Abstract
In this report, cell-penetrating streptavidin (CPS) is introduced to exploit the full power of streptavidin–biotin biotechnology in cellular uptake. For this purpose, transporters, here cyclic oligochalcogenides (COCs), are covalently attached to lysines of wild-type streptavidin. This leaves all four biotin binding sites free for at least bifunctional delivery. To maximize the standards of the quantitative evaluation of cytosolic delivery, the recent chloroalkane penetration assay (CAPA) is coupled with automated high content (HC) imaging, a technique that combines the advantages of fluorescence microscopy and flow cytometry. According to the resulting HC-CAPA, cytosolic delivery of CPS equipped with four benzopolysulfanes was the best among all tested CPSs, also better than the much smaller TAT peptide, the original cell-penetrating peptide from HIV. HaloTag-GFP fusion proteins expressed on mitochondria were successfully targeted using CPS carrying two different biotinylated ligands, HaloTag substrates or anti-GFP nanobodies, interfaced with peptide nucleic acids, flipper force probes, or fluorescent substrates. The delivered substrates could be released from CPS into the cytosol through desthiobiotin–biotin exchange. These results validate CPS as a general tool which enables unrestricted use of streptavidin–biotin biotechnology in cellular uptake.
Introduction
Streptavidin1−4 is a 52 kDa β-barrel tetramer that binds one biotin per monomer with exceptionally high affinity because the barrels close upon binding (Figure 1A). The advantages of such encapsulation combined with tetravalency, stability, and ease of use empower the high versatility of biotin–streptavidin technology. For cellular uptake, streptavidin has been used to noncovalently couple biotinylated substrates to biotinylated transporters such as cell-penetrating peptides (CPPs)5 and other dynamic covalent systems,6 including cell-penetrating poly(disulfide)s,7,8 related disulfide-containing systems,9,10 and cyclic oligochalcogenides (COCs, Figure 1B).11−15 However, this approach is limited to the reliable delivery of only one functionality because it is difficult to control the interfacing of more than two different ligands with the streptavidin tetramer. This limitation is occasionally overcome by covalent linking of fluorophores to proteins.14 A more powerful solution would be cell-penetrating streptavidin (CPS) with covalently attached transporters and all four binding sites free to harness the full power of streptavidin–biotin technology for bifunctional delivery (Figure 1C) with, for example, retention-using-selective-hooks (RUSH)-like16 spatiotemporal control (Figure 1D).
Figure 1.
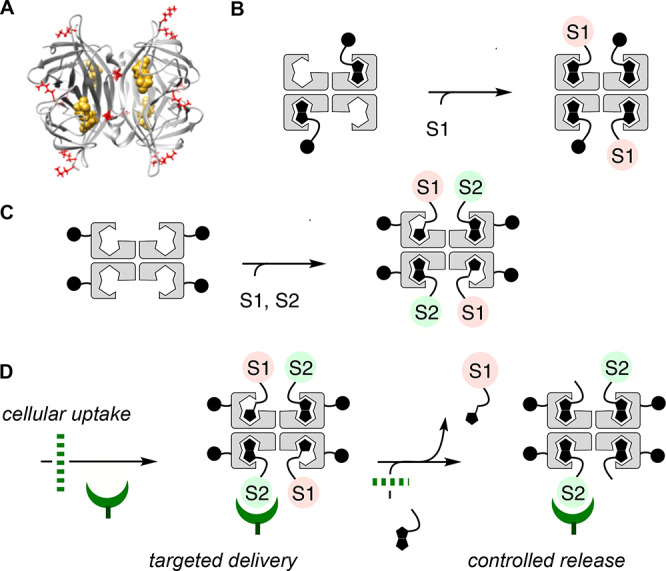
(A) Molecular model of wild-type streptavidin tetramer with bound biotins (yellow) and lysine residues (red, front view) used to (C) covalently attach transporters. (B) Conventional use of streptavidin to interface biotinylated transporters with one biotinylated substrate. (C) Cell-penetrating streptavidin (CPS) with all biotin binding sites free to interface with two different biotinylated substrates S1 and S2 for bifunctional uptake with (D) spatiotemporal control.
Covalent protein modification has been used previously for cellular uptake. Examples include supercharging of proteins by either addition of positive or removal of negative charges17 or simple covalent conjugation to possible new transporters such as boronic acids or halogen-bond donors.5−10,18
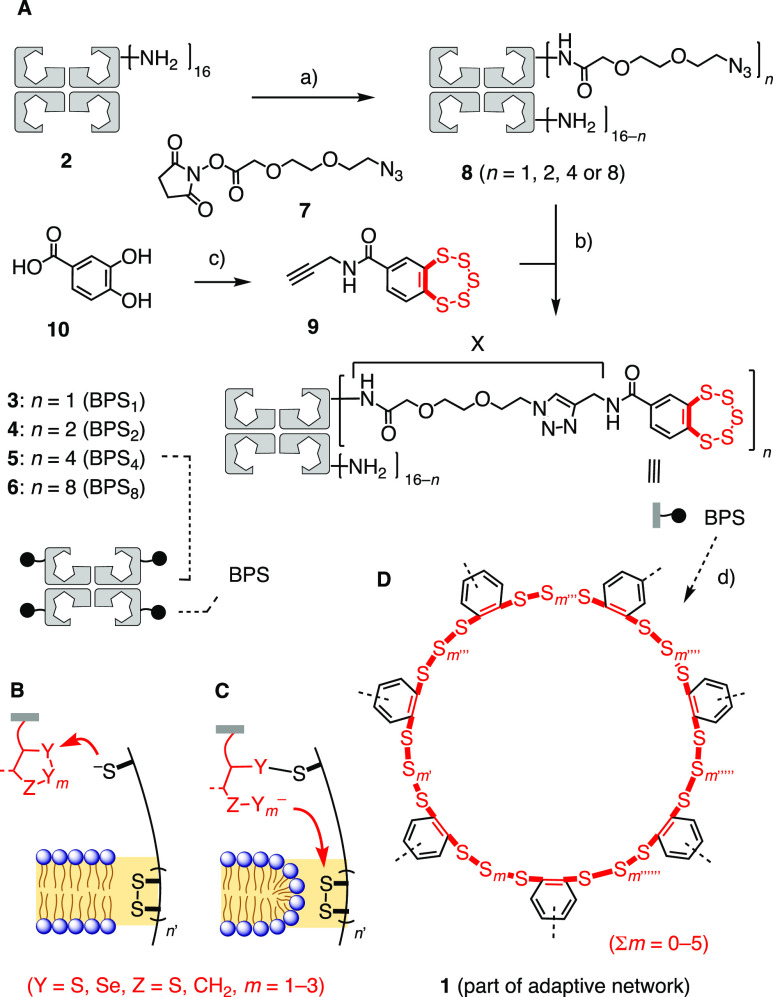
To elaborate on the idea of CPS, we selected COCs as transporters. COCs such as asparagusic acid (AA),11,12 epidiketodithiopiperazines,13 diselenolipoic acid (DSL),14 or the most recent benzopolysulfanes (BPS)15 are currently being explored to access increasingly unorthodox dynamic covalent oligochalcogenide exchange chemistry on the way into the cytosol. Such thiol-mediated uptake of COCs11−15 and related transporters7−10 has allowed delivery of not only small molecules but also larger substrates such as DNA,9b antibodies,9c quantum dots,8 other nanoparticles,9c liposomes, and polymersomes11d to the cytosol without significant capture within endosomes. Mechanistic hypotheses envision COCs as molecular walkers, walking along disulfide tracks in membrane proteins12,14,19,20 through the transient micellar pores known from CPPs but also from disulfide-rich scramblases or voltage-gated ion channels (Figures 2B and 2C).14,20,21 Driving the growing impact of dynamic covalent chemistry to cellular uptake6−14 to the extreme, BPS have been hypothesized to act by forming adaptive networks of rare sulfur species such as macrocycles 1 containing up to 19 sulfur atoms for cells to select from (Figure 2D).15 BPS are known to occur in marine natural products,22 have attracted early attention in total synthesis,23 and appeared top in recent library screens to reverse cognitive defects in mouse models.24
Figure 2.
