1. Our results substantiate, however, that B1 is a major and widespread component of awnlessness in winter wheat, particularly for the European countries of origin, but probably even on a global scale (Mackay et al., 2014; DeWitt et al., 2020; Huang et al., 2020).
Fig. 3.
Effect of the B1 (Tipped 1) locus on awnedness in wheat. (A) Awnedness dependent on Tipped 1 (marker D993093) with the wild-type b1 and the awnlessness-conferring B1 alleles. (B) Frequency of b1/B1 (D993993) in different countries of origin.
Fine-mapping of Tipped 1
We narrowed Tipped 1 down to a region at ~698.5 Mbp on chromosome 5A, close to two strongly associated markers recently reported by Mackay et al. (2014) who investigated this trait in a wheat MAGIC population (Fig. 2C; Supplementary Fig. S1). Our two most strongly associated markers flank the candidate gene TraesCS5A01G542800 at 698.531 Mbp, and downstream there is no annotated gene for ~100 kb, followed by a non-significant marker. In the other direction, the association signal also drops off rapidly, with the next two markers being much more weakly associated.
We therefore assessed the five genes TraesCS5A01G542400 to TraesCS5A01G542800 for polymorphisms in genotypes carrying the wild-type b1 allele or presumed to carry the B1 inhibitor allele. No polymorphisms were found in TraesCS5A01G542400 and TraesCS5A01G542500, including their promoter regions. In contrast, TraesCS5A01G542600 and TraesCS5A01G542700 carry non-synonymous polymorphisms in the coding regions, as well as polymorphisms including InDels >10 bp in introns (Supplementary Fig. S2). No polymorphism was found in the coding region of TraesCS5A01G542800, but a 25 bp InDel was identified in the promoter at position –346 bp relative to the start codon, with awnless B1 genotypes carrying the deletion. All polymorphisms formed one haplotype, i.e. followed the same pattern, with one allele in the b1 genotypes and another in the presumed B1 plants. TraesCS5A01G542600 is annotated as a sugar and other transporter, TraesCS5A01G542700 as a universal stress protein family gene with a protein kinase domain, and TraesCS5A01G542800 had no functional annotation. TraesCS5A01G542800 is a rather short gene, consisting of a single exon of 366 bp, and has two homoeologs on chromosomes 4D (TraesCS4D02G476700LC) and 4B (not annotated), of which the latter lacks the C-terminal region. A BLAST search against Arabidopsis, rice, and maize yielded several genes with sequence similarity, which revealed TraesCS5A01G542800 to be a C2H2 zinc finger, having the zinc finger domain as well as the EAR motif at the C-terminal end (Supplementary Fig. S3).
Misexpression reveals a C2H2 zinc finger as the candidate for Tipped 1
For Tipped 1, the awned wild-type b1 allele is recessive and the awn inhibitor B1 allele is dominant. If the mutant version, B1, was a hypomorphic allele, b1 would have to act as an activator of awn formation, in which case it is more likely that b1 would be dominant or the phenotype intermediate with incomplete dominance, as dominance would require a threshold level of activity not reached in heterozygous plants (Supplementary Fig. S4). If, in contrast, B1 was a hypermorphic allele, b1 would have to be a repressor of awn formation, with an increase of this activity resulting in a dominant mode of action with complete repression of awns. This may be caused either by an allele with increased activity compared with the wild-type allele, or by a gene product with normal function, but produced at increased amounts. In addition, B1 could be a neomorphic allele; that is, an allele that has gained a novel function not related to that of the wild-type allele or a novel expression pattern. While the former would be caused by mutations in the gene’s coding region, the latter would require changes in the regulatory region resulting in expression in tissues or developmental stages in which the gene is not normally expressed. Notably, mutant screenings of awnless cultivars have yielded awned mutants (Rakszegi et al., 2010, Dhaliwal et al., 2015), which is less likely for a hypomorphic allele that would have to restore its functionality, but is consistent with B1 being a hypermorphic or a neomorphic allele. Mutations that disrupt the gene’s function would render the misexpressed transcript non-functional and reverse the phenotype, and, in the case of redundancy, without any other effect. Thus, misexpression, either overexpression or ectopic expression, appeared as a likely molecular cause for B1, and we therefore examined the expression of the candidate genes in wheat spikes.
