Abstract
Long non-coding RNA (lncRNA) LINC00473 plays a carcinogenic role in a variety of different tumor types. Nevertheless, the mechanisms through which LINC00473 regulates the radiosensitivity of esophageal squamous cell carcinoma (ESCC) cells remains elusive. In the present study, reverse transcription-quantitative PCR was used to quantify the expression of LINC00473, microRNA (miRNA/miR)-497-5p and cell division cycle 25A (CDC25A) in ESCC tissues. The association between LINC00473 expression and the clinicopathological characteristics of patients with ESCC was also assessed. Furthermore, Cell Counting kit-8 and colony formation assays were carried out to monitor the proliferation of ESCC cells exposed to X-ray radiation. A dual-luciferase reporter assay was also conducted to analyze the interaction between LINC00473 and miR-497-5p, as well as the interaction between CDC25A and miR-497-5p. The findings of the present study demonstrated that in ESCC tissues and cells, the expression levels of LINC00473 and CDC25A were significantly upregulated, while the expression of miR-497-5p was downregulated. The high expression level of LINC00473 was associated with a higher T stage, lymph node metastasis stage and a lower tumor differentiation grade in patients with ESCC. Following irradiation, transfection with miR-497-5p mimics reduced the promoting effect of LINC00473 overexpression on ESCC cell proliferation, and partially impeded the resistance of ESCC cells to X-ray radiation induced by LINC00473 overexpression. Moreover, transfection with miR-497-5p inhibitors partially alleviated the inhibitory effects of LINC00473 knockdown on cellular proliferation, and partly reversed the sensitivity of cells to X-ray irradiation induced by LINC00473 knockdown. Furthermore, it was confirmed that miR-497-5p was able to bind LINC00473 and the 3′-untranslated region of CDC25A. On the whole, the findings of the present study demonstrate that LINC00473 reduces the radiosensitivity of ESCC cells by modulating the miR-497-5p/CDC25A axis.
Keywords: LINC00473, microRNA-497-5p, cell division cycle 25A, esophageal squamous cell carcinoma, radiosensitivity
Introduction
Esophageal carcinoma is one of the most life-threatening malignancies worldwide (1,2). Esophageal squamous cell carcinoma (ESCC) is a major histological type of esophageal carcinoma, and accounts for ~70-80% of esophageal cancer cases in China (3). Radiotherapy is one of the preferred treatment options for patients with advanced ESCC (4). However, the radioresistance of ESCC hinders the radiotherapeutic effect, which can result in treatment failure (5). Therefore, the detailed mechanisms responsible for the radioresistance of ESCC cells require urgent elucidation.
Long non-coding RNAs (lncRNAs) are a group of highly conserved transcripts of >200 nucleotides, with no protein coding ability (6-8). An increasing number of studies have documented the involvement of lncRNAs in regulating the biological behaviors of cancer cells (9,10). lncRNAs can be categorized as oncogenes and/or tumor suppressors depending on the specific malignancy (11,12), and the aberrant expression of lncRNAs can be used to evaluate the tumorigenesis, progression and relapse of malignant tumors. For example, LINC01133 can be used to predict the progression of ESCC with high sensitivity and specificity (13), and the upregulation of lncRNA HOX transcript antisense RNA indicates a poor prognosis and early-onset metastasis in patients with ESCC (14). Furthermore, lncRNAs have been demonstrated to modulate the radiosensitivity of ESCC cells. Specifically, lncRNA FAM201A induces the radioresistance of ESCC cells, which results in a reduced overall survival time (15); in addition, LINC00473 may reduce the radiotherapeutic efficacy in patients with ESCC (16). However, the underlying mechanisms through which LINC00473 regulates the radiosensitivity of ESCC cells remain to be elucidated.
MicroRNAs (miRNAs or miRs) are a class of small non-coding RNAs of 18-25 nucleotides in length that specifically bind to the 3-untranslated region (3′-UTR) of their target genes, resulting in translation inhibition or mRNA degradation. In this manner, miRs are able to regulate the expression of multiple genes (17,18). The abnormal expression of miRs has been closely associated with the development and progression of tumors. miRs have thus been shown to play pivotal roles in the biological functions of cancer cells, such as differentiation, the stress response, proliferation, apoptosis and sensitivity to irradiation (19). Various miRs of the miR-15/16 family, including miR-15a-3p, miR-16-1-3p and miR-497, exhibit anticancer behaviors (20-26). For instance, miR-497 has been shown to suppress the malignant behavior of papillary thyroid cancer cells by reversely modulating the expression of Yes associated protein 1 (YAP1) (22); it has also been shown to inhibit the angiogenesis and metastasis of hepatocellular carcinoma by impeding the actions of vascular endothelial growth factor A and astrocyte elevated gene-1 (23). Notably, miR-497 can influence the DNA damage response (DDR) and facilitate the radiosensitivity of prostate cancer cells by inhibiting the expression of Wee1-like protein kinase (24). Nonetheless, the exact role and mechanisms of action of miR-497-5p in the radio-resistance of ESCC cells remain elusive.
Cell division cycle 25A (CDC25A), a specific protein phosphatase, is a key regulator of the cell cycle (27). The human CDC25A protein possesses 524 amino acid residues and has 2 distinctive regions; the N-terminal domain and the C-terminal catalytic domain (28). CDC25A is also involved in regulating the radiosensitivity of a variety of malignancies, including prostate and colorectal cancer (29,30), although the CDC25A-associated mechanisms of action in ESCC remain unclear.
LINC00473, miR-497-5p and CDC25A conduct crucial roles in the development of malignancies, and bioinformatics analyzes suggest that binding sites may exist between them. Based on these predictions, the present study aimed to elucidate the effectss of LINC00473 on the radiosensitivity of ESCC cells via miR-497-3p/CDC25A, the results of which may aid in the optimization of radiotherapeutic regimens for patients with ESCC.
Materials and methods
Tissue collection
All patients provided written informed consent for the collection and use of their tissue samples. The present study was approved by the Ethics Review Committee of the Third People's Hospital of Linyi (Linyi, China). In total, 46 pairs of ESCC and adjacent-normal tissues were collected from the Department of Oncology at the Third People's Hospital of Linyi between April, 2016 and March, 2018. All tumors were pathologically confirmed as ESCC, and no other malignant tumors were identified. None of the patients had received neoadjuvant therapy (chemotherapy or radiotherapy) prior to surgery.
Bioinformatics prediction analysis
The StarBase database (starBase, v2.0, http://starbase.sysu.edu.cn/) was employed to identify potential miR-lncRNA binding partners. TargetScan (http://www.targetscan.org/vert_72/) was then used to determine the downstream target genes of these miRs.
Cells and cell culture
Human ESCC cell lines (KYSE-30, KYSE-180, KYSE-150 and TE-5) and a normal esophageal epithelial cell line (Het-1A) were procured from the Shanghai Institute of Biochemistry and Cell Biology. All the cell lines were cultured in DMEM (HyClone; GE Healthcare life Sciences) containing 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin (all Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C (5% CO2). The media were replaced at 3-4-day intervals, and the cells were sub-cultured using the trypsinization method (0.25%; Amerecesco, Inc.).