(A) Synthesis of CPSs 3–6: (a) PBS, pH 7.4, rt, 2 h; (b) CuSO4·5H2O, BTTAA, sodium ascorbate, aminoguanidine hydrochloride, PBS, pH 7.4, rt, 1 h; (c, d) ref (15). (B–D) Working hypotheses for the modes of action of COCs as molecular walkers (B) first exchanging with exofacial thiols and (C) then walking along disulfide tracks and transient micellar pores into cells and of (D) BPS as an adaptive network including macrocycles as large as 1.
The objective of this study was to create a general tool which warrants unrestricted availability of streptavidin–biotin technology in cellular uptake. With COCs as a timely example of freely variable transporters, bifunctional delivery with spatiotemporal16 controllability is tackled as a functional challenge of biological relevance (Figure 1D). Specifically, HC-CAPA, a new combination of the chloroalkane penetration assay (CAPA)25,26 with automated high content fluorescence microscopy,27 is introduced first to secure direct quantitative functional evidence for cytosolic delivery of CPS. On the basis of these results, compatibility of CPS with targeted delivery, the interfacing of nanobodies,8 peptide nucleic acids (PNAs),28 and mechanosensitive fluorescent flipper probes29 with HaloTag-GFP fusion proteins,25,30 and their controlled release through intracellular desthiobiotin–biotin exchange are demonstrated.3,4,16,29a
Results and Discussion
Design and Synthesis of CPS
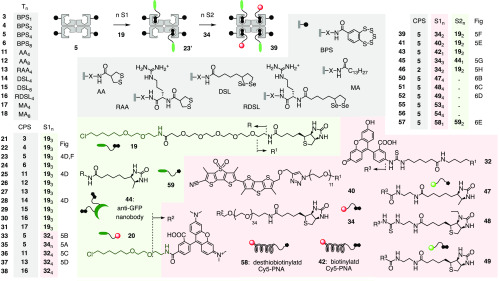
Wild-type streptavidin tetramer 2 contains 16 lysine residues (Figures 1A and 2A),1−4 which were partially derivatized to give CPS with different numbers of COCs. We chose to chemically modify amines rather than carboxylates to preclude misleading CPP-like contributions5−8,21 to the uptake activity of CPS due to increased net positive charge.26,17 The synthesis of CPSs 3–6 is shown as an example (Figures 2A and 3, Schemes S7 and S8). The reaction of 2 and a controlled amount of activated ester 7 gave modified streptavidin 8 with up to ca. eight azide groups. The degree of functionalization was estimated using a colorimetric amine assay (Figure S1). The obtained key intermediate 8 underwent copper-catalyzed alkyne–azide cycloaddition reaction (CuAAC) quantitatively with alkynylated transporter 9 under mild conditions to give CPSs with ca. one (3, BPS1), two (4, BPS2), four (5, BPS4) or eight covalently linked BPSs (6, BPS8, Figure 2A). BPS 9 was prepared by multistep synthesis from catechol 10 following previously reported procedures.15 CuAAC between pre-CPS 8 and other alkynylated transporters and controls afforded CPSs covalently linked with four or eight AAs (11, AA4; 12, AA8), four AA-arginines (13, RAA4), four or eight DSLs (14, DSL4; 15, DSL8), four DSL-arginines (16, RDSL4), and four or eight myristic acids (17, MA4; 18, MA8, Figure 3, Schemes S1, S7, and S8). An overview of CPS with different transporters, CPS complexes with different substrates, and controls described in the following is provided in Figure 3. A comprehensive summary of all structures used can be found in Figure S39, Supporting Information.
Figure 3.
Structure of monomers, CPS with n transporters T, CPS complexes with n substrates S1 and S2, and controls made and used in this study with representative assembly of 39 and indication of pertinent figures (Fig) reporting on activity. n is the stoichiometry used for complex preparation (35: n = 2–4, see text); 47 and 49 are a mixture of m/p-carboxyfluorescein isomers.
Cytosolic Delivery of CPS Evidenced by HC-CAPA
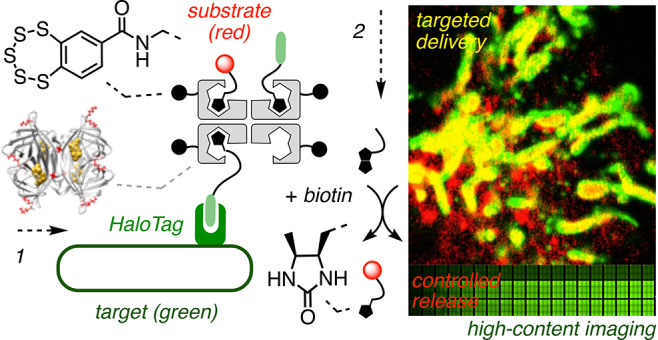
CAPA has been introduced recently by the Kritzer group to quantify cytosolic delivery reliably based on function.25 This is important because the commonly used fluorescence intensity-based assays are often significantly affected by the environment2,11−15 and, in general, demonstrate neither cytosolic location nor intact functionality.25,26 CAPA operates with HGM cells, e.g., HeLa cells that have a fusion protein of HaloTag and GFP (Figure 4F, green) expressed on the outer surface of mitochondria (Figure 4A).25,30 Delivered chloroalkanes, such as 19, react in cytosol with HaloTags (Figure 4B) and thus inhibit reaction with the subsequently added fluorescent chloroalkane 20 (Figure 4C and 4H vs 4I, red). Thus, the delivery of chloroalkanes can be quantified from the fluorescence intensity of 20 on mitochondria. To secure quantitative data of the highest possible quality, we adapted CAPA to an image-based assay using automated high-content fluorescence microscopy (referred to also as high-content screening (HCS) or high-content analysis (HCA)),27 preserving the power of flow cytometry to record and analyze thousands of cells in a short period of time without losing the subcellular spatial resolution of fluorescence imaging. For HC-CAPA, the mitochondria with HaloTags were first segmented applying a top-hat transform to the unmodifiable fluorescence of the GFP (Figure 4F). Then this mask was used to collect the intensity of 20 only in the region of mitochondria, thus excluding any possible false positives from off-target signals (Figure 4G, Figures S2–S5).
Figure 4.
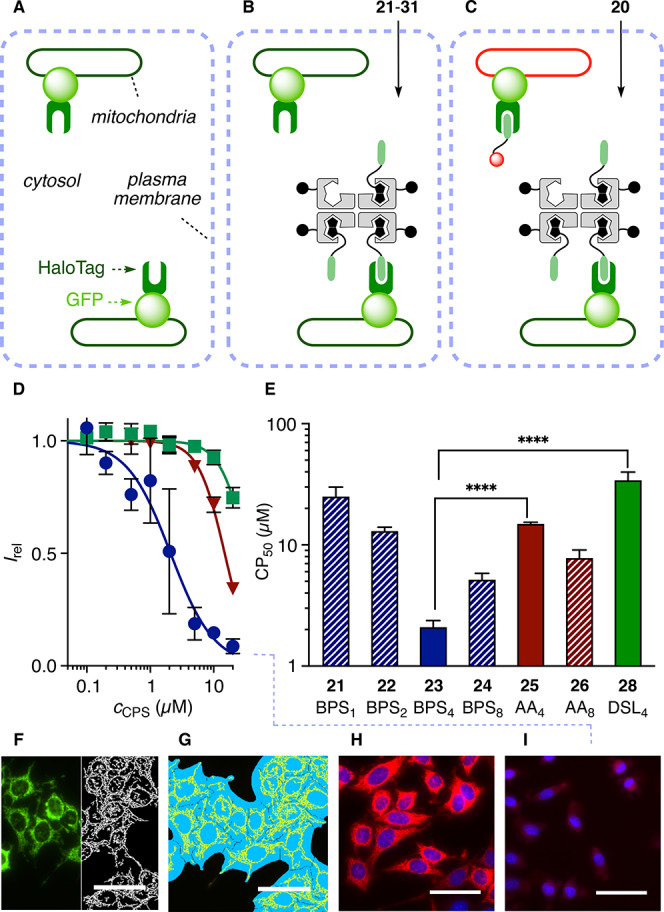
(A–C) Schematic representation of CAPA: (A) HGM cells were incubated with (B) 21–31 and then (C) 20. (D) HC-CAPA dose–response curves for 23 (●), 25 (▼), and 28 (■). (E) CP50 values of 21–26 and 28. Statistical significance (**** p < 0.0001) was estimated by the t test using GraphPad Prism. (F–I) Representative images for HC-CAPA: (F) top-hat transform (white) of the image generated from the emission of GFP (green); (G) cell body (light blue) and mitochondria (yellow) masks applied to selectively extract from the mitochondria the emission intensity in the rhodamine channel; (H) rhodamine channel (red) from labeling with 20 without incubation with transporters and (I) after incubation with 20 μM 23 (nuclei in blue; Hoechst 33342, poststaining, scale bars 50 μm).