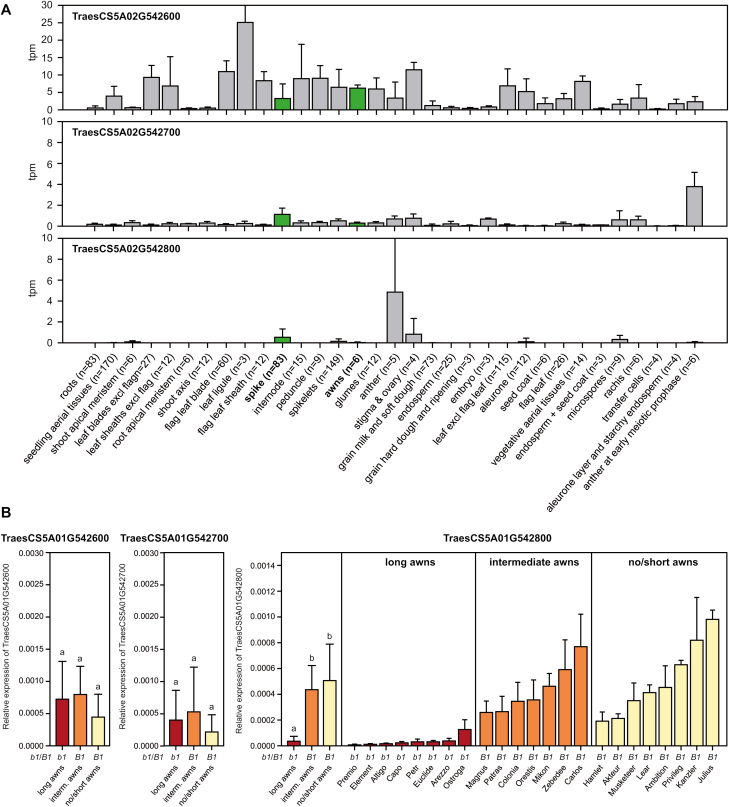
Using available gene expression data in wheat, we found that all three candidate genes are expressed in spikes, as well as in awns (Fig. 4A). Both TraesCS5A01G542600 and TraesCS5A01G542700 appear to be more universally expressed, while TraesCS5A01G542800 showed a more specific expression, being restricted to tissues within spikes.
Fig. 4.
Expression analysis of candidate genes. (A) Expression of the three candidate genes TraesCS5A02G542600, TraesCS5A02G542700, and TraesCS5A02G542800 in different tissues. Data are taken from the Wheat Expression Browser expVIP (Ramírez-González et al., 2018; Borrill et al., 2016). (B) Expression of the three candidate genes in developing spikes of cultivars with long awns, intermediate long awns, or no/short awns. Samples were taken from at least 10 plants per genotype. The whiskers represent the SD and different letters indicate significant differences between the three groups. The presumed allelic state at Tipped 1 (D993993) is indicated underneath.
We next assessed the expression of the three candidate genes in developing, whole wheat spikes at around the stage when the second stem node is visible, using at least 10 plants per genotype. Expression was assessed in cultivars with no or short awns carrying B1, those with intermediate awns, and from wild-type b1 plants with long awns. qPCR revealed no significant difference in expression for TraesCS5A01G542600 and TraesCS5A01G542700 (Fig. 4B; Supplementary Fig. S5). In contrast, for TraesCS5A01G542800, there was a significantly (P<0.01) higher expression in plants with no or short awns carrying B1 compared with wild-type b1 plants. Plants with intermediate long awns have been described as half-awned and considered to be allelomorphic with B1 and b1 [i.e. to carry another allele termed b1a (Watkins and Ellerton, 1940)]. Cultivars with intermediate long awns were found to have expression levels similar to the short-awned genotypes, indicating that they carry the same allele. The different effects of this B1 allele on awn length (debated previously) probably depend on the allelic state at B2 and Hd, and potentially other QTL. Taken together, our results strongly suggest the C2H2 zinc finger protein TraesCS5A01G542800 as the gene underlying the major awnedness inhibitor Tipped 1 in wheat, with misexpression of this gene resulting in a reduction of awn length.