Transfection
The empty pcDNA vector (NC), pcDNA-LINC00473 (LINC00473), short hairpin (sh)RNA negative control (sh-NC), shRNAs targeting LINC00473 (sh-LINC00473), miR control (miR-NC), miR-497-5p mimic and miR-497-5p inhibitor were designed and constructed by Shanghai GenePharma Co., Ltd. KYSE-30 and TE-5 cells were seeded into a 24-well plate (3×105 cells/well) and cultured for 24 h at 37°C (5% CO2), prior to transfection using Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.). All transfections were performed using a final concentration of 60 nM of miR-497-5p mimics and 100 nM of miR-497-5p inhibitor and sh-LINC00473. Reverse transcription-quantitative (RT-q)PCR was conducted to confirm transfection efficiency 24 h post-transfection, and the subsequent experiments were then performed.
Ionizing irradiation
Transfected ESCC cells were harvested in the logarithmic phase, and irradiated using a linear accelerator (Varian Medical Systems) at room temperature. The cells were exposed to various doses of radiation (0, 2, 4, 6 and 8 Gy) for 24-96 h prior to subsequent analysis. For the time-course experiment, cells were irradiated with X-rays at a dose of 6 Gy, and were then collected for immediate RT-qPCR analysis; RT-qPCR was conducted every 3 h within a 24 h period.
RT-qPCR
Total RNA was isolated from the ESCC tissues and cultured cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.); 1 μg total RNA was reverse transcribed into cDNA using the SuperScript™ First Strand cDNA System (Invitrogen; Thermo Fisher Scientific, Inc.), and qPCR was subsequently performed. SYBR-Green Premix Ex Taq II (Takara) and ABI 7500 real-time PCR system (Applied Biosystems) was applied for qPCR. The qPCR cycling conditions were as follows: 95°C for 2 min, then 40 cycles at the conditions of 95°C for 15 sec, 60°C for 15 sec, and 68°C for 20 sec. The relative expression levels of LINC00473 and miR-497-5p were calculated using the 2−ΔΔCq method (31). The primers sequences were as follows: LINC00473 forward, 5′-GATGGAAAGGAGGGAAGG-3′ and reverse, 5′-CACAGTGGGTCCAGGGTT-3′; miR-497-5p forward, 5′-CCTTCAGCAGCACACTGTGG-3′ and reverse, 5′-CAGTGCAGGGTCCGAGGTAT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; and β-actin forward; 5′-TTCGAGCAAGAGATGGCCA-3′ and reverse, 5′-TACATGGTGGTGCCGCC-3′.
Western blot analysis
The ESCC tissues and cells were lysed with RIPA buffer (Beyotime Institute of Biotechnology) supplemented with protease inhibitors. Following high-speed centrifugation (10,000 × g, 4°C, 5 min), the supernatants were collected and the proteins were denatured in a water bath at 100°C for 10 min; the protein concentrations were determined using the bicinchoninic acid method. A total of 10 μl total protein extracts were loaded into each well, and were separated by SDS-PAGE using 12% gels, and then transferred to polyvinylidene fluoride membranes. The membrane was washed once in PBS with 0.2% Tween-20 (PBST) and then blocked with 5% non-fat milk for 1 h at room temperature. After washing in TBST solution, the membranes were incubated overnight with primary antibodies against CDC25A (cat. no. ab79252; Abcam, 1:1,000) and β-actin (cat. no. ab20272; Abcam; 1:100) on a shaking platform at 4°C. The membranes were then rinsed with TBS-T solution and incubated with goat anti-rabbit IgG H&L (cat. no. ab150077; Abcam; 1:1,000) at room temperature for 1 h. Following a final wash with TBS-T (3 times), signals were developed with Hypersensitive ECL Chemiluminescent Substrate (Hubei Biossci Biotechnology Co, Ltd.) and exposed to X-ray films.
Cell counting kit-8 (CCK-8) assay
The KYSE-30 and TE-5 cells were harvested in the logarithmic phase and adjusted to a density of 1×104/ml; 100 μl cell suspension was then inoculated into the wells of a 96-well plate as appropriate, and cultured at 37°C for 24 h. Subsequently, 10 μl enhanced CCK-8 solution (Hubei Biossci Biotechnology Co, Ltd.) was added and the plate was returned to the incubator for a further 1 h. The absorbance (OD value) of each well was determined at 450 nm using a plate reader (Multiscan FC; Thermo Fisher Scientific, Inc.) at 24, 48, 72 and 96 h.
Colony formation assay
Transfected ESCC cells were seeded into 6-well plates at a density of 1×103 cells/well and irradiated at a specified single dose (0, 2, 4, 6 or 8 Gy). The culture medium was then discarded and the cells were carefully washed twice with PBS. Following incubation at 37°C (5% CO2) for 2 weeks, the cells were fixed with 10% methanol, stained with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA) for 15 min and dried at room temperature. Subsequently, the number of colonies was recorded under a light microscope (Olympus Corporation).
Dual-luciferase reporter gene assay
Wild-type (WT) or mutant-type (MUT) LINC00473 was subcloned into a pGL3 Basic vector (Promega Corporation). The cells were then seeded into a 24-well plate at a density of 5×103 cells/well, and the Dual-Luciferase® Reporter Assay System (Promega Corporation) was used to determine luciferase activity. miR-497-5p or miR-NC were co-transfected with WT or MUT reporter vectors into KYSE-30 and TE-5 cells, respectively, using Lipofectamine® 3000. Luciferase activity was assessed at 48 h post-transfection. The results were normalized through comparison with Renilla luciferase activity.
Statistical analysis
All statistical analyzes were conducted using SPSS 20.0 (IBM Corp.), and all data are presented as the means ± standard deviation. Whether the data are normally distributed or not was examined using the Kolmogorov-Smirnov test. For normally distributed data, an unpaired or paired t-test was used to compare the data between 2 groups. Comparisons among ≥3 groups were conducted with one-way ANOVA. If the data exhibited significant differences, Tukey's post hoc test was then performed to compare the data between groups. For data that were not normally distributed, comparisons between 2 groups were performed by a paired sample Wilcoxon signed-rank test. Pearson's correlation coefficient was used to evaluate the correlation between the expression levels of the genes in the ESCC samples. A Chi-squared test was used to analyze the association between the expression of LINC00473 and the patient clinicopathological characteristics. P<0.05 was considered to indicate a statistically significant difference.
Results
Association between the expression levels of LINC00473, miR-497-5p and CDC25A in ESCC tissues
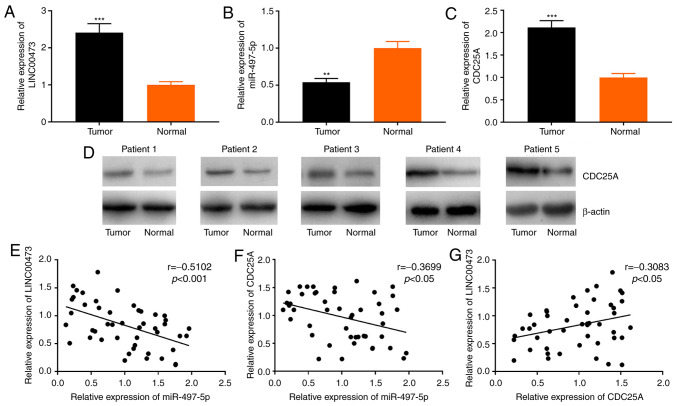
RT-qPCR was conducted to determine the association between the expression levels of LINC00473, miR-497-5p and CDC25A mRNA in 46 paired ESCC and adjacent-normal tissue samples. The expression levels of LINC00473 and CDC25A mRNA were significantly higher, while miR-497-5p expression was significantly lower in ESCC tissues, compared with those in the adjacent normal tissues (Fig. 1A-C). Additionally, western blot analysis was used to assess CDC25A protein expression in the ESCC and adjacent tissues of 5 randomly selected patient samples; the results indicated that CDC25A was also upregulated at the protein level (Fig. 1D). LINC00473 expression was also found to inversely correlate with that of miR-497-5p (Fig. 1E; r=-0.5102, P<0.001). Moreover, miR-497-5p expression negatively correlated with CDC25A expression (Fig. 1F; r=−0.3699, P<0.05), while LINC00473 expression positively correlated with CDC25A expression (Fig. 1G; r=0.3083, P<0.05). These data suggest a possible regulatory association among LINC00473, miR-497-5p and CDC25A.