The newly devised HC-CAPA was then used to assess cytosolic delivery of CPS-chloroalkane complexes 21–31, prepared by mixing the corresponding CPS with biotinylated chloroalkane 19 (Figure 3, Scheme S9). The absence of unbound 19 was assured using a substoichiometric 3 equiv of ligand per CPS and by meticulous purification using centrifugal filters. Results from HC-CAPA were quantified as CP50, which is the CPS concentration needed for 50% inhibition of HaloTagging by rhodamine 20. The best CP50 = 2.1 ± 0.3 μM was found for BPS423, with the onset of activity detectable already in the nanomolar range (Figures 4D (●), 4E, and 4I). This activity exceeded the CP50 = 3.1 ± 0.5 μM reported for chloroalkane-tagged Tat peptide, the original CPP from the HIV virus.25 Although direct quantitative comparison of these two values has to be made with due caution considering the different methods used for data collection, it perfectly illustrates the high efficiency of cytosolic delivery with BPS423. BPS together with a large 52 kDa protein outperforming this small undecapeptide in cytosolic delivery is particularly interesting considering that uptake efficiencies in general decrease rapidly with increasing substrate size.5,8,9,11
The presence of fewer and more transporters in BPS121, BPS222, and BPS824 reduced activity (Figure 4E). The second best COC was AA with CP50 = 8 ± 1 μM for AA826 and CP50 = 14.9 ± 0.5 μM for AA425, followed by DSL428 with CP50 = 34 ± 6 μM (Figures 4D–I). The presence of more transporters in DSL829 rather deteriorated the poor performance of DSL under these conditions. Insertion of guanidinium cations between the protein and COCs in RAA427 and RDSL430 was also ineffective in improving the transport activity, thus excluding contributions from CPP-like mechanisms7−11 (CP50 = 20 ± 1 μM for 27, >35 μM for 30). Decreasing activities from BPS423 to BPS824 but not from AA425 to AA826 suggested that the higher hydrophobicity of BPS starts to hinder thiol-mediated uptake in the presence of more than four transporters per streptavidin, possibly due to decreasing solubility in water, aggregation, and even precipitation (Figure 4E). The best activity found with an intermediate number of BPS, or “Goldilocks” behavior,31 implied that the right balance between sufficient hydrophilicity and number of transporters is rather subtle. Goldilocks behavior further supported that simple passive diffusion does not account for the top activity of BPS423. Corroborative experimental support for this important conclusion was secured with myristyl control MA431, which is as hydrophobic as BPS but completely inactive (Figure S6, Table S3, and Scheme S9).
We previously reported the mechanistic insights in support of thiol-mediated uptake of BPS, including partial inhibition with Ellman’s reagent which inactivates cell surface thiols, insensitivity toward endocytosis inhibitors (cytochalasin B, wortmannin, chlorpromazine, or methyl-β-cyclodextrin), the formation of complex adaptive networks of extreme sulfur species in the presence of thiols and disulfides (up to heptamers 1, Figure 1D), and stability in PBS buffer for >2 weeks.15 In the current context focusing on cell-penetrating streptavidin, which is a general and unrestricted availability of biotin–streptavidin biotechnology for cellular uptake independent of the transporter used, repetition of these mechanistic studies on BPS-mediated uptake was less relevant and off topic. Nevertheless, retention on and thiol-meditated release from thiol-affinity columns, arguably the most compelling test for thiol-mediated uptake,14a,15 was confirmed for intact BPS423 (Figure S7). As previously noted with AA,12 the presence of 10% serum in Leibovitz’s media did not affect the activity much, resulting in a less than doubled CP50 of BPS423.
Targetless Delivery of CPS into Various Cells
The delivery of four biotinylated fluoresceins 32(14) in BPS433 into unmodified HeLa cells afforded confocal laser scanning microscopy (CLSM) images with diffusely emitting cells, including their nuclei (Figures 5B and S8).15 Similar images were obtained with biotinylated rhodamine 34(2) in BPS435 inside C2C12, HeLa, MCF7, or MDCK cells (Figures 5A and S9; n34 = 2). Weaker fluorescence was observed in CLSM images of AA436 in HeLa cells (Figure 5C). The added cations in RAA437 and RDSL438 did not help to improve the activity significantly, confirming that CPP-like mechanisms7−11 are negligible for COC-mediated uptake (Figures 5D and S8). Contrary to BPS433, images of AA436 and RAA437 contain bright punctate patterns on a diffuse background, which usually indicate contributions from endolysosomes and perhaps also precipitating aggregates besides emission from the cytosol. With flow cytometry, such contributions from endosomal capture are recorded as false positives. The here introduced HC-CAPA is powerful because all eventual false positives25,26 are eliminated without losses in fast acquisition of statistically relevant high-content data.
Figure 5.
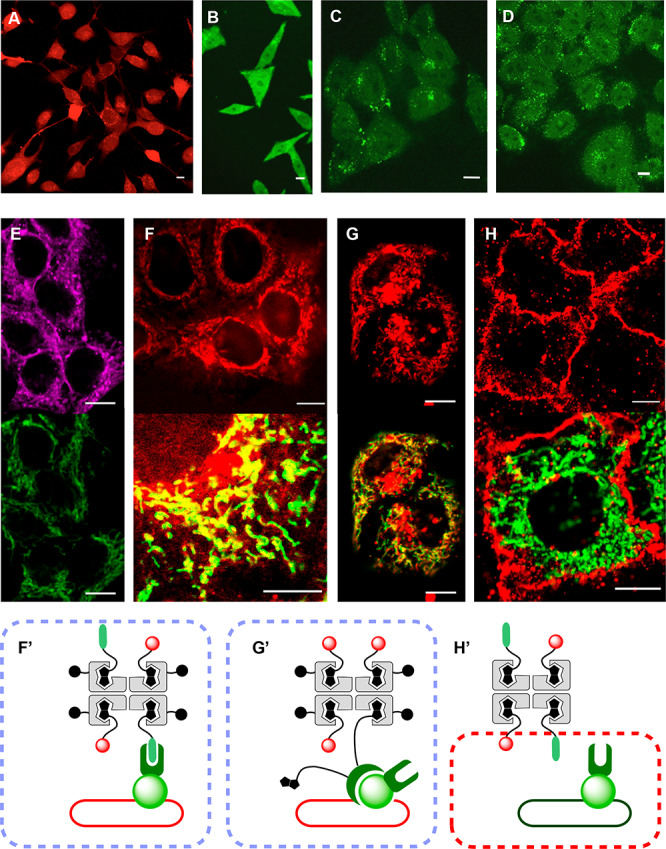
CLSM images of (A) C2C12 cells after incubation with fluorescently labeled BPS435 (n34 = 2, 10 μM, 2 h), of HeLa cells after incubation with (B) BPS433, (C) AA436, and (D) RAA437 (all 10 μM, 8 h), and of HGM cells after incubation with (E) BPS441 (emission top, 40; bottom, GFP; 5 μM, 2 h), (F) 39 (top, 34; bottom, 34 (red) + GFP (green), 5 μM, 2 h), (G) 45 (top, 34; bottom, 34 (red) + GFP (green), 2.5 μM, 2 h), and (H) 46 (top, 40; bottom, 40 (red) + GFP (green), 5 μM, 2 h) in Leibovitz’s media; complex stoichiometry indicates the ratios of preparation. (F′–H′) Design of experiments F–H. Scale bars: 10 μm.