This misexpression might be caused by the 25 bp InDel in the promotor region at position –346 relative to the start codon (Supplementary Fig. S2). Awnless B1 genotypes such as Cadenza carry the deletion, which would mean that removal of this putative regulatory region results in overexpression or ectopic expression and, conversely, that the 25 bp region contains elements that restrict the expression of Tipped 1 (Supplementary Table S3). Sequencing showed this InDel to coincide with the awn phenotype and the presumed b1/B1 genotype in another 26 wheat cultivars (Supplementary Table S4). However, the haplotype structure with several linked polymorphisms does not allow us to conclude that this InDel is causative for the altered expression. Alternatively, another of the identified polymorphisms in the promoter region may cause the observed misexpression or another polymorphism in a regulatory region not assessed here. While further upstream is a region rich in transposable elements, delimiting what might classically be assumed as a promoter, the regulatory elements disrupted in B1 might be further away from the gene itself, in either direction. For example, another 276 bp long InDel is located 575 bp downstream of the gene, again with Chinese Spring having the insertion and Cadenza the deletion. Notably, Huang et al. (2020) also identified the 25 bp InDel in the promoter region and reported it to be highly predictive of awn inhibition, whereas DeWitt et al. (2020) reported sequence variation in the promoter region to not be predictive for the phenotype but rather a 30 bp deletion ~4 kb downstream of the gene. Thus, further research is required to uncover the source behind the observed misexpression of B1.
Discussion
Awns are a characteristic feature of many cereals and, in wheat, both awned and awnless cultivars exist. In rice, most wild species and most Oryza sativa ssp. indica lines produce awns, whereas in O. sativa ssp. japonica awns are partially or completely suppressed as a result of domestication and breeding (Toriba and Hirano, 2014). Genetic studies have indicated multiple genes to be involved in the formation and/or elongation of awns. Awn-1 (An-1) is required for awn elongation and encodes a basic helix–loop–helix protein that regulates cell division and is strongly expressed at the apex of the lemma primordia (Luo et al., 2013). The identification of additional genes suggested that different developmental processes are involved in awn formation (Toriba and Hirano, 2014). In barley, the dominant Hooded (K) replaces the awn by an extra flower of inverse polarity on the lemma. This homeotic transformation is caused by a 305 bp duplication in intron 4 of the homeobox gene HvKnox3, altering the strength and pattern of expression in the lemma (Müller et al., 1995). The barley short awn 2 (lks2) is a natural variant from Eastern Asia that shortens awns by about half and was found to encode a SHORT INTERNODES (SHI) family transcription factor (You et al., 2012). However, the awn inhibitors of wheat do not appear to be orthologs of the awn development genes identified in rice and barley (Yoshioka et al., 2017).
Using a combination of fine-mapping in a large diversity panel and expression analysis, we identified the C2H2 zinc finger TraesCS5A01G542800 as the likely candidate underlying the major awnedness inhibitor Tipped 1 in wheat, the same candidate gene suggested by DeWitt et al. (2020) and Huang et al. (2020). It contains a zinc finger that enables binding to the target sequence and in the C-terminal region an ethylene-responsive element-binding factor (ERF)-associated amphiphilic repression (EAR) motif (Laity et al., 2001). The EAR motif functions in active repression of transcription and is highly conserved in transcriptional regulators with known function as negative regulators across a range of developmental processes (Ohta et al., 2001; Hiratsu et al., 2002; Payne et al., 2004; Kagale and Rozwadowski, 2011). Based on homology, it appears likely that the B1 (Tipped 1) candidate TraesCS5A01G542800 functions as a transcriptional repressor.
While there was no polymorphism within the gene, we found increased expression of TraesCS5A01G542800 in spikes of B1 plants compared with fully awned wild-type b1 genotypes. This indicates that misexpression of this transcriptional repressor may contribute to B1, which is in line with its dominant nature. This misexpression might either directly or indirectly lead to the reduction of awn length and might be caused by polymorphisms in a regulatory region of the gene. Epigenetic modifications are another possible explanation, but appear less likely given the stability of the phenotype. Thus, while we have identified the misexpression of B1, its basis requires further research.