Figure 1.
Association between the expression levels of LINC00473, miR-497-5p and CDC25A. (A-C) LINC00473, miR-497-5p and CDC25A miRNA expression was detected in ESCC tissues by RT-qPCR. (D) CDC25A expression in the cancerous and adjacent tissues of 5 patients with ESCC was detected by western blotting. (E) Expression of LINC00473 inversely correlated with that of miR-497-5p in 46 cases of ESCC. (F) miR-497-5p expression negatively correlated with that of CDC25A. (G) The expression of CDC25A mRNA positively correlated with that of LINC00473. **P<0.01 and ***P<0.001 vs. normal tissue. CDC25A, cell division cycle 25A; ESCC, esophageal squamous cell cancer; miR, microRNA.
LINC00473 expression is associated with multiple patho- logical indicators in patients with ESCC
The association between the expression of LINC00473 and the clinicopathological indexes of patients with ESCC was also analyzed. High expression levels of LINC00473 in tumor tissues were found to be significantly associated with local lymph node metastasis, a low degree of differentiation and a higher T stage in patients with ESCC; however, they were not associated with sex, age, tumor size or a history of smoking (Table I). These findings suggest that high expression of LINC00473 may be involved in the progression of ESCC.
Table I.
Associations between LINC00473 expression and the clinicopathological characteristics of patients with esophageal squamous cell cancer.
| Clinicopathological indicators | No. of patients | Relative expression level of LINC00473
|
P-value | |
|---|---|---|---|---|
| High | Low | |||
| All cases | 46 | 24 | 22 | |
| Age, years | ||||
| ≥60 | 35 | 19 | 16 | 0.609 |
| <60 | 11 | 5 | 6 | |
| Sex | ||||
| Male | 34 | 16 | 18 | 0.242 |
| Female | 12 | 8 | 4 | |
| Smoking | ||||
| Smoker | 29 | 14 | 15 | 0.489 |
| Never smoked | 17 | 10 | 7 | |
| Tumor size (d/cm) | ||||
| >3 cm | 25 | 15 | 10 | 0.246 |
| ≥3 cm | 21 | 9 | 12 | |
| T classification | ||||
| T1 + T2 | 20 | 7 | 13 | 0.041 |
| T3 + T4 | 26 | 17 | 9 | |
| Lymph node metastasis | ||||
| N0-N1 | 17 | 5 | 12 | 0.018 |
| N2-N3 | 29 | 19 | 10 | |
| Tumor differentiation | ||||
| Poor/medium | 24 | 16 | 8 | 0.039 |
| High | 22 | 8 | 14 | |
LINC00473 expression is increased and miR-497-5p expression is decreased in ESCC cells exposed to X-ray radiation
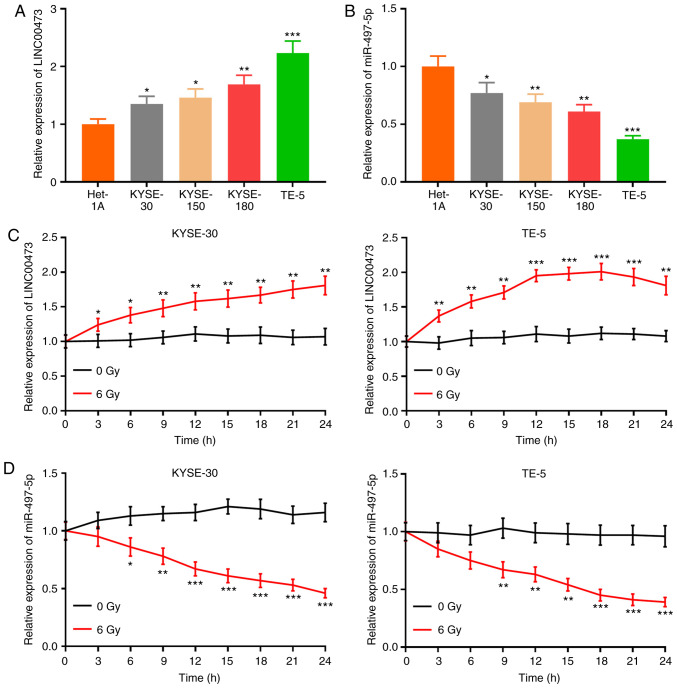
RT-qPCR was used to quantify the expression levels of LINC00473 and miR-497-5p in 4 ESCC cell lines (KYSE-30, KYSE-150, KYSE-180 and TE-5). Compared with Het-1A cells, the expression of LINC00473 was significantly higher in the ESCC cell lines, while the expression of miR-497-5p was notably decreased (Fig. 2A and B). The KYSE-30 and TE-5 cells were irradiated with 6 Gy X-rays, and the expression of LINC00473 was detected every 3 h by RT-qPCR. The results revealed that the expression of LINC00473 in the irradiated KYSE-30 and TE-5 cells was significantly higher than that in the control group (0 Gy) (Fig. 2C and D), and that following irradiation, the expression of miR-497-5p was markedly suppressed (Fig. 2E and F). These results indicated that the expression of LINC00473 and miR-497-5p was inversely affected by X-ray irradiation.
Figure 2.
LINC00473 expression is increased, while miR-497-5p expression is decreased following X-ray irradiation. RT-qPCR was performed to detect the expression of (A) LINC00473 and (B) miR-497-5p in 4 esophageal squamous cell cancer cell lines (KYSE-30, KYSE-150, KYSE-180 and TE-5) and a normal esophageal epithelial cell line (Het-1A). (C) LINC00473 and (D) miR-497-5p expression in KYSE-30 and TE-5 cells was detected every 3 h following X-ray irradiation (6 Gy). *P<0.05, **P<0.01 and ***P< 0.001 vs. Het-1A cells or control (0 Gy). miR, microRNA.