Targeted Delivery of Fluorophores, Nanobodies, and PNA
Using the best CPS, that is, BPS45, we explored the simultaneous delivery of two functions, here a reporter and a targeting unit. BPS45 was loaded with biotinylated chloroalkane 19 and rhodamine 34 (Figures 3 and 5F′, Scheme S12). Incubation of HeLa cells expressing HaloTags on mitochondria with the resulting complex 39 afforded CLSM images with fluorescently labeled mitochondria (Figure 5F, top). The diffuse emission from cytosol and nuclei observed without targeting (Figures 5A and 5B) disappeared, thus confirming that delivery of complex 39 to the HaloTag on the surface of mitochondria is as specific as expected. A good colocalization with GFP emission demonstrated the targeted delivery of the complex to HaloTags (Figures 5F, bottom, S10A, and S10B). HaloTag targeting was substrate independent; the substitution of rhodamine 34 with flipper 40(29) gave similar images for delivery of the respective complex 41 to HaloTags on mitochondria (Figures 3, 5E, and S11). This result was of interest for the fluorescence imaging of precisely localized membrane tension within living cells.29 Targeted delivery of biotinylated and Cy5-labeled 18-mer PNA 42(28) in complex 43 was also accomplished in HGM cells (vide infra, Figure S13).
Biotinylated anti-GFP nanobodies844 were probed as alternative targeting units in complex 45, recognizing GFP instead of HaloTag of the fusion protein on mitochondria. The resulting images showed good colocalization with GFP together with less important endolysosomal capture and/or precipitates (red dots, Figures 5G and S12). These off-targeted, endolysosomal capture and/or precipitates probably originate from the larger oligomers of complex 45 cross-linked through the multivalent anti-GFP nanobodies844 with more than one biotin (Figures 3 and 5G′). Shifts in uptake mechanism from cytosolic delivery to endosomal capture have been observed frequently with increasing substrate size, although much more for CPPs5 than for thiol-mediated uptake.8,9,11 To minimize these side effects, complex 45 was prepared by incubation of BPS45 with 3 equiv of fluorophores 34 before incubation with the multivalent nanobodies 44, a procedure that should in turn increase formation of the targetless CPS side product 35 loaded with four fluorophores 34 only (Figure 3, n34 = 4). However, eventual contributions from off-targeted delivery with CPS 35 were very minor (diffuse red as in Figures 5A, 5G, and S12). Considering this situation, the targeted delivery mediated by anti-GFP nanobodies844 to GFP expressed on the surface of mitochondria was remarkably efficient and supported, with red fluorescence originating from ligand 34, that the majority of complex 45 reaches the cytosol in intact functional form (yellow, Figures 5G, 5G′, and S12). The control complex 46 without COCs was unable to penetrate cells and showed no colocalization with GFP (Figures 5H, S10C, and S10D).
Controlled Intracellular Release of Substrates from CPS
Once delivered in the cells, the substrate would ideally be detached from transporter and target at a well-defined time16 to ensure and follow its proper function on demand (Figure 1D). To elaborate on the timed release of substrates from CPS, fluorescein as a responsive model substrate was equipped with desthiobiotin 47, N-ethylbiotin 48, and biotin 49 (Figures 3 and 6, Schemes S4 and S13). Desthiobiotin (Kd = 10–11 M)3 and N-ethylbiotin (Kd = 10–9 M)4 bind to streptavidin but with reduced affinity compared to biotin (Kd = 10–15 M).3,4 The corresponding complexes 50–52 were readily accessible by incubation with BPS45 (Figures 3 and 6).
Figure 6.
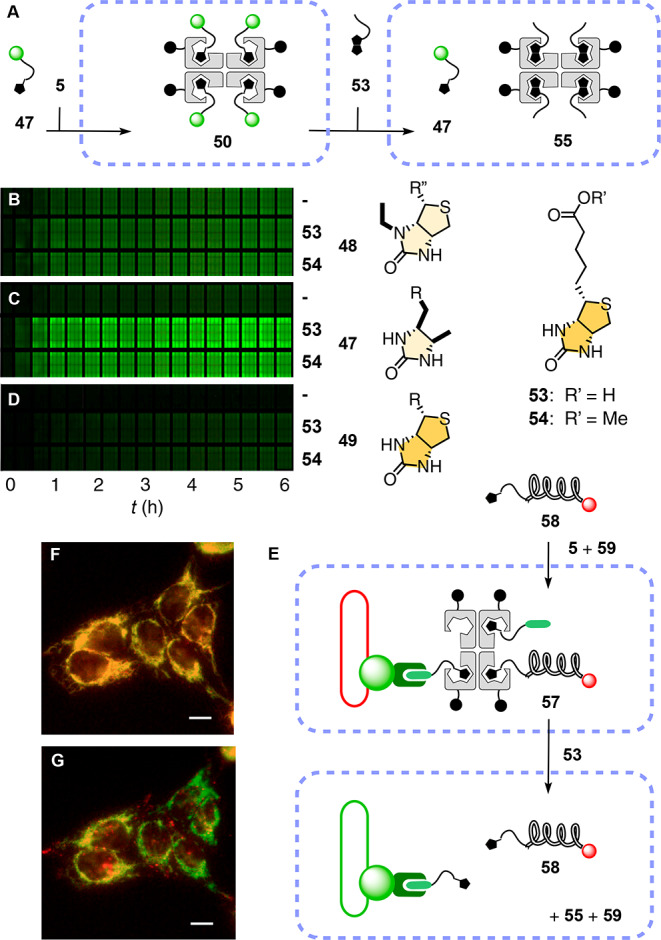
(A) Delivery of 47 with BPS45 followed by addition of biotin 53 to release 47 from complex 50 in the cytosol. (B–D) Original image of the multiwell plate to monitor the time course of substrate release in HeLa cells by automated high-content fluorescence microscopy. Thousands of cells preincubated with (B) 51, (C) 50, or (D) 52 (10 μM, 6 h in Leibovitz’s medium) were treated with 53 (middle), 54 (bottom; 40 μM each), or none (top) and imaged every 12 min (left to right; shown are every other images). (E–G) Spatiotemporal control of (F) the delivery of 58 with 5 and 59 to the mitochondria of HGM cells (5 μM, 2 h incubation in Leibovitz’s medium) and (G) the release of 58 after addition of 53 (58 (red) + GFP (green), 40 μM). Scale bars: 10 μm.
Desthiobiotin 47 was first delivered in complex 50; then biotin 53, a nontoxic vitamin, or its methyl ester 54 was added at a 40 μM concentration as recommended by the RUSH technology16 to diffuse into the cell, exchange, and release substrate 47 in the cytosol (Figure 6A). As streptavidin strongly quenches the fluorescence of bound carboxyfluorescein,2 intracellular desthiobiotin–biotin exchange was detectable by fluorescence recovery of the released substrate 47, whereas the biotin–streptavidin side product 55 or 56 passes undetected.
Quantitative release kinetics were obtained by automated high-content fluorescence microscopy of wells containing thousands of cells with an increasing incubation time with biotin 53 and its methyl ester 54 (Figures 6B–D and S14). Identical t50 = 17 min obtained for controlled release of desthiobiotin 47 from CPS 50 with 53 and 54 demonstrated that passive diffusion of the protonatable carboxylate 53 into the cell is not rate limiting (Figures 6C and S14). In clear contrast, the release of N-ethylbiotin 48 from complex 51 was spontaneous and not affected by the presence of either 53 or 54 (Figure 6B). The lower fluorescence intensity observed with 48 (Figure 6B) compared with 47 (Figure 6C) suggested that 48 dissociated from CPS already during incubation time, diffused out of cells, and was washed away before the start of the acquisition (Figure S14C). Similar spontaneous release of desthiobiotin 47 was slow on the time scale of biotin-controlled release (Figures 6C and S14B). This difference in spontaneous release was in agreement with the poorer binding affinity of N-ethylbiotin 48 (Kd = 10–9 M)4 compared to desthiobiotin 47 (Kd = 10–11 M).3 On the other hand, the controlled release of 49 from 52 by biotin–biotin exchange was negligible even after an extended incubation period (Figure 6D), consistent with much stronger binding of biotin 49 compared to desthiobiotin 47. Insufficient release of biotin 49 and excessive release of N-ethylbiotin 48 added up again in Goldilocks behavior,31 identifying desthiobiotin 47 with neither too strong nor too weak binding as the ligand of choice for controlled release within cells.