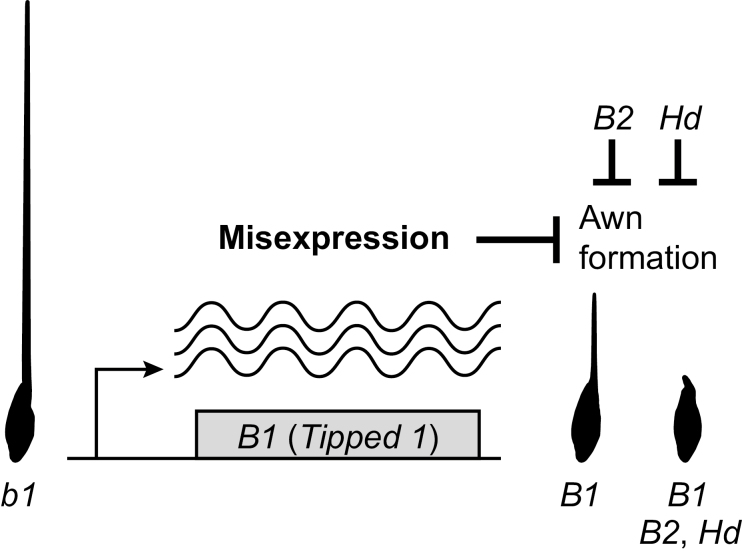
Our model suggests that through misexpression B1 acts on downstream target(s), which reduces the activity of the awn development pathway to result in the formation of intermediate long awns (Fig. 5). This remaining activity may be due to an incomplete penetrance, in which case B1 may be able to fully repress awns if expressed at sufficiently high levels. In combination with B2 and/or Hd, this residual activity is further diminished, resulting in no or only short awns. B2 and Hd may act in parallel to B1, but the epistatic interactions reported for these three awn inhibitor genes (Yoshioka et al., 2017) rather suggest that they act in the same pathway or that their gene products act together. Interestingly, it has recently been shown that the TOPLESS family of co-repressors, that are involved in the regulation of development, hormone signaling, and stress response, interact with the EAR motif (Ke et al., 2015). This, in combination with other results of genetic interactions, co-complex formation, and physical interaction, led to a model where EAR repressors coordinate development and environmental response by regulating expression of their targets via recruitment and action of chromatin-remodeling factors (Kagale and Rozwadowski, 2011).
Fig. 5.
Model for the evolution of awnlessness in wheat by misexpression of B1 (Tipped 1) resulting in a reduction of awn length. B1 in combination with B2 and/or Hooded (Hd) leads to awnless plants.
Taken together, our results show that the awnedness inhibitor locus B1 (Tipped 1) is widespread and probably the major determinant of awnlessness in wheat globally. We identified a C2H2 zinc finger protein with an EAR motif characteristic for transcriptional repressors as a candidate for Tipped 1, and show that its misexpression is consistent with the reduction of awn length in B1 plants. Importantly, these findings lend further support to the results of two very recent studies that also identified the C2H2 zinc finger protein as a candidate underlying the B1 locus. Further characterization of this gene and identification of the other known awnedness inhibitors B2 and Hd will shed further light on the molecular mechanisms underlying the expression of this morphological characteristic in wheat.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Fine-mapping of Tipped 1.
Fig. S2. Polymorphisms in candidate genes.
Fig. S3. Alignment of protein sequences.
Fig. S4. Schematic presentation of B1 function.
Fig. S5. Expression of candidate genes.
Table S1. Summary statistics for awnedness.
Table S2. Marker-explained variance.
Table S3. Analysis of the promotor InDel.
Table S4. Presence or absence of the InDel.