LINC00473 plays a crucial role in reducing the radiosensitivity of ESCC cells
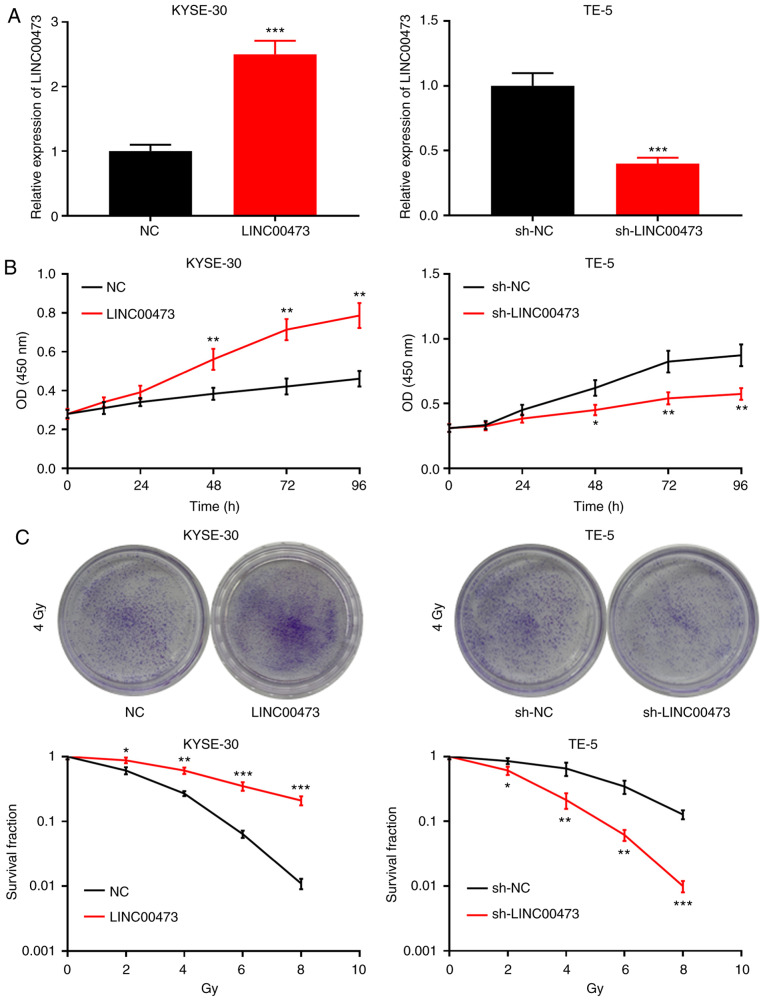
The results of the aforementioned experiments illustrated that the expression of LINC00473 was upregulated in the irradiated ESCC cells. The expression level of LINC00473 was lowest in the KYSE-30 cells, and highest in the TE-5 cells; therefore, to further investigate the role of LINC00473 in the radiosensi-tivity of ESCC cells, KYSE-30 and TE-5 cells were selected to construct LINC00473 overexpression and knockdown models, respectively. The KYSE-30 cells were transfected with a LINC00473 overexpression plasmid, and the TE-5 cells with LINC00473 shRNA; RT-qPCR indicated the successful establishment of each model (Fig. 3A). CCK-8 assay was subsequently performed to assess the proliferative capacity of each cell line; the results revealed that LINC00473 over-expression significantly increased KYSE-30 cell proliferation at 48, 72 and 96 h, while LINC00473 knockdown inhibited the proliferation of TE-5 cells, demonstrating that LINC00473 enhanced the proliferative ability of the ESCC cells (Fig. 3B). A colony formation assay was then performed, and the results indicated that LINC00473 overexpression and knockdown increased and decreased the number of colony-forming units, respectively (Fig. 3C). Collectively, it was thus concluded that LINC00473 enhances the proliferative ability and reduces the sensitivity of ESCC cells to irradiation.
Figure 3.
LINC00473 enhances the viability of esophageal squamous cell cancer cells and promotes resistance to radiotherapy. (A) KYSE-30 cells were transfected with pcDNA-NC and pcDNA-LINC00473, and TE-5 cells were transfected with sh-NC and sh-LINC00473. RT-qPCR was conducted to confirm transfection efficiency. (B) KYSE-30 and TE-5 cell viability was assessed using the Cell Counting kit-8 assay at 0, 24, 48 and 96 h. (C) Following X-ray irradiation (0, 2, 4, 6 and 8 Gy), the colony number was counted using a colony formation assay. *P<0.05, **P<0.01 and ***P<0.001 vs. NC. sh, short hairpin (RNA); NC, negative control.
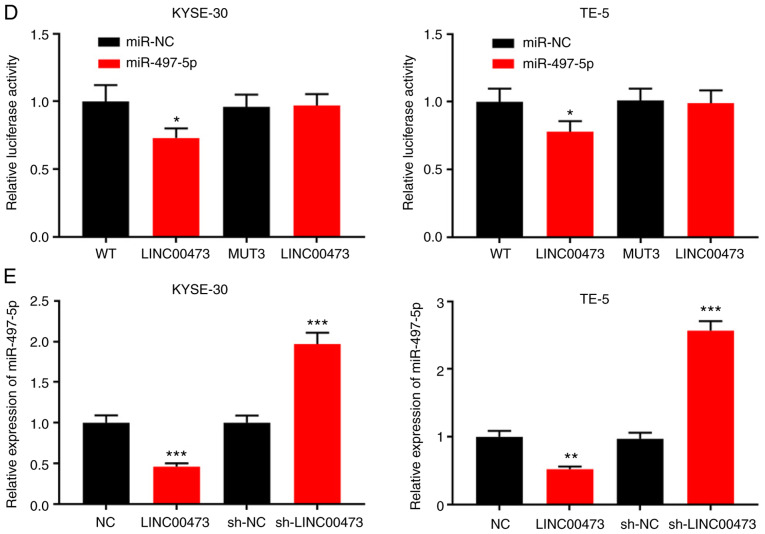
LINC00473 directly interacts with and negatively regulates the expression of miR-497-5p
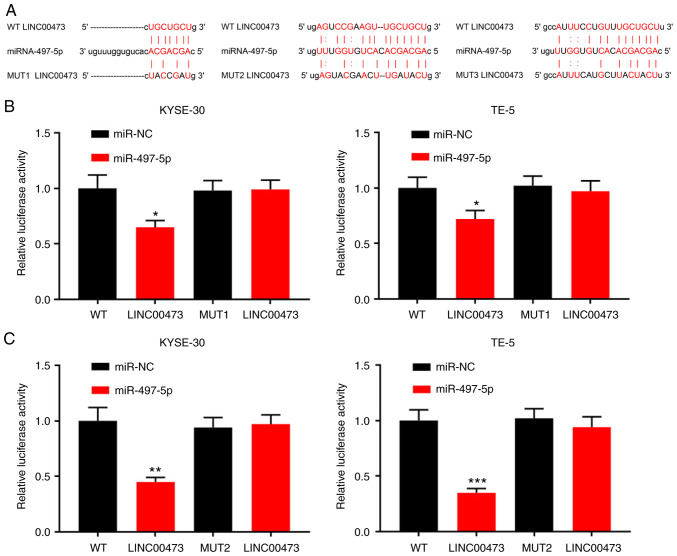
Bioinformatics analysis (starBase, v2.0, http://starbase.sysu.edu.cn/) predicted miR-497-5p to be a candidate target of LINC00473, and the potential binding site is illustrated in Fig. 4A. Using a dual-luciferase assay, following the transfection of miR-497 mimics into ESCC cells (Figs. S1 and S2), miR-497-5p overexpression was shown to markedly decrease the luciferase activity of pGL3-LINC00473-WT, whereas it did not significantly affect that of pGL3-LINC00473-MUT (Fig. 4B-D). The results suggested that of the three predicted binding fragments, the first and third had similar binding capacities, and the second exhibited the most statistically significant difference (Fig. 4B-D). In addition, the KYSE-30 and TE-5 cells were transfected with pcDNA-LINC00473 or sh-LINC00473. The results suggested that compared with the control group, the expression of miR-497-5p was notably inhibited by LINC00473 overexpression and increased by LINC00473 knockdown, respectively (Fig. 4E). Thus, LINC00473 can directly interact with miR-497-5p and negatively regulate its expression.
Figure 4.