To finally combine targeted delivery and controlled release, PNA was selected as a prototypical example of a useful reagent that has been limited in scope by its cellular permeability.28 In complex 57, CPS 5 was loaded with desthiobiotinylated 18-mer PNA 58 and chloroalkane 59 (Figures 3 and 6E, Schemes S5, S6, and S13). The good colocalization (yellow) of Cy5 attached to PNA 58 (red) and GFP (green) demonstrated targeted delivery of complex 57 to HaloTags on the mitochondria of HGM cells in intact form and with excellent selectivity (Figures 6F and S15). Subsequent addition of biotin 53 induced PNA release, detectable as weakened emission of Cy5 on the mitochondria due to dilution of the liberated PNA 58 into the cytosol (Figures 6G, 6E, and S15). These results thus supported the compatibility of CPS delivery with spatiotemporal control using biotin analogs.16
Conclusion
This study introduces CPS as a privileged scaffold for cellular uptake, simple and robust, with biotin–streptavidin technology made fully available for general bifunctional delivery with spatiotemporal control. HC-CAPA, the recent CAPA25 combined with automated high-content fluorescence microscopy27 is devised as the method of choice to quantify cytosolic delivery; the currently emerging benzopolysulfanes15 are validated as COCs of choice for cytosolic delivery. Their performance supports the integration of increasingly unorthodox32 oligochalcogenide exchange chemistry to find new ways to enter into cells and encourages studies on mechanistic hypotheses, i.e., molecular walkers and dynamic-covalent adaptive networks.14,15,19−22
Acknowledgments
This paper is dedicated to George Whitesides on the occasion of his 80th birthday. We thank E. Bartolami, A. Goujon, R. Martinent, and K. Strakova for contributions to CPS synthesis, H. Riezman, N. Jiménez-Rojo, and S. Vossio for help with plasmid amplification, J. A. Kritzer (Tufts) for providing materials, the NMR, the MS, and the Bioimaging platforms for services, and the University of Geneva, the NCCR Chemical Biology, the NCCR Molecular Systems Engineering, and the Swiss NSF for financial support. J.L.A. acknowledges a Curie fellowship (740288).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.9b13621.
Materials and methods; small-molecule synthesis; cell lines and plasmids; CPS synthesis; combining CAPA with automated HC microscopy; cellular uptake of CPS; targeted delivery; supporting references; NMR spectra; characterization of PNA derivatives; summary of structures (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- a Wilson M. E.; Whitesides G. M. Conversion of a Protein to a Homogeneous Asymmetric Hydrogenation Catalyst by Site-Specific Modification with a Diphosphinerhodium(I) Moiety. J. Am. Chem. Soc. 1978, 100, 306–307. 10.1021/ja00469a064. [DOI] [Google Scholar]; b Schreiber C. L.; Smith B. D. Molecular Conjugation Using Non-Covalent Click Chemistry. Nat. Rev. Chem. 2019, 3, 393–400. 10.1038/s41570-019-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Dundas C. M.; Demonte D.; Park S. Streptavidin–Biotin Technology: Improvements and Innovations in Chemical and Biological Applications. Appl. Microbiol. Biotechnol. 2013, 97, 9343–9353. 10.1007/s00253-013-5232-z. [DOI] [PubMed] [Google Scholar]; d Sun X.; Montiel D.; Li H.; Yang H. Plug-and-Go” Strategy to Manipulate Streptavidin Valencies. Bioconjugate Chem. 2014, 25, 1375–1380. 10.1021/bc500296p. [DOI] [PubMed] [Google Scholar]; e Heinisch T.; Ward T. R. Artificial Metalloenzymes Based on the Biotin–Streptavidin Technology: Challenges and Opportunities. Acc. Chem. Res. 2016, 49, 1711–1721. 10.1021/acs.accounts.6b00235. [DOI] [PubMed] [Google Scholar]; f Dubacheva G. V.; Araya-Callis C.; Geert Volbeda A.; Fairhead M.; Codee J.; Howarth M.; Richter R. P. Controlling Multivalent Binding through Surface Chemistry: Model Study on Streptavidin. J. Am. Chem. Soc. 2017, 139, 4157–4167. 10.1021/jacs.7b00540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber H. J.; Marek M.; Schindler H.; Kaiser K. Biotin–Fluorophore Conjugates with Poly(Ethylene Glycol) Spacers Retain Intense Fluorescence after Binding to Avidin and Streptavidin. Bioconjugate Chem. 1997, 8, 552–559. 10.1021/bc970087m. [DOI] [PubMed] [Google Scholar]
- a Ding Y.; Williams N. H.; Hunter C. A. A Synthetic Vesicle-to-Vesicle Communication System. J. Am. Chem. Soc. 2019, 141, 17847–17853. 10.1021/jacs.9b09102. [DOI] [PubMed] [Google Scholar]; b Sadhu K. K.; Eierhoff T.; Römer W.; Winssinger N. Photoreductive Uncaging of Fluorophore in Response to Protein Oligomers by Templated Reaction in Vitro and in Cellulo. J. Am. Chem. Soc. 2012, 134, 20013–20016. 10.1021/ja310171s. [DOI] [PubMed] [Google Scholar]; c Osuna Gálvez A.; Bode J. W. Traceless Templated Amide-Forming Ligations. J. Am. Chem. Soc. 2019, 141, 8721–8726. 10.1021/jacs.9b03597. [DOI] [PubMed] [Google Scholar]
- Ying L.-Q.; Branchaud B. P. Design of a Reversible Biotin Analog and Applications in Protein Labeling, Detection, and Isolation. Chem. Commun. 2011, 47, 8593–8595. 10.1039/c1cc12738a. [DOI] [PubMed] [Google Scholar]
- a Juanes M.; Lostalé-Seijo I.; Granja J. R.; Montenegro J. Supramolecular Recognition and Selective Protein Uptake by Peptide Hybrids. Chem. - Eur. J. 2018, 24, 10689–10698. 10.1002/chem.201800706. [DOI] [PubMed] [Google Scholar]; b Kuan S. L.; Ng D. Y. W.; Wu Y.; Förtsch C.; Barth H.; Doroshenko M.; Koynov K.; Meier C.; Weil T. pH Responsive Janus-like Supramolecular Fusion Proteins for Functional Protein Delivery. J. Am. Chem. Soc. 2013, 135, 17254–17257. 10.1021/ja4084122. [DOI] [PubMed] [Google Scholar]; c Li S.-D.; Huang L. Targeted Delivery of Antisense Oligodeoxynucleotide and Small Interference RNA into Lung Cancer Cells. Mol. Pharmaceutics 2006, 3, 579–588. 10.1021/mp060039w. [DOI] [PubMed] [Google Scholar]; d Lee H.; Kim T. H.; Park T. G. A Receptor-Mediated Gene Delivery System Using Streptavidin and Biotin-Derivatized, Pegylated Epidermal Growth Factor. J. Controlled Release 2002, 83, 109–119. 10.1016/S0168-3659(02)00166-9. [DOI] [PubMed] [Google Scholar]
- Ulrich S. Growing Prospects of Dynamic Covalent Chemistry in Delivery Applications. Acc. Chem. Res. 2019, 52, 510–519. 10.1021/acs.accounts.8b00591. [DOI] [PubMed] [Google Scholar]