Acknowledgements
This research was funded by the Deutsche Forschungsgemeinschaft under grant number WU 658/1-1. AP was funded under the Designing Future Wheat programme (BBS/E/C/000I0220) of the UK Biotechnology and Biological Sciences Research Council. The authors gratefully acknowledge IPK-Genebank (Gatersleben, Germany) for wheat accessions. The authors would like to thank the International Wheat Genome Sequencing Consortium (www.wheatgenome.org) for providing pre-publication access to IWGSC RefSeq v1.0.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Boeven PH, Longin CF, Würschum T. 2016. A unified framework for hybrid breeding and the establishment of heterotic groups in wheat. Theoretical and Applied Genetics 129, 1231–1245. [DOI] [PubMed] [Google Scholar]
- Borrill P, Ramirez-Gonzalez R, Uauy C. 2016. expVIP: a customizable RNA-seq data analysis and visualization platform. Plant Physiology 170, 2172–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DA, Owen BD, Steacy G, Crowle WL, Mtimuni JP. 1977. Nutritive value of whole crop silage made from seven cereal cultivars. Canadian Journal of Animal Science 57, 537–542. [Google Scholar]
- DeWitt N, Guedira M, Lauer E, et al. 2020. Sequence-based mapping identifies a candidate transcription repressor underlying awn suppression at the B1 locus in wheat. New Phytologist 225, 326–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaliwal AK, Mohan A, Sidhu G, Maqbool R, Gill KS. 2015. An ethylmethane sulfonate mutant resource in pre-green revolution hexaploid wheat. PLoS One 10, e0145227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum R, Zaltzman L, Burgert I, Fratzl P. 2007. The role of wheat awns in the seed dispersal unit. Science 316, 884–886. [DOI] [PubMed] [Google Scholar]
- Gilmour AR, Gogel B, Cullis B, Thompson R. 2009. ASReml user guide release 3.0. Hemel Hempstead, UK: VSN International Ltd. [Google Scholar]
- Harper JL, Lovell PH, Moore KG. 1970. The shapes and sizes of seeds. Annual Review of Ecology, Evolution, and Systematics 1, 327–356. [Google Scholar]
- Hiratsu K, Ohta M, Matsui K, Ohme-Takagi M. 2002. The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Letters 514, 351–354. [DOI] [PubMed] [Google Scholar]
- Huang D, Zheng Q, Melchkart T, et al. 2020. Dominant inhibition of awn development by a putative zinc-finger transcriptional repressor expressed at the B1 locus in wheat. New Phytologist 225, 340–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Schnurbusch T. 2016. Costs and benefits of awns. Journal of Experimental Botany 67, 2533–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Rozwadowski K. 2011. EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6, 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karren DB, Goonewardene LA, Bradley JA. 1994. The effect of feed type on mouth lesions in slaughter cattle. Canadian Journal of Animal Science 74, 571–573. [Google Scholar]
- Ke J, Ma H, Gu X, Thelen A, Brunzelle JS, Li J, Xu HE, Melcher K. 2015. Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors. Science Advances 1, e1500107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laity JH, Lee BM, Wright PE. 2001. Zinc finger proteins: new insights into structural and functional diversity. Current Opinion in Structural Biology 11, 39–46. [DOI] [PubMed] [Google Scholar]
- Li H, Vikram P, Singh RP, et al. 2015. A high density GBS map of bread wheat and its application for dissecting complex disease resistance traits. BMC Genomics 16, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Liu H, Zhou T, et al. 2013. An-1 encodes a basic helix–loop–helix protein that regulates awn development, grain size, and grain number in rice. The Plant Cell 25, 3360–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C-Y, Gao L-Y, Li N, Li X-H, Ma W-J, Appels R, Yan Y-M. 2012. Proteomic analysis of albumins and globulins from wheat variety Chinese Spring and its fine deletion line 3BS-8. International Journal of Molecular Sciences 13, 13398–13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay IJ, Bansept-Basler P, Barber T, et al. 2014. An eight-parent multiparent advanced generation inter-cross population for winter-sown wheat: creation, properties, and validation. G3 4, 1603–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh RA, Yamazaki Y, Dubcovsky J, Rogers J, Morris C, Appels R, Xia XC.2013. https://shigen.nig.ac.jp/wheat/komugi/genes/download.jsp Catalogue of Gene Symbols for Wheat.