Interaction between LINC00473 and miR-497-5p. (A) Binding sites between LINC00473 and miR-497-5p were predicted by bioinformatics analysis. (B-D) KYSE-30 and TE-5 cells were co-transfected with miR-NC or miR-497-5p and pGL3-LINC00473-WT or pGL3-LINC00473-MUT, and luciferase activity was detected using a luciferase reporter gene assay. Of the three predicted binding fragments, the first and third had similar binding capacities, and the second exhibited the most statistically significant difference. (E) KYSE-30 and TE-5 cells were transfected with pcDNA-NC or pcDNA-LINC00473, and sh-NC or sh-LINC00473, respectively, and the expression of miR-497-5p was detected by RT-qPCR. *P<0.05, **P<0.01 and ***P<0.001 vs. NC and sh-NC. miR, microRNA; sh, short hairpin (RNA); NC, negative control; WT, wildtype; MUT, mutant.
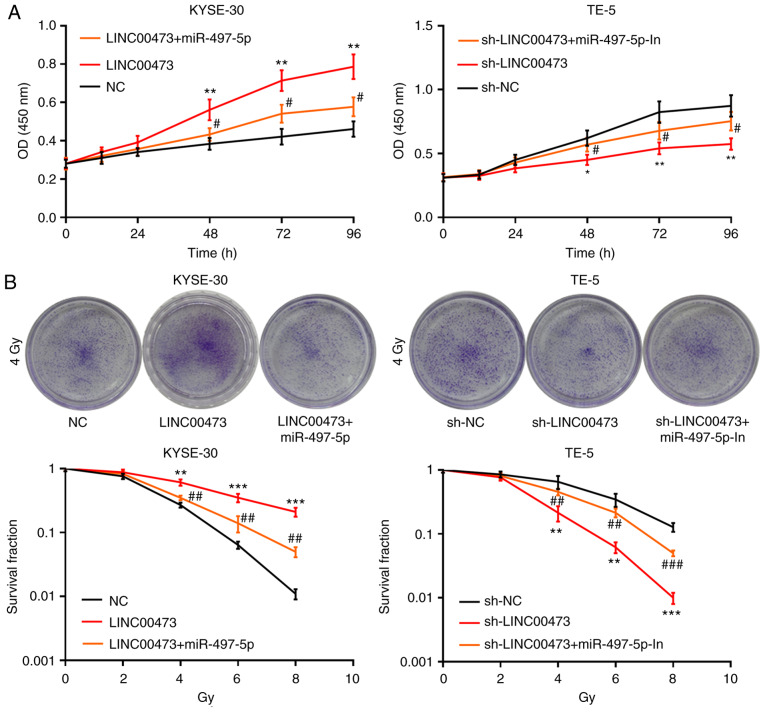
LINC00473 regulates the radiosensitivity of ESCC cells via miR-497-5p
To investigate the potential mechanisms responsible for the radiosensitivity in ESCC cells induced by LINC00473, the KYSE-30 cells were transfected with pcDNA-NC, pcDNA-LINC00473 or pcDNA-LINC00473-miR-497-5p mimics; TE-5 cells were concurrently transfected with sh-NC, sh-LINC00473 or sh-LINC00473-miR-497-5p inhibitors (Figs. S1 and S2). The inhibitory effects of LINC00473 knockdown on ESCC cell viability were weakened by transfection with miR-497-5p inhibitors, while transfection with miR-497-5p mimics attenuated the promoting effects of LINC00473 on cell proliferation (Fig. 5A). Moreover, transfection with miR-497-5p mimics was shown to partially counteract the radioresistance of ESCC cells (2, 4, 6 and 8 Gy) induced by LINC00473, while transfection with miR-497-5p inhibitors exerted the opposite effect (Fig. 5B). Collectively, these data indicate that LINC00473 reduces the radiosensitivity of ESCC cells via miR-497-5p.
Figure 5.
LINC00473 influences the viability and radiosensitivity of esophageal squamous cell cancer cells by regulating miR-497-5p. Following pcDNA-NC, pcDNA-LINC00473 or pcDNA-LINC00473/miR-497-5p mimic transfection into KYSE-30 cells, and sh-NC, sh-LINC00473 or sh-LINC00473/miR-497-5p inhibitor transfection into TE-5 cells, cellular viability/proliferation was detected using the Cell Counting Kit-8 assay (A). Following X-ray irradiation (0, 2, 4, 6 and 8 Gy) for 2 weeks, the survival ratio of colony-forming cells was detected using a colony formation assay (B). *P<0.05, **P<0.01 and ***P<0.001, NC vs. LINC00473 and sh-NC vs. sh-LINC0473. #P<0.05, ##P<0.01 and ###P<0.001, LINC00473 vs. LINC00473 + miR-497-5p and sh-LINC00473 vs. sh-LINC00473 + miR-497-5p inhibitors. miR, microRNA; NC, negative control; sh, short hairpin (RNA).
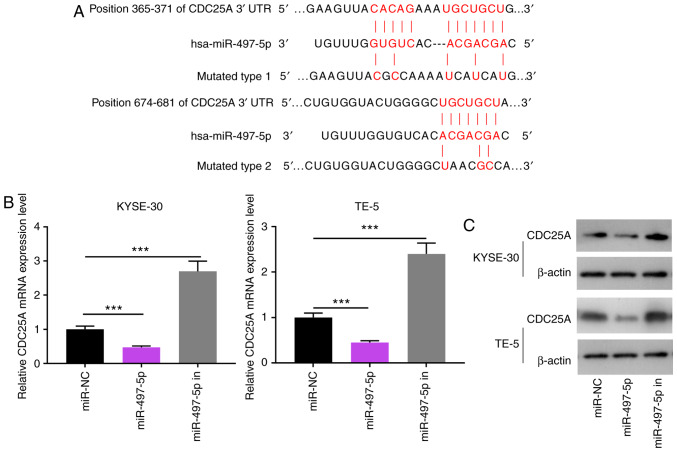
miR-497-5p directly targets the 3′-UTR of CDC25A
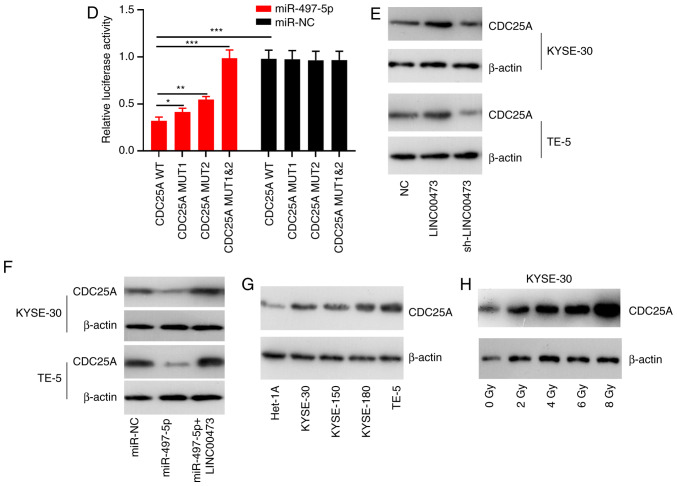
TargetScan predicted that CDC25A was one of the candidate target genes for miR-497-5p, and the speculative binding sites are displayed in Fig. 6A. The results of RT-qPCR and western blot analysis revealed that the mRNA and protein levels of CDC25A were notably decreased in the KYSE-30 and TE-5 cells following transfection with miR-497-5p mimics, respectively; on the other hand, miR-497-5p inhibitors induced the upregulation of CDC25A expression in ESCC cells at both the mRNA and protein levels (Fig. 6B and C). The results of the luciferase reporter assay indicated that miR-497-5p could specifically bind to the 3′UTR of CDC25A (Fig. 6D). The results of western blot analysis revealed that the expression of CDC25A was markedly elevated in the KYSE-30 and TE-5 cells following LINC00473 overexpression, but was reduced after LINC00473 was knocked down (Fig. 6E). Additionally, LINC00473 overexpression reversed the inhibitory effect on CDC25A expression induced by miR-497-5p (Fig. 6F). Furthermore, the expression level of CDC25A was markedly upregulated in the ESCC cell lines, compared with the Het-1A cells (Fig. 6G), and its expression in ESCC cells was induced by irradiation (Fig. 6H). Collectively, these data confirm that CDC25A is a downstream gene of miR-497-5p, and that its expression is directly and inversely regulated by miR-497-5p, though indirectly and positively modulated by LINC00473.