- a Bang E.-K.; Gasparini G.; Molinard G.; Roux A.; Sakai N.; Matile S. Substrate-Initiated Synthesis of Cell-Penetrating Poly(disulfide)s. J. Am. Chem. Soc. 2013, 135, 2088–2091. 10.1021/ja311961k. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Qian L.; Fu J.; Yuan P.; Du S.; Huang W.; Li L.; Yao S. Q. Intracellular Delivery of Native Proteins Facilitated by Cell-Penetrating Poly(disulfide)s. Angew. Chem., Int. Ed. 2018, 57, 1532–1536. 10.1002/anie.201711651. [DOI] [PubMed] [Google Scholar]; c Yuan P.; Mao X.; Wu X.; Liew S. S.; Li L.; Yao S. Q. Mitochondria-Targeting, Intracellular Delivery of Native Proteins Using Biodegradable Silica Nanoparticles. Angew. Chem., Int. Ed. 2019, 58, 7657–7661. 10.1002/anie.201901699. [DOI] [PubMed] [Google Scholar]; d Liu Y.; Jia Y.; Wu Q.; Moore J. S. Architecture-Controlled Ring-Opening Polymerization for Dynamic Covalent Poly(Disulfide)s. J. Am. Chem. Soc. 2019, 141, 17075–17080. 10.1021/jacs.9b08957. [DOI] [PubMed] [Google Scholar]; e Zhang X.; Waymouth R. M. 1,2-Dithiolane-Derived Dynamic, Covalent Materials: Cooperative Self-Assembly and Reversible Cross-Linking. J. Am. Chem. Soc. 2017, 139, 3822–3833. 10.1021/jacs.7b00039. [DOI] [PubMed] [Google Scholar]; f Zhang Q.; Deng Y. X.; Luo H. X.; Shi C. Y.; Geise G. M.; Feringa B. L.; Tian H.; Qu D. H. Assembling a Natural Small Molecule into a Supramolecular Network with High Structural Order and Dynamic Functions. J. Am. Chem. Soc. 2019, 141, 12804–12814. 10.1021/jacs.9b05740. [DOI] [PMC free article] [PubMed] [Google Scholar]; g Okamoto Y.; Kojima R.; Schwizer F.; Bartolami E.; Heinisch T.; Matile S.; Fussenegger M.; Ward T. R. A Cell-Penetrating Artificial Metalloenzyme Regulates a Gene Switch in a Designer Mammalian Cell. Nat. Commun. 2018, 9, 1943. 10.1038/s41467-018-04440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E.; Bartolami E.; Matile S.; Gonzalez-Gaitan M. Efficient Delivery of Quantum Dots into the Cytosol of Cells Using Cell-Penetrating Poly(disulfide)s. J. Am. Chem. Soc. 2017, 139, 10172–10175. 10.1021/jacs.7b02952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Meng X.; Li T.; Zhao Y.; Wu C. CXC-Mediated Cellular Uptake of Miniproteins: Forsaking “Arginine Magic.. ACS Chem. Biol. 2018, 13, 3078–3086. 10.1021/acschembio.8b00564. [DOI] [PubMed] [Google Scholar]; b Shu Z.; Tanaka I.; Ota A.; Fushihara D.; Abe N.; Kawaguchi S.; Nakamoto K.; Tomoike F.; Tada S.; Ito Y.; Kimura Y.; Abe H. Disulfide-Unit Conjugation Enables Ultrafast Cytosolic Internalization of Antisense DNA and SiRNA. Angew. Chem., Int. Ed. 2019, 58, 6611–6615. 10.1002/anie.201900993. [DOI] [PubMed] [Google Scholar]; c Zhou J.; Sun L.; Wang L.; Liu Y.; Li J.; Li J.; Li J.; Yang H. Self-Assembled and Size-Controllable Oligonucleotide Nanospheres for Effective Antisense Gene Delivery through an Endocytosis-Independent Pathway. Angew. Chem., Int. Ed. 2019, 58, 5236–5240. 10.1002/anie.201813665. [DOI] [PubMed] [Google Scholar]
- a Torres A. G.; Gait M. J. Exploiting Cell Surface Thiols to Enhance Cellular Uptake. Trends Biotechnol. 2012, 30, 185–190. 10.1016/j.tibtech.2011.12.002. [DOI] [PubMed] [Google Scholar]; b Oupický D.; Li J. Bioreducible Polycations in Nucleic Acid Delivery: Past, Present, and Future Trends. Macromol. Biosci. 2014, 14, 908–922. 10.1002/mabi.201400061. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Du S.; Liew S. S.; Li L.; Yao S. Q. Bypassing Endocytosis: Direct Cytosolic Delivery of Proteins. J. Am. Chem. Soc. 2018, 140, 15986–15996. 10.1021/jacs.8b06584. [DOI] [PubMed] [Google Scholar]; d Aubry S.; Burlina F.; Dupont E.; Delaroche D.; Joliot A.; Lavielle S.; Chassaing G.; Sagan S. Cell-Surface Thiols Affect Cell Entry of Disulfide-Conjugated Peptides. FASEB J. 2009, 23, 2956–2967. 10.1096/fj.08-127563. [DOI] [PubMed] [Google Scholar]; e Hashim P. K.; Okuro K.; Sasaki S.; Hoashi Y.; Aida T. Reductively Cleavable Nanocaplets for siRNA Delivery by Template-Assisted Oxidative Polymerization. J. Am. Chem. Soc. 2015, 137, 15608–15611. 10.1021/jacs.5b08948. [DOI] [PubMed] [Google Scholar]; f Balakirev M.; Schoehn G.; Chroboczek J. Lipoic Acid-Derived Amphiphiles for Redox-Controlled DNA Delivery. Chem. Biol. 2000, 7, 813–819. 10.1016/S1074-5521(00)00030-2. [DOI] [PubMed] [Google Scholar]; g Brülisauer L.; Kathriner N.; Prenrecaj M.; Gauthier M. A.; Leroux J.-C. Tracking the Bioreduction of Disulfide-Containing Cationic Dendrimers. Angew. Chem., Int. Ed. 2012, 51, 12454–12458. 10.1002/anie.201207070. [DOI] [PubMed] [Google Scholar]; h Drake C. R.; Aissaoui A.; Argyros O.; Thanou M.; Steinke J. H. G.; Miller A. D. Examination of The Effect of Increasing the Number of Intra-Disulfide Amino Functional Groups on the Performance of Small Molecule Cyclic Polyamine Disulfide Vectors. J. Controlled Release 2013, 171, 81–90. 10.1016/j.jconrel.2013.02.014. [DOI] [PubMed] [Google Scholar]; i Schneider A. F. L.; Wallabregue A. L. D.; Franz L.; Hackenberger C. P. R. Targeted Subcellular Protein Delivery Using Cleavable Cyclic Cell-Penetrating Peptides. Bioconjugate Chem. 2019, 30, 400–404. 10.1021/acs.bioconjchem.8b00855. [DOI] [PubMed] [Google Scholar]; j Bode S. A.; Wallbrecher R.; Brock R.; van Hest J. C. M.; Löwik D. W. P. M. Activation of Cell-Penetrating Peptides by Disulfide Bridge Formation of Truncated Precursors. Chem. Commun. 2014, 50, 415–417. 10.1039/C3CC46826G. [DOI] [PubMed] [Google Scholar]
- a Gasparini G.; Sargsyan G.; Bang E.-K.; Sakai N.; Matile S. Ring Tension Applied to Thiol-Mediated Cellular Uptake. Angew. Chem., Int. Ed. 2015, 54, 7328–7331. 10.1002/anie.201502358. [DOI] [PubMed] [Google Scholar]; b Tirla A.; Rivera-Fuentes P. Peptide Targeting of an Intracellular Receptor of the Secretory Pathway. Biochemistry 2019, 58, 1184–1187. 10.1021/acs.biochem.9b00029. [DOI] [PubMed] [Google Scholar]; c Li X.; Hou Y.; Meng X.; Ge C.; Ma H.; Li J.; Fang J. Selective Activation of a Prodrug by Thioredoxin Reductase Providing a Strategy to Target Cancer Cells. Angew. Chem., Int. Ed. 2018, 57, 6141–6145. 10.1002/anie.201801058. [DOI] [PubMed] [Google Scholar]; d Chuard N.; Gasparini G.; Moreau D.; Lörcher S.; Palivan C.; Meier W.; Sakai N.; Matile S. Strain-Promoted Thiol-Mediated Cellular Uptake of Giant Substrates: Liposomes and Polymersomes. Angew. Chem., Int. Ed. 2017, 56, 2947–2950. 10.1002/anie.201611772. [DOI] [PubMed] [Google Scholar]; e Singh R.; Whitesides G. M. Degenerate Intermolecular Thiolate-Disulfide Interchange Involving Cyclic Five-Membered Disulfides Is Faster by ∼ 103 Than That Involving Six- or Seven-Membered Disulfides. J. Am. Chem. Soc. 1990, 112, 6304–6309. 10.1021/ja00173a018. [DOI] [Google Scholar]