- Money D, Gardner KM, Migicovsky Z, Schwaninger H, Zhong GY, Myles S. 2015. LinkImpute—fast and accurate genotype imputation for non-model organisms. G3 5, 2383–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzo R, Giunta F. 2002. Awnedness affects grain yield and kernel weight in near-isogenic lines of durum wheat. Australian Journal of Agricultural Research 53, 1285–1293. [Google Scholar]
- Müller KJ, Romano N, Gerstner O, Garcia-Maroto F, Pozzi C, Salamini F, Rohde W. 1995. The barley Hooded mutation caused by a duplication in a homeobox gene intron. Nature 374, 727–730. [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. 2001. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. The Plant Cell 13, 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne T, Johnson SD, Koltunow AM. 2004. KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 131, 3737–3749. [DOI] [PubMed] [Google Scholar]
- Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M. 2009. Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Molecular Biology 10, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepho HP, Möhring J. 2007. Computing heritability and selection response from unbalanced plant breeding trials. Genetics 177, 1881–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2014. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rakszegi M, Kisgyörgy BN, Tearall K, Shewry PR, Láng L, Phillips A, Bedö Z. 2010. Diversity of agronomic and morphological traits in a mutant population of bread wheat studied in the Healthgrain program. Euphytica 174, 409–421. [Google Scholar]
- Ramírez-González RH, Borrill P, Lang D, et al. 2018. The transcriptional landscape of hexaploid wheat across tissues and cultivars. Science 361, eaar6089.30115782 [Google Scholar]
- Rebetzke GJ, Bonnett DG, Reynolds MP. 2016. Awns reduce grain number to increase grain size and harvestable yield in irrigated and rainfed spring wheat. Journal of Experimental Botany 67, 2573–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambussi EA, Bort J, Guiamet JJ, Nogués S, Araus JL. 2007. The photosynthetic role of ears in C3 cereals: metabolism, water use efficiency and contribution to grain yield. Critical Reviews in Plant Sciences 26, 1–16. [Google Scholar]
- Teich AH. 1982. Interaction of awns and environment on grain yield in winter wheat (Triticum aestivum L.). Cereal Research Communications 10, 11–15. [Google Scholar]
- Toriba T, Hirano H-Y. 2014. The DROOPING LEAF and OsETTIN2 genes promote awn development in rice. The Plant Journal 77, 616–626. [DOI] [PubMed] [Google Scholar]
- Utz HF, Melchinger AE, Schön CC. 2000. Bias and sampling error of the estimated proportion of genotypic variance explained by quantitative trait loci determined from experimental data in maize using cross validation and validation with independent samples. Genetics 154, 1839–1849. [PMC free article] [PubMed] [Google Scholar]
- Wallsten J, Nadeau E, Bertilsson J, Martinsson K. 2008. Voluntary intake and diet selection by dairy heifers fed ensiled whole-crop barley and oats harvested at different stages of maturity. Livestock Science 122, 94–98. [Google Scholar]
- Watkins AE, Ellerton S. 1940. Variation and genetics of the awn in Triticum. Journal of Genetics 40, 243–270. [Google Scholar]
- Weyhrich RA, Carver BF, Martin BC. 1995. Photosynthesis and water-use efficiency of awned and awnletted near-isogenic lines of hard red winter wheat. Crop Science 35, 172–176. [Google Scholar]
- Williams E, Piepho HP, Whitaker D. 2011. Augmented p-rep designs. Biometrical Journal 53, 19–27. [DOI] [PubMed] [Google Scholar]
- Würschum T, Boeven PHG, Longin CFH, Leiser WL. 2015. Multiply to conquer: copy number variations at Ppd-B1 and Vrn-A1 facilitate global adaptation in wheat. BMC Genetics 16, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würschum T, Langer SM, Longin CFH, Tucker MR, Leiser WL. 2017. A modern Green Revolution gene for reduced height in wheat. The Plant Journal 92, 892–903. [DOI] [PubMed] [Google Scholar]
- Yoshioka M, Iehisa JCM, Ohno R, Kimura T, Enoki H, Nishimura S, Nasuda S, Takumi S. 2017. Three dominant awnless genes in common wheat: fine mapping, interaction and contribution to diversity in awn shape and length. PLoS One 12, e0176148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You T, Yamashita Y, Kanamori H, et al. 2012. A SHORT INTERNODES (SHI) family transcription factor gene regulates awn elongation and pistil morphology in barley. Journal of Experimental Botany 63, 5223–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.