Figure 6.
Interaction between miR-497-5p and CDC25A. Binding sites between miR-497-5p and CDC25A were predicted using bioinformatics analysis. (A) Reverse transcription-quantitative PCR was used to quantify the expression of CDC25A miRNA in KYSE-30 and TE-5 cells transfected with miR-NC or miR-497-5p. (B) RT-qPCR analysis of CDC25A expression in KYSE-30 and TE-5 cells transfected with miR-497 mimics or miR-497 inhibitors. (C) Western blot analysis was performed to detect CDC25A expression in KYSE-30 and TE-5 cells transfected with miR-497 mimics or miR-497 inhibitors. (D) The luciferase activity of KYSE-30 and TE-5 cells co-transfected with miR-497-5p and pGL3-miR-497-5p-WT or pGL3-miR-497-5p-MUT. (E) Western blot analysis was performed to detect CDC25A expression in KYSE-30 and TE-5 cells transfected with pcDNA-LINC00473 or sh-LINC00473. (F) Western blot analysis was performed to detected CDC25A expression in KYSE-30 and TE-5 cells transfected with miR-497-5p alone or miR-975-5p+pcDNA-LINC00473. (G) Western blot analysis was performed to detect CDC25A expression in four esophageal squamous cell cancer cell lines (KYSE-30, KYSE-150, KYSE-180 and TE-5) and a normal esophageal epithelial cell line (Het-1A). (H) Western blot analysis was conducted to detect CDC25A expression in KYSE-30 cells under different doses of irradiation. *P<0.05, **P<0.01 and ***P<0.001. miR, microRNA; CDC25A, cell division cycle 25A; NC, negative control; WT, wild-type; MUT, mutant.
Discussion
Radiotherapy is extensively used for the treatment of advanced malignant tumors, including ESCC (32,33). Nevertheless, the radioresistance of ESCC cells, which is caused by a series of factors, can result in treatment failure (34). Consistent with the findings of a previous study, the results of the present study revealed significantly higher expression levels of LINC00473 in ESCC tissues and cells, compared with paired-adjacent tissues and normal esophageal epithelial cells, respectively (16). In the present study, a high LINC00473 expression in tumor tissues was significantly associated with local lymph node metastasis, a low degree of differentiation and a higher T stage in patients with ESCC. Through further experimentation, LINC00473 was confirmed to reduce the radiosensitivity of ESCC cells by modulating the miR-497-5p/CDC25A axis.
lncRNAs are regarded as key regulators of the proliferation, apoptosis, metastasis and radioresistance of multiple types of tumor cells, including ESCC cells (6-8,35). lncRNAs may act as potential indicators to monitor the progression of malignant tumors (36); for example, LINC00657 promotes the proliferation, migration and invasion, but reduces the radiosensitivity of ESCC cells, and its high expression level is associated with poor prognosis (35). LINC00473 promotes the progression of pancreatic cancer by upregulating the expression of programmed death-ligand 1 (37). LINC00473 knockdown has also been reported to induce the radiosensitization of ECSS cells (16); this is consistent with the results of the present study, which also revealed that X-ray exposure significantly increased the expression of LINC00473 in ESCC cells, compared with non-irradiated cells. A CCK-8 assay also revealed that LINC00473 knockdown largely blocked the viability of ESCC cells, while LINC00473 overexpression enhanced proliferative capacity. Different doses of X-ray irradiation also significantly increased the colony-forming potential of cells overexpressing LINC00473, which was impeded by LINC00473 knockdown. Taken together, these results indicate that LINC00473 enhances the proliferative and colony-forming abilities of ESCC cells, and induces radioresistance.
miRNAs are able to regulate the malignant phenotype of human tumor cells, including their proliferation, invasiveness and resistance to X-rays (17-19,38,39). A growing number of studies have suggested that numerous members of the miR-15/16 family possess an antitumor role in human malignant tumors (20,40). For example, miR-15b can enhance the sensitivity of colorectal cancer cells to radiotherapy, which can be used as a valuable marker for prognosis and treatment outcome (41). It has been demonstrated that miRNAs regulate sensitivity to radiotherapy through the DNA injury mechanism (42). miRNAs are involved in DNA injury in almost all cell types, including its detection, the transmission of injury signals, DNA repair, cell cycle activation and inducing apoptosis (43). This makes miRNAs a potential target for the early detection of radioresistance, which may improve the response of cells to radiotherapy. miR-497 interferes with the cell cycle, DNA synthesis and function, and activates DDR to induce apoptosis (44). In the present study, miR-497-5p was expressed at low levels in ESCC tissues and cells; thus, examining the potential mechanisms of miR-497-5p may provide possible treatment options for patients with ESCC. Common binding sites between LINC00473 and miR-497-5p were subsequently predicted using an online bioinformatics database (StarBase, v2.0, http://starbase.sysu.edu.cn/), and luciferase reporter gene assays confirmed that LINC00473 could sponge miR-497-5p. In addition, LINC00473 overexpression was shown to inhibit the expression of miR-497-5p, while LINC00473 knockdown increased the expression level of miR-497-5p. The viability of ESCC cells was also markedly reduced by the transfection of miR-497-5p mimics into cells overexpressing LINC00473, while an miR-497-5p inhibitor produced opposing effects. Furthermore, colony formation assays revealed that miR-497-5p inhibition reverses the enhanced radiosensitivity caused by LINC00473 knockdown. Thus, it can be concluded that LINC00473 reduces the radiosensitivity of ESCC cells by regulating the expression of miR-497-5p.
Previous studies have highlighted the significance of CDC25A in numerous cellular behaviors, including sensitivity to radiotherapy (45). For example, miR-339-5p enhances ESCC radiosensitivity by inhibiting CDC25A (46). In the present study, binding sites between miR-497-5p and CDC25A were identified, and CDC25A expression was markedly downregulated both at the miRNA and protein levels in ESCC cells transfected with miR-497-5p mimics. miR-497-5p also decreased the luciferase activity of WT CDC25A, but not the mutant type. Furthermore, LINC00473 overexpression in ESCC cells elevated the expression of CDC25A. Hence, CDC25A was concluded to be directly and reversely regulated by miR-497-5p, as well as positively and indirectly by LINC00473.