- Abegg D.; Gasparini G.; Hoch D. G.; Shuster A.; Bartolami E.; Matile S.; Adibekian A. Strained Cyclic Disulfides Enable Cellular Uptake by Reacting with the Transferrin Receptor. J. Am. Chem. Soc. 2017, 139, 231–238. 10.1021/jacs.6b09643. [DOI] [PubMed] [Google Scholar]
- Zong L.; Bartolami E.; Abegg D.; Adibekian A.; Sakai N.; Matile S. Epidithiodiketopiperazines: Strain-Promoted Thiol-Mediated Cellular Uptake at the Highest Tension. ACS Cent. Sci. 2017, 3, 449–453. 10.1021/acscentsci.7b00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Chuard N.; Poblador-Bahamonde A. I.; Zong L.; Bartolami E.; Hildebrandt J.; Weigand W.; Sakai N.; Matile S. Diselenolane-Mediated Cellular Uptake. Chem. Sci. 2018, 9, 1860–1866. 10.1039/C7SC05151D. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Bartolami E.; Basagiannis D.; Zong L.; Martinent R.; Okamoto Y.; Laurent Q.; Ward T. R.; Gonzalez-Gaitan M.; Sakai N.; Matile S. Diselenolane-Mediated Cellular Uptake: Efficient Cytosolic Delivery of Probes, Peptides, Proteins, Artificial Metalloenzymes and Protein-Coated Quantum Dots. Chem. - Eur. J. 2019, 25, 4047–4051. 10.1002/chem.201805900. [DOI] [PubMed] [Google Scholar]; c Laurent Q.; Sakai N.; Matile S. The Opening of 1,2-Dithiolanes and 1,2-Diselenolanes: Regioselectivity, Rearrangements, and Consequences for Poly(disulfide)s, Cellular Uptake and the Pyruvate Dehydrogenase Complex. Helv. Chim. Acta 2019, 102, e1800209. 10.1002/hlca.201800209. [DOI] [Google Scholar]
- Cheng Y.; Zong L.; López-Andarias J.; Bartolami E.; Okamoto Y.; Ward T. R.; Sakai N.; Matile S. Cell-Penetrating Dynamic-Covalent Benzopolysulfane Networks. Angew. Chem., Int. Ed. 2019, 58, 9522–9526. 10.1002/anie.201905003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Boncompain G.; Divoux S.; Gareil N.; de Forges H.; Lescure A.; Latreche L.; Mercanti V.; Jollivet F.; Raposo G.; Perez F. Synchronization of Secretory Protein Traffic in Populations of Cells. Nat. Methods 2012, 9, 493–498. 10.1038/nmeth.1928. [DOI] [PubMed] [Google Scholar]; b Terai T.; Kohno M.; Boncompain G.; Sugiyama S.; Saito N.; Fujikake R.; Ueno T.; Komatsu T.; Hanaoka K.; Okabe T.; Urano Y.; Perez F.; Nagano T. Artificial Ligands of Streptavidin (ALiS): Discovery, Characterization, and Application for Reversible Control of Intracellular Protein Transport. J. Am. Chem. Soc. 2015, 137, 10464–10467. 10.1021/jacs.5b05672. [DOI] [PubMed] [Google Scholar]
- a Kuan S. L.; Stöckle B.; Reichenwallner J.; Ng D. Y. W.; Wu Y.; Doroshenko M.; Koynov K.; Hinderberger D.; Müllen K.; Weil T. Dendronized Albumin Core–Shell Transporters with High Drug Loading Capacity. Biomacromolecules 2013, 14, 367–376. 10.1021/bm301531c. [DOI] [PubMed] [Google Scholar]; b McNaughton B. R.; Cronican J. J.; Thompson D. B.; Liu D. R. Mammalian Cell Penetration, SiRNA Transfection, and DNA Transfection by Supercharged Proteins. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 6111–6116. 10.1073/pnas.0807883106. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Mix K. A.; Lomax J. E.; Raines R. T. Cytosolic Delivery of Proteins by Bioreversible Esterification. J. Am. Chem. Soc. 2017, 139, 14396–14398. 10.1021/jacs.7b06597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Andersen K. A.; Smith T. P.; Lomax J. E.; Raines R. T. Boronic Acid for the Traceless Delivery of Proteins into Cells. ACS Chem. Biol. 2016, 11, 319–323. 10.1021/acschembio.5b00966. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Jakka S. R.; Govindaraj V.; Mugesh G. A Single Atom Change Facilitates the Membrane Transport of Green Fluorescent Proteins in Mammalian Cells. Angew. Chem., Int. Ed. 2019, 58, 7713–7717. 10.1002/anie.201902347. [DOI] [PubMed] [Google Scholar]; c Pelegri-O’Day E. M.; Maynard H. D. Controlled Radical Polymerization as an Enabling Approach for the Next Generation of Protein-Polymer Conjugates. Acc. Chem. Res. 2016, 49, 1777–1785. 10.1021/acs.accounts.6b00258. [DOI] [PubMed] [Google Scholar]; d Hou Y.; Zhou Y.; Wang H.; Sun J.; Wang R.; Sheng K.; Yuan J.; Hu Y.; Chao Y.; Liu Z.; Lu H. Therapeutic Protein PEPylation: The Helix of Nonfouling Synthetic Polypeptides Minimizes Antidrug Antibody Generation. ACS Cent. Sci. 2019, 5, 229–236. 10.1021/acscentsci.8b00548. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Velonia K. Protein - Polymer Amphiphilic Chimeras: Recent Advances and Future Challenges. Polym. Chem. 2010, 1, 944–952. 10.1039/b9py00362b. [DOI] [Google Scholar]
- Beld J.; Woycechowsky K.; Hilvert D. Small-Molecule Diselenides Catalyze Oxidative Protein Folding in Vivo. ACS Chem. Biol. 2010, 5, 177–182. 10.1021/cb9002688. [DOI] [PubMed] [Google Scholar]
- a Alvadia C.; Lim N. K.; Clerico Mosina V.; Oostergetel G. T.; Dutzler R.; Paulino C. Cryo-EM Structures and Functional Characterization of the Murine Lipid Scramblase TMEM16F. eLife 2019, 8, e44365. 10.7554/eLife.44365. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wu J.; Yan Z.; Li Z.; Qian X.; Lu S.; Dong M.; Zhou Q.; Yan N. Structure of the Voltage-Gated Calcium Channel Cav1.1 at 3.6 Å Resolution. Nature 2016, 537, 191–196. 10.1038/nature19321. [DOI] [PubMed] [Google Scholar]; c Jing S.; Trowbridge I. S. Identification of the Intermolecular Disulfide Bonds of the Human Transferrin Receptor and its Lipid-Attachment Site. EMBO J. 1987, 6, 327–331. 10.1002/j.1460-2075.1987.tb04758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Ali Khan H.; Mutus B. Protein Disulfide Isomerase: A Multifunctional Protein with Multiple Physiological Roles. Front. Chem. 2014, 2, 70. 10.3389/fchem.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Schmidt D.; Jiang Q. X.; MacKinnon R. Lipid-Dependent Gating of a Voltage-Gated Potassium Channel. Nature 2006, 444, 775–779. 10.1038/nature05416. [DOI] [PubMed] [Google Scholar]; b Sakai N.; Matile S. Anion-Mediated Transfer of Polyarginine Across Liquid and Bilayer Membranes. J. Am. Chem. Soc. 2003, 125, 14348–14356. 10.1021/ja037601l. [DOI] [PubMed] [Google Scholar]