Although previous studies have reported that LINC000473 in ESCC tissues and cells is significantly upregulated, and the upregulation of LINC00473 indicated radioresistance and a poor prognosis of patients with ESCC (16,47), the findings of the present study highlight a unique lncRNA-regulated mechanism in ESCC, namely that LINC00473 facilitates the radioresistance of ESCC cells through miR-497-5p. These results provide a novel theoretical basis for the diagnosis and treatment of ESCC. There are several limitations in the present study. Firstly, the conclusion of the present study is only based on in vitro experiments, and animal models are required to further validate the results. Secondly, more data are required to prove that in ESCC, miR-497-5p regulated the radiosensitivity of cancer cells by suppressing CDC25A.
Supplementary Data
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The data used to support the findings of this study are available from the corresponding author upon request.
Authors' contributions
XW, WHL and YLW designed the present study. WHL, HYQ, WQW and JX performed the experiments, and analyzed and interpreted the experimental data. WHL, YLW, XW contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All patients provided written informed consent for the collection and use of their tissue samples. Tissues were collected during surgery following approval from the Ethics Review Committee of Third People's Hospital of Linyi (Linyi, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Wang W, Wu D, He X, Hu X, Hu C, Shen Z, Lin J, Pan Z, He Z, Lin H, Wang M. CCL18-induced HOTAIR upregulation promotes malignant progression in esophageal squamous cell carcinoma through the miR-130a-5p-ZEB1 axis. Cancer Lett. 2019;460:18–28. doi: 10.1016/j.canlet.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Gao J, Wang Y, Yang J, Zhang W, Meng K, Sun Y, Li Y, He QY. RNF128 promotes invasion and metastasis via the EGFR/MAPK/MMP-2 pathway in esophageal squamous cell carcinoma. Cancers (Basel) 2019;11:E840. doi: 10.3390/cancers11060840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao XH, Wang D, Wang F, Zhu SC. Comparison of the effect of postoperative radiotherapy with surgery alone for esophagus squamous cell carcinoma patients: A meta-analysis. Medicine (Baltimore) 2018;97:e13168. doi: 10.1097/MD.0000000000013168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo J, Wang W, Tang Y, Zhou D, Gao Y, Zhang Q, Zhou X, Zhu H, Xing L, Yu J. mRNA and methylation profiling of radioresistant esophageal cancer cells: The involvement of Sall2 in acquired aggressive phenotypes. J Cancer. 2017;8:646–656. doi: 10.7150/jca.15652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, Sun X, Chi C, Liu Y, Lin C, Xie D, Shen X, Lin X. Upregulation of long noncoding RNA LINC00152 promotes proliferation and metastasis of esophageal squamous cell carcinoma. Cancer Manag Res. 2019;11:4643–4654. doi: 10.2147/CMAR.S198905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang J, Li J, Li Y, Lu Z, Che Y, Mao S, Lei Y, Zang R, Zheng S, Liu C, et al. Interferon-inducible lncRNA IRF1-AS represses esophageal squamous cell carcinoma by promoting interferon response. Cancer Lett. 2019;459:86–99. doi: 10.1016/j.canlet.2019.05.038. [DOI] [PubMed] [Google Scholar]
- 9.Wang YY, Yan L, Yang S, Xu HN, Chen TT, Dong ZY, Chen SL, Wang WR, Yang QL, Chen CJ. Long noncoding RNA AC073284.4 suppresses epithelial-mesenchymal transition by sponging miR-18b-5p in paclitaxel-resistant breast cancer cells. J Cell Physiol. 2019;234:23202–23215. doi: 10.1002/jcp.28887. [DOI] [PubMed] [Google Scholar]
- 10.Liu B, Cao W, Ma H. Knockdown of lncRNA LSINCT5 suppresses growth and metastasis of human glioma cells via up-regulating miR-451. Artif Cells Nanomed Biotechnol. 2019;47:2507–2515. doi: 10.1080/21691401.2019.1626404. [DOI] [PubMed] [Google Scholar]
- 11.Yang L, Ye Y, Chu J, Jia J, Qu Y, Sun T, Yin H, Ming L, Wan J, He F. Long noncoding RNA FEZF1-AS1 promotes the motility of esophageal squamous cell carcinoma through Wnt/β-catenin pathway. Cancer Manag Res. 2019;11:4425–4435. doi: 10.2147/CMAR.S196004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang W, Zhou R, Mao L, Deng C, Dang X. Esophageal cancer related gene-4 inhibits the migration and proliferation of oral squamous cell carcinoma through BC200 lncRNA/MMP-9 and -13 signaling pathway. Cell Signal. 2019;62:109327. doi: 10.1016/j.cellsig.2019.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Yang XZ, He QJ, Cheng TT, Chi J, Lei ZY, Tang Z, Liao QX, Zhang H, Zeng LS, Cui SZ. Predictive value of LINC01133 for unfavorable prognosis was impacted by alcohol in esophageal squamous cell carcinoma. Cell Physiol Biochem. 2018;48:251–262. doi: 10.1159/000491724. [DOI] [PubMed] [Google Scholar]
- 14.Song W, Zou SB. Prognostic role of lncRNA HOTAIR in esophageal squamous cell carcinoma. Clin Chim Acta. 2016;463:169–173. doi: 10.1016/j.cca.2016.10.035. [DOI] [PubMed] [Google Scholar]
- 15.Chen M, Liu P, Chen Y, Chen Z, Shen M, Liu X, Li X, Li A, Lin Y, Yang R, et al. Long noncoding RNA FAM201A mediates the radiosensitivity of esophageal squamous cell cancer by regulating ATM and mTOR expression via miR-101. Front Genet. 2018;9:611. doi: 10.3389/fgene.2018.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen W, Zhang Y, Wang H, Pan T, Zhang Y, Li C. LINC00473/miR-374a-5p regulates esophageal squamous cell carcinoma via targeting SPIN1 to weaken the effect of radiotherapy. J Cell Biochem. 2019;120:14562–14572. doi: 10.1002/jcb.28717. [DOI] [PubMed] [Google Scholar]
- 17.Ruggieri V, Russi S, Zoppoli P, La Rocca F, Angrisano T, Falco G, Calice G, Laurino S. The role of MicroRNAs in the regulation of gastric cancer stem cells: A meta-analysis of the current status. J Clin Med. 2019;8:E639. doi: 10.3390/jcm8050639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rong D, Lu C, Zhang B, Fu K, Zhao S, Tang W, Cao H. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol Cancer. 2019;18:25. doi: 10.1186/s12943-019-0958-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, Xu P, Sun G, Xu J, Lv J, Xu Z. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang T, Hou J, Li Z, Zheng Z, Wei J, Song D, Hu T, Wu Q, Yang JY, Cai JC. miR-15a-3p and miR-16-1-3p negatively regulate Twist1 to repress gastric cancer cell invasion and metastasis. Int J Biol Sci. 2017;13:122–134. doi: 10.7150/ijbs.14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gajera M, Desai N, Suzuki A, Li A, Zhang M, Jun G, Jia P, Zhao Z, Iwata J. MicroRNA-655-3p and microRNA-497-5p inhibit cell proliferation in cultured human lip cells through the regulation of genes related to human cleft lip. BMC Med Genomics. 2019;12:70. doi: 10.1186/s12920-019-0535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng H, Dong H, Feng J, Tian H, Zhang H, Xu L. miR-497 inhibited proliferation, migration and invasion of thyroid papillary carcinoma cells by negatively regulating YAP1 expression. Onco Targets Ther. 2018;11:4711–4721. doi: 10.2147/OTT.S164052. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Yan JJ, Zhang YN, Liao JZ, Ke KP, Chang Y, Li PY, Wang M, Lin JS, He XX. MiR-497 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting VEGFA and AEG-1. Oncotarget. 2015;6:29527–29542. doi: 10.18632/oncotarget.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatano K, Kumar B, Zhang Y, Coulter JB, Hedayati M, Mears B, Ni X, Kudrolli TA, Chowdhury WH, Rodriguez R, et al. A functional screen identifies miRNAs that inhibit DNA repair and sensitize prostate cancer cells to ionizing radiation. Nucleic Acids Res. 2015;43:4075–4086. doi: 10.1093/nar/gkv273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Ye Z, Mei D, Gu H, Zhang J. Long noncoding RNA DLX6-AS1 promotes tumorigenesis by modulating miR-497-5p/FZD4/FZD6/Wnt/β-catenin pathway in pancreatic cancer. Cancer Manag Res. 2019;11:4209–4221. doi: 10.2147/CMAR.S194453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X, Wang L, Liu W, Li F. MicroRNA-497-5p inhibits proliferation and invasion of non-small cell lung cancer by regulating FGF2. Oncol Lett. 2019;17:3425–3431. doi: 10.3892/ol.2019.9954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen T, Huang S. The role of Cdc25A in the regulation of cell proliferation and apoptosis. Anticancer Agents Med Chem. 2012;12:631–639. doi: 10.2174/187152012800617678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Jiang M, Cui M, Dong J, Li Y, Xiao H, Fan S. MiR-365 enhances the radiosensitivity of non-small cell lung cancer cells through targeting CDC25A. Biochem Biophys Res Commun. 2019;512:392–398. doi: 10.1016/j.bbrc.2019.03.082. [DOI] [PubMed] [Google Scholar]
- 29.Mao A, Zhao Q, Zhou X, Sun C, Si J, Zhou R, Gan L, Zhang H. MicroRNA-449a enhances radiosensitivity by downregulation of c-Myc in prostate cancer cells. Sci Rep. 2016;6:27346. doi: 10.1038/srep27346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang MY, Wang JY, Chang HJ, Kuo CW, Tok TS, Lin SR. CDC25A, VAV1, TP73, BRCA1 and ZAP70 gene overexpression correlates with radiation response in colorectal cancer. Oncol Rep. 2011;25:1297–1306. doi: 10.3892/or.2011.1193. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Chen NB, Qiu B, Zhang J, Qiang MY, Zhu YJ, Wang B, Guo JY, Cai LZ, Huang SM, Liu MZ, et al. Intensity-modulated radiotherapy versus three-dimensional conformal radiotherapy in definitive chemoradiotherapy for cervical esophageal squamous cell carcinoma: Comparison of survival outcomes and toxicities. Cancer Res Treat. 2020;52:31–40. doi: 10.4143/crt.2018.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, Yi J, Tao L, Huang G, Chu X, Song H, Chen L. Wnt signaling induces radioresistance through upregulating HMGB1 in esophageal squamous cell carcinoma. Cell Death Dis. 2018;9:433. doi: 10.1038/s41419-018-0466-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C, Wang X, Wang X, Han C, Wang P, Pang Q, Chen J, Sun X, Wang L, Zhang W, et al. A multicenter phase III study comparing Simultaneous Integrated Boost (SIB) radiotherapy concurrent and consolidated with S-1 versus SIB alone in elderly patients with esophageal and esophagogastric cancer-the 3JECROG P-01 study protocol. BMC Cancer. 2019;19:397. doi: 10.1186/s12885-019-5544-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Y, Wang J, Pan S, Yang T, Sun X, Wang Y, Shi X, Zhao X, Guo J, Zhang X. LINC00657 played oncogenic roles in esophageal squamous cell carcinoma by targeting miR-615-3p and JunB. Biomed Pharmacother. 2018;108:316–324. doi: 10.1016/j.biopha.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 36.Tong YS, Wang XW, Zhou XL, Liu ZH, Yang TX, Shi WH, Xie HW, Lv J, Wu QQ, Cao XF. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14:3. doi: 10.1186/1476-4598-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou WY, Zhang MM, Liu C, Kang Y, Wang JO, Yang XH. Long noncoding RNA LINC00473 drives the progression of pancreatic cancer via upregulating programmed death-ligand 1 by sponging microRNA-195-5p. J Cell Physiol. 2019;234:23176–23189. doi: 10.1002/jcp.28884. [DOI] [PubMed] [Google Scholar]
- 38.Chen GZ, Zhu HC, Dai WS, Zeng XN, Luo JH, Sun XC. The mechanisms of radioresistance in esophageal squamous cell carcinoma and current strategies in radiosensitivity. J Thorac Dis. 2017;9:849–859. doi: 10.21037/jtd.2017.03.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong S, Yin H, Dong C, Sun K, Lv P, Meng W, Ming L, He F. Predictive value of plasma MicroRNA-216a/b in the diagnosis of esophageal squamous cell carcinoma. Dis Markers. 2016;2016:1857067. doi: 10.1155/2016/1857067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.You C, Liang H, Sun W, Li J, Liu Y, Fan Q, Zhang H, Yue X, Li J, Chen X, Ba Y. Deregulation of the miR-16-KRAS axis promotes colorectal cancer. Sci Rep. 2016;6:37459. doi: 10.1038/srep37459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhan D, Ji T, Li M, Yao Y, Jia J, Yi H, Qiao M, Xia J, Zhang Z, Ding H, et al. Enhancement of sensitivity to chemo/radiation therapy by using miR-15b against DCLK1 in colorectal cancer. Stem Cell Reports. 2018;11:1506–1522. doi: 10.1016/j.stemcr.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu H, Gatti RA. MicroRNAs: New players in the DNA damage response. J Mol Cell Biol. 2011;3:151–158. doi: 10.1093/jmcb/mjq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao L, Lu X, Cao Y. MicroRNA and signal transduction pathways in tumor radiation response. Cell Signal. 2013;25:1625–1634. doi: 10.1016/j.cellsig.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soriano A, Paris-Coderch L, Jubierre L, Martínez A, Zhou X, Piskareva O, Bray I, Vidal I, Almazán-Moga A, Molist C, et al. MicroRNA-497 impairs the growth of chemoresistant neuroblastoma cells by targeting cell cycle, survival and vascular permeability genes. Oncotarget. 2016;7:9271–9287. doi: 10.18632/oncotarget.7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishioka K, Doki Y, Shiozaki H, Yamamoto H, Tamura S, Yasuda T, Fujiwara Y, Yano M, Miyata H, Kishi K, et al. Clinical significance of CDC25A and CDC25B expression in squamous cell carcinomas of the oesophagus. Br J Cancer. 2001;85:412–421. doi: 10.1054/bjoc.2001.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo A, Zhou X, Shi X, Zhao Y, Men Y, Chang X, Chen H, Ding F, Li Y, Su D, et al. Exosome-derived miR-339-5p mediates radio-sensitivity by targeting Cdc25A in locally advanced esophageal squamous cell carcinoma. Oncogene. 2019;38:4990–5006. doi: 10.1038/s41388-019-0771-0. [DOI] [PubMed] [Google Scholar]
- 47.He Z. LINC00473/miR-497-5p regulates esophageal squamous cell carcinoma progression through targeting PRKAA1. Cancer Biother Radiopharm. 2019;34:650–659. doi: 10.1089/cbr.2019.2875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.