- Jiang C.-S.; Müller W. E. G.; Schröder H. C.; Guo Y.-W. Disulfide- and Multisulfide-Containing Metabolites from Marine Organisms. Chem. Rev. 2012, 112, 2179–2207. 10.1021/cr200173z. [DOI] [PubMed] [Google Scholar]
- Behar V.; Danishefsky S. J. Total Synthesis of the Novel Benzopentathiepin Varacinium Trifluoroacetate: The Viability of ″Varacin-Free Base″. J. Am. Chem. Soc. 1993, 115, 7017–7018. 10.1021/ja00068a087. [DOI] [Google Scholar]
- Xu J.; Chatterjee M.; Baguley T. D.; Brouillette J.; Kurup P.; Ghosh D.; Kanyo J.; Zhang Y.; Seyb K.; Ononenyi C.; Foscue E.; Anderson G. M.; Gresack J.; Cuny G. D.; Glicksman M. A.; Greengard P.; Lam T. L.; Tautz L.; Nairn A. C.; Ellman J. A.; Lombroso P. J. Inhibitor of the Tyrosine Phosphatase STEP Reverses Cognitive Deficits in a Mouse Model of Alzheimer’s Disease. PLoS Biol. 2014, 12, e1001923. 10.1371/journal.pbio.1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peraro L.; Deprey K. L.; Moser M. K.; Zou Z.; Ball H. L.; Levine B.; Kritzer J. A. Cell Penetration Profiling Using the Chloroalkane Penetration Assay. J. Am. Chem. Soc. 2018, 140, 11360–11369. 10.1021/jacs.8b06144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Deprey K.; Becker L.; Kritzer J.; Plückthun A. Trapped! A Critical Evaluation of Methods for Measuring Total Cellular Uptake versus Cytosolic Localization. Bioconjugate Chem. 2019, 30, 1006–1027. 10.1021/acs.bioconjchem.9b00112. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Méndez-Ardoy A.; Lostalé-Seijo I.; Montenegro J. Where in the Cell Is Our Cargo? Methods Currently Used to Study Intracellular Cytosolic Localisation. ChemBioChem 2019, 20, 488–498. 10.1002/cbic.201800390. [DOI] [PubMed] [Google Scholar]; c Lacroix A.; Vengut-Climent E.; de Rochambeau D.; Sleiman H. F. Uptake and Fate of Fluorescently Labeled DNA Nanostructures in Cellular Environments: A Cautionary Tale. ACS Cent. Sci. 2019, 5, 882–891. 10.1021/acscentsci.9b00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Abraham V. C.; Taylor D. L.; Haskins J. R. High Content Screening Applied to Large-Scale Cell Biology. Trends Biotechnol. 2004, 22, 15–22. 10.1016/j.tibtech.2003.10.012. [DOI] [PubMed] [Google Scholar]; b Eggert U. S.; Mitchison T. J. Small Molecule Screening by Imaging. Curr. Opin. Chem. Biol. 2006, 10, 232–237. 10.1016/j.cbpa.2006.04.010. [DOI] [PubMed] [Google Scholar]; c Moreau D.; Gruenberg J. Automated Microscopy and High Content Screens (Phenotypic Screens) in Academia Labs. Chimia 2016, 70, 878–882. 10.2533/chimia.2016.878. [DOI] [PubMed] [Google Scholar]
- a Saarbach J.; Sabale P. M.; Winssinger N. Peptide Nucleic Acid (PNA) and Its Applications in Chemical Biology, Diagnostics, and Therapeutics. Curr. Opin. Chem. Biol. 2019, 52, 112–124. 10.1016/j.cbpa.2019.06.006. [DOI] [PubMed] [Google Scholar]; b Zhao X.-L.; Chen B.-C.; Han J.-C.; Wei L.; Pan X.-B. Delivery of Cell-Penetrating Peptide-Peptide Nucleic Acid Conjugates by Assembly on an Oligonucleotide Scaffold. Sci. Rep. 2015, 5, 17640. 10.1038/srep17640. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Wolfe J. M.; Fadzen C. M.; Choo Z.-N.; Holden R. L.; Yao M.; Hanson G. J.; Pentelute B. L. Machine Learning to Predict Cell-Penetrating Peptides for Antisense Delivery. ACS Cent. Sci. 2018, 4, 512–520. 10.1021/acscentsci.8b00098. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Dragulescu-Andrasi A.; Rapireddy S.; He G.; Bhattacharya B.; Hyldig-Nielsen J. J.; Zon G.; Ly D. H. Cell-Permeable Peptide Nucleic Acid Designed to Bind to the 5′-Untranslated Region of E-Cadherin Transcript Induces Potent and Sequence-Specific Antisense Effects. J. Am. Chem. Soc. 2006, 128, 16104–16112. 10.1021/ja063383v. [DOI] [PubMed] [Google Scholar]; e Turner J. J.; Ivanova G. D.; Verbeure B.; Williams D.; Arzumanov A. A.; Abes S.; Lebleu B.; Gait M. J. Cell-Penetrating Peptide Conjugates of Peptide Nucleic Acids (PNA) as Inhibitors of HIV-1 Tat-Dependent Trans-Activation in Cells. Nucleic Acids Res. 2005, 33, 6837–6849. 10.1093/nar/gki991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Goujon A.; Straková K.; Sakai N.; Matile S. Streptavidin Interfacing as a General Strategy to Localize Fluorescent Membrane Tension Probes in Cells. Chem. Sci. 2019, 10, 310–319. 10.1039/C8SC03620A. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Goujon A.; Colom A.; Straková K.; Mercier V.; Mahecic D.; Manley S.; Sakai N.; Roux A.; Matile S. Mechanosensitive Fluorescent Probes to Image Membrane Tension in Mitochondria, Endoplasmic Reticulum and Lysosomes. J. Am. Chem. Soc. 2019, 141, 3380–3384. 10.1021/jacs.8b13189. [DOI] [PubMed] [Google Scholar]
- a Los G. V.; Encell L. P.; McDougall M. G.; Hartzell D. D.; Karassina N.; Zimprich C.; Wood M. G.; Learish R.; Ohana R. F.; Urh M.; Simpson D.; Mendez J.; Zimmerman K.; Otto P.; Vidugiris G.; Zhu J.; Darzins A.; Klaubert D. H.; Bulleit R. F.; Wood K. V. HaloTag: A Novel Protein Labeling Technology for Cell Imaging and Protein Analysis. ACS Chem. Biol. 2008, 3, 373–382. 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]; b Long M. J. C.; Poganik J. R.; Aye Y. On-Demand Targeting: Investigating Biology with Proximity-Directed Chemistry. J. Am. Chem. Soc. 2016, 138, 3610–3622. 10.1021/jacs.5b12608. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Chambers J. E.; Kubánková M.; Huber R. G.; López-Duarte I.; Avezov E.; Bond P. J.; Marciniak S. J.; Kuimova M. K. An Optical Technique for Mapping Microviscosity Dynamics in Cellular Organelles. ACS Nano 2018, 12, 4398–4407. 10.1021/acsnano.8b00177. [DOI] [PubMed] [Google Scholar]
- a Behr J.-P.; Kirch M.; Lehn J.-M. Carrier-Mediated Transport through Bulk Liquid Membranes: Dependence of Transport Rates and Selectivity on Carrier Properties in a Diffusion-Limited Process. J. Am. Chem. Soc. 1985, 107, 241–246. 10.1021/ja00287a043. [DOI] [Google Scholar]; b Vargas Jentzsch A.; Emery D.; Mareda J.; Metrangolo P.; Resnati G.; Matile S. Ditopic Ion Transport Systems: Anion-π Interactions and Halogen Bonds at Work. Angew. Chem., Int. Ed. 2011, 50, 11675–11678. 10.1002/anie.201104966. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Cotelle Y.; Sakai N.; Matile S. Unorthodox Interactions at Work. J. Am. Chem. Soc. 2016, 138, 4270–4277. 10.1021/jacs.5b13006